The imbalance between energy intake and expenditure is the underlying cause of the current obesity and diabetes pandemics. Central to these pathologies is the fat depot: white adipose tissue (WAT) stores excess calories, and brown adipose tissue (BAT) consumes fuel for thermogenesis using tissue-specific uncoupling protein 1 (UCP1)1, 2. BAT was once thought to have a functional role only in rodents and human infants, but it has been recently shown that in response to mild cold exposure, adult human BAT consumes more glucose per gram than any other tissue3. In addition to this non-shivering thermogenesis, human BAT may also combat weight gain by becoming more active in the setting of increased whole-body energy intake4-7. This phenomenon of BAT-mediated diet-induced thermogenesis has been observed in rodents8 and suggests that human BAT could be utilized as a safe treatment for obesity and metabolic dysregulation9. In this study, we isolated anatomically defined neck fat from adult human volunteers and compared its gene expression, differentiation capacity, and basal oxygen consumption to different mouse adipose depots. While the properties of human neck fat vary substantially, some human samples share many similarities with classical/constitutive rodent BAT.
Initial examinations of the principal human neck fat depots revealed brown adipocytes with classic multilocular adipocytes, numerous mitochondria, and a rich investment with capillaries and sympathetic neurons10 as well as higher expression of UCP110,11. Critical molecular and functional studies are lacking, however, because the human brown adipocytes are distributed heterogeneously, and their precise sites are unknown. To gain a specific description of where BAT is located and its gene expression and functional profiles, we resected adipose tissue from the necks of 31 individuals undergoing surgery (Supplementary Tables 1 and 2 and Supplementary Methods online). Samples were collected from up to five different depots (Fig. 1a), from superficial to most deep: subcutaneous, subplatysmal, carotid sheath, longus colli, and prevertebral. Superficial and deeper fat had the classical histological (Fig. 1b) and ultrastructural (Supplementary Fig. 1a–d) features of rodent white and brown fat, respectively12.
Figure 1. Anatomical localization of adult human neck BAT.
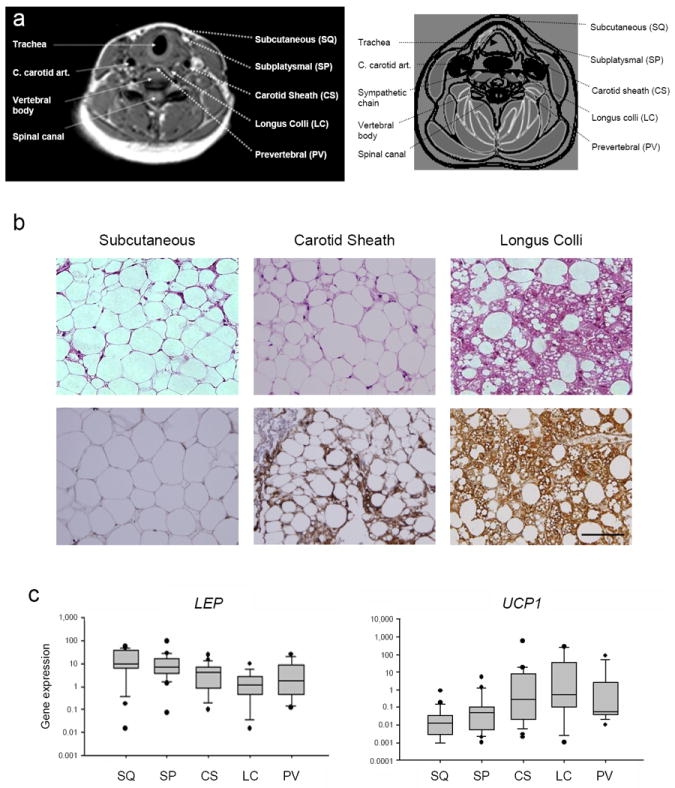
(a) The five different sites from which adipose tissue was resected are shown. On the left side are standard anatomical landmarks in italics: trachea, common carotid artery (C. carotid art.), sympathetic chain, vertebral body, and spinal canal. On the right side are the fat depots. The superficial depots comprise the subcutaneous (SQ) and subplatysmal (SP) regions and deep depots comprise the carotid sheath (CS), longus colli (LC), and prevertebral (PV) regions. The left side shows the axial cross-sectional MRI image of a healthy volunteer at the level of the fifth cervical vertebra (“C5”). Dashed white arrows point to the location of the landmarks and adipose tissue depots. The right side is a schematic diagram of the MRI image. Skeletal muscle is gray; air in the trachea is white; blood vessels are black; fascial planes are separated by black lines; and landmarks are identified using dashed black arrows. (b) Human neck fat from the subcutaneous (left), carotid sheath (center), and longus colli (right) depots from a representative volunteer was stained with hematoxylin and eosin (upper panels) or immunohistochemically for UCP1 (lower panels). Scale bar, 100 μm. (c) From 18 individuals undergoing routine neck surgery, gene expression levels of WAT-associated LEP and BAT-associated UCP1 are plotted on a logarithmic scale. The gray boxes enclose the 25th and 75th percentiles, and the internal line is the median. The whiskers are the 10th and 90th percentiles, and the black circles are outliers. Non-parametric ANOVA for both genes had a P = 0.002.
