Abstract
During early stages of B-lineage differentiation in bone marrow, signals emanating from IL-7 receptor and pre-B cell receptor (pre-BCR) are thought to synergistically induce proliferative expansion of progenitor cells. Paradoxically, loss of pre-BCR signaling components is associated with leukemia in both mice and humans. Exactly how progenitor B cells perform the task of balancing proliferative burst dependent on IL-7 with the termination of IL-7 signals and the initiation of LC gene rearrangement remains to be elucidated. In this report, we provide genetic and functional evidence that the cessation of IL-7 response of pre-B cells is controlled via a cell-autonomous mechanism that operates at a discreet developmental transition inside Fraction C’ (Large Pre-BII) marked by transient expression of c-Myc. Our data indicates that pre-BCR cooperates with IL-7R in expanding pre-B cell pool, but it is also critical to control differentiation program shutting off c-Myc gene in large pre-B cells.
INTRODUCTION
B lymphopoiesis progresses through a developmental program that links the ordered rearrangement of immunoglobulin gene segments with cellular expansion and differentiation events (1–6). The nomenclature of developing B cell subsets is based on expression of cell surface markers and the rearrangement status of the IgH and IgL loci (7). The earliest B-lineage progenitors are contained within the Fraction A (Fr. A) of bone marrow (also termed “Pro-B”). These cells begin the rearrangement of IgH chain genes and differentiate into Fr. B and C distinguished by a pattern of expression of CD24 and BP.1 (Fr. B and C are collectively termed “Pre-BI”). The completion of IgH rearrangement and the expression of µHC protein on the cell surface with the surrogate light chain (LC) proteins (VpreB and λ5) to form the pre-B cell receptor (pre-BCR) marks the transition to Fr. C’ (also termed “Large Pre-BII”) marked by high levels of CD24 and CD25. Fr. C’ cells are large and undergo rapid proliferative expansion critically dependent on IL-7 and the pre-BCR. Subsequently, however, the pre-BCR induces differentiation of C’ cells into Fr. D (also termed “Small Pre-BII”), which cease to proliferate, upregulate RAG-1/-2 genes, and begin the rearrangement of LC gene loci (3, 4).
The exact mechanism that controls these transitions remains incompletely understood. Intriguingly, the loss of pre-BCR signaling components results not only in a developmental arrest of pre-B cells, but in both mice and humans also leads to development of spontaneous pre-B cell leukemias (8–11). In this context, the pre-BCR signaling is initiated by tyrosine phosphorylation of ITAM sequences in Igα and Igβ (CD79a/CD79b) subunits followed by recruitment and activation of Syk tyrosine kinase and the assembly of the SLP65/BLNK signalosome (4, 12, 13). On the other hand, IL-7 initiates signaling events by heterodimerization of the IL-7Rα chain and γ chain, leading to trans-phosphorylation of JAK3 and JAK1, phosphorylation of the IL-7Rα chain, and recruitment of STAT proteins, STAT5A and STAT5B (14). This permits STATs to dimerize and translocate to the nucleus, where they act as transcription factors for a number of target genes. The IL-7Rα chain also serves for direct recruitment and activation of the p85 subunit of phosphotidylinositol-3-OH kinase (PI3-K) that is responsible for many downstream survival and proliferation related events (15).
Thus, while signals emanating from both IL-7R and the pre-BCR synergistically regulate proliferative expansion of early stage B lineage cells by promoting c-Myc expression and their survival (16), paradoxically, the pre-BCR complex is also critical for cell cycle exit of large pre-B cells and their differentiation into small pre-B cells, as the loss of pre-BCR signaling results in an arrest in differentiation and leads to pre-B cell lymphoblastic leukemia characterized by expression of c-Myc (17, 18). In this report, we have used a fluorescently tagged c-Myc gene “knock-in” approach to track transient expression of c-Myc protein in developing B cells. Strikingly, using this approach we have discovered a previously unrecognized developmental stage of large pre-B cells. We present functional and biochemical evidence that during large pre-B cell differentiation, the ability of cells to respond to IL-7 receptor stimulation is controlled in a cell-autonomous manner at a new developmental transition we term C’-1 to C’-2.
MATERIALS AND METHODS
Mice
c-MyceGFP mice were previously described (19). Rag-2−/− and Rag-1−/− mice were a gift from M. White (Washington University). Mice were maintained in the specific-pathogen-free facility in accordance with institutional policies.
