Abstract
Background
Neuroblastoma (NB), a childhood neoplasm arising from neural crest cells, is characterized by a diversity of clinical behaviors ranging from spontaneous remission to rapid tumor progression and death. In addition to genetic abnormalities, recent studies have indicated that epigenetic aberrations also contribute to NB pathogenesis. However, the epigenetic mechanisms underlying the pathogenesis of NB are largely unknown.
Methods
Inhibition of euchromatic histone-lysine N-methyltransferase 2 (EHMT2) was evaluated through the measurement of H3K9Me2 levels. Cell proliferation was examined by the use of cell counting in human NB cell lines (LA1-55n, IMR-5 and NMB). The RNA expression of EHMT2, MYCN, and p21 was measured by real-time PCR. The expression of PCNA, MYCN, p53, cyclinD1, H3, H3K27M2, and H3K9Me2 was examined by Western blot analysis. In vitro invasion and the effects of the EHMT2 inhibitor (BIX-01294) were assessed in the Transwell chamber assay. Caspase-3 and -8 activities were measured by a Caspase-Glo assay kit. The level of global DNA methylation was measured by liquid chromatography-mass spectroscopy.
Results
BIX-01294, a specific inhibitor of EHMT2 (a key enzyme for histone H3 dimethylation at lysine-9), specifically decreases global H3K9Me2 level but not H3K27Me2. The inhibition of EHMT2 decreased proliferation of NB cells and induced apoptosis by increasing caspase 8/caspase 3 activity. BIX-01294 inhibited NB cell mobility and invasion. This was accompanied with a decreased expression of the MYCN oncogene. Inhibition of EHMT2 enhanced a doxorubicin induced inhibitory effect on cell proliferation. Finally EHMT2 inhibition modulated global DNA methylation levels in NB cells.
Conclusion
Our results demonstrate that histone lysine methylation is involved in cell proliferation, apoptosis, cell invasion, and global DNA methylation in human NB cells. Further understanding of this mechanism may provide insight into the pathogenesis of NB progression and lead to novel treatment strategies.
Keywords: EMHT2, Histone lysine methyltransferase, Cell proliferation, Global DNA methylation, Caspase, Phenotype, Neuroblastoma
Introduction
Neuroblastoma (NB) is the most common extracranial pediatric solid tumor and is characterized by its broad spectrum of clinical behavior [1, 2]. Although some patients have high cure rates, approximately 45% of the patients are presented with a widely disseminated and high-risk disease which remains difficult to cure. NB originates from the neural crest precursor cells as a result of genetic and epigenetic alterations that disrupt the normal developmental program. It has been reported that a CpG island methylator phenotype (CIMP) was a powerful prognostic factor, independent of age and stage in NB [3, 4]. Our group and others have shown that a poor outcome is associated with the hypermethylation of a number of tumor suppressor genes including RASSF1A, CASP8, and HIN-1 [5–7]. In preclinical studies, we demonstrated that NB tumor growth was impaired with agents that inhibit DNA methyltransferase and histone deacetylase, demonstrating the important role the epigenome plays in NB tumor growth [8–10].
Histone lysine methyltransferase EHMT2 is a key enzyme for histone H3 dimethylation at lysine-9 (H3K9me2), which is an epigenetic mark of gene suppression [11–13]. EHMT2 is highly expressed in human cancer cells and plays a key role in promoting cancer invasion and metastasis. The RNAi-mediated knockdown of EHMT2 in highly invasive lung cancer cells inhibited cell migration and invasion in vitro as well as metastasis in vivo [14]. The suppression of EHMT2 by knockdown inhibited cell growth in prostate cancer cells and led to morphologically senescent cells with telomere abnormalities [15]. These studies indicated that EHMT2 is likely required for the maintenance of the malignant phenotype. However the involvement of EHMT2 in the regulation of the NB phenotype and cell proliferation remains unknown. In this study, we investigated the effect of the inhibition of EHMT2 on NB proliferation and apoptosis, We also determined the effect of EHMT2 inhibition on cell invasion and global DNA methylation in NB cells.
Methods
NB cell culture
MYCN amplified NB cell lines, LA1-55n, IMR5 and NMB, were kindly provided by Dr. Susan L. Cohn at University of Chicago and the cell lines used in this study have been described previously[8, 16, 17]. They were grown at 5% CO2 in RPMI 1640 (Invitrogen, Carlsbad, CA) and supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Invitrogen), L-Glutamine, and antibiotics as previously described [10].
Cell treatment
NB cells were treated with either EHMT2 inhibitor BXI-01294 (EMD Millipore, Billerica, MA), doxorubicin (EMD Millipore), or in combination. The cells were also treated with 1 μM staurosporin (Sigma-Aldrich, St. Louis, MO) for 1 day and samples were used as a positive control for caspase activation.
Cell proliferation assay
The cell number was determined by the use of a hemocytometer. Trypan blue staining was used to differentiate between dead and live cells. LA1-55n, IMR-5 and NMB cells were plated in 6-well plates and cultured overnight. BIX-01294 was added and cells were incubated for 24 h and 48 h at the indicated concentrations.
