Abstract
To elucidate the putative function of a gene, effective tools are required for genetic characterization that facilitate its inactivation, deletion or modification on the bacterial chromosome. In the present study, the nucleotide sequence of the Escherichia coli/Aggregatibacter actinomycetemcomitans shuttle vector pYGK was determined, allowing us to redesign and construct a new shuttle cloning vector, pJT4, and promoterless lacZ transcriptional/translational fusion plasmids, pJT3 and pJT5. Plasmids pJT4 and pJT5 contain the origin of replication necessary to maintain shuttle vector replication. In addition, a new suicide vector, pJT1, was constructed for the generation of scarless and markerless deletion mutations of genes in the oral pathogen A. actinomycetemcomitans. Plasmid pJT1 is a pUC-based suicide vector that is counter-selectable for sucrose sensitivity. This vector does not leave antibiotic markers or scars on the chromosome after gene deletion and thus provides the option to combine several mutations in the same genetic background. The effectiveness of pJT1 was demonstrated by the construction of A. actinomycetemcomitans isogenic qseB single deletion (ΔqseB) mutant and lsrRK double deletion mutants (ΔlsrRK). These new vectors may offer alternatives for genetic studies in A. actinomycetemcomitans and other members of the HACEK (Haemophilus spp., A. actinomycetemcomitans, Cardiobacterium hominis, Eikenella corrodens, and Kingella kingae) group of Gram-negative bacteria.
1. Introduction
Aggregatibacter actinomycetemcomitans is a Gram-negative, facultative bacterium, and non-motile coccobacillus of the Pasteurellaceae familly that is found in the oral cavity, and it is implicated in the etiology of aggressive and chronic periodontitis, but also is associated with extra-oral infections such as infective endocarditis, soft tissue abscesses, meningitis, pneumonia, septicemia, urinary tract infections, and osteomyelitis (Haffajee and Socransky, 1994; Henderson et al., 2003; Paturel et al., 2004; Nørskov-Lauritsen and Kilian, 2006; Rahamat-Langendoen et al., 2011 Wang et al., 2010; Hyvärinen et al., 2012). A. actinomycetemcomitans expresses a variety of virulence factors to survive in the oral cavity and causes tissue inflammation, tissue destruction, immune suppression and bone resorption (Han et al., 2006; Rogers et al.,2007). The molecular mechanisms by which A. actinomycetemcomitants successfully colonizes and persists in the oral cavity, and its ability to disseminate to other organs of the body have not been well defined. In part, this is due to the limited repertoire of molecular genetic tools that are available to manipulate A. actinomycetemcomitans.
Several molecular biological approaches have been reported for studies of A. actinomycetemcomitans gene function including conjugation-based transposon-insertion mutagenesis (Thomson et al., 1999) which was applied for the mutagenesis of the katA gene and the characterization of genes involved in the tight adherence (tad) locus Kachlany et al., 2000; Kachlany et al., 2001a; Kachlany et al., 2001b; Planet et al., 2003). Gene inactivation by a single recombinant event to insert an antibiotic marker carried on a plasmid vector has been performed using mobilizable vectors such as pVT1461 (R6K ori) to inactivate recA (Mintz et al., 2002), and vectors such as pUC19 that do not replicate in A. actinomycetemcomitans have been used to inactivate the lsrB gene (Shao et al., 2007a). Gene replacement by an antibiotic marker, generated by double recombination events, using plasmids based on the pUC replicon and containing the counter-selectable marker sacB for sucrose-sensitivity (Reyrat et al., 1998) have also been reported to produce genetic deletions of orfA (Schaeffer et al., 2008) and ihfB (Kolodrubetz et al., 2010). Finally, the Cre/loxP recombinase system, which represents a step forward in the generation of markerless deletion mutations in A. actinomyctemcomitans, has been applied to generate a deletion mutation of the SPA-a gene cluster, and the mutant strain was complemented by the reinsertion of the SPA-a gene cluster. In addition, a deletion mutation of the pagBC genes was performed using this methodology (Fujise et al., 2004). However, these approaches may have limitations for the construction of multiple deletion mutations arising the antibiotic markers or the scars (i.e, the loxP sequence left in the genome after the recombination event between two loxP sequences) generated by the Cre recombinase system. For example, the loxP sequences remaining in the chromosome may be substrates for future recombination events during the construction of new deletion mutations in the same genetic background unless loxP with different spacer sequences are used (Lee and Saito, 1998)
A. actinomycetemcomitans is genetically hetereogeneous and comprises strains of six clonal populations expressing distinct serotypes that exhibit variation in natural competence for DNA uptake Kittichotirat et al., 2011). Serotypes a, d and e are naturally competent (Fujise et al., 2004), but serotypes b, c and f contain an insertional inactivation of the comM gene that results in the inability of these strains to be readily transformed (Mena and Chen, 2007; Kittichotirat et al., 2011 ). This is important since serotype b and c strains are commonly associated with human oral infections. The Cre/loxP recombinase system has been successfully used in naturally competent A. actinomycetemcomitans strains, but its efficacy in non-natural competent strains has not yet been reported. In an attempt to overcome the limitation presented by non-naturally competent strains, Bhattacharjee et al. (2007) reported that inducible expression of the tfox gene from a plasmid restored the natural competent phenotype in A. actinomycetemcomitans strains except serotype f. While this system has utility, the approach also has several limitations in that the DNA used for the transformation must contain a 9-bp uptake sequence (USS), it relies on selection by antibiotic resistance, and the replicating plasmid containing tfoX must be eliminated before the performing genetic studies. In general, there remains a strong need for additional genetic tools to manipulate A. actinomycetemcomitans that are effective in naturally competent as well as non-naturally competent strains.
Plasmid pYGK (Brogan et al., 1996) is an A. actinomycetemcomitans-E. coli shuttle vector that was derived from plasmid pYG10 extracted from A. pleuropneumoniae 80-8141 Lalonde et al., 1989). It has been used as a transcriptional reporter plasmid and cloning vector for complementation studies in A. actinomyctemcomitans James et al., 2006; Novak et al., 2010; Shao et al., 2007a; Shao et al., 2007b). However, the nucleotide sequence of pYGK has not been determined, nor has its origin of replication been identified which has limited its further development. In the present study, we report the nucleotide sequence of pYGK and define the minimal origin of replication required to maintain the plasmid in A. actinomycetemcomitans and E. coli. This allowed us to redesign and construct three new pYGK-derived plasmids including the cloning vector pJT4 and the transcriptional/translational reporter plasmids pJT3 and pJT5. In addition, we describe the construction of pJT1, a suicide plasmid that facilitates the generation of scarless and markerless chromosomal deletion mutations in A. actinomycetemcomitans. The utility of each plasmid was demonstrated in the serotype c strain A. actinomycetemcomitans 652.
2. Materials and methods
2.1 Bacterial strains, plasmids, and media
The bacterial strains and plasmids used in this study are listed in Table 1. LB (Luria-Bertani) broth, LB agar (LB broth plus 1.5% agar), brain heart infusion (BHI) broth, BHI agar, TYE (1% tryptone, and 0.5% yeast extract) broth, and TYE agar (all from DIFCO) were routinely used for propagation and plating of bacteria. SOC medium (2% tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4 and 20 mM glucose) (Hanahan, 1983) was used for the recovery post-transformation bacterial cells. A. actinomycetemcomitans strain 652 (serotype c) that was grown at 37°C under microaerophilic conditions. When required the medium was supplemented with 10 % sucrose, 1 mM IPTG, 25 μg ml−1 kanamycin (Km), 12.5 μg ml−1 tetracycline, 100 μg ml−1 ampicillin (Ap), or 50 μg ml−1 spectinomycin (Sp).
Table 1.