Flow cytometry
Single-cell suspensions were stained with antibodies to AA4.1, B220, CD43, CD127, CD132, C-Kit, CXCR4 and SLC/pre-BCR (BD Pharmingen), and CD24, CD25 (eBioscience) BP.1 (Biolegend), and pSTAT5 and pFoxO1/3a (Cell Signal), and IgM (Southern Biotech), according to standard protocols. Cell sorts were performed on FACS Aria II (Becton Dickinson). Intracellular stains were performed by fixing the cells in 2% PFA for 15 minutes followed by washing with permeabilization buffer (PBS+2% FBS and 0.1% Saponin).
OP-9 cell cultures
Sorted B cell subsets were cultured in the presence of 10ng/ml IL-7 in DMEM-10 media in 96 well flat bottom plates with a layer of 104 OP-9 cells and analyzed as indicated.
Quantitative RT-PCR analysis
Sorted cell subsets were harvested in Trizol (Invitrogen), RNA was extracted, and cDNA was generated using SuperScript First-Strand RT system (Invitrogen) according to manufacturer’s instructions. RT-PCR primers are listed in Supplementary Table I. Quantitative real-time PCR was performed with Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) and a Stratagene Mx3000P.
PCR Analysis of Igκ Rearrangements
C’-1 and C’-2 cells were sorted via FACS and DNA was extracted. PCR reactions were performed for Vκ-Jκ and IL-2 as a loading control (Supplementary Table I), and PCR products were separated and analyzed on a Phosphorimager after Southern blotting using Jκ probe using standard protocols.
Auto-reconstitution assays
c-MyceGFP mice were sublethally irradiated (500 Rads) and sacrificed at the time points indicated, bone marrow was harvested, and B cell populations were analyzed by flow cytometry.
Statistical analysis
Data are expressed throughout as mean ± SD. Datasets were compared using the 2-tailed unpaired Student t test. Differences were considered statistically significant (*) when P < .05, and (**)P < .005.
RESULTS AND DISCUSSION
c-Myc marks a novel stage inside the Fr. C’ (Large-Pre-BII) subset of precursor B cells
Precursor B cells have to coordinate significant proliferative burst dependent on IL-7 with the termination of IL-7 signals to permit light chain gene rearrangement (1–6, 20). However, IL-7 receptor is still present on small pre-B cells (14), and despite some evidence that IL-7R+ large pre-B cells may migrate away from IL-7 producing stroma (6) the mechanism by which IL-7 receptor signaling is terminated remains unclear. In order to precisely determine the stage of pre-B cell development at which the large Fr. C’ cells cease to proliferate we decided to directly visualize c-Myc protein abundance in live cells ex vivo. To this end, we used a mouse model in which eGFP reporter is engineered to replace the start codon of Myc in the endogenous locus (c-MyceGFP) in which c-Myc is still functionally expressed (19). This model is uniquely suited for these analyses because c-Myc expression in early lymphoid progenitors is thought to be primarily regulated by a post-translational mechanism (21).
