Abstract
Viruses are usually thought to form parasitic associations with hosts, but all members of the family Polydnaviridae are obligate mutualists of insects called parasitoid wasps. Phylogenetic data founded on sequence comparisons of viral genes indicate that polydnaviruses in the genus Bracovirus (BV) are closely related to pathogenic nudiviruses and baculoviruses. However, pronounced differences in the biology of BVs and baculoviruses together with high divergence of many shared genes make it unclear whether BV homologs still retain baculovirus-like functions. Here we report that virions from Microplitis demolitor bracovirus (MdBV) contain multiple baculovirus-like and nudivirus-like conserved gene products. We further show that RNA interference effectively and specifically knocks down MdBV gene expression. Coupling RNAi knockdown methods with functional assays, we examined the activity of six genes in the MdBV conserved gene set that are known to have essential roles in transcription (lef-4, lef-9), capsid assembly (vp39, vlf-1), and envelope formation (p74, pif-1) during baculovirus replication. Our results indicated that MdBV produces a baculovirus-like RNA polymerase that transcribes virus structural genes. Our results also supported a conserved role for vp39, vlf-1, p74, and pif-1 as structural components of MdBV virions. Additional experiments suggested that vlf-1 together with the nudivirus-like gene int-1 also have novel functions in regulating excision of MdBV proviral DNAs for packaging into virions. Overall, these data provide the first experimental insights into the function of BV genes in virion formation.
Author Summary
Microorganisms form symbiotic associations with animals and plants that range from parasitic (pathogens) to beneficial (mutualists). Although numerous examples of obligate, mutualistic bacteria, fungi, and protozoans exist, viruses are almost always considered to be pathogens. An exception is the family Polydnaviridae, which consists of large DNA viruses that are obligate mutualists of insects called parasitoid wasps. Prior studies show that polydnaviruses in the genus Bracovirus evolved approximately 100 million years ago from a group of viruses called nudiviruses, which are themselves closely related to a large family of insect pathogens called baculoviruses. Polydnaviruses are thus of fundamental interest for understanding the processes by which viruses can evolve into mutualists. In this study we characterized the composition of virus particles from Microplitis demolitor bracovirus (MdBV) and conducted functional experiments to assess whether BV genes share similar functions with related essential baculovirus replication genes. Our results indicate that several genes in MdBV retain ancestral functions, but select other genes have novel functions unknown from baculoviruses. Our results also provide the first experimental data on the function of polydnavirus replication genes and enhance understanding of the similarities between these viruses and their pathogenic ancestors.
Introduction
Microorganisms form associations with metazoan hosts that range from beneficial symbiosis (mutualists) to parasitic (pathogens). Mutualists serve as important sources of evolutionary innovation for hosts, while pathogens often acquire genes from hosts or other organisms that facilitate their own survival and cause disease. Although most research on obligate mutualists focuses on bacteria, several fungi and protozoans are also known to form beneficial partnerships [1]–[3]. Viruses in contrast are almost always thought to form parasitic associations [4]–[6]. A notable exception to this is the family Polydnaviridae, which consists entirely of large DNA viruses that are obligate mutualists of insects called parasitoid wasps [7], [8]. Polydnaviruses (PDVs) thus provide an opportunity for understanding the adaptations involved in the evolution of viruses into mutualists from pathogenic ancestors.
Parasitoid wasps reproduce by laying eggs into other insects (hosts) that their progeny consume [9]. The Polydnaviridae consists of two genera: the Bracovirus (BV) associated with ca. 20,000 species of wasps in the family Braconidae, and the Ichnovirus (IV) associated with ca. 18,000 species of wasps in the family Ichneumonidae [10]. Each wasp species carries a genetically unique PDV that persists in all cells as an integrated provirus. Viral replication only occurs in pupal and adult stage female wasps in a type of cell in the ovary called calyx cells. Virions from calyx cells are released via cell lysis and accumulate to high density in the lumen of the reproductive tract to form calyx fluid. Virions are also enveloped and contain multiple, circular, double-stranded DNAs of large aggregate size (190–600 kbp) that encode many virulence genes. Most PDV-carrying wasps parasitize larval stage Lepidoptera (moths) by depositing eggs containing the proviral genome plus a quantity of virions. These virions rapidly infect host cells, which is followed by expression of virulence genes that immunosuppress and alter the development of hosts in ways that are essential for survival of the wasp's progeny [11].
The origin and genomic organization of IVs is unclear [11]. In contrast, BV genomes exhibit features unlike any other known viruses [12]–[14]. As proviruses, their genomes consist of two types of DNA domains: those that contain genes with predicted roles in replication, and others that contain the virulence genes that become packaged into virions. Remarkably, these domains reside in different locations in the wasp genome [12], [13]. In addition, while genes with predicted roles in replication are transcribed in calyx cells, their transmission is entirely vertical and independent of any viral DNA replication or encapsidation [7], [11], [12]. Virulence gene-containing domains are likewise transmitted vertically. However, they also are amplified, excised from the wasp genome into circular forms and packaged into virions during replication in calyx cells [7], [15]–[20]. In all other cells of the wasp including the germ line both the replication and virulence genes of the proviral genome are silent [11]. BVs cause no apparent disease in wasps because almost no virulence genes are expressed in wasp cells and lytic replication is restricted to only calyx cells [11], [21]. In contrast, BVs cause severe disease in the hosts wasps parasitize because virions systemically infect the host insect and all of the virulence genes virions deliver are expressed [11], [21], [22]. The disease symptoms BVs cause in the host, however, are also essential for development of the wasp's offspring. Thus, BVs depend on wasps for genetic transmission, while wasps depend on BVs for parasitism of hosts.
Genes in the proviral genome of BVs with predicted roles in replication were identified because they exhibit homology with core genes from two other types of arthropod-infecting viruses: nudiviruses and their sister taxon the Baculoviridae [7], [12]. Like BVs, nudiviruses and baculoviruses replicate in cell nuclei and package large, circular ds-DNA genomes into enveloped virions. Unlike the developmentally-linked and tissue-specific replication of BVs, however, baculoviruses are virulent pathogens, which establish systemic infections in insects by undergoing lytic replication in virtually all cells of the infected host and expressing a variety of virulence genes [23]. Nudiviruses produce either systemic, lytic infections or latent infections [23], [24]. More than 60 baculoviruses have been sequenced and a survey of a subset (13) of these genomes indicates that all share 31 genes, which are collectively referred to as the baculovirus core gene set [24], [25] (Figure 1). Functional studies of model species like Autographa californica multinucleopolyhedrosis virus (AcMNPV) indicate about half of these genes have essential roles in viral replication [24]. These include genes with roles in replicating the viral genome, several genes that code for virion structural components, and four genes that code for subunits of a novel RNA polymerase, which selectively transcribes viral genes because it recognizes unique promoter sequences (Figure 1) [24]. Six nudivirus genomes have been sequenced and each contains 20 genes with homology to structural, replication and transcription components of the baculovirus core gene set [23] (Figure 1). The actual function of these genes, however, is unknown beyond inferences from baculoviruses. Data from three braconid wasps, Cotesia congregata, Chelonus inanitus, and Microplitis demolitor, indicate they lack recognizable homologs of most baculovirus core genes with roles in viral DNA replication (Figure 1), which suggests that, unlike baculoviruses, replication of BV DNAs packaged into virions is regulated by machinery from the wasp [7]. However, BVs do encode homologs of several baculovirus/nudivirus-like structural genes plus the four subunits of a baculovirus/nudivirus-like RNA polymerase [7], [12]. Each of these genes is also transcribed in ovaries when BV virions are produced. Together, these genes form a conserved gene set likely present in all BV genomes (Figure 1). However, we refrain from referring to these as “core” genes because of the small number of BV genomes currently available and their non-discrete organization in wasp genomes [7]. Other predicted members of a conserved BV gene set include a baculovirus/nudivirus-like sulfhydryl oxidase (ac92), 11 nudivirus-like genes unknown from baculoviruses, and 11 novel genes [7].
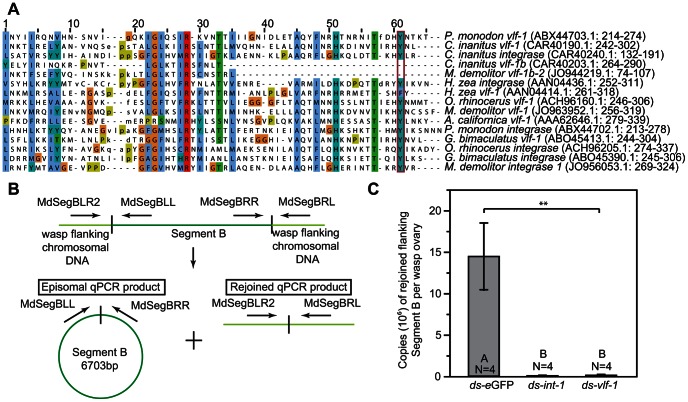
Figure 1. BV conserved gene products detected in MdBV virions.
The predicted conserved gene set for BVs (gray boxes) is shown to the right relative to the predicted core genes for nudiviruses (black boxes) and baculoviruses (hatched boxes) as previously detailed [7]. Black circles in hatched boxes represent baculovirus core gene products that are detected in mature occlusion-derived baculovirus virions [55]. Two independently prepared protein samples for MdBV virions were analyzed and are indicated as replicate 1 and 2 in the figure. Each gray box for the two replicates with a number or asterisk (*) indicates that peptides matching this conserved gene product were detected. The number in each gray box indicates the total number of unique peptides that matched the product. Gray boxes with asterisks indicate that the MdBV proviral genome contains two or more homologs of nudivirus/baculovirus genes and peptides matching multiple homologs were identified (see Table S1). Gray boxes with no number or asterisk indicate that no peptides were identified that matched the product.
Since BV-carrying braconids are monophyletic [26], these data overall indicate that BVs evolved from an ancestral nudivirus-wasp association. Fossil calibrations estimate this association arose 100 million years ago (Mya), while the last common ancestor of BVs, nudiviruses, and baculoviruses existed approximately 312 Mya [27]. Given these timelines and the pronounced differences that exist today between BVs and baculoviruses, it is not surprising many of the genes they share have diverged to the point that homology is difficult to recognize outside of essential residues or functional domains. Indeed, algorithms like BLAST cannot detect homology between BV and baculovirus genes, while identity between predicted BV and more closely related nudivirus proteins ranges between 19–41% [7]. Such divergence, however, also begs the question of whether baculovirus-like genes in BV proviral genomes retain baculovirus-like functions. Here, we used proteomic, RNA interference (RNAi), and functional assays to characterize selected members of the conserved gene set of Microplitis demolitor bracovirus (MdBV) in the wasp M. demolitor. Our results indicate that six genes with hypothesized roles in replication exhibited conserved functions relative to baculoviruses. Our data also identified novel functions for two genes in excision of viral DNAs for packaging into virions.