Flow cytometry for analysis of cell cycle
LA1-55n cells were harvested at the completion of the respective BIX-01294 treatments. The cells were washed with a phosphate buffered saline (PBS, pH 7.4) twice then subsequently xed with 70% ethyl alcohol for 15 min on ice. The cells were then centrifuged at 2000 rpm to obtain pellets and the residual alcohol was aspirated. Cells were then digested with DNase-free RNase A (2 mg/ml) for 30 min at 37°C. Before ow cytometric analysis, cells were resuspended in 1 ml of 10 mg/ml propidium iodide (PI) (Sigma-Aldrich) for staining cellular DNA as previously described [15]. Cellular DNA content was then analyzed using an Epics XL-MCL Flow Cytometer (Beckman Coulter, Fullerton, CA).
Cell invasion assay
The in vitro invasive properties of LA1-55n cells were evaluated using BD Matrigel Biocoat invasion chambers (BD Biosciences, Bedford, MA). Briefly, after transwells and inserts were warmed up, 1 × 105/ml LA155n cells in 0.5 ml of media with 1% FBS were added to the upper surface of the filter for each assay and 0.75 ml of media with 10% FBS was used in the well as a chemoattractant. After 2 days, non-invaded cells in the upper chamber were removed with cotton swabs and invaded cells were fixed with 100% methanol for 2 min and then stained with 1% Toluidine blue in 1% borax solution for 2 min. For each insert, at least five random microscopic fields (200× magnification) were counted. Experiments were carried out in triplicates. The percentage of invasion was calculated by the number of cells invading through the Matrigel membrane divided by the number of cells which migrated through the control membrane.
cDNA synthesis and SYBR green real-time PCR
RNA was isolated from untreated and BIX-01294-treated LA1-55n cells using Trizol reagent (Invitrogen). Reverse transcription was performed using Superscript III (Invitrogen) according to the manufacturer’s instructions. SYBR green real-time PCR reactions were set up containing 1X Power SYBR Green Master Mix (Applied Biosystems, Foster City, CA), 250 nM forward and reverse primers in a 20 μl reaction. All assays were carried out in a 96-well format. Real-time fluorescent detection of PCR products was performed with a StepOne Plus Real-Time PCR System (Applied Biosystems) using the following thermocycling conditions: 1 cycle of 95°C for 10 min; 40 cycles of 95°C for 30 s, and 60°C for 1 min. The primer sequences for EHMT2 are sense: GCTCCGACGTGTGGTTTGC; antisense: CGATTTCCCACCCCAAGTC. The other primer sequences are as previously described [18]. The sequences of the primers for Real-time PCR were designed using the Primer Express software (Applied Biosystems). β-actin was used as an endogenous control for gene expression. For data analysis, the comparative method (ΔΔCt) was used to calculate the relative quantities of a nucleic acid sequence.
Western blot analysis
Total protein from LA1-55n cells were extracted by lysing the cells in cell lysis buffer (RIPA buffer: 20mM Tris-HCl–pH 7.5, 150mM NaCl, 1mM EDTA, 1mM EGTA, 1% IGEPAL, 2.5mM sodium pyrophosphate, 1mM β-glycerophosphate) containing protease and phosphatase inhibitor cocktails (Sigma-Aldrich). Protein concentrations were determined by using the Bradford protein assay kit (Bio-Rad, Hercules, CA). Equal amounts of total protein (10–25 μg) from cells were subjected to SDS-PAGE. Proteins were then transferred to a nitrocellulose membrane for 90 min at 100 V. Membranes were blocked for 1 h at room temperature in a Tris-buffered saline (TBS) containing 5% nonfat powdered milk. The membranes were then probed with a primary antibody in TBS with 2.5% nonfat powdered milk at concentrations from 1:500 to 1:20,000 dilutions and pre-incubated for overnight according to the manufacturer’s instructions for each antibody. In all cases, a secondary antibody labeled with horseradish peroxidase (GE Healthcare, Piscataway, NJ) was used at concentrations from 1:2,000 to 1:20,000 for 1 h at room temperature, and immuno-reactive bands were detected by using SuperSignal West Pico Chemi-luminescent Substrate (Pierce, Rockford, IL) and recorded on photosensitive film. The primary antibodies used for this study include: anti-cyclinD1 (Sigma-Aldrich), anti-PCNA (Proteintech,Chicago, IL), anti-p53 (Santa Cruz, Santa Cruz, CA), anti-MycN (Santa Cruz, Santa Cruz), anti-H3K9me2, H3K27Me2 (Abcam, Cambridge, MA), H3 (Abcam) and anti- β-actin (Santa Cruz).
Caspase assays
Caspase 3 and 8 activities in LA1-55n cells were measured using Caspase-Glo assay kit (Promega, Madison, WI). Briefly, the cells were plated in white-walled 96-well plates (Corning Incorporated, Lowell, MA), and incubated for 24 h. For caspase inhibition assay, caspase-8 inhibitor II, and negative control were added 30 min prior to adding BIX-01294. BIX-01294 was added into the medium, and incubated for 24h. The proluminescent substrate containing DEVD or LETD (EMD Millipore), which are cleaved by caspase 3 or caspase 8 respectively, was added to each well, and incubated at room temperature for 1 h. The luminescence of each sample was measured in a plate-reading luminometer as directed by the luminometer manufacturer (Promega).