The microanatomical distinctions persisted at the level of gene expression. Non-parametric ANOVA demonstrated a significant difference among the depots for expression of WAT-associated leptin gene (LEP), which was more enriched in superficial fat (P = 0.002) (Fig. 1c and Supplementary Table 3). The opposite was seen with UCP1 gene expression (P = 0.002). Specifically, in two of the deeper fat depots, carotid sheath and longus colli, UCP1 expression was 12- to 72-fold higher than the two superficial depots, subcutaneous and subplatysmal (P = 0.03 or lower for all four pair-wise comparisons). The PV site also had more UCP1 than that of subcutaneous fat (P = 0.006). Therefore, although there was great variability among adult human neck fat in terms of UCP1 expression, we consistently found BAT to be most abundant near the carotid sheath and longus colli muscles. These deep locations may have functional significance: the longus colli depot is adjacent to the sympathetic chain, which mediates the rapid neuronal response to cold13, and the carotid sheath BAT envelops the carotid arteries, permitting effective heating of the cerebral vasculature14.
Having established the anatomical location of human neck BAT, we determined its likely developmental lineage. It has recently been shown with mouse tissue that brown adipocytes are not all the same. The large and constitutively present interscapular BAT derives from a lineage common to skeletal muscle and is termed classical/constitutive BAT15-17, while other brown adipocytes can be induced to grow within predominantly white depots and have been termed brite/beige/inducible BAT16-19. In order to provide more than one perspective on the relationship between lineage marker genes and human neck fat, we used three different, complementary approaches: (a) paired comparisons between the subcutaneous and deep fat in each person to give a basic description of which genes are found in which sites; (b) cluster analysis to view how the gene expression patterns relate to each other; and (c) factor analysis to identify a potential underlying framework that accounts for the correlations seen among the genes’ expression patterns.
From 13 pairs of superficial and deep human neck adipose tissue, expression levels of 12 mouse marker genes were compared. In the subcutaneous depots, LEP (P = 0.001) and HOXC9 (P = 0.03), both markers of WAT1, 15, were higher, while in the deeper depots, UCP1 (P = 0.001) and the classical/constitutive-associated markers ZIC115, 18, 20-22 (P = 0.002) and LHX815, 18, 20-22 (P = 0.02) were significantly elevated (Fig. 2a, Supplementary Fig. 2, and Supplementary Table 4). Cluster analysis (Fig. 2b) reinforced this patterning and showed that there were three distinct groups of marker genes in human neck fat: one that is down-regulated in deeper depots (SHOX2, LEP, HOXC9); one that is not changed (TNFRSF9 – also known as CD137, TMEM26, TBX1, MPZL2 – also known as EVA1, EBF3, FBXO31); and one that is up-regulated (UCP1, ZIC1, LHX8). Factor analysis (Table 1) provided a similar categorization, with some slight differences. In this case, the best markers for the groupings were MPZL2, HOXC9, EBF3, FBXO31, and LEP (“white” component); TNFRSF9, TMEM26, and SHOX2 (“brite/beige” component); and UCP1, LHX8, and ZIC1 (“brown” component).
Figure 2. Adult human neck BAT lineage tracing.
