Abstract
Ameloblasts, the cells responsible for making enamel, modify their morphological features in response to specialized functions necessary for synchronized ameloblast differentiation and enamel formation. Secretory and maturation ameloblasts are characterized by the expression of stage-specific genes which follows strictly controlled repetitive patterns. Circadian rhythms are recognized as key regulators of development and diseases of many tissues including bone. Our aim was to gain novel insights on the role of clock genes in enamel formation and to explore the potential links between circadian rhythms and amelogenesis. Our data shows definitive evidence that the main clock genes (Bmal1, Clock, Per1 and Per2) oscillate in ameloblasts at regular circadian (24h) intervals both at RNA and protein levels. This study also reveals that two markers of ameloblast differentiation i.e. amelogenin (Amelx; a marker of secretory ameloblasts) and kallikrein-related peptidase 4 (Klk4, a marker of maturation ameloblasts) are downstream targets of clock genes. Both, Amelx and Klk4 show 24h oscillatory expression patterns and their expression levels are up-regulated after Bmal1 over-expression in HAT-7 ameloblast cells. Taken together, these data suggest that both the secretory and the maturation stage of amelogenesis might be under circadian control. Changes in clock genes expression patterns might result in significant alterations of enamel apposition and mineralization.
Keywords: Clock genes, enamel, amelogenin, kallikrein-related peptidase 4, odontoblasts
Introduction
Circadian rhythms are self-sustained endogenous oscillations occurring over a 24 hours (hr) period. These biological rhythms are involved in many physiological processes. Though there is a site in the suprachiasmatic nucleus of the brain that is considered the “master clock”, “peripheral clocks” have been found in several tissues in the body. Peripherals clocks may respond to the environmental light-dark cycles of an organism but can persist oscillating even after the light-dark stimulus is removed. The relationship between the “master clock” and the “peripheral clocks” has not been made clear [1, 2].
Several genes, named “clock genes” have been identified as core maintainers of circadian rhythms. The main mammalian clock genes include Circadian Locomotor Output Cycles Kaput (Clock), Brain and Muscle Aryl Hydrocarbon Receptor Nuclear Translocation (ARNT)-like (Bmal1), Period 1 (Per1), Period 2 (Per2), Period 3 (Per3), and Cryptochromes (Cry1) and Cry2. Additionally, nuclear receptor subfamily 1, group D, member 1 and 2 (Nr1d1 and Nr1d2), RAR-Related Orphan Receptor Alpha (Rora), and Albumin D-binding protein (Dbp) also play a key role in circadian rhythms biology by regulating the expression of the main mammalian clock genes [3]. Transcription of the clock genes oscillates over an approximate 24h period and their output signals induce rhythms of gene expression that create repetitive patterns in physiological processes. This method of circadian control involves binding of clock genes to the promoter region of clock-controlled genes (CCG) [4]. Binding to the promoter of target genes (CCG) occurs via three specific DNA sequences called E-, D-, RRE- box [3]. In a more indirect way, clock genes may also bind to intermediate clock-controlled genes, such as key transcription factors, which then influence expression of downstream target genes involved in cell proliferation or cell differentiation processes [1].
Clock genes regulate circadian rhythms and metabolic functions in mammals but have also been implicated in mineralized tissue development, including bone formation [5]. More recent studies have indicated that Cry2 and Per2 affect distinct pathways in the regulation of bone volume. More specifically, Cry2 influences mostly the osteoclastic cellular component of bone and Per2 acts on osteoblast’s parameters [6].
Similar to bone, teeth are formed incrementally and previous studies suggest the existence of molecular clocks in enamel forming ameloblasts [7, 8] and in dentin forming odontoblasts [9]. Indeed, several lines of evidence support the idea that ameloblasts and odontoblasts are under circadian control. First, there are daily growth patterns found in enamel and dentin that are produced during the secretory stage and seem to follow circadian rhythms. These daily growth lines, called cross-striations in enamel, mark the amount of matrix deposited each 24h by the ameloblast cells [10]. Second, it has been demonstrated that odontoblasts show circadian rhythms with regard to collagen synthesis and secretion [9]. It has also been suggested that these rhythms may be responsible for the circadian incremental lines observed in dentin [9]. Third, in addition to the circadian matrix deposition during secretory stage, ameloblasts alternate between two functionally and morphologically distinct cell forms every 8h in rats during maturation stage i.e. the smooth- and ruffle-ended ameloblasts [11]. Ruffle-ended ameloblasts transport calcium and phosphate ions in the enamel matrix, whereas when smooth-ended ameloblasts degrade proteins of the matrix [11, 12]. Although the above observations strongly suggest that dental tissue formation is under circadian control, no clear evidence for a “dental” circadian clock exists. It is also still unclear how circadian control affects ameloblast and odontoblast functions and dental tissue formation resulting in fully mineralized enamel and dentin.
We recently reported evidence that clock genes are differentially expressed during tooth development, mainly by ameloblasts and odontoblasts [8, 13]. Furthermore, we provided preliminary evidence that the clock gene Nr1d1 is expressed and oscillates in 24h intervals in an ameloblast cell line (HAT-7) [14]. We also found that key ameloblast markers such as amelogenin may be under the control of Nr1d1 [14]. Consistently, we have showed that the total amount of enamel secreted proteins follows daily biological rhythms [8]. However, a detailed analysis of clock genes circadian expression in ameloblasts and evaluation of their multi-level control in ameloblast genes is still missing. This research was undertaken to increase our knowledge on the role of clock genes in enamel formation.