Results
MdBV virions contain conserved viral gene products
BV replication in calyx cells begins with amplification of a portion of the proviral genome, which is followed by the de novo assembly and packaging of virions in nuclei [20], [28]. Calyx cells then lyse which releases virions into the lumen. In the case of MdBV, prior studies establish the timing of these events and the chronology of replication gene expression during the pupal and adult stages of M. demolitor [7]. MdBV packages multiple circular, double-stranded DNA segments into virions but each individual virion contains only a single viral DNA [14], [29]. The sequence of these DNAs as episomes and their wasp-viral boundary sequences when integrated into the genome of M. demolitor are known [14], [29], [30]. Prior studies document that these DNAs are specifically amplified in M. demolitor calyx cells, followed by excision from flanking DNA and circularization [7]. Flanking wasp DNA at the site of excision is then rejoined to form an ‘empty locus’ [7].
Given this background, we first conducted a proteomic analysis of MdBV virions to determine whether predicted conserved structural components were present. To accomplish this, we produced two independent samples of calyx fluid with the second sample further purified on a sucrose gradient that produces morphologically pure and intact virions [31]. Following separation on SDS-PAGE gels, proteins were in-gel trypsin digested and analyzed using an Orbitrap Elite mass spectrometer. Mass spectra were then compared to our previously generated M. demolitor ovary transcriptome database [7] to identify proteins present. We present our findings relative to the predicted core/conserved gene sets for baculoviruses, nudiviruses, and BVs in Figure 1 and Table S1. Four proteins (38K, VP39, VLF-1, AC92) detected in MdBV virions were homologs of baculovirus capsid or capsid/envelope components. VP39 was the most accurately detected of these proteins based on the total number of unique peptides identified, which corresponded with vp39 also being the most abundant viral gene transcript detected in ovaries during MdBV replication [7] (Table S1). We also detected several proteins related to envelope components of baculovirus occlusion-derived virus. These included variants of ODV-E66 and ODV-E56 ( = PIF-5) plus the infectivity factors P74, PIF-1 through -6 and envelope component VP91 (Figure 1, Table S1). Seven MdBV virion proteins corresponded to genes in the BV conserved gene set for which homologs are known from all or some nudiviruses but are unknown from baculoviruses (Figure 1). These included the product of the integrase-1 (int-1) gene, which is structurally related to vlf-1, plus products of several nudivirus-like genes of unknown function (HzNVORF9-1 and -2, 64, 94, 106, PMV Hypothetical Protein). We also detected products of four conserved genes or gene families unique to BVs (17A, 35A, 97A) (Figure 1, Table S1). In contrast, we did not detect any proteins in virions that corresponded to the helicase gene or the RNA polymerase subunits (lef-4, lef-8, lef-9, p47) (Figure 1).
MdBV conserved genes are efficiently knocked down by RNAi
The preceding data showed that MdBV virions contain BV conserved gene products but provided no experimental evidence for their function. We therefore selected six genes from MdBV for functional studies. Our choices included two predicted subunits of a baculovirus-like RNA polymerase (lef-4, lef-9), 2), two homologs of baculovirus capsid genes (vp39, and vlf-1), and 3) two homologs of baculovirus envelope genes (p74 and pif-1). Each of these genes is a member of the BV conserved gene set because orthologs are likely present in all other BVs studied to date (see Figure 1). Each is also a member of the baculovirus core gene set with prior studies from AcMNPV or other isolates providing experimental evidence for the function of each [24]. In addition, we selected one nudivirus-like gene (int-1), unknown from baculoviruses, for which no functional studies have been conducted. As previously noted, sequence divergence between members of the BV conserved gene set and corresponding baculovirus/nudivirus core genes is high. Identities of the above gene products from MdBV with corresponding predicted proteins from the closest known nudivirus relative, Heliothis zea nudivirus-1 (HzNV-1), were: lef-4 (25%), lef-9 (32%), vp39 (19%), vlf-1 (28%), p74 (26%), pif-1 (28%), and int-1 (30%).
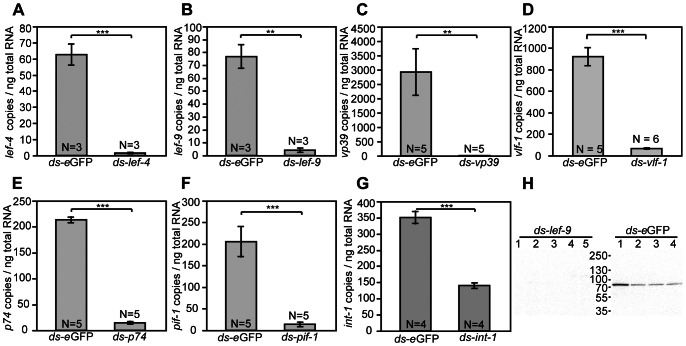
The genes we selected reside in the MdBV proviral genome and each is transcribed in ovary calyx cells during replication [7]. However, conventional knock out techniques used to characterize baculovirus gene function are untenable because the DNA domains where these genes reside are not replicated and packaged into MdBV virions. We thus assessed whether RNAi could be used to knock down transcription of these genes in M. demolitor. Since MdBV replication begins in the pupal stage of the wasp, we developed methods for injecting gene-specific dsRNAs into wasp larvae after they emerged from a host caterpillar and spun a cocoon. We then compared the abundance of each targeted transcript in newly emerged adult wasps by qPCR relative to treatment with a non-specific dsRNA (ds-eGFP). Our results showed that we reduced transcript abundance on average 60–99% for each gene we targeted (Figure 2). Using an antibody we generated to MdBV LEF-9, we also confirmed that knockdown at the transcript level resulted in knockdown of the corresponding protein (Figure 2).
Figure 2. RNAi knockdown of six MdBV putative replication genes.
M. demolitor larvae were injected with double-stranded RNA (dsRNA) specific for (A) lef-4, (B) lef-9, (C) vp39, (D) vlf-1, (E) p74, (F) pif-1, (G) int-1. Control larvae were injected with ds-eGFP. The ovaries from individual, newly emerged adults wasps were then dissected and total RNA isolated. The bars in each graph compare copy number of each target gene per ng of total RNA in wasps treated with dsRNA specific for the target gene versus ds-eGFP (control). Error bars represent one standard error from the mean. The number of individuals examined for each treatment is indicated by the N value at the bottom of each bar. Statistical significance is indicated by asterisks: *, p<0.05; **, p<0.001; ***, p<0.0001; N. S., not significant. (H) Knockdown of lef-9 depletes LEF-9 protein. M. demolitor larvae were injected with ds-lef-9 as in (B). Total protein from the ovaries of a newly emerged adult wasp was loaded into lanes of an SDS-PAGE gel followed by immunoblot analysis using an MdBV anti-LEF-9 antibody (1∶5000). The left lanes show results from 5 individual M. demolitor females treated with ds-lef-9 (knockdown), while the lanes on the right show results from 4 females treated with ds-eGFP (control). The predicted size of MdBV LEF-9 is 74.4 kDa.
Before initiating any functional experiments, we further verified our approach by examining the effects of dsRNA dose, time required after treatment for target transcript degradation, and specificity. Using vlf-1 as an example, our results showed that injection of 50 ng to 5 µg of dsRNA per wasp larva yielded a similar level of knockdown (Figure S1A). Our results also indicated that injection of vlf-1 dsRNA into wasp larvae did not significantly reduce transcript abundance until day 3 of the pupal stage, which suggested that 2–3 days were required before an RNAi effect was observed (Figure S1B). We examined specificity of knockdown in two ways. Since vlf-1 and int-1 are homologous genes, we verified that int-1 dsRNA, which strongly knocked down int-1 (Fig. 2G), did not affect transcript abundance of vlf-1 via off-target effects [32] (Figure S2A). We also generated a second vlf-1 dsRNA that did not overlap the dsRNA used for the data presented in Figure 2D to verify that it had a similar knockdown effect on vlf-1 transcript abundance (Figure S2B).
MdBV LEF-4 and LEF-9 are RNA polymerase subunits
Baculovirus RNA polymerases consist of four subunits (LEF-4, LEF-8, LEF-9, P47), which transcribe baculovirus genes with roles in virion formation [33]. These subunits are categorized as ‘early’ genes because they are transcribed before DNA replication and transcription of structural ‘late’ and ‘very late’ genes commences. As noted above, baculovirus RNA polymerases selectively transcribe late and very late viral genes because they recognize unique promoter elements with the consensus sequence (A/G/T)TAAG absent from host insect genes [34], [35]. The conserved gene set of BVs contains homologs of each RNA polymerase subunit. Expression data from M. demolitor and Cotesia congregata also indicate these subunits are transcribed in ovaries earlier than predicted structural genes, while sequence analysis has identified baculovirus late gene promoter elements upstream of the start codon of several predicted BV structural genes [7], [12]. Thus, if the BV RNA polymerase subunits form a functionally similar enzyme as baculoviruses, RNAi knockdown of one or more subunits should compromise transcription of MdBV structural genes but not wasp genes.
