Abstract
Nucleosome organization at promoter regions plays an important role in regulating gene activity. Genome-wide studies in yeast, flies, worms, mammalian embryonic stem cells and transformed cell lines have found well-positioned nucleosomes flanking a nucleosome depleted region (NDR) at transcription start sites. This nucleosome arrangement depends on DNA sequence (cis-elements) as well as DNA binding factors and ATP-dependent chromatin modifiers (trans-factors). However, little is understood about how the nascent embryonic genome positions nucleosomes during development. This is particularly intriguing since the embryonic genome must undergo a broad reprogramming event upon fusion of sperm and oocyte. Using four stages of early embryonic zebrafish development, we map nucleosome positions at the promoter region of 37 zebrafish hox genes. We find that nucleosome arrangement at the hox promoters is a progressive process that takes place over several stages. At stages immediately after fertilization, nucleosomes appear to be largely disordered at hox promoter regions. At stages after activation of the embryonic genome, nucleosomes are detectable at hox promoters, with positions becoming more uniform and more highly occupied. Since the genomic sequence is invariant during embryogenesis, this progressive change in nucleosome arrangement suggests that trans-factors play an important role in organizing nucleosomes during embryogenesis. Separating hox genes into expressed and non-expressed groups shows that expressed promoters have better positioned and occupied nucleosomes, as well as distinct NDRs, than non-expressed promoters. Finally, by blocking the retinoic acid-signaling pathway, we disrupt early hox gene transcription, but observe no effect on nucleosome positions, suggesting that active hox transcription is not a driving force behind the arrangement of nucleosomes at the promoters of hox genes during early development.
Introduction
The nucleosome is comprised of an octamer histone core wrapped nearly 1.7 times by approximately 147 bp of DNA that represents the basic unit of eukaryotic chromatin [1]. While packaging of nucleosomes into a higher order structure enables the compaction of chromatin into the nucleus, it also limits access to various DNA binding factors, thereby placing an accessibility constraint on all DNA-dependent processes (e.g. replication, transcription) [2]. Nucleosome arrangements on genomic DNA are defined both in terms of positioning (how precisely a nucleosome resides at a particular site in all cells of a population) and occupancy (how frequently a specific position is bound by a nucleosome). In particular, nucleosome positioning and occupancy at transcription start sites (TSSs) is thought to impact gene expression. Accordingly, genome-wide nucleosome mapping studies in yeast have revealed a nucleosome-depleted region (NDR) upstream of most TSSs [3]–[7] that likely permits access by the transcription machinery. However, some yeast promoters appear to be occupied by nucleosomes that are actively removed in response to inducing signals [8]–[10]. Such promoters display higher transcriptional plasticity and are more responsive to signaling pathways, than are promoters with pronounced NDRs, suggesting that nucleosome positioning represents a mechanism to achieve regulated gene expression in yeast [11]. Nucleosome positioning may play an even greater role in the regulation of gene expression in metazoans since regulatory DNA sequences are invariant among all cells of a multi-cellular organism, but only a subset of cells may express a specific gene. Indeed, while many promoters in flies [12]–[14], worms [15], [16], fish [17], and humans [18], [19] display NDRs upstream of TSSs, many other promoters are occupied by nucleosomes [20] and inductive signals cause nucleosome rearrangements at such promoters (e.g. nucleosome occupancy is greatly increased in the region immediately upstream of repressed promoters upon T-lymphocyte stimulation [19] and NDRs form at androgen-responsive enhancers in prostate cells [21]). This suggests that nucleosomes need to be rearranged at many metazoan promoters prior to transcription and, accordingly, there is an overall bias towards expressed promoters having a more pronounced NDR [12], [18], [19].
Nucleosome positioning is partially encoded by the DNA sequence and experimental studies have identified sequences that favor (e.g. dinucleotide repeats [22], [23] and G+C rich regions [5], [24]) or disfavor (e.g. dA:dT tracts [3], [5], [18], [25], [26]) nucleosome binding. More recently, experimentally derived nucleosome position information has been used to design theoretical models for the purpose of predicting nucleosome positioning de novo. These models are reasonably successful at predicting nucleosome positions in yeast [24], [27]–[29], but are less successful in C. elegans [7] or in human cells [20]. In particular, the models appear less accurate at predicting nucleosome positioning at metazoan regulatory regions (including promoters [7], [20]). Notably, regulatory regions have higher G+C content in metazoans than in yeast and are therefore more likely to be bound by nucleosomes [20]. As discussed above, such nucleosomes are actively removed in cells where the corresponding promoter is expressed, possibly accounting for the observed discrepancies between predicted and actual nucleosome positioning. Nucleosomes may be repositioned from such G+C rich promoter regions by a variety of mechanisms, including competition with sequence-specific transcription factors [30], [31] or the RNA Polymerase II complex [13], [14], [19], [32], [33], as well as by the action of ATP-dependent nucleosome remodelers (reviewed in [34]). It is also worth noting that regions defined as NDRs are not necessarily completely devoid of nucleosomes [33], [35], but may represent sites with less robust nucleosomes, perhaps because they contain histone variants such as H2.AZ or H3.3 that are less stably bound to DNA [36]. Such nucleosomes are more easily displaced and might therefore make promoters more responsive to inductive signals, but would also make them more sensitive to DNase-based methods used to map nucleosome organization. Taken together, work to date suggests that active processes control nucleosome positioning at many promoters and that this is an important regulatory mechanism for inducible and cell-specific gene expression in metazoans.
Nucleosome organization has been analyzed in blastula stage O. latipes (medaka fish [17]) embryos, as well as in samples of mixed stage D. melanogaster [13] and C. elegans [15], [37] embryos. In spite of metazoan embryos consisting of multiple cell types, these experiments nevertheless detected well-organized nucleosomes. In particular, many promoters reveal a nucleosome arrangement with pronounced nucleosomes flanking the TSS. One nucleosome is observed downstream of the TSS in the coding sequence (+1 nucleosome) and a second upstream of the TSS (−1 nucleosome) with an intervening NDR observed immediately upstream of the TSS. This represents a canonical arrangement in most embryonic cells regardless of tissue type, stage of development or level of transcription. However, it is not clear that such a pattern is truly fixed throughout embryogenesis since chromatin structure appears to be remodeled during embryonic development. For instance, the hox genes, which encode homeodomain-containing transcription factors essential for development of all metazoans [38], [39] and that are arranged into several genomic clusters, have been observed to decondense coincident with their expression during mouse embryogenesis [40], [41] – a process that can be mimicked by using retinoic acid (RA; an endogenous inducer of hox gene expression) to treat murine ES cells [42]. Chromatin rearrangements at the hox clusters have also been observed during mouse embryogenesis using 4C technology [43]. Hence, while the canonical arrangement of a +1 nucleosome at the TSS preceded by an upstream NDR has been observed at hox promoters in human cell lines [44], it is unclear if chromatin remodeling during embryonic development generates nucleosome profiles that differ from the canonical organization. Indeed a time course of nucleosome organization, and its refinement in response to inductive signals, has not been reported for any metazoan embryo.
We have mapped nucleosomes near the TSS (herein referred to as ‘promoter’) of 37 zebrafish hox genes under different conditions. We first examined nucleosome arrangements at the TSS of all 37 genes at various stages of embryogenesis and find relatively poorly positioned and weakly occupied nucleosomes at 2 hpf and 4 hpf. Notably, no hox genes are expressed at these stages of development and we do not observe NDRs at these time points. At the 6 hpf and 9 hpf time points nucleosomes become better organized. The progressive nature of nucleosome positioning on the invariant sequence of hox promoters through early development suggests an important role for trans-factors in positioning nucleosomes at hox promoters. More detailed analyses revealed that promoters of genes expressed at these stages have better nucleosome organization and occupancy with an NDR immediately upstream of the TSS. Non-expressed promoters have nucleosomes that are less organized and lack an NDR at early stages, suggesting that NDR formation correlates with gene expression. However, blocking hox gene transcription by disruption of the RA signaling pathway results in no change in nucleosome positioning or NDR formation, indicating that transcription does not drive nucleosome organization at hox promoters. Our data therefore indicate that trans-factors act at hox promoters during embryogenesis to dynamically rearrange nucleosomes independently of hox gene transcription.
Materials and Methods
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Animal Experiments of the University of Massachusetts (A-1565).
Fish Care
Ekkwill (EK) embryos were collected through natural matings and staged using morphological criteria for two, four, six, and nine hours post fertilization (hpf) as defined by Kimmel et al [45].
Drug Treatments
Retinoic acid (RA): 2 cell embryos (∼45 minutes post-fertilization) were treated with 100 nM RA diluted in fish-water (5 mM NaCl, 0.17 mM KCL, 0.33 mm CaCl2, 0.33 mM MgSO4, and 0.004% methylene blue). Embryos remained in RA-treated water until they were harvested (2 hpf RA embryos were treated for ∼1 hour, 4 hpf embryos ∼3 hours etc.). Diethylaminobenzaldehyde (DEAB): 4–8 cell embryos (∼1–1.25 hours post fertilization) were treated with 10 uM DEAB diluted in fish-water. Embryos remained in DEAB-water until the developmental stage harvested. Drug concentrations were chosen based on embryonic survival to limit embryonic death.
Embryo Processing and Nucleosome Cross-linking
Embryos were collected and the chorion was removed using 10 mg/ml Pronase. Embryos were then washed with Fish ringers (0.1 M NaCl, 3 mM KCl, 3 mM CaCl2, 2.4 mM NaHCO3) and mechanically dissociated by pipetting. Cells were washed once with PBS, resuspended in 1% formaldehyde in PBS and incubated for 10 minutes at 27°C. The reaction was quenched with equal volume of 1M glycine and cells were spun down at 5000 g.