Materials and Methods
Cell culture, synchronization and transfection studies
HAT-7 [15] cells are maintained in DMEM/F-12 (Sigma, St. Louis, MO) supplemented with 100 units/ml penicillin G, 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA) and 10% (v/v) FBS (foetal bovine serum; SAFC Biosciences, Lenexa, KS). For synchronization study, cells are exposed for 2h to serum-free medium containing 15μM forskolin (FSK; Calbiochem, La Jolla, CA), as previously described [16]. After that, cells are changed to regular culture medium. All time intervals calculations are based at the indicated zeitgeber (ZT- is an event that provides the sets of a biological clock) and ZT0 is considered 2 hours after medium changing. Cells are first harvested at 2 hours after medium changing (ZT0), and then are collected every 4 hours for 2 days, for a total of 13 time points. RNAs and proteins are isolated and analyzed by real-time RT-PCR and western blot, respectively.
For transfection studies, HAT-7 cells are plated at 70% confluence in 6-well plates. After changing to serum-free DMEM/F-12 without antibiotics for overnight, 2 μg of Bmal1-pCMV or Cry1-pCMV (kindly provided by Dr. Lei Yin, Department of Molecular and Integrative Physiology, University of Michigan, Ann Arbor, MI) or empty pCMV plasmid is transfected into HAT-7 by Lipofectamine 2000 (Invitrogen), following the manufacturer’s instructions. After 4h, the medium is replaced with DMEM/F-12 supplemented with 10% (v/v) FBS and cultured for an additional 44 h.
RNA isolation and RT-PCR
Total RNA is isolated from the HAT-7 cells by using TRIzol (Invitrogen), and 2 μg of RNA is reverse transcribed with TaqMan reverse transcription reagents (Applied Biosystems, Branchbury, NJ), following the manufacturer’s recommendations. The resulting cDNA is then amplified by RT-PCR or real time RT-PCR (qRT-PCR) using AmpliTaq Gold DNA Polymerase (Applied Biosystems). The RT-PCR products are subcloned into pGEM-T Easy vector (Promega, Madison, WI) and confirmed by sequencing. For RNA quantification, qRT-PCR amplifications are performed at 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s using specific primers for the house keeping gene β-actin (Actb), the four key clock genes Bmal1, Clock, Per1, Per2 as well as the ameloblast specific genes i.e. Amelogenin (Amelx) and Kallikrein-related peptidase 4 (Klk4). The PCR primers are based on published cDNA sequences (Table 1). The relative expression levels for each gene are calculated based on the expression levels of Actb and the differences are presented in graphs using the 2-DDCT method. p-values are calculated using two-sample t-test.
Table 1.
Primers sequences for RT-PCR
| Gene name | 5’- Sequence -3’ | Product size (bp) | GenBank Number | |
|---|---|---|---|---|
| Actb | Forward | AAGTACCCCATTGAACACGG | 257 | NM_031144 |
| Reverse | ATCACAATGCCAGTGGTACG | |||
| Amelx | Forward | TACCACCTCATCCTGGGAGC | 164 | NM_001271074 |
| Reverse | CTGTTGAGACAGCACAGGGA | |||
| Bmal1 | Forward | CCAAGAAAGTATGGACACAGACAAA | 81 | NM_024362 |
| Reverse | GCATTCTTGATCCTTCCTTGGT | |||
| Clock | Forward | CAAAATGTCACGAGCACTTAATGC | 84 | NM_021856 |
| Reverse | ATATCCACTGCTGGCCTTTGG | |||
| Cry1 | Forward | TCAGTTGGGAAGAAGGGATG | 212 | NM_198750 |
| Reverse | TTTTGCAGGGAAGCCTCTTA | |||
| Enam | Forward | GAAAGAACTGCTGGCCTGAC | 211 | NM_001106001 |
| Reverse | GCCCTCCATTGTTAGTTGGA | |||
| Klk4 | Forward | CTGGGGTACCTCATCCTTGA | 208 | NM_001004101 |
| Reverse | CCACGGTGTAGGAGTCCTGT | |||
| Per1 | Forward | AAACCTCTGGCTGTTCCTACCA | 74 | NM_001034125 |
| Reverse | AATGTTGCAGCTCTCCAAATACC | |||
| Per2 | Forward | ATGCTCGCCATCCACAAGA | 72 | NM_031678 |
| Reverse | GCGGAATCGAATGGGAGAAT |
Western blot analysis
Cells are washed twice with PBS before being lysed for 20 min on ice in RIPA lysis buffer. Total proteins (50 μg) per lane are separated by SDS–PAGE, and transferred to a polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA). The membrane is blocked with 5% nonfat milk, incubated overnight at 4°C with the primary antibody against BMAL1 (1:250) (Novus, NB300-596), CLOCK (1:250) (Thermo, PA1-520), PER1 (1:100) (Abcam, ab3443), PER2 (1:100) (Lifespan, LS-C2836), AMELX (1:500) and KLK4 (1:500) (both AMELX and KLK4 antibodies are gifts from Dr. James Simmer’s laboratory at the University of Michigan, Ann Arbor, USA). Anti-Tubulin antibody (Abcam, ab4074) is used to determine the loading control. The membranes are incubated for 1h at room temperature with a horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibody, after washing. Bound antibodies are visualized by enhanced chemiluminescence (ECL) detection system. All bands are measured by densitometry and normalized to tubulin (means±standard error of three or four experiments) using the ImageJ software.