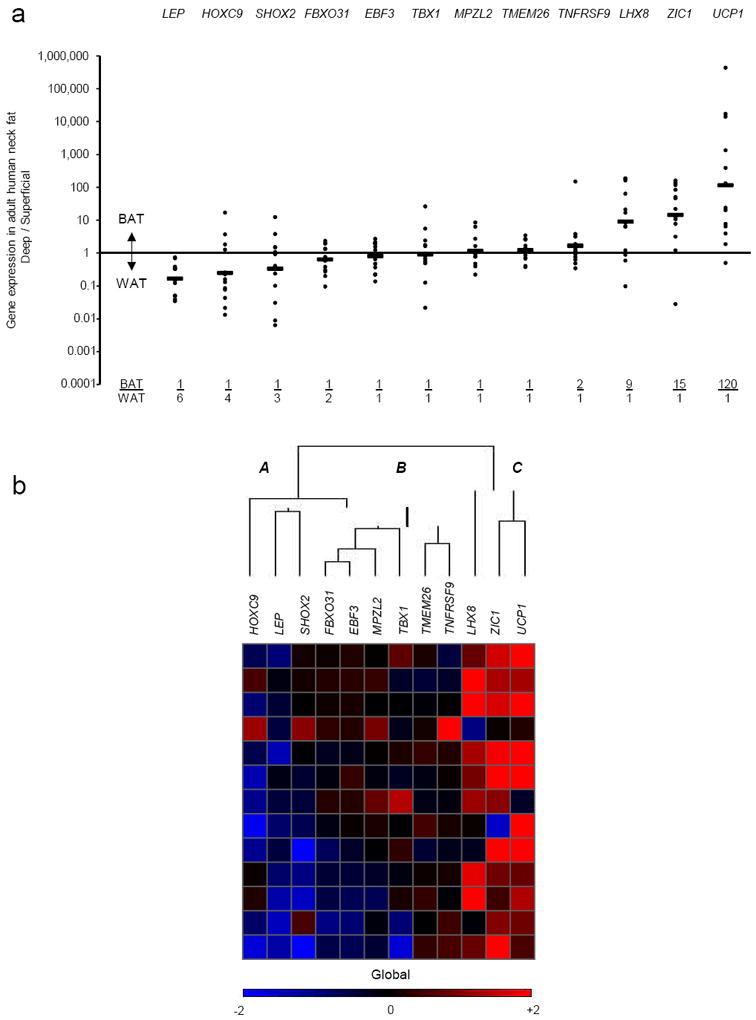
(a) Shown for 13 individuals undergoing routine neck surgery is the ratio of the gene expression found in the deep tissue expressing the highest level of UCP1 (“BAT”) vs. that found in each individual’s SQ fat (“WAT”) for each of the 12 lineage markers. Each closed circle represents a single person. The geometric mean for each gene is represented by the horizontal bars and is also shown at the bottom of the graph as the BAT/ WAT ratio. (b) For the cluster analysis, each color in the figure represents the log10 ratio of the (BAT gene expression) / (WAT gene expression) of a particular gene in each volunteer. Each column contains data from a specific gene, and each row contains data from a single person. Red squares represent high ratios and blue squares represent low ratios. The values in the heat map range between −2 and +2 (i.e. expression ratios between 100-fold in one direction to 100-fold in the other). For visualization purposes, values outside of that range are replaced by either −2 or +2, as appropriate. The dendrogram reflects the degree of correlation of the genes assessed by the hierarchical clustering. Letters “A,”, “B,” and “C” represent the groupings of markers that were, respectively, down-regulated in deeper depots (SHOX2, LEP, HOXC9); not changed (TNFRSF9, TMEM26, TBX1, MPZL2, EBF3, FBXO31); and up-regulated (UCP1, ZIC1, LHX8).
Table 1.
Factor analysis showing Varimax rotated components of human BAT lineage marker genesa
| Gene | R1 | R2 | R3 | Communality |
|---|---|---|---|---|
| UCP1 | +0.131 | +0.780 | −0.051 | 0.629 |
| LHX8 | +0.306 | +0.665 | +0.211 | 0.580 |
| ZIC1 | −0.217 | +0.630 | +0.368 | 0.579 |
| TNFRSF9 | −0.250 | −0.033 | +0.812 | 0.724 |
| TMEM26 | +0.189 | +0.246 | +0.792 | 0.724 |
| SHOX2 | +0.256 | −0.468 | +0.634 | 0.686 |
| TBX1 | +0.067 | +0.313 | −0.154 | 0.127 |
| MPZL2 | +0.805 | +0.258 | +0.107 | 0.727 |
| HOXC9 | +0.817 | +0.109 | −0.159 | 0.704 |
| EBF3 | +0.908 | +0.081 | +0.103 | 0.842 |
| FBXO31 | +0.945 | +0.225 | +0.015 | 0.944 |
| LEP | +0.814 | −0.320 | −0.039 | 0.767 |
|
| ||||
| Variance (%) | 33.5 | 17.2 | 16.2 | |
| Cumulative Variance (%) | 67 | |||
Factor loadings >0.500 are shown in bold
The three complementary marker analyses indicate that deep human neck brown adipocytes most closely resemble cells from the classical/constitutive BAT lineage in the mouse. This designation is supported by anatomical studies showing this depot is found in human infants and persists even into the eighth decade of life 23. Functionally, this may also indicate that human neck BAT may have the same capacity for high rates of energy expenditure seen in rodent interscapular BAT24. Of note, these results represent the combined signal from whole tissue biopsies and not individual cells. Given the mixed composition of human BAT5-7, 10, 19, 25, our findings are also consistent with the presence of some brite/beige adipocytes in the neck depot, since beige marker TNFRSF918 showed a nonsignificant, two-fold enrichment in the deeper tissue and clustered the closest to UCP1 after brown markers ZIC1 and LHX8. That some genes previously designated as markers of a brown lineage in mice (EBF3, MPZL2, FBXO31)19 were not higher in the deeper human neck depots or associated closely with UCP1 reflect the possibility that adipose tissue actually comprises multiple mini-organs both in the mouse22 and human26, with distinct developmental and functional characteristics. A clear definition of what these cell types are, their origins, and their gene signatures is still a work in progress.