Nuclei Purification
Protocol was adapted from Dennis et al 2007 [46]. Cell pellets were resuspended by pipetting vigorously in sucrose buffer (0.3 M sucrose, 2 mM MgAc2, 3 mM CaCl2, 1% Triton X-100, 500 uM DTT, 1× complete protease inhibitor Roche: 11873580001, and 10 mM HEPES at pH 7.8) and incubated for 30 minutes on ice. Cells were pipetted vigorously again and diluted 1∶1 with GB buffer (25% glycerol, 5 mM MgAc2, 0.1 mM EDTA, 500 uM DTT, 1× complete protease inhibitor Roche: 11873580001, and 10 mM HEPES at pH 7.8). Nuclei were purified by layering on an equal volume of GB and spun at 1000 g for 10 minutes at 4°C.
MNase Digestion and Chromatin Purification
Protocol was adapted from Yuan et al 2005 [3]. Isolated nuclei were resuspended and washed once in Reaction buffer (50 mM NaCl, 10 mM Tris pH 7.4, 5 mM MgCl2, 1 mM CaCl2, 1 mM β-mercaptoethanol, 500 uM spermidine and 500 uM DTT) followed by resuspension in reaction buffer with a titrated amount of MNase (5–20 units/ml, Worthington: LS004797) and incubated at 37°C for 10 minutes. Reactions were terminated with 50 mM EDTA and placed on ice. Samples were then diluted in water and treated with 1× RNase cocktail (Ambion: AM2286) and 200 mM NaCl (to remove RNA and reverse crosslinks) and incubated at 55°C for 2 hours. 2 ul proteinase K (20 mg/ml) was added and samples were placed at 65°C overnight. Chromatin was extracted using phenol:chloroform followed by ethanol precipitation. Samples were visualized by gel electrophoresis and samples containing a 80–90% mono-nucleosome DNA (faint tri-nucleosome band visible) were used for tiling array hybridization. Mono-nucleosome sized fragments were gel extracted using the Qiagen Gel Extraction kit (28706).
Array Build and Hybridization
Zebrafish genome v7 sequence of the seven hox clusters was masked for repetitive sequence using the Sanger Institute’s Zebrafish RepeatMasker (http://www.sanger.ac.uk/Projects/D_rerio/fishmask.shtml). The resulting sequences were used to construct a 144 k feature array of 50 bp probes positioned every 20 bp designed using Agilent eArray web software (https://earray.chem.agilent.com/earray/GEO: GPL16536). Isolated mono-nucleosome sized fragments were hybridized to the hox array using protocols adapted from Agilent protocols substituting COT DNA for salmon sperm DNA (Mammalian ChIP-on-chip Protocol G4481-90010). Arrays were scanned using either an Axon 4000B or Agilent’s High-Resolution C Scanner.
Array Analysis and Nucleosome Positioning
Probe sequences were remapped to Zv9 and the distance from the center of a probe to the TSS of the nearest hox gene was calculated. Log2 ratios were calculated based on normalized r-processed and g-processed signals from the Agilent chip for each probe. Mean signal from two replicates for each sample was assigned to each probe location. Signals were tallied using a 30 bp sliding window with a step of 10 bp for each window. A Lowess fitting line (f = 0.05) was plotted to show the trend of the aggregated signals. Nucleosome spacing was calculated based on the predicted di- and mono-nucleosome sized fragments identified from gel images, represented in figure S2 in File S1. Our observations indicate that the di-nucleosome band is 320–360 bp, the mono-nucleosome band 150–175 bp and the linker is 20–60 bp, indicating that the peak-to-peak distance between neighboring nucleosomes is 170–210 bp. This distance was used in the text when comparing observed peak distances in the aggregate nucleosome plots. Signals for expressed and non-expressed genes were compared using a two-sided non-paired Wilcoxon rank sum test to calculate the significance of the difference between the two gene sets (GEO: GSE43757 ).
hox Expression
hox gene expression was determined using both Agilent and Affymetrix Zebrafish expression arrays. Only genes found to be expressed by both platforms were included in the RA and WT expression groups. Agilent Arrays: RNA was isolated from retinoic acid treated and untreated WT zebrafish embryos at 2 hpf, 4 hpf, 6 hpf, and 9 hpf embryos using Trizol (Invitrogen#15596-026) following standard procedures. RNA was processed and hybridized to Agilent Zebrafish (V3) Gene Expression Microarrays (G2519F-026437) essentially as outlined in Agilent protocols. Since no hox genes are reported to be expressed maternally, the 2 hpf WT embryonic sample was taken to represent baseline and signal above this baseline was taken to represent expression (GEO: GSE43756 ). Affymetrix Arrays: RNA was isolated from retinoic acid treated embryos at 4 hpf, 6 hpf, and 9 hpf while RNA from untreated embryos was collected at 9 hpf. RNA was processed and hybridized to Zebrafish Genechip Arrays (900487) by the UMass Genomic Core facility using standard Affymetrix protocols. CEL files from Affymetrix were normalized using invariantset probe set and background corrected by mas5 using expresso from the R affy package. Present/absent calls were calculated using mas5 call from R affy package with default parameters (GEO: GSE43755).
QPCR
DEAB-treated embryos were collected at 9 hpf and RNA was extracted using Trizol. cDNA was synthesized using the Superscript III RT First strand cDNA synthesis kit priming with oligo dT (18080-051). hox gene cDNA was quantified by QPCR using the Qiagen QuantiFast SYBR Green PCR kit (204054) on an ABI 7300 thermocycler. hox expression was normalized to a beta-actin control. Data represents 3 technical replicates.
Primers
hoxb1a: FWD-5′-ACC TAC GCT GAC TTA TCG GCC TCT CAA GG
RVS-5′-CTC AAG TGT GGC AGC AAT CTC CAC ACG
hoxb7a: FWD-5′-CCA TCC GAA TCT ACC CAT GGT GAG CGC
RVS-5′-TCT CGA TAC GCC GCC GTC TTG AAA GG
hoxb1b: FWD-5′-GGT TCG TTC AGC AAG TAT CAG GTC TCC CC
RVS-5′-TCT CAA GTT CCG TGA GCT GCT TGG TGG
hoxb5b: FWD-5′-CCT AAC CCA GGA CCA GTG CAA GAC GG
RVS-5′-CGT TCC GTC AAA CAC AGA GCG TGC G
hoxb6b: FWD-5′-AGT GCA AGA CGG ACT GCA CAG AAC AGG
RVS-5′-CGT TCC GTC AAA CAC AGA GCG TGC G
hoxc8a: FWD-5′-AGC AAG AGG CCA CCT TAG CGC AAT ACC
RVS-5′-CTT CAA TAC GGC GCT TGC GTG TGA GG
hoxc9a: FWD-5′-CGG AGA CTG TTT GGG CTC GAA CGG A
RVS-5′-ACC TCA TAT CGC CGG TCT CTT GTG AGG T
Beta-Actin: FWD-5′-ATA CAC AGC CAT GGA TGA GGA AAT CC
RVS-5′-GGT CGT CCA ACA ATG GAG GGG AAA A
Transcription Start Sites and Genes Included in Study
For this study we used the Embryonic Transcriptome TSSs determined in Pauli et al [47]. Genes with multiple TSSs were left out of this study. This resulted in the inclusion of 37 of the 44 known Zebrafish hox genes (Table 1).
Table 1. hox gene expression during zebrafish embryogenesis.
| 9 hpf WT non-expressed | 9 hf WT expressed | 6 hpf RA treated uninduced | 6 hpf RA treated induced | RA-only |
| hoxa4a | hoxb1a | hoxa9a | hoxa4a | hoxa4a |
| hoxa5a | hoxb7a | hoxa11a | hoxa5a | hoxa5a |
| hoxa9a | hoxb5b | hoxa13a | hoxb1a | hoxb5a |
| hoxa11a | hoxb6b | hoxa9b | hoxb5a | hoxc1a |
| hoxa13a | hoxc8a | hoxa11b | hoxb5b | hoxc4a |
| hoxa9b | hoxc9a | hoxa13b | hoxb6b | hoxc5a |
| hoxa11b | hoxb2a | hoxc1a | ||
| hoxa13b | hoxb4a | hoxc4a | ||
| hoxb2a | hoxb6a | hoxc5a | ||
| hoxb4a | hoxb7a | |||
| hoxb5a | hoxb9a | |||
| hoxb6a | hoxb13a | |||
| hoxb9a | hoxb8b | |||
| hoxb13a | hoxc6a | |||
| hoxb8b | hoxc8a | |||
| hoxc1a | hoxc9a | |||
| hoxc4a | hoxc10a | |||
| hoxc5a | hoxc11a | |||
| hoxc6a | hoxc12a | |||
| hoxc10a | hoxc13a | |||
| hoxc11a | hoxc6b | |||
| hoxc12a | hoxc12b | |||
| hoxc13a | hoxd4a | |||
| hoxc6b | hoxd9a | |||
| hoxc12b | hoxd10a | |||
| hoxd4a | hoxd11a | |||
| hoxd9a | hoxd12a | |||
| hoxd10a | hoxd13a | |||
| hoxd11a | ||||
| hoxd12a | ||||
| hoxd13a |
Results
To investigate nucleosome organization at hox promoters during embryogenesis, we used zebrafish (Danio rerio) embryos from 2, 4, 6, and 9 hours post fertilization (hpf). These time points were chosen since zygotic gene expression is initiated at 3–4 hpf in the zebrafish [48]. Hence, 2 hpf and 4 hpf embryos consist of a relatively uniform population of largely undifferentiated cells in which hox genes are not transcribed, while in 6 hpf and 9 hpf embryos some cell populations have begun to differentiate and hox gene transcription is being initiated. Nucleosome densities were determined by micrococcal nuclease (MNase) digestion of cross-linked chromatin isolated from staged embryos (adapted from [46]). Mononucleosome sized fragments were gel-purified and hybridized to an Agilent custom DNA array tiled with 50 bp oligonucleotides positioned every 20 bp across the seven zebrafish hox clusters. Randomly fragmented mononucleosome sized genomic DNA (gDNA) was co-hybridized as a control. The nucleosomal signal was expressed as a ratio of the MNase digested fragments to the random gDNA fragments. Nucleosome densities were averaged for 37 zebrafish hox genes (Table 1) from −600 bp to +600bp relative to the annotated transcription start site (TSS). Two separate MNase digestions were carried out for each time point and we find that the results are highly reproducible (r2 values range from 0.70 to 0.93; S1 in File S1).