Immunohistochemistry and Immunocytofluorescence
For immunohistochemistry, male C57BL/6J mice (4 weeks of age) are purchased from Jackson Laboratory (Bar Harbor, ME). The animal use was approved by the Animal Welfare Committee of the University of Michigan. Serial sections (7 μm thick) of mandible are prepared. endogenous peroxidase activity is blocked with 2% hydrogen peroxide in methanol for 20 min. Sections are blocked for 1h with 10% goat serum in PBS and then incubated for 1h with Rabbit anti-BMAL1 (1:500) or rabbit anti-PER2 (1:200). After that, the rabbit IgG Vectastain ABC kit (Vector Laboratories, Burlingame, CA) is used following manufacturer’s instruction. All slides are developed in parallel using peroxidase substrate kit DAB (SK-4100, Vector Laboratories), and the reaction is stopped before detection of nonspecific staining in control pre-immune serum-treated sections. For the 12-hour interval experiments, mice are sacrificed, at 8 am and 8 pm, and incisors are dissected out and then embedded in paraffin and sectioned.
For immunocytofluorescence, HAT-7 cells are cultured on slide before fixation. After permeabilization by Triton X-100 and blocking by 3% bovine serum albumin (BSA), cells are incubated with Rabbit anti-BMAL1 (1:500) or rabbit anti-PER2 (1:200) for 1 hour at RT. Next, a fluorescent Alexa Fluor 594 goat anti-mouse (Invitrogen, A-11012) secondary antibody is applied to all sections followed by DAPI counter staining. Sections are then mounted with Prolong Gold antifade reagent (Invitrogen). All sections are examined and then photographed on an Olympus microscope. As negative controls, both omission of primary antibodies and omission of secondary antibodies are used. Normal rabbit IgG instead of the primary antibodies is also used for determination of nonspecific binding of antibodies.
Statistics
We used the Fast Fourier Transform (FFT) method to determine how well a 24h sinusoidal wave period fits the clock gene expression time course data. To calculate the significance of this period, we calculate the probability of obtaining FFT power values higher than the power value obtained with the circadian rhythmic expression (power of reference). For this purpose, we produce 10000 random permutations of the time course data and obtain the maximum power value associated with each permutation. A period is considered statistically significant if the number of power values obtained by the random permutations is greater than the power of reference with a significance level of 1%.
We also used Student’s unpaired t-test for statistical analysis of our qRT-PCR data. Each experiment is performed at least twice, and the representative data are presented as means ± S.D. from at least three independent replicates.
Results
Clock RNAs follow circadian rhythms in HAT-7 cell line
We first used qRT-PCR to evaluate if clock genes RNAs exhibit a circadian rhythm in HAT-7 cells, an ameloblast cell line, after cell cycle synchronization. Circadian rhythms are evaluated usually in 6 consecutive 4-hours intervals (full 24h period) as previously described [17]. We extended our analysis to two consecutive 24h periods as an additional confirmation of the existence of circadian rhythms in ameloblasts. Rhythmic expression patterns are found for two consecutive days for all clock RNAs evaluated here in HAT-7 cells (Fig. 1A-D). Bmal1 (Fig. 1A) and Clock (Fig. 1B) RNAs show clear 24h cycles of expression having the highest expression levels at ZT20. Per1 (Fig. 1C) and Per2 (Fig. 1D) RNAs also exhibit 24h oscillations being at the nadir of expression between ZT12 and ZT10. The expression levels of clock genes RNAs follow similar oscillatory patterns during the second consecutive day after synchronization, confirming the presence of a 24h cycle.
Fig. 1.
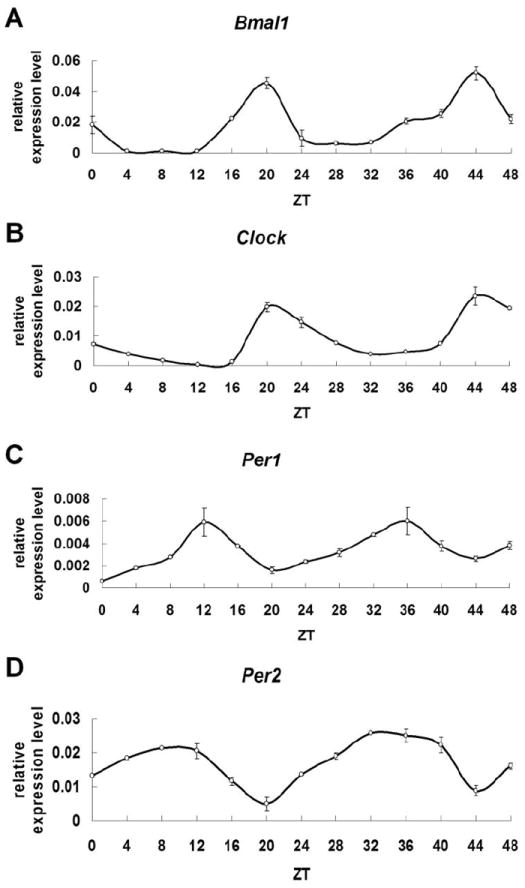
Clock gene RNAs follow circadian rhythms in HAT-7 cell line. A) Bmal1 RNA highest expression levels are observed at ZT20. B) Clock RNA highest expression levels are found at ZT 20. C) Per1 RNA shows highest expression levels at ZT12. D) Per2 shows the strongest expression levels at ZT10-ZT12. Data are means ± S.D. from at least three independent replicates.
