Abstract
MicroRNAs (miRNAs) play important roles in the regulation of immune responses. There is evidence that miRNAs also participate in the pathogenesis of multiple sclerosis (MS), but how the miRNAs regulate the pathogenesis of MS is still under investigation. The identification of new members of the miRNA family associated with the pathogenesis of MS could facilitate early diagnosis and treatment. Here we show that the level of miRNA let-7e is significantly upregulated in experimental autoimmune encephalomyelitis (EAE), an animal model of MS using miRNA array and quantitative real-time PCR. The expression of let-7e was mainly in CD4+ T cells and infiltrated mononuclear cells of central nervous system, and highly correlated with the development of EAE. We found that let-7e silencing in vivo inhibited encephalitogenic Th1 and Th17 cells and attenuated EAE, with reciprocal increase of Th2 cells; overexpression of let-7e enhanced Th1 and Th17 cells and aggravated EAE. We also identified IL-10 as one of the functional targets of let-7e. Together, we propose that let-7e is a new miRNA involved in the regulation of encephalitogenic T-cell differentiation and the pathogenesis of EAE.
Keywords: MicroRNA, mir-let-7e, EAE/MS, Th1/Th2 cells, Cell differentiation
Introduction
Multiple sclerosis (MS) is a complex neurological disorder characterized by demyelination and axonal loss with unknown etiology. The pathogenesis starts with increased migration of autoreactive lymphocytes across the blood-brain barrier [1-3]. Experimental autoimmune encephalomyelitis (EAE) is a commonly used and well established animal model with many similarities to human MS including episodes of relapsing and remitting paralysis. IFN-γ-producing Th1 and IL-17-producing Th17 have been recognized as the main effector T cells that elicit the pathogenesis of the disease with different pathological phenotypes; and the evidences also showed that Th1 cells facilitate the entry of Th17 cells to the CNS during the development of the disease [4, 5]. The reciprocal balance between the effector T cells and their cytokines controls the development of the disease. Accumulation of IL-4-producing Th2 or a shift from Th1-type to Th2-type immune response affords protection against the disease. Therefore, it is tremendously interesting and important to identify new drugs that suppress the pathogenic effector T cells and enhance the protective immune response [6-10]. Other protective factors, such as cytokine IL-10 and regulatory T (Treg) cells play a negative role in the development of the disease. IL-10 is critical in the development of the disease by suppressing Th1 while promoting Th2 responses. Mice transgenic for IL-10 are resistant to the development of EAE [11-13].
MiRNAs are a class of ~22-nt non-coding RNAs that regulate gene expression at the posttranscriptional level. They bind to the 3′-UTR of the target mRNAs and lead to degradation or translational inhibition of the target mRNAs. They are estimated to regulate up to one third of all human genes, play critical roles in multiple biological processes, such as regulation of immune cell differentiation, immunity, and for the correct functioning of central nervous system, thus emerge as new tools for diagnosis and therapy of several human diseases [14-17]. The involvement of miRNAs in the pathogenesis of neurodegenerative diseases, such as Alzheimer’s and Parkinson’s disease, has been demonstrated [18, 19]. Recently, miRNA expression was detected in the context of MS. MiR-155 and miR-326 are among the most upregulated miRNAs in active MS lesions and blood compared with control specimens; both of them can promote the development of encephalitogenic Th1 and Th17 cells; deletion or inhibition of either one could significantly attenuate the severity of the disease [20-22]. In this report, we show that miRNA let-7e (let-7e) is dramatically upregulated in EAE-sensitive mice but significantly downregulated in EAE-resistant mice. We further demonstrate that let-7e promotes the development of Th1 and Th17 cells; however, inhibition of let-7e shifts the immune response to a Th2 profile and attenuates the severity of the disease. Therefore, we demonstrate that let-7e was critically involved in the pathogenesis of EAE. Our results suggest that let-7e is a new member of miRNA family in MS pathogenesis.
Results
Let-7e expression positively correlates with the development of EAE
We have previously shown that knocking out CD44 in C57BL/6 mice (WT) leads to resistance to EAE (Fig. 1A and reference [23]). In the CD44 knockout (KO) mice, the development of Th1 and Th17 cells were significantly inhibited while the development of Th2 and Treg cells was significantly enhanced; the immune response was dramatically shifted from Th1 to Th2. We showed that the role of CD44 was dependent on the signaling following the interaction between osteopontin and CD44, which modulated the DNA methylation of ifnγ, il4, il17, and foxp3 promoter [23, 24]. The original purpose of the current study was to explore the miRNA regulation on such CD44-dependant DNA methylation. To that end, miRNA profiles were first determined using miRNA array. The level of miRNA expression from naïve CD4+ T cells was set as baseline. The fold change of each miRNA expression from encephalitogenic CD4+ T cells from EAE mice was calculated based on the baseline (Fig. 1B and 1C). The results showed that out of 609 targets, 122 targets were positive for WT but negative for KO CD4+ T cells; 113 targets were negative for WT but positive for KO CD4+ T cells; 374 targets were either positive or negative in both WT and KO CD4+ T cells (Fig. 1D). When we used two-fold change as a cut-off, it was noted that there were 18 targets with over two-fold change exclusively in WT CD4+ T cells; there were 8 targets over two-fold change exclusively in KO CD4+ T cells; and there were 15 targets over two-fold change in both WT and KO CD4+ T cells (Fig.1E).
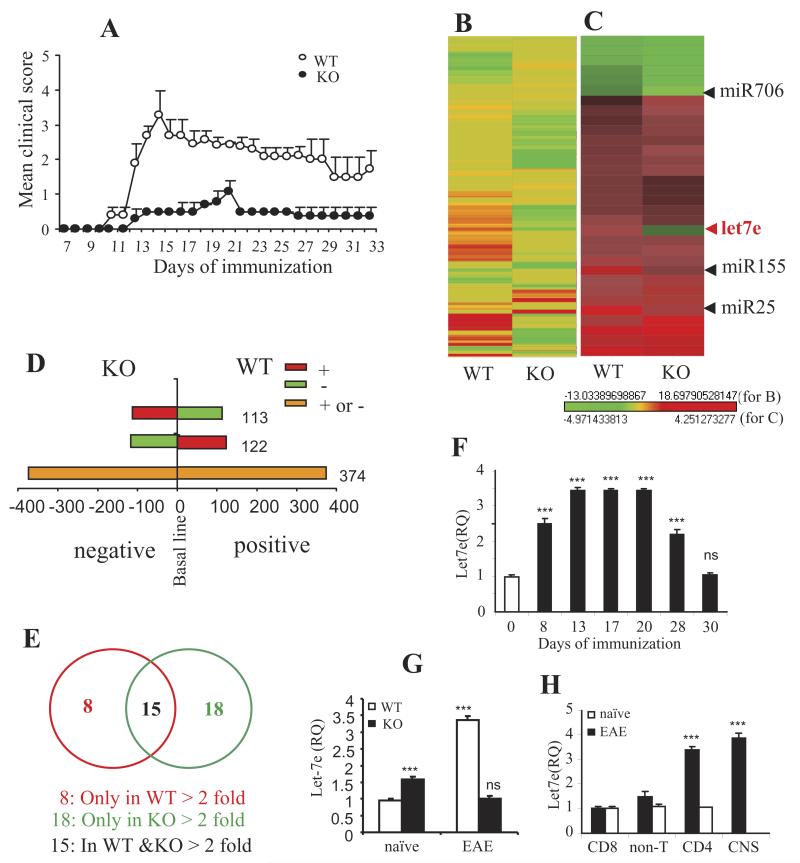
Figure 1. Upregulation of let-7e in WT and CD44 KO EAE mice.