Nucleosome Organization at hox Promoters is Dynamic during Embryogenesis
MNase digests revealed that mononucleosome fragments are 150–175 bp and dinucleosome fragments are 320–360 bp in zebrafish (Fig. S2 in File S1), indicating that linker regions range from 20–60 bp. This is similar to results seen for other fish species [17]. Based on these observations, the expected distance between two nucleosome peaks is 170–210 bp.
Our analysis revealed that nucleosomes are poorly occupied and positioned in 2 hpf and 4 hpf embryos (Fig. 1A and B). In particular, we are unable to identify any peaks that correspond to the predicted size of a nucleosome at these stages. Instead peaks have low amplitudes and are broad, indicating low occupancy and a lack of uniform positioning in the promoter region. At 6 hpf, nucleosome peaks begin to appear roughly +60, +260 and +480 bp from the TSS (+1, +2, and +3 nucleosomes respectively in Fig. 1C). The spacing of these peaks (200 bp and 220 bp respectively) indicates a nucleosomal unit of ∼150 bp of protected sequence separated by a linker fragment of ∼60 bp – values that correspond to those expected based on our gel analysis. We note that the amplitudes of the peaks in this region remain modest at 6 hpf, suggesting either that nucleosome occupancy is limited in all embryonic cells, or that nucleosomes are becoming more highly occupied in only a subset of cells. As in 2 hpf and 4 hpf embryos, nucleosomes upstream of the TSS are loosely positioned in 6 hpf embryos. At 9 hpf, nucleosome peaks are observed at roughly −450, −290, −170, +115, and +250 bp (−3, −2, −1, +1, and +2 nucleosomes respectively in Fig. 1D). The amplitude of the nucleosome peaks is greater at 9 hpf than 6 hpf. In particular, the amplitude of the +1 peak increases relative to the other peaks, indicating that nucleosome occupancy increases at this position. We interpret the change in nucleosome occupancy and positioning from 6 hpf to 9 hpf to mean that nucleosomes are less uniformly positioned at 6 hpf and take on more uniform positions by 9 hpf. However, the distances between the −3/−2, −2/−1 and +1/+2 peaks (150 bp, 120 bp, and 130 bp respectively) are closer than the expected distance between nucleosomes, possibly due to nucleosomes occupying different positions between expressed and non-expressed genes, as explored further below. Our results suggest that the arrangement of nucleosomes at hox promoters is established gradually during zebrafish embryogenesis.
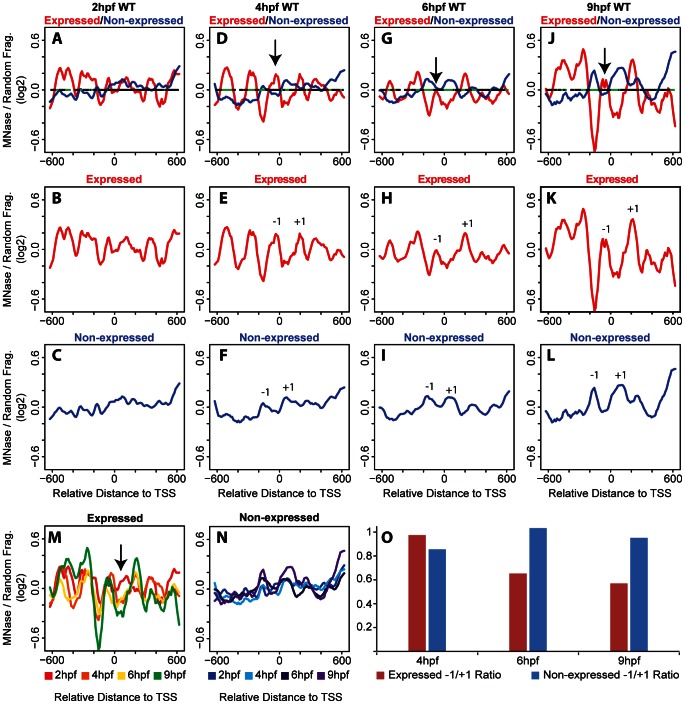
Figure 1. Nucleosome positioning is progressive during early embryonic development.
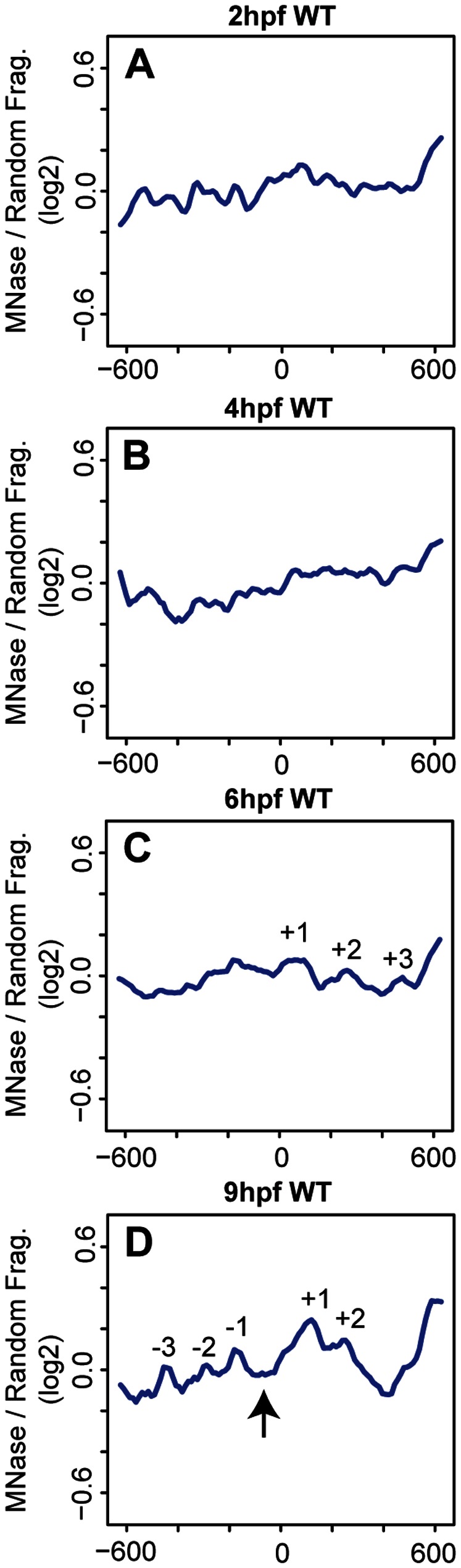
(A–D) Average nucleosome density for 37 zebrafish hox promoters was calculated as the log2 ratio of MNase digested to randomly fragmented genomic DNA for positions −600 to +600 relative to the TSS (TSS is set as 0 on X-axis) at 2 hpf (A), 4 hpf (B), 6hpf (C) and 9hpf (D). Detectable nucleosome peaks are numbered in panels C (at positions +60, +260 and +480, separated by 200 bp and 220 bp respectively) and D (at positions −450, −290, −170, +155, and +250 bp, separated by 150 bp, 120 bp, 290 bp, and 130 bp respectively). Arrow in panel D indicates a nucleosome depleted region (NDR) formed between the −1 and +1 nucleosomes.
Several groups have reported a nucleosome-depleted region (NDR) flanked by −1 and +1 nucleosomes upstream of the TSS in many metazoan genes (including hox genes) regardless of their expression state [12]–[19], [44]. In many of these reports, the size of the NDR corresponds to approximately one nucleosome. At 2 hpf, 4 hpf, and 6 hpf, nucleosomes around the TSS are too disordered to observe an NDR structure, but we observe an NDR at 9 hpf, where the +1 and −1 nucleosome peaks sit ∼290 bp apart (arrow in Fig. 1D). This is equivalent to an NDR of ∼130 bp, slightly shorter than one nucleosome length. There is also reduced nucleosome density around +400 bp at 9 hpf (Fig. 1D), but the significance of this observation is unclear. Hence, our data indicate that an NDR slightly shorter than one nucleosome is present at 9 hpf.