Liquid chromatography-mass spectroscopy (LC/MS)
Analysis of total cytosine methylation was performed by LC/MS as described previously [19]. Briefly, genomic DNA from BIX-01294 treated and untreated LA1-55n cells was hydrolyzed to nucleosides by adding 5U nuclease P1 (Sigma-Aldrich) at 37°C for 2 hs, 0.002 units of venom phosphodiesterase I (Sigma-Aldrich) at 37°C for 2 hrs, and 0.5 units of alkaline phosphatase at 37°C for 1 h. Stock solutions of 2′-deoxycytidine and 5-methyl-2′-deoxycytidine were prepared in water. An eight-point stock mixture of a standard was carefully prepared to give an exact known concentration ratio of 2′-deoxycytidine to 5-methyl-2′-deoxycytidine. The concentration of 2′-deoxycytidine and 5-methyl-2′-deoxycytidine in each sample was calculated from the standard curve. Each DNA sample was analyzed in triplicate. 25 μl (80ng) of sample was injected into the LC and run through an Atlantis DC18 silica column (Waters Corporation, Milford, MA), and identification of 2′-deoxycytidine and 5-methyl-2′-deoxycytidine was obtained by mass spectra of chromatographic peaks.
Statistical analyses
Statistical analyses were performed using a two-tailed Student’s t test. A p value of < 0.05 was considered statistically significant.
Results
Effect of EHMT2 inhibition on LA1-55n cell proliferation
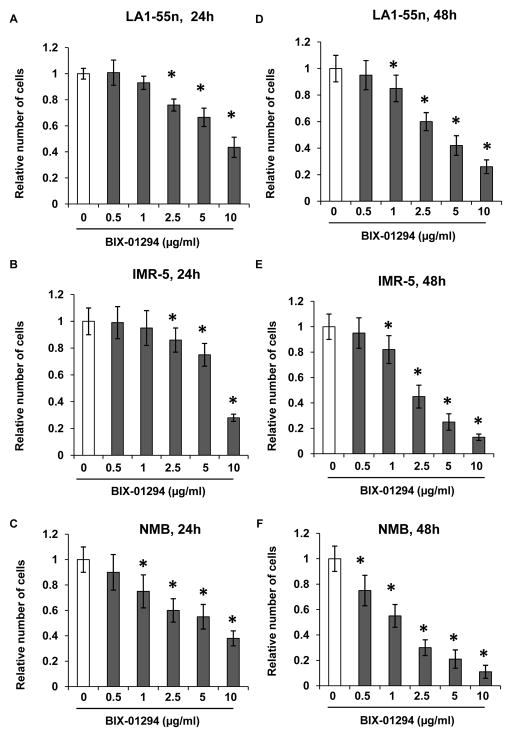
To test if EHMT2 regulates NB cell proliferation, LA1-55n, IMR-5 and NMB NB cells were cultured for 24 h and 48 h in media containing various concentrations of BIX-01294, the specific EHMT2 inhibitor. Trypan blue exclusion analysis revealed that EHMT2 inhibition suppressed LA1-55n cell proliferation in a dose and time dependent manner. For 24 h treatment, at concentrations ranging from 2.5 μg/ml to 10 μg/ml, BIX-01294 significantly inhibited LA1-55n, IMR-5 and NMB NB cell proliferation as shown in Figure 1A, 1B and 1C (p<0.01). BIX-01294 treatment resulted in decreased cell number of NMB cells even at the concentration of 1 μg/ml. However there is no significant change of cell number for LA1-55n and IMR-5 after treatment with BIX-01294 at concentration of 1 μg/ml, indicating that NMB cells are much more sensitive in response to BIX-01294 treatment.
Figure 1. Effect of EHMT2 inhibition on cell proliferation.
LA1-55n (A), IMR-5 (B) and NMB (C) cells were treated with BIX-01294 for 24 h at the concentration of 0.5, 1, 2.5, 5 and 10 μg/ml Proliferation was assessed by trypan blue exclusion to differentiate between dead and live cells. LA1-55n (D), IMR-5 (E) and NMB (F) cells were treated with BIX-01294 for 48 h at the concentration of 0.5, 1, 2.5, 5 and 10 μg/ml. Proliferation was assessed by trypan blue exclusion to differentiate between dead and live cells. * represents a statistical significance (p<0.01) in the difference between the vehicle- and BIX-01294-treated cells.
For 48 h treatment, three NB cells exhibited the inhibitory effect of BIX-01294 on cell proliferation at concentrations ranging from 1 μg/ml to 10 μg/ml. Moreover, BIX-01294 treatment resulted in decreased cell number of NMB cells at the concentration of 0.5 μg/ml. However there is no significant change of cell number for LA1-55n and IMR-5 after treatment with BIX-01294 at concentration of 0.5 μg/ml.
Effect of EHMT2 inhibition on the H3K9Me2 level in LA1-55n cells
To determine if BIX-01294 induced the inhibitory effect of NB cell proliferation via EHMT2, Western blot analysis was performed in LA1-55n cells using antibody against H3K9Me2, since EHMT2 is a key enzyme for histone H3 dimethylation at lysine-9 (H3K9me2). As shown in Figure 2A and 2B, global level of H3K9Me2 was decreased in LA1-55n cells treated with EHMT2 inhibitor (BIX-01294), as compared to control. However, global level of H3K27Me2 was not altered in LA1-55n cells treated with BIX-01294 as shown in Figure 2A and 2C, suggesting that BIX-01294 specifically targets EHMT2, leading to decreased level of H3K9Me2. To test if decreased level of H3K9me2 was due to an altered level of EHMT2 expression, real-time PCR was performed. As shown in Figure 2D, expression of EHMT2 was not significantly decreased in BIX-01295-treatedLA1-55n compared to control, indicating that BIX-01294-induced decreased level of H3K9Me2 is via inhibition of EHMT2 activity.