Strains and plasmids used in this work
| Strain or plasmid | Derived, relevant genotype or characteristicsa | Source or reference |
|---|---|---|
| Escherichia coli | ||
| XL1-Blue MRF′ | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB. lacIqZΔM15 Tn10 (Tcr)] | Stratagene |
| Aggregatibacter actinomycetemcomitans | ||
| 652 | Wild type, serotype c, | Laboratory stock |
| 652-JR37 | 652 ΔqseB | This study |
| 652-TE47 | 652 ΔlsrRK | This study |
| Plasmids | ||
| pUC18 | Ampr; cloning vector | (Yanisch-Perron et al., 1985) |
| pBAD-HA-lacZ | Ampr, expression vector | Invitrogen |
| pYGK | Kmr, cloning shuttle vector | Brogan et al., 1996 |
| pYGK-lacZ | Kmr, promoterless lacZ reporter vector | James et al., 2006 |
| pUC4K | Kmr, cloning shuttle vector | (Viera and Messing, 1982) |
| Suicide vectors | ||
| pVT1461 | Spr; template for amplifying the spectinomycin gene | (Mintz et al., 2002) |
| pRE112 | Cmr: template for amplifying the sacB | (Edwards et al., 1998) |
| pJT1 | Spr; suicide vector | This study |
| Derived from suicide vector pJT1 | ||
| pDJR37 | Spr; flanking region to qseB | This study |
| pATE47 | Spr; flanking region to lsrRK | This study |
| Derived from reporter vector pYGK-lacZ | ||
| pJT3 | Kmr, promoterless lacZ reporter vector | This study |
| pJT4 | Kmr, cloning vector | This study |
| pJT5 | Kmr, promoterless lacZ reporter vector | This study |
| pJT6 | Kmr, cloning vector | This study |
| pJT12 | Kmr, promoter lsrR (255-bp)-lacZ | This study |
| Derived from pJT3 | ||
| pATE23 | Kmr, promoter lsrR (255-bp)-lacZ | Torres-Escobar et al., 2013 |
| Derived from pJT5 | ||
| pATE65 | Kmr, promoter lsrR (255-bp)-lacZ | This study |
Ampr ampicillin resistance, Kmr kanamycin resistance, Tcr tetracyclin resistance, Spr Spectinomycin resistance.
2.2 DNA procedures
DNA manipulations were carried out as described (Sambrook and Russell, 2001). Transformation of E. coli and A. actinomycetemcomitans was done by electroporation (Bio-Rad) [2 mm cuvette, voltage (V) 1800, capacitance (μF) 25, resistance (ohm) 200]. For replicative plasmids the ~2μg was used for electroporation whereas for non-replicative plasmids, ~20 μg was used. For this study, smooth colony strains of A. actinomycetemcomitans were used. However, we previously inactivated luxS in both a smooth strain and a clinical isolate via electroporation (Demuth et al., 2011) and observed little difference in transformation efficiency. Transformant cells containing plasmids were selected on LB agar plates supplemented with appropriated antibiotics. Plasmid DNA was isolated using the QIAprep Spin Miniprep Kit (Qiagen). Restriction enzymes were used as recommended by the manufacturer (New England Biolabs). All PCR products were amplified with the Platinum PCR Super Mix High Fidelity used as recommended by the manufacturer (Invitrogen). All primers used in this study (Integrated DNA Technology) were flanked with restriction enzyme recognition sites (underlined in the primer sequences) and are shown in Table 2. Primer sequences were designed based on the genome sequence of A. actinomycetemcomitans D11S-1 strain available from the Pathosystems Resource Integration Center (http://patricbrc.vbi.vt.edu). All nucleotide substitutions in the recombinant plasmids were performed using a Quick-Change site-directed mutagenesis kit (Stratagene). All constructs, and the presence of a desired mutation were verified by DNA sequencing (University of Louisville Facilities).
Table 2.
Oligonucleotides used in this work
| Related product and oligonucletide | Sequence (5′ →3′) |
|---|---|
| MCS assembly | |
| MDJR-16F | AGCTTGTGCTAGCATCTGGTACCCCACCCGGGTGGAAATTGGGCCCAATTAATGGCGGCCGCTACGTCGTAGTA |
| MDJR-17R | CTGGAAGTACTACGACGTAGCGGCCGCCATTAATTGGGCCCAATTTCCACCCGGGTGGGGTACCAGATGCTAGCACA |
| MDJR-18F | CTTCCAGCTACTCGAGTTATTCGAACTAGTATTCGTTACTGCAGCCAATATTGCGCGCTTATTGCTGAGCTCCAGCTTCGT |
| MDJR-19R | CAAGAGATCTACGAAGCTGGAGCTCAGCAATAAGCGCGCAATATTGGCTGCAGTAACGAATACTAGTTCGAATAACTCGAGTAG |
| MDJR-20F | AGATCTCTTGTTACACGCGTCTTATTCTGTCGACCTTCCTTTAGGATCCATTGAGTTACCTAGGGGTTATGCTAGCTAG |
| MDJR-21R | AATTCTAGCTAGCATAACCCCTAGGTAACTCAATGGATCCTAAAGGAAGGTCGACAGAATAAGACGCGTGTAA |
| Ptrc promoter | |
| MDJR-22F | CATTCTGAAATGAGCTGTTGACAATTAATCATCCGGCTCGTATAATGTGTGGAATTGTGAGCGGATAACAATTTCACACAGGAAACA |
| MDJR-23R | GATCTGTTTCCTGTGTGAAATTGTTATCCGCTCACAATTCCACACATTATACGAGCCGGATGATTAATTGTCAACAGCTCATTTCAGAATGAGCT |
| sacB gene | |
| MDJR-24F | AAGGAGACAAGATCTATGAACATCAAAAAGTTTGCAAAACAAGC |
| MDJR-27R | AACTAGCGAACGCGTTCAAAAGGTTAGGAATACGGTTAGC |
| pUC ori | |
| MDJR-34F | GCGGTAATACGCGTATCCACAGAATCAGGGGATAACG |
| MDJR-35R | TAAGCATTGTCGACTGTCAGACCAAGTTTACTCATATATAC |
| Spectinomycin gene | |
| MDJR-28F | GTCGTACGGTCGACTTTCGTTCGTGAATACATGTTATAATAAC |
| MDJR-29R | GTAATCTGGCTAGCCCTAGGCAAGGGTTTATTGTTTTCTAAAATC |
| qseB deletion | |
| MDJR-49F NotI | |
| MDJR-82R XhoI | GCCGTTACCCTCGAGAGGATCATCTTCGATTAATAAAATCCGC |
| MDJR-83F XhoI | GTTCACGGCCTCGAGTATGCACTGGGAAAAAATGATGAAACTG |
| MDJR-84R PstI | GTCCGCTTTCCTGCAGGACACCAACTTTAGTCATGAAAGAC |
| PCR verification | |
| MDJR-63F | CCAAGTGGAAATCGAACGCAAAGCCTGGC |
| MDJR-61R | CGTACGTTTTGCCACTTGTTCACCGATTTGTTTCAGG |
| lsrRK double deletion | |
| ATE-86F NotI | CGTCAACATAATTATTCCCTCCGCGGCCGCTTATTTCGACCGC |
| ATE-87R XhoI | CGTTTCTTGATTATCCGTCTCGAGCATTAATTCGTTC |
| ATE-88F XhoI | GGCACCAGGGTTGTAGCTCGAGCGAACGATAAAAAATCG |
| ATE-89R PstI | GATGCAATTTTTGTCTGCAGCATATATACCTC |
| PCR-verification | |
| ATE-62F | GAGCTGATGGGCTTTATTCG |
| ATE-102R | CAAGATCTCAGCCGTTTCATCACC |
| Upstream region lsrR | |
| lsrR255 f-3 | GTGAGTTTGGGTACCCATCTTTTACCTGCCG |
| lsrR255 r-6 | CTTGATTATCCG TGGATCCCATTAATTCGTTC |
| lsrRK operon | |
| lsrR F-109 | CCGGCGATAATTTTCATGGATCCCAGCGTGGATTTCC |
| lsrK R-110 | GGAACCGGTGGCATAATCTAGAAAGTGCGGTGGG |
| crp gene | |
| crp F-151 | GCCGAATCAGGGTACCAACCAGCAATGGC |
| crp R-152 | CCCGACAGGTTCTCTAGAGTTTGTAACGATGC |
| Nucleotide sequence of pYGK-lacZ | |
| ATE9R | CGGAAACCAGGCAAAGCGCCATTC |
| ATE21R | CCCCTTGTATTACTGTTTATGTAAGC |