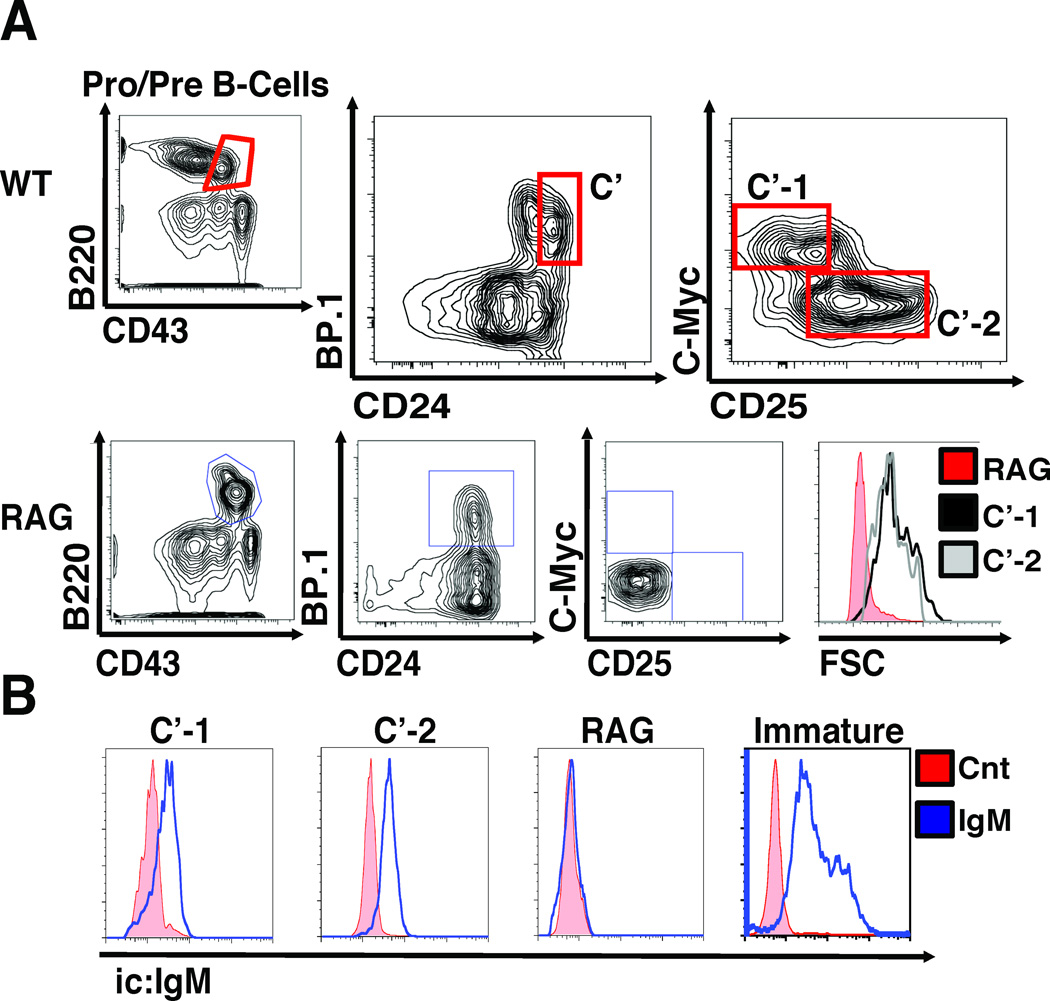
Indeed, our analyses of the distribution of differentiation markers including AA4.1, B220, c-kit, CD43, CD24, CD25, BP.1, and c-MyceGFP in bone marrow cells from c-MyceGFP/eGFP and control mice showed that expression of c-MyceGFP marked a distinct stage of developing pre-B cells within Fr. C’ (Large Pre-BII) (Fig.1A, Supplemental Figure S1A). Importantly, analyses of the expression of µHC genes showed that virtually all c-MyceGFP+ cells express intracellular µHC protein (Fig.1B) and SLC components (Supplemental Figure S1B) which are the defining traits of Fr. C’ cells. Consistent with a view that c-MyceGFP+ cells comprise Fr. C’, they appear large (Fig.1A) and express c-kit (Supplemental Figure S1C). Strikingly, however, c-MyceGFP+ cells completely lack expression of CD25, a conventional marker of C’ cells. Therefore, expression of c-Myc protein appears to mark a previously unrecognized stage within the Fr. C’ large pre-B cells, which we further refer to as C’-1. On the other hand, our data clearly indicates that large pre-B cells marked by expression of CD25 no longer contain c-Myc protein (Fig.1A). We hypothesized that this latter subset, we further refer to as C’-2, may encompass a stage that acquires the ability to rearrange LC genes.
Figure 1. Phenotypic classification of a novel c-Myc-positive stage of large pre-B cells.
(A) Flow cytometry analysis of c-Myc, CD25 expression, and cell size in bone marrow cells from c-MyceGFP/eGFP and RAG mice gated as B220+/CD43+ cells of Fr. A (CD24−/BP.1−), B (CD24+/BP.1−), C (CD24med/BP.1+/CD25−) and C’ (CD24+/BP.1+/CD25−); note the absence of C’-1 (c-Myc+/CD25−) and C’-2 (c-Myc−/CD25+) cells in RAG mice. Data is representative of over 20 independent experiments (n>30). (B) Intracellular µHC staining. Fr. C’-1, and C’-2 bone marrow cells were sorted by FACS from c-MyceGFP/eGFP mice, labeled with anti-µHC or isotype control, and analyzed by flow cytometry, as indicated. Sorted B220+/IgD− immature B cells from spleen of WT mice and B220+/CD43+ cells from bone marrow of Rag-1-deficient mice were used as a positive and a negative control, respectively. Data shown is representative of two independent experiments (n=4).