Expressed and Non-expressed Promoters Display Distinct Nucleosome Profiles
We note that hox gene expression is initiated by the 6 hpf and 9 hpf time points, raising the possibility that nucleosome arrangements may be distinct at promoters of transcribed genes relative to promoters of genes which are not transcribed at these stages. To examine this possibility, we first used microarray analysis to identify all hox genes that become expressed during the stages analyzed here and find that six hox genes are transcribed by 9 hpf (Table 1). We next examined the nucleosome arrangement surrounding the TSS of the 31 non-expressed genes compared to the six genes expressed at 9 hpf. At 2 hpf, promoters of non-expressed genes do not reveal readily apparent nucleosomes (Fig. 2C). However, nucleosomes become progressively more apparent at non-expressed promoters as embryogenesis progresses (Fig. 2F, I) and by 9hpf several well-positioned and well-occupied nucleosomes are detected (Fig. 2L). We note that while there are clear differences in amplitude, nucleosome positioning remains relatively constant across the stages analyzed (Fig. 2N). Since 31 of 37 promoters belong to the non-expressed group, it is expected that the nucleosome profile at non-expressed promoters will closely parallel the profile seen when all promoters are averaged together. While this is indeed the case (compare Fig. 2C, F, I, L to Fig. 1A–D), it is noteworthy that there are also clear differences. For instance, nucleosomes can be seen surrounding the TSS at 4 hpf at non-expressed promoters (−1 and +1 in Fig. 2F), but such nucleosomes are not observed at 4 hpf when all promoters are averaged (Fig. 1B). Furthermore, the −1 nucleosome is better occupied in non-expressed promoters at 9 hpf (Fig. 2L) than when all promoters are averaged (Fig. 1D). These observations suggest that although the number of expressed promoters is small, they must have a distinct nucleosome profile from non-expressed promoters. This turns out to be the case, as can be seen in Fig. 2B, E, H, K. Indeed, promoters of expressed genes reveal relatively well-defined nucleosomes already at 2 hpf (Fig. 2B) and these are further refined by 4 hpf (Fig. 2E), and remain as such at 6 hpf (Fig. 2H) and 9 hpf (Fig. 2K). In addition to being detected earlier than nucleosome peaks at non-expressed promoters, peaks at expressed promoters are also narrower and have higher amplitudes, suggesting that nucleosomes are better positioned and more highly occupied at expressed promoters. As noted for non-expressed promoters, nucleosome positioning also remains relatively constant at expressed promoters across the stages analyzed here (Fig. 2M). One exception is at 2hpf, when nucleosome density is higher near the TSS than at later stages (arrow in Fig. 2M), perhaps indicating that nucleosomes are evicted or repositioned from the TSS upon initiation of gene activation.
Figure 2. Nucleosome organization differs between expressed and non-expressed promoters.
(A–L) Average nucleosome density was calculated as in figure 1 for expressed (red line in panels A, B, D, E, G, H, J, K) and non-expressed (blue lines in panels A, C, D, F, G, I, J, L) promoters at 2 hpf (A–C), 4 hpf (D–F), 6 hpf (G–I) and 9 hpf (J−L). Nucleosome densities at expressed and non-expressed promoters were compared using a Wilcoxon Ranked Sum test and statistically significant differences (p<0.05) are illustrated in green on the horizontal line in panels A, D, G, J. Arrows in D, G, and J indicate the −1 nucleosome. (M, N) Overlay of profiles for expressed (M) and non-expressed (N) promoters at all time points. Arrow in M indicates region where 2 hpf time point (red line) has greater nucleosome density than later time points. (O) Change in occupancy of the −1 nucleosome was calculated as a ratio of density at the −1 nucleosome to density at the +1 nucleosome for expressed (red bars) and non-expressed (blue bars) promoters at 4 hpf, 6 hpf and 9 hpf.
A closer examination reveals additional differences in nucleosome positioning at promoters of expressed versus non-expressed hox genes. These differences are observed most readily when the profiles are overlayed as in figures 2 A, D, G and J. In particular, in the region surrounding the TSS (−300 to +300), non-expressed promoters display peaks at −160 and +70, while expressed promoters display peaks at −270,−50 and +200. Notably, the −1 nucleosome in expressed promoters (arrow in Fig. 2 D, G, J) appears to be dynamic, as it is reduced at 6 hpf and 9 hpf (when hox genes are expressed) relative to 4 hpf (when hox genes are not expressed). This is particularly clear when the amplitude of the −1 nucleosome peak is compared to the amplitudes of the adjacent peaks. Expressing the amplitude of the −1 nucleosomes as a ratio to the +1 nucleosomes reveals that the −1 nucleosome in expressed promoters at 6 hpf and 9 hpf is reduced by 35% and 43%,respectively (Fig. 2O), while the −1 nucleosome remains unchanged in the non-expressed promoters. The net result is a reduction in nucleosome density between the −270 and +200 peaks in the expressed promoters at stages when hox genes are expressed. While this is consistent with previous reports of NDRs forming at expressed promoters, the region is not devoid of nucleosomes since a peak persists at the TSS at 6 hpf and 9 hpf. It is possible that this peak represents a less stable nucleosome or that it reflects the fact that not all cells in the embryo express these hox genes, but our experiments cannot distinguish between these possibilities.
In an attempt to determine the significance of the observed differences between expressed and non-expressed promoters, we employed a two-sided Wilcoxon rank sum test. The results of this test are indicated on the horizontal line in figure 2A, D, G, J where regions with a statistically significant difference in nucleosome density between expressed and non-expressed promoters are indicated in green. As can be seen, the greatest difference between the two conditions is centered near the TSS at 6 hpf and 9 hpf, although other regions (most notably the region −200 to −600 in 4 hpf embryos) also show significant differences. We conclude that nucleosomes are detectable earlier at promoters of expressed hox genes and that these nucleosomes are better positioned and more highly occupied than nucleosomes at promoters of non-expressed hox genes. We further conclude that nucleosome occupancy changes as hox genes become expressed such that nucleosome density decreases near the TSS, although we do not observe the formation of a region truly depleted of nucleosomes. Hence, hox promoters may fall into the class of promoters where a nucleosome positioned upstream of the TSS must be actively removed prior to transcription, thereby providing additional regulation and permitting high transcriptional plasticity.
Disruption of Retinoic Acid Signaling Blocks hox Transcription, but does not Affect Nucleosome Organization
As mentioned, the retinoic acid (RA) signaling pathway is an activator of hox gene expression and plays a role in chromatin rearrangements at the hox clusters in both cell lines and mouse embryos [40]–[42]. To test if the RA signaling pathway plays a role in the nucleosome positioning observed in our experiments, we treated embryos with diethylaminobenzaldehyde (DEAB), a compound that blocks the RA synthesis pathway by inhibiting retinaldehyde dehydrogenase (RALDH) [49]. DEAB has also previously been shown to affect hindbrain development, particularly hox gene expression, in zebrafish embryos [50]. DEAB treatment was begun at the 2–4 cell stage in order to prevent initiation of hox transcription and embryos were collected at 9 hpf to determine transcript levels and nucleosome organization of the six active hox genes. RT-qPCR analysis revealed that transcription of the six hox genes was maximally blocked by 10 uM DEAB, with higher DEAB concentrations not providing further blockade (Fig. 3). Plotting average nucleosome profiles for all 37 hox genes from DEAB-treated embryos revealed no change from untreated embryos (Fig. 4 A). When hox genes are divided into expressed and non-expressed groups, nucleosomes in DEAB-treated embryos are again positioned very similarly to untreated embryos (Fig. 4B, compare to Fig. 2J). Overlaying nucleosome traces for expressed and non-expressed genes from DEAB and untreated embryos confirms the similarity (Fig. 4C, D). Hence, while the six genes expressed at these stages are RA sensitive and blocking RA synthesis disrupts their transcription, no detectable change in nucleosome organization is observed in the absence of RA signaling. We conclude that RA-induced transcription is not driving changes in nucleosome organization at the promoter regions of hox genes during zebrafish embryogenesis.
Figure 3. DEAB treatment blocks hox transcription.
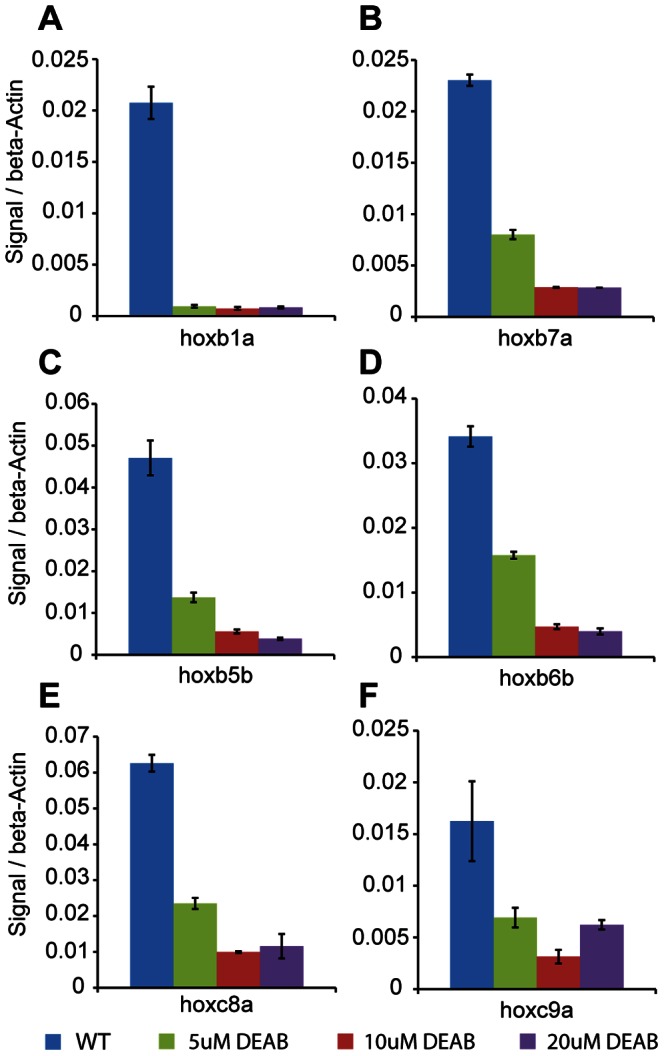
(A–F) Zebrafish embryos were left untreated (blue bars) or treated with 5 uM (green bars), 10 uM (red bars) or 20 uM (purple bars) DEAB and harvested at 9 hpf. Transcript levels for hoxb1a (A), hoxb7a (B), hoxb5b (C), hoxb6b (D), hoxc8a (E) and hoxc9a (F) were determined by quantitative RT-PCR and normalized to β-actin. Error bars indicate standard deviations of 3 technical replicates.