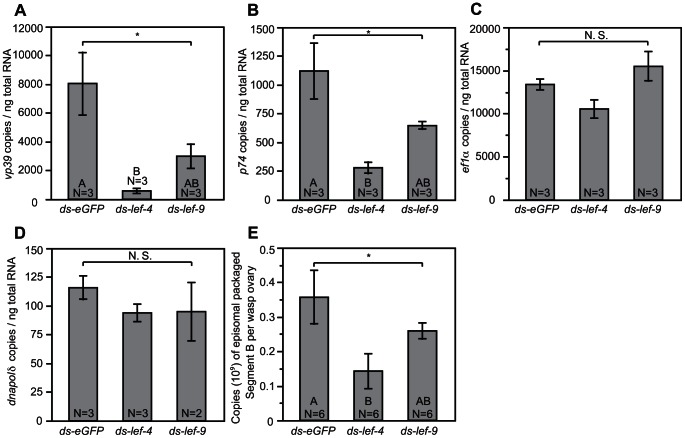
As shown above (Figure 2A, B), we knocked down lef-4, a predicted 5′ capping enzyme [36], [37] and lef-9, a predicted RNA polymerase subunit that forms part of the catalytic cleft [38], [39]. We then measured transcript abundance of two predicted MdBV structural genes (vp39 and p74), and two typical insect genes expressed in ovaries (elongation factor 1 alpha (ef1α) and DNA polymerase delta subunit (dnapolδ)). Our results showed that knockdown of lef-4 and lef-9 significantly reduced transcript abundance of vp39 and p74 while having no effect on ef1α and dnapolδ (Figure 3A–D). Given this outcome and the putative role of vp39 and p74 as structural genes we reasoned that reduced expression of vp39 and p74 could also result in production of fewer virions on average than control females. We therefore estimated viral titer by using episomal MdBV DNA segment B as a marker and a previously developed qPCR assay that includes a DNAse step to remove all non-encapsidated DNA before isolating DNA from ovary homogenates [7]. In this manner, the copy number of episomal segment B in virions, which protect the packaged DNA, could be determined. These results showed that knockdown of lef-4 and lef-9 significantly reduced the titer of DNAse-protected segment B relative to ds-eGFP-treated controls (Figure 3E).
Figure 3. Knockdown of lef-4 and lef-9 reduces expression of MdBV structural genes.
M. demolitor larvae were injected with ds-eGFP, ds-lef-4 or ds-lef-9 as shown in Figure 2. The ovaries from individual, newly emerged adults wasps were then dissected and total RNA isolated followed by measurement of copy number per ng of total RNA of: (A) vp39, (B) p74, (C) ef1α, (D) dnapolδ. (E) Copy number of DNase-protected MdBV episomal genomic segment B in the ovaries of newly emerged M. demolitor adult females. The ovaries from individual, newly emerged adults wasps were dissected and treated with DNAse. DNA was then isolated and total copy number of episomal genomic segment B determined. Error bars, N values, and statistical significance are indicated as defined in Figure 2.
VP39 and VLF-1 likely encode structural components of viral nucleocapsids
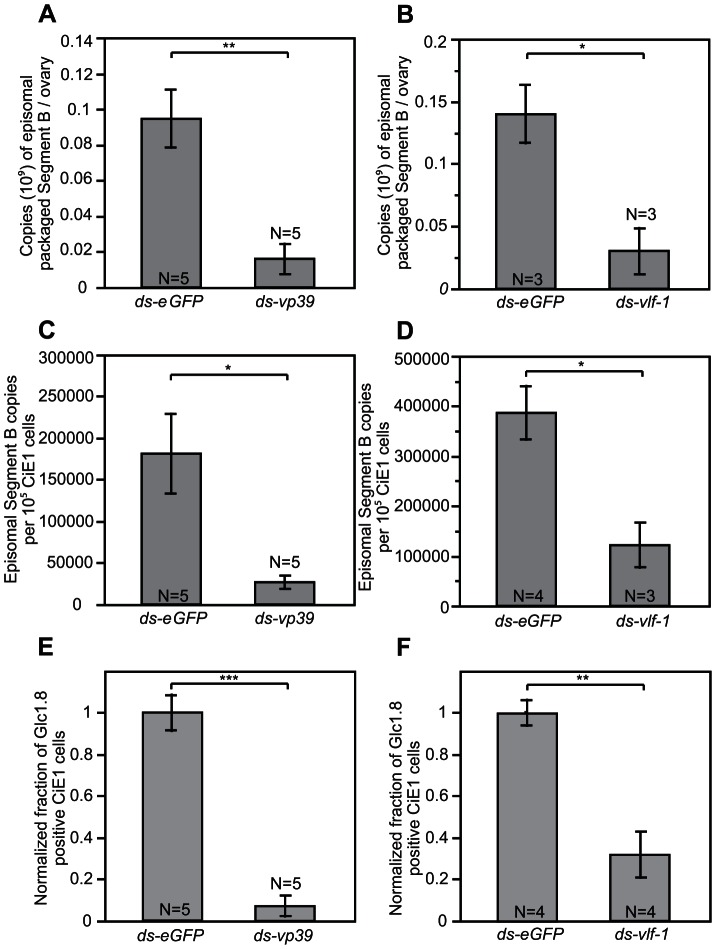
In baculoviruses, VP39 is a major capsid protein while VLF-1 is a structural component, and is also functionally required for capsid production and very late gene expression [24], [40]–[42]. After knocking down vp39 and vlf-1 in M. demolitor (Figure 2C, D), we first assessed whether either treatment affected virion structural integrity by measuring the DNase sensitivity of packaged genomic DNAs as described above. These assays indicated that the abundance of DNase-protected segment B declined by 83% and 78% in vp39 and vlf-1 knockdown samples respectively relative to the control (Figure 4A, B). We also used the non-overlapping dsRNA, ds-vlf-1-2 in these assays, which produced the same result as ds-vlf-1 (Figure S2C).
Figure 4. Knockdown of vp39 and vlf-1 increases DNAse sensitivity and reduces infectivity.
M. demolitor larvae were injected with ds-eGFP, ds-vp39 or ds-vlf-1 as shown in Figure 2. (A) Copy number of DNase-protected MdBV episomal genomic segment B in the ovaries of newly emerged M. demolitor adult females pretreated with ds-eGFP or ds-vp39. (B) Copy number of DNase-protected MdBV episomal genomic segment B in the ovaries of newly emerged M. demolitor adult females pretreated with ds-eGFP or ds-vlf-1. (C) Copy number of MdBV episomal genomic segment B in CiE1 cells infected with MdBV from wasps pretreated with ds-eGFP or ds-vp39. (D) Copy number of MdBV episomal genomic segment B in CiE1 cells infected with MdBV from wasps pretreated with ds-eGFP or ds-vlf-1. (E) Normalized fraction of CiE1 cells expressing the gene product GLC1.8 on their surface after infection with MdBV from wasps pretreated with ds-eGFP or ds-vp39. (F) Normalized fraction of CiE1 cells expressing the gene product GLC1.8 on their surface after infection with MdBV from wasps pretreated with ds-eGFP or ds-vlf-1. Error bars, N values, and statistical significance are indicated as defined in Figure 2.
We then examined the effects of vp39 and vlf-1 knockdown on the ability of MdBV to infect cells from the moth Chrysodeixis includens, which is a host for M. demolitor. For these assays, we used CiE1 cells, which is a continuous, hemocyte-like cell line established from C. includens that is highly permissive to MdBV infection [30], [43]. We determined by qPCR that 2–4 copies of episomal DNA segment B were present per CiE1 cell when cultures were infected at an estimated MOI of 100 with MdBV from control wasps treated with ds-eGFP (Figure 4C, D). In contrast, copy number was 85.2% and 69.6% less when cells were infected with the same amount of calyx fluid from vp39 and vlf-1 knockdown wasps. We also assessed infection using the MdBV gene product GLC1.8, which is an excellent marker because it is rapidly expressed on the surface of CiE1 cells and is easily visualized immunocytochemically [43], [44]. Normalizing the control samples, we observed that vp39 knockdown reduced the fraction of cells stained for GLC1.8 to less than 10%, while vlf-1 knockdown reduced this fraction to 26.8% (Figure 4E,F).
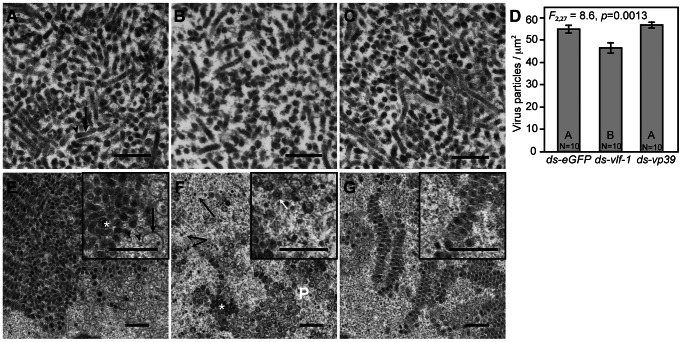
These results could be explained by vp39 and vlf-1 knockdown either adversely affecting virion formation, which would result in calyx fluid containing a lower titer of virus, or causing structural defects that do not reduce virion density but nonetheless compromise function. We therefore examined virion morphology in calyx fluid by transmission electron microscopy (TEM). We previously documented that MdBV virions in calyx fluid consist of a single barrel-shaped nucleocapsid surrounded by a highly elongate envelope [29], [31]. By counting the number of virions in randomly selected fields of view from treatment and control wasp sections, we determined that calyx fluid from a vlf-1 knockdown wasp contained a slightly lower concentration of virions than observed in a control wasp, whereas a vp39 knockdown wasp did not (Figure 5A–D).
Figure 5. Electron microscopy analysis shows defects in MdBV morphogenesis after vlf-1 and vp39 knockdown.
M. demolitor larvae were injected with ds-eGFP, ds-vp39 or ds-vlf-1 as shown in Figure 2. (A) Image of calyx fluid in the lumen of the reproductive tract of a newly emerged adult female pretreated with ds-eGFP. Each virion consists of one electron dense nucleocapsid (arrowhead) surrounded by a single elongate envelope (arrow). (B) Image of calyx fluid from a wasp pretreated with ds-vlf-1. Note the lower density of virions present relative to (A). (C) Image of calyx fluid from a wasp pretreated with ds-vp39. Scale bars in A-C equal 500 nm. (D) Graph showing that virion density in calyx fluid from wasps treated with ds-vlf-1 was significantly lower than in wasps treated with ds-eGFP or ds-vp39. (E) Image showing a portion of a calyx cell nucleus from a wasp pretreated with ds-eGFP. The left side of the image shows a large array of assembled MdBV virions. Virions in the process of assembly are visible in the lower right of the image. The insert in the upper right shows virion assembly at higher magnification. Note that assembled virions (*), de novo forming envelopes, empty capsids (arrowhead) and electron dense nucleocapsids in the process of being surrounded by envelopes (arrow). (F) Image showing a portion of a calyx cell nucleus from a wasp pretreated with ds-vlf-1. Unlike normal calyx cells, few virions are assembled into arrays (*). Most of these virions have envelopes that are rounded. Numerous electron dense nucleocapsids without envelopes (arrows) and rounded envelopes without electron dense nucleocapsids are visible. The insert in the upper right shows these defects at higher magnification. It also shows that some rounded envelopes contain empty capsids (arrow). (G) Image showing a portion of a calyx cell nucleus from a wasp pretreated with ds-vp39. Arrays of assembled virions are visible but they are much smaller than the arrays observed in calyx cell nuclei from control wasps. The insert in the upper right shows virions with elongate envelope and electron dense nucleocapsid similar to virions from control wasps. Scale bar for images and inset images E–G equal 500 nm.