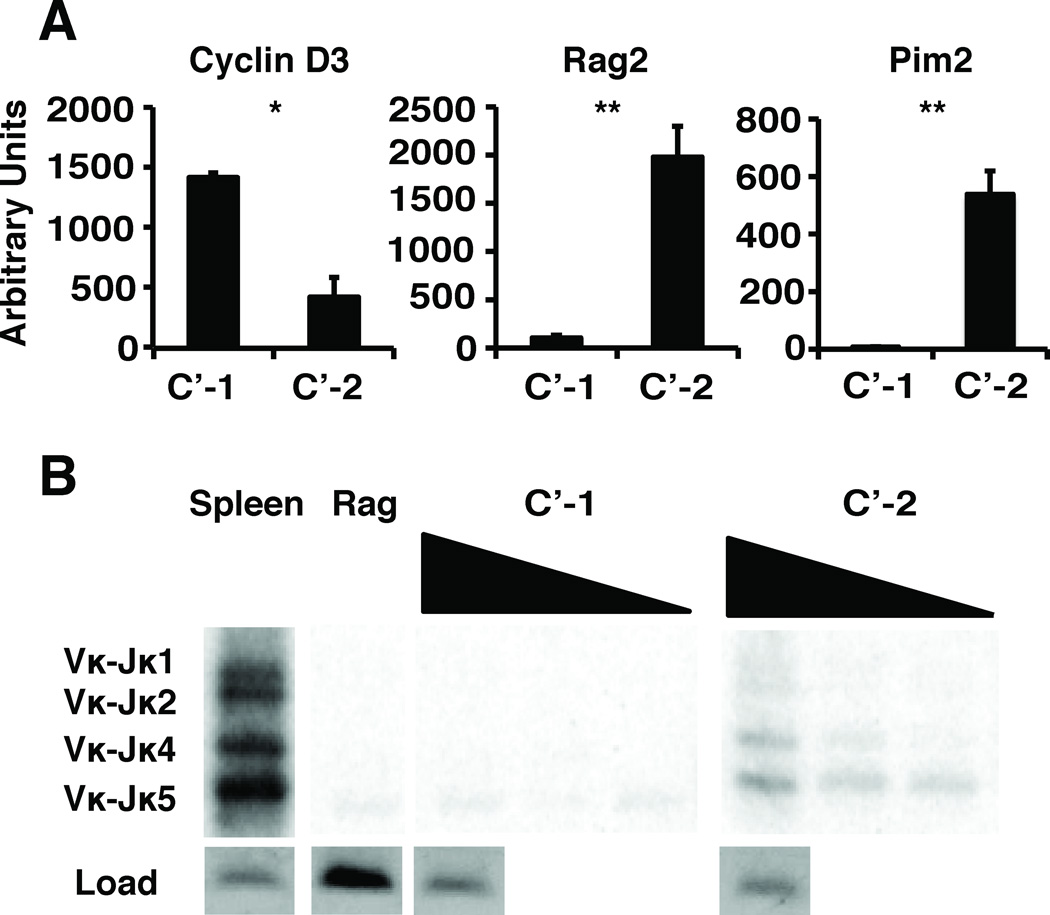
In agreement with this view, our analyses show that C’-1 express high levels of Ccnd3 gene transcripts encoding cyclin D3 that are virtually absent in C’-2 cells (Fig.2A). In sharp contrast, C’-2 cells have abundant expression of RAG2 and double-stranded DNA break marker, Pim2, and appear to actively rearrange their Ig LC genes as indicated by the presence of recombined V-Jκ chain gene segments (Fig.2A,B). Moreover, our intracellular staining analyses with phosphospecific antibodies revealed significantly higher levels of pFoxO1/3a and pSTAT5 in C’-1 cells, consistent with a scenario that these cells are responding to IL-7 in vivo (Supplemental Figure S1D). Taken together, we interpret these data as indicating that the assembly and expression of the pre-BCR complex promotes a discrete stage of pre-B cell development marked by expression of c-Myc protein (C’-1), which is distinct from the large but non-proliferative CD25+ stage (C’-2). Therefore, it appears that pre-BCR initially contributes to C’ cell proliferation during a discrete C’-1 stage but subsequently may induce differentiation to a non-proliferative C’-2 stage at which rearrangements of Ig LC genes are initiated.