Figure 4. DEAB treatment has little effect on nucleosome organization at hox promoters.
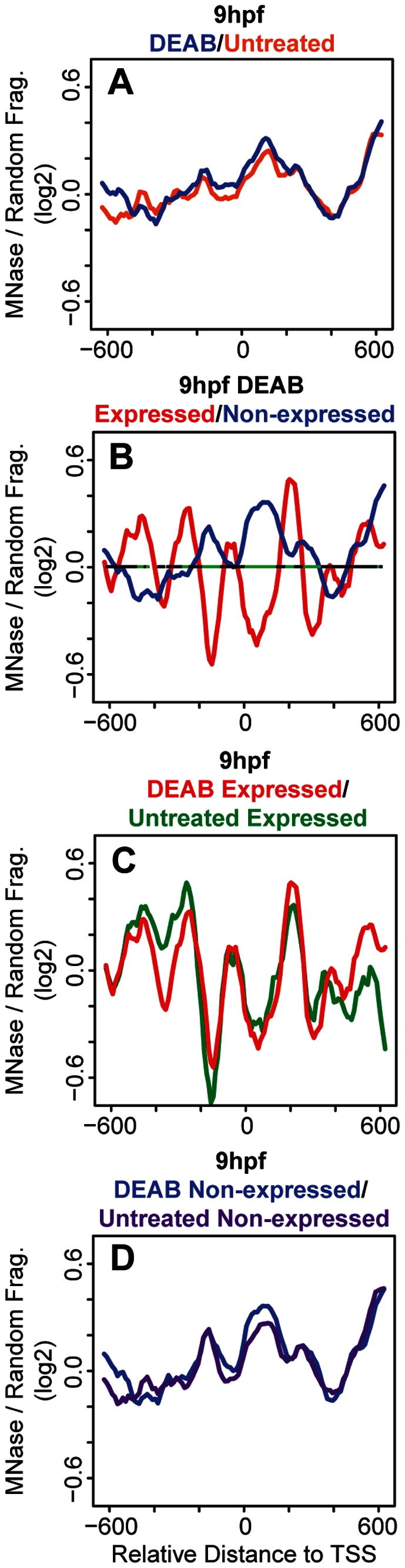
(A–D) Average nucleosome density was calculated as in figure 1. (A) Overlay of average nucleosome profiles for 37 hox promoters from DEAB-treated (blue line) and untreated (orange line) embryos at 9 hpf. (B) Overlay of nucleosome profiles for expressed (red line) and non-expressed (blue line) promoters in DEAB-treated embryos at 9 hpf. Nucleosome densities at expressed and non-expressed promoters were compared using a Wilcoxon Ranked Sum test and statistically significant differences (p<0.05) are illustrated in green on the horizontal line in panel B. (C) Overlay of nucleosome profiles for expressed promoters from DEAB-treated (red line) and untreated (green line) embryos at 9 hpf. (D) Overlay of nucleosome profiles for non-expressed promoters from DEAB-treated (blue line) and untreated (purple line) embryos at 9 hpf.
Retinoic Acid Treatment does not Affect Nucleosome Organization at hox Promoters
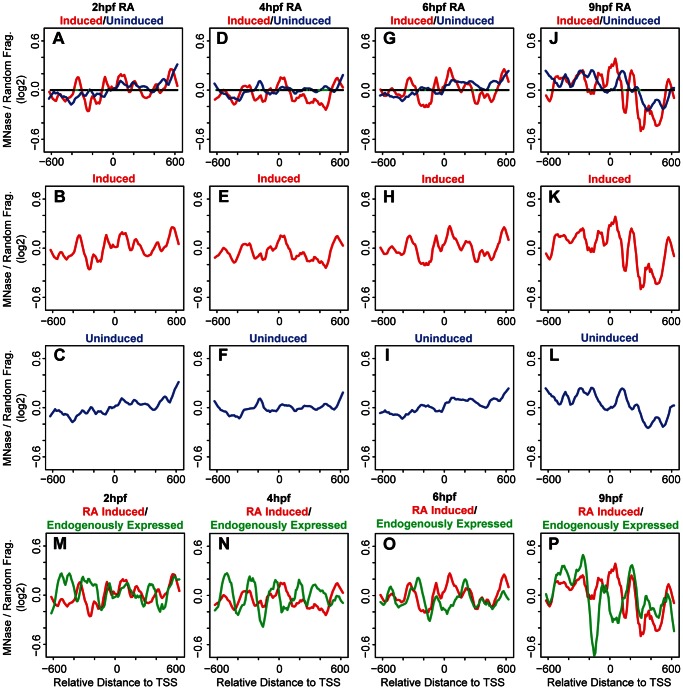
We next examined if addition of exogenous RA affects nucleosome organization at hox promoters. Embryos were treated with RA starting at the 2-cell stage and collected at 2 hpf, 4 hpf, 6 hpf and 9 hpf. We initially examined average nucleosome organization at all 37 hox promoters. We find the nucleosome profiles of RA-treated embryos to be similar to the profiles of untreated embryos, although there are some minor differences when overlayed (Fig. 5A–D). Using microarray analysis we identified nine hox genes whose expression is induced in RA treated embryos (Table 1). We next used this information to compare nucleosome organization at RA-induced and uninduced hox promoters. Promoters of genes not induced by RA do not display detectable nucleosomes until 9 hpf (Fig. 6C, F, I, L). As expected, this is similar to the non-expressed promoters in untreated embryos (Fig. 2C, F, I, L), although it is somewhat more difficult to detect individual nucleosomes in RA treated embryos and there may be additional nucleosomes forming in the region of −200 to −600 at 9 hpf (Fig. 6L). RA-induced promoters (Fig. 6B, E, H, K) show better positioned and more highly occupied nucleosomes than uninduced promoters (Fig. 6C, F, I, L) as can be seen when profiles of the two groups are overlayed (Fig. 6A, D, G, J). However, there are essentially no regions with statistically significant differences between RA-induced and uninduced promoters. This finding is in contrast to the changes in nucleosome organization we observed when comparing expressed and unexpressed promoters in untreated embryos (Fig. 2A, D, G, J) and suggests that although RA induces transcription of several hox genes, it does not drive their nucleosome organization to mimic that of endogenously expressed genes. Indeed, when the nucleosome profiles of RA-induced promoters (from Fig. 6B, E, H, K) are overlayed on the profile of endogenously expressed promoters (from Fig. 2B, E, H, K) it is clear that the profiles differ (Fig. 6 M–P). In particular, while nucleosomes are depleted in the region from −100 to −200 in both sets of promoters at 4, 6, and 9 hpf, this depletion is less pronounced at RA-induced promoters and depletion in the region from 0 to +100 is not observed at all at RA-induced promoters.
Figure 5. Exogenous RA has little effect on nucleosome organization at hox promoters.
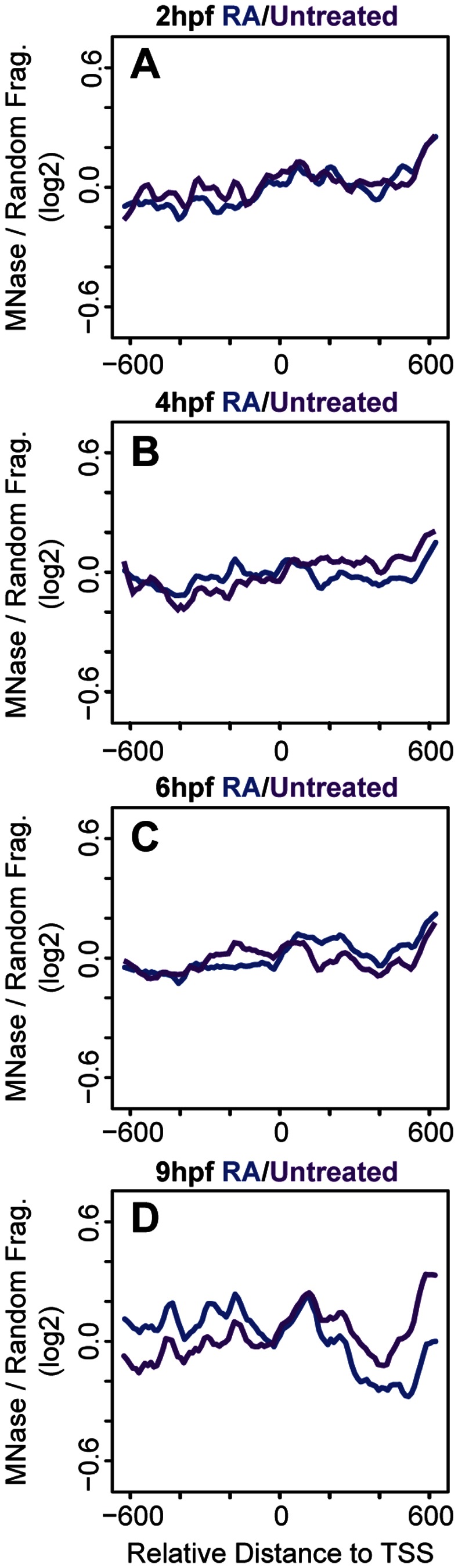
(A–D) Average nucleosome density was calculated as in figure 1 for 37 hox promoters from RA-treated embryos. Overlay of nucleosome profiles for 37 hox promoters from RA-treated (blue line) and untreated (purple line) embryos at 2 hpf (A), 4 hpf (B), 6 hpf (C) and 9 hpf (D).