| ATE22F | GATTGGTGGCGACGACTCCTGGA |
| ATE30R | CGAAAATCGCACCTGCTTCC |
| ATE31F | GCCTGTTTTTTGTCGGTTGG |
| ATE34F | GGCGGTAGGTTTTTAGTGTTGG |
| ATE35R | CCTATTCAATGTATTGGGGATC |
| Mapping oriT region | |
| ATE-38F | GTCGACCTGCAGGGGACTAGTGGTACCGGGCCCGGAAAGCC |
| ATE-39R | CGCTGAGGTCTGCTAGCCTCGTGAAGAAGG |
| ATE-40F | GGTTGAAACTAGTAAGCCTATTATCTAGATTTG |
| ATE-41R | CTTGACCCCCGACCGCTAGCTTTCAGCGGTC |
| ATE-107F | GAGTCGACCTGCAGGCATGCTCTAGATGGTTGAAAC |
| ATE-108R | CCCTCTGACTTGGGGATCCTTGATGTTCATGCCGCC |
| ATE-115 | CAGGCGGCATGAACATCAAGTCGACGGGGGGGGGGGAAAGCCACGTTGTG |
| ATE-116 | CACAACGTGGCTTTCCCCCCCCCCCGTCGACTTGATGTTCATGCCGCCTG |
| ATE-117 | CTTCACGAGGTACCAGAGGGCCCTCAGCGGATCCCCCCCCCCCTCTAGATGGTTGAAACTAAA |
| ATE-118 | TTTAGTTTCAACCATCTAGAGGGGGGGGGGGATCCGCTGAGGGCCCTCTGGTACCTCGTGAAG |
| ATE-177F | GCCCTTTTGC AATCTAGAGGTCAAAA AGGAAACTCG |
| ATE-178R | CCAATGCTTTAGGTCGACAAAGAAAAAGGGATCG |
| Kanamycin marker | |
| ATE-36F | GATACTAGTTTTCTAGAGGTGAGCCTG |
| ATE-37R | GCAAAGGCTAGCTAGTAATGCAAGAGATTGCG |
| ATE-105F | TAACGTCGTGATTCGACGGATCCGGGGGG |
| ATE-106R | GCAAAATTGATTCGACTCTAGAGGGGGGG |
| ATE-179F | TAACGTCGTGATTCGACGTCGACGGGGGG |
2.3 Nucleotide sequence and analysis of the pYGK-lacZ shuttle vector
The initial plasmid sequence was obtained by using primers designed based on the lacZ and aph(3′)Ia genes contained in pYGK-lacZ. Subsequently, the sequencing of the whole plasmid was performed using new primers designed on the new sequence revealed. Sequencing was carried out at the University of Louisville Core Sequencing Facility. The sequence was submitted to GenBank with the accession numbers, JX826404 for pYGK and JX826405 for pYGK-lacZ. ORFs were predicted with the ORF finder tools (National Center for Biotechnology Information). The DNA and putative protein sequences were compared to the public sequence databases with the Blast program.
2.4 Construction of the physical map of the pYGKlacZ shuttle vector
The pYGK-lacZ is a pYG53-derived plasmid that contains the lacZ and aph(3′)Ia genes from E. coli C600 strain (Brogan et al., 1996) and pUC4K (Vieira and Messing, 1982), respectively. The pYG53 is in turn a pYG10-derived plasmid (Lalonde et al., 1989). To construct the physical map of pYGK-lacZ, purified plasmid DNA was digested with XbaI, PstI, EcoRI, HindIII KpnI BamHI, or various combinations of these enzymes. Digested DNAs were analyzed by conventional agarose gel electrophoresis in 1 % agarose gels (Bio-rad).
2.5 Construction of pJT4 and pJT6 shuttle plasmids
The construction of pJT4 was performed in several steps that involved the construction of precursor plasmids pJT4-A and pJT4-B, described below. The fragment containing the putative origin of replication, two ORFs as well as the cluster of genes involved in the mobility of pYGK-lacZ, was PCR amplified using primer set ATE-107F and ATE-108R. The 2484-bp PCR product was digested with XbaI-BamHI and ligated with the 1214 bp XbaI-BamHI-digested fragment containing the aph(3′)Ia from pYGK-lacZ. This latter fragment was amplified with the primer set ATE-105F and ATE-106R to create pJT4-A. Next, the unique BamHI site in pJT4-A plasmid was replaced with a SalI site by site-directed mutagenesis using primer set ATE-115F and ATE-116R, to create pJT4-B. Three additional unique restriction sites, KpnI, ApaI and BamHI sites, were then introduced downstream of the mobA gene in the pJT4-B by site-directed mutagenesis using primer set ATE-117F and ATE-118R, the resulting plasmid was designated pJT4. The fragment containing the putative origin of replication and the two ORFs of pYGK-lacZ, was PCR amplified using primer set ATE-177F and ATE-178R. The 820-bp PCR product was digested with XbaI-SalI and ligated with the 1214 bp XbaI-SalI-digested fragment containing the aph(3′)Ia from pYGK-lacZ. This latter fragment was amplified with the primer set ATE-179F and ATE-106R to create pJT6.
2.6 Construction of pJT3, pJT5, pATE12 and pATE65, transcriptional/translational lacZ gene reporter plasmids
Plasmid pJT3 was constructed from pYGK-lacZ by excising a 3167-bp BamHI-XbaI fragment and replacing it with a 3537-bp BamHI-XbaI fragment containing the promoterless lacZ gene and the transcriptional terminator rrnBT1 and rrrnBT2, amplified from pBAD-lacZ (Invitrogene) using the primer set ATE-10F and ATE-12R. This terminator product was also cloned into BamHI-XbaI-digested pJT4 to create pJT5. The promoter region of A. actinomycetemcomitans lsrR was then PCR amplified with the primer set lsrR255-f3 and lsrR255-r6. The 255-bp PCR product was digested with KpnI-BamHI and cloned into KpnI-BamHI-digested pYGK-lacZ and pJT5 to create pATE12 and pATE65 respectively.
2.7 Construction of pATE52, pATE60 plasmids
The promoter region and structural lsrRK genes was amplified from A. actinomycetemcomitans genomic DNA using primer sets lsrR-f109 and lsrK-r110, and the resulting 3090 bp fragment was digested with BamHI-XbaI and cloned into BamHI-XbaI-digested pJT4 (see Table 2) to create pATE52. Similarly, the promoter region and structural crp gene was amplified from A. actinomycetemcomitans genomic DNA using primer sets crp-F151 and crp-r152 and the resulting 1120-bp product was digested with KpnI-XbaI and cloned into KpnI-XbaI-digested pJT4 (see Table 2) to create pATE60.
2.8 Growth kinetics
A single colony of A. actinomycetemcomitans harboring each recombinant plasmid was independently inoculated into 10 ml of BHI media supplemented with 25 μg ml−1 Km and was grown standing for 24 hour at 37°C. The next day, the overnight culture (optical density at 600 nm [OD600nm] of 0.6) was diluted at a 1:30 ratio to inoculate 12 ml of BHI (12 ml in 15-ml conical centrifuge tubes) with 25 μg ml−1 Km and grown standing at 37°C. For the first hours of growth, an aliquot of 1.1 ml was taken from each culture until 9 h. Additional aliquots were taken from each culture at the 24 h (1 ml to read the OD600nm and 0.1 ml for the β-galactosidase activity assay. β-galactosidase activity was also determined for each aliquot as described below.