Figure 2. Cell cycle and LC gene rearrangement analysis of C’-1 and C’-2 stage cells.
(A) qRT-PCR analysis of FACS sorted cells was performed to detect RAG2 (RAG2), PIM2 (PIM2) and ccnd3 (Cyclin D3) transcripts. Data is a composite of 3 independent experiments (n=6). (B) Detection of LC gene (Vκ-Jκ) rearrangements in genomic DNA isolated from FACS sorted cells, as indicated. IL-2 gene was used as a loading control, WT splenic DNA was used as a positive control, and Rag-1-deficient pro-B cell DNA was used as a negative control. Data shown is representative of 4 independent experiments (n=4).
Cell-autonomous control of IL-7 driven proliferation in novel subsets of large pre-B cells
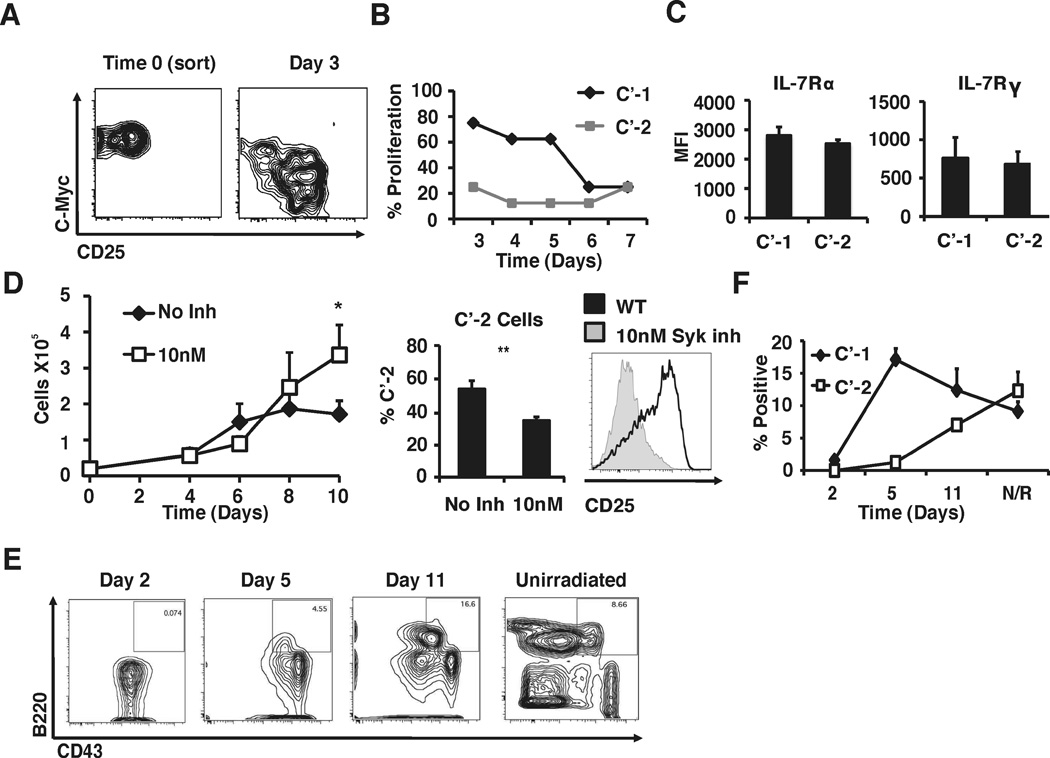
Given these results, we hypothesized that pro-proliferative signals emanating from IL-7R in large pre-B cells may be suppressed at the C’-1/C’-2 transition in a cell-autonomous manner. To test this hypothesis, we analyzed the ability of isolated C’-1 and C’-2 cells to respond with proliferation to IL-7 treatment in vitro. Despite similar levels of surface expression of IL-7Rα and IL-7Rγ chains C’-1 cells proliferate in response to IL-7 in OP9 cultures in vitro, while C’-2 cells completely fail to proliferate (Fig.3A–C). Remarkably, C’-1 cells appear to spontaneously differentiate into C’-2 cells in vitro with the accompanying loss of their ability to proliferate in response to IL-7. Moreover, blocking pre-BCR signaling by treatment with a specific Syk kinase inhibitor leads to an arrest at the C’-1 to C’-2 transition in vitro with an accompanying increase in cell proliferation (Fig.3D). Thus, while the generation of C’-2 stage cells is virtually blocked by Syk inhibitor in this system, developmentally arrested C’-1 cells remain viable and retain their proliferative properties (Fig.3D). We interpret these results as suggesting that expression of c-Myc marks C’-1 stage cells that respond to IL-7R signals with proliferation, but later differentiate into C’-2 stage cells that no longer proliferate in response to IL-7.