We also used standard Fast Fourier analysis to predict the phase and its associated power value for clock RNA expression. 10000 random permutations of the time series data are used to calculate the probability of obtaining the power value by chance (significance level of 1%). Results reconfirm that clock genes RNAs exhibit 24h period rhythms as shown here for Bmal1 (Fig. 2A) and Per2 (Fig. 2B).
Fig. 2.
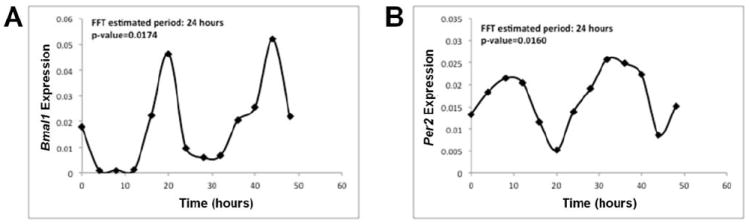
Prediction of expression rhythm phase by Fast Fourier analysis is done using time series data of Bmla1 and Per2. 10000 random permutations of the time series data are used to calculate the probability of obtaining the power value by chance (significance level of 1%). The p-value is shown in the figure.
Clock proteins follow circadian rhythms in HAT-7 cell line
We then examine whether the corresponding clock proteins are also following a circadian cycle pattern in HAT-7 cells. Western blot results (Fig. 3A) indicate that all four clock proteins exhibit circadian patterns similar to the observed RNAs oscillations. After analysis using ImageJ software, our data reveals that the highest levels of BMAL1 proteins expression are found at ZT4 (Fig. 3B). CLOCK expression reaches its highest level at ZT0 (Fig. 3C). PER1 shows strong expression levels at ZT12 (Fig. 3D) and PER2 expression peak is observed at ZT8 (Fig. 3E).
Fig. 3.
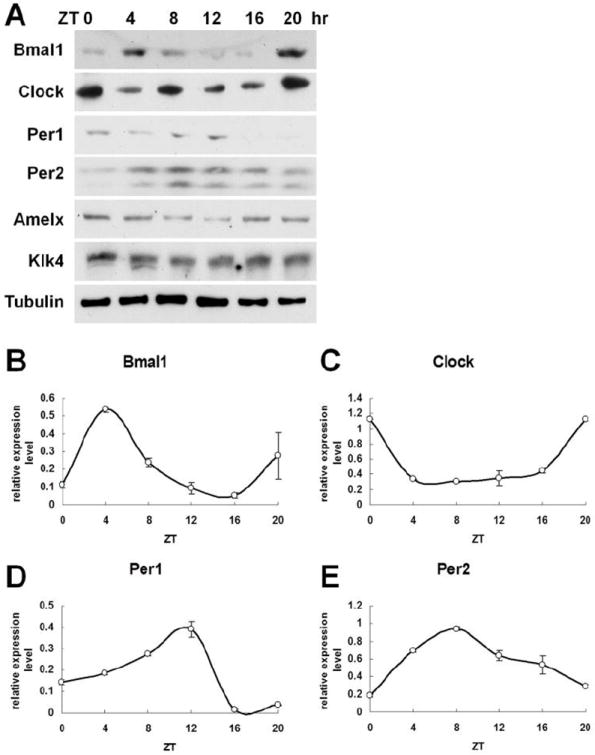
Clock gene proteins exhibit circadian rhythms in HAT-7 cell line. A) Western blot data demonstrate that the expression levels of clock proteins BMAL1, CLOCK, PER1, PER2 and of two ameloblast-specific proteins, AMELX and KLK4, follow circadian rhythms. After analyzed using ImageJ software, results show that B) BMAL1 protein expression levels arrive to zenith at ZT4; C) CLOCK protein expression reaches the highest levels at ZT0; D) PER1 proteins show strong expression levels at ZT12; E) Both PER2 detected protein isoforms exhibit the highest expression peak at ZT8 after synchronization. Data are presented as means± S.D. from three independent replicates.
Clock proteins cellular localization shifts in HAT-7 cells and in mouse ameloblasts
To evaluate if clock proteins shifts from the cytoplasm into the nucleus of cells (where actually clock proteins activate or suppress the expression of CCG) also follow circadian patterns, we used immunocytofluorescence in HAT-7 cells isolated every 4h after cell cycle synchronization. Interestingly, we found that BMAL1 and PER2 proteins have opposite intracellular distribution in HAT-7 cells (Fig. 4A). BMAL1 is mostly expressed in the cytoplasm at ZT0 but shifts into the nucleus at ZT12 (Fig. 4A, left). In contrast, PER2 is observed in the nucleus at ZT0 and moves into the cytoplasm at ZT12 (Fig. 4A, right). We also detected BMAL1 and PER2 on mice incisor serial sections by immunohistochemistry. When BMAL1 is strongly expressed in the cytoplasm of ameloblasts (Fig. 4B and 4D), PER2 is mostly expressed in the nucleus of ameloblasts (Fig. 4C and 4E). We then tested BMAL1 and PER2 protein localization in mouse incisor ameloblasts at 8 am and 8 pm by immunohistochemistry (Fig. 4F and 4G). BMAL1 is mostly expressed in the cytoplasm of ameloblasts at 8 am and in the nucleus at 8 pm (Fig. 4F). In contrast, PER2 is mostly found in the nucleus at 8 am and in the cytoplasm of ameloblasts at 8 pm (Fig. 4G). These data reveal that BMAL1 and PER2 intracellular distribution shifts in opposite directions during a 24h interval.