We then examined MdBV morphogenesis in calyx cell nuclei. Early studies of BVs show that calyx cells exhibit a progression of development with smaller, younger cells being situated closer to the ovarioles and older, large cells being closer to the lumen of the ovary [45]. In turn, young cells show no evidence of BV replication, while old calyx cells contain an abundance of assembled virions in their nuclei [45], [46]. In control wasps, we observed that MdBV morphogenesis began with the de novo appearance of short membrane profiles in calyx cell nuclei. This was followed by the formation of nucleocapsids near virogenic stroma, which is where DNA packaging also occurs in baculoviruses. Assembled MdBV virions then formed large aggregations with a layered crystalline structure (Figure 5E). At this stage, virions were rod-shaped and of uniform length. The envelope surrounding each nucleocapsid was also not as elongated as seen for virions in calyx fluid (see Figure 5A versus 5E). Calyx cells from vlf-1 knockdown wasps in contrast exhibited an abundance of membrane profiles that appeared to be envelope progenitors (Figure 5F). Rather than elongating, these envelopes were spherical and either lacked capsids entirely or contained empty capsids (Figure 5F). A number of electron dense and empty capsids were also observed with no envelope (Figure 5F). Lastly, almost no aggregations of rod-shaped, electron dense virions were present in calyx cells from vlf-1 knockdown wasps. Calyx cells from vp39 knockdown wasps showed no distinct alterations in virion assembly, but aggregations of rod shaped, electron dense virions were consistently much smaller than those observed in control wasps (Figure 5G).
Knockdown of p74 and pif-1 reduces the fraction of host cells that express the marker GLC1.8
p74 and the pif genes are known as per os infectivity factors because their loss in baculoviruses such as AcMNPV disables oral infection of host insects by occlusion derived virus [47]–[49]. Each is also a component of the occlusion derived virus envelope where they form a complex with one another [50]. Unlike baculoviruses, BV virions never infect host insects orally but instead are injected into the hemocoel by wasps where they bind to host cells such as hemocytes via fusion of the envelope with the plasma membrane [51]. Nucleocapsids then travel through the cytoplasm to nuclear pores where they release their DNA into the nucleus to initiate transcription of virulence genes like glc1.8 [51], [52]. Given the differences in the known functions of per os infectivity factors in baculoviruses relative to the biology of BVs we asked whether the p74 and pif-1-like genes of MdBV still play a role in infectivity by knocking down each (Figure 2E, F) and then conducting the same assays in CiE1 cells as described above. Our results revealed no differences between knockdown and control wasps in the copy number of DNA segment B in CiE1 cells at 24 h post-infection (Figure 6A). However, the fraction of CiE1 cells expressing GLC1.8 on their surface was dramatically lower using virus from p74 and pif-1 knockdown wasps (Figure 6B).
Figure 6. Knockdown of p74 and pif-1 reduces the fraction of host cells that express GLC1.8.
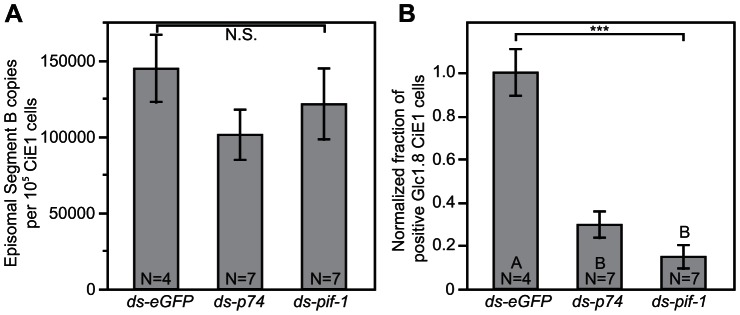
M. demolitor larvae were injected with ds-eGFP, ds-p74 or ds-pif-1 as shown in Figure 2. (A) Copy number of MdBV episomal genomic segment B in CiE1 cells infected with MdBV from wasps pretreated with ds-eGFP, ds-p74, or ds-pif-1. (B) Normalized fraction of CiE1 cells expressing the gene product GLC1.8 on their surface after infection with MdBV from wasps pretreated with ds-eGFP, ds-p74, or ds-pif-1. Error bars, N values, and statistical significance are indicated as defined in Figure 2.
Knockdown of VLF-1 and INT-1 disables proviral DNA excision
As noted above, all baculoviruses encode a vlf-1 gene while nudiviruses and BVs also encode related integrase genes (known as vlf-1 or vlf-1a, vlf-1b-1 and -2 or HzNVORF140, and int-1 and -2 or HzNVORF144) [7], [12]. Although VLF-1 is a structural component of baculovirus virions, it along with nudivirus integrase genes are members of the tyrosine (Tyr) recombinase family, which includes several enzymes that mediate the excision and integration of genetic elements [53]. As noted above, elimination of vlf-1 from AcMNPV disables capsid formation while mutation of the conserved Tyr residue required for integrase activity in other Tyr recombinase family members produces non-infectious virus [40]. Whether baculovirus VLF-1 possesses any integrase activity, however, remains unstudied in all likelihood because baculovirus genomes persist as episomes in infected host cells and are unknown to integrate. In contrast, a key feature of BVs is their persistence in wasps as integrated proviruses that amplify, excise and package a portion of the genome when replicating in calyx cells. We therefore assessed whether vlf-1 and/or int-1 homologs from MdBV regulate proviral DNA excision.
We had previously determined that the MdBV proviral genome encodes three distinct vlf-1 genes (named vlf-1, vlf1b-1, -2) and two integrase genes (int-1, -2) that are all transcribed in ovaries during replication [7]. Phylogenetic analysis further suggested the integrase genes of BVs likely arose from duplication of vlf-1 in the nudivirus ancestor, which was then followed by duplication of each gene in M. demolitor [7]. Alignment of vlf-1 and integrase family members from MdBV, select nudiviruses, AcMNPV, and Chelonus inanitus bracovirus (CiBV) showed that MdBV vlf-1 and int-1 both retain a typical active site Tyr residue for predicted integrase activity, whereas other M. demolitor family members do not (Figure 7A). We thus knocked down vlf-1 and int-1 (Figure 2D, G), and then isolated DNA from newly emerged adult wasp ovaries to determine whether proviral DNAs had excised from the wasp genome as normally occurs. This was accomplished using MdBV segment B as a marker and qPCR assays that measured copy number of the rejoined ‘empty locus’ that only forms if proviral DNA segment B was excised from the wasp genome (Figure 7B). Copy number of the empty locus in ovaries from control wasps treated with ds-eGFP was 14.4×106, which indicated a high level of excision of DNA segment B from calyx cells. In contrast, we detected almost no copies of the empty locus in wasps treated with ds-int-1 and ds-vlf-1 (Figure 7C).
Figure 7. Knockdown of vlf-1 and int-1 disables excision of genomic segment B from the M. demolitor genome.
(A) Alignment of MdBV vlf-1 and integrase family members to family members from select nudiviruses (Paenaeus monodon nudivirus (PmNV), Heliothis zea nudivirus (HzNV-1), Gryllus bimaculatus nudivirus (GbNV), Oryctes rhinoceros nudivirus (OrNV)), the baculovirus Autographa californica multinnucleopolyhedrosis virus (AcMNPV), and Chelonus inanitus bracovirus (CiBV). The alignment shows only the portion of each where the predicted conserved tyrosine (Y) residue required for recombinase activity by other tyrosine recombinase family members is located (box). Select other conserved residues are highlighted with other background colors. (B) Schematic illustrating the qPCR assays to quantify copy number of MdBV genomic segment B (6307 bp) in M. demolitor ovaries [7], [14], [30]. Episomal segment B is specifically amplified using the primers MdSegBLL and MdSegBRR, which can only generate a product if segment B is circularized. Copy number of the rejoined empty locus in M. demolitor is specifically amplified using the primers MdSegBLR2 and MdSegBRL, which can only generate a product if segment B has been excised and the wasp flanking domains are rejoined. (C) Copy number of the rejoined empty locus from excision of segment B in the ovaries of newly emerged M. demolitor adult females pretreated with ds-eGFP, ds-int-1 or ds-vlf-1. The ovaries from individual, newly emerged adults wasps were dissected, followed by DNA isolation and qPCR analysis as outlined in (B). Error bars, N values, and statistical significance are indicated as defined in Figure 2.
Discussion
Phylogenetic data strongly support that BVs evolved from a nudivirus ca. 100 Mya, and that nudiviruses and baculoviruses shared a more ancient common viral ancestor ca. 200 Mya earlier [12], [26], [27]. Detailed studies of AcMNPV and select other species also provide important insights into the function of baculovirus core genes. In contrast, the hypothesized function of baculovirus core gene homologs in BVs (and nudiviruses) is founded on inferences from the baculovirus literature and/or expression patterns in wasp ovaries during replication. Thus, the primary goal of this investigation was to assess whether RNAi methods could be used to disrupt BV gene expression, and then to use these methods with a subset of genes to determine whether their roles in replication were consistent with or differed from baculoviruses.
Prefacing these functional studies, we conducted a proteomic analysis of purified MdBV virions to assess whether BV conserved genes that are homologs of baculovirus structural components were present. We also did this to compare MdBV virions to virions from CcBV and CiBV, which are the only other BVs for which any proteomic data are available [12], [54]. We detected all of the baculovirus-like capsid and envelope components previously identified in CcBV and CiBV as well as two additional baculovirus-like conserved genes not detected in CcBV or CiBV. These included AC92, which is associated with baculovirus nucleocapsids, and PIF-3, which is associated with baculovirus occlusion-derived virus envelopes [55]. We also detected four nudivirus-like (HzNVORF9-1 and -2, HzNVORF106, PmV) and three novel conserved gene products (17A, 35A, 97A) in MdBV virions identified in CiBV virions. In contrast, we did not detect three novel gene products (27B, 30B, 97B) reported from CiBV, yet did detect three nudivirus-like gene products (INT-1, HZNVORF64, HZNVORF94) not reported from CiBV virions.