Figure 3. Functional and precursor-product relationship analysis of C’-1 and C’-2 cells.
(A) C’-1 cells were sorted via FACS and then co-cultured with OP-9 stromal cells supplemented with IL-7. After 3 days in culture, cells were analyzed by flow cytometry to assess differentiation into C’-2 cells. Data shown is representative of 5 independent experiments (n=8). (B) Freshly isolated C’-1 and C’-2 cells were directly FACS sorted at 10 cells/well into microtiter plates and co-cultured with OP-9 stroma in the presence of 10ng/ml IL-7. Wells were scored daily to determine IL-7-induced proliferation by microscopic observation. Data is representative of 2 independent experiments (n=4). (C) Flow cytometry analysis of IL-7Rα and cγ in C’-1 and C’-2 cells. MFI is shown, data is composite of 3 independent experiments (n=6). (D) Analysis of pre-B cell proliferation in the presence of Syk kinase inhibitor (BAY61-3606). B220+/CD43+/CD25− cells were FACS sorted and cultured in the presence of indicated concentrations of IL-7, with or without BAY61-3606 (10nM). Cells were counted every 2 days and analyzed for CD25 expression on day 10. Data is composite of 4 independent experiments (n=6). (E) Autoreconstitution analysis of pre-B cell repopulation kinetics. c-MyceGFP/eGFP mice were sublethally irradiated (500 rads) and pro/pre-B cell populations were analyzed as indicated. (F) Percent of C’-1 and C’-2 cells were measured as well as total B220+/CD43+ populations. Data is representative of 4 independent experiments (n>10).
In addition to IL-7 and pre-BCR, other environmental cues such as cytokines induced by stress or tissue injury can exert progenitor cell proliferation, differentiation, and modulate c-Myc protein abundance in progenitor cells (22–24). To further address the precursor/product relationship between the C’-1 and the C’-2 subsets, we treated c-MyceGFP/eGFP mice with low doses of γ-radiation inducing ablation of B-lineage cells and activation of quiescent progenitor cells. Analyses of bone marrow at 24 h post irradiation showed an absence of virtually all B-lineage cells in irradiated mice (Fig.3E). Strikingly, analyses of the repopulation kinetics revealed that development of C’-1 cells preceded C’-2 cells by 3–5 days (Fig.3F) consistent with the view that the former subset represents an earlier stage as compared to the latter.
Taken together, these observations indicate that expression of c-Myc protein in C’ large pre-B cells is subject to pre-BCR dependent regulation. Thus, the pre-BCR may function to suppress signals emanating from IL-7R and/or, conceivably, is involved in the recruitment of downstream effectors necessary for the suppression of c-Myc expression and the induction of C’-1 cell differentiation to the non-proliferative C’-2 stage. In this context, our analyses revealed virtually identical levels of IL-7Rα and IL-7Rγ chains on the surface of C’-1 and C’-2 cells (Fig.3C). Therefore, the more mature C’-2 cells maintain IL-7R expression, but are unresponsive to IL-7.