To determine if adult human neck BAT also has a functional profile similar to classical/constitutive rodent BAT, we compared the expression of genes associated with rodent WAT and BAT differentiation, function, and thermogenesis18, 20, 27, 28. From five mice, levels were measured in five different depots: interscapular BAT (iBAT), inguinal subcutaneous WAT, perigonadal WAT, mesenteric WAT, and perirenal WAT. These five depots were analyzed together with the deep neck fat from three individuals with high UCP1 expression and hence likely to be most purely BAT. The associated WAT from the individuals’ subcutaneous neck depots was used as a comparator. Human BAT had an expression pattern that was very similar to mouse iBAT (Fig. 3a and Supplementary Tables 3 and 5), including the high expression of genes involved with mitochondrial biogenesis and thermogenesis (UCP1, PPARGC1A - also known as PGC1α, DIO2) and low levels of NRIP1, also known as RIP140, a nuclear corepressor whose reduction leads to increased oxidative metabolism and mitochondrial biogenesis29. Human SQ WAT was strikingly different, more like mouse mesenteric and perigonadal depots.
Figure 3. Adult human neck BAT expresses functional genes similar to mouse interscapular BAT, can be grown ex vivo from multiple adipose tissue depots, and has an unstimulated energy expenditure similar to that of mouse interscapular BAT.
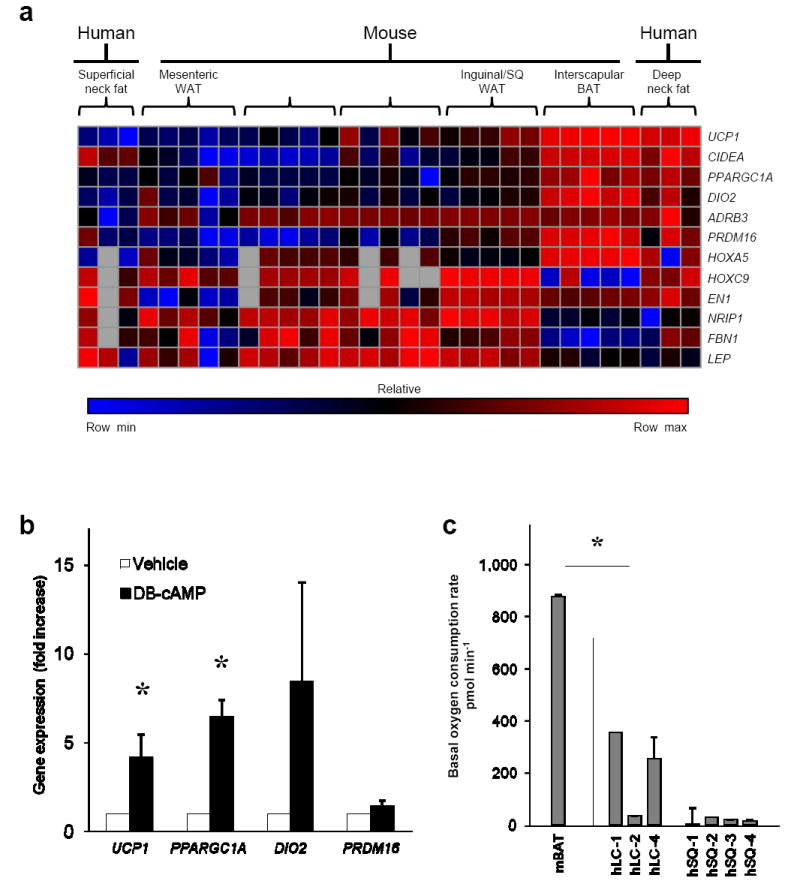
(a) Gene expression of BAT- and WAT-associated genes was measured in five different mouse adipose tissue depots taken from five C57Bl/6 male mice, 12 weeks-old. Clustering of the genes from the mouse depots was done with the samples of human deep neck fat that had the highest expression of UCP1, along with their associated subcutaneous fat. The heat map represents the relative abundance of each gene across all 31 samples. The values were normalized within each row using a linear color scale. The highest value in each row is colored bright red; the lowest is colored bright blue; those near the midpoint between these values are colored black. Missing values are colored gray. (b) Preadipocytes from the stromal vascular fraction of both superficial and deep neck adipose tissue fat depots from four individuals (eight different sites in total), were differentiated into mature adipocytes and then treated for 4 hours with 500 μM dibutyryl-cAMP (DB-cAMP) or vehicle. Shown is the fold increase in expression of four different BAT-associated genes, UCP1, PPARGC1A, DIO2, and PRDM16, compared to treatment with vehicle for the same duration. *, P < 0.05; errors bars are s.e.m. (c) Adipose tissue from mouse interscapular BAT or human cervical adipose tissue from four individuals’ subcutaneous (SQ) and longus colli (LC) depots. Labeling is as follows: “m” or “h” for mouse and human, respectively; neck fat depot; and number referring to the particular individual. *, P < 0.05; errors bars are s.e.m.