(A) EAE was induced by s.c. immunization of MOG35-55 peptide in WT and KO mice. The mean clinical scores are shown. (B and C) The heatmap of miRNA array for the CD4+ T cells of WT and KO mice from day 15 of EAE. Naïve CD4+ T cells from WT or KO were used as background control in the array. Heatmap from (B) a total of 609 probes and (C)for the highlight of let-7e, along with some additional miRNAs. (D) Distribution of total 609 miRNAs in WT and KO mice. (E) Distribution of miRNAs with >2 fold change of expression level in WT and KO mice. (F–H) miRNAs quantitation using QPCR. RNU1A was used for endogenous control. (F)let-7e in CD4+ T cells of WT mice from different days following immunization; (G)let-7e in the CD4+ T cells of WT and KO mice from day 15 of EAE (including the validation for the miRNA array); (H) let-7e in the different cell subsets of WT mice on day 15 of EAE. ***: p<0.001, ns: not significant, versus (G and H) WT naïve control or (F) day 0 (ANOVA test). (F-H) Data are shown as mean ± SEM of n = 10 per group (A) or n= 5 (F-H) and are representative of three independent experiments.
Table 1 shows a select list of miRNAs that showed significant differences between CD4+ T cells from WT and CD44 KO mice. A prominent member was miR let-7e that was upregulated in WT CD4+ T cells but downregulated in KO CD4+ T cells (fold change: 2.7 vs. −1). Figure 1C and Table 1 also show additional miRs such as miR-155 whose expression was increased in both WT and KO CD4+ T cells (positive); nevertheless, the extent of the increase was much lower in KO CD4+ T cells (fold change: 3.21 vs. 1.95). We also noted that miR-706 was downregulated in both WT and KO cells with much stronger decrease in KO cells, while miR-25 was upregulated in both groups with WT having a stronger increase (Table 1).
Table 1.
miRNA expression fold changea)
| ProbeSet name | WT | CD44 KO |
|---|---|---|
| mmu-miR-685 | −3.4955 | −1.4525 |
| mmu-let-7i | −3.1613 | −1.8551 |
| mmu-miR-762 | −2.6765 | −1.3745 |
| mmu-miR-466j | −2.3856 | −1.189 |
| mmu-miR-706 | −1.5624 | −4.9714 |
| mmu-let-7a | −1.1254 | −1.1154 |
| mmu-let-7b | 2.0063 | 1.2563 |
| mmu-let-7c | 1.1509 | −1.0778 |
| mmu-let-7d | 1.4022 | 1.0277 |
| mmu-let-7f | 1.8187 | 1.6543 |
| mmu-let-7g | −1.1315 | 1.0693 |
| mmu-miR-466e-3p | 2.0531 | 1.3378 |
| mmu-miR-194 | 2.0801 | 1.293 |
| mmu-let-7e | 2.6918 | −1.0018 |
| mmu-miR-29a | 2.8452 | 4.2513 |
| mmu-miR-155 | 3.212 | 1.9518 |
| mmu-miR-25 | 3.8236 | 2.6011 |
| mmu-miR-326 | 1.0322 | −1.0401 |
| mmu-miR-106a | 1.6841 | 1.552 |
| mmu-miR-106b | 3.5431 | 3.4794 |
| mmu-miR-146a | 2.474 | 1.6428 |
Selected miRNAs in miRNA array assay. The full lists of miRNAs whose expression in CD4+ T cells can be found in the ArrayExpress database under accession number E-MEXP-3594.
Because we noted contrasting expression of let-7e between WT and KO cells, we further examined let-7e expression at different stages of the EAE disease and in the different cell populations. We found that the expression of let-7e started to increase at an early stage (day 8) and continued to peak at the height of the disease (day 13 to day 20); the expression had reduced on the recession (day 28) and returned to the normal level on the remission (day 30) of the disease (Fig. 1F). MiRNA analysis using QPCR revealed that let-7e was significantly increased in the encephalitogenic WT CD4+ T cells but its expression was dramatically inhibited in KO CD4+ T cells, even lower than the expression in the naïve CD4+ T cells (Fig. 1G). In WT EAE mice, let-7e was predominantly expressed in the CD4+ T cells and CNS-MNCs but not in CD8+ T cells and non-T cells (Fig. 1H). Thus, our results indicated a positive correlation between the expression of let-7e and the development of the disease and suggested a possible role of let-7e on the differentiation of encephalitogenic CD4+ T cells.
Let-7e regulates Th1-Th2 differentiation of encephalitogenic CD4+ T cells
To examined the effect of let-7e on the Th1-Th2 differentiation, splenocytes from EAE-induced WT and KO mice on day 15 were prepared and transfected with the miRNA oligonucleotides that mimic endogenous let-7e (labeled as pre-miR) or inhibit the activity of endogenous let-7e (labeled as anti-miR) or serve as the negative control (labeled as mock). After re-stimulation with MOG35-55 peptide for 24 h, the production of IFN-γ and IL-4 in the CD4+ T cells were assayed. We found that overexpression of let-7e enhanced IFN-γ production and mutually inhibited IL-4 production in encephalitogenic CD4+ T cells, while downregulation of let-7e inhibited IFN-γ production and enhanced IL-4 production (Fig. 2A, panel WT and KO). In another study, naïve CD4+ T cells from unimmunized WT mice were transfected with pre-miR, anti-miR or mock and incubated in the Th1- or Th2-polarization condition with the stimulation of mAbs anti-CD3 and anti-CD28. We found that overexpression of let-7e promoted Th1 polarization and inhibited Th2 polarization, while downregulation of let-7e inhibited Th1 polarization and enhanced Th2 differentiation (Fig. 2A, panel TH1 and TH2). Meanwhile, part of the cultured cells was examined for the expression of let-7e. We found that cells from WT EAE mice or Th1 polarization expressed significantly higher level of let-7e than those from KO EAE mice or Th2 polarization; and pre-miR increased the let-7e level and antimiR decreased the let-7e level in all groups (Fig. 2B). Together, our results indicated that let-7e regulates Th1-Th2 differentiation of encephalitogenic CD4+ T cells.