One model proposes that pre-BCR signaling upregulates the transcription factor IRF4 that in turn promotes higher expression of the chemokine receptor CXCR4 on pre-B cells and their movement, directed by the chemokine CXCL12, away from stromal cells expressing IL-7 thereby attenuating IL-7 signaling (6). Notably, IRF4 also induces germline transcription of Ig LC loci and, consequently, their accessibility for recombination (6, 25, 26). However, in our hands both C’-1 and C’-2 stage cells show similar levels of expression of surface CXCR4 (Supplemental Figure S1E), even though the developmental transition from C’-1 to C’-2 is accompanied by dramatic increase in IRF4 production (Supplemental Figure S1F). Moreover, our analyses of expression of critical transcription factors involved in pre-B cell differentiation indicate that C’-2 cells contain significantly higher levels of Irf4 and Izkf1 gene (encoding Ikaros) as compared to C’-1 (Supplemental Figure S1F) consistent with the view that these factors negatively regulate pre-B-cell proliferation by directly suppressing c-Myc expression (3). Notably, our data also reveals a significant increase in the Socs-1 gene expression at the transition from C’-1 to C’-2 (Supplemental Figure S1F). These results indicate that the increase in Socs-1 expression we find in C’-2 cells could, conceivably, contribute to downregulation of STAT5 signaling in this population as suggested by previously published studies (27). We note, however, that Socs-1 expression levels continue to dramatically rise in Fr. D cells (Supplemental Figure S1F).
Thus, whatever the mechanism, we show here the first evidence of existence of two distinct subsets of Fr. C’ cells distinguished by their ability to respond to IL-7. Our data clearly indicates that termination of IL-7 signaling is a cell-autonomous process critically dependent on signals emanating from the pre-BCR. Strikingly, pharmacological inhibition of the pre-BCR signaling leads to a dramatic enhancement of proliferation in response to IL-7 that coincides with a developmental arrest at the C’-1 stage expressing c-Myc. We propose that these results help explain observations of spontaneous leukemia upon loss of pre-BCR in mice and humans (8–10).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Chyi Hsieh, Jerry Lio, Jeff Bednarski, and Beth Helmink for discussion and experimental help.
Footnotes
This work was supported by National Institutes of Health grants R01AI061077, R01AI073718 (W. Swat), DK083756 and DK043351 (R.J. Xavier), AI047829 (B. Sleckman), K01 DK078318 (D. Bhattacharya), Leukemia & Lymphoma Society Scholar Award (W. Swat), and Special Fellowship Award (D.B. Graham).
REFERENCES
- 1.Oda A, Ono T, Yamamoto M, Goitsuka R, Kitamura D. PKC eta directs induction of IRF-4 expression and Ig kappa gene rearrangement in pre-BCR signaling pathway. Int Immunol. 2008;20:1417–1426. doi: 10.1093/intimm/dxn101. [DOI] [PubMed] [Google Scholar]
- 2.Geier JK, Schlissel MS. Pre-BCR signals and the control of Ig gene rearrangements. Semin Immunol. 2006;18:31–39. doi: 10.1016/j.smim.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Ma S, Pathak S, Mandal M, Trinh L, Clark MR, Lu R. Ikaros and Aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol Cell Biol. 2010;30:4149–4158. doi: 10.1128/MCB.00224-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandal M, Powers SE, Ochiai K, Georgopoulos K, Kee BL, Singh H, Clark MR. Ras orchestrates exit from the cell cycle and light-chain recombination during early B cell development. Nat Immunol. 2009;10:1110–1117. doi: 10.1038/ni.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson K, Reddy KL, Singh H. Molecular pathways and mechanisms regulating the recombination of immunoglobulin genes during B-lymphocyte development. Adv Exp Med Biol. 2009;650:133–147. doi: 10.1007/978-1-4419-0296-2_11. [DOI] [PubMed] [Google Scholar]
- 6.Johnson K, Hashimshony T, Sawai CM, Pongubala JM, Skok JA, Aifantis I, Singh H. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 2008;28:335–345. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 7.Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 8.Jumaa H, Bossaller L, Portugal K, Storch B, Lotz M, Flemming A, Schrappe M, Postila V, Riikonen P, Pelkonen J, Niemeyer CM, Reth M. Deficiency of the adaptor SLP-65 in pre-B-cell acute lymphoblastic leukaemia. Nature. 2003;423:452–456. doi: 10.1038/nature01608. [DOI] [PubMed] [Google Scholar]