The ability to grow new functional brown adipocytes is likely to be essential for utilizing BAT thermogenesis to treat metabolic dysregulation30. We isolated preadipocytes from the stromal vascular fraction of human neck adipose tissue and differentiated them ex vivo over ten days in conditions used to induce brown adipogenesis in mouse preadipocytes17. To reproduce the effects of cold-mediated adrenergic stimulation, we treated the differentiated human adipocytes with 500 μM dibutyryl-cAMP. Compared with vehicle control, mature adipocytes derived from SVF of neck adipose tissue responded to dibutyryl-cAMP with significant increases in several BAT genes, including UCP1 (P = 0.04) and PPARGC1A (P = 0.01)(Fig. 3b and Supplementary Table 6). These increases in gene expression demonstrates that as with stromal vascular fraction from supraclavicular sites31, the in vitro differentiated cells from neck fat possess the capacity to respond to adrenergic stimulation with expression of genes needed for thermogenesis and are therefore bona fide brown adipocytes. That PRDM16 did not increase as much reflects its role as a cell fate determining factor20 rather than a mediator of sympathetically-mediated thermogenesis.
To compare the energy expenditure of mouse and human fat, we isolated samples of neck fat from four individuals’ superficial and deep depots and compared the unstimulated oxygen consumption rate (OCR) in whole 10 mg sections with mouse iBAT (Fig. 3c). The OCR in mouse iBAT was 874 pmol/min and in the deeper fat next to the longus colli in two individuals was 252 and 354 pmol/min, nearly 50% of mouse iBAT. In contrast, the basal OCR of human subcutaneous WAT was nearly two orders of magnitude lower than both longus colli and mouse iBAT (P = 0.001).
In summary, we more precisely define the anatomical regions where adult human BAT can be found in the principal neck fat depots. Adult human neck BAT possesses molecular signatures of classical/constitutive BAT. With further work it should ultimately be distinguishable by a small number of genes that will permit rapid screening of interventions designed to increase BAT mass and energy expenditure. Most notably, functional human BAT can be induced to grow ex vivo from precursor cells present in neck fat depots, and its unstimulated energy expenditure is similar to mouse BAT.
Online Methods
Human Study Population
This study followed institutional guidelines and was approved by the Human Studies Institutional Review Boards of Beth Israel Deaconess Medical Center, Joslin Diabetes Center, and Massachusetts General Hospital. Individuals were identified either by Dr. White prior to anterior cervical spine surgery or by Dr. Hasselgren prior to thyroidectomy, and written informed consented was obtained by other study staff prior to surgery. All people undergoing thyroidectomies had TSH values within the normal range. There were two independent cohorts: for anatomical localization and comparison to mouse adipose tissue depots, neck fat from18 individuals was studied. For lineage tracing, neck fat from13 different people was studied. Healthy volunteers for the MRI imaging were recruited via electronic advertisements.
MRI
Studies were carried out using a 3.0 Tesla Siemens Allegra MRI System equipped for echo planar imaging with quadrature head coil. The adult human subject lay supine in the scanner with the head immobilized using cushioned supports. Two sets of structural images were collected using a T1-weighted MPRAGE sequence (TR/TE = 2.73/3.19 ms, flip angle = 7°, FOV = 256 × 256 mm; slice thickness = 1.33 mm).
Light and Electron Microscopy
For light microscopy, we placed freshly resected brown and white fat into 4% PBS-buffered formalin (Sigma), and processed as described17,32. Immunohistochemical assays were done with the use of polyclonal goat IgG to UCP1 (Santa Cruz Biotechnology, sc6528) at 1:50 dilution. For light and transmission electron microscopy, fat was fixed in 2.5% glutaraldehyde in 0.1M phosphate buffer, pH 7.4, osmicated, and embedded in plastic (Araldite) then processed as described in the Joslin Advanced Microscopy Core33.