Proteomic data must be interpreted cautiously when investigating viral structural proteins, because of the potential for non-integral proteins to become non-specifically associated with virions during assembly [24]. Nonetheless, the baculovirus literature combined with our detection of the capsid and envelope proteins in Figure 1 suggest these baculovirus-like gene products are likely structural components of MdBV virions. While no functional data from nudiviruses exist, we speculate for the same reason that products of the nudivirus-like conserved genes HzNVorf 9-1, 9-2, -64, -94, -106, PmV hypothetical protein and novel conserved genes 17a, 35a, and 97a, are also structural proteins. Our proteomic data did not identify any peptides corresponding to MdBV conserved genes with predicted roles in DNA replication (helicase) or transcription (lef-4, lef-8, lef-9, p47), which at minimum indicates our samples were not contaminated with some non-integral products transcribed in calyx cells [7]. However, it is notable that we consistently detect the products of the int-1, vlf-1b-1 and vlf-1b-2 genes, which could suggest that similar to baculovirus vlf-1 they too are capsid components or are packaged into capsids together with episomal DNA.
Because of the unique biology of BVs and limited genetic data available for their associated wasps, the options available for studying gene function are obviously constrained. RNAi is a potentially powerful method for studying BV gene function, but its efficacy in insects is also patchy with examples of successful use being more prevalent in some taxa (beetles (Coleoptera), mosquitoes (Diptera)) [56], [57] than others (moths (Lepidoptera) [58]). We thus were very careful in validating our RNAi approach for knocking down MdBV genes in M. demolitor before initiating any functional studies. Our analysis of the ovary transcriptome indicated that all genes of the siRNAi pathway are present in M. demolitor and transcribed [7]. Results presented in this study further show that ds-RNA injection into late larval stage M. demolitor effectively and specifically knocks down the genes we targeted. While we present the outcome of a number of validation experiments using vlf-1 as an example, we have conducted similar experiments with other MdBV conserved genes, which all showed the same trends.
With knockdown methods established, we used the baculovirus literature and our proteomic data to select six genes in the MdBV conserved gene set with hypothesized roles in viral transcription (lef-4, lef-9), capsid assembly (vp39, vlf-1), and envelope formation (p74, pif-1). Our rationale for selecting these genes was also driven by the strength of the functional literature for each in baculoviruses, which provided in most cases clear expectations for what a conserved phenotype should be for MdBV. Our results with lef-4 and lef-9 strongly support that MdBV produces a baculovirus-like RNA polymerase that preferentially transcribes structural genes. We also note that knockdown of lef-4 more strongly disabled structural gene expression than knockdown of lef-9. This could reflect that as a capping enzyme the effect of knocking down of lef-4 was further enhanced by degradation of cap-lacking transcripts. The transcription of reporter virus structural genes vp39 and p74 was not completely abolished for either lef-4 or lef-9 knockdowns. Despite detecting no LEF-9 protein after knockdown on immunoblots, this could reflect incomplete knockdown, and the presence of enough RNA polymerase subunit proteins to produce some functional viral RNA polymerase holoenzyme. Alternatively, viral structural genes may also be transcribed in part by wasp RNA polymerase II. Activity of replication gene transcription compared to relative silence of BV genes that are ultimately packaged into virions suggests that replication genes are transcribed from ancestral viral RNA polymerase promoters whereas virulence genes are not.
The phenotypic effects we observe in response to vlf-1 knockdown are consistent with this protein being both a structural component and a product required for virion assembly. The defects in morphogenesis of MdBV virions we observe, however, differ somewhat from the defects observed with AcMNPV where knockout of vlf-1 resulted in formation of tubular structures that appeared to be aberrant capsids that fail to package DNA [40]. The technical approaches to these studies, however, resulted in observations associated with budded virus production and precluded examination of potential defects associated with formation of occlusion-derived virus (see below). In contrast, the severe defects we observed in the assembly of MdBV virions suggest vlf-1 may be important in both DNA packaging and proper association of capsids with envelopes. Like vlf-1, knockdown of vp39 greatly increased the sensitivity of packaged DNA to DNAse treatment while also reducing infectivity. For both genes, dsRNA treatment did not completely abolish DNAse protection or infectivity of virus particles, which we presume is due to incomplete knockdown of transcript levels. Unlike vlf-1 though, knockdown of vp39 did not cause any obvious morphological defects in virion assembly with the exception that virion aggregations in calyx cells were much smaller owing possibly to a reduction in VP39 for production of capsids. Given these observations, we are unclear why knockdown of vp39 did not reduce virion density in calyx fluid.
Unlike BVs, which produce only one virion type, AcMNPV and most other baculoviruses produce two virion phenotypes named occlusion-derived virus and budded virus [24]. Occlusion-derived virus is embedded in a protein matrix called an occlusion body and is the type that initiates a midgut infection when ingested by a new host. Budded virus in contrast is non-occluded and is the form of the virus that disseminates from the midgut and other cells to systemically infect the insect. Occlusion-derived and budded virus capsids contain the same structural proteins [55] but their envelopes differ greatly with the former assembling de novo in host cell nuclei and containing products of several core genes including per os infectivity factors. Budded virus in contrast acquires an envelope when budding through host cell plasma membranes, which contains only one or two viral proteins (GP64, F) [55]. Although never occluded, the de novo assembly of BVs in calyx cell nuclei together with the envelope components detected in their virions (see above) indicate that BV particles structurally share more features with the occlusion-derived phenotype of baculoviruses. On the other hand, while per os infectivity factors are required for infectivity and binding of the occlusion-derived phenotype of AcMNPV to midgut cells, they are not required for infection of cultured cells or host larvae when injected into the hemocoel [49], [59]. We thus were unclear what effect, if any, knockdown of p74 or pif-1 might have on infectivity given that BVs infect host insects only when injected into the hemocoel by wasps. Our results reveal no defects in the copy number of MdBV DNA detected in CiE1 cells after infection with a high MOI. Yet, knockdown of each gene resulted in a large decline in the fraction of infected cells that expressed the marker gene GLC1.8. These findings are interesting because they suggest the loss of per os infectivity factors from the MdBV envelope results in improper translocation of MdBV to host cell nuclei where transcription of glc1.8 and other virulence genes occurs. No such activity has previously been associated with per os infectivity factors in baculoviruses but intriguingly GP64, the envelope fusion protein of budded virus, has been implicated in affecting baculovirus translocation to host cell nuclei [60].
In addition to targeting six baculovirus-like conserved genes, we also examined the function of nudivirus-like int-1 because this gene and vlf-1 are both tyrosine recombinase family members and BV replication requires the excision of proviral genomic DNAs from the wasp genome for packaging into virions. The near complete inhibition of empty locus formation after vlf-1 and int-1 knockdown suggests a role for both in proviral DNA excision. At this time we have little understanding of the recombination reactions VLF-1 and INT-1 potentially mediate, although detailed investigation of other tyrosine recombinases suggest they perform recombination events by establishing a synapse between their cognate binding sites and performing two consecutive strand exchanges [61], [62]. If on the same molecule recombination between two binding sites leads to excision of a circular intermediate [63]. Analysis of tyrosine-recombinase structural properties also suggest the mechanism of recombination requires binding of four enzyme monomers, which suggests the possibility for involvement of both vlf-1 and int-1 in proviral DNA excision [53]. Additionally, proviral DNA segments that become encapsidated possess direct repeats at the sequence boundaries that abut the flanking wasp DNA, which have been previously hypothesized to contain binding sites for recombinases that mediate excision [18], [19], [30]. Lastly detection of both VLF-1 and INT-1 in virions suggests these factors may also have important functions in parasitized hosts given recent findings that all DNA segments packaged into MdBV virions rapidly integrate into the genome of infected host cells [30].
Taken together, our results provide the first experimental insights into the function of a subset of MdBV genes. We fully recognize that additional experiments will be needed to more fully characterize the function of the individual genes we targeted, but by examining several key genes at once we provide evidence that: 1) RNAi can be used to knock down a number of MdBV genes, and 2) the baculovirus-like genes we targeted exhibit conserved functions despite divergence from nudiviruses more than 100 Mya. At the same time our results with vlf-1 and int-1 also identify novel functions unknown from baculoviruses but essential to the biology of BVs. With these results in hand, we are now positioned to undertake both more detailed experiments on these genes as well as studies on nudivirus-like and novel genes in the BV conserved gene set for which expectations about function are less clear.
The arms race between wasps and the hosts wasps parasitize likely underlies the high rates of speciation of BV-carrying braconids [26], [64]. The genetic mechanisms guiding host range evolution in contrast are largely unknown. One hypothesis would be that PDV virion structure has undergone rapid adaptation in response to the different lepidopteran host species each wasp species parasitizes, which could result in high variation in BV virion structure. The similarities thus far found in BV conserved genes together with the functional insights provided here, however, strongly suggest that BV gene functions will be conserved across isolates. Thus, differences in the sequence of BV genes may affect whether a given isolate can infect a given host species but we think it unlikely large differences will be found in the structure of BV virions across isolates. In contrast, the literature already indicates that the virulence genes BVs package into virions vary greatly among isolates associated with phylogenetically distant species of wasps. Thus, we would expect that differences in the virulence genes BVs deliver to hosts strongly affect host range by impacting the ability of wasp offspring to successfully develop. Finally, conservation in the function of the MdBV RNA polymerase and structural proteins suggest that key features in the evolution of BVs into mutualists do not involve radical changes in virion structural products but rather in how: 1) transcription of early factors required for DNA replication and viral transcription are regulated so that replication only occurs in calyx cells, 2) the genome is organized so that only some portions are amplified and packaged into virions, and 3) virulence genes are kept silent in wasps.
Materials and Methods
Ethics statement
All studies were approved by the Biological Safety and Animal Care and Use Committee of the University of Georgia and were performed in compliance with relevant institutional policies, National Institutes of Health regulations, Association for the Accreditation of Laboratory Animal care guidelines, and local, state, and federal laws.
Insects and developmental staging of wasp larvae
M. demolitor parasitizes several species of larval stage Lepidoptera including Chrysodeixis ( = Pseudoplusia) includens. Both species were reared at 27°C with a 16 h-light:8 h-dark photoperiod. M. demolitor has an 11 day developmental period described in detail elsewhere [7]. For this study, M. demolitor females were allowed to parasitize C. includens larvae, and wasp offspring were then allowed to develop in the host for 6 days. On day 7, wasp larvae emerge from hosts and spin a cocoon within several hours. Cocoons are slightly asymmetrical with the anterior end generally being more elevated and pointed than the posterior end that contains the wasp abdomen. Nine-12 hours after spinning their cocoon, wasps pupate and develop for four more days until emerging as an adult. Adult wasps were then maintained in constant dark at 18°C.