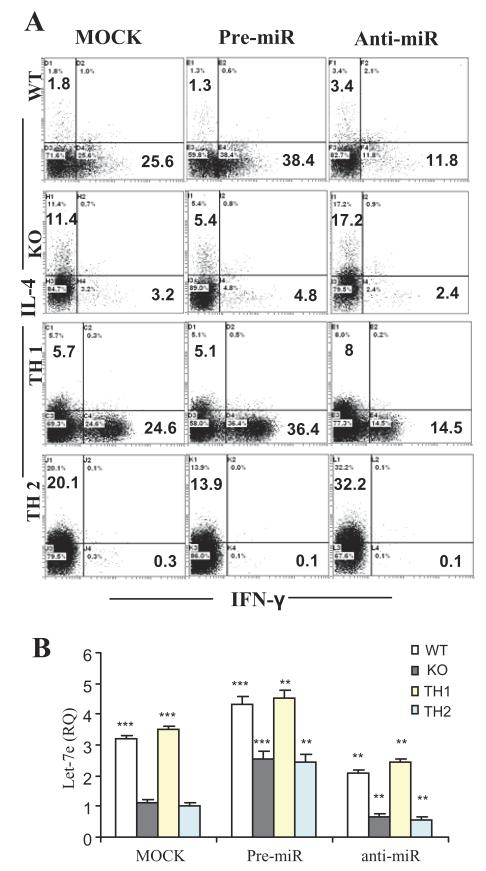
Figure 2. Let-7e regulates Th1-Th2 differentiation.
(A) Splenocytes from WT or CD44 KO EAE–induced mice on day 15 following the immunization were transfected with pre-miR, anti-miR or mock oligonucleotides, and restimulated with MOG35-55 peptide (30 μg/ml) for 24 h (panel WT and KO). Naive CD4+ T cells with the transfection of the oligonucleotides were polarized to Th1 or Th2 under the stimulation of anti-CD3 and anti-CD28 antibodies (panel TH1 and TH2). Intracellular production of IL-4 and IFN-γ in CD4+ T cells was measured using flow cytometry. (B) At the end of the culture, part of cells was used for quantitation of let-7e by QPCR. **: p<0.01, ***: p<0.001, versus respective mock control in each group (Student’s t-test). Data are shown as mean ± SEM of n = 5 and are representative of three independent experiments.
Overexpression or knockdown of let-7e affects the development of EAE
In order to know whether the expression of let-7e can influence the development of EAE, we developed the lentivirus vectors that can, in vivo, increase the expression levels of let-7e (labeled as LV-pre) or inhibit the activity of endogenous let-7e (labeled as LV-anti) or serve as the negative control (labeled as LV-ctrl) (Fig. 3A). We introduced 30 ×106 recombinant lentivirus into WT mice through vein injection 7 days before the immunization. The data showed that LV-pre-infected mice developed severe EAE, whereas LV-anti-infected mice developed significantly milder EAE in comparison to that of LV-ctrl-infected mice (Fig. 3B). In another experiment, we introduced the lentivirus into WT mice 5 days after immunization. The results showed a similar effect on the EAE development as described above (data not shown).
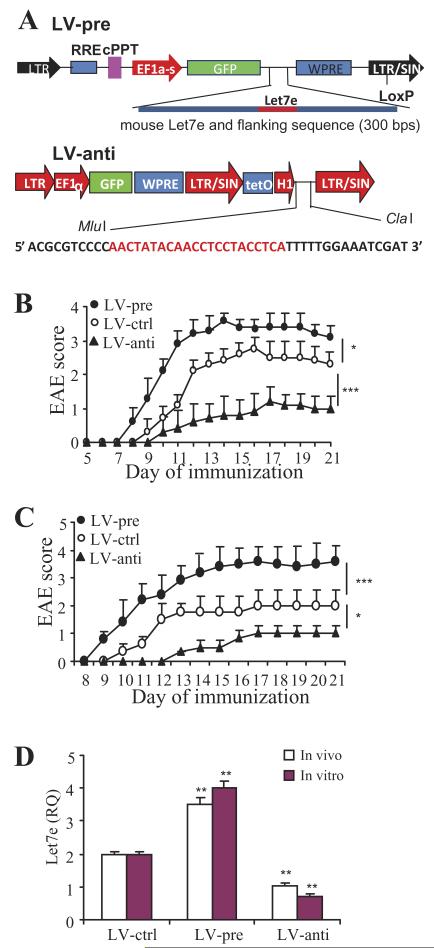
Figure 3. Silencing let-7e in vivo ameliorates the development of EAE.
(A) Lentivirus vectors overexpress (LV-pre) or inhibit (LV-ctrl) let-7e expression. The sequence of specific inhibitor of miR-let-7e is shown. (B) WT mice were injected intravenously with 30 × 106 lentivirus 7 days before MOG35-55 immunization; (C)splenocytes from virus-infected EAE mice on day 8 were restimulated with MOG35-55 for 3 days; CD4+ T cells were then isolated and transferred into naïve WT mice; the mean clinical scores were presented. (D) The expression of let-7e in the transferred cells was detected by QPCR. *: p<0.02, **: p<0.01, ***: p<0.007, versus LV-ctrl. (B and C) The mean clinical scores is shown (n = 10, Mann-Whitney test). (D) Data are shown as mean ± SEM of n = 5 (ANOVA test) and (B-D) are representative of three independent experiments.
To confirm whether let-7e directly affects the encephalitogenesis of effector CD4+ T cells, splenocytes from the virus-infected EAE mice on day 8 were restimulated with MOG35-55 for 3 days; CD4+ T cells were then isolated and transferred into naïve WT mice. The results showed that CD4+ T cells from LV-pre-infected mice caused severe EAE, whereas CD4+ T cells from LV-anti-infected mice triggered significantly milder EAE in comparison to that of LV-ctrl-infected mice (Fig. 3C). In another experiment, splenocytes from uninfected EAE WT mice on day 8 were infected with LV-pre, LV-anti, or LV-ctrl in vitro, and restimulated with MOG35-55 for 3 days; CD4+ T cells were then isolated and transferred into naïve WT mice. The results were similar to the above data using adoptive transfer (data not shown).
We examined the level of let-7e in CD4+ T cells used for the transfer. Consistent with the previous finding (Fig. 1F), CD4+ T cells on day 8 of EAE showed increased expression of let-7e. The virus infection in vivo or in vitro equally affected the expression of let-7e (Fig. 3D). These results showed that when compared with the LV-controls, LV-pre significantly increased the expression of let-7e; whereas LV-anti significantly inhibited the expression of let-7e (Fig. 3D). All together, the above results indicated that overexpression of let-7e enhanced the encephalitogenesis of the effector CD4+ T cells and induced severe EAE; whereas downregulation of let-7e played the opposite role.