Gene Expression for Anatomical Localization of Adult Human Neck Fat
Freshly resected fat from the neck was placed immediately into RNAlater (Qiagen). We extracted total cellular RNA from tissue using an RNeasy minikit (Qiagen) according to instructions. Quantity and purity were assessed by ultraviolet absorbance at 260 and 280 nm. cDNA was prepared from 6 ng/μL of RNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the manufacturer’s instructions. 6 μL (36 ng) of cDNA was used in a 20 μL PCR using TaqMan® Gene Expression Assays with a FAM dye label for the following genes (Supplementary Tables 3 and 5): uncoupling protein 1 (UCP1), type 2 deiodinase (DIO2), β3-adrenergic receptor (ADRB3), peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PPARGC1A, also known as PGC1α), cell death-inducing DNA fragmentation factor alpha-like effector A (CIDEA), PRD1-BF1-RIZ1 homologous domain containing 16 (PRDM16), receptor-interacting protein 140 (NRIP1, also known as RIP140); fibrillin 1 (FBN1); engrailed 1 (EN1); homeobox A5 (HOXA5); homeobox C9 (HOXC9); and leptin (LEP). Quantitative RT-PCR assays were run in duplicates and quantitated in the ABI Prism 7900 sequence detection system. The values were normalized to the expression of TATA-binding protein (TBP) in each sample, and results were expressed as ratios in arbitrary units.
Gene Expression and Cluster Analysis of Markers of Adipocyte Lineage
From 13 individuals undergoing routine neck surgery, we resected fat was and prepared cDNA as above using primer sequences for quantitative real-time PCR (Supplementary Table 4). Assays were run in duplicates and quantified in the ABI Prism 7900 sequence detection system using SYBR® Green as previously described32 for the following genes: tumor necrosis factor receptor superfamily, member 9 (TNFRSF9, also known as CD137); early B-cell factor 3 (EBF3); myelin protein zero-like 2 (MPZL2, also known as EVA1); F-box protein 31 (FBXO31); LIM homeobox 8 (LHX8); short stature homeobox 2 (SHOX2); T-box 1 (TBX1); transmembrane protein 26 (TMEM26); Zic family member 1 (ZIC1). We normalized values to the expression of 18S ribosomal RNA in each sample and expressed results as ratios in arbitrary units. WAT was defined as the subcutaneous fat, and BAT was defined as the sample of deep fat that had the highest expression of UCP1. We clustered genes via pairwise complete-linkage hierarchical clustering according to the city-block distance between rows. The clustering visualization was done using GenePattern34.
Heat Map for Mouse-Human Comparison
All animals were housed and utilized according to the institutional guidelines stipulated by the Joslin Diabetes Center Institutional Animal Care and Usage Committee. We resected mouse fat depots from C57Bl/6 male mice, 12 weeks old (Taconic). Human and mouse RNA was isolated and measured as above. After normalization to TBP, the gene expression levels were log-transformed. We generated the heat map using GenePattern34 using the relative abundance of each gene across all 31 samples.
Ex Vivo Differentiation
For isolation of pre-adipocytes, we isolated the stromal vascular fraction (SVF) from (1) superficial fat (pooled subcutaneous and subplatysmal) and (2) fat located in the deeper neck regions (pooled: carotid sheath, longus colli, prevertebral). Freshly resected fat from the neck was collected, minced and subsequently digested using collagenase 1 (2 mg/mL in phosphate-buffered saline with addition of 3.5% bovine serum albumin; Worthington Biochemical Corporation), and the SVF was isolated as previously described17,35. SVF cells were plated and grown until 90% confluence in growth medium supplemented with 10% fetal bovine serum in the presence of 5 ng/mL human basic fibroblast growth factor (Sigma-Aldrich), 10 ng/mL human epidermal growth factor, 10 ng/mL platelet-derived growth factor-BB (both from PeproTech), and 10 ng/mL murine leukemia inhibitory factor (EMD Millipore)17,36. We seeded cells at 15k cells per 48-well and grew them for two days until confluent. They were then differentiated for ten days in growth medium without growth factors and supplemented with 2% FBS and adipogenic induction cocktail (50 μM indomethacin, 0.5 μM insulin, 33 μM biotin, 17 μM pantothenate, 0.1 μM dexamethasone, 2 nM liothyronine, 540 μM isobutylmethylxanthine). Ten days later, we treated the mature adipocytes with 500 μM dibutyryl-cAMP or vehicle for 4 h, and then isolated RNA as described32.