Proteomic analysis of MdBV virions
Virus was collected from M. demolitor ovaries. For the first replicate, 100 whole ovaries were crushed in PBS and the debris was removed by centrifugation. Then, the supernatant containing the virus was spun down at 20,000×g for 5 minutes and washed with PBS 3 times to collect virus particles. For replicate 2, calyx fluid was dissected from 100 dissected ovaries and resuspended in PBS. Virions were then isolated on a sucrose gradient as previously described [31]. Both virus samples were electrophoresed on either a 4–20% or 12.5% Tris-Glycine gel (Lonza). For each sample, the entire lane was cut into four pieces to separate proteins by size. In-gel trypsin digestion was performed for each gel slice, by overnight incubation with trypsin (20 µg/ml) in 20 mM ammonium bicarbonate. Tryptic fragments were extracted with 50% acetonitrile and 0.1% TFA and vacuum dried. Samples (4 µl) were analyzed by an Orbitrap Elite mass spectrometer, coupled to an Easy-nLC II Liquid Chromatography (LC) instrument (Thermo Fisher Scientific). Samples were desalted and pre-concentrated on a C18 Easy LC pre-column (100 um internal diameter (ID) ×2 cm, 5 µm particle packing (PP)). Peptides were eluted from a reverse-phase column (75 um ID ×10 cm, 3 µm PP) with a gradient of 10–35% B for 70 min, 35–95% B for 10 min, 95% B for 5 min (A = 0.1% formic acid in water, B = 0.1% formic acid in acetonitrile) at 300 nl/min. Nanospray ionization was performed with a spray voltage of 2 kV, with a capillary temperature of 200°C. The Orbitrap mass analyzer was used to provide resolutions of 120,000 and 30,000 for MS and MS/MS analyses, respectively. Briefly, a cycle of one full-scan mass spectrum (300–2000 m/z) was performed, followed by continuous cycles of CID and HCD MS/MS spectra acquisitions of the 2 or 5 most abundant peptide ions throughout the LC separation until the candidate ions were exhausted. Data were acquired using Xcalibur software (v2.2, Thermo Fisher Scientific).
Proteins were identified by searching against a custom database “Md” consisting of translated open reading frames (ORFs) greater than 33 amino acids in size from transcripts described in [7] using the Mascot v2.3 algorithm (Matrix Science Inc.). Transcripts can be accessed through NCBI accession numbers JO913492 through JO979916 and JR139425 through JR139430. Data were visualized with ProteomeDiscoverer v1.3 (Thermo Fisher Scientific). Peptides with scores greater than the identity score (p<0.05) were considered significant matches. Only ORFs that were matched by at least two peptide spectra were considered positive identifications.
RNAi assays and quantification of target gene expression
To target individual genes for RNAi knockdown, gene-specific primers were designed with added T7 promoter adaptors to amplify a 300–400 bp template for double-stranded RNA (dsRNA) synthesis (Table S2). cDNA from adult wasp ovaries was amplified using standard PCR and the resulting products were used as template in the MegaScript RNAi Kit (Ambion). Larval stage wasp cocoons were marked within 15 h of spinning and were subsequently injected within 1–3 h. As M. demolitor is protandrous with males developing faster than females, the cocoons selected for injections were biased towards later emergence times and thus female wasps. Cocoons were affixed to double-sided tape, and oriented so that the posterior ends were facing in the same direction. Cocoons were pierced with a minuten pin in the abdomen region and wasps were injected through the cocoon with a glass needle directly into the abdomen. Approximately 0.5–1 µl of 2–4 µg/µl dsRNA was injected into each individual. All control wasps were injected with a non-specific dsRNA probe homologous to the bacterial eGFP gene. Wasps were sampled within 24 h of emerging as adults, and ovaries were removed and separated using opthalmic scissors. One ovary half was snap-frozen at −80°C for RNA extraction, and the other was used to assay RNAi phenotypes as described below.
Extraction of total RNA was performed using the QIAGEN RNeasy kit following the standard kit protocol with a 20 min on-column DNAse treatment and elution in 30 µl of RNAse-free H2O. RNA concentration was measured using a Nanodrop Spectrophotometer and cDNA was synthesized from a quantity of total RNA normalized across samples. First-strand cDNA synthesis was performed using Invitrogen reagents including the Superscript III enzyme and oligo(dT) primers as outlined by the manufacturer (Invitrogen).
We used quantitative PCR (qPCR) to detect differences in expression of target genes after dsRNA treatment. An absolute standard curve was generated via PCR amplification of the corresponding cDNA for each gene of interest using specific primers (Table S2). Each product was cloned into pSC-A-amp/kan, and after propagating and isolating each plasmid from minipreps, its identity was confirmed by sequencing. Standard curves were generated followed by determination of copy numbers from serially diluted amounts (102 to 107 copies) of each plasmid standard. qPCR was performed on a Rotor-Gene Q using the Rotor-Gene SYBR Green PCR Kit with 1 µM primers and 1 µl of undiluted cDNA per 10 µl reaction (QIAGEN). After 5 minutes of denaturation at 95°C, a two-step amplification cycle with 95°C for 5 sec denaturation and 60°C for 20 sec of annealing and extension was used for 45 cycles. Melting curve analyses were performed to ensure that amplified products were specific for the gene of interest. At least three independently acquired biological replicates were analyzed per stage for each gene, with each sample internally replicated 4 times.
A polyclonal antibody against Lef-9 was generated by PCR amplifying and cloning a portion of MdBV lef-9 into pET-30-EK-LIC (Novagen) as previously outlined [65]. Briefly, 31% of the lef-9 coding sequence was amplified and cloned to create an expression product of 22.9 kDa. This construct was confirmed by DNA sequencing, and then expressed in Escherichia coli BL21 (DE3) cells grown in 6 L Luria Broth with 50 µg/ml kanamycin at 37°C. The cultures were then induced with 0.025 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for an additional 24 h at 37°C. Bacterial cells were harvested by centrifugation, lysed, and the insoluble recombinant protein was purified from the cell pellet using PerfectPro Ni-NTA (QIAGEN) agarose beads under denaturing conditions. After analysis by SDS–PAGE and immunoblotting using an anti-His monoclonal antibody, the identity of the recombinant protein was validated by mass spectrometry. A polyclonal antibody was then produced by a commercial service (Pacific Immunology) which generated antisera in rabbits by immunizing with ca. 500 µg of affinity purified truncated LEF-9. Antiserum was then purified by nitrocellulose-based immunoaffinity chromatography as previously outlined [65], [66]. The resulting antibody was then used in immunoblotting experiments with control and lef-9 knockdown wasps by explanting ovaries, separating ovary extracts on 5–20% SDS-PAGE gradient gels and transferring to PVDF (Immobilon). LEF-9 was then visualized using a goat anti-mouse secondary antibody and the ECL Advance kit as previously described [65], [67].
PCR-based quantification of viral titer, proviral DNA excision, and TEM
The titer of MdBV virions with DNAse-protected episomes of MdBV segment B was quantified by qPCR. One half of an ovary pair for each wasp individual was homogenized with a pestle in 100 µl of DNase buffer from the Roche HighPure RNA Isolation Kit, and NP40 was added to a final concentration of 1% to solubilize wasp cells and virus particle envelopes. After 20 min of gentle rocking, 1 µl of TURBO DNase from the Ambion DNA free kit was added and samples were incubated at 37°C for 40 min to digest all free wasp and episomal viral DNAs. After the addition of EDTA (10 mM) to inactivate the DNase, 250 µg of proteinase K (Roche) and 2% sarcosyl were added to samples, followed by incubation at 62°C for 1 h and by phenol∶chloroform extraction and ethanol precipitation in the presence of 0.3 M sodium acetate, pH 5.2. DNA pellets were resuspended in 30 µl of 10 mM Tris-Cl pH 8.5 and diluted to 1 ng/µl for use as template. Segment B specific primers flanking the point of circularization in the viral segment were used to amplify circularized viral DNA using qPCR as described above and previously [7] (Figure 7B, Table S2). Per-ovary copy numbers were calculated by multiplying the qPCR estimate of copy number for a half ovary by two, and by the dilution factor and elution volume. At least 3 independently acquired biological replicates were performed for each treatment with samples internally replicated 4 times. Quantification of viral segment excision was performed on genomic DNAs extracted from the entire tissue from an ovary half with the proteinase K, sarcosyl, and phenol/chloroform extraction method described above, without a DNAse step. Primers specific for the empty locus for Segment B were used in qPCR to quantify copy number (Figure 7B, Table S2). TEM was performed as in [31].
Infectivity assays
The CiE1 cell line was maintained as in [43]. The infectivity of virus preparations was measured by counting the number of Segment B viral genome copies in CiE1 cells after 24 hours of incubation [43]. Virus was collected from 24 h old wasps treated dsRNA by puncturing a half ovary in 50 µl of PBS and allowing the virus to dissolve. The entire contents of the droplet were added to a microcentrifuge tube containing 200 ul of Sf900 media with 5% fetal bovine serum and 1% antibiotics. This solution was filtered through a 0.45 µm membrane to remove any bacteria and cellular debris. Fifty µl of each virus preparation, corresponding to 0.1 wasp equivalents or an estimated MOI of 100 for control samples [29] was added to wells in a 24-well cell culture plate containing 105 CiE1 cells per well. Virus particles were incubated with cells for 2 h at room temperature, followed by removal of virus-containing media and the addition of 500 µl of fresh media. CiE1 cells were incubated for an additional 22 hours at 26°C. DNA was isolated from cells following the protocol for quantification of viral titer above without prior DNase treatment. All samples were diluted to a concentration of 50 ng/µl for qPCR amplification of circularized Segment B as described above.
Successful viral gene expression, translation and export of protein products in host cells was quantified by counting the percentage of cells displaying the MdBV protein GLC1.8 on their surface. CiE1 cells were infected with virus preparations as described above, and after 24 hours of total incubation were fixed with 3.7% paraformaldehyde and stained with a murine monoclonal antibody specific for GLC1.8 followed by goat anti-mouse Alexa-fluor 568 secondary antibody (Molecular Probes) as described [44]. One hundred CiE1 cells were counted from a randomly selected field of view using an epifluorescent, phase-contrast microscope (Leica DM IRB).