Let-7e regulates IL-4, IL-10, IL-17, and IFN-γ production in encephalitogenic CD4+ T cells
On the day 15 of EAE, splenic CD4+ T cells continued to overexpress let-7e following the LV-pre infection; whereas the LV-anti infection suppressed the expression of let -7e (Fig. 4A). We also examined the cytokine production in the encephalitogenic CD4+ T cells. After re-stimulation with MOG35-55, production of IFN-γ and IL-17 was dramatically increased in CD4+ T cells with let-7e overexpression while production of IL-4 and IL-10 was dramatically inhibited; on the contrary, production of IFN-γ and IL-17 was dramatically inhibited in CD4+ T cells with let-7e downregulation while production of IL-4 and IL-10 was dramatically increased (Fig 4B). Similar pattern was also found in the culture supernatant and sera (Fig 4C). CNS-MNCs were also prepared at the same time and assessed for the protein level of IL-4, IL-10, L-17, and IFN-γ in the culture supernatants. The results were similar to that of the splenic CD4+ T cells (data not shown). Overall, the results showed that the overexpression of let-7e enhanced IFN-γ and IL-17 production while the inhibiting IL-4 and IL-10 production. In contrast, downregulation of let-7e resulted in opposite effects. These results demonstrated that let-7e plays a critical role in the differentiation of Th1, Th2, Th17 cells as well as the regulation of IL-10 production.
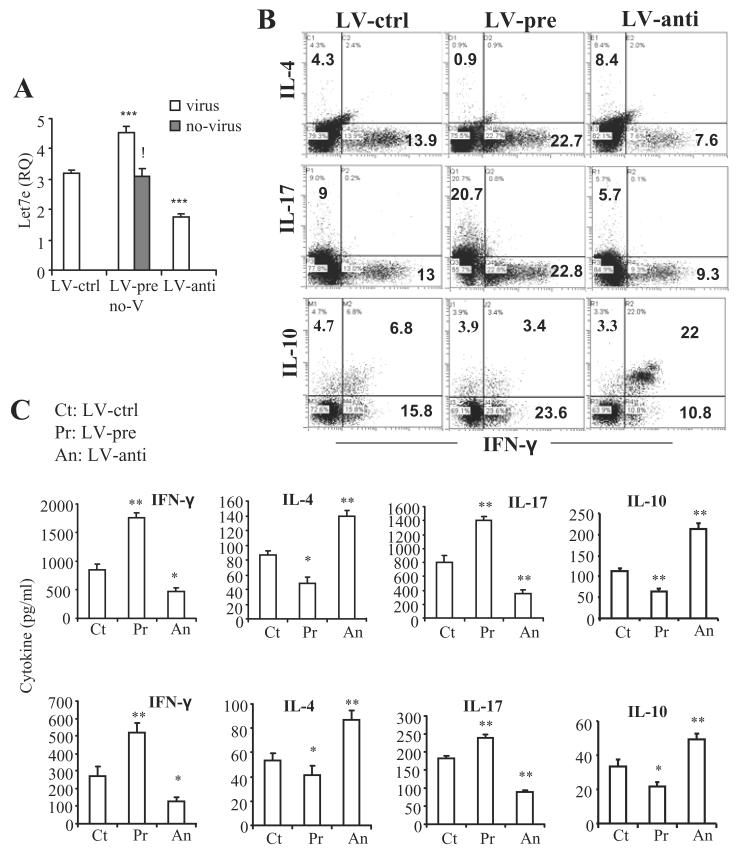
Figure 4. Modulation of let-7e level in vivo regulates the development of encephalitogenic Th1, Th2 and Th17 cells as well as IL-10 production.
(A) Splenic CD4+ T cells were prepared on day 15 of immunization from the virusinfected WT EAE mice as described in Figure 3B and subject to quantitation of let-7e by QPCR. The gray bar shows the quantitation of let-7e in the splenic CD4+ T cells from uninfected WT EAE mice (labeled as no-V in the histogram). ***: p<0.001, versus LV-ctrl (ANOVA test).!:p<0.001, between LV-pre and WT (Student’s t-test). (B and C) Splenocytes from the same mice were re-stimulated with MOG35-55 peptide (30 μg/ml) for 24 h. Production of IL-4, IL-10, IL-17 and IFN-γ in CD4+ T cells, culture supernatants as well as serum were measured. (B) Dot plots of the intracellular staining; (C) cytokines in the culture supernatants (top row) and serum (bottom row). *: p<0.02, **: p<0.01, versus LV-ctrl (Student’s t-test or ANOVA test). . Data are shown as mean ± SEM of n = 5 (top row) or 10 (bottom row and A) and are representative of three independent experiments.
IL-10 is the functional target of let-7e
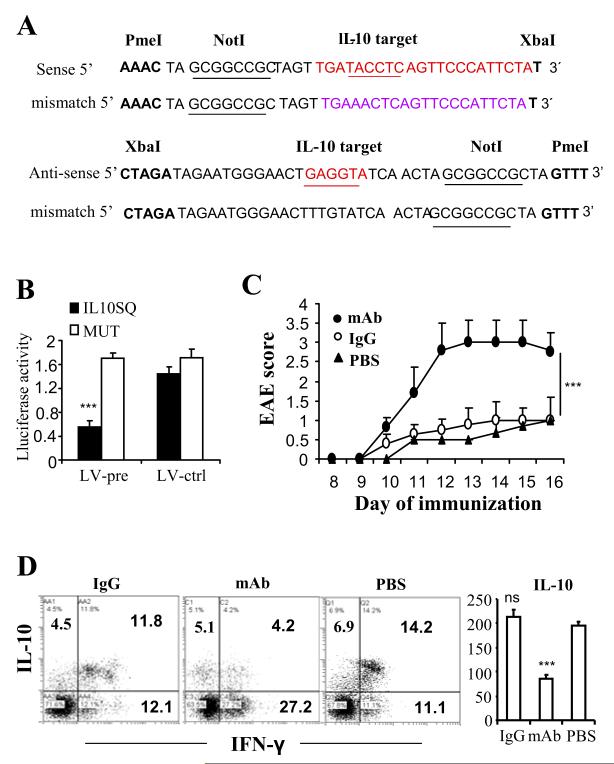
We found that immunoregulatory gene, IL-10, has the putative binding site in the 3′-UTR of let-7e. The luciferase assay showed that IL-10 could specifically and directly bind to let-7e (Fig. 5A and 5B). Our results showed that LV-pre enhanced the level of let-7e and correspondingly inhibited the protein level of IL-10; whereas LV-anti inhibited the level of let-7e and enhanced the protein level of IL-10 (Fig. 4B and 4C). To further determine whether IL-10 is the functional target of let-7e and associated with the development of EAE, we performed the IL-10 neutralization study by administering the mice with anti-IL-10 neutralizing antibody. WT mice were infected with LV-anti 7 days before the MOG35-55 immunization for the induction of EAE as described above. On different days of EAE, mice were given i.p. the antibody, isotype Ig, or the same amount of PBS (labeled as mAb, IgG, PBS, respectively). The results showed that the anti-IL-10 antibody treatment caused an aggravated disease (Fig. 5C). On day 15, splenocytes were restimulated with MOG35-55 for 24 h and production of IL-10 in the CD4+ T cells was detected by flow cytometry after intracellular staining. The results showed that IL-10 production in CD4+ T cells was significantly inhibited by the anti-IL-10 antibody treatment while IFN-γ production in CD4+ T cells was significantly enhanced (Fig. 5D). Also, data on production of IL-10 in the culture supernatants yielded a pattern similar to the above (Fig. 5D). These results demonstrated that neutralization of IL-10 could reverse the protective effect of LV-anti on the development of EAE and therefore, IL-10 is the functional target of let-7e.