Figure 2. Effect of EHMT2 inhibition on H3K9Me2 level.
(A) LA1-55n cells were treated with BIX-01294 for 24 h at the concentration of 5 μg/ml. The effect of EHMT2 inhibition on the expression of H3K9me2, H3K27Me2, and H3 proteins in LA1-55n cells was determined by Western blot using antibody against H3K9Me2, H3K27Me2, and H3. β-actin was used as an endogenous control. (B) Summary data show BIX-01294 treatment at concentration of 5 μg/ml decreased the H3K9Me level. (C) Summary data show BIX-01294 treatment at concentration of 5 μg/ml does not affect the expression level of H3K27Me2. (D) LA1-55n cells were treated with BIX-01294 at the concentration of 2.5 μg/ml for 24 h. Total RNA was isolated, and real-time PCR was performed to determine the expression of EHMT2. β-actin was used as an endogenous control.
Effect of EHMT2 inhibition on LA1-55n cell cycle progression
To determine if inhibition of LA1-55n cell proliferation is due to an increase in cell cycle arrest, LA1-55n cells were plated and cultured for 24 h in media containing 10% FBS, and cells were treated with BIX-01294 or vehicle for 24 h. Cells were stained with propidium iodide to study the cell cycle progression. As shown in Figure 3A, EHMT2 inhibition showed no significant change in cell cycle progression.
Figure 3. Effect of EHMT2 inhibition on cell cycle progression.

(A) Cell cycle distribution was determined by flow cytometric analysis. LA1-55n cells were treated with either vehicle or BIX-01294 (2.5 μg/ml) for 24 h, and subjected to flow cytometric analysis. (B) The expression of cyclin D1, p53 and PCNA was determined by Western blot analysis in LA1-55n cells in the presence or absence of BIX-01294 (2.5 μg/ml). β-actin was used as a loading control. (C) p21 expression was examined by real-time PCR in LA1-55n cells in the presence or absence of BIX-01294 (2.5 μg/ml). β-actin was used as an endogenous control.
To determine if EHMT2 inhibition altered the expression of cell proliferation related genes, protein expression of p53, Cyclin D1 and PCNA genes was analyzed by Western blot. As shown in Figure 3B, there was no marked difference in expression of CCND1, p53, or PCNA between control and BIX-01294 treated NB cells. In addition, p21 RNA expression was not induced in LA1-55n cells treated with BIX-01294 (Figure 3C).
Effect of EHMT2 inhibition on motility and invasive abilities of LA1-55n cells
To determine if EHMT2 inhibition affect motility and invasion of LA1-55n cells, a transwell chamber assay was performed. As shown in Figure 4A, the number of migrating and invading LA1-55n cells were both lower in BIX-01294-treated cells than those in the non-treated cells (17±2.1 vs 12±2.01, p<0.01; 9±1.8 vs 3.8±1.2, p<0.01). Moreover, the index of invading cells was lower in BIX-01294 treated groups compared to control groups (53% vs 32%, p<0.01). The data suggested that inhibition of EHMT2 significantly suppressed the motility and invasive ability of LA1-55n cells.
Figure 4. Effect of EHMT2 inhibition on cell invasion and MYCN expression.
(A) Cells were cultured on BD Matrigel invasion chambers for 48 h. Cells were treated with BIX-01294 at a concentration of 1 μg/ml for 48 h in the chambers. Invaded cells were stained and the number of invaded cells in each field was counted under microscopic fields at 200× magnification. Experiments were carried out in triplicate. * p< 0.05 compared with control group. (B) LA1-55n cells were treated with BIX-01294 at a concentration of 5 μg/ml for 24 h. The expression of MYCN was examined by real-time PCR in the vehicle and BIX-01294 treated LA1-55n cells. (C) LA1-55n cells were treated with BIX-01294 at concentration of 5 μg/ml for 48 h. The expression of MYCN was examined by real-time PCR in the vehicle and BIX-01294 treated LA1-55n cells. * represents a statistical significance (p<0.01) between the vehicle- and BIX-01294-treated cells. (D) LA1-55n cells were treated with BIX-01294 at concentration of 1, 2.5, and 5 μg/ml for 24 h. The protein level of MYCN was examined by Western blot analysis in the cell lysis from vehicle and BIX-01294 treated LA1-55n cells. β-actin was used as an endogenous control.
To determine the correlation between the expression of MYCN and invasive ability of NB cells, MYCN expression was examined by real-time PCR in LA1-55n cells treated with BIX-01294. As shown in Figure 4B and 4C, the expression of MYCN was significantly decreased in BIX-01294- treated cells compared to control by 50% (24 h treatment), and 40% (48 h treatment) respectively (p<0.01). To determine whether protein level of MYCN was also decreased in response to BIX-01294 treatment, Western blot analysis was performed in LA1-55n cells treated with vehicle and BIX-10294. As shown in Figure 4D, EHMT2 inhibition resulted in decreased MYCN protein level at a dose dependent manner.