Figure 6. Exogenous RA does not affect nucleosome organization at hox promoters.
(A–L) Average nucleosome density was calculated as in figure 1 for expressed (red line in panels A, B, D, E, G, H, J, K) and non-expressed (blue lines in panels A, C, D, F, G, I, J, L) promoters at 2 hpf (A–C), 4 hpf (D–F), 6 hpf (G–I) and 9 hpf (J–L). Nucleosome densities at induced and uninduced promoters were compared using a Wilcoxon Ranked Sum test and statistically significant differences (p<0.05) are indicated in green on the horizontal line in panels A, D, G and J. (M–P) Overlay of nucleosome profiles for expressed promoters in untreated (green line) and RA-treated (red line) embryos at 2 hpf (M), 4 hpf (N), 6 hpf (O) and 9 hpf (P).
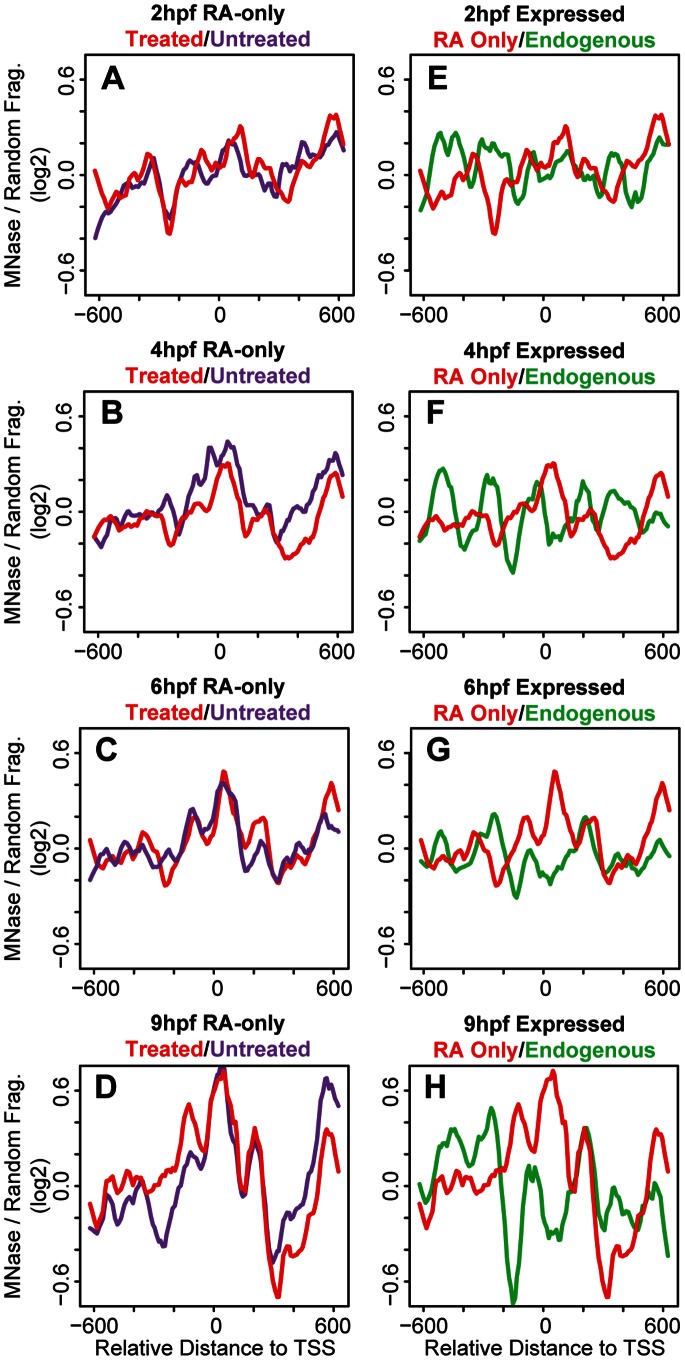
We note that three hox genes are shared between the group of endogenously expressed genes and the group of RA-induced genes (Table 1). To better isolate the effects of RA, we created a third group of promoters that are only induced by RA (Table 1; RA-only). Overlays of the nucleosome profiles of the six RA-only promoters from RA-treated embryos on the profiles of the same promoters from untreated embryos, reveal the nucleosome profiles to be similar (Fig. 7A–D). Hence, while RA induces the expression of these six hox genes, it has no effect on nucleosome organization at their promoters. Furthermore, the nucleosome organization at induced RA-only promoters is clearly distinct from that of endogenously expressed promoters (Fig. 7 E–H). Taken together, the results of our DEAB and RA treatments demonstrate that RA regulates hox gene transcription, but does not drive nucleosome organization at hox promoters during early zebrafish development.
Figure 7. Exogenous RA does not affect nucleosome positioning at induced promoters.
(A–H) Average nucleosome density was calculated as in figure 1 for RA-only genes in RA-treated embryos (red line in panels A–H), for RA-only genes in untreated embryos (purple line in panels A–D), and for endogenously expressed genes in untreated embryos (green line in panels E–H).
Discussion
While nucleosomes have been mapped in several different systems, little is known about nucleosome organization in a developing vertebrate embryo. Initial analyses of nucleosome organization focused on yeast and cultured cells that represent relatively uniform populations and that, while responsive to some stimuli, in many cases have relatively limited developmental potential. In contrast, developing embryos are multicellular and contain diverse cell types that represent a range of developmental potentials. Recent studies have analyzed nucleosome arrangements in C. elegans [15], [35] and D. melanogaster [13] embryos using mixtures of embryonic stages. However, this strategy limits the ability to detect changes in chromatin structure at specific developmental stages. Here we use staged zebrafish embryos to analyze the nucleosome arrangement at hox promoters during vertebrate embryogenesis. We find that nucleosomes are poorly organized at early stages, but become better organized by 6 hpf and 9 hpf. These latter stages correspond to the time when hox genes first become expressed in the embryo. Comparing expressed and non-expressed genes, we observe several differences in nucleosome organization at the promoter regions. First, we observe increased nucleosome occupancy at expressed promoters when compared to non-expressed promoters. Interestingly, the increased amplitude is observed in most of the nucleosomes in the promoter region, with exception of the −1 nucleosome. We find that occupancy of the −1 nucleosome decreases at 6 hpf and 9 hpf at expressed promoters. Second, we detect changes in the spacing between the −1 and +1 nucleosomes of expressed and non-expressed promoters. The larger spacing is most evident at 6 hpf and 9 hpf in the expressed promoters and coincides with a likely NDR. Due to this change in spacing, nucleosomes also appear out of phase between the expressed and non-expressed promoters. Finally, though hox transcription is dependent on RA signaling, we find that blocking RA signaling does not cause changes in nucleosome organization at the expressed promoters, suggesting that nucleosome arrangement is independent of RA-induced transcription. The fact that nucleosome organization is dynamic, but genomic sequence is invariant, during embryogenesis, also suggests that trans-factors play a role in dynamically positioning nucleosomes at the promoters of hox genes in the developing embryo.
The Role of Transcription in Nucleosome Organization at hox Promoters
Transcription has been shown previously to correlate with specific nucleosome profiles at some TSSs in metazoans [12], [18], [19]. Indeed, in our bulk nucleosome plots at 9 hpf, when hox transcription is initiated, nucleosomes appear to be better positioned as compared to bulk nucleosome positions at 2 hpf−6 hpf (Fig. 1A–D). Grouping the hox genes into expressed and non-expressed promoters revealed that nucleosomes at expressed promoters are better positioned and have increased occupancy when compared to nucleosomes at non-expressed promoters (Fig. 2A, D, G, J). While these data suggest that transcription may have a direct effect on the nucleosome arrangement at hox promoters, we find that blocking RA signaling represses hox transcription (Fig. 3) with no changes in the nucleosome profile (Fig.4). We note that our DEAB protocol was designed to prevent initiation of hox transcription and that we may have observed a different effect if hox gene transcription had been allowed to initiate prior to being inactivated. Hence, our data suggest that the nucleosome profile at hox promoters is independent of RA-induced hox transcription. We see further support for this conclusion when embryos are treated with RA. Though exogenous RA induces hox transcription, RA-induced genes do not recapitulate the nucleosome positions observed at endogenously expressed promoters (Fig. 6) and display little change from nucleosome positions observed in untreated embryos (Fig. 7), again suggesting that the nucleosome profile at hox promoters is independent of hox transcription.
