Abstract
An evaluation of 70 accessions of ash gourd germplasm grown at National Bureau of Plant Genetic Resources, New Delhi, India during Kharif season (2010) showed natural occurrence of a yellow stunt disease in three accessions (IC554690, IC036330 and Pusa Ujjwal). A set of begomovirus specific primers used in PCR gave expected amplicon from all the symptomatic plants; however no betasatellite was detected. Complete genome of the begomovirus (DNA-A and DNA-B), amplified through rolling circle amplification, was cloned and sequenced. The begomovirus under study shared high sequence identities to different isolates of Tomato leaf curl New Delhi virus (ToLCNDV) and clustered with them. Among those isolates, the DNA-A and DNA-B of the present begomovirus isolate showed highest 99.6 and 96.8 % sequence identities, respectively with an isolate reported on pumpkin from India (DNA-A: AM286433, DNA-B: AM286435). Based on the sequence analysis, the begomovirus obtained from ash gourd was considered as an isolate of ToLCNDV. Thus, the present findings constitute the first report of occurrence of a new yellow stunt disease in ash gourd from India and demonstrated the association of ToLCNDV with the symptomatic samples. Occurrence of ToLCNDV in ash gourd germplasm not only adds up a new cucurbitaceous host of this virus but also raises the concern about the perpetuation of this virus in absence of its main host tomato and thus has an epidemiological relevance for understanding the rapid spread of this virus in tomato and other hosts in Indian sub-continent.
Keywords: Benincasa hispida, Begomovirus, Tomato leaf curl New Delhi virus, Rolling circle amplification
Ash gourd (Benincasa hispida), also known as wax gourd, white gourd or winter melon; the only member of the genus Benincasa belonging to family Cucurbitaceae, is an important vegetable crop in India. In northern India and Pakistan, the vegetable is widely used to prepare a popular candy called Petha. In Ayurvedic remedies, it is believed to increase appetite and its fresh juice is used to cure kidney stones.
Begomoviruses (family Geminiviridae) are characterized by their tween-quasiisometric virion of 20 × 30 nm and are exclusively transmitted by whitefly (Bemisia tabaci) in a persistent manner. Genome of begomoviruses is either bipartite i.e. consisted of two circular single stranded DNA molecules designated as DNA-A and DNA-B or monopartite having only one component homologous to that of DNA-A of bipartite begomoviruses. In India, first begomovirus disease of cucurbits was recorded as yellow vein mosaic disease of pumpkin during 1955. Later, it was established that Squash leaf curl China virus was responsible for the disease [9]. Begomovirus disease problem in cucurbits emerged in India during 1980s onward. Only two begomovirus species, Squash leaf curl China virus (SLCCNV) and Tomato leaf curl New Delhi virus (ToLCNDV) are known to affect different cucurbits in India [8].
In case of ash gourd, a yellow leaf disease was reported from Thailand and partial characterization of the associated virus indicated presence of a begomovirus with the disease [13]. However, the exact identity of the begomovirus associated with such disease was not confirmed due to unavailability of full length sequence of the begomovirus, which is essential for nomenclature of any begomovirus. Another begomovirus associated disease of ash gourd was reported from central Thailand where the infected plants exhibited yellow leaf curl symptom [14]. An isolate of SLCCNV was reported to be associated with such disease. Prior to this report, neither any begomovirus associated disease nor was any begomovirus reported from ash gourd in India.
In the evaluation experiment on 70 accessions of ash gourd germplasm, during Kharif season (2010) at National Bureau of Plant Genetic Resources, New Delhi, India, seven plants of three accessions (IC554690, IC036330 and Pusa Ujjwal) showed a yellow stunt disease. The leaves of the diseased plants became small, rough and brittle and exhibited yellowing, puckering and vein thickening symptoms (Fig. 1a). Overall the infected plants appeared severely stunted. Considerable population of whiteflies (Bemisia tabaci) was noticed on the field, however, the number of adult whiteflies was more (average 30–50 per plant) in the symptomatic plants than the asymptomatic one (average 5–15 per plant). Based on the symptoms and the association of more whiteflies with the diseased plants, a begomovirus infection was suspected. Therefore, attempt was made to detect the begomovirus through PCR using begomovirus specific primers PAL1v722/PAL1c1960 [2]. The PCR conditions for amplification were denaturation at 94 °C for 3 min, followed by 30 cycles each consists of denaturation for 30 s at 94 °C, annealing for 30 s at 52 °C and synthesis for 80 s at 72 °C with final extension for 10 min at 72 °C. All the seven symptomatic samples yielded an expected 1.2 kb amplicon (Fig. 1b), whereas no such amplification was obtained from asymptomatic plants, thus, indicating the association of a begomovirus with symptomatic samples. To detect the presence of betasatellite, amplification was carried out with universal betasatellite primers β01/β02 [1], however, the reaction did not showed any positive result even with repeated attempts.