Effect of EHMT2 inhibition on caspase activity and doxorubicin-induced inhibitory effect of cell proliferation
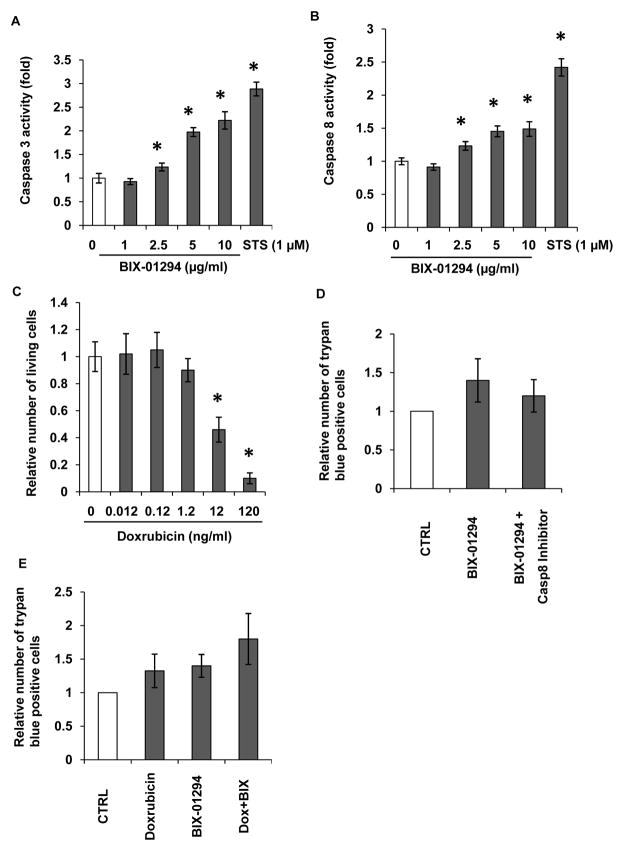
To determine if EHMT2 inhibition attenuated cell proliferation through the apoptotic pathway, caspase 3/7 and 8 activities were measured. As shown in Figure 5A, caspase 3/7 were activated after treatment with BIX-01294 in a dose-dependent manner. At a concentration of 2.5 μg/ml, BIX-01294 activated caspase 3/7 activity by 130% (p<0.01), At concentration of 10 μg/ml, BIX-01294 activated caspase 3/7 activity by 220% (p<0.01), As a positive control, STS at concentration of 1 μM activated caspase 3/7 activity by 280% compared to untreated cells (p<0.01). Activated caspase 8 was also seen in LA1-55n cells treated with BIX-01294. At concentrations ranging from 2.5 μg/ml to 10 μg/ml, BIX-01294 activated caspase 8 activity by 130% to 150% (p<0.01) as shown in Figure 5B. To determine if EHMT2 inhibition enhanced a doxorubicin induced inhibitory effect on cell proliferation, we initially determine the dose effect of doxorubicin on cell proliferation. As shown in Figure 5C, at concentrations above 12 ng/ml, doxorubicin resulted in a marked decrease in cell number, indicating that doxorubicin inhibited cell proliferation in a dose-dependent manner. To further determine the effect of EHMT2 inhibition on cell death, 40 μM of LETD caspase 8 inhibitor was added to the media 30 min prior to BIX-01294 treatment, and cells were incubated in10 μM of BIX-01294 for 24 h. The dead cells were counted by trypan blue exclusion. As shown in Figure 5D, EHMT2 inhibition significantly increases number of cell death. However, inhibition of caspase 8 activity by LETD attenuated BIX-01294-induced inhibitory effect of cell proliferation. Next we determined if EHMT2 inhibition enhanced a doxorubicin-induced inhibitory effect on cell proliferation. As shown in Figure 5E, BIX-01294 treatment at concentration of 5 μg/ml in combination with doxorubicin (12 ng/ml) markedly increased cell death as compared with either single treatment.
Figure 5. Effect of EHMT2 inhibition on caspase activity.
(A) Caspase 3 activities were measured in vehicle or BIX-01294 treated LA1-55n cells. The LA1-55n cells were treated with BIX-01294 at the concentrations of 1, 2.5, 5 and 10 μg/ml. Staurosporin (STS) at the concentration of 1 μM was used as a positive control for activation of caspase activity. (B) Caspase 8 activities were measured in vehicle or BIX-01294 treated LA1-55n cells. The LA1-55n cells were treated with BIX-01294 at the concentrations of 1, 2.5, 5 and 10 μg/ml. Staurosporin (STS) at the concentration of 1 μM was used as a positive control for activation of caspase activity. * represents a statistical significant difference (p<0.01) between the vehicle- and BIX-01294-treated cells. (C) LA1-55n cells were treated with various concentrations of doxorubicin (0.012, 0.12, 1.2, 12, 120 ng/ml) and proliferation was assessed by trypan blue exclusion. (D) 40 μM of LETD caspase 8 inhibitor was added to the media 30 min prior to BIX-01294 treatment, and cells were incubated in10 μM of BIX-01294 for 24 h. And trypan blue exclusion was performed to determine the relative number of dead cells. (E) LA1-55n cells were treated with either 12 ng/ml doxorubicin, 5 μg/ml BIX-01294 or in combination with 5 μg/ml BIX-01294 for 24 h. Trypan blue exclusion was performed to determine the percentage of dead cells.