Statistical analyses
JMP v10 was used for all statistical analyses. For qPCR analyses, the number of copies of a gene or DNA product were averaged for all technical replicates within a biological replicate. For functional assays, means were calculated from experimental values derived from biological replicates. Each biological replicate represented an individual wasps' ovary. Differences between means of biological replicates were tested using a t-test assuming equal variances or ANOVA. Differences between means for experiments with more than two treatments were distinguished using Tukey's HSD test at the p<0.05 significance level.
Supporting Information
BV conserved gene-related proteins detected in MdBV virions. Conserved gene names and locus identifier in the M. demolitor ovary transcriptome are listed to the left [7]. Number of unique peptides from the two proteomic replicates are indicated in the middle columns relative to transcript abundance for each gene that was previously determined as reads per kilobase per million reads mapped (RPKM) in whole adult ovaries [7].
(PDF)
Primers used for dsRNA synthesis template amplification, qPCR assays of expression or segment abundance, and protein expression.
(PDF)
Knockdown of vlf-1 occurs over a range of ds- vlf-1 quantities and is detectable 2 days post-treatment. (A) M. demolitor larvae were injected with ds-eGFP (500 ng) or 50 ng, 500 ng or 5 µg of ds-vlf-1. The ovaries from individual, newly emerged adults wasps were then dissected and total RNA isolated. The bars in each graph compare copy number of vlf-1 per ng of total RNA in wasps treated with ds-eGFP and each dose of ds-vlf-1. (B) Effect of time post-injection of ds-vlf-1 on transcript knockdown. Larvae were injected with ds-vlf-1 as described in Fig. 2. Ovaries were then dissected from 2 day old pupae (stage 2), 3 day old pupae (stage 3), 1 day, 3 day and 5 day old adult wasps and total RNA extracted. Level of knockdown is presented as % inhibition relative to ovaries from 1 day adult wasp pretreated with ds-eGFP. Error bars, N values, and statistical significance are indicated as defined in Figure 2.
(PDF)
ds- int-1 has no effect on vlf-1 while ds- vlf-1-2 has a similar knockdown effect as ds- vlf-1 . M. demolitor larvae were injected with ds-eGFP, ds-int-1, or ds-vlf-1-2 which is specific for the vlf-1 gene but does not overlap ds-vlf-1 used in assays shown in Figure 2. The ovaries from individual, newly emerged adults wasps were then dissected and total RNA isolated. (A) The bars in the graph compare copy number of vlf-1 per ng of total RNA in wasps treated with ds-eGFP versus ds-int-1. (B) The bars in the graph compare copy number of vlf-1 per ng of total RNA in wasps treated with ds-eGFP versus ds-vlf-1-2. (C) Copy number of DNase-protected MdBV episomal genomic segment B in the ovaries of newly emerged M. demolitor adult females pretreated with ds-eGFP or ds-vlf-1-2. Error bars, N values, and statistical significance are indicated as defined in Figure 2.
(PDF)
Acknowledgments
We thank Mary Ard at the UGA Electron Microscopy Lab for electron microscopy sample preparation and microscope operation. We also thank Dennis Phillips and Chau-Wen Chou at the UGA Proteomics and Mass Spectrometry core facility for technical assistance with proteomic analysis of MdBV virions.
Funding Statement
This work was supported by the National Institutes of Health (F32 AI096552) (GRB), US National Science Foundation (IOS-1261328) (MRS) and US Department of Agriculture (2009-35302-05250) (MRS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Corradi N, Bonfante P (2012) The arbuscular mycorrhizal symbiosis: origin and evolution of a beneficial plant infection. PLoS Pathog 8: e1002600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moran NA (2006) Symbiosis. Curr Biol 16: R866–871. [DOI] [PubMed] [Google Scholar]
- 3. Husseneder C (2010) Symbiosis in subterranean termites: a review of insights from molecular studies. Environ Entomol 39: 378–388. [DOI] [PubMed] [Google Scholar]
- 4. Moreira D, Lopez-Garcia P (2009) Ten reasons to exclude viruses from the tree of life. Nat Rev Microbiol 7: 306–311. [DOI] [PubMed] [Google Scholar]
- 5. Villarreal LP (2009) Persistence pays: how viruses promote host group survival. Curr Opin Microbiol 12: 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Villarreal LP, Witzany G (2010) Viruses are essential agents within the roots and stem of the tree of life. J Theor Biol 262: 698–710. [DOI] [PubMed] [Google Scholar]
- 7. Burke GR, Strand MR (2012) Deep sequencing identifies viral and wasp genes with potential roles in replication of Microplitis demolitor bracovirus. J Virol 86: 3293–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strand MR (2010) Polydnaviruses. In: Asgari S, Johnson KN, editors. Insect Virology. Norwich, UK: Caister Academic Press. pp. 171–197. [Google Scholar]
- 9. Pennacchio F, Strand MR (2006) Evolution of developmental strategies in parasitic Hymenoptera. Annu Rev Entomol 51: 233–258. [DOI] [PubMed] [Google Scholar]
- 10.Strand MR, Drezen JM (2012) Family polydnaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus taxonomy: ninth report of the international committee on taxonomy of viruses. Amsterdam: Elsevier. pp. 237–248.
- 11. Strand MR, Burke GR (2012) Polydnaviruses as symbionts and gene delivery systems. PLoS Pathog 8: e1002757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bezier A, Annaheim M, Herbiniere J, Wetterwald C, Gyapay G, et al. (2009) Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science 323: 926–930. [DOI] [PubMed] [Google Scholar]
- 13. Desjardins CA, Gundersen-Rindal DE, Hostetler JB, Tallon LJ, Fadrosh DW, et al. (2008) Comparative genomics of mutualistic viruses of Glyptapanteles parasitic wasps. Genome Biol 9: R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Webb BA, Strand MR, Dickey SE, Beck MH, Hilgarth RS, et al. (2006) Polydnavirus genomes reflect their dual roles as mutualists and pathogens. Virology 347: 160–174. [DOI] [PubMed] [Google Scholar]
- 15. Albrecht U, Wyler T, Pfister-Wilhelm R, Gruber A, Stettler P, et al. (1994) Polydnavirus of the parasitic wasp Chelonus inanitus (Braconidae): characterization, genome organization and time point of replication. J Gen Virol 75: 3353–3363. [DOI] [PubMed] [Google Scholar]
- 16. Annaheim M, Lanzrein B (2007) Genome organization of the Chelonus inanitus polydnavirus: excision sites, spacers and abundance of proviral and excised segments. J Gen Virol 88: 450–457. [DOI] [PubMed] [Google Scholar]
- 17. Marti D, Grossniklaus-Burgin C, Wyder S, Wyler T, Lanzrein B (2003) Ovary development and polydnavirus morphogenesis in the parasitic wasp Chelonus inanitus. I. Ovary morphogenesis, amplification of viral DNA and ecdysteroid titres. J Gen Virol 84: 1141–1150. [DOI] [PubMed] [Google Scholar]
- 18. Savary S, Beckage N, Tan F, Periquet G, Drezen JM (1997) Excision of the polydnavirus chromosomal integrated EP1 sequence of the parasitoid wasp Cotesia congregata (Braconidae, Microgastinae) at potential recombinase binding sites. J Gen Virol 78: 3125–3134. [DOI] [PubMed] [Google Scholar]
- 19. Savary S, Drezen JM, Tan F, Beckage NE, Periquet G (1999) The excision of polydnavirus sequences from the genome of the wasp Cotesia congregata (Braconidae, microgastrinae) is developmentally regulated but not strictly restricted to the ovaries in the adult. Insect Mol Biol 8: 319–327. [DOI] [PubMed] [Google Scholar]
- 20. Wyler T, Lanzrein B (2003) Ovary development and polydnavirus morphogenesis in the parasitic wasp Chelonus inanitus. II. Ultrastructural analysis of calyx cell development, virion formation and release. J Gen Virol 84: 1151–1163. [DOI] [PubMed] [Google Scholar]
- 21. Burke GR, Strand MR (2012) Polydnaviruses of parasitic wasps: domestication of viruses to act as gene delivery vectors. Insects 3: 91–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreau SJM, Huguet E, Drezen JM (2009) Polydnaviruses as tools to deliver wasp virulence factors to impair lepidopteran host immunity. In: Reynolds S, editor. Insect Infection and Immunity: Evolution, Ecology, and Mechanisms. Oxford, UK: Oxford University Press. pp. 137–158. [Google Scholar]
- 23.Jehle JA (2010) Nudiviruses. In: Asgari S, Johnson KN, editors. Insect Virology. Norwich, UK: Caister Academic Press. pp. 153–170. [Google Scholar]
- 24.Rohrmann GF (2011) Baculovirus molecular biology. Bethesda: National Library of Medicine, National Center for Biotechnology information.