Bioenergetics
Studies were carried as described previously37. We collected adipose tissue samples from individuals during surgery and from C57Bl/6 male mice, 12-14 weeks old. Both sets of tissues were rinsed with unbuffered KHB media containing 111 mM NaCl, 4.7 mM KCl, 2 mM MgSO4, 1.2 mM Na2HPO4, 0.5 mM carnitine, 2.5 mM glucose, and 10 mM sodium pyruvate, cut into pieces (~10 mg), washed extensively, and then each piece placed in a single well of a XF24-well Islet Flux plate (#101174-100; Seahorse Bioscience) and covered with a customized screen that allows for free perfusion while minimizing tissue movement. We added KHB (450 μL) to each well and studied samples in an XF24 extracellular flux analyzer machine using the following protocol: oxygen concentration was measured over time periods of 2 min at 6-min intervals, consisting of a 2-min mixing period and a 4-min waiting period. Basal OCR was measured for each sample in triplicate. Each reported OCR value was an average of five independent pieces per tissue per experiment for mice and 2-3 for individuals.
Statistical Analyses
We analyzed the data using JMP® Pro 9.0.0 software (SAS Institute, Inc., Cary, NC). All P values presented are two-tailed, and values less than 0.05 were considered to indicate statistical significance. Comparison of the gene expression in different anatomical depots was done using the nonparametric Wilcoxon sign-ranks test. We generated principal components using log-transformed gene expression values via the principal components method with diagonals = 1 and did orthogonal rotation using the Varimax method. Values are factor loadings, the correlation coefficient of the relationship between the components produced and the individual adipose tissue genes. R1, R2, and R3 represent the first, second, and third rotated components extracted.
Supplementary Material
Acknowledgments
This work was supported by US National Institutes of Health (NIH) grants DK087317, DK081604, DK046200, DK077097, RR025757, and P30 DK036836 from the National Institute of Diabetes and Digestive and Kidney Diseases, the Clinical Translational Science Award UL1RR025758 to Harvard University and BIDMC from the National Center for Research Resources, Harvard Catalyst ∣ The Harvard Clinical and Translational Science Center (NIH Award #UL1 RR 025758 and financial contributions from Harvard University and its affiliated academic health care centers), as well as the Harvard Stem Cell Institute and Eli Lilly Foundation.
We thank Y.I.. Chen and K.K Kwong for the MRI imaging; K. Chaudhary for his assistance in figure design; M. Mittleman, H. Keenan, and E. Wilker for their biostatistical advice; J. Skupien for his advice on factor analysis; H. Sacks for his insights into the role of brown fat in warming blood; C. Cahill at the Joslin Diabetes Center Microscopy Core for his expertise in histology; the teams of operating room nurses for their assistance in tissue collection; and C.R. Kahn for his input in preparing this manuscript. Finally, we are grateful to our volunteers for their commitment to the studies.
Competing Financial Interests: Drs. Cypess and Tseng are recipients of a sponsored research grant from Chugai Pharmaceutical Co., Ltd, through Joslin Diabetes Center.
Footnotes
Author contributions: Experimental design: A.M.C., A.P.W., R.X., Y-H.T.; manuscript writing: A.M.C., A.P.W., C.V., T.J.S., C.A.S., Y-H.T.; immunohistochemistry: T.L.H.; human gene expression profiling: A.M.C., A.P.W., R.X., C.A.S., C. R.-T., L.S.W., C.S., A.T.C., L.N.D., L.H., N.T., A. G., A.R.H., A. G., P.-O. H., M.A.M., M.M.; mouse gene expression profiling: C.A.S., N.T., M.A.M; brown adipogenesis: T.J.S.; bioenergetics: C.V., C.A.S.; all authors contributed to editing the manuscript.