Effect of EHMT2 inhibition on global DNA methylation
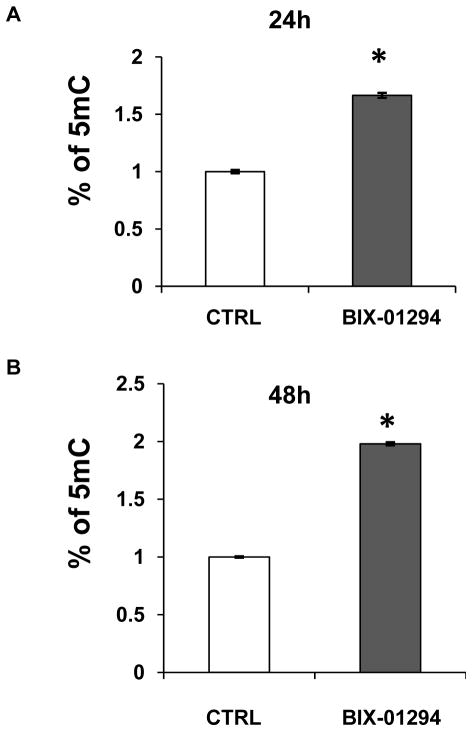
As a histone modifier, BIX-01294 modulated histone modification by inhibiting EHMT2 activity. To determine if the interplay between histone modification and DNA methylation occurred, LC/MS analysis was performed to determine the percentage of cytosine methylation in vehicle-treated and BIX-01294-treated LA1-55n cells. Using a standard curve average of 5-methylcytosine, the level of 5-methylcytosine was increased significantly (p<0.01) as compared to controls by 1.7 and 1.9 fold following 24 h and 48 h of BIX-01294 treatment (Figure 6A, 6B), suggesting that EHMT2 inhibition affected the pattern of DNA methylation in LA1-55n NB cells.
Figure 6. Effect of EHMT2 inhibition on global DNA methylation level.
Stock solutions of 2′-deoxycytidine and 5-methyl-2′-deoxycytidine were prepared in water. An eight-point stock mixture of a standard was carefully prepared to give an exact known concentration ratio of 2′-deoxycytidine and 5-methyl-2′-deoxycytidine. DNA methylation was determined by LC/MS. Genomic DNA was isolated from control and LA1-55n cells treated with 5 μg/ml BIX-01294 for 24h (A) or 48 h (B) respectively. DNA was then subjected to digestion with nuclease P1, venom phosphodiesterase I, and alkaline phosphatase respectively. The concentration of 2′-deoxycytidine and 5-methyl-2′-deoxycytidine in each sample was calculated from the standard curve. Each DNA sample was analyzed in triplicate. * represents the statistical significant difference (p<0.01) between the vehicle- and BIX-01294-treated cells.
Discussion
There are several lines of evidence indicating that the level of EHMT2 expression is associated with a malignant phenotype in a variety of cancer types [12, 14, 20]. Highly expressed EHMT2 has been reported in metastatic prostate cancer, lymphomas, and aggressive breast cancer [21]. In lung cancer, EHMT2 represents a metastasis promoter by silencing the cell adhesion molecular Ep-CAM [14]. Although the role of EHMT2 in NB progression has not been characterized, another well-known histone mark, trimethylation of histone 3 on lysine 27 (H3K27Me3), associated with gene silencing, was recently studied [22]. H3K27Me3 is mediated by the lysine methyltransferase EZH2, which is the enzymatically active component of the Polycomb Repressor Complex 2 (PRCR2) [21, 23]. RNAi-mediated knockdown of EZH2 or pharmacological inhibition of EZH2 inhibited cell growth and induced neurite extension with an increase in expression of tumor suppressors CLU,NGFR, and RUNX3, suggesting that EZH2 plays an essential role in maintenance of an undifferentiated phenotype in NB tumors [22]. We postulated that EHMT2 may also serve an important role in the growth regulation of NB cells. In this study, BIX-01294 induced an inhibitory effect on cell proliferation. At 2.5 μg/ml of BIX-01294, there is no marked cell arrest by flow cytometry. Because it has been reported that EHMT2 is involved in p21 promoter activity [11], we determined the expression level of p21 in BIX-01294 treated LA1-55n cells vs vehicle-treated cells. No marked induction of p21 expression was seen. In addition, the expression of cell proliferation related proteins of CCND1, p53, and PCNA was not changed between vehicle and BIX-01294 treated NB cells suggesting that the EHMT2 mediated inhibitory effect of gene expression such as p21 is cell type specific.
Next we evaluated the role of EHMT2 in cellular apoptosis. We found that the induction of apoptosis in NB cells played a role in regulation of cell proliferation. Caspase 3 and 8 activities were significantly increased within 24 h after treatment with the EHMT2 inhibitor. Studies using caspase 8 specific inhibitors further revealed the role of EHMT2 in regulation of cell growth via apoptotic pathways. Our studies are consistent with a previous report showing that knockdown of EHMT2 by siRNA resulted in the suppression of growth of cancer cells and possibly caused apoptotic cell death in bladder and lung cancer cells [20].