Figure 5. IL-10 is the functional target of miR-let-7e.
(A) Matched or mutant sequence of IL-10 3′-UTR binding site for the construction of the dual-luciferase vectors. (B) The relative luciferase activity. (C) WT mice were infected with LV-anti 7 days before the MOG35-55 immunization as described in Materials and Methods. On day 5, 6, 10 and 13, mice were injected i.p. with anti-IL10 mAb, isotype control, or PBS. The mean clinical scores is shown. (D) Splenocytes were prepared on day 15 and re-stimulated with MOG35-55 (30 μg/ml) for 24 h. Production of IL-10 and IFN-γ in CD4+ T cells were measured after intracellular staining. Representative dot plots of three experiments are shown. The culture supernatants were detected for IL-10 and results were presented as histogram (pg/ml). ***: p<0.001, versus MUT control (B, ANOVA test) or PBS (C, Mann-Whitney test and D, Student’s t-test or ANOVA test), ns: not significance between IgG and PBS. (B-D) Data are shown as mean ± SEM of n = 3 (B), 10 (C), or 5 (D) and are representative of three independent experiments.
Let-7e targeted IL-13
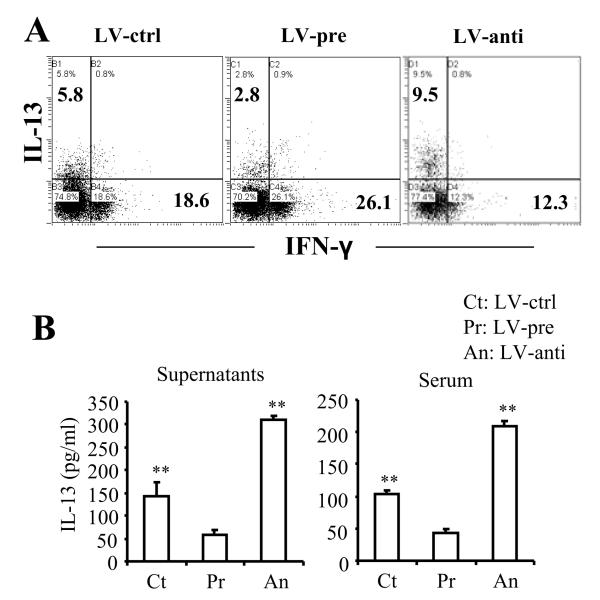
Although we showed that IL-10 was the target of let-7e, to further corroborate that let-7e overexpression or knockdown was functional in vivo, we tested the effect on another known functional target of let-7e such as IL-13 [25]. This target was also selected because previous studies have shown that induction of IL-13 protects against EAE [26-28]. To investigate whether overexpression or suppression of let-7e by the lentivirus transfection could affect IL-13 production, WT mice were infected with LV-pre or LV-anti, 7 days before the MOG35-55 immunization, for the induction of EAE as described above. On day 15, splenocytes were restimulated with MOG35-55 for 24 h and production of IL-13 in the CD4+ T cells was detected by flow cytometry after intracellular staining. The results showed that LV-pre significantly inhibited IL-13 production in CD4+ T cells with enhancement of IFN-γ production; whereas LV-anti significantly enhanced the IL-13 production in CD4+ T cells with suppression of IFN-γ production (Fig. 6A). Also, data on the production of IL-13 in the culture supernatants and serum yielded a pattern similar to the above (Fig. 6B). These results demonstrated that the LV vector systems used to generate let-7e overexpression and knockdown was functional in vivo because it did lead to repression and derepression of IL-13, a known let-7e target.
Figure 6. Let-7e targets IL-13 in EAE.
Splenocytes from the virus-infected WT EAE mice as described in Fig. 3B were prepared on day 15 of the immunization and re-stimulated with MOG35-55 peptide (30 μg/ml) for 24 h. Production of IL-13 and IFN-γ in CD4+ T cells, culture supernatants as well as serum were measured as in Figure 5. (A) Dot plots of the intracellular staining; (B) cytokines in the culture supernatants and serum. (B) Data are shown as mean ± SEM of n = 5 and are representative of three independent experiments. **: p<0.01, versus LV-pre (ANOVA test).
Discussion
The precise role of miRNAs in the regulation of the pathogenesis of MS is unclear. In the current study, we demonstrate for the first time that the miRNA let-7e was significantly upregulated in experimental EAE as well as in multiple sclerosis patients. Importantly, we found that silencing of let-7e in vivo inhibited encephalitogenic Th1 and Th17 cells and attenuated EAE, with reciprocal increase of Th2 cells, while overexpression of let-7e enhanced Th1 and Th17 cells and aggravated EAE. Our studies also identified IL-10 as the functional target of let-7e inasmuch as neutralization of IL-10 in vivo led to reversal of the protective effect of LV-anti on the development of EAE. Together, our studies demonstrated that let-7e plays a crucial role in the regulation of encephalitogenic T-cell differentiation and the pathogenesis of EAE and MS.
The let-7 miRNA, a founding member of miRNA gene family, was first identified in Caenorhabditis elegans and is conserved in invertebrates and vertebrates including humans, and controls the timing of cell cycle exit and terminal differentiation in C. elegans [29, 30]. Recent studies suggested that let-7 functions as a tumor suppressor in certain types of cancer cells through targeting RAS, IMP-1, HMGA2, and BACH1 [31-33]; for example, let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2 [34]. In the nervous system, it was reported that overexpression of let-7e negatively modulated Casp3 expression and protected against neuronal apoptosis following anoxia/reoxygenation injury [35]. In the immune response, it has been reported that let-7e regulates the endotoxin sensitivity and tolerance of macrophages through targeting TLR4 [36]; let-7a and let-7e play proinflammatory roles in experimental asthma by targeting IL-13 [25]. Lately, it is reported that let-7b, let-7c and let-7f are involved in HIV infection by targeting IL-10 [37]. Thus, our studies are novel in establishing for the first time that let-7e modulates Th differentiation and the development of EAE by targeting IL-10.