- 25. Herniou EA, Olszewski JA, Cory JS, O'Reilly DR (2003) The genome sequence and evolution of baculoviruses. Annu Rev Entomol 48: 211–234. [DOI] [PubMed] [Google Scholar]
- 26. Murphy N, Banks JC, Whitfield JB, Austin AD (2008) Phylogeny of the parasitic microgastroid subfamilies (Hymenoptera: Braconidae) based on sequence data from seven genes, with an improved time estimate of the origin of the lineage. Mol Phylogenet Evol 47: 378–395. [DOI] [PubMed] [Google Scholar]
- 27. Theze J, Bezier A, Periquet G, Drezen JM, Herniou EA (2011) Paleozoic origin of insect large dsDNA viruses. Proc Natl Acad Sci U S A 108: 15931–15935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Webb BA, Strand MR (2005) The biology and genomics of polydnaviruses. In: Gilbert L, Iatrou I, Gill SS, editors. Comprehensive molecular insect science. Boston: Elsevier. pp. 323–360. [Google Scholar]
- 29. Beck MH, Inman RB, Strand MR (2007) Microplitis demolitor bracovirus genome segments vary in abundance and are individually packaged in virions. Virology 359: 179–189. [DOI] [PubMed] [Google Scholar]
- 30. Beck MH, Zhang S, Bitra K, Burke GR, Strand MR (2011) The encapsidated genome of Microplitis demolitor bracovirus integrates into the host Pseudoplusia includens . J Virol 85: 11685–11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Strand MR, McKenzie DI, Grassl V, Dover BA, Aiken JM (1992) Persistence and expression of Microplitis demolitor polydnavirus in Pseudoplusia includens . J Gen Virol 73: 1627–1635. [DOI] [PubMed] [Google Scholar]
- 32. Ma Y, Creanga A, Lum L, Beachy PA (2006) Prevalence of off-target effects in Drosophila RNA interference screens. Nature 443: 359–363. [DOI] [PubMed] [Google Scholar]
- 33. Guarino LA, Xu B, Jin J, Dong W (1998) A virus-encoded RNA polymerase purified from baculovirus-infected cells. J Virol 72: 7985–7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rankin C, Ooi BG, Miller LK (1988) Eight base pairs encompassing the transcriptional start point are the major determinant for baculovirus polyhedrin gene expression. Gene 70: 39–49. [DOI] [PubMed] [Google Scholar]
- 35. Xing K, Deng R, Wang J, Feng J, Huang M, et al. (2005) Analysis and prediction of baculovirus promoter sequences. Virus Res 113: 64–71. [DOI] [PubMed] [Google Scholar]
- 36. Jin J, Dong W, Guarino LA (1998) The LEF-4 subunit of baculovirus RNA polymerase has RNA 5′-triphosphatase and ATPase activities. J Virol 72: 10011–10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gross CH, Shuman S (1998) RNA 5′-triphosphatase, nucleoside triphosphatase, and guanylyltransferase activities of baculovirus LEF-4 protein. J Virol 72: 10020–10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lu A, Miller LK (1994) Identification of three late expression factor genes within the 33.8- to 43.4-map-unit region of Autographa californica nuclear polyhedrosis virus. J Virol 68: 6710–6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD (2006) Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell 127: 941–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vanarsdall AL, Okano K, Rohrmann GF (2006) Characterization of the role of very late expression factor 1 in baculovirus capsid structure and DNA processing. J Virol 80: 1724–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McLachlin JR, Miller LK (1994) Identification and characterization of vlf-1, a baculovirus gene involved in very late gene expression. J Virol 68: 7746–7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang S, Miller LK (1999) Activation of baculovirus very late promoters by interaction with very late factor 1. J Virol 73: 3404–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Johnson JA, Bitra K, Zhang S, Wang L, Lynn DE, et al. (2010) The UGA-CiE1 cell line from Chrysodeixis includens exhibits characteristics of granulocytes and is permissive to infection by two viruses. Insect Biochem Mol Biol 40: 394–404. [DOI] [PubMed] [Google Scholar]
- 44. Beck M, Strand MR (2005) Glc1.8 from Microplitis demolitor bracovirus induces a loss of adhesion and phagocytosis in insect high five and S2 cells. J Virol 79: 1861–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stoltz DB, Vinson SB (1977) Baculovirus-like particles in the reproductive tracts of female parasitoid wasps. II. The genus Apanteles . Can J Microbiol 23: 28–37. [DOI] [PubMed] [Google Scholar]
- 46. Stoltz DB, Vinson SB, MacKinnon EA (1976) Baculovirus-like particles in the reproductive tracts of female parasitoid wasps. Can J Microbiol 22: 1013–1023. [DOI] [PubMed] [Google Scholar]
- 47. Faulkner P, Kuzio J, Williams GV, Wilson JA (1997) Analysis of p74, a PDV envelope protein of Autographa californica nucleopolyhedrovirus required for occlusion body infectivity in vivo. J Gen Virol 78: 3091–3100. [DOI] [PubMed] [Google Scholar]
- 48. Kikhno I, Gutierrez S, Croizier L, Croizier G, Ferber ML (2002) Characterization of pif, a gene required for the per os infectivity of Spodoptera littoralis nucleopolyhedrovirus. J Gen Virol 83: 3013–3022. [DOI] [PubMed] [Google Scholar]
- 49. Ohkawa T, Washburn JO, Sitapara R, Sid E, Volkman LE (2005) Specific binding of Autographa californica M nucleopolyhedrovirus occlusion-derived virus to midgut cells of Heliothis virescens larvae is mediated by products of pif genes Ac119 and Ac022 but not by Ac115. J Virol 79: 15258–15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peng K, van Oers MM, Hu Z, van Lent JW, Vlak JM (2010) Baculovirus per os infectivity factors form a complex on the surface of occlusion-derived virus. J Virol 84: 9497–9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Strand MR (1994) Microplitis demolitor polydnavirus infects and expresses in specific morphotypes of Pseudoplusia includens haemocytes. J Gen Virol 75: 3007–3020. [DOI] [PubMed] [Google Scholar]
- 52. Bitra K, Zhang S, Strand MR (2011) Transcriptomic profiling of Microplitis demolitor bracovirus reveals host, tissue and stage-specific patterns of activity. J Gen Virol 92: 2060–2071. [DOI] [PubMed] [Google Scholar]
- 53. Cambray G, Guerout AM, Mazel D (2010) Integrons. Annu Rev Genet 44: 141–166. [DOI] [PubMed] [Google Scholar]
- 54. Wetterwald C, Roth T, Kaeslin M, Annaheim M, Wespi G, et al. (2010) Identification of bracovirus particle proteins and analysis of their transcript levels at the stage of virion formation. J Gen Virol 91: 2610–2619. [DOI] [PubMed] [Google Scholar]
- 55. Hou D, Zhang L, Deng F, Fang W, Wang R, et al. (2012) Comparative proteomics reveal fundamental structural and functional differences between the two progeny phenotypes of a baculovirus. J Virol 87: 829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shin SW, Kokoza VA, Raikhel AS (2003) Transgenesis and reverse genetics of mosquito innate immunity. J Exp Biol 206: 3835–3843. [DOI] [PubMed] [Google Scholar]
- 57. Bai H, Zhu F, Shah K, Palli SR (2011) Large-scale RNAi screen of G protein-coupled receptors involved in larval growth, molting and metamorphosis in the red flour beetle. BMC Genomics 12: 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Terenius O, Papanicolaou A, Garbutt JS, Eleftherianos I, Huvenne H, et al. (2011) RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol 57: 231–245. [DOI] [PubMed] [Google Scholar]
- 59. Li X, Song J, Jiang T, Liang C, Chen X (2007) The N-terminal hydrophobic sequence of Autographa californica nucleopolyhedrovirus PIF-3 is essential for oral infection. Arch Virol 152: 1851–1858. [DOI] [PubMed] [Google Scholar]
- 60. Katou Y, Ikeda M, Kobayashi M (2006) Abortive replication of Bombyx mori nucleopolyhedrovirus in Sf9 and High Five cells: defective nuclear transport of the virions. Virology 347: 455–465. [DOI] [PubMed] [Google Scholar]
- 61. Nunes-Duby SE, Kwon HJ, Tirumalai RS, Ellenberger T, Landy A (1998) Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res 26: 391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Duyne GD (2002) A structural view of tyrosine recombinase. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: ASM Press. pp. 93–117. [Google Scholar]
- 63. Collis CM, Hall RM (1992) Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol Microbiol 6: 2875–2885. [DOI] [PubMed] [Google Scholar]
- 64. Smith MA, Rodriguez JJ, Whitfield JB, Deans AR, Janzen DH, et al. (2008) Extreme diversity of tropical parasitoid wasps exposed by iterative integration of natural history, DNA barcoding, morphology, and collections. Proc Natl Acad Sci U S A 105: 12359–12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Eum J-H, Bottjen RC, Pruijssers AJ, Clark KD, Strand MR (2010) Characterization and kinetic analysis of protein tyrosine phosphatase-H2 from Microplitis demolitor bracovirus. Insect Bioch Mol Biol 40: 690–698. [DOI] [PubMed] [Google Scholar]
- 66. Robinson PA, Anderton BH, Loviny TL (1988) Nitrocellulose-bound antigen repeatedly used for the affinity purifcaiotn of specific polyclonal antibodies for screening DNA expression libraries. J Immunol Methods 108: 115–122. [DOI] [PubMed] [Google Scholar]
- 67. Bitra K, Suderman RJ, Strand MR (2012) Polydnavirus Ank proteins bind NF-kappaB homodimers and inhibit processing of Relish. PLoS Pathog 8: e1002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BV conserved gene-related proteins detected in MdBV virions. Conserved gene names and locus identifier in the M. demolitor ovary transcriptome are listed to the left [7]. Number of unique peptides from the two proteomic replicates are indicated in the middle columns relative to transcript abundance for each gene that was previously determined as reads per kilobase per million reads mapped (RPKM) in whole adult ovaries [7].
(PDF)
Primers used for dsRNA synthesis template amplification, qPCR assays of expression or segment abundance, and protein expression.
(PDF)
Knockdown of vlf-1 occurs over a range of ds- vlf-1 quantities and is detectable 2 days post-treatment. (A) M. demolitor larvae were injected with ds-eGFP (500 ng) or 50 ng, 500 ng or 5 µg of ds-vlf-1. The ovaries from individual, newly emerged adults wasps were then dissected and total RNA isolated. The bars in each graph compare copy number of vlf-1 per ng of total RNA in wasps treated with ds-eGFP and each dose of ds-vlf-1. (B) Effect of time post-injection of ds-vlf-1 on transcript knockdown. Larvae were injected with ds-vlf-1 as described in Fig. 2. Ovaries were then dissected from 2 day old pupae (stage 2), 3 day old pupae (stage 3), 1 day, 3 day and 5 day old adult wasps and total RNA extracted. Level of knockdown is presented as % inhibition relative to ovaries from 1 day adult wasp pretreated with ds-eGFP. Error bars, N values, and statistical significance are indicated as defined in Figure 2.
(PDF)
ds- int-1 has no effect on vlf-1 while ds- vlf-1-2 has a similar knockdown effect as ds- vlf-1 . M. demolitor larvae were injected with ds-eGFP, ds-int-1, or ds-vlf-1-2 which is specific for the vlf-1 gene but does not overlap ds-vlf-1 used in assays shown in Figure 2. The ovaries from individual, newly emerged adults wasps were then dissected and total RNA isolated. (A) The bars in the graph compare copy number of vlf-1 per ng of total RNA in wasps treated with ds-eGFP versus ds-int-1. (B) The bars in the graph compare copy number of vlf-1 per ng of total RNA in wasps treated with ds-eGFP versus ds-vlf-1-2. (C) Copy number of DNase-protected MdBV episomal genomic segment B in the ovaries of newly emerged M. demolitor adult females pretreated with ds-eGFP or ds-vlf-1-2. Error bars, N values, and statistical significance are indicated as defined in Figure 2.
(PDF)