Doxorubicin is a well known apoptotic inducer in NB [24–26]. Doxorubicin-induced apoptosis in caspase-8-deficient neuroblastoma cells (SK-N-SH) is mediated through direct action on mitochondria [25]. In this study, we found that BIX-01294 enhanced the doxorubicin-induced inhibitory effect on cell proliferation in LA1-55n cells. We have previously reported that methylation of the caspase 8 promoter is higher in N-type tumorigenic LA1-55n cells than in S-type non-tumorigenic LA1-5s NB cells. The level of caspase 8 expression was upregulated in LA1-55n cells treated with 5-Aza-dC, a DNA methyltransferase inhibitor, suggesting that expression of caspase 8 is regulated by DNA methylation[18]. Although the expression of caspase 8 is relatively lower in LA1-55n cells due to its promoter methylation, the basal level of caspase 8 was sufficiently activated by BIX-01294 treatment, which was confirmed by enhanced activation of caspase 8. Moreover, the BIX-01294-induced cell death was attenuated by a caspase 8 inhibitor.
Although EHMT2 plays an essential role in histone modification leading to dysregulation of epigenetic pathways in cancer progression, little is known about how DNA methylation is mediated by EHMT2. In murine embryonic stem cells, EHMT2 is required for de novo DNA methylation and establishment of proviral silencing [27]. In this study, we demonstrated that EHMT2 inhibition resulted in modulation of global DNA methylation as analyzed by LC/MS. Inhibition of EHMT2 is capable of altering the distribution of DNA methylation, which is associated with changes in NB phenotype, including down regulation of MYCN expression and decrease in cell invasive ability. Our studies indicated that a small compound, BIX-01294, which specifically inhibits EHMT2 enzymatic activity is not only able to modulate histone modification by reducing H3K9Me2 levels as previously described [28, 29], but also alters genome wide DNA methylation. Interestingly, DNA hypermethylation profiles are associated with glioma subtypes and EZH2 mRNA expression [30]. According to our knowledge, this is the first report to show that EHMT2 inhibition mediates changes in malignant phenotype and global DNA methylation in NB. That is also consistent with our previous studies showing that an interplay between histone methylation and DNA methylation occurred [18], and DNA methylation and histone modifications act in concert to form an appropriate epigenome leading to modulation of cancer phenotype [31]. Although the molecular mechanisms underlying enhanced global DNA level regulated by BIX-01294 is unknown, there are some evidences showing that beside DNA methyltransferase, some other proteins including Tet 1, 2 and 3, AID, and APOBEC1 are also involved in modulation of global DNA methylation levels related to epigenetic reprogramming [32–35]. The extent of both DNA hypomethylation and hypermethylation in NB cells is likely to reflect distinctive biological and clinical characteristics, and understanding of the epigenetic reprogramming mechanisms that occur in NB should provide important new information about the relationship between NB phenotype and epigenetic state.
Taken together, our results indicate that the inhibition of EHMT2 is capable of inhibiting NB proliferation by enhancing caspase activities, leading to NB cells undergoing apoptosis. In addition, inhibition of EHMT2 modulated the phenotype and global DNA methylation in NB cells. Understanding the role of the interaction between histone modification and DNA methylation and how they function at the molecular level may shed light on the mechanisms involved in the development and progression of NB.
Acknowledgments
This work was supported in part by Children’s Neuroblastoma Cancer Foundation (QY), National Institutes of Health grants R01 HL075187 (JUR) and R01 HL059435 to (JUR).
Footnotes
Competing Interests: The authors declare that they have no competing interests.
References
- 1.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 2.Cohn SL, Pearson AD, London WB, Monclair T, Ambros PF, Brodeur GM, et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abe M, Ohira M, Kaneda A, Yagi Y, Yamamoto S, Kitano Y, et al. CpG island methylator phenotype is a strong determinant of poor prognosis in neuroblastomas. Cancer research. 2005;65:828–834. [PubMed] [Google Scholar]
- 4.Abe M, Watanabe N, McDonell N, Takato T, Ohira M, Nakagawara A, et al. Identification of genes targeted by CpG island methylator phenotype in neuroblastomas, and their possible integrative involvement in poor prognosis. Oncology. 2008;74:50–60. doi: 10.1159/000139124. [DOI] [PubMed] [Google Scholar]
- 5.Yang Q, Kiernan CM, Tian Y, Salwen HR, Chlenski A, Brumback BA, et al. Methylation of CASP8, DCR2, and HIN-1 in neuroblastoma is associated with poor outcome. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:3191–3197. doi: 10.1158/1078-0432.CCR-06-2846. [DOI] [PubMed] [Google Scholar]
- 6.Yang Q, Zage P, Kagan D, Tian Y, Seshadri R, Salwen HR, et al. Association of epigenetic inactivation of RASSF1A with poor outcome in human neuroblastoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:8493–8500. doi: 10.1158/1078-0432.CCR-04-1331. [DOI] [PubMed] [Google Scholar]
- 7.Buckley PG, Das S, Bryan K, Watters KM, Alcock L, Koster J, et al. Genome-wide DNA methylation analysis of neuroblastic tumors reveals clinically relevant epigenetic events and large-scale epigenomic alterations localized to telomeric regions. International journal of cancer Journal international du cancer. 2011;128:2296–2305. doi: 10.1002/ijc.25584. [DOI] [PubMed] [Google Scholar]