It is known that IL-10 is a negative regulator of EAE by suppressing Th1 and Th17 and promoting Th2 cells [11-13, 38]. Our study showed that modulation of expression level of let-7e directly affected IL-10 at the protein level and simultaneous Th1-Th2-Th17 differentiation. It is known that various kinds of immune cells express IL-10, such as monocytes/macrophages, neutrophils, mast cells, basophils, NK cells and effector T-cell subsets, including Th1, Th2, Th9, Th17 and Treg cells [39]. Our results showed the IL-10 expression by CD4+ T cells in two populations: CD4+IFN-γ−and CD4+IFN-γ+cells. The majority of CD4+IL-10+ cells were also CD4+IFN-γ+ cells. The differential production of IL-10 caused by the modulation of let-7e was mainly and significantly restricted to CD4+IFN-γ+ cells. While overexpression of let-7e repressed IL-10 production and augmented IFN-γ production in the culture supernatants and sera, similar pattern was seen in CD4+IFN-γ+ IL-10+ and CD4+IFN-γ+ IL-10− (Th1) cells. It was not clear whether these were two different populations or they could convert from each other due to the plasticity of the effector T cells. We also showed that neutralization of IL-10 successfully reversed the therapeutic effects of LV-anti in EAE, which clearly demonstrated that IL-10 was the functional target of let-7e and IL-10 was the negative regulator of EAE.
IL-13 was originally identified as a Th2 cytokine; however, it is now known that IL-13 is produced by a variety of cell types, such as natural killer T cells, natural killer cells, eosinophils, basophils [40], CD4+CD25+ Treg cells [26] and macrophages [27]. Due to the plasticity of effector T cells, Th1 cells could also produce IL-13 in response to the chronic antigen stimulation [40]. It has been demonstrated that IL-13 is a negative regulator of the development of EAE [26-28]. Our study examined the IL-13 expression by CD4+ T cells and showed that IFN-γ− CD4+ T cells are the main producing cells in CD4+ T-cell population. Our results showed that modulation of let-7e could target IL-13 and result in a pattern similar to the targeting of IL-10. Overexpression of let-7e repressed IL-13 production and augmented IFN-γ production whereas suppression of let-7e enhanced IL-13 production and inhibited IFN-γ production from the encephalitogenic CD4+ T cells. These results indicated that let-7e can also target IL-13 and thereby regulate pathogenesis of EAE. It should be noted that IL-10 is still a critical target of let-7e in our study because its neutralization reversed the protective effect of LV-anti on the development of EAE. Because let-7e has numerous targets and uses different targets in different models [31-33, 35-37], we are currently identifying additional targets of let-7e in EAE model.
The anti-inflammatory properties of IL-10 play a central role in infection by limiting the immune response to pathogens and thereby preventing damage to the host. However, regulation of IL-10 expression is of much complexity. Recently, an increasing interest in how IL-10 expression is regulated in different immune cells has revealed some of the molecular mechanisms involved at the levels of signal transduction, epigenetics, transcription factor binding and gene activation [39]. Since it is a negative regulator of EAE, understanding the specific molecular events that regulate the production of IL-10 are important for the design of new strategies of immune intervention. It has been shown that miR-106a modulates the IL-10 expression [41]. However, we did not find any difference in the levels of miR-106a between WT and KO mice (fold change: 1.7 vs. 1.6). Thus, we suggest that miR-106a may not be involved in the regulation of IL-10 in EAE.
It has been demonstrated that miR-155 and miR-326 affected the development of encephalitogenic Th1, Th2 and Th17 cells and the pathogenesis of EAE [20-22, 42, 43]. In our study, we observed significantly increased levels of miR-155 in WT-EAE mice when compared with WT-naïve mice; the level of miR-155 in CD44 KO-EAE mice was also increased but the degree of increase was significantly lower than WT-EAE mice (fold change: 3.21 vs. 1.95). Meanwhile, miR-326 was positively increased in WT-EAE mice and decreased in KO-EAE mice (fold change: 1.03 vs. -1.04) as was the pattern of let-7e (fold change: 2.7 vs. -1). These results suggested that multiple miRNAs may be involved in the differentiation of encephalitogenic T cells and the pathogenesis of EAE.
It has been demonstrated by us that CD44 reciprocally regulates the differentiation of encephalitogenic Th1/Th17 and Th2/Treg cells through epigenetic modulation involving DNA methylation of cytokine gene promoters, thereby controlling the development of EAE [23]. In the CD44 KO mice, the development of Th1 and Th17 cells were significantly inhibited while the development of Th2 and Treg cells was significantly enhanced; the immune response was dramatically shifted from Th1-type to Th2-type. We also showed that the role of CD44 depends on the signaling pathway involving the interaction between osteopontin-CD44 [23]. It has been reported that Raf kinase inhibitory protein (RKIP) regulates let-7e expression; the latter can target HMGA2 and BACH1, thereby regulating the osteopontin expression [32]. We speculate that CD44 could modulate the levels of RKIP thereby influencing let-7e expression. The related study is undergoing.
Material and Methods
Mice and reagents
Wild-type C57BL/6 (WT or CD44+/+) mice were purchased from the National Cancer Institute. CD44 knockout (CD44 KO or CD44−/−) mice were generated at Amgen Institute (Canada) and kindly provided to us by Dr. Tak Mak. These mice are on C57BL/6 background and have been extensively characterized in our previous studies [44, 45]. Mice were housed in the University of South Carolina Animal Facility. Animal procedures were performed according to NIH guidelines under protocols approved by the Institute Animal Care and Use Committee of the university. MOG35-55 peptide was purchased from NeoMPS. Incomplete Freund’s adjuvant (IFA) and desiccated Mycobacterium tuberculosis H37RA were purchased from DIFCO. Pertussis toxin (PTX) was purchased from List Biological Laboratories. All antibodies and isotype controls were purchased from Biolegend. PMA and ionomycin were purchase from Sigma.
Lentivirus-mediated overexpression or inhibition of miR-let-7e
A 300 base pair genomic sequence spanning the mouse miR-let-7e coding region and the flanking regions from 5′ or 3′ on either end was cloned into lentiviral vector pWPT-GFP downstream of EF1α-short promoter (named as LV-pre in the text). A sequence complementary to let-7e was cloned into pLVTHM vector under the control of H1 promoter to obtain let-7e inhibition vector (named as LV-anti in the text). A sequence encoding mutant let-7e was also cloned for vector control (named as LV-ctrl in the text). Virus was produced in HEK293T cells as previously described [46]. Splenocytes or CD4+ T cells were infected at MOI of 10 by spinning at 2,000 rpm for 60 min at room temperature. For direct in vivo administration of lentiviruses, mice were injected 30 × 106 viruses intravenously.