2.9 β-galactosidase assays
β-galactosidase (β-gal) activity was qualitatively assessed on BHI agar plates that were supplemented with 50 μg ml−1 5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside (X-gal). Quantitative evaluation of β-gal activity was carried out using permeabilized cells incubated with o-nitrophenyl-β-D-galactopyranoside (ONPG) substrate (Sigma) as previously described (Miller, 1972). Average values (± the standard deviations) for activity units were routinely calculated from three independent assays using GraphPad Prisma v5 software.
2.10 Determination of plasmid stability
The plasmid stability of strain A. actinomycetemcomitans harboring pATE3 or pJT4 or pJT5 or pJT6 was performed essentially as described previously by Torres-Escobar et al. (2010) except that A. actinomycetemcomitans harboring each recombinant plasmid was determined with BHI media supplemented with 25 μg ml−1 Km.
2.11 Construction of the suicide vector pJT1, for the generation of scarless and markerless deletion mutations
The construction of the pJT1 suicide vector was performed in several steps that involved the construction of precursor plasmids pJT-A, pJT-B, and pJT-C, described below. A 240 bp DNA fragment containing a multiple cloning site (MCS) flanked by EcoRI and HindIII overhanging sequences was obtained by annealing three sets of complementary single-stranded 80-b oligonucleotides, (MDJR-16F and MDJR-17R), (MDJR-18F and MDJR-19R) and (MDJR-20F and MDJR-21R). The 240 bp synthetic DNA was cloned into EcoRI-HindIII-digested pUC18 (Yanisch et al., 1985) to create pJT-A. A 87-bp DNA fragment containing the synthetic trc promoter (Ptrc) (Amann et al., 1983) with by SacI and BglII sites was obtained by annealing two complementary single-stranded 80 base oligonucleotides, (MDJR-22F and MDJR-23R). The resulting 87 bp synthetic DNA was cloned into SacI-BglII-digested pJT-A to create pJT-B. Finally, the sacB gene was PCR amplified from pRE112 (Edwards et al., 1998) using primer set MDJR-24F and MDJR-27R. The 1552 bp PCR product was digested with BglII-MluI and cloned into BglII-MluI-digested pJT-B to create pJT-C. pJT-C was then digested with NheI-MluI to produce a 1777 bp fragment containing the MCS-Ptrc -sacB region. To produce pJT-1, this fragment was simultaneously ligated with two other fragments; an 857 bp fragment containing the pUC replication origin that was PCR amplified from pUC18 using primer set MDJR-34F and MDJR-35R, and a 1191 bp fragment containing the spectinomycin resistance marker that was PCR amplified from pVT1461 (Mintz et al., 2002) using primer set MDJR-28F and MDJR-29R.
2.12 Generation of A. actinomycetemcomitans markerless deletion mutants
The construction of A. actinomycetemcomitans harboring a markerless deletion mutation of qseB gene was carried out by amplifying the upstream and downstream flanking regions of qseB with primer sets (MDJR-49F and MDJR-82R) and (MDJR-83F and MDJR-84R; Table 2). The respective 1630-bp and 1890-bp PCR products were digested with NotI-XhoI, and XhoI-PstI and cloned adjacently (joined by the XhoI restriction site) into NotI-PstI-digested pJT1 suicide vector to create pDJR37 (Table 1). A similar approach was used to generate the A. actinomycetemcomitans markerless double-deletion mutant in lsrRK by constructing and using the suicide plasmid pATE47 (Table 1). Primer sets used to amplify the flanking regions of lsrRK were (ATE-86F and ATE-87R) and (ATE-88F and ATE-89R) (Table 2). Each recombinant suicide plasmid (~20 μg) was introduced individually into A. actinomycetemcomitans by electroporation. Electroporated cells were incubated in 0.5 ml of SOC broth standing for 5 h at 37°C in microaerophilic conditions (anaerobic jar). Bacterial cells with single recombinant event were selected onto BHI agar containing 50 μg ml−1 spectinomycin (Sp) at 37°C under microaerophilic conditions. Ten spectinomycin-resistant (Spr) colonies were randomly selected and subcultured daily for 24 h at 37°C for three consecutive days, except that the final cultures were grown in presence of 1mM IPTG (IPTG-induced Ptrc-sacB). To select for bacteria that had undergone a second recombination event, the culture was diluted 10-fold and spread onto TYE agar supplemented with 1 mM IPTG and 10% sucrose, and grown at 37°C in microaerophilic conditions. One thousand sucrose-resistant (Sucr) colonies were replica plated onto TYE agar supplemented with sucrose and onto BHI agar supplemented with spectinomycin. Spectinomycin-sensitive (SpS) colonies were selected to perform PCR for the deletion mutation of the target genes, using the primer sets for qseB deletion (MDJR-63F and MDJR61R) or for lsrRK deletion (ATE-62F and ATE-102R). SpS colonies that were PCR positive were selected for further analysis.
3. Results
3.1 Nucleotide sequence of the shuttle vector pYGKlacZ
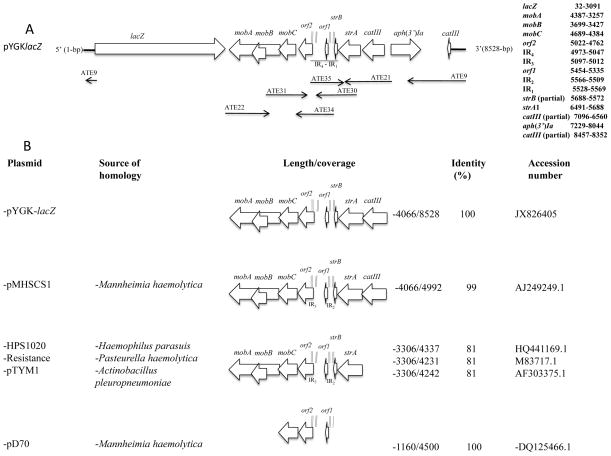
The complete nucleotide sequence of pYGK was determined as described in Materials and Methods and a map of the plasmid containing the lacZ gene of E. coli C600 is shown in Fig. 1A. Through comparative analysis with the nucleotide/protein sequences in the GenBank database, four regions were identified on plasmid. As shown in Fig. 1B, pYGK contains several open reading frames (orfs) that are conserved in other organisms of the Pasteurellaceae and Enterobacteriaceae (Kehrenberg and Schwarz, 2002; Matter et al., 2008; Kang et al., 2009). The first region harbors genes encoding antibiotic resistance markers such as catIII, which encodes chloramphenicol acetyltransferase. This gene is truncated in pYGK as a result of the previous cloning by Brogan et al. (1996) of a 1252-bp DNA fragment harboring the aph(3′)-Ia gene (encoding aminoglycoside phosphotransferase [Km]) from pUC4K. This region of pYGK also contains strA, encoding aminoglycoside 3′-phosphotranferase [APH0(3″)-Ib], and a truncated strB gene that encodes a portion of aminoglycoside 6′-phosphotranferase (APH0(6)-Id). The second region comprises three ORFs encoding proteins involved in plasmid mobilization (MobA, MobB and MobC) that share high sequence similarity to related genes identified in other organisms in the family Pasteurellaceae (Matter et al., 2008). The mobC and mobA genes overlap by four bases. In addition, mobB resides completely within mobA, but is translated in a different reading frame. The third region revealed two ORFs (orf1 and orf2) encoding proteins with no significant homologies to amino acid sequences available in the Gene-Bank database. The fourth region contained a potential origin of replication that shares 100% identity to the 1160 bp region containing the origin of replication of plasmid pD70 from Mannheimia hemolytica serotype 1 (Briggs and Tatum, 2005). In addition, two putative oriT sequences, four inverted repeat (IR1-4) sequences, and 2 direct repeat (DR) sequences that reside within the IR1 (Briggs and Tatum, 2005) were identified (see Table 3). The entire nucleotide sequence of pYGK is 5469 bp with position 1 defined as the guanine residue that corresponds to the first nucleotide of the unique restriction site EcoRI. The length of pYGK containing lacZ as shown in Figure 1A is 8528 bp.