Fig. 4.
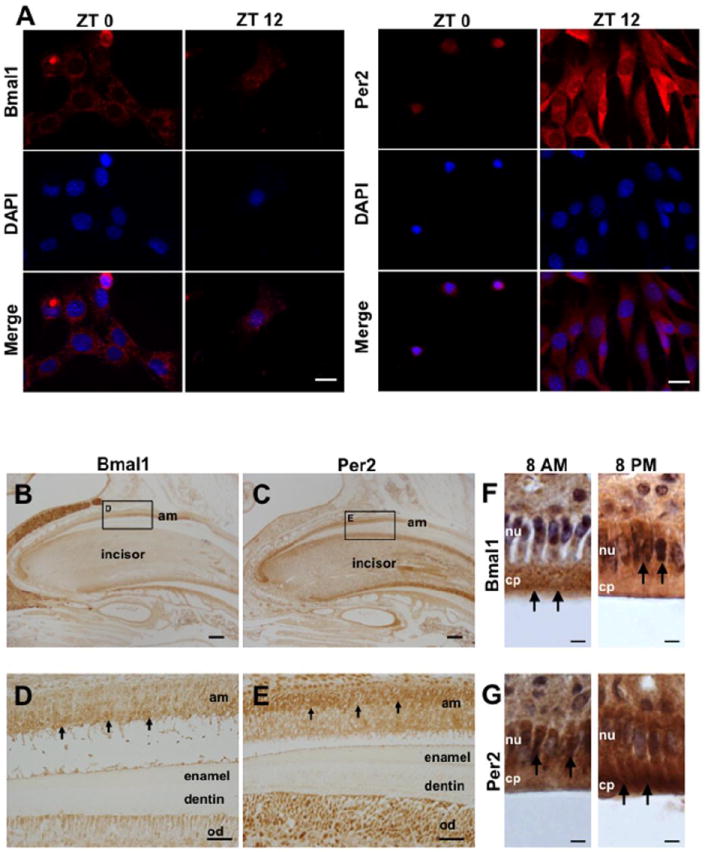
Cellular localization of clock proteins in HAT-7 during a 24h period. A) Immunocytofluorescence staining reveals that BMAL1 proteins are mostly localized in the cytoplasm at ZT0 and then shift into the nucleus at ZT12 (left). In contrast, PER2 is localized in the cell nucleus at ZT0 and then it shifts to the cytoplasm at ZT12 (right). B) IHC results show that BMAL1 (C) and PER2 (D) are expressed in ameloblasts (black arrows). On serial sections, BMAL1 is mostly expressed in the cytoplasm (D), and PER2 is localized mainly in the nucleus (E). Most of BMAL1 proteins are localized in the cytoplasm at 8 am and then in the nucleus of ameloblasts at 8 pm (F). PER2 proteins are found in the nucleus at 8 am and in the cytoplasm of ameloblasts at 8 pm. Am, ameloblast; od, odontoblast; nu, nucleus; cp, cytoplasm. Scale bars = 20 μm in A, 200 μm in B and C, 40 μm in D and E, 10 μm in F and G.
Ameloblast stage-specific genes exhibit circadian rhythms in HAT-7 cells
We then tested if the ameloblast stage-specific genes, Amelx and Klk4, also follow circadian rhythms. QRT-PCR analysis reveals that Amelx RNA is strongly expressed at ZT0 and ZT24 (Fig. 5A). Klk4 RNA also shows high expression levels at around the ZT24 time point (Fig. 5B). Western blot results confirm circadian patterns for both AMELX and KLK4 proteins. AMELX is highly expressed at ZT0 (Fig. 4A and 5C) and KLK4 is highly expressed at ZT4 (Fig. 4A and 5D).
Fig. 5.