- 9.Hendriks RW, Kersseboom R. Involvement of SLP-65 and Btk in tumor suppression and malignant transformation of pre-B cells. Semin Immunol. 2006;18:67–76. doi: 10.1016/j.smim.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Nakayama J, Yamamoto M, Hayashi K, Satoh H, Bundo K, Kubo M, Goitsuka R, Farrar MA, Kitamura D. BLNK suppresses pre-B-cell leukemogenesis through inhibition of JAK3. Blood. 2009;113:1483–1492. doi: 10.1182/blood-2008-07-166355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nahar R, Ramezani-Rad P, Mossner M, Duy C, Cerchietti L, Geng H, Dovat S, Jumaa H, Ye BH, Melnick A, Muschen M. Pre-B cell receptor-mediated activation of BCL6 induces pre-B cell quiescence through transcriptional repression of MYC. Blood. 2011;118:4174–4178. doi: 10.1182/blood-2011-01-331181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herzog S, Storch B, Jumaa H. Dual role of the adaptor protein SLP-65: organizer of signal transduction and tumor suppressor of pre-B cell leukemia. Immunol Res. 2006;34:143–155. doi: 10.1385/IR:34:2:143. [DOI] [PubMed] [Google Scholar]
- 13.Martensson IL, Almqvist N, Grimsholm O, Bernardi AI. The pre-B cell receptor checkpoint. FEBS Lett. 2010;584:2572–2579. doi: 10.1016/j.febslet.2010.04.057. [DOI] [PubMed] [Google Scholar]
- 14.Milne CD, Paige CJ. IL-7: a key regulator of B lymphopoiesis. Semin Immunol. 2006;18:20–30. doi: 10.1016/j.smim.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Werner M, Hobeika E, Jumaa H. Role of PI3K in the generation and survival of B cells. Immunol Rev. 2010;237:55–71. doi: 10.1111/j.1600-065X.2010.00934.x. [DOI] [PubMed] [Google Scholar]
- 16.Peschon JJ, Morrissey PJ, Grabstein KH, Ramsdell FJ, Maraskovsky E, Gliniak BC, Park LS, Ziegler SF, Williams DE, Ware CB, Meyer JD, Davison BL. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med. 1994;180:1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoelbl A, Schuster C, Kovacic B, Zhu B, Wickre M, Hoelzl MA, Fajmann S, Grebien F, Warsch W, Stengl G, Hennighausen L, Poli V, Beug H, Moriggl R, Sexl V. Stat5 is indispensable for the maintenance of bcr/abl-positive leukaemia. EMBO Mol Med. 2010;2:98–110. doi: 10.1002/emmm.201000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, Strasser A, Busslinger M. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol. 2010;11:171–179. doi: 10.1038/ni.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang CY, Bredemeyer AL, Walker LM, Bassing CH, Sleckman BP. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur J Immunol. 2008;38:342–349. doi: 10.1002/eji.200737972. [DOI] [PubMed] [Google Scholar]
- 20.Ochiai K, Maienschein-Cline M, Mandal M, Triggs JR, Bertolino E, Sciammas R, Dinner AR, Clark MR, Singh H. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat Immunol. 2012 doi: 10.1038/ni.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vervoorts J, Luscher-Firzlaff J, Luscher B. The ins and outs of MYC regulation by posttranslational mechanisms. J Biol Chem. 2006;281:34725–34729. doi: 10.1074/jbc.R600017200. [DOI] [PubMed] [Google Scholar]
- 22.de Alboran IM, O'Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- 23.Kelly K, Cochran BH, Stiles CD, Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983;35:603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- 24.Maclean KH, Keller UB, Rodriguez-Galindo C, Nilsson JA, Cleveland JL. c-Myc augments gamma irradiation-induced apoptosis by suppressing Bcl-XL. Mol Cell Biol. 2003;23:7256–7270. doi: 10.1128/MCB.23.20.7256-7270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma S, Turetsky A, Trinh L, Lu R. IFN regulatory factor 4 and 8 promote Ig light chain kappa locus activation in pre-B cell development. J Immunol. 2006;177:7898–7904. doi: 10.4049/jimmunol.177.11.7898. [DOI] [PubMed] [Google Scholar]
- 26.Ma S, Pathak S, Trinh L, Lu R. Interferon regulatory factors 4 and 8 induce the expression of Ikaros and Aiolos to down-regulate pre-B-cell receptor and promote cell-cycle withdrawal in pre-B-cell development. Blood. 2008;111:1396–1403. doi: 10.1182/blood-2007-08-110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corfe SA, Rottapel R, Paige CJ. Modulation of IL-7 thresholds by SOCS proteins in developing B lineage cells. J Immunol. 2011;187:3499–3510. doi: 10.4049/jimmunol.1100424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.