Our findings raise the question as to what role RA signaling plays in hox transcription if it does not affect nucleosome organization. Given the complexity of eukaryotic chromatin structure, it is possible that RA affects chromatin structure at a level distinct from the nucleosome. For instance, previous studies detected chromatin changes at the HoxB and HoxD clusters using fluorescent in situ hybridization [40]–[42]. Hox loci were observed to decondense during mouse embryogenesis in correlation with hox gene transcription and this process was recapitulated by RA-treatment of ES cells. It is therefore possible that RA affects chromatin at the level of the 30 nm fiber without affecting the positioning of individual nucleosomes. It is also possible that RA affects hox expression by promoting histone modifications that are supportive of transcription. Indeed, RA receptors are known to recruit histone-modifying enzymes [51]. Lastly, RA may simply recruit components of the transcription machinery, again via RA receptors, to hox promoters. The fact that RA induces hox transcription without affecting nucleosome organization could also be taken to indicate that many nucleosome arrangements are permissive for transcription. However, it is important to note that the exogenously applied RA is likely in significant excess relative to endogenous levels and this may permit over-riding of a nucleosome arrangement that would not otherwise support transcription. In summary, we propose that an RA-independent mechanism promotes a nucleosome arrangement that is permissive for transcription, but that RA is required for actual transcription. A transcription-independent mechanism for nucleosome organization is also supported by our observation that an NDR forms at non-expressed promoters by 9 hpf. Since genes in this group will become expressed at later stages of embryogenesis, it is possible that this NDR forms in preparation for subsequent transcriptional activation.
A Likely Role for Trans-factors in Nucleosome Organization at hox Promoters
Nucleosome positioning has been shown to result from the combination of intrinsic characteristics of DNA sequence, such as base pair composition (cis-elements), and from factors that interact with DNA, such as transcription factors and ATP-dependent chromatin modifiers (trans-factors). However, the relative contribution of each mechanism remains unclear. A recent study addressed how cis-elements and trans-factors influence nucleosome positioning in yeast. By using YACs to transfer large DNA fragments between divergent yeast strains, analysis of nucleosome organization in the native strain could be compared to nucleosome organization on the YAC in the new host strain [52]. This analysis revealed that inter-nucleosome spacing and positioning of the +1 nucleosome was altered upon transfer to the new host strain. Since sequence remains constant between the YAC and native yeast strain, these findings suggest that trans-factors play a more important role in nucleosome positioning than cis-elements. Similarly, we find that nucleosome organization changes during embryogenesis, but since the underlying sequence is invariant during development, trans-factors also likely play a role in nucleosome positioning during embryogenesis. We note that the changes in nucleosome organization that we observe correlate with important transitions during embryonic development. In particular, 2 hpf and 4 hpf embryos display relatively disordered nucleosomes at promoter regions (Fig. 1A, B), while at 6 hpf and 9 hpf nucleosomes are readily identified (Fig. 1C, D). This change coincides with activation of the zygotic genome at the maternal zygotic transition (MZT), which occurs in a time window at approximately 3–4 hpf. Our data do not reveal whether there is a causal relationship between this transition and the observed nucleosome rearrangement. However, since we observe better nucleosome positioning after the MZT, it is plausible that trans-factors (such as transcription factors and ATP-dependent chromatin remodelers) become expressed at the MZT and subsequently regulate nucleosome arrangements at hox promoters.
NDR Formation at hox Promoters during Embryogenesis
Nucleosome depleted regions (NDRs) were initially identified at promoters in yeast, but have subsequently been identified in other cell types. In most cases, NDRs are readily observed in bulk analyses of promoters regardless of whether the promoters are active or not. Indeed, previous analyses of bulk hox promoters in human cell lines identified an NDR upstream of the TSS [44]. Accordingly, when we average nucleosome positions for all 37 zebrafish hox genes, we observe an NDR as soon as −1 and +1 nucleosomes are resolved at the TSS (9 hpf, Fig. 1D). The NDR observed in the bulk plot at 9 hpf is ∼130 bp, while the NDRs observed at expressed promoters at 6 hpf and 9 hpf are ∼100 bp and ∼110 bp respectively and the NDR observed at non-expressed promoters at 9 hpf is ∼85 bp, suggesting an average NDR size of ∼100 bp. This is relatively similar to NDRs observed in other genome-wide nucleosome mapping studies, including fish, where NDR lengths vary somewhat, but are ∼150 bp.
Though the NDRs observed in our study are similar to other bulk studies, they are smaller than the NDR previously observed at human hox promoters, which was reported to be ∼500 bp [44]. We suspect the difference in NDR lengths between the two studies is due to differences between zebrafish embryos and human cell lines. First, the embryo is made up of a heterogeneous population of cell types, while cell lines represent a homogeneous population. The heterogeneity of cell types in the embryo might lead to variable nucleosome occupancy. For instance, cells in the embryo that do not express a given hox gene might have a nucleosome positioned upstream of the TSS, thereby reducing the size of the NDR observed when signals from all cells in the embryo are averaged. Indeed, a previous study found nucleosomes to be differentially positioned at the serum albumin enhancer in a tissue specific manner in mouse [53]. Such variable nucleosome occupancy presumably does not occur in cell lines since they represent a homogeneous population of cells that would all have similar nucleosome positions. Interestingly, if some cells in the embryo lacked the −1 nucleosome, then the NDR of these promoters would expand to 310 bp and 320 bp at 6 hpf and 9 hpf respectively, making it more similar to the NDR observed at hox promoters in human cell lines. Second, the difference in NDR length may be due to differences between fish and humans. For instance, divergence of regulatory sequences in the promoters as well as divergence in the trans-factors responsible for nucleosome positioning may lead to different sized NDRs. Support for this possibility comes from the analysis of NDRs in evolutionary divergent yeast species, which were found to have different sized NDRs at orthologous promoters [52].
Our data do not address how NDRs form, but we consider several possibilities. First, NDRs could form in a competitive process. Evidence exists for competition between nucleosomes and trans-factors for binding to specific sequences [54], [55]. Once a trans-factor is bound, positioning of nucleosomes would be restricted to other available sites in a process similar to that suggested by the “barrier model”. The barrier model is driven by trans-factors interacting with DNA and providing a barrier that blocks free nucleosome diffusion, creating well-ordered and positioned nucleosomes [6], [56]. Hence, binding of trans-factors at expressed hox promoters would create more uniform nucleosome positions as well as increased amplitude of nucleosome peaks, while the lack of trans-factor binding at non-expressed genes would lead to lower occupancy and less well-positioned nucleosomes. Such competition has been observed at the CLN2 promoter in yeast where binding sites for three sequence specific transcription factors are needed for NDR formation. In the absence of these binding sites, the CLN2 promoter has increased nucleosome occupancy [54]. Meis and Pbx proteins, which bind elements in many hox promoters and are involved in the regulation of hox transcription, have been suggested to act as pioneer transcription factors capable of binding nucleosome-occupied DNA [57] and may impact nucleosome binding at hox promoters. Since RA-receptors may be bound to DNA even in the absence of RA-signaling [58], [59], RARs may play a similar role by binding RA response elements. However, our analyses have failed to identify an enrichment of binding sites for any known trans-factor in the NDR regions of hox promoters. Second, NDR formation could be an active process mediated throughout embryogenesis by ATP-dependent remodelers. ATP-dependent SWI2/SNF2 complexes, which slide nucleosomes through DNA sequence, have been previously shown to regulate hox genes [60], [61]. Many of these factors do not bind DNA directly and would therefore need to be recruited to hox promoters by DNA binding factors such as the Meis and Pbx factors mentioned above.
Nucleosome Occupancy and Histone Modifications at hox Promoters are Temporally Coincident
The accessibility of genomic DNA is regulated not only by nucleosome positioning, but also by post-translational modifications made to the N-termini of histone tails, that in turn affect chromatin structure. For instance, histone H3 lysine 4 tri-methylation (H3K4me3) by trithorax group proteins and histone H3 lysine 27 tri-methylation (H3K27me3) by polycomb group proteins, associate with active and inactive promoters, respectively [62]. A recent study mapped H3K4me3 and H3K27me3 marks throughout the zebrafish genome at 2.5 hpf (pre-MZT), as well as at 4.5 hpf (post-MZT), and detected chromatin marks only post-MZT [63]. Notably, this coincides with the time point where we first observe well-defined nucleosomes. This temporal coincidence of emerging well-positioned nucleosomes and detectable histone modifications suggests that histones may become modified as soon as they are deposited at a promoter. While the significance of this observation is unclear, it is noteworthy that hox promoters are bivalently marked with both H3K4me3 and H3K27me3 at this stage [63]. Bivalency is thought to act as a developmental control, poising developmentally important genes for rapid activation at the appropriate stage of embryogenesis [64]. Indeed, the inability to deposit H3K27me3 marks leads to misregulated hox gene expression and homeotic transformations in Drosophila [65]. Hence, it is possible that recently deposited nucleosomes at hox promoters must be rapidly modified in order to ensure proper regulation of hox genes.
Supporting Information
File with Figures S1 and S2. Figure S1 Comparison of biological replicates used for calculation of nucleosome densities. Data from two biological replicates were plotted against each other for untreated embryos at 2 hpf (A), 4 hpf (B), 6 hpf (C), 9 hpf (D), as well as for RA-treated embryos at 2 hpf (E), 4 hpf (F), 6 hpf (G), 9 hpf (H) and for DEAB-treated embryos at 9 hpf (I). R2 values are indicated in the top right quadrant of each panel. Figure S2 Representative MNase digestion. Cross-linked genomic DNA from 4 hpf embryo was left untreated (lane 2) or treated for 10 minutes at 37°C with serially diluted concentrations of micrococcal nuclease (MNase) increasing from 0.5 units/ml −8 units/ml (lanes 3–6) and separated by agarose gel electrophoresis. Lanes 1 and 7 contain size ladders.
(PDF)
Acknowledgments
We are grateful to John Holik, Marta Radman-Livaja, Ben Carone, Oliver Rando, Simon Xi and Zhiping Weng for advice regarding tiling array analysis and calculation of nucleosome positions, as well as to members of the Sagerström lab for helpful discussions.