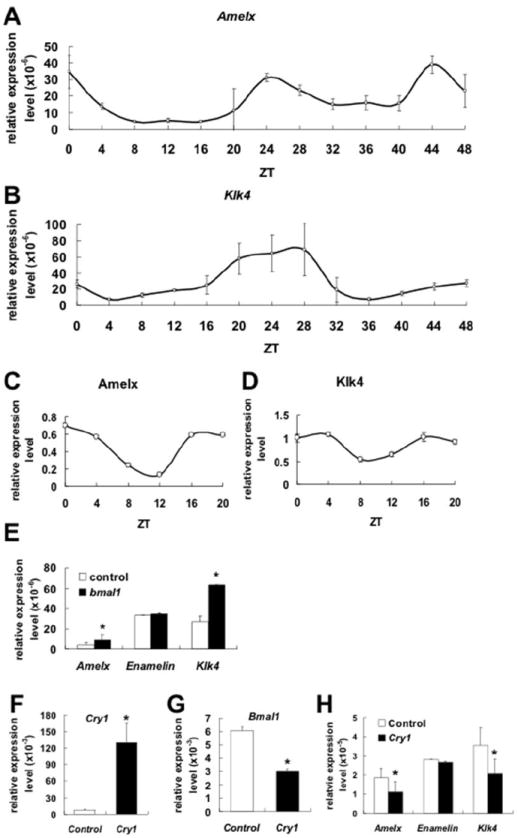
Ameloblast-specific genes expression exhibits circadian rhythms in HAT-7. A) Real-time qRT-PCR reveals that Amelx, a marker of secretory amelobalsts, RNA expression follows a circadian pattern in HAT-7 cells after cell cycle synchronization. B) Klk4, a marker of maturation ameloblsts, also exhibits RNA circadian pattern of expression. At the protein levels, after ImageJ analysis, AMELX shows the highest protein expression levels at ZT0 (C), and KLK4 protein shows the highest level of expression at ZT4 (D). Klk4 relative expression levels are slightly higher than Amelx relative expression levels in HAT-7 cells but both genes follow clear circadian patterns. Over-expression of Bmal1 cDNA results in up-regulation of Amexl and Klk4 RNAs expression in HAT-7 (E). In contrast, Enamelin (which is another ameloblast-specific gene) RNA expression remains unchanged. Over-expression of Cry1 (F) results in down-regulation of Bmal1 (G). Amelx and Klk4 expression levels also are significantly down-regulated (H). The representative data are presented as means ± S.D. from at least three independent replicates where data from cells transfected with Bmla1 or Cry1 were compared with cells transfected with empty vector. Statistical significance (*) was considered when p<0.05.
Bmal1 up-regulates Amelx and Klk4 RNAs steady state levels
Over-expression of Bmal1 in HAT-7 is performed to evaluate if Amelx, Enamelin and Klk4, which are ameloblast stage-specific genes, are regulated by clock genes. QRT-PCR results demonstrate that Amelx and Klk4 RNAs levels are clearly up-regulated after Bmal1 over-expression (Fig. 5E). In contrast, Enamelin RNA expression levels remain unchanged (Fig. 5E). We are also performed Cry1 over-expression to down-regulate Bmal1 in HAT-7 cells (Fig. 5F). After Cry1 over-expression, Bmal1 expression levels dropped to 50% compared to control group (transfection with empty vector) (Fig. 5G). Amelx and Klk4 RNAs levels are also found down-regulated, similar to Bmal1 RNA, in HAT-7 cell line (Fig. 5H). Enamelin RNA expression is not affected by Cry1 transfection and Bmal1 down-regulation (Fig. 5H).
Discussion
Although previous research from our group [8, 13, 14] and others [7, 18] has suggested that enamel formation might be controlled by circadian rhythms little is known regarding the mechanisms of clock genes control in amelogenesis. Here, we conducted a series of studies to clarify how clock genes and their products may guide ameloblast function by controlling ameloblast-specific genes at RNA and protein expression levels. Our data supports a key role of clock genes in ameloblast functions.
To the best of our knowledge, this is the first study that shows that all four main clock genes (Clock, Bmal1, Per1, and Per2) are oscillating in ameloblasts (Fig. 1, 2). We confirm this result both at RNA and protein levels over two successive daily cycles. We also use a Fast Fourier analysis that shows a perfect, statistically significant, 24h oscillatory expression pattern (Fig. 2). These data in combination with our previous reports [8, 13, 14] strongly suggest that clock genes expression is a functional part of amelogenesis.
This study also reveals that clock genes exhibit distinct patterns in the cycle of transcription-translation, adding complexity to the ameloblast clock. At the RNA level, both groups of clock genes, Bmal1/Clock (the positive regulators of circadian rhythms) and Per1/Per2 (the negative regulators of the core clock feedback mechanism), show a perfectly repetitive pattern of oscillation with 8 hours difference in the expression peak between the two groups (Fig. 1). This data is consistent with previous observations in other systems [19-21]. However, it is worthy to note that Per2 peak of expression is much “smoother” than the peak of expression for all other three clock genes suggesting that Per2 RNAs may have a longer half life in ameloblasts. At the protein level, all four proteins (BMAL1, CLOCK, PER1 and PER2) were also found to oscillate at regular 24h intervals. Of interest, BMAL1 and PER1 proteins peaks of expression were observed about 4 hours later than CLOCK and PER2 peaks, suggesting different post-transcriptional modifications.
Finally, immuno fluorescence localization of all four proteins in synchronized ameloblast cells and in vivo tooth sections showed differential localization patterns. Unexpectedly, nucleus localization of clock proteins does not coincide with the high expression peaks observed at the protein levels by western blot. In HAT-7 cells, clock proteins are translocated into the nucleus when their relative expression levels decline. Several levels of control of the circadian clock have been reported including the circadian control of nucleus translocation [22-24]. Our study further highlights the complexity of clock genes regulation in ameloblasts and opens novel directions for future research.