Figure 1.
Schematic representation of plasmid pYGK-lacZ and its comparative analysis. A) The transcriptional orientation of open reading frames in pYGKlacZ is indicated by open arrows. Thin arrows below the gene name show the sequence revealed and localization of the primers. B) A BLAST search using a 4066-bp DNA fragment from pYGK-lacZ identified related plasmid sequences in Mannheimia haemolytica, Haemophilus parasuis, Pasteurella haemolytica and Actinobacillus pleuropneumoniae. The orientation of gene transcription is indicated by open arrows and percent nucleotide identity is indicated on the right. The inverted repeat sequences (IR) are indicated with double smaller vertical lines.
Table 3.
Invert, direct and putative oriT sequences of pYGK.
| Invert repeat 1 | 5′ TCTTTTTCCCTAGCTT-N9-AAGCTAGGGAAAAAGA 3′ 3′ AGAAAAAGGGATCGAA-N9-TTCGATCCCTTTTTCT 5′ |
| Invert repeat 2 | 5′ GAAACGATTCTAT-N18-ATAGAATCGTTTC 5′ 3′ CTTTGCTAAGATA-N18-TATCTTAGCAAAG 3′ |
| Invert repeat 3 | 5′ TTTTT-N6-AAAAA 3′ 3′ AAAAA-N6-TTTTT 3′ |
| Imperfect Invert repeat 4 | 5′ CACAACCTTTCGACAACCTTTCGAGCACCCATTGAC-N3-CTCAATAGGTAGTCGAAAAGTTACCGAAAGGTTATG 3′ 3′ GTGTTGGAAAGCTGTTGGAAAGCTCGTGGGTAACTG-N3-GAGTTATCCATCAGCTTTTCAATGGCTTTCCAATAC 5′ |
| Direct repeat 1 | 5′ CGAAA-N6-CGAAA 3′ 3′ GCTTT-N6-GCTTT 3′ |
| Direct repeat 2 | 5′ ACAACCTTTCGACAACCTTTCGA 3′ 3′ TGTTGGAAAGCTGTTGGAAAGCT 5′ |
| oriT 1 | 5′ CCGAAAG 3′ 3′ GGCTTTC 5′ |
| oriT 2 | 5′ CCGAAAG 3′ 3′ GGCTTTC 5′ |
3.2 Mapping the replication origin of pYGK
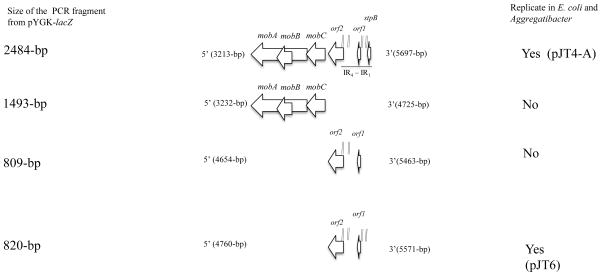
A series of four fragments of 2484 bp, 1493 bp, 820 and 809 bp (see Fig 2) flanked with appropriate restriction enzymes sites were amplified (as described in Material and Methods) from pYGK and each was individually ligated with a 1252 bp fragment (flanked with restriction sites) containing a kanamycin resistance marker. Each ligation was introduced into E. coli XLI-blue MRF′ strain by electroporation and the cells were spread onto LB agar supplemented with 20 μg ml−1 kanamycin and incubated at 37 °C. Kmr colonies were detected in transformants harboring the 2484 bp or 820 bp fragments. This recombinant plasmid, designated pJT4-A and pJT6, respectively, also replicated in A. actinomycetemcomitans 652. No recombinants were recovered from ligations using the 1493 bp or 809 bp fragments, indicating that the 820-bp (containing the two ORF plus the four IR sequences) that resides into the 2484-bp fragment is enough for supporting the replication of this plasmid in both E. coli and A. actinomycetemcomitans cells and that the mob genes region is unnecessary for pYGK replication.
Figure 2.
Mapping the pYGK origin of replication. A) Schematic representation of DNA fragments harboring different regions of the pYGK origin of replication and mapping of the minimal region required for plasmid replication in A. actinomycetemcomitans and E. coli.
3.3 Improvement of the shuttle plasmid pYGK-lacZ
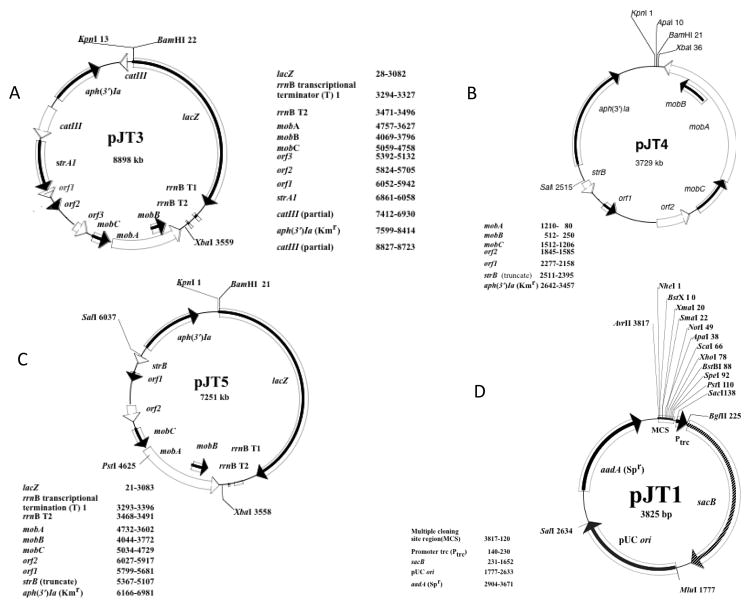
pYGK-lacZ is a transcriptional reporter plasmid that has been successfully used to characterize the leukotoxin promoter of A. actinomycetemcomitans (Brogan et al., 1996). To improve this vector and to minimize potential toxic effects arising from run-on transcription from strong promoters fused to lacZ, the lacZ gene from E. coli C600 was replaced with the promoterless lacZ gene from pBAD-lacZ that includes the strong transcriptional terminator sequences T1 and T2 derived from the rrnB gene (Orosz et al., 1991). The resulting plasmid was named pJT3 (Fig. 3A). The utility of the pJT3 transcriptional/translational reporter plasmid was recently validated through the determination of the minimal regulatory region of the A. actinomycetemcomitans lsrCADBFG and lsrRK operons (Torres-Escobar et al., 2013) as well as ygiW-qseBC operon (Juarez-Rodriguez et al., unpublished results). Since pJT4-A contains a replication origin that is required for maintenance in both E. coli and A. actinomycetemcomitans, further modifications of this plasmid were performed through deletion/insertion of new restriction sites (see Material and Methods) with the aim to conserve the same unique restriction sites KpnI and BamHI previously mapped on pYGK-lacZ. The resulting new cloning vector was designated pJT4 (Fig. 3B) and this plasmid lacks all sequences of pYGK that are not required for replication or selection. To validate its utility as shuttle cloning vector, the A. actinomycetemcomitans lsrRK operon and crp gene were cloned individually into pJT4 (see Material and Methods) and the resulting recombinant plasmids pATE52 and pATE60, respectively, were selected for in E. coli and analyzed by restriction digestion. Subsequently, these plasmids were introduced by electroporation into A. actinomycetemcomitans 652, re-isolated from recombinant cells and shown to be identical to the input plasmid by restriction analysis (data not shown). These results confirm that the pJT4-derivative plasmids replicate and are maintained in both E. coli and A. actinomycetemcomitans cells. Finally, the lacZ gene from pBAD-lacZ was introduced into pJT4 resulting in the transcriptional/translational reporter plasmid pJT5 (Fig. 3C). To demonstrate the utility of pJT5, the 255 bp A. actinomycetemcomitans lsrR promoter was cloned into the plasmid to generate pATE65. pATE65 was introduced by electroporation into A. actinomycetemcomitans 652 and β-galactosidase activity produced by this strain was compared to a similar strain harboring pATE12 (pYGK-lacZ fused to the lsrR promoter) and pATE23 (pJT3, lsrR promoter-lacZ) (Torres-Escobar et al., 2013). The lsrR promoter expressed similar levels of activity (see Table 4), indicating that pJT5 functions as a transcriptional/translational reporter plasmid for studies of gene expression in A. actinomycetemcomitans. Plasmid pJT3, pJT4, pJT5 and pJT6 were stably maintained for 50 or more generations in A. actinomycetemcomitans under selective conditions and A. actinomycetemcomitans transformant cells [OD600nm of 0.6 (1.15×109 colony forming, CFU, units ml−1)] produced the same amount of each recombinant plasmid (~0.75 μg ml−1) as pYGK (not shown). pJT4 and pJT5 are both almost 2 kbp smaller than the pYGK plasmids from which they were derived.