EAE induction, adoptive transfer, intracellular staining, and cytokine measurement
EAE was induced by s.c. immunization of MOG35-55 peptide in WT and KO mice as we described previously [23]. For adoptive transfer of EAE, splenocytes were prepared from EAE-induced WT mice (virus-infected or uninfected) on day 8 after the immunization, and re-stimulated with MOG35-55 peptide (30μg/ml) for 3 days without or with the lentivirus infection, respectively. CD4+ T cells were then purified with magnetic beads (Stemcell), and 3 × 106 CD4+ T cells were intravenously transferred into naïve WT recipient mice. Mice were given 200 ng at the day of transfer and 400 ng of pertussis toxin at day 2 post transfer. Mice were monitored and scored daily for disease progression as described previously [23].
CNS-infiltrating mononuclear cells (CNS-MNCs) from pooled spinal cord and brain were prepared with Percoll gradient (70%/30%) centrifugation after perfusion with 30 ml heparin-PBS. Splenocytes or CNS-infiltrating MNCs were stimulated with 30μg/ml of MOG35-55 for 24 h, followed by stimulation with 50 ng/ml PMA and 1 μg /ml ionomycin in the presence of 2 μm monensin for 4 h. Production of IL-4, IL-10, IL-17, and IFN-γ in the CD4+ T cells were detected by flow cytometry after intracellular staining (Beckman Coulter, CXP FC500); the cytokines in the culture supernatants were detected using sandwich ELISA or multiplexed microsphere cytokine immunoassay (Bio-Plex Cytokine Assay kit, Bio-Rad), The cytokines in the sera were measured using the same methods as for the culture supernatants.
MicroRNA array and quantitative real-time PCR
MicroRNA array in Affymetrix platform was performed with 869 probes for human PBL and 609 probes for mouse CD4+ T cells. CD4+ T cells from naïve mice and PBL from normal individuals were used as background control. Quantitative real-time PCR (QPCR) was applied for the validation and amplification of the test samples. Relative fold expression values were calculated based on the expression of RNU1A gene as endogenous control. Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MEXP-3594.
Th1-Th2 polarization and oligonucleotide transfection
Naïve CD4+ T cells were polarized under Th1 or Th2 -polarizing condition [23]. Some cells were transfected with Pre-miR let-7e, anti-miR let-7e and mock oligonucleotides (Applied Biosystems, named as Pre-miR, anti-miR or mock in the text) by Lipofectamine RNAiMAX (Invitrogen) as per the manufacturer’s protocol.
Luciferase reporter assay
HEK293 cells (ATCC) were transfected with lentivirus LV-ctrl or LV-pre; the latter became high miR-let-7e-expressing cells. To develop the Dual-Luciferase reporter vectors, wild-type (named as IL10SQ in the text) or mutated 3′-UTR sequences (named as MUT in the text) of mouse IL-10 that overlap the binding site for miR-let-7e were cloned into pmirGLO Dual-Luciferase miRNA Target Expression vector (Promega). The constructs were introduced into HEK293 cells by Fugene HD (Roche). Cell extracts were prepared 24 h later and luciferase activity was measured with Dual-Glo Luciferase Assay system.
Statistical analysis
Data were presented as mean ± SEM and analyzed for significance using 2-tailed Student’s t-test (unequal variance type) or the Mann-Whitney U test. In case of multiple testing, One-way ANOVA test followed by Newman-Keuls Multiple Comparision Test was applied. Comparisons were considered significant at p≤0.05.
Acknowledgments
This work was supported in part by NIH grants P01AT003961, R01MH094755, P20RR032684 to PN; R01ES019313 to PN and MN; R01AT006888 and VA Merit Award BX001357 to MN; R21HL106325 to DF; and the Research Development Fund from University of South Carolina to HG.
Footnotes
Conflict of interest The authors declare no financial or commercial conflict of interest.
Reference
- 1.Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372:1502–1517. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- 2.Goverman J. Autoimmune T-cell responses in the central nervous system. Nat Rev Immunol. 2009 doi: 10.1038/nri2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasper LH, Shoemaker J. Multiple sclerosis immunology: The healthy immune system vs the MS immune system. Neurology. 74(Suppl 1):S2–8. doi: 10.1212/WNL.0b013e3181c97c8f. [DOI] [PubMed] [Google Scholar]
- 4.Jager A, Dardalhon V, Sobel RA, Bettelli E, Kuchroo VK. Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. J Immunol. 2009;183:7169–7177. doi: 10.4049/jimmunol.0901906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Connor RA, Prendergast CT, Sabatos CA, Lau CW, Leech MD, Wraith DC, Anderton SM. Cutting edge: Th1 cells facilitate the entry of Th17 cells to the central nervous system during experimental autoimmune encephalomyelitis. J Immunol. 2008;181:3750–3754. doi: 10.4049/jimmunol.181.6.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bettelli E, Korn T, Oukka M, Kuchroo VK. Induction and effector functions of T(H)17 cells. Nature. 2008;453:1051–1057. doi: 10.1038/nature07036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aharoni R, Teitelbaum D, Leitner O, Meshorer A, Sela M, Arnon R. Specific Th2 cells accumulate in the central nervous system of mice protected against experimental autoimmune encephalomyelitis by copolymer 1. Proc Natl Acad Sci U S A. 2000;97:11472–11477. doi: 10.1073/pnas.97.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber MS, Prod’homme T, Youssef S, Dunn SE, Rundle CD, Lee L, Patarroyo JC, et al. Type II monocytes modulate T cell-mediated central nervous system autoimmune disease. Nat Med. 2007;13:935–943. doi: 10.1038/nm1620. [DOI] [PubMed] [Google Scholar]
- 9.Steinman L. A brief history of T(H)17, the first major revision in the T(H)1/T(H)2 hypothesis of T cell-mediated tissue damage. Nat Med. 2007;13:139–145. doi: 10.1038/nm1551. [DOI] [PubMed] [Google Scholar]
- 10.Qin X, Guo BT, Wan B, Fang L, Lu L, Wu L, Zang YQ, et al. Regulation of Th1 and Th17 cell differentiation and amelioration of experimental autoimmune encephalomyelitis by natural product compound berberine. J Immunol. 185:1855–1863. doi: 10.4049/jimmunol.0903853. [DOI] [PubMed] [Google Scholar]
- 11.Dai H, Ciric B, Zhang GX, Rostami A. Interleukin-10 plays a crucial role in suppression of experimental autoimmune encephalomyelitis by Bowman-Birk inhibitor. J Neuroimmunol. 245:1–7. doi: 10.1016/j.jneuroim.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L, Yuan S, Cheng G, Guo B. Type I IFN promotes IL-10 production from T cells to suppress Th17 cells and Th17-associated autoimmune inflammation. PLoS One. 6:e28432. doi: 10.1371/journal.pone.0028432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bettelli E, Nicholson LB, Kuchroo VK. IL-10, a key effector regulatory cytokine in experimental autoimmune encephalomyelitis. J Autoimmun. 2003;20:265–267. doi: 10.1016/s0896-8411(03)00048-9. [DOI] [PubMed] [Google Scholar]