To elucidate the potential down-stream clock genes targets in ameloblasts, we have undertaken extensive promoter analysis of potential clock controlled genes (CCG). This analysis predicted clock binding sites on the promoters of several ameloblast specific genes including amelogenin (a marker of secretory ameloblasts) and klk4 (a marker of maturation ameloblasts) [14]. To confirm these predictions, we have undertaken circadian assays and transient transfection experiments in HAT-7 cells. Our data shows regular 24h oscillations of ameloblast stage-specific genes (amelogenin and Klk4). In contrast, enamelin, another ameloblast-specific gene, does not show any circadian oscillations. This is in accordance with the fact that enamelin promoter lacks potential binding sites for clock proteins. Only 6-10% of genes in a given tissue are CCG [25-28]. To complete the list of CCG in ameloblasts it may be necessary to undertake whole genome RNA arrays at regular time intervals.
This study also reveals that the expression of both Amelx and Klk4 RNA is in phase with Clock/Bmal1 expression. Accordingly, Bmal1 over-expression (a positive regulator of the circadian clock) results in the up-regulation of amelogenin and Klk4 RNAs, suggesting that clock genes regulate amelogenin and Klk4 expression at the transcriptional level. Consistently, Cry1 over-expression (a negative regulator of the circadian clock) results in the down-regulation of amelogenin and Klk4 RNAs. These findings are also consistent with previous work from our group [14]. Additional levels of control that imply indirect transcriptional control (through the control of transcription factors controlled by clock genes), as we have previously suggested [14], or post-transcriptional and/or translational and post-translational mechanisms may also co-exist during amelogenesis. Further investigation is needed to clarify the mechanisms of transcriptional control of amelogenin and Klk4 by clock genes.
The appositional growth of enamel is characterized by enamel matrix proteins (mainly amelogenin) production and secretion into the enamel matrix. Appositional growth is a linear event that follows circadian (24h) cycles. These circadian cycles are evident in humans as growth lines, called cross striations, traversing enamel prisms [29, 30]. Measurements in rat enamel have also clearly showed that enamel volume and size increase follows a linear pattern of growth per unit of time across the secretory stage [11]. The in vitro circadian RNA pattern for amelogenin reported here is in accordance with these previous observations in humans and rodents. Consistently, our recently published data generated in Dr. Charles Smith’s laboratory (University of Montreal, Canada) shows amelogenin circadian protein secretion variation in rat enamel matrix [8]. Similarly, unpublished data from our group shows higher RNA expression at 10pm versus 10am in mouse teeth suggesting an overall higher Amelx RNA and protein expression/secretion in phase with the wake status of rodents. Additional studies are needed to clarify the complex controls of the observed circadian cycles of expression for clock genes and their CCG in ameloblasts.
Similar to enamel apposition, enamel maturation also occurs perfectly linearly over time suggesting the involvement of a clock mechanism in the regulation of the process[11]. Klk4 plays a key role in the process of enamel maturation [31] and is considered as the main marker of mature ameloblasts [32]. Of interest, similar to amelogenin, Klk4 expression follows circadian rhythms in synchronized HAT-7 cells. Although at this time we do not know if circadian Klk4 expression does relate with precise functions in mature ameloblasts that follow a biological clock, it is tempted to speculate that removal of enamel proteins from the enamel matrix also follows a circadian pattern similar to their secretion.
Since the enamel secretion and maturation might be dependent on circadian clock, genetic polymorphisms in clock genes could modify their rhythms and produce differences in enamel characteristics. This could account for individual differences in enamel shape, thickness, or hardness among the general population. By gaining a better understanding of how enamel is made, this information could be also applied to efforts in the enamel regeneration field.
Highlights.
Several clock proteins show circadian oscillatory expression patterns in ameloblast cells
BMAL1 and PER2 proteins localization shifts from the nucleus to the cytoplasm in regular 12h intervals in ameloblasts
CRY1 and BMAL1 proteins regulate Amelx and Klk4 RNA and protein levels in ameloblasts
Our data suggest that amelogenesis might be under circadian control
Acknowledgments
This research was funded by funds provided by the Department of Orthodontics and Pediatric Dentistry, School of Dentistry, University of Michigan and NIH grant DE018878 to PP.