Figure 3.
Physical maps of transcriptional/translation reporter plasmid pJT3 (A), the cloning vector pJT4 (B), the transcriptional/translation reporter plasmid pJT5 (C) and the suicide vector pJT1 (D).
Table 4.
| Strain/plasmid | Derived from | Miller units |
|---|---|---|
| Aa 652 | 0 | |
| Aa 652/pYGK-lacZ | 0 | |
| Aa 652/pATE12(PlsrR255-lacZ) | pYGK-lacZ | 916±31 |
| Aa 652/pJT3 | 0 | |
| Aa 652/pATE23(PlsrR255-lacZ) | pJT3 | 930±36 |
| Aa 652/pJT5 | 0 | |
| Aa 652/pATE65(PlsrR255-lacZ) | pJT5 | 928±28 |
3.4 pJT1, a scarless-markerless suicide plasmid
The suicide plasmid pJT1 (Fig. 3D) carries the pUC origin of replication and does not replicate in A. actinomycetemcomitans. In addition, pJT1 is a sucrose-sensitive suicide vector containing the Bacillus subtilis sacB gene encoding levane saccharase. This gene is lethal in most Gram-negative bacteria in the presence of sucrose (Gay et al., 1983) and thus provides strong selection for isolation of organisms that have undergone both recombination events required for gene deletion mutagenesis. The expression of sacB is under the control of the trc promoter (Ptrc), and thus is inducible in the presence of isopropyl-β-D-thiogalactopyranoside (IPTG). In its present form, pJT1 also contains a spectinomycin resistance gene and a multiple cloning site (MCS) with thirteen unique restriction endonuclease sites for cloning the DNA fragments flanking the target gene to be deleted or inactivated. However, as described in the Materials and Methods, pJT1 was constructed from individual fragments containing a multiple cloning site (MCS), trc promoter and sacB; the pUC origin of replication; and an antibiotic resistance marker. The use of these separate functional cassettes readily facilitates additional modifications to develop new vectors with different selectable markers, origins or reporter genes.
3.5 Construction of the A. actinomycetemcomitans deletion mutants
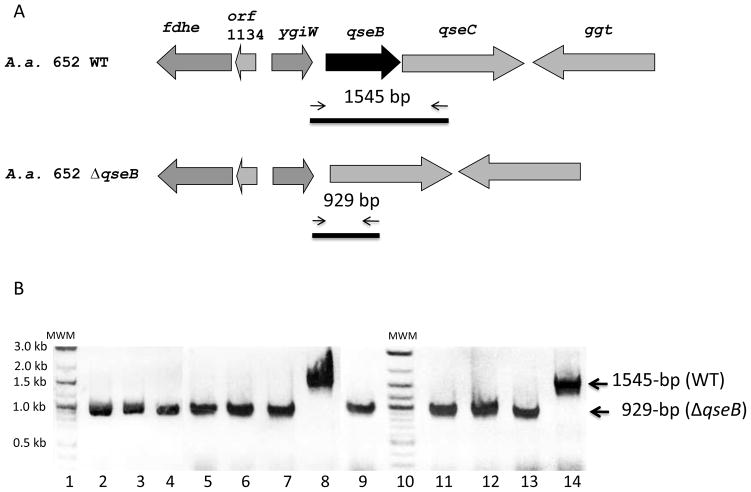
To demonstrate the utility of pJT1, we generated markerless deletion mutants of the lsrRK operon and the qseB gene of A. actinomycetemcomitans. DNA fragments flanking qseB or lsrRK were cloned individually into pJT1 (as stated in Materials and Methods) to create the suicide plasmids pDJR37 and pATE47, respectively. Twenty micrograms of each plasmid were introduced into A. actinomycetemcomitans 652 cells by electroporation. Transformed bacterial cells with the suicide plasmid integrated in the chromosome by a single homologous recombination event were selected on medium containing 50 μg ml−1 spectinomycin. The transformation efficiencies were about 10 CFU μg−1 DNA using 1×1010 electroporated cells. Ten Spr colonies were randomly chosen and sub-cultured daily for three consecutive days. To identify gene deletion mutants that had undergone a second event of recombination, cells were diluted and spread on TYE agar with 1mM IPTG and 10% sucrose. Approximately 1,000 Sucr colonies were replica plated onto agar medium containing sucrose in the presence and absence of spectimomycin. Twenty one and twenty SucR-SpS colonies were obtained with pDJR37 and pATE47, respectively. About 85 % of SucR-SpS colonies contained the expected modified allele, as shown in Figure 4B. Deletion of qseB was confirmed in ten independent SucR-SpS colonies by the generation of a 929 bp amplicon. A similar PCR approach was used to confirm deletion of lsrRK (data not shown). Subsequently, markerless deletion mutagenesis using pJT1 was used successfully to generate deletion mutants lacking qseC, qseBC, and ygiW (Juarez-Rodrigues et al., unpublished results) and lsrR, lsrK, and crp (Torres-Escobar et al., 2013). The transformation efficiency of each of these suicide plasmids was similar to that reported above. In addition, the yield of clones that contained the expected modified allele was similar to that obtained in the experiment described above.
Figure 4.
Deletion of the qseB gene in A. actinomycetemcomitans 652. (A). Schematic representation of the ygiW-qseB-qseC locus. The transcriptional orientation of genes is indicated by open arrows. Thin arrows represent primers used for PCR confirmation of the deletion of qseB. (B). PCR products obtained with the A. actinomycetemcomitans ΔqseB mutant strains. Deletion of the qseB gene was confirmed by PCR amplification of a 929-bp fragment with primer set MDJR-63F and MDJR-61R in ten randomly selected SucR and SpS colonies obtained with pDJR37 suicide vector as described in Material and Methods. The amplification profile used was: 94°C for 2 min for 1 cycle and then 94°C for 30 s, 58°C for 30 s, and 68°C for 2 min for 35 cycles. MWM, molecular weight marker.