Fig. 1.
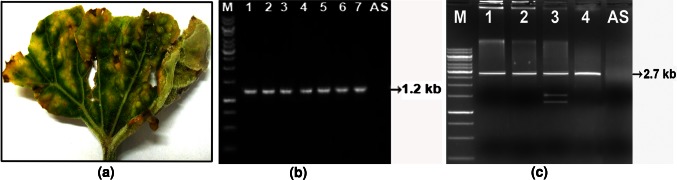
a Symptom of yellowing, puckering and vein thickening in ash gourd leaf. b Detection of begomovirus through PCR. Lane 1–7: symptomatic sample yielded a 1.2 kb amplicon which was not present in asymptomatic samples. c RCA product digested with BglI (lane 1), EcoRV (lane 2), PstI (lane 3) and XbaI. M 1 kb molecular weight marker, AS asymptomatic sample. (Color figure online)
Full length genome of the begomovirus was isolated through rolling circle amplification (RCA) using ϕ29 DNA polymerase (Thermo Scientific, USA) following the standard protocol [15]. The RCA products digested with BglI, EcoRV and XbaI produced c.a. 2.7 kb fragments corresponding to the unit genome-length of begomovirus, while PstI enzyme produced a c.a. 2.7 kb fragment along with one c.a. 1.5 kb and one c.a. 1.2 kb fragments (Fig. 1c). The 2.7 kb fragments generated separately by PstI (PstAG) and XbaI (XbaAG) were purified using Wizard SV Gel and PCR Clean-up System (Promega Corporation, Madison, WI, USA) and ligated to PstI and XbaI—linearized pUC18 vector, respectively. The ligation mixture was used to transform Escherichia coli strain DH5α. Vectors containing inserts of the expected size were identified in miniprep screening by size estimation. The sequences of PstAG (2,738 nucleotides) and XbaAG (2,694 nucleotides) were obtained in forward and reverse directions from commercial facility (Chromous Biotech, India) and the sequences were deposited in the nucleotide sequence database with the accession numbers JN208136 and JN208137, respectively. The sequences of PstAG and XbaAG were initially searched for similarity using BLASTn Program (http://www.ncbi.nlm.nih.gov/BLAST) and the sequences which showed high scores were selected for further analysis. The genome organization was determined by analyzing those sequences using ORF Finder (www.ncbi.nlm.nih.gov/gorf/gorf.html). Sequence identity matrix was generated using Bioedit Sequence Alignment Editor (version 5.0.9) [4]. After multiple alignment, phylogenetic analysis was done in MEGA 4.0 software [19] using the default parameters of Maximum Parsimony, and the Bootstrapped consensus dendrogram was generated with 1,000 replication (Fig. 2).
Fig. 2.
Phylogenetic analysis of the DNA-A (a) and DNA-B (b) of ToLCNDV associated with yellow stunt disease of ash gourd, showing distinct relationships to isolates of ToLCNDV. ToLCBV-Tomato leaf curl Bangalore virus, ToLCKeV-Tomato leaf curl Kerala virus, ToLCPNV-Tomato leaf curl Pune virus, ToLCJoV-Tomato leaf curl Joydebpur virus, ToLCRV-Tomato leaf curl Ranchi virus, ToLCGV-Tomato leaf curl Gujarat virus, ToLCKV-Tomato leaf curl Karnataka virus. The Maximum Parsimonious tree was constructed using MEGA 4.0 software. Percent bootstrap values are indicated in the node
Sequences of PstAG and XbaAG showed >90 and >86 % sequence identities with DNA-A and DNA-B sequences of different isolates of ToLCNDV, respectively. Among the different isolates of ToLCNDV, the DNA-A (PstAG) and DNA-B (XbaAG) of the present isolate showed highest 99.6 and 96.8 % sequence identity with that of pumpkin isolate (DNA-A: AM286433, DNA-B: AM286435) reported from New Delhi. A total of 33 begomovirus isolates were taken for phylogenetic relationship study which revealed that the present isolate formed cluster with different isolates of ToLCNDV with respect to both DNA-A and DNA-B sequences. Based on sequence analysis, it is evident that the present isolate of begomovirus, associated with yellow stunt disease of ash gourd, is an isolate of ToLCNDV and hence, the descriptor for this isolate is proposed as ToLCNDV-[India:New Delhi:Ash gourd:2011] (ToLCNDV-[IN:ND:Ash:11]).
The genome organization of the DNA-A and DNA-B of ToLCNDV-[IN:ND:Ash:11] was similar to that of all other Old World bipartite begomoviruses. DNA-A has four open reading frames (ORFs) on the complementary strand (AC1; 1,499–2,584, AC2; 1,177–1,596, AC3; 1,047–1,457, and AC4; 2,251–2,427) and two ORFs on the viral sense strand (AV1; 280–1,053 and AV2; 120–458). The AV1 (CP) ORF was 774 nt long encoding 28.6 kDa coat protein. AV2 was 339 nt long encoding 12.5 kDa protein. The AC1 was 1,086 nt long encoding 40.2 kDa replication associated protein (Rep). AC2 was 420 nt long encoding (15.5 kDa) transcription activator protein. The AC3 was 411 nt long encoding 15.2 kDa replication enhancer protein. The AC4 was 177 nt long encoding 6.5 kDa protein. DNA-B contains two ORFs, one on the complementary strand (BC1; 1,306–2,151) and the other on sense strand (BV1; 442–1,248) coding for a movement protein (31.3 kDa) and a nuclear shuttle protein (29.8 kDa), respectively.
ToLCNDV is an emerging problem in various agricultural crops in India, Pakistan, Bangladesh and Thailand [8]. It was reported for the first time in India from tomato and further its infectivity study demonstrated that both DNA-A and DNA-B are essential for symptom development [10]. ToLCNDV is known to infect tomato in India since nearly two decades, but during the last one decade, its host range has increased enormously to various crops such as potato [21], papaya [12], eggplant [11] okra [23] and several cucurbitaceous vegetables like bottle gourd, bitter gourd, cucumber, ivy gourd, long melon, pumpkin, ridge gourd and watermelon in northern India and chayote in north-western India [7, 16, 17, 20]. In Pakistan, besides tomato, ToLCNDV was also reported on crops like chilli [5], bitter gourd [18], and on weed Eclipta prostrata [3]. In Thailand, ToLCNDV has been reported to infect bottle gourd, cucumber and muskmelon [6]. The frequency of new strains of ToLCNDV emerging in several agricultural crops and non-crop species indicated that the virus species has diverse virulent strains in the field and thus pose a serious threat to vegetable cultivation particularly in Indian sub-continent.
In the present study, we report the occurrence of a severe yellow stunt disease in ash gourd germplasm from India and documented complete sequence of an isolate of ToLCNDV associated with such disease of ash gourd for the first time. The use of such susceptible germplasm accessions in breeding programme is a serious matter of concern as it may lead to introduction of susceptible genes into cultivated varieties and such event has already been reported in many other crops [22]. Occurrence of ToLCNDV in cucurbitaceous crops was not recorded before 1980s. Presently ToLCNDV is a serious concern for most of the cucurbitaceous vegetable crops. These crops are mainly grown during Kharif season and their harvesting time often coincides with the sowing time of tomato in northern India. Thus on the epidemiological point of view the natural occurrence of ToLCNDV in newer cucurbitaceous host strongly supports its perpetuation in absence of main host and its subsequent spread to tomato.
Acknowledgments
The authors wish to thank Director, National Bureau of Plant Genetic Resources, New Delhi, India for providing necessary facilities to carry out the study.
References
- 1.Briddon RW, Bull SE, Mansoor S, Amin I, Markham PG. Universal primers for the PCR-mediated amplification of DNA β molecule associated with some monopartite begomoviruses. Mol Biotechnol. 2002;20:315–318. doi: 10.1385/MB:20:3:315. [DOI] [PubMed] [Google Scholar]
- 2.Chowda Reddy RV, Colvin J, Muniyappa V, Seal S. Diversity and distribution of begomoviruses infecting tomato in India. Arch Virol. 2005;150:845–867. doi: 10.1007/s00705-004-0486-5. [DOI] [PubMed] [Google Scholar]
- 3.Haider MS, Tahir M, Latif S, Briddon RW. First report of Tomato leaf curl New Delhi virus infecting Eclipta prostrata in Pakistan. Plant Pathol. 2006;55(2):285. doi: 10.1111/j.1365-3059.2005.01278.x. [DOI] [Google Scholar]
- 4.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 5.Hussain M, Mansoor S, Iram S, Zafar Y, Briddon RW. First report of Tomato leaf curl New Delhi virus affecting chilli pepper in Pakistan. Plant Pathol. 2004;53:794–795. doi: 10.1111/j.1365-3059.2004.01073.x. [DOI] [Google Scholar]
- 6.Ito T, Sharma P, Kittipakorn K, Ikegami M. Complete nucleotide sequence of a new isolate of Tomato leaf curl New Delhi virus infecting cucumber, bottle gourd and muskmelon in Thailand. Arch Virol. 2008;153:611–613. doi: 10.1007/s00705-007-0029-y. [DOI] [PubMed] [Google Scholar]
- 7.Mandal B, Mandal S, Sohrab SS, Pun KB, Varma A. A new yellow mosaic disease of chayote in India. Plant Pathol. 2004;53:797. doi: 10.1111/j.1365-3059.2004.01075.x. [DOI] [Google Scholar]
- 8.Mandal B. Emerging geminiviral diseases and their management. In: Sharma P, Gaur Rajarshi K, Ikegami M, editors. Emergence of begomovirus diseases in cucurbits in India. New York: Nova Science Publishers Inc; 2010. pp. 167–181. [Google Scholar]
- 9.Muniyappa V, Maruthi MN, Babitha CR, Colvin J, Briddon RW, Rangaswamy KT. Characterization of Pumpkin yellow vein mosaic virus from India. Ann Appl Biol. 2003;142:323–331. doi: 10.1111/j.1744-7348.2003.tb00257.x. [DOI] [Google Scholar]
- 10.Padidam M, Beachy RN, Fauquet CM. Tomato leaf curl geminivirus from India has a bipartite genome and coat protein is not essential for infectivity. J Gen Virol. 1995;76(Pt 1):25–35. doi: 10.1099/0022-1317-76-1-25. [DOI] [PubMed] [Google Scholar]
- 11.Pratap D, Kashikar AR, Mukherjee SK. Molecular characterization and infectivity of a Tomato leaf curl New Delhi virus variant associated with newly emerging yellow mosaic disease of eggplant in India. Virol J. 2011;8:305. doi: 10.1186/1743-422X-8-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raj SK, Snehi SK, Khan MS, Singh R, Khan AA. Molecular evidence for association of Tomato leaf curl New Delhi virus with leaf curl disease of papaya (Carica papaya L.) in India. Australas Plant Dis Notes. 2008;3:152–155. doi: 10.1071/DN08059. [DOI] [Google Scholar]
- 13.Samretwanich K, Chiemsombat P, Kittipakorn K, Ikegami M. Yellow leaf disease of cantaloupe and wax gourd from Thailand caused by Tomato leaf curl virus. Plant Dis. 2000;84:200. doi: 10.1094/PDIS.2000.84.2.200C. [DOI] [PubMed] [Google Scholar]
- 14.Sawangjit S. The complete nucleotide sequence of Squash leaf curl China virus-[wax gourd] and its phylogenetic relationship to other geminiviruses. Sci Asia. 2009;35:131–136. doi: 10.2306/scienceasia1513-1874.2009.35.131. [DOI] [Google Scholar]
- 15.Singh MK, Haq QMR Singh K, Mandal B, Varma A. Molecular characterization of Tobacco leaf curl Pusa virus, a new monopartite begomovirus associated with Tobacco leaf curl disease in India. Virus Genes. 2011;43:296–306. doi: 10.1007/s11262-011-0631-7. [DOI] [PubMed] [Google Scholar]
- 16.Sohrab SS, Mandal B, Ali A, Varma A. Chlorotic curly stunt: a severe begomovirus disease of bottle gourd in northern India. Indian J Virol. 2010;21:56–63. doi: 10.1007/s13337-010-0002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sohrab SS, Mandal B, Ali A, Varma A. Molecular diagnosis of emerging begomovirus diseases in cucurbits occurring in northern India. Indian J Virol. 2006;17:88–95. [Google Scholar]
- 18.Tahir M, Haider MS. First report of Tomato leaf curl New Delhi virus infecting bitter gourd in Pakistan. Plant Pathol. 2005;54:807–808. doi: 10.1111/j.1365-3059.2005.01215.x. [DOI] [Google Scholar]
- 19.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 20.Tiwari AK, Sharma PK, Khan MS, Snehi SK, Raj SK, Rao GP. Molecular detection and identification of tomato leaf curl New Delhi virus isolate causing yellow mosaic disease in bitter gourd (Momordica charantia), a medicinally important plant in India. Med Plants. 2010;2:117–123. [Google Scholar]
- 21.Usharani KS, Surendranath B, Paul-Khurana SM, Garg ID, Malathi VG. Potato leaf curl-a new disease of potato in northern India caused by a strain of Tomato leaf curl New Delhi virus. Plant Pathol. 2004;53:235–236. doi: 10.1111/j.0032-0862.2004.00959.x. [DOI] [Google Scholar]
- 22.Varma A, Mandal B, Singh MK. Global emergence and spread of whitefly (Bemisia tabaci) transmitted geminiviruses. In: Thompson WMO, editor. The whitefly, Bemisia tabaci (homoptera: aleyrodidae) interaction with geminivirus-infected host plants. 1. India: Springer link; 2011. pp. 205–292. [Google Scholar]
- 23.Venkataravanappa V, Lakshminarayana RCN, Salil J, Krishna RM. Molecular characterization of distinct bipartite begomovirus infecting bhendi (Abelmoschus esculentus L.) in India. Virus Genes. 2012;44(3):522–535. doi: 10.1007/s11262-012-0732-y. [DOI] [PubMed] [Google Scholar]