Footnotes
Conflict of interest: The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brown SA, Schibler U. The ins and outs of circadian timekeeping. Curr Opin Genet Dev. 1999;9:588–94. doi: 10.1016/s0959-437x(99)00009-x. [DOI] [PubMed] [Google Scholar]
- 2.Barclay NL, Eley TC, Mill J, Wong CC, Zavos HM, Archer SN, Gregory AM. Sleep quality and diurnal preference in a sample of young adults: associations with 5HTTLPR, PER3, and CLOCK 3111. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:681–90. doi: 10.1002/ajmg.b.31210. [DOI] [PubMed] [Google Scholar]
- 3.Ukai-Tadenuma M, Kasukawa T, Ueda HR. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat Cell Biol. 2008;10:1154–63. doi: 10.1038/ncb1775. [DOI] [PubMed] [Google Scholar]
- 4.Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 5.Fu L, Patel MS, Bradley A, Wagner EF, Karsenty G. The molecular clock mediates leptin-regulated bone formation. Cell. 2005;122:803–15. doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 6.Maronde E, Schilling AF, Seitz S, Schinke T, Schmutz I, van der Horst G, Amling M, Albrecht U. The clock genes Period 2 and Cryptochrome 2 differentially balance bone formation. PLoS One. 2010;5:e11527. doi: 10.1371/journal.pone.0011527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antoine D, Hillson S, Dean MC. The developmental clock of dental enamel: a test for the periodicity of prism cross-striations in modern humans and an evaluation of the most likely sources of error in histological studies of this kind. J Anat. 2009;214:45–55. doi: 10.1111/j.1469-7580.2008.01010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simmer JP, Papagerakis P, Smith CE, Fisher DC, Rountrey AN, Zheng L, Hu JC. Regulation of dental enamel shape and hardness. J Dent Res. 2010;89:1024–38. doi: 10.1177/0022034510375829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohtsuka M, Shinoda H. Ontogeny of circadian dentinogenesis in the rat incisor. Arch Oral Biol. 1995;40:481–5. doi: 10.1016/0003-9969(95)00002-7. [DOI] [PubMed] [Google Scholar]
- 10.Shellis RP. Utilization of periodic markings in enamel to obtain information on tooth growth. J Hum Evol. 1998;35:387–400. doi: 10.1006/jhev.1998.0260. [DOI] [PubMed] [Google Scholar]
- 11.Smith CE. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9:128–61. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- 12.Josephsen K, Takano Y, Frische S, Praetorius J, Nielsen S, Aoba T, Fejerskov O. Ion transporters in secretory and cyclically modulating ameloblasts: a new hypothesis for cellular control of preeruptive enamel maturation. Am J Physiol Cell Physiol. 2010;299:1299–307. doi: 10.1152/ajpcell.00218.2010. [DOI] [PubMed] [Google Scholar]
- 13.Zheng L, Papagerakis S, Schnell SD, Hoogerwerf WA, Papagerakis P. Expression of clock proteins in developing tooth. Gene Expr Patterns. 2011;11:202–6. doi: 10.1016/j.gep.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Athanassiou-Papaefthymiou M, Kim D, Harbron L, Papagerakis S, Schnell S, Harada H, Papagerakis P. Molecular and circadian controls of ameloblasts. Eur J Oral Sci. 2011;119(Suppl 1):35–40. doi: 10.1111/j.1600-0722.2011.00918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawano S, Morotomi T, Toyono T, Nakamura N, Uchida T, Ohishi M, Toyoshima K, Harada H. Establishment of dental epithelial cell line (HAT-7) and the cell differentiation dependent on Notch signaling pathway. Connect Tissue Res. 2002;43:409–12. doi: 10.1080/03008200290000637. [DOI] [PubMed] [Google Scholar]
- 16.Atanasov AG, Leiser D, Roesselet C, Noti M, Corazza N, Schoonjans K, Brunner T. Cell cycle-dependent regulation of extra-adrenal glucocorticoid synthesis in murine intestinal epithelial cells. Faseb J. 2008;22:4117–25. doi: 10.1096/fj.08-114157. [DOI] [PubMed] [Google Scholar]
- 17.Kowalska E, Ripperger JA, Hoegger DC, Bruegger P, Buch T, Birchler T, Mueller A, Albrecht U, Contaldo C, Brown SA. Feature Article: NONO couples the circadian clock to the cell cycle. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1213317110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacruz RS, Hacia JG, Bromage TG, Boyde A, Lei Y, Xu Y, Miller JD, Paine ML, Snead ML. The circadian clock modulates enamel development. J Biol Rhythms. 2012;27:237–45. doi: 10.1177/0748730412442830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–5. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 20.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS. PERIOD2∷LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–46. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondratov RV, Chernov MV, Kondratova AA, Gorbacheva VY, Gudkov AV, Antoch MP. BMAL1-dependent circadian oscillation of nuclear CLOCK: posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 2003;17:1921–32. doi: 10.1101/gad.1099503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon I, Lee J, Chang SH, Jung NC, Lee BJ, Son GH, Kim K, Lee KH. BMAL1 shuttling controls transactivation and degradation of the CLOCK/BMAL1 heterodimer. Mol Cell Biol. 2006;26:7318–30. doi: 10.1128/MCB.00337-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamanini F, Yagita K, Okamura K, Horst GTJVD. Methods in Enzymology. In: Young MW, editor. Circadian Rhythms. 1. Waltham: Academic Press; 2005. pp. 418–435. [DOI] [PubMed] [Google Scholar]
- 25.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–92. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 26.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 27.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 28.Bozek K, Relogio A, Kielbasa SM, Heine M, Dame C, Kramer A, Herzel H. Regulation of clock-controlled genes in mammals. PLoS One. 2009;4:e4882. doi: 10.1371/journal.pone.0004882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moggi-Cecchi J. Questions of growth. Nature. 2001;414:595–7. doi: 10.1038/414595a. [DOI] [PubMed] [Google Scholar]
- 30.Guatelli-Steinberg D, Reid DJ, Bishop TA, Larsen CS. Anterior tooth growth periods in Neandertals were comparable to those of modern humans. Proc Natl Acad Sci U S A. 2005;102:14197–202. doi: 10.1073/pnas.0503108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Y, Papagerakis P, Yamakoshi Y, Hu JC, Bartlett JD, Simmer JP. Functions of KLK4 and MMP-20 in dental enamel formation. Biol Chem. 2008;389:695–700. doi: 10.1515/BC.2008.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simmer JP, Hu Y, Lertlam R, Yamakoshi Y, Hu JC. Hypomaturation enamel defects in Klk4 knockout/LacZ knockin mice. J Biol Chem. 2009;284:19110–21. doi: 10.1074/jbc.M109.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]