4. Discussion
To continue to make progress in understanding the genetic and molecular basis of A. actinomycetemcomitans pathogenicity and virulence, new molecular tools and genetic techniques, and/or improvements on the currently available vectors are needed to manipulate the A. actinomycetemcomitans genome. In this study, we focused on the construction of vectors that will facilitate gene cloning, gene deletion and the analysis of gene expression in A. actinomycetemcomitans. To accomplish this, the complete sequence of the A. actinomycetemcomitans-E. coli shuttle vector pYGK was determined in order to further characterize the plasmid and improve its stability by eliminating sequences that are non-essential for its replication. Analysis of the nucleotide sequence of pYGK revealed that the original parent plasmid pYG10, isolated from A. pleuropneumoniaes 80-8141 obtained from a clinical case of swine pleuropneumoniae (Lalonde et al., 1989), is a mobile plasmid that mediates antibiotic resistance. The nucleotide sequence also allowed us to identify elements that may represent the origin of replication required for maintenance of the plasmid in both A. actinomycetemcomitans and E. coli. Indeed, the overall structure of the pYGK repeat region was similar to the 720 bp minimal replication unit identified by Briggs and Tatum, (2005) in plasmid pD70 from Mannheimia haemolytica. Similar 720 bp origins also exist in plasmid pIG1 from Pasteurella multocida (Gene Bank accession No. U57657.1). Briggs and Tatum, (2005) determined that pD70-3′Δ400oriKmr can be replicated in the Pasteurella multocida NADC TT94 strain, but they did not determine if the 720 bp origin of pD70 was functional in E. coli. Our results indicate that a 809 bp fragment containing the repeat region of pYGK (but excluding IR1) was insufficient to support replication in either A. actinomycetemcomitans or E. coli. Similarly, a 1493 bp fragment harboring only the mobilization genes was unable to support replication, whereas a 820 bp fragment containing the two ORF’s plus the four IR sequences as well as a 2484 bp fragment containing both the cluster of mobilization genes and inverted repeat sequences allowed replication. Therefore, this fragment was subsequently used in the construction of cloning and lacZ reporter vectors. These new vectors, pJT4 and pJT5, contain only the sequences required for plasmid replication and selection and eliminate over 1,600 bp of pYGK that are not necessary for these essential functions. This will facilitate the cloning of larger inserts and may increase the stability and transformation efficiency of these constructs.
We were also interested in developing news tools to facilitate gene replacement or gene deletion mutagenesis approaches in A. actinomycetemcomitans. The pJT1 suicide vector was designed for the generation of chromosomal non-polar, scarless and markerless gene deletions or gene modifications in A. actinomycetemcomitans. pJT1 carries strong selectable markers including a spectinomycin-resistance marker that is efficiently expressed in A. actinomycetemcomitans to select for plasmid insertion into the target locus and the sacB gene under the control of the Ptrc promoter, which provides a strong counter-selectable marker that facilitates the positive selection of mutants that have undergone the replacement of the wild type allele by the mutated allele, and have lost the integrated plasmid. Using an approach that has been used successfully in Salmonella to generate markerless deletion mutations (Kang et al., 2002) we have used pJT1 to generate several deletion mutants and in frame deletions that target the removal of specific functional domains of the protein encoded by the target gene. The mutant strains that were obtained are free of antibiotic markers and scars and thus can readily be subjected to additional rounds of mutagenesis without encountering the limitations of current approaches for generating multiple mutations in A. actinomycetemcomitans. Furthermore, pJT1 deletion mutagenesis was carried out using a serotype c strain which is not naturally competent, suggesting that the pJT1 vector will be useful for mutagenesis in both naturally and non-naturally competent strains of A. actinomycetemcomitans.
In summary, characterizing the A. actinomycetemcomitans-E. coli shuttle vector pYGK has allowed us to design new cloning, lacZ reporter and suicide vectors that should facilitate genetic studies and mutagenesis approaches in A. actinomycetemcomitans. Given the similarity of the pYGK origin of replication with other plasmids from Mannheimia and Haemophilus species, these vectors may be more generally applicable for genetic studies in other HACEK organisms as well.
Highlights.
The nucleotide sequence of the shuttle vector pYGK facilitated contruction of new pYGK-derived plasmids.
The minimal origin of replication of the shuttle vector pYGK was identified.
The mob genes region is unnecessary for pYGK replication.
A new scarless-markerless suicide plasmid, pJT1 was constructed.
The effectiveness of pJT1 was demonstrated by the construction of ΔqseB and ΔlsrRK mutants.
Acknowledgments
This research was supported by the Public Health Service grant RO1DE14605 from the NIDCR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Amann E, Brosius J, Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983;25:167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharjee MK, Fine DH, Figurski DH. tfoX (sxy)-dependent transformation of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Gene. 2007;399:53–64. doi: 10.1016/j.gene.2007.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briggs RE, Tatum FM. Generation and molecular characterization of new temperature-sensitive plasmids intended for genetic engineering of Pasteurellaceae. Appl Environ Microbiol. 2005;71:7187–95. doi: 10.1128/AEM.71.11.7187-7195.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brogan JM, Lally ET, Demuth DR. Construction of pYGK, an Actinobacillus actinomycetemcomitans-Escherichia coli shuttle vector. Gene. 1996;169:141–2. doi: 10.1016/0378-1119(95)00792-x. [DOI] [PubMed] [Google Scholar]
- 5.Demuth DR, Novak EA, Shao H. Alternative autoinducer-2 quorum sensing response circuits; impact on microbial community development. In: Kolenbrander Paul E., editor. Oral Microbial Communities: Genomic Inquiry and Interspecies Communication. ASM Press; 2011. pp. 263–282. [Google Scholar]
- 6.Edwards RA, Keller LH, Schifferli DM. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene. 1998;207:149–157. doi: 10.1016/s0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 7.Fujise O, Lakio L, Wang Y, Asikainen S, Chen C. Clonal distribution of natural competence in Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 2004;19:340–2. doi: 10.1111/j.1399-302x.2004.00157.x. [DOI] [PubMed] [Google Scholar]
- 8.Fujise O, Wang Y, Chen W, Chen C. Adherence of Aggregatibacter actinomycetemcomitans via serotype-specific polysaccharide antigens in lipopolysaccharides. Oral Microbiol Immunol. 2008;23:226–33. doi: 10.1111/j.1399-302X.2007.00416.x. [DOI] [PubMed] [Google Scholar]
- 9.Gay P, Le Coq D, Steinmetz M, Ferrari E, Hoch JA. Cloning structural gene sacB, which codes for exoenzyme levansucrase of Bacillus subtilis; expression of the gene in Escherichia coli. J Bacteriol. 1983;153:1424–1431. doi: 10.1128/jb.153.3.1424-1431.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haffajee AD, Socransky SS. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78–111. doi: 10.1111/j.1600-0757.1994.tb00020.x. [DOI] [PubMed] [Google Scholar]
- 11.Han X, Kawai T, Eastcott JW, Taubman MA. Bacterial-responsive B lymphocytes induce periodontal bone resorption. J Immunol. 2006;176:625–31. doi: 10.4049/jimmunol.176.1.625. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 13.Henderson B, Nair SP, Ward JM, Wilson M. Molecular pathogenicity of the oral opportunistic pathogen Actinobacillus actinomycetemcomitans. Annu Rev Microbiol. 2003;57:29–55. doi: 10.1146/annurev.micro.57.030502.090908. [DOI] [PubMed] [Google Scholar]