Reference List
- 1.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 2.Richard D, Picard F. Brown fat biology and thermogenesis. Front Biosci. 2011;16:1233–1260. doi: 10.2741/3786. [DOI] [PubMed] [Google Scholar]
- 3.Orava J, et al. Different metabolic responses of human brown adipose tissue to activation by cold and insulin. Cell Metab. 2011;14:272–279. doi: 10.1016/j.cmet.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Wijers SL, Saris WH, Marken Lichtenbelt WD. Individual thermogenic responses to mild cold and overfeeding are closely related. J Clin Endocrinol Metab. 2007;92:4299–4305. doi: 10.1210/jc.2007-1065. [DOI] [PubMed] [Google Scholar]
- 5.van Marken Lichtenbelt WD, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 6.Cypess AM, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saito M, et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cannon B, Nedergaard J. Nonshivering thermogenesis and its adequate measurement in metabolic studies. J Exp Biol. 2011;214:242–253. doi: 10.1242/jeb.050989. [DOI] [PubMed] [Google Scholar]
- 9.Vernochet C, McDonald ME, Farmer SR. Brown adipose tissue: a promising target to combat obesity. Drug News Perspect. 2010;23:409–417. doi: 10.1358/dnp.2010.23.7.1487083. [DOI] [PubMed] [Google Scholar]
- 10.Zingaretti MC, et al. The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue. FASEB J. 2009;23:3113–3120. doi: 10.1096/fj.09-133546. [DOI] [PubMed] [Google Scholar]
- 11.Svensson PA, et al. Gene expression in human brown adipose tissue. Int J Mol Med. 2011;27:227–232. doi: 10.3892/ijmm.2010.566. [DOI] [PubMed] [Google Scholar]
- 12.Cinti S. Transdifferentiation properties of adipocytes in the Adipose Organ. Am J Physiol Endocrinol Metab. 2009;297:E977–986. doi: 10.1152/ajpendo.00183.2009. [DOI] [PubMed] [Google Scholar]
- 13.Morrison SF. 2010 Carl Ludwig Distinguished Lectureship of the APS Neural Control and Autonomic Regulation Section: Central neural pathways for thermoregulatory cold defense. J Appl Physiol. 2011;110:1137–1149. doi: 10.1152/japplphysiol.01227.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith RE. Thermoregulatory and adaptive behavior of brown adipose tissue. Science. 1964;146:1686–1689. doi: 10.1126/science.146.3652.1686. [DOI] [PubMed] [Google Scholar]
- 15.Timmons JA, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci U S A. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulz TJ, et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci U S A. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrovic N, et al. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J Biol Chem. 2010;285:7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seale P, et al. Transcriptional Control of Brown Fat Determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walden TB, Petrovic N, Nedergaard J. PPARalpha does not suppress muscle-associated gene expression in brown adipocytes but does influence expression of factors that fingerprint the brown adipocyte. Biochem Biophys Res Commun. 2010;397:146–151. doi: 10.1016/j.bbrc.2010.05.053. [DOI] [PubMed] [Google Scholar]
- 22.Walden TB, Hansen IR, Timmons JA, Cannon B, Nedergaard J. Recruited vs. nonrecruited molecular signatures of brown, “brite,” and white adipose tissues. Am J Physiol Endocrinol Metab. 2012;302:E19–E31. doi: 10.1152/ajpendo.00249.2011. [DOI] [PubMed] [Google Scholar]
- 23.Heaton JM. The distribution of brown adipose tissue in the human. J Anat. 1972;112:35–39. [PMC free article] [PubMed] [Google Scholar]
- 24.Foster DO, Frydman ML. Nonshivering thermogenesis in the rat. II. Measurements of blood flow with microspheres point to brown adipose tissue as the dominant site of the calorigenesis induced by noradrenaline. Can J Physiol Pharmacol. 1978;56:110–122. doi: 10.1139/y78-015. [DOI] [PubMed] [Google Scholar]
- 25.Virtanen KA, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 26.Tchkonia T, et al. Identification of depot-specific human fat cell progenitors through distinct expression profiles and developmental gene patterns. Am J Physiol Endocrinol Metab. 2007;292:E298–E307. doi: 10.1152/ajpendo.00202.2006. [DOI] [PubMed] [Google Scholar]
- 27.Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto Y, et al. Adipose depots possess unique developmental gene signatures. Obesity (Silver Spring) 2010;18:872–878. doi: 10.1038/oby.2009.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leonardsson G, et al. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc Natl Acad Sci U S A. 2004;101:8437–8442. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravussin E, Galgani JE. The implication of brown adipose tissue for humans. Annu Rev Nutr. 2011;31:33–47. doi: 10.1146/annurev-nutr-072610-145209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee P, Swarbrick MM, Zhao JT, Ho KK. Inducible brown adipogenesis of supraclavicular fat in adult humans. Endocrinology. 2011;152:3597–3602. doi: 10.1210/en.2011-1349. [DOI] [PubMed] [Google Scholar]
For the Online Methods
- 32.Tseng YH, et al. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454:1000–1004. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pisania A, et al. Quantitative analysis of cell composition and purity of human pancreatic islet preparations. Lab Invest. 2010;90:1661–1675. doi: 10.1038/labinvest.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 35.Tchkonia T, et al. Fat depot-specific characteristics are retained in strains derived from single human preadipocytes. Diabetes. 2006;55:2571–2578. doi: 10.2337/db06-0540. [DOI] [PubMed] [Google Scholar]
- 36.Steenhuis R, Pettway GJ, Ignelzi MA. Cell surface expression of stem cell antigen-1 (Sca-1) distinguishes osteo-, chondro-, and adipoprogenitors in fetal mouse calvaria. Calcif Tissue Int. 2008;82:44–56. doi: 10.1007/s00223-007-9083-4. [DOI] [PubMed] [Google Scholar]
- 37.Kiefer FW, et al. Retinaldehyde dehydrogenase 1 regulates a thermogenic program in white adipose tissue. Nat Med. 2012;18:918–925. doi: 10.1038/nm.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