Funding Statement
This work was supported by National Institutes of Health grant NS038183 to CGS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kornberg RD, Lorch Y (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98: 285–294. [DOI] [PubMed] [Google Scholar]
- 2. Widom J (1998) Structure, dynamics, and function of chromatin in vitro. Annual review of biophysics and biomolecular structure 27: 285–327. [DOI] [PubMed] [Google Scholar]
- 3. Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, et al. (2005) Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630. [DOI] [PubMed] [Google Scholar]
- 4. Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, et al. (2007) Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576. [DOI] [PubMed] [Google Scholar]
- 5. Lee W, Tillo D, Bray N, Morse RH, Davis RW, et al. (2007) A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39: 1235–1244. [DOI] [PubMed] [Google Scholar]
- 6. Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, et al. (2008) A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18: 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, et al. (2009) The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Almer A, Rudolph H, Hinnen A, Horz W (1986) Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. The EMBO journal 5: 2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fedor MJ, Kornberg RD (1989) Upstream activation sequence-dependent alteration of chromatin structure and transcription activation of the yeast GAL1-GAL10 genes. Molecular and cellular biology 9: 1721–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD (2004) Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36: 900–905. [DOI] [PubMed] [Google Scholar]
- 11. Tirosh I, Barkai N (2008) Two strategies for gene regulation by promoter nucleosomes. Genome Res 18: 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mito Y, Henikoff JG, Henikoff S (2005) Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet 37: 1090–1097. [DOI] [PubMed] [Google Scholar]
- 13. Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, et al. (2008) Nucleosome organization in the Drosophila genome. Nature 453: 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, et al. (2010) Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143: 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, et al. (2008) A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res 18: 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ercan S, Lubling Y, Segal E, Lieb JD (2011) High nucleosome occupancy is encoded at X-linked gene promoters in C. elegans. Genome Res 21: 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sasaki S, Mello CC, Shimada A, Nakatani Y, Hashimoto S, et al. (2009) Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites. Science 323: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ozsolak F, Song JS, Liu XS, Fisher DE (2007) High-throughput mapping of the chromatin structure of human promoters. Nat Biotechnol 25: 244–248. [DOI] [PubMed] [Google Scholar]
- 19. Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, et al. (2008) Dynamic regulation of nucleosome positioning in the human genome. Cell 132: 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tillo D, Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, et al. (2010) High nucleosome occupancy is encoded at human regulatory sequences. PLoS One 5: e9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Andreu-Vieyra C, Lai J, Berman BP, Frenkel B, Jia L, et al. (2011) Dynamic nucleosome-depleted regions at androgen receptor enhancers in the absence of ligand in prostate cancer cells. Mol Cell Biol 31: 4648–4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ioshikhes I, Bolshoy A, Derenshteyn K, Borodovsky M, Trifonov EN (1996) Nucleosome DNA sequence pattern revealed by multiple alignment of experimentally mapped sequences. Journal of molecular biology 262: 129–139. [DOI] [PubMed] [Google Scholar]
- 23. Thastrom A, Lowary PT, Widlund HR, Cao H, Kubista M, et al. (1999) Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. Journal of molecular biology 288: 213–229. [DOI] [PubMed] [Google Scholar]
- 24. Peckham HE, Thurman RE, Fu Y, Stamatoyannopoulos JA, Noble WS, et al. (2007) Nucleosome positioning signals in genomic DNA. Genome Res 17: 1170–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Iyer V, Struhl K (1995) Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. The EMBO journal 14: 2570–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Suter B, Schnappauf G, Thoma F (2000) Poly(dA.dT) sequences exist as rigid DNA structures in nucleosome-free yeast promoters in vivo. Nucleic acids research 28: 4083–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ioshikhes IP, Albert I, Zanton SJ, Pugh BF (2006) Nucleosome positions predicted through comparative genomics. Nature genetics 38: 1210–1215. [DOI] [PubMed] [Google Scholar]
- 28. Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, et al. (2006) A genomic code for nucleosome positioning. Nature 442: 772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yuan GC, Liu JS (2008) Genomic sequence is highly predictive of local nucleosome depletion. PLoS computational biology 4: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shim EY, Woodcock C, Zaret KS (1998) Nucleosome positioning by the winged helix transcription factor HNF3. Genes & development 12: 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, et al. (2008) A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Molecular Cell 32: 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, et al. (2008) NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes & development 22: 1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N (2010) High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res 20: 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hargreaves DC, Crabtree GR (2011) ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res 21: 396–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xi Y, Yao J, Chen R, Li W, He X (2011) Nucleosome fragility reveals novel functional states of chromatin and poises genes for activation. Genome Res 21: 718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jin C, Felsenfeld G (2007) Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 21: 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnson SM, Tan FJ, McCullough HL, Riordan DP, Fire AZ (2006) Flexibility and constraint in the nucleosome core landscape of Caenorhabditis elegans chromatin. Genome Res 16: 1505–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lewis EB (1978) A gene complex controlling segmentation in Drosophila. Nature 276: 565–570. [DOI] [PubMed] [Google Scholar]
- 39. Krumlauf R (1994) Hox genes in vertebrate development. Cell 78: 191–201. [DOI] [PubMed] [Google Scholar]
- 40. Chambeyron S, Da Silva NR, Lawson KA, Bickmore WA (2005) Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development 132: 2215–2223. [DOI] [PubMed] [Google Scholar]
- 41. Morey C, Da Silva NR, Perry P, Bickmore WA (2007) Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development 134: 909–919. [DOI] [PubMed] [Google Scholar]
- 42. Chambeyron S, Bickmore WA (2004) Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev 18: 1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, et al. (2011) The dynamic architecture of Hox gene clusters. Science 334: 222–225. [DOI] [PubMed] [Google Scholar]
- 44. Kharchenko PV, Woo CJ, Tolstorukov MY, Kingston RE, Park PJ (2008) Nucleosome positioning in human HOX gene clusters. Genome Res 18: 1554–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310. [DOI] [PubMed] [Google Scholar]
- 46. Dennis JH, Fan HY, Reynolds SM, Yuan G, Meldrim JC, et al. (2007) Independent and complementary methods for large-scale structural analysis of mammalian chromatin. Genome research 17: 928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, et al. (2012) Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 22: 577–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schier AF, Talbot WS (2005) Molecular genetics of axis formation in zebrafish. Annu Rev Genet 39: 561–613. [DOI] [PubMed] [Google Scholar]
- 49. Perz-Edwards A, Hardison NL, Linney E (2001) Retinoic acid-mediated gene expression in transgenic reporter zebrafish. Dev Biol 229: 89–101. [DOI] [PubMed] [Google Scholar]
- 50. Maves L, Kimmel CB (2005) Dynamic and sequential patterning of the zebrafish posterior hindbrain by retinoic acid. Dev Biol 285: 593–605. [DOI] [PubMed] [Google Scholar]
- 51. Perissi V, Jepsen K, Glass CK, Rosenfeld MG (2010) Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet 11: 109–123. [DOI] [PubMed] [Google Scholar]
- 52. Hughes AL, Jin Y, Rando OJ, Struhl K (2012) A functional evolutionary approach to identify determinants of nucleosome positioning: a unifying model for esishing the genome-wide pattern. Mol Cell 48: 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McPherson CE, Shim EY, Friedman DS, Zaret KS (1993) An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell 75: 387–398. [DOI] [PubMed] [Google Scholar]
- 54. Bai L, Ondracka A, Cross FR (2011) Multiple sequence-specific factors generate the nucleosome-depleted region on CLN2 promoter. Mol Cell 42: 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Anderson JD, Widom J (2000) Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J Mol Biol 296: 979–987. [DOI] [PubMed] [Google Scholar]
- 56. Fu Y, Sinha M, Peterson CL, Weng Z (2008) The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet 4: e1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, et al. (2004) Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell 14: 465–477. [DOI] [PubMed] [Google Scholar]
- 58. Koide T, Downes M, Chandraratna RA, Blumberg B, Umesono K (2001) Active repression of RAR signaling is required for head formation. Genes Dev 15: 2111–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mahony S, Mazzoni EO, McCuine S, Young RA, Wichterle H, et al. (2011) Ligand-dependent dynamics of retinoic acid receptor binding during early neurogenesis. Genome Biol 12: R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, et al. (2001) Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell 8: 531–543. [DOI] [PubMed] [Google Scholar]
- 61. Li M, Belozerov VE, Cai HN (2010) Modulation of chromatin boundary activities by nucleosome-remodeling activities in Drosophila melanogaster. Mol Cell Biol 30: 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G (2007) Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745. [DOI] [PubMed] [Google Scholar]
- 63. Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, et al. (2010) Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464: 922–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, et al. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. [DOI] [PubMed] [Google Scholar]
- 65. Pengelly AR, Copur O, Jackle H, Herzig A, Muller J (2013) A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339: 698–699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
File with Figures S1 and S2. Figure S1 Comparison of biological replicates used for calculation of nucleosome densities. Data from two biological replicates were plotted against each other for untreated embryos at 2 hpf (A), 4 hpf (B), 6 hpf (C), 9 hpf (D), as well as for RA-treated embryos at 2 hpf (E), 4 hpf (F), 6 hpf (G), 9 hpf (H) and for DEAB-treated embryos at 9 hpf (I). R2 values are indicated in the top right quadrant of each panel. Figure S2 Representative MNase digestion. Cross-linked genomic DNA from 4 hpf embryo was left untreated (lane 2) or treated for 10 minutes at 37°C with serially diluted concentrations of micrococcal nuclease (MNase) increasing from 0.5 units/ml −8 units/ml (lanes 3–6) and separated by agarose gel electrophoresis. Lanes 1 and 7 contain size ladders.
(PDF)