- 14.Hyvärinen K, Mäntylä P, Buhlin K, Paju S, Nieminen MS, Sinisalo J, Pussinen PJ. A common periodontal pathogen has an adverse association with both acute and stable coronary artery disease. Atherosclerosis. 2012;223:478–484. doi: 10.1016/j.atherosclerosis.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 15.James D, Shao H, Lamont RJ, Demuth DR. The Actinobacillus actinomycetemcomitans ribose binding protein RbsB interacts with cognate and heterologous autoinducer 2 signals. Infect Immun. 2006;74:4021–9. doi: 10.1128/IAI.01741-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang HY, Dozois CM, Tinge SA, Lee TH, Curtiss R., 3rd Transduction-mediated transfer of unmarked deletion and point mutations through use of counterselectable suicide vectors. J Bacteriol. 2002;184:307–12. doi: 10.1128/JB.184.1.307-312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang M, Zhou R, Liu L, Langford PR, Chen H. Analysis of an Actinobacillus pleuropneumoniae multi-resistance plasmid, pHB0503. Plasmid. 2009;61:135–139. doi: 10.1016/j.plasmid.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Kachlany SC, Planet PJ, Bhattacharjee MK, Kollia E, DeSalle R, Fine DH, Figurski DH. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J Bacteriol. 2000;182:6169–6176. doi: 10.1128/jb.182.21.6169-6176.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kachlany SC, Planet PJ, DeSalle R, Fine DH, Figurski DH. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. 2001a;9:429–437. doi: 10.1016/s0966-842x(01)02161-8. [DOI] [PubMed] [Google Scholar]
- 20.Kachlany SC, Planet PJ, Desalle R, Fine DH, Figurski DH, Kaplan JB. flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol Microbiol. 2001b;40:542–554. doi: 10.1046/j.1365-2958.2001.02422.x. [DOI] [PubMed] [Google Scholar]
- 21.Kehrenberg C, Schwarz S. Nucleotide sequence and organization of plasmid pMVSCS1 from Mannheimia varigena: identification of a multiresistance gene cluster. J Antimicrob Chemother. 2002;49:383–6. doi: 10.1093/jac/49.2.383. [DOI] [PubMed] [Google Scholar]
- 22.Kittichotirat W, Bumgarner RE, Asikainen S, Chen C. Identification of the pangenome and its components in 14 distinct Aggregatibacter actinomycetemcomitans strains by comparative genomic analysis. PLoS One. 2011;6:e22420. doi: 10.1371/journal.pone.0022420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolodrubetz D, Phillips L, Burgum A. Repression of aerobic leukotoxin transcription by integration host factor in Aggregatibacter actinomycetemcomitans. Res Microbiol. 2010;161:541–548. doi: 10.1016/j.resmic.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lalonde G, Miller JF, Tompkins LS, O’Hanley P. Transformation of Actinobacillus pleuropneumoniae and analysis of R factors by electroporation. Am J Vet Res. 1989;50:1957–60. [PubMed] [Google Scholar]
- 25.Lee G, Saito I. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene. 1998;216:55–65. doi: 10.1016/s0378-1119(98)00325-4. [DOI] [PubMed] [Google Scholar]
- 26.Matter D, Rossano A, Sieber S, Perreten V. Small multidrug resistance plasmids in Actinobacillus porcitonsillarum. Plasmid. 2008;59:144–52. doi: 10.1016/j.plasmid.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Mena J, Chen C. Identification of strain-specific DNA of Actinobacillus actinomycetemcomitans by representational difference analysis. Oral Microbiol Immunol. 2007;22:429–32. doi: 10.1111/j.1399-302X.2007.00371.x. [DOI] [PubMed] [Google Scholar]
- 28.Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 29.Mintz KP, Brissette C, Fives-Taylor PM. A recombinase A-deficient strain of Actinobacillus actinomycetemcomitans constructed by insertional mutagenesis using a mobilizable plasmid. FEMS Microbiol Lett. 2002;206:87–92. doi: 10.1111/j.1574-6968.2002.tb10991.x. [DOI] [PubMed] [Google Scholar]
- 30.Nørskov-Lauritsen N, Kilian M. Reclassification of Actinobacillus actinomycetemcomitans, Haemophilus aphrophilus, Haemophilus paraphrophilus and Haemophilus segnis as Aggregatibacter actinomycetemcomitans gen.nov., comb. nov., Aggregatibacter aphrophilus comb. nov. and Aggregatibacter segnis comb. nov., and emended description of Aggregatibacter aphrophilus to include V factor-dependent and V factor-independent isolates. Int J Syst Evol Microbiol. 2006;56:2135–46. doi: 10.1099/ijs.0.64207-0. [DOI] [PubMed] [Google Scholar]
- 31.Novak EA, Shao H, Daep CA, Demuth DR. Autoinducer-2 and QseC control biofilm formation and in vivo virulence of Aggregatibacter actinomycetemcomitans. Infect Immun. 2010;78:2919–26. doi: 10.1128/IAI.01376-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orosz A, Boros I, Venetianer P. Analysis of the complex transcription termination region of the Escherichia coli rrnB gene. Eur J Biochem. 1991;201:653–9. doi: 10.1111/j.1432-1033.1991.tb16326.x. [DOI] [PubMed] [Google Scholar]
- 33.Paturel L, Casalta JP, Habib G, Nezri M, Raoult D. Actinobacillus actinomycetemcomitans endocarditis. Clin Microbiol Infect. 2004;10:98–118. doi: 10.1111/j.1469-0691.2004.00794.x. [DOI] [PubMed] [Google Scholar]
- 34.Planet PJ, Kachlany SC, Fine DH, DeSalle R, Figurski DH. The widespread colonization island of Actinobacillus actinomycetemcomitans. Nat Genet. 2003;34:193–198. doi: 10.1038/ng1154. [DOI] [PubMed] [Google Scholar]
- 35.Rahamat-Langendoen JC, van Vonderen MG, Engström LJ, Manson WL, van Winkelhoff AJ, Mooi-Kokenberg EA. Brain abscess associated with Aggregatibacter actinomycetemcomitans: case report and review of literature. J Clin Periodontol. 2011;38:702–6. doi: 10.1111/j.1600-051X.2011.01737.x. [DOI] [PubMed] [Google Scholar]
- 36.Reyrat JM, Pelicic V, Gicquel B, Rappuoli R. Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infect Immun. 1998;66:4011–4017. doi: 10.1128/iai.66.9.4011-4017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers JE, Li F, Coatney DD, Rossa C, Bronson P, Krieder JM, Giannobile WV, Kirkwood KL. Actinobacillus actinomycetemcomitans lipopolysaccharide-mediated experimental bone loss model for aggressive periodontitis. J Periodontol. 2007;78:550–8. doi: 10.1902/jop.2007.060321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 39.Schaeffer LM, Schmidt ML, Demuth DR. Induction of Aggregatibacter actinomycetemcomitans leukotoxin expression by IS1301 and orfA. Microbiology. 2008;154:528–38. doi: 10.1099/mic.0.2007/012195-0. [DOI] [PubMed] [Google Scholar]
- 40.Shao H, James D, Lamont RJ, Demuth DR. Differential interaction of Aggregatibacter (Actinobacillus) actinomycetemcomitans LsrB and RbsB Proteins with Autoinducer 2. J Bacteriol. 2007a;189:5559–5565. doi: 10.1128/JB.00387-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shao H, Lamont RJ, Demuth DR. Autoinducer 2 is required for biofilm growth of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Infect Immun. 2007b;75:4211–4218. doi: 10.1128/IAI.00402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomson VJ, Bhattacharjee MK, Fine DH, Derbyshire KM, Figurski DH. Direct selection of IS903 transposon insertions by use of a broad-host-range vector: isolation of catalase-deficient mutants of Actinobacillus actinomycetemcomitans. J Bacteriol. 1999;181:7298–307. doi: 10.1128/jb.181.23.7298-7307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torres-Escobar A, Juárez-Rodríguez MD, Gunn BM, Branger CG, Tinge SA, Curtiss R., 3rd Fine-tuning synthesis of Yersinia pestis LcrV from runaway-like replication balanced-lethal plasmid in a Salmonella enterica serovar Typhimurium vaccine induces protection against a lethal Y. pestis challenge in mice. Infect Immun. 2010;78:2529–43. doi: 10.1128/IAI.00005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torres-Escobar A, Juárez-Rodríguez MD, Lamont RJ, Demuth DR. Transcriptional Regulation of Aggregatibacter actinomycetemcomitans lsrACDBFG and lsrRK operons and their role in biofilm formation. J Bacteriol. 2013;195:56–65. doi: 10.1128/JB.01476-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–68. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 46.Wang CY, Wang HC, Li JM, Wang JY, Yang KC, Ho YK, Lin PY, Lee LN, Yu CJ, Yang PC, Hsueh PR. Invasive infections of Aggregatibacter (Actinobacillus) actinomycetemcomitans. J Microbiol Immunol Infect. 2010;43:491–7. doi: 10.1016/S1684-1182(10)60076-X. [DOI] [PubMed] [Google Scholar]
- 47.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]