- 14.Coolen M, Bally-Cuif L. MicroRNAs in brain development and physiology. Curr Opin Neurobiol. 2009;19:461–470. doi: 10.1016/j.conb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 16.Taganov KD, Boldin MP, Baltimore D. MicroRNAs and immunity: tiny players in a big field. Immunity. 2007;26:133–137. doi: 10.1016/j.immuni.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Mattes J, Collison A, Foster PS. Emerging role of microRNAs in disease pathogenesis and strategies for therapeutic modulation. Curr Opin Mol Ther. 2008;10:150–157. [PubMed] [Google Scholar]
- 18.Eacker SM, Dawson TM, Dawson VL. Understanding microRNAs in neurodegeneration. Nat Rev Neurosci. 2009;10:837–841. doi: 10.1038/nrn2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Junker A, Krumbholz M, Eisele S, Mohan H, Augstein F, Bittner R, Lassmann H, et al. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain. 2009;132:3342–3352. doi: 10.1093/brain/awp300. [DOI] [PubMed] [Google Scholar]
- 21.O’Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T-cell development. Immunity. 33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 23.Guan H, Nagarkatti PS, Nagarkatti M. CD44 Reciprocally regulates the differentiation of encephalitogenic Th1/Th17 and Th2/regulatory T cells through epigenetic modulation involving DNA methylation of cytokine gene promoters, thereby controlling the development of experimental autoimmune encephalomyelitis. J Immunol. 186:6955–6964. doi: 10.4049/jimmunol.1004043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guan H, Nagarkatti PS, Nagarkatti M. Role of CD44 in the differentiation of Th1 and Th2 cells: CD44-deficiency enhances the development of Th2 effectors in response to sheep RBC and chicken ovalbumin. J Immunol. 2009;183:172–180. doi: 10.4049/jimmunol.0802325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polikepahad S, Knight JM, Naghavi AO, Oplt T, Creighton CJ, Shaw C, Benham AL, et al. Proinflammatory role for let-7 microRNAS in experimental asthma. J Biol Chem. 285:30139–30149. doi: 10.1074/jbc.M110.145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ochoa-Reparaz J, Rynda A, Ascon MA, Yang X, Kochetkova I, Riccardi C, Callis G, et al. IL-13 production by Treg cells protects against experimental autoimmune encephalomyelitis independently of autoantigen. J Immunol. 2008;181:954–968. doi: 10.4049/jimmunol.181.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cash E, Minty A, Ferrara P, Caput D, Fradelizi D, Rott O. Macrophage-inactivating IL-13 suppresses experimental autoimmune encephalomyelitis in rats. J Immunol. 1994;153:4258–4267. [PubMed] [Google Scholar]
- 28.Young DA, Lowe LD, Booth SS, Whitters MJ, Nicholson L, Kuchroo VK, Collins M. IL-4, IL-10, IL-13, and TGF-beta from an altered peptide ligand-specific Th2 cell clone down-regulate adoptive transfer of experimental autoimmune encephalomyelitis. J Immunol. 2000;164:3563–3572. doi: 10.4049/jimmunol.164.7.3563. [DOI] [PubMed] [Google Scholar]
- 29.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 30.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 31.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 32.Yun J, Frankenberger CA, Kuo WL, Boelens MC, Eves EM, Cheng N, Liang H, et al. Signalling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer. Embo J. 30:4500–4514. doi: 10.1038/emboj.2011.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, et al. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–2591. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- 34.Dong Q, Meng P, Wang T, Qin W, Qin W, Wang F, Yuan J, et al. MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PLoS One. 5:e10147. doi: 10.1371/journal.pone.0010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng G, Yuan Y, He Q, Wu W, Luo BY. MicroRNA let-7e regulates the expression of caspase-3 during apoptosis of PC12 cells following anoxia/reoxygenation injury. Brain Res Bull. 86:272–276. doi: 10.1016/j.brainresbull.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 36.Androulidaki A, Iliopoulos D, Arranz A, Doxaki C, Schworer S, Zacharioudaki V, Margioris AN, et al. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31:220–231. doi: 10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swaminathan S, Suzuki K, Seddiki N, Kaplan W, Cowley MJ, Hood CL, Clancy JL, et al. Differential Regulation of the Let-7 Family of MicroRNAs in CD4+ T Cells Alters IL-10 Expression. J Immunol. 188:6238–6246. doi: 10.4049/jimmunol.1101196. [DOI] [PubMed] [Google Scholar]
- 38.Bettelli E, Das MP, Howard ED, Weiner HL, Sobel RA, Kuchroo VK. IL-10 is critical in the regulation of autoimmune encephalomyelitis as demonstrated by studies of IL-10- and IL-4-deficient and transgenic mice. J Immunol. 1998;161:3299–3306. [PubMed] [Google Scholar]
- 39.Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 40.Motomura Y, Kitamura H, Hijikata A, Matsunaga Y, Matsumoto K, Inoue H, Atarashi K, et al. The transcription factor E4BP4 regulates the production of IL-10 and IL-13 in CD4+ T cells. Nat Immunol. 12:450–459. doi: 10.1038/ni.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma A, Kumar M, Aich J, Hariharan M, Brahmachari SK, Agrawal A, Ghosh B. Posttranscriptional regulation of interleukin-10 expression by hsa-miR-106a. Proc Natl Acad Sci U S A. 2009;106:5761–5766. doi: 10.1073/pnas.0808743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen D, McKallip RJ, Zeytun A, Do Y, Lombard C, Robertson JL, Mak TW, et al. CD44-deficient mice exhibit enhanced hepatitis after concanavalin A injection: evidence for involvement of CD44 in activation-induced cell death. J Immunol. 2001;166:5889–5897. doi: 10.4049/jimmunol.166.10.5889. [DOI] [PubMed] [Google Scholar]
- 45.McKallip RJ, Fisher M, Do Y, Szakal AK, Gunthert U, Nagarkatti PS, Nagarkatti M. Targeted deletion of CD44v7 exon leads to decreased endothelial cell injury but not tumor cell killing mediated by interleukin-2-activated cytolytic lymphocytes. J Biol Chem. 2003;278:43818–43830. doi: 10.1074/jbc.M304467200. [DOI] [PubMed] [Google Scholar]
- 46.Fan D, Qiu S, Overton CD, Yancey PG, Swift LL, Jerome WG, Linton MF, et al. Impaired secretion of apolipoprotein E2 from macrophages. J Biol Chem. 2007;282:13746–13753. doi: 10.1074/jbc.M611754200. [DOI] [PubMed] [Google Scholar]