- 8.Yang QW, Liu S, Tian Y, Salwen HR, Chlenski A, Weinstein J, et al. Methylation-associated silencing of the thrombospondin-1 gene in human neuroblastoma. Cancer research. 2003;63:6299–6310. [PubMed] [Google Scholar]
- 9.Yang Q, Tian Y, Liu S, Zeine R, Chlenski A, Salwen HR, et al. Thrombospondin-1 peptide ABT-510 combined with valproic acid is an effective antiangiogenesis strategy in neuroblastoma. Cancer research. 2007;67:1716–1724. doi: 10.1158/0008-5472.CAN-06-2595. [DOI] [PubMed] [Google Scholar]
- 10.Yang Q, Liu S, Tian Y, Hasan C, Kersey D, Salwen HR, et al. Methylation-associated silencing of the heat shock protein 47 gene in human neuroblastoma. Cancer research. 2004;64:4531–4538. doi: 10.1158/0008-5472.CAN-04-0956. [DOI] [PubMed] [Google Scholar]
- 11.Kim JK, Esteve PO, Jacobsen SE, Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic acids research. 2009;37:493–505. doi: 10.1093/nar/gkn961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes & development. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes & development. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen MW, Hua KT, Kao HJ, Chi CC, Wei LH, Johansson G, et al. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer research. 2010;70:7830–7840. doi: 10.1158/0008-5472.CAN-10-0833. [DOI] [PubMed] [Google Scholar]
- 15.Kondo Y, Shen L, Ahmed S, Boumber Y, Sekido Y, Haddad BR, et al. Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PloS one. 2008;3:e2037. doi: 10.1371/journal.pone.0002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foley J, Cohn SL, Salwen HR, Chagnovich D, Cowan J, Mason KL, et al. Differential expression of N-myc in phenotypically distinct subclones of a human neuroblastoma cell line. Cancer research. 1991;51:6338–6345. [PubMed] [Google Scholar]
- 17.Cohn SL, Salwen H, Quasney MW, Ikegaki N, Cowan JM, Herst CV, et al. Prolonged N-myc protein half-life in a neuroblastoma cell line lacking N-myc amplification. Oncogene. 1990;5:1821–1827. [PubMed] [Google Scholar]
- 18.Yang Q, Tian Y, Ostler KR, Chlenski A, Guerrero LJ, Salwen HR, et al. Epigenetic alterations differ in phenotypically distinct human neuroblastoma cell lines. BMC cancer. 2010;10:286. doi: 10.1186/1471-2407-10-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah MY, Vasanthakumar A, Barnes NY, Figueroa ME, Kamp A, Hendrick C, et al. DNMT3B7, a truncated DNMT3B isoform expressed in human tumors, disrupts embryonic development and accelerates lymphomagenesis. Cancer research. 2010;70:5840–5850. doi: 10.1158/0008-5472.CAN-10-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho HS, Kelly JD, Hayami S, Toyokawa G, Takawa M, Yoshimatsu M, et al. Enhanced expression of EHMT2 is involved in the proliferation of cancer cells through negative regulation of SIAH1. Neoplasia. 2011;13:676–684. doi: 10.1593/neo.11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutation research. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Wang C, Liu Z, Woo CW, Li Z, Wang L, Wei JS, et al. EZH2 mediates epigenetic silencing of neuroblastoma suppressor genes CASZ1, CLU, RUNX3 and NGFR. Cancer research. 2011 doi: 10.1158/0008-5472.CAN-11-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 24.Hopkins-Donaldson S, Yan P, Bourloud KB, Muhlethaler A, Bodmer JL, Gross N. Doxorubicin-induced death in neuroblastoma does not involve death receptors in S-type cells and is caspase-independent in N-type cells. Oncogene. 2002;21:6132–6137. doi: 10.1038/sj.onc.1205879. [DOI] [PubMed] [Google Scholar]
- 25.Rebbaa A, Chou PM, Emran M, Mirkin BL. Doxorubicin-induced apoptosis in caspase-8-deficient neuroblastoma cells is mediated through direct action on mitochondria. Cancer chemotherapy and pharmacology. 2001;48:423–428. doi: 10.1007/s002800100375. [DOI] [PubMed] [Google Scholar]
- 26.Friesen C, Fulda S, Debatin KM. Induction of CD95 ligand and apoptosis by doxorubicin is modulated by the redox state in chemosensitive- and drug-resistant tumor cells. Cell death and differentiation. 1999;6:471–480. doi: 10.1038/sj.cdd.4400512. [DOI] [PubMed] [Google Scholar]
- 27.Leung DC, Dong KB, Maksakova IA, Goyal P, Appanah R, Lee S, et al. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5718–5723. doi: 10.1073/pnas.1014660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang Y, Zhang X, Horton JR, Upadhyay AK, Spannhoff A, Liu J, et al. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nature structural & molecular biology. 2009;16:312–317. doi: 10.1038/nsmb.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Molecular cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 30.Zheng S, Houseman EA, Morrison Z, Wrensch MR, Patoka JS, Ramos C, et al. DNA hypermethylation profiles associated with glioma subtypes and EZH2 and IGFBP2 mRNA expression. Neuro-oncology. 2011;13:280–289. doi: 10.1093/neuonc/noq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu S, Tian Y, Chlenski A, Salwen HR, Lu Z, Raj JU, et al. Valproic acid shows a potent antitumor effect with alteration of DNA methylation in neuroblastoma. Anti-cancer drugs. 2012 doi: 10.1097/CAD.0b013e32835739dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139:15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- 33.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes & development. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petersen-Mahrt S. DNA deamination in immunity. Immunological reviews. 2005;203:80–97. doi: 10.1111/j.0105-2896.2005.00232.x. [DOI] [PubMed] [Google Scholar]