Transcriptional coregulators play key roles in the epigenetic regulation of gene expression. This review highlights recent in vivo studies that reveal wide-ranging roles for the corepressors NCoR1 and SMRT in developmental and homeostatic processes, including metabolism, inflammation, and circadian rhythms. The authors discuss the emerging roles of NCoR1 and SMRT in disease pathways spanning from genomic stability and cancer to metabolic diseases such as type 2 diabetes.
Keywords: transcription factor, cofactor, development, epigenetics, metabolism
Abstract
Epigenetic regulation of gene expression is strongly influenced by the accessibility of nucleosomal DNA or the state of chromatin compaction. In this context, coregulators, including both coactivators and corepressors, are pivotal intermediates that bridge chromatin-modifying enzymes and transcription factors. NCoR1 (nuclear receptor corepressor) and SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) are among the best-characterized corepressors from a molecular point of view. These coregulators have conserved orthologs in lower organisms, which underscores their functional importance. Here we summarize the results from recent in vivo studies that reveal the wide-ranging roles of NCoR1 and SMRT in developmental as well as homeostatic processes, including metabolism, inflammation, and circadian rhythms. We also discuss the potential implications of NCoR1 and SMRT regulation of pathways ranging from genomic stability and carcinogenesis to metabolic diseases such as type 2 diabetes.
Epigenetic regulation by coregulators
Chromatin plays a central role in determining the transcriptional activity of genes. Epigenetic modifications, such as DNA methylation and histone modifications, directly modulate chromatin compaction and thus the accessibility of coding regions to the transcriptional machinery (Gardner et al. 2011). One of the best-characterized types of histone tail modifications is acetylation, which, like histone methylation, phosphorylation, ADP-ribosylation, ubiquitination, and sumoylation, impacts chromatin compaction and function. It is commonly accepted that acetylation of lysine residues on histones generally correlates with a decondensed and more accessible state of chromatin, resulting in transcriptional activation of associated genes. Therefore, histone acetyltransferases (HATs) usually stimulate gene transcription, whereas histone deacetylases (HDACs) repress gene expression (Fig. 1A).
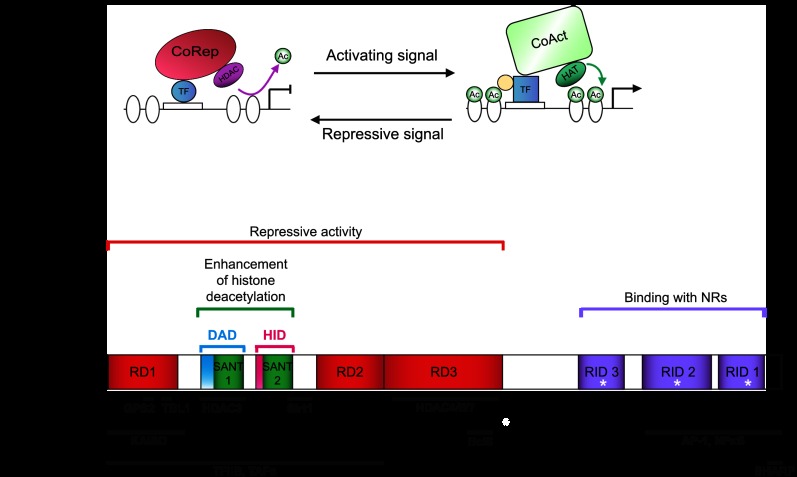
Figure 1.
(A) Illustration of corepressor/coactivator exchange. Corepressor (e.g., NCoR1/SMRT; CoRep)/coactivator (CoAct) exchange results in the differential recruitment of enzymes acting on histone (white ovals) acetylation (Ac). The transcription factor in its repressive form induces a locked chromatin state through interaction with CoReps and recruitment of HDACs. The CoActs and HATs are docked when the transcription factor is active due to a conformational change induced by post-transcriptional modifications or ligand binding (e.g., NRs), schematized by a yellow dot. (B) Scheme of the functional domains of SMRT and NCoR1. Different domains are similarly arranged and conserved in both corepressors. The regions interacting with some cofactors (first row) and transcription factors (second row) are indicated. The white star represents the CoRNR box, a leucine-rich, helical motif responsible for the interaction with NRs. Based on the work of Glass and Rosenfeld (2000), Hu and Lazar (2000), Jepsen and Rosenfeld (2002), Yu et al. (2003), and Watson et al. (2012b). See the text for detailed description.
Transcriptional coregulators, or cofactors, are the main actors in the plot of epigenetic regulation (Perissi et al. 2010), as they recruit chromatin-modifying enzymes. Coregulators interact with transcription factors that bind in a sequence-specific way to the regulatory regions of genes. Thus, they can be considered as intermediates between transcription factors and chromatin-modifying enzymes, determining the final transcriptional output. Indeed, the same transcription factors can have opposite effects, depending on the nature of the coregulator to which they are bound, with coactivators in general stimulating and corepressors inhibiting transcription. Generally, the switch from corepressor to coactivator binding goes hand in hand with activation of the transcription factor. A good illustration of this concept is the fact that nuclear receptors (NRs), upon ligand binding, release their corepressors in order to recruit coactivators (Glass and Rosenfeld 2000). Coregulators often act as a docking platform or scaffold for the binding of additional regulatory proteins, which is exemplified by the fact that corepressors and coactivators recruit HDACs and HATs, respectively.
The present review is focused on the role of the NR corepressor (NCoR or NCoR1) and the silencing mediator of retinoic acid and thyroid hormone receptor (SMRT, also known as NCoR2), in physiology and homeostasis. For a more detailed discussion of how NCoR1/SMRT mechanistically regulate transcription, we refer the interested reader to some recent reviews (Perissi et al. 2010; Watson et al. 2012b).
NCoR1 and SMRT corepressors—gene and protein
NCoR1 and SMRT were both discovered in a quest to explain repression by NRs (Chen and Evans 1995; Horlein et al. 1995), as they were found to bind thyroid hormone receptor (TR) and retinoic acid receptor (RAR) and inhibit their respective target genes. Although described as prototypical NR corepressors, later studies showed that they could also associate with non-NR transcription factors (Jepsen and Rosenfeld 2002), such as FOXP1 (Jepsen et al. 2008).
HDAC3 appears to be the main enzyme responsible for the repressive activity of SMRT and NCoR1 because it is the protein that associates in the most stable and reproducible way with both corepressors (Perissi et al. 2010). Indeed, in the context of repression by TR, the recruitment of HDAC3 is essential for repression (Ishizuka and Lazar 2003, 2005). In addition, the catalytic deacetylase activity of HDAC3 requires interaction with NCoR1 or SMRT (Guenther et al. 2001; You et al. 2013). However, NCoR1 and SMRT can also recruit other deacetylases in a context-specific manner, such as HDAC4 (Fischle et al. 2002), HDAC5 and HDAC7 (Kao et al. 2000), Sirt1 (Picard et al. 2004), or HDAC1, through interaction with mSin3 (mammalian Sin3), another corepressor (Heinzel et al. 1997; Nagy et al. 1997). Interestingly, the interaction with class II HDACs, like HDAC4, HDAC5, or HDAC7, enables the corepressors to become nutrient-sensitive, as phosphorylation by AMP-activated kinase (AMPK) controls the nuclear/cytoplasmic shuttling of class II HDACs, contrary to HDAC3 (Mihaylova et al. 2011). In addition to their association with HDACs, NCoR1/SMRT can bind components of the basal transcriptional machinery, such as transcription factor IIB (TFIIB) and template-activating factors (TAFs), potentially blocking them in a nonfunctional conformation, hindering the initiation complex (Muscat et al. 1998; Wong and Privalsky 1998). Although corepressor complexes are heterogeneous, context-specific, and of transient nature, some partners are regularly found in stoichiometric association with NCoR1/SMRT, as they are essential for their repressive function. In addition to HDAC3, these partners include the proteins G protein pathway suppressor (GPS2) and transducing β-like 1 (TBL1) and its homolog, TBL-related 1 (TBLR1), which all together form the core repression complex (Oberoi et al. 2011). In addition to binding with repressive transcription factors, NCoR1/SMRT can also recruit SMRT/HDAC1-associated repressor protein (SHARP), a repressor protein that can bind RNA and also contains motifs that are conserved in corepressors, such as the receptor interaction domain (RID) motif (Shi et al. 2001; Ariyoshi and Schwabe 2003; Mikami et al. 2012; see below).
The genes for NCoR1 and SMRT map to chromosomes 17 and 12 (11 and 5 in mice), respectively, and contain multiple exons. Alternative splicing events generate several mRNAs for each corepressor, which in turn translate into distinct proteins that contain different RIDs or N-terminal-interacting regions (Fig. 1B; Seol et al. 1996; Goodson et al. 2005; see below). Hence, this repertoire of corepressor isoforms constitutes an additional level of regulation for NCoR1/SMRT, which changes in a species-, context-, or tissue-dependent manner (Goodson et al. 2011).
Both Ncor1 and Ncor2 encode extremely large proteins composed of ∼2500 amino acids (molecular mass of ∼270 kDa), a characteristic that facilitates protein interactions and makes working with them challenging. The homology between the two corepressors is evidenced by the high level of overall amino acid sequence identity (∼40%) (Ordentlich et al. 1999; Park et al. 1999). Therefore, it is no surprise that NCoR1 and SMRT share a similar overall structure containing conserved functional domains. Both proteins are rather unstructured in nature, as few regions are intrinsically folded. This is typical for proteins that act as a hub or platform for interactions with multiple partners (Watson et al. 2012b). The functional domains of NCoR1 and SMRT are composed of (Fig. 1B):
The repression domains (RDs)—These three domains, which retain high homology between NCoR1 and SMRT, are responsible for the repressive activity of the corepressors (Li et al. 1997). They contain highly conserved sequences for interaction with members of the core repression complex as well as with HDAC4 or Sirt1.
The SANT-like domains (the name coming from their presence in Swi3, Ada2, NCoR1, and TFIIB)—These synergistically promote histone deacetylation. One of the SANT domains is part of the deacetylase activation domain (DAD), the region of the corepressor responsible for recruitment and activation of HDAC3 (Guenther et al. 2001). The other SANT domain, the histone interaction domain (HID), directly binds histone tails, preferentially when they are deacetylated, and enhances repression (Yu et al. 2003).
The RIDs—These three domains are able to bind to NRs (Downes et al. 1996; Li et al. 1997; Webb et al. 2000; Cohen et al. 2001) mainly by interacting with their ligand-binding domains (LBDs) in the absence of ligands. RIDs contain an isoleucine-rich domain, named the CoRNR box. Generally, the RIDs of the corepressors bind the same region of the NRs as coactivators do, meaning that their respective binding to the NR is mutually exclusive.
Besides these well-characterized domains, other parts of the corepressors mediate the interaction with transcription factors.
Phylogenetic studies reveal that NCoR1 and SMRT proteins are conserved throughout evolution. NCoR1 and SMRT homologs have been identified in Danio rerio, Xenopus laevis, Drosophila melanogaster, and Caenorhabditis elegans, displaying conserved SANT domains (see below). In addition, the SET3 complex in Saccharomyces cerevisiae has been identified as a functional homolog of the NCoR1/SMRT complex (Pijnappel et al. 2001). Indeed, it contains a SANT domain protein called Snt1, which binds the closest homolog of HDAC3, named Hos2, and Sif2, the homolog of TBL1. The SET3 complex inhibits the sporulation gene program by repressing meiosis.
Physiological role of NCoR1 and SMRT
Lower species and germline knockout mouse models illustrate a role in development
Clues about the in vivo functions of NCoR1/SMRT first pointed toward their important role in development. The characterization of SMRTER, the Drosophila NCoR1/SMRT homolog, demonstrated the evolutionarily conserved mode of action for these corepressors (Tsai et al. 1999). SMRTER possesses a domain repetition similar to NCoR1 and SMRT and binds the ecdysone receptor (EcR), a NR that responds to ecdysone hormone fluctuations to regulate molting. The Drosophila homolog also mediates repression by recruitment of the dSin3 complex and associated HDACs. Developmental defects and lethality arise when the interaction between SMRTER and EcR is disrupted. In addition, the SMRTER/Ebi (homolog of TBL1) complex was discovered to mediate the cross-talk between the epidermal growth factor receptor (EGFR) and Notch signaling pathways during eye development, which determines the differentiation of photoreceptor cells (Tsuda et al. 2006). The findings in these fly studies mirrored the central role of SMRT/HDAC1 in the mammalian Notch pathway, where Notch activation frees the Notch effector CBF1/RBP-Jκ {the mammalian homolog of Drosophila suppressor of Hairless [Su(H)]} from the repression exerted by the corepressor complex (Kao et al. 1998).
In mammals, NCoR1 and SMRT seem to have nonredundant roles because mice with germline mutations of either gene are embryonic-lethal. Characterization of NCoR1−/− mice highlighted its role in CNS development (Jepsen et al. 2000). Impaired erythropoiesis and a developmental arrest at the double-negative stage of T lymphocytes are also a consequence. These defects were mainly attributed to the disruption of RAR and TR signaling and resulted in the death of most embryos at embryonic day 15.5 (E15.5). SMRT also appeared critical for forebrain development by regulating RAR-dependent differentiation of neural stem cells (Jepsen et al. 2007) as well as for heart growth by interacting with a forkhead family member, FoxP1 (Jepsen et al. 2008). The lethality of the SMRT−/− mouse before E16.5 was caused by heart defects.
The NCoR1/SMRT association with RAR was found also to be crucial for proper development in other nonmammalian organisms. In X. laevis, RAR repression by SMRT is necessary for head development (Koide et al. 2001). Moreover, whereas the lethality of NCoR1−/−/SMRT−/− mouse models underscored their important role in mid-embryogenesis, early embryogenesis is also regulated by the interaction of these corepressors with RAR, as evidenced by work in X. laevis and D. rerio (Koide et al. 2001; Xu et al. 2009; Linney et al. 2011). Both NCoR1 and SMRT are expressed in the early zebrafish embryo, where they repress retinoid signaling. NCoR1 was shown to be crucial for the early patterning of the anterior–posterior axis of the zebrafish hindbrain, since interfering with NCoR1 mimics the developmental defects seen with abnormal RA levels (Xu et al. 2009). Also, SMRT is present as early as the eight-cell stage of zebrafish embryogenesis, which could suggest that it keeps important genes repressed from the time of fertilization until RA appears (Linney et al. 2011). Recently, GEI-8, the C. elegans homolog of NCoR1/SMRT, was also shown to control worm development and homeostasis. Loss of function of gei-8 results in developmental defects and impairs growth, movement, gonadogenesis, cholinergic neurotransmission, and mitochondrial metabolism (Yamamoto et al. 2011; Mikolas et al. 2013).
In conclusion, the requirement of NCoR1/SMRT repression during development seems to be conserved, but they may operate at different stages of embryogenesis or larval development depending on the species.
Domain- and tissue-specific mouse models reveal a role in homeostasis
In this section, we describe results obtained with domain- and tissue-specific gain-of-function or loss-of-function mutants that circumvent the lethality of germline NCoR1−/−/SMRT−/− mouse models and help to precisely define their functions in homeostasis (Table 1).
Table 1.
Summary of the different mouse models discussed in this review
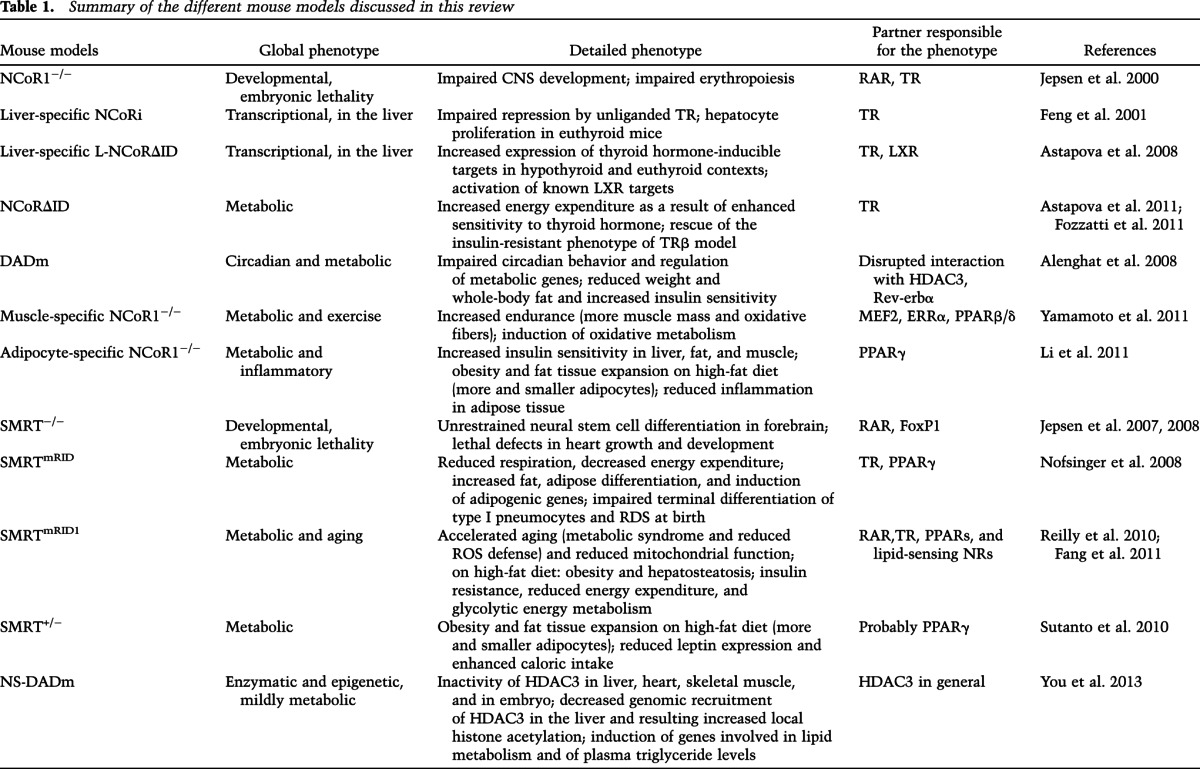
Liver-specific NCoRi, L-NCoRΔID, and NCoRΔID mice
In combination, these three mouse models allowed researchers to establish how NCoR1 is implicated in thyroid hormone signaling. Thyroid hormone is important in growth, differentiation, and development as well as metabolic homeostasis (Song et al. 2011). Thyroid hormone modulates serum lipid profiles and enhances fat oxidation in the liver. Thyroid hormone also favors mitochondrial uncoupling and biogenesis and regulates glucose metabolism, as low levels of, or resistance to, thyroid hormone correlate with insulin resistance. These effects of thyroid hormone on lipid and glucose metabolism occur mainly through its impact on the liver, while in fat tissue, thyroid hormone reduces lipid storage and favors fat oxidation.
NCoRi stands for a version of NCoR1 in which the RDs are deleted, meaning that binding of NCoRi to TR is “sterile” because no repression will ensue in the absence of the RDs. A liver-specific knock-in of NCoRi blocks the basal repression of endogenous TR targets by competing with endogenous NCoR1 for binding to TR, providing in vivo evidence that NCoR1 mediates the repression by unliganded TR (Feng et al. 2001). Furthermore, the increase in hepatocyte proliferation in NCoRi mice indicated possible implications of NCoR1 in the control of cell proliferation.
L-NCoR1ΔID mice have a liver-specific deletion of the two RIDs that are essential for the interaction of NCoR1 with TR (Astapova et al. 2008). In both the absence and the presence of thyroid hormone, expression of thyroid hormone-inducible targets is increased, which confirms the important contribution of NCoR1 to repress the nonliganded TR. Moreover, it introduces the concept that NCoR1 controls the amplitude of the response to thyroid hormone levels and determines individual or tissue-specific responses to similar levels of thyroid hormone. In parallel, studies in this mouse model also demonstrate that NCoR1 participates in liver X receptor (LXR) signaling, as the expression of LXR targets was induced in the liver of L-NCoR1ΔID mice.
In NCoR1ΔID mice, the generalized abrogation of the interaction between NCoR1 and TR increased the sensitivity of multiple tissues to thyroid hormone, typified by increased energy expenditure (Astapova et al. 2011). Crossing of these mice with TRβ mutant mice, a model of thyroid hormone resistance, rescued the resistant phenotype, demonstrating the role of NCoR1 in central and peripheral thyroid hormone resistance in vivo (Fozzatti et al. 2011).
DADm mice
A whole-body mutation in the DAD domain of NCoR1 was shown to impede binding and activation of HDAC3, resulting in impaired regulation of clock genes and circadian behavior (Alenghat et al. 2008). The involvement of NCoR1 in diurnal regulation was in line with the fact that the repression of the Bmal1 component of the clock is mediated by the Rev-erbα/NCoR1 complex (Yin and Lazar 2005; Yin et al. 2007; Cho et al. 2012; Solt et al. 2012). Rhythmicity in mammals is patterned by the regulation of the clock genes by two negative interlocked feedback loops. Rev-erbα is involved in one of these loops—Bmal1 activates the expression of Rev-erbα, which in turn represses Bmal1 (Yin et al. 2010). The metabolic phenotype of the DADm mice is characterized by an altered oscillatory pattern of metabolic genes, lower body weight, decreased whole-body fat mass, and insulin sensitization (Alenghat et al. 2008). Moreover, this mouse model also revealed a role for NCoR1 in the induction of thyroid hormone-mediated autophagy in the liver (Sinha et al. 2012).
Muscle-specific NCoR1−/−
NCoR1-specific deletion in the muscle enhanced exercise endurance, a consequence of both increased muscle mass and a shift to more oxidative muscle fibers (Yamamoto et al. 2011), which contained both more and hyperactive mitochondria. NCoR1 favors the development of larger muscle, possibly through interaction with MEF2, a key myogenic transcription factor. Furthermore, the muscle fiber type shift and the induction of oxidative metabolism are achieved in this model by the absence of NCoR1's repressive interactions with peroxisome proliferator-activated receptors β/δ (PPARβ/δ) and/or ERRα (Yamamoto et al. 2011; Perez-Schindler et al. 2012). Most importantly, this study indicated a dynamic regulation of NCoR1 activity, whose expression was reduced in conditions in which fatty acid oxidation was solicited, such as long-term fasting, high-fat feeding, and endurance exercise. Further explanation about NCoR1 regulation is given below.
Adipocyte-specific NCoR1−/−
The phenotypic features of adipocyte-specific NCoR1−/− mice were mainly ascribed to the derepression of PPARγ, the master regulator of adipocyte differentiation (Li et al. 2011). Consequently, adipocyte-specific NCoR1-deficient mice became more obese when fed a high-fat diet. These mice had a larger quantity of smaller adipocytes than are usually seen in obesity, which is normally characterized by adipocyte hypertrophy. Consistent with the insulin-sensitizing role of PPARγ (Heikkinen et al. 2007), glucose tolerance and insulin sensitivity of liver, muscle, and fat, however, were significantly improved. Adipose-specific NCoR1−/− mice also displayed reduced macrophage infiltration and diminished adipose tissue inflammation. As an additional proof of the derepression of PPARγ, rosiglitazone, a thiazolidinedione PPARγ agonist used to treat type 2 diabetes, was unable to further improve insulin sensitivity in these mice, meaning that the PPARγ transcriptional program was already maximally induced. The assessment of the phosphorylation status of PPARγ at Ser273 constituted a possible mechanism to explain the insulin sensitization of the adipocyte-specific NCoR1−/− mice. Cyclin-dependent kinase 5 (CDK5) was previously shown to phosphorylate PPARγ at this residue, inducing insulin resistance. NCoR1 depletion in adipose tissue enriched the unphosphorylated form of PPARγ, which has insulin-sensitizing actions; as a consequence, the mice were refractory to the activation of CDK5 with TNFα, meaning that NCoR1 directly facilitates phosphorylation of PPARγ by CDK5.
SMRTmRID mice
Homozygous mutations in the RID1 and RID2 of SMRT specifically disrupt its interactions with NRs (Nofsinger et al. 2008). Mainly as a result of derepression of TR, these mutant mice exhibited reduced respiration and energy expenditure and were insulin-resistant with increased hepatic glucose output. These mice were also fatter, as a result of the induction of adipogenic genes, consequent to the derepression of PPARγ. This translated into a lowered adipogenic set point, meaning a reduced threshold for adipose differentiation. In addition, pulmonary development was severely compromised in SMRTmRID mice. These mice die from acute respiratory distress syndrome (RDS) just after birth, resulting from impaired terminal differentiation of type I pneumocytes (Pei et al. 2011). This phenotype is rescued by anti-thyroid drugs, and thus SMRT repression governs the thyroid hormone-dependent differentiation program of pneumocytes I through the transcription factor Klf2.
SMRTmRID1 mice
In these mice, mutations are restricted to one of the RIDs. On the one hand, this model can be seen as a loss of function because it impairs interactions with transcription factors through the absence of the RID1. Alternatively, these mice can also reflect a gain of function when considering that the interactions with lipid-sensing NRs, such as the PPARs, acting through the unopposed actions of RID2, may be reinforced or at least favored. Indeed, NRs binding to RID2 of SMRT face less competition for SMRT binding in SMRTmRID1 mice, as several other NRs are unable to bind SMRT anymore. The gain-of-function model was invoked to underscore the role of SMRT in aging in SMRTmRID1 mice (Reilly et al. 2010). Expression of endogenous SMRT is induced with age in tissues with high OXPHOS activity, such as the muscle. Accordingly, SMRTmRID1 mice age in an accelerated way, as PPAR activity was potentially inhibited. Notably, SMRTmRID1 mice developed characteristics of the metabolic syndrome, such as hyperlipidemia and insulin resistance. Genes involved in fatty acid catabolism and oxidative metabolism were repressed, thereby resulting in reduced mitochondrial function. Finally, these mice were more sensitive to oxidative stress because of reduced expression of genes responsible for reactive oxygen species (ROS) defense.
The loss-of-function phenotype, revealed by disrupted interactions through RID1, seemed to manifest when SMRTmRID1 mice were exposed to an environmental stressor such as high-fat feeding (Fang et al. 2011). Such mice became obese and showed signs of hepatosteatosis, accompanied by increased serum cholesterol and triglyceride levels. Moreover, the SMRTmRID1 mice were insulin-resistant, with reduced energy expenditure and lowered mitochondrial function, thereby shifting whole-body (and, in particular, adipose cell) metabolism toward glycolysis. Importantly, PGC-1α expression was decreased and adipocyte-associated inflammation was accentuated in SMRTmRID1 mice fed a high-fat diet. These phenotypic observations suggested that RAR, TR, and PPAR signaling pathways were likely impaired.
NS-DADm
Bearing a single-amino-acid mutation in the DAD domains of both NCoR1 and SMRT, NS-DADm mice provide the in vivo proof of concept that the interaction with the SMRT/NCoR1 corepressors is absolutely required for the deacetylase activity of HDAC3 in multiple adult tissues (e.g., liver, heart, and skeletal muscle) as well as the embryo (You et al. 2013). As a result of the NS-DAD mutation, the level of histone acetylation in the liver was elevated to a level comparable with that measured in the liver-specific HDAC3 knockout animals (Knutson et al. 2008). Likewise, in the liver, the recruitment of HDAC3 to NCoR1/SMRT-recruiting genomic regions was decreased, which elegantly demonstrates the pivotal role of NCoR1 and SMRT in mediating the epigenetic cues of HDAC3.
In contrast to the lethality seen in SMRT, NCoR1, and HDAC3 germline mutant mice (Jepsen et al. 2000, 2007; Bhaskara et al. 2008), the viability of the NS-DADm mice implies that the critical properties of these proteins are not linked to the enzymatic activation of HDAC3 by the corepressors. Furthermore, HDAC3 seems to exert a nonenzymatic action on liver fat metabolism, as NS-DADm mice with a selective defect in HDAC3 deacetylase activity display far less pronounced triglyceride and cholesterol accumulation than liver-specific HDAC3 knockout mice (Knutson et al. 2008; You et al. 2013).
Regulation of NCoR1/SMRT
Transcription
In our laboratory, we established how NCoR1 expression ties into metabolic homeostasis (Yamamoto et al. 2011). Low-glucose or high-fatty-acid levels were identified to decrease NCoR1 expression, while insulin and high-glucose levels increase its expression (Fig. 2A). Hence, NCoR1 levels are reduced in conditions favoring fatty acid oxidation in vitro and in vivo. In other words, NCoR1 activity is induced when glycolysis is favored over fat oxidation as energy source. These observations also demonstrate a logical and physiological link between the muscle- and adipocyte-specific NCoR1−/− models (Saltiel 2011); when glucose is available, NCoR1 discriminates against oxidative metabolism and fatty acid utilization to favor the use of glucose in the muscle. At the same time, adipogenesis is kept repressed. In case of the consumption of a high-fat diet, NCoR1 levels adapt by going down, thus allowing muscle to switch to oxidative metabolism and adipose tissue to store the excess fat.
Figure 2.
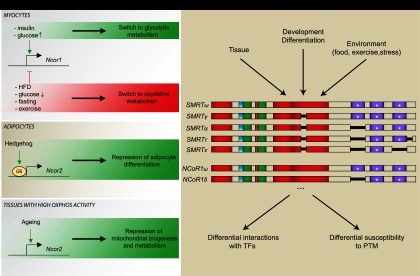
(A) Transcriptional regulation of NCoR1/SMRT. Metabolic cues direct Ncor1 transcription in myocytes, whereas hedgehog signaling controls the expression of Ncor2, the gene coding for SMRT, in adipocytes. SMRT expression also increases upon aging. (B) Regulation through alternative splicing. A nonexhaustive and schematic representation of NCoR1/SMRT isoforms (based on Goodson et al. 2011). Depending on the context (i.e., tissue, developmental stage, and environment), alternative splicing can generate NCoR1/SMRT isoforms with different affinity for binding partners (transcription factors) and responsiveness to post-translational modifications (PTM). Color codes of the NCoR1 and SMRT domains are identical as in Figure 1B.
Another illustration of the importance of NCoR1/SMRT expression as a dynamic regulatory mechanism was highlighted by its identification, together with the hedgehog pathway, in an RNAi screen to identify players in obesity in D. melanogaster (Pospisilik et al. 2010). When translated to a mammalian context, Gli, the effector of the hedgehog pathway, occupied the SMRT promoter. Consequently, SMRT was induced upon the activation of the hedgehog pathway to repress adipogenesis (Pospisilik et al. 2010). This was in line with earlier studies on limb development, where SMRT had already been identified as a Gli target (Vokes et al. 2008).
Alternative splicing
As mentioned previously, the domain composition of corepressor isoforms can direct their repressive activity toward specific transcription factors and their associated target genes in a tissue- or developmental stage-specific way (Fig. 2B; Goodson et al. 2005). A case in point, the NCoR1δ isoform favors adipocyte differentiation of 3T3-L1 preadipocytes, while NCoR1ω represses adipogenesis, as NCoR1δ lacks one of the RIDs contained in NCoR1ω (Goodson et al. 2011). Interestingly, the relative amount of these NCoR1 isoforms varies throughout the differentiation process, diversifying the spectrum of action of NCoR1 (Goodson et al. 2011). NCoR1 isoforms can also be differentially sensitive to post-translational modifications, such as mitogen-activated protein (MAP) kinase phosphorylation (Jonas et al. 2007).
Post-translational modifications
Two major post-translational modifications are reported to interrupt the repression by the NCoR1/SMRT corepressors and trigger their exit out of the nucleus; namely, phosphorylation and ubiquitination (Fig. 3; Perissi et al. 2010).
Figure 3.
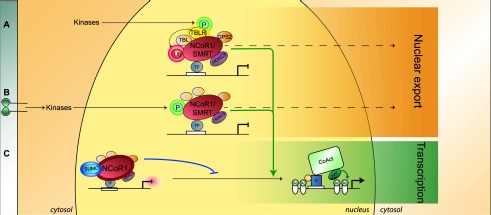
Post-translational regulation of NCoR1/SMRT. (A) Ubiquitination of NCoR1/SMRT (red sphere with halo; Ub) by TBLR, following phosphorylation of TBLR by upstream pathways, leads to their nuclear export and proteasomal degradation. (B) Growth factor and cytokine receptors (EGFR, IL-βR, etc.; see details in the text) signal through downstream kinases, such as MAP3K, AKT, and Cdk2, which phosphorylate NCoR1/SMRT (green sphere with halo; P). This triggers their export from the nucleus, enabling the docking of coactivators, as such derepressing transcription. (C) Conversely, sumoylation of NCoR1 (blue oval; SUMO) stabilizes the complex and enhances repressive activity. The color code for the members of the core repression complex (HDAC3, TBL1, TBLR1, and GPS2) is conserved throughout the figures.
Ubiquitination by TBLR1 targets SMRT/NCoR1 for proteasomal degradation and favors the exchange of corepressors for coactivators (Perissi et al. 2004, 2008). This usually occurs following the phosphorylation of TBLR1 itself by various kinase cascades. Interestingly, TBLR1, which belongs to the core repression complex, is thus involved in both repression with NCoR1/SMRT and the regulation of the dismissal of these corepressors, thus achieving a dual role. This may mean that repression and formation of the core repression complex are intrinsically transient because the complex is prone to dissociate sooner or later (Perissi et al. 2010).
The inhibitor of the NF-κB kinase subunit α (IKKα), MAP kinase kinase kinase 1 (MAP3K1), and AKT were all found to phosphorylate NCoR1 and/or SMRT, resulting in their differential cellular localization (Hong et al. 1998; Hong and Privalsky 2000; Baek et al. 2002; Hermanson et al. 2002; Hoberg et al. 2004; Perissi et al. 2010). As an example, EGF induces the phosphorylation of SMRT through the activation of MAP3K1, while MAP3K1 phosphorylates NCoR1 following interleukin-1β (IL-1β) signaling (Jonas and Privalsky 2004; Perissi et al. 2010). Increased SMRT phosphorylation and its aberrant cytosolic localization correlate with IKKα hyperactivation, which results in the enhanced expression of Notch target genes in the context of colon cancer (Fernandez-Majada et al. 2007a). Interestingly, SMRT is capable of homodimerization to stabilize the corepression complex; SMRT dimerization is abrogated when ERK2, a component of the MAP kinases cascade, phosphorylates SMRT (Varlakhanova et al. 2011).
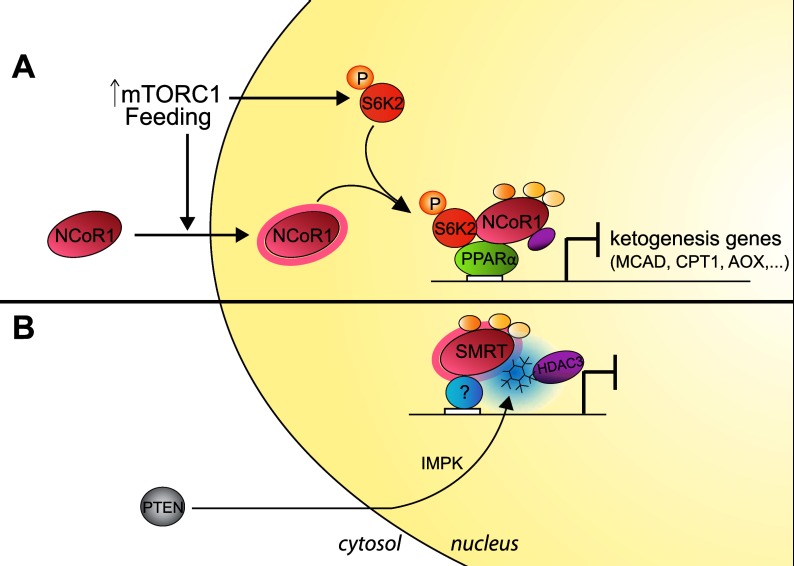
mTOR is another signaling pathway that affects NCoR1 activity and energy homeostasis in the liver (Fig. 4A; Sengupta et al. 2010). The mTOR signaling pathway adapts cellular activities to environmental signals and to the energy status of the cell (Laplante and Sabatini 2012). PPARα, the main transcriptional activator of ketogenesis, is repressed in livers of aged mice due to hyperactive mTORC1 signaling, thereby blunting ketogenesis during fasting (Sengupta et al. 2010). mTORC1 may achieve this effect by regulating the subcellular localization of NCoR1, the predominant corepressor of PPARα. Indeed, in the context of activation of mTORC1 signaling, suppression of NCoR1 restored ketogenesis, suggesting that mTORC1 hyperactivation during aging reduces the reactivity (dynamics or flexibility) of NCoR1 toward the nutrient status and increases nuclear NCoR1 retention upon fasting (Sengupta et al. 2010). Recent data have identified S6 kinase 2 (S6K2) as being the mTORC1 effector responsible for the repression of PPARα targets (Kim et al. 2012). S6K2, which is phosphorylated by mTORC1 in nutrient-affluent conditions, interacts with and enhances NCoR1 nuclear localization, stabilizing the PPARα/corepressor complex, ultimately silencing PPARα target genes. In line with this, S6K2 was found to be strongly associated with NCoR1 in nuclei of ob/ob hepatocytes, confirming the direct effect of nutrient availability and mTORC1 signaling onto NCoR1 intracellular localization. Together, these data establish a role for NCoR1 in the regulation of PPARα activity and ketogenesis by bridging them with mTOR signaling in the contexts of obesity and aging (Sengupta et al. 2010; Kim et al. 2012). The fact that NCoR1 also suppresses oxidative metabolism in the muscle (Yamamoto et al. 2011) is concordant with this role in ketogenesis, as ketogenesis is critically dependent on fatty acid oxidation in the liver. These findings also resonate well with the implication of SMRT in the accelerated aging, sensitivity to oxidative stress, and development of the metabolic syndrome displayed by the SMRTmRID1 mouse (Reilly et al. 2010).
Figure 4.
Post-translational regulation of NCoR1/SMRT. (A) mTORC1 activation in livers during aging or feeding alters the interaction of S6K2 with NCoR1 and changes its subcellular localization. Ketogenic target genes of PPARα are thus silenced depending on environmental input. (B) PTEN, a negative component of the PI3K pathway, acts concomitantly with the IPMK to form Ins(1,4,5,6)P4, which stabilizes and activates the SMRT/HDAC3 complex.
However, these observations of a homeostatic role of NCoR1 seem to be in apparent contradiction with its role in early neural development and in glioma cells, where NCoR1 phosphorylation by AKT was thought to trigger its export out of the nucleus (Hermanson et al. 2002; Park et al. 2007; Perissi et al. 2010). Indeed, as mentioned above, phosphorylation and ubiquitination of NCoR1/SMRT were commonly considered to be the two mechanisms that dismiss the corepressors from the nucleus (Perissi et al. 2010). Thus, the fact that the repressive activity of NCoR1 is enhanced by glucose, insulin, and mTORC1 activation (Sengupta et al. 2010; Yamamoto et al. 2011; Kim et al. 2012), which usually are associated with enhanced AKT phosphorylation, seems in conflict with AKT phosphorylation being a transcriptional derepression mechanism that would inhibit NCoR1 action (Hermanson et al. 2002; Park et al. 2007; Perissi et al. 2010). However, in line with the concept of AKT shutting down NCoR1-mediated repression, phosphatase and tensin homolog (PTEN), a known negative regulator of AKT signaling, was recently proposed to indirectly enhance the repressive activity of these corepressors (Fig. 4B; Watson et al. 2012a). Together with inositol polyphosphate multikinase (IPMK), PTEN contributes to the formation of D-myo-inositol-tetrakisphosphate [Ins(1,4,5,6)P4], a molecule that has been found to act as a glue for the SMRT/HDAC3 complex, thus being essential for the deacetylase activity of the complex.
The multiplicity of possible phosphorylation sites—many potentially not yet mapped—within the sequence of these corepressors might be one explanation for these apparent contradictory effects. Other post-translational modifications could contribute to some of these differences; namely, sumoylation of NCoR1 (Fig. 3) enhances the repressive activity of NCoR1 (Tiefenbach et al. 2006), while prolyl-isomerization mediated by the prolyl isomerase Pin1 (consequently to the phosphorylation of SMRT by Cdk2) affects the stability of SMRT (Stanya et al. 2008; Ryo et al. 2009). Future studies should therefore characterize not only these distinct phosphorylation sites, but also the role of other NCoR1/SMRT post-translational modifications. Furthermore, it should be explored whether context-specific differences in signaling—i.e., early development versus adult homeostasis—can explain some of these differences.
Functional integration of the roles of NCoR1 and SMRT—a systems view
Integrating the wealth of information obtained through the phenotypic analysis of these different models allows one to appreciate the pleiotropic functions that are influenced by NCoR1/SMRT.
Development
One of the most important functions of NcoR1/SMRT concerns their role in cell fate determination, cell differentiation, and lineage progression. NCoR1 and SMRT are both required for the maintenance of neural stem cells (Hermanson et al. 2002; Jepsen et al. 2007) and thus determine the development of neural lineages and the CNS. Additionally, NCoR1 takes part in embryonic hematopoiesis, and SMRT takes part in heart formation. Their homologs in lower organisms also confirm their crucial implication in neural development, as they influence forebrain and eye development (Tsuda et al. 2006; Xu et al. 2009), with crucial roles as early as the eight-cell stage embryo (Linney et al. 2011). Even earlier in evolution, the homolog gei-8 is critical in multiple tissues (neurons, muscle, and intestine) and various stages of C. elegans development (Yamamoto et al. 2011; Mikolas et al. 2013).
In adult mice, NCoR1/SMRT also controls cell fate decisions. This was first illustrated by their role in adipocyte differentiation in vivo, as the SMRTmRID, SMRTmRID1, and adipocyte-specific NCoR1−/− mouse models all show significant adipose tissue phenotypes, which have been linked to PPARγ derepression (Nofsinger et al. 2008; Fang et al. 2011; Li et al. 2011). In line with this, SMRT seems to keep the lid on terminal adipocyte differentiation of the 3T3-L1 fibroblast cell line until the appropriate signals appear; this repressive effect was traced to the interaction of SMRT with the transcription factors C/EBPβ and KAISO (Raghav et al. 2012). Additionally, SMRT also governs cell fate decisions in the lung through the regulation of a set of TR targets as discussed above (Pei et al. 2011). The increased muscle mass and oxidative fiber type switch in muscle-specific NCoR1−/− mice is a further indication of a role for NCoR1 in cell fate determination (Yamamoto et al. 2011). This function of NCoR1 in muscle fiber type determination seems conserved throughout evolution, given that gei-8 also determines worm muscle function (Yamamoto et al. 2011) and regulates transcription of muscle-specific genes (Mikolas et al. 2013).
NCoR1 and SMRT both have nonredundant functions during vertebrate development, as germline mutants of both corepressors are lethal. In line with this, mouse mutants for HDAC3, the enzymatic effector of the NCoR1/SMRT corepressor complex, are also not viable (Bhaskara et al. 2008). In this context, it is interesting to note that NS-DADm and SMRTmRID mice are viable, which, respectively, means that lethality is not dependent on the interaction between NCoR1/SMRT and HDAC3 and that SMRT–NR interactions are dispensable during development (Alenghat et al. 2008; Nofsinger et al. 2008; You et al. 2013).
Metabolic homeostasis—lipid, glucose, mitochondria, and circadian regulation
Strikingly, each of the NCoR1 and SMRT mouse models cited above shows metabolic abnormalities, highlighting the fact that NCoR1 and SMRT are essential for metabolic homeostasis (Fig. 5). Through interacting with NRs, such as the PPARs and ERRs, both corepressors modulate energy expenditure and oxidative metabolism by repressing mitochondrial biogenesis and function, as illustrated by the phenotypes seen in the muscle-specific NCoR1−/−, NCoR1ΔID, SMRTmRID, and SMRTmRID1 mouse models (Nofsinger et al. 2008; Reilly et al. 2010; Astapova et al. 2011; Yamamoto et al. 2011).
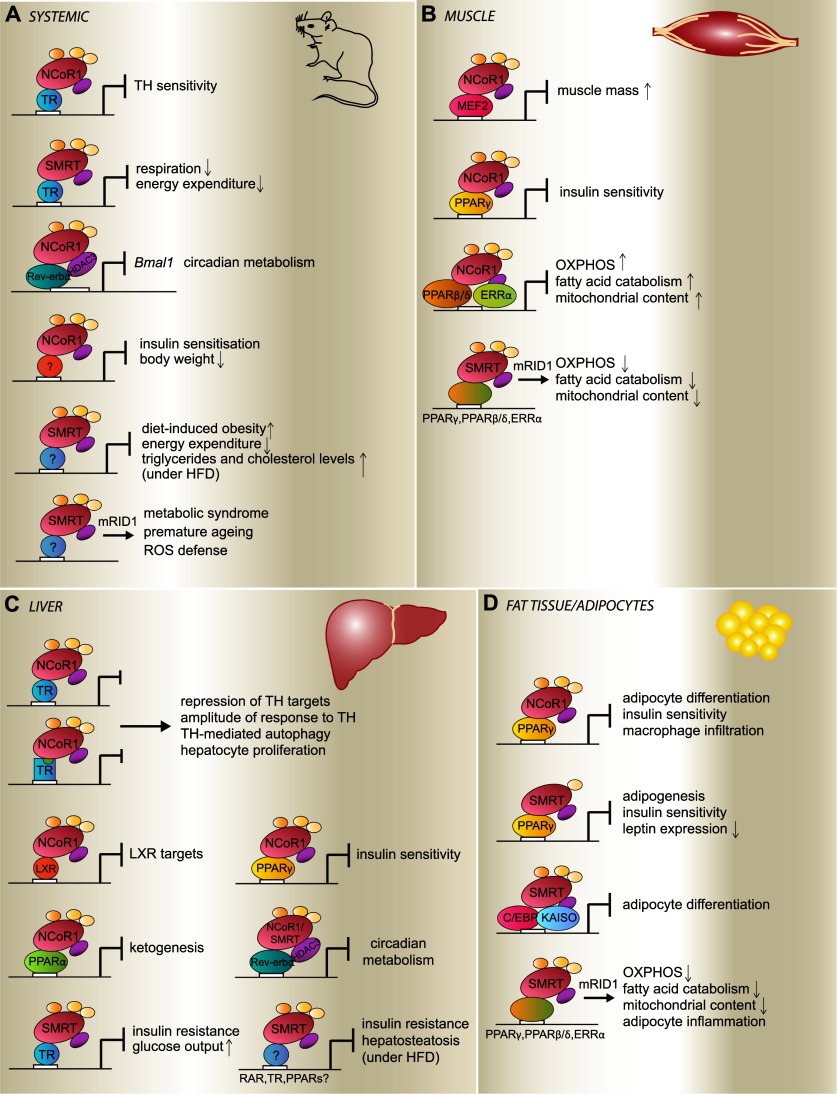
Figure 5.
Metabolic actions of NCoR1 and SMRT. Scheme of the functional consequences of NCoR1 and SMRT action in the entire mouse (A), in muscle (B), in liver (C), and in adipocytes (D). The phenotypes displayed by the mutant mice are listed, as these are logically inexistent in wild-type mice when NCoR1/SMRT achieves proper repression. Conversely, gain-of-function phenotypes of the SMRTmRID1 mice are introduced by an arrow at the bottom of the panels. Details are found in the text. The transcription factors thought to mediate the effects are indicated. (TH) Thyroid hormone. Liganded TR is indicated by a square with a green circle.
Glucose homeostasis is also modulated by SMRT and NCoR1, notably by regulating the sensitivity to thyroid hormone, as reflected by studies of the liver-specific NCoRi, L-NCoRΔID, and NCoRΔID mice (Astapova et al. 2008, 2011; Feng et al. 2011). Moreover, PPARγ and PPARβ/δ also influence insulin sensitivity and glucose homeostasis, as exposed by studies in the muscle- and adipocyte-specific NCoR1−/− mice as well as in SMRTmRID and SMRTmRID1 mouse lines (Nofsinger et al. 2008; Reilly et al. 2010; Fang et al. 2011; Li et al. 2011; Yamamoto et al. 2011). The predominant impact of NCoR1 (and PPARγ) on insulin sensitivity was perhaps nowhere better highlighted by the fact that thiazolidinediones were unable to further improve insulin sensitivity despite a striking induction of obesity in the adipocyte-specific NCoR1−/− mice (Li et al. 2011).
Likewise, these corepressors were also shown to impact lipid homeostasis, as several of the above models develop obesity and features of the metabolic syndrome. Indeed, phenotypes such as fat accumulation, enhanced adipocyte differentiation, deregulated serum lipid profiles, or liver steatosis were observed in the above-listed mutant models, which disrupted the inhibitory interactions of NCoR1/SMRT with NRs, such as PPARγ, PPARβ/δ, TR, Rev-erbα, etc. In line with this, SMRT+/− mice became more obese on a high-fat diet compared with control mice, an effect not observed when these mice were fed a chow diet (Sutanto et al. 2010). Interestingly, SMRT+/− mice showed an increase in the number of smaller subcutaneous adipocytes, which are known to be more insulin-sensitive (Hallakou et al. 1997), and a decrease in leptin expression, resulting in an increased caloric intake upon high-fat feeding. As discussed in the frame of regulation by post-translational modifications, NCoR1 impacts ketogenesis through its interaction with PPARα (Sengupta et al. 2010). It is worth noting that the identification of SMRTER in a Drosophila screen for genes involved in obesity confirms a long-standing implication of this corepressor family in lipid homeostasis (Pospisilik et al. 2010). Similarly, gei-8 in C. elegans impacts mitochondrial homeostasis (Yamamoto et al. 2011) and the transcription of metabolic genes related to lipid, sugar, and amino acid metabolism (Mikolas et al. 2013).
Furthermore, NCoR1 seems to occupy a pivotal role in the link between metabolism and circadian rhythm as the mediator for the repression of Bmal1 by the key circadian regulator Rev-erbα (Alenghat et al. 2008). Also, the recruitment of HDAC3 to genes involved in lipid metabolism and thus hepatic lipogenesis follows a diurnal pattern (Feng et al. 2011). Consistent with this, mice with a liver-specific mutation of Rev-erbα are characterized by hepatosteatosis, reminiscent of the phenotype of the liver-specific HDAC3−/− mice (Knutson et al. 2008; Feng et al. 2011; discussed below). Together, these findings indicate a critical function for NCoR1, and perhaps SMRT, in the circadian regulation of metabolism.
Whereas all of the above clearly underscore that the pleiotropic metabolic and circadian actions of both corepressors are partially explained by their intersection with multiple transcriptional pathways, NCoR1 and SMRT also cross-talk with other cofactors. Strikingly, within this context, there exist strong parallels between the SMRT mutant mouse models and the liver-specific HDAC3−/− mice. Like the SMRT mutant mouse models, the liver HDAC3−/− mice develop nonalcoholic fatty liver disease as a result of derepression of lipid and cholesterol synthesis genes (Knutson et al. 2008). This phenotype could be rescued by antagonists of either PPARγ or mTOR signaling, indicating that HDAC3 regulates hepatic lipid metabolism through PPARγ and mTOR (Knutson et al. 2008). Although NS-DADm mice are devoid of HDAC3 deacetylase activity, these mice did not display the severe metabolic impairments that typified HDAC3 gene deletion in the liver and heart (Knutson et al. 2008; Montgomery et al. 2008; Sun et al. 2011, 2012), suggesting the existence of metabolic defects that are independent of the loss of the deacetylase activity of HDAC3 (You et al. 2013).
The alterations observed after muscle NCoR1 deletion, such as enhanced oxidative metabolism, fiber type switching, and exercise endurance, are reminiscent of the changes that typify PGC-1α overexpression and/or activation (Lin et al. 2002; Arany et al. 2008; Yamamoto et al. 2011). The repression that SMRT exerts on oxidative stress resistance genes during the aging process (Reilly et al. 2010) goes hand in hand with this affirmation, as PGC-1α enhances ROS defense (St-Pierre et al. 2006). In summary, the NCoR1/SMRT (or HDAC3) corepressors seem to oppose the action of coactivators, such as PGC-1α, highlighting the yin-yang relationship between corepressors and coactivators on a physiological level.
Inflammation
Regulation of inflammation is another major function of NCoR1/SMRT. Both NCoR1 and SMRT repress proinflammatory genes in macrophages, many of which are NF-κB targets (Fig. 6A; Pascual et al. 2005). While innate inflammatory stimuli, such as LPS, cause NCoR1 clearance from promoters and activation of the expression of these inflammatory genes, the anti-inflammatory effects of PPARγ and LXRs ligands were shown to prevent NCoR1 dismissal (Pascual et al. 2005; Ghisletti et al. 2007). This mechanism involves ligand-dependent sumoylation of PPARγ and LXRs on their ligand-binding site, which in turn inhibits NCoR1 ubiquitination or phosphorylation induced by LPS, normally leading to its clearance from promoters in proinflammatory conditions. In other words, sumoylated PPARγ or LXRs are able to inhibit inflammatory responses by binding to NCoR1 and keeping it docked on the promoter region of NF-κB target genes even if proinflammatory signals are heightened. While this mechanism appeared to be a general strategy underpinning NR transrepression of proinflammatory genes, the differential sumoylation of LXRs and PPARγ, although similar in nature, enable distinct transrepression pathways to be solicited in a signal- or gene-specific manner. Moreover, NCoR1 dismissal after Toll-like receptor (TLR) activation can be triggered by various signaling pathways, such as p65/IKKɛ kinase or calmodulin kinase (CaMKII or CaMK2), depending on whether TLR4 or TLR-2 is stimulated, respectively (Huang et al. 2009). The highly flexible and combinatorial mode through which this enables NCoR1 to regulate inflammatory responses is remarkable.
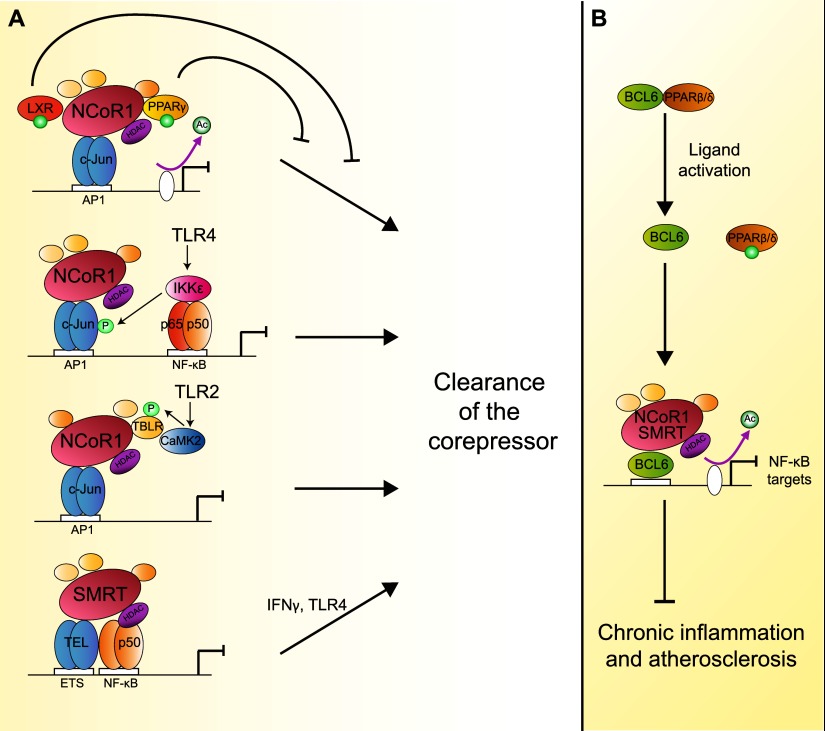
Figure 6.
Anti-inflammatory roles NCoR1/SMRT. (A) Through binding with c-Jun, NCoR1 docks to AP1-binding sites, silencing inflammatory genes. Liganded and sumoylated PPARγ/LXR maintain NCoR1 repression by counteracting the nuclear export and proteasomal degradation of NCoR1 (Pascual et al. 2005; Ghisletti et al. 2007). In response to the activation of TLR in the context of innate immunity, specific downstream kinase pathways (e.g., IKKɛ and CamK2) trigger the clearance of NCoR1 (Huang et al. 2009). Activation of TLR4 or interferon γ (IFNγ) signaling can clear SMRT from ETS sites. (B) Ligand activation of PPARβ/δ dissociates it from the transcriptional repressor BCL6. BCL6 mediated recruitment of NCoR1/SMRT to the promoter of NF-κB target genes, then counters sustained inflammation in the context of atherosclerosis.
In the context of TLR4-inducible target genes, both corepressors can be recruited independently, as the transcription factors mediating their anchorage in the promoter region are different (Ghisletti et al. 2009). NCoR1 is apparently recruited by interacting with unphosphorylated c-JUN bound to activator protein 1 (AP1) target sites, while SMRT binds to the ETS leukemia (TEL) protein and the p50 subunit of NF-κB. This differential anchorage defines specific sets of targets for NCoR1 and SMRT repression. However, these sets are overlapping, meaning that, for a subset of TLR4-inducible target genes, both NCoR1 and SMRT are required on the promoter at the same time and in a mutually dependent way to mediate repression. Moreover, as discussed above, separate mechanisms are responsible for the dismissal of NCoR1 and SMRT.
Another angle of the overall contribution of NCoR1/SMRT to the inflammation process has been exposed by studying their respective cistromes in macrophages (Fig. 6B; Barish et al. 2012). In fact, part of the anti-inflammatory role of SMRT and NCoR1 involves its binding to B-cell lymphoma 6 (BCL6) protein, a central transcription factor that represses TLR-initiated responses in macrophages. NCoR1/SMRT antagonize the NF-κB-driven proinflammatory program by also mediating the repression exerted by BCL6, which makes both corepressors major players that inhibit chronic inflammation and atherosclerosis (Lee et al. 2003; Barish et al. 2012). Interestingly, BCL6 was found to mediate the anti-inflammatory actions of ligand-activated PPARβ/δ in macrophages (Lee et al. 2003). Unliganded PPARβ/δ is bound to BCL6, hindering its anti-inflammatory potential. Ligand activation of PPARβ/δ releases BCL6 and reduces inflammation-related atherosclerosis (Lee et al. 2003; Takata et al. 2008; Bishop-Bailey and Bystrom 2009).
The multiplicity of distinct signaling pathways and mechanisms that take part in the regulation of inflammation by NCoR1/SMRT suggests that these proteins have evolved as central integrators of pro- and anti-inflammatory signals.
Cancer
Given its involvement in the control of metabolism and inflammation, it is no surprise that NCoR1/SMRT are involved in the development of cancer. NCoR1 and SMRT have previously been linked to several kinds of leukemia as well as glioblastoma multiforme and colorectal and endometrial carcinoma (Fernandez-Majada et al. 2007a,b; Karagianni and Wong 2007; Park et al. 2007; Kashima et al. 2009; Perissi et al. 2010). In these cases, their implication was explained by aberrant interactions with mutated proteins or the deregulated expression and subcellular localization of NCoR1/SMRT. In the context of breast cancer, a decrease in the stability and levels of SMRT was directly associated with acquired tamoxifen resistance, as SMRT mediates repression by the estrogen receptor (Ryo et al. 2009). During the progression of prostate cancer, the decreased recruitment of SMRT alters the response of androgen receptor to androgens, which may also explain resistance to hormonal treatments (Liao et al. 2003; Yoon and Wong 2006; Godoy et al. 2012).
The role of NCoR1/SMRT in cell proliferation was already suggested by work performed on their functional homolog in yeast, Snt1, which is part of the SET3 complex (Pijnappel et al. 2001) and is known to be involved in cell cycle regulation. In line with this, HDAC3 deletion in mouse embryonic fibroblasts led to apoptosis because of DNA damage and disruption of DNA repair pathways (Bhaskara et al. 2008). Thus, these findings revealed roles of the NCoR1/SMRT complexes in S-phase progression and genomic stability (Bhaskara et al. 2008). More precisely, the NCoR1/SMRT complex is responsible for the maintenance of acetylation and methylation patterns during S phase, which are essential for DNA repair and genomic stability (Bhaskara et al. 2010). Furthermore, deletion of the chromosomal region containing NCoR1 or down-regulation of NCoR1 expression are common in human hepatocellular carcinoma. Consistent with this hypothesis, SMRT and HDAC3 were shown to be involved in cell cycle regulation in 3T3-L1 fibroblasts (Fajas et al. 2002; Raghav et al. 2012). These observations functionally link HDAC3-containing corepressor complexes involved in cell cycle regulation with adipocyte differentiation and metabolic control. In combination, these studies in different species suggest that cell cycle regulation is one of the ancestral roles of NCoR1/SMRT complexes, which has evolved in higher organisms to encompass the formation of heterochromatin to fine-tune gene expression patterns in a cell- and context-specific way.
Perspectives
The corepressors NCoR1/SMRT have been proven to be crucial hubs in the complex network of transcriptional regulation during development and homeostasis across evolution. The multiplicity of the physiological pathways, which these corepressors influence on an organismal level, is a hallmark of their wide-ranging regulatory roles. Despite their structural similarity, NCoR1 and SMRT use mechanisms such as alternative splicing and differential post-translational modifications to diversify their actions depending on the developmental stage, tissue, metabolic milieu, and signaling context. Thus, while NCoR1 and SMRT are partially both similar and redundant, they have specific and divergent signaling actions that are complex and finely tuned. Future studies should further clarify the precise mechanisms by which NCoR1/SMRT achieve this diversity. One important aspect is to establish the role of NCoR1 and SMRT as bona fide metabolic sensors, which involves their regulation by upstream regulatory factors and their integration into established nutrient-sensitive signaling pathways, such as those controlled by insulin, mTORC1, and AMPK. Other work should elucidate the downstream signaling and transcriptional networks governed by NCoR1 and SMRT and should involve the characterization of additional temporally and spatially controlled NCoR1/SMRT mouse models. These studies could also be facilitated by establishing the role of these corepressors in simple model organisms, such as C. elegans and D. melanogaster, as corepressor signaling is evolutionary conserved. Finally, human genetic studies are required to further validate the role of NCoR1 and SMRT pathways for human biology with the ultimate hope that their signaling pathways could be exploited as potential drug targets to prevent or treat disease states that involve altered function of these corepressors.
Acknowledgments
J.A. is the Nestlé Chair in Energy Metabolism. The work in our laboratory is supported by the École Polytechnique Fédérale de Lausanne, the EU Ideas program (ERC-2008-AdG-231138), the NIH (R01HL106511-01A and R01AG043930), the Velux Stiftung, the Swiss National Science Foundation (31003A-124713, 310030-143748, and CRSII3-136201).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.214023.113.
References
- Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, Bucan M, Ahima RS, Kaestner KH, Lazar MA 2008. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature 456: 997–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany Z, Foo SY, Ma Y, Ruas JL, Bommi-Reddy A, Girnun G, Cooper M, Laznik D, Chinsomboon J, Rangwala SM, et al. 2008. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1α. Nature 451: 1008–1012 [DOI] [PubMed] [Google Scholar]
- Ariyoshi M, Schwabe JW 2003. A conserved structural motif reveals the essential transcriptional repression function of Spen proteins and their role in developmental signaling. Genes Dev 17: 1909–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astapova I, Lee LJ, Morales C, Tauber S, Bilban M, Hollenberg AN 2008. The nuclear corepressor, NCoR, regulates thyroid hormone action in vivo. Proc Natl Acad Sci 105: 19544–19549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astapova I, Vella KR, Ramadoss P, Holtz KA, Rodwin BA, Liao XH, Weiss RE, Rosenberg MA, Rosenzweig A, Hollenberg AN 2011. The nuclear receptor corepressor (NCoR) controls thyroid hormone sensitivity and the set point of the hypothalamic–pituitary–thyroid axis. Mol Endocrinol 25: 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG 2002. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-κB and β-amyloid precursor protein. Cell 110: 55–67 [DOI] [PubMed] [Google Scholar]
- Barish GD, Yu RT, Karunasiri MS, Becerra D, Kim J, Tseng TW, Tai LJ, Leblanc M, Diehl C, Cerchietti L, et al. 2012. The Bcl6-SMRT/NCoR cistrome represses inflammation to attenuate atherosclerosis. Cell Metab 15: 554–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara S, Chyla BJ, Amann JM, Knutson SK, Cortez D, Sun ZW, Hiebert SW 2008. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell 30: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara S, Knutson SK, Jiang G, Chandrasekharan MB, Wilson AJ, Zheng S, Yenamandra A, Locke K, Yuan JL, Bonine-Summers AR, et al. 2010. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell 18: 436–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop-Bailey D, Bystrom J 2009. Emerging roles of peroxisome proliferator-activated receptor-β/δ in inflammation. Pharmacol Ther 124: 141–150 [DOI] [PubMed] [Google Scholar]
- Chen JD, Evans RM 1995. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377: 454–457 [DOI] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. 2012. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485: 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen RN, Brzostek S, Kim B, Chorev M, Wondisford FE, Hollenberg AN 2001. The specificity of interactions between nuclear hormone receptors and corepressors is mediated by distinct amino acid sequences within the interacting domains. Mol Endocrinol 15: 1049–1061 [DOI] [PubMed] [Google Scholar]
- Downes M, Burke LJ, Bailey PJ, Muscat GE 1996. Two receptor interaction domains in the corepressor, N-CoR/RIP13, are required for an efficient interaction with Rev-erbAα and RVR: Physical association is dependent on the E region of the orphan receptors. Nucleic Acids Res 24: 4379–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajas L, Egler V, Reiter R, Hansen J, Kristiansen K, Debril MB, Miard S, Auwerx J 2002. The retinoblastoma–histone deacetylase 3 complex inhibits PPARγ and adipocyte differentiation. Dev Cell 3: 903–910 [DOI] [PubMed] [Google Scholar]
- Fang S, Suh JM, Atkins AR, Hong SH, Leblanc M, Nofsinger RR, Yu RT, Downes M, Evans RM 2011. Corepressor SMRT promotes oxidative phosphorylation in adipose tissue and protects against diet-induced obesity and insulin resistance. Proc Natl Acad Sci 108: 3412–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Jiang Y, Meltzer P, Yen PM 2001. Transgenic targeting of a dominant negative corepressor to liver blocks basal repression by thyroid hormone receptor and increases cell proliferation. J Biol Chem 276: 15066–15072 [DOI] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA 2011. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331: 1315–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Majada V, Aguilera C, Villanueva A, Vilardell F, Robert-Moreno A, Aytes A, Real FX, Capella G, Mayo MW, Espinosa L, et al. 2007a. Nuclear IKK activity leads to dysregulated notch-dependent gene expression in colorectal cancer. Proc Natl Acad Sci 104: 276–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Majada V, Pujadas J, Vilardell F, Capella G, Mayo MW, Bigas A, Espinosa L 2007b. Aberrant cytoplasmic localization of N-CoR in colorectal tumors. Cell Cycle 6: 1748–1752 [DOI] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E 2002. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell 9: 45–57 [DOI] [PubMed] [Google Scholar]
- Fozzatti L, Lu C, Kim DW, Park JW, Astapova I, Gavrilova O, Willingham MC, Hollenberg AN, Cheng SY 2011. Resistance to thyroid hormone is modulated in vivo by the nuclear receptor corepressor (NCOR1). Proc Natl Acad Sci 108: 17462–17467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner KE, Allis CD, Strahl BD 2011. Operating on chromatin, a colorful language where context matters. J Mol Biol 409: 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Ogawa S, Pascual G, Lin ME, Willson TM, Rosenfeld MG, Glass CK 2007. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARγ. Mol Cell 25: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, Rosenfeld MG, Glass CK 2009. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev 23: 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev 14: 121–141 [PubMed] [Google Scholar]
- Godoy AS, Sotomayor PC, Villagran M, Yacoub R, Montecinos VP, McNerney EM, Moser M, Foster BA, Onate SA 2012. Altered corepressor SMRT expression and recruitment to target genes as a mechanism that change the response to androgens in prostate cancer progression. Biochem Biophys Res Commun 423: 564–570 [DOI] [PubMed] [Google Scholar]
- Goodson ML, Jonas BA, Privalsky ML 2005. Alternative mRNA splicing of SMRT creates functional diversity by generating corepressor isoforms with different affinities for different nuclear receptors. J Biol Chem 280: 7493–7503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson ML, Mengeling BJ, Jonas BA, Privalsky ML 2011. Alternative mRNA splicing of corepressors generates variants that play opposing roles in adipocyte differentiation. J Biol Chem 286: 44988–44999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA 2001. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol 21: 6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallakou S, Doare L, Foufelle F, Kergoat M, Guerre-Millo M, Berthault MF, Dugail I, Morin J, Auwerx J, Ferre P 1997. Pioglitazone induces in vivo adipocyte differentiation in the obese Zucker fa/fa rat. Diabetes 46: 1393–1399 [DOI] [PubMed] [Google Scholar]
- Heikkinen S, Auwerx J, Argmann CA 2007. PPARγ in human and mouse physiology. Biochim Biophys Acta 1771: 999–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, et al. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387: 43–48 [DOI] [PubMed] [Google Scholar]
- Hermanson O, Jepsen K, Rosenfeld MG 2002. N-CoR controls differentiation of neural stem cells into astrocytes. Nature 419: 934–939 [DOI] [PubMed] [Google Scholar]
- Hoberg JE, Yeung F, Mayo MW 2004. SMRT derepression by the IκB kinase α: A prerequisite to NF-κB transcription and survival. Mol Cell 16: 245–255 [DOI] [PubMed] [Google Scholar]
- Hong SH, Privalsky ML 2000. The SMRT corepressor is regulated by a MEK-1 kinase pathway: Inhibition of corepressor function is associated with SMRT phosphorylation and nuclear export. Mol Cell Biol 20: 6612–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, Wong CW, Privalsky ML 1998. Signaling by tyrosine kinases negatively regulates the interaction between transcription factors and SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) corepressor. Mol Endocrinol 12: 1161–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, et al. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377: 397–404 [DOI] [PubMed] [Google Scholar]
- Hu X, Lazar MA 2000. Transcriptional repression by nuclear hormone receptors. Trends Endocrinol Metab 11: 6–10 [DOI] [PubMed] [Google Scholar]
- Huang W, Ghisletti S, Perissi V, Rosenfeld MG, Glass CK 2009. Transcriptional integration of TLR2 and TLR4 signaling at the NCoR derepression checkpoint. Mol Cell 35: 48–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka T, Lazar MA 2003. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol Cell Biol 23: 5122–5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka T, Lazar MA 2005. The nuclear receptor corepressor deacetylase activating domain is essential for repression by thyroid hormone receptor. Mol Endocrinol 19: 1443–1451 [DOI] [PubMed] [Google Scholar]
- Jepsen K, Rosenfeld MG 2002. Biological roles and mechanistic actions of co-repressor complexes. J Cell Sci 115: 689–698 [DOI] [PubMed] [Google Scholar]
- Jepsen K, Hermanson O, Onami TM, Gleiberman AS, Lunyak V, McEvilly RJ, Kurokawa R, Kumar V, Liu F, Seto E, et al. 2000. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell 102: 753–763 [DOI] [PubMed] [Google Scholar]
- Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim HJ, Glass CK, Hermanson O, Rosenfeld MG 2007. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature 450: 415–419 [DOI] [PubMed] [Google Scholar]
- Jepsen K, Gleiberman AS, Shi C, Simon DI, Rosenfeld MG 2008. Cooperative regulation in development by SMRT and FOXP1. Genes Dev 22: 740–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas BA, Privalsky ML 2004. SMRT and N-CoR corepressors are regulated by distinct kinase signaling pathways. J Biol Chem 279: 54676–54686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas BA, Varlakhanova N, Hayakawa F, Goodson M, Privalsky ML 2007. Response of SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) and N-CoR (nuclear receptor corepressor) corepressors to mitogen-activated protein kinase kinase kinase cascades is determined by alternative mRNA splicing. Mol Endocrinol 21: 1924–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T 1998. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev 12: 2269–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Downes M, Ordentlich P, Evans RM 2000. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev 14: 55–66 [PMC free article] [PubMed] [Google Scholar]
- Karagianni P, Wong J 2007. HDAC3: Taking the SMRT-N-CoRrect road to repression. Oncogene 26: 5439–5449 [DOI] [PubMed] [Google Scholar]
- Kashima H, Horiuchi A, Uchikawa J, Miyamoto T, Suzuki A, Ashida T, Konishi I, Shiozawa T 2009. Up-regulation of nuclear receptor corepressor (NCoR) in progestin-induced growth suppression of endometrial hyperplasia and carcinoma. Anticancer Res 29: 1023–1029 [PubMed] [Google Scholar]
- Kim K, Pyo S, Um SH 2012. S6 kinase 2 deficiency enhances ketone body production and increases peroxisome proliferator-activated receptor α activity in the liver. Hepatology 55: 1727–1737 [DOI] [PubMed] [Google Scholar]
- Knutson SK, Chyla BJ, Amann JM, Bhaskara S, Huppert SS, Hiebert SW 2008. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J 27: 1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide T, Downes M, Chandraratna RA, Blumberg B, Umesono K 2001. Active repression of RAR signaling is required for head formation. Genes Dev 15: 2111–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM 2012. mTOR signaling in growth control and disease. Cell 149: 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Chawla A, Urbiztondo N, Liao D, Boisvert WA, Evans RM, Curtiss LK 2003. Transcriptional repression of atherogenic inflammation: Modulation by PPARδ. Science 302: 453–457 [DOI] [PubMed] [Google Scholar]
- Li H, Leo C, Schroen DJ, Chen JD 1997. Characterization of receptor interaction and transcriptional repression by the corepressor SMRT. Mol Endocrinol 11: 2025–2037 [DOI] [PubMed] [Google Scholar]
- Li P, Fan W, Xu J, Lu M, Yamamoto H, Auwerx J, Sears DD, Talukdar S, Oh D, Chen A, et al. 2011. Adipocyte NCoR knockout decreases PPARγ phosphorylation and enhances PPARγ activity and insulin sensitivity. Cell 147: 815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G, Chen LY, Zhang A, Godavarthy A, Xia F, Ghosh JC, Li H, Chen JD 2003. Regulation of androgen receptor activity by the nuclear receptor corepressor SMRT. J Biol Chem 278: 5052–5061 [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, et al. 2002. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature 418: 797–801 [DOI] [PubMed] [Google Scholar]
- Linney E, Perz-Edwards A, Kelley B 2011. Identification and characterization of a functional zebrafish smrt corepressor (ncor2). Gene 486: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, Shaw RJ 2011. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 145: 607–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami S, Kanaba T, Ito Y, Mishima M 2012. NMR assignments of SPOC domain of the human transcriptional corepressor SHARP in complex with a C-terminal SMRT peptide. Biomol NMR Assign doi: 10.1007/s12104-012-9424-8 [DOI] [PubMed] [Google Scholar]
- Mikolas P, Kollarova J, Sebkova K, Saudek V, Yilma P, Kostrouchova M, Krause MW, Kostrouch Z 2013. GEI-8, a homologue of vertebrate nuclear receptor corepressor NCoR/SMRT, regulates gonad development and neuronal functions in Caenorhabditis elegans. PLoS ONE 8: e58462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RL, Potthoff MJ, Haberland M, Qi X, Matsuzaki S, Humphries KM, Richardson JA, Bassel-Duby R, Olson EN 2008. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest 118: 3588–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscat GE, Burke LJ, Downes M 1998. The corepressor N-CoR and its variants RIP13a and RIP13Δ1 directly interact with the basal transcription factors TFIIB, TAFII32 and TAFII70. Nucleic Acids Res 26: 2899–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89: 373–380 [DOI] [PubMed] [Google Scholar]
- Nofsinger RR, Li P, Hong SH, Jonker JW, Barish GD, Ying H, Cheng SY, Leblanc M, Xu W, Pei L, et al. 2008. SMRT repression of nuclear receptors controls the adipogenic set point and metabolic homeostasis. Proc Natl Acad Sci 105: 20021–20026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberoi J, Fairall L, Watson PJ, Yang JC, Czimmerer Z, Kampmann T, Goult BT, Greenwood JA, Gooch JT, Kallenberger BC, et al. 2011. Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery. Nat Struct Mol Biol 18: 177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordentlich P, Downes M, Xie W, Genin A, Spinner NB, Evans RM 1999. Unique forms of human and mouse nuclear receptor corepressor SMRT. Proc Natl Acad Sci 96: 2639–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Schroen DJ, Yang M, Li H, Li L, Chen JD 1999. SMRTe, a silencing mediator for retinoid and thyroid hormone receptors-extended isoform that is more related to the nuclear receptor corepressor. Proc Natl Acad Sci 96: 3519–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DM, Li J, Okamoto H, Akeju O, Kim SH, Lubensky I, Vortmeyer A, Dambrosia J, Weil RJ, Oldfield EH, et al. 2007. N-CoR pathway targeting induces glioblastoma derived cancer stem cell differentiation. Cell Cycle 6: 467–470 [DOI] [PubMed] [Google Scholar]
- Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK 2005. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ. Nature 437: 759–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L, Leblanc M, Barish G, Atkins A, Nofsinger R, Whyte J, Gold D, He M, Kawamura K, Li HR, et al. 2011. Thyroid hormone receptor repression is linked to type I pneumocyte-associated respiratory distress syndrome. Nat Med 17: 1466–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Schindler J, Summermatter S, Salatino S, Zorzato F, Beer M, Balwierz PJ, van Nimwegen E, Feige JN, Auwerx J, Handschin C 2012. The corepressor NCoR1 antagonizes PGC-1α and estrogen-related receptor α in the regulation of skeletal muscle function and oxidative metabolism. Mol Cell Biol 32: 4913–4924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG 2004. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116: 511–526 [DOI] [PubMed] [Google Scholar]
- Perissi V, Scafoglio C, Zhang J, Ohgi KA, Rose DW, Glass CK, Rosenfeld MG 2008. TBL1 and TBLR1 phosphorylation on regulated gene promoters overcomes dual CtBP and NCoR/SMRT transcriptional repression checkpoints. Mol Cell 29: 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Jepsen K, Glass CK, Rosenfeld MG 2010. Deconstructing repression: Evolving models of co-repressor action. Nat Rev Genet 11: 109–123 [DOI] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L 2004. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature 429: 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M, Rigaut G, Seraphin B, Aasland R, Stewart AF 2001. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev 15: 2991–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospisilik JA, Schramek D, Schnidar H, Cronin SJ, Nehme NT, Zhang X, Knauf C, Cani PD, Aumayr K, Todoric J, et al. 2010. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of brown versus white adipose cell fate. Cell 140: 148–160 [DOI] [PubMed] [Google Scholar]
- Raghav SK, Waszak SM, Krier I, Gubelmann C, Isakova A, Mikkelsen TS, Deplancke B 2012. Integrative genomics identifies the corepressor SMRT as a gatekeeper of adipogenesis through the transcription factors C/EBPβ and KAISO. Mol Cell 46: 335–350 [DOI] [PubMed] [Google Scholar]
- Reilly SM, Bhargava P, Liu S, Gangl MR, Gorgun C, Nofsinger RR, Evans RM, Qi L, Hu FB, Lee CH 2010. Nuclear receptor corepressor SMRT regulates mitochondrial oxidative metabolism and mediates aging-related metabolic deterioration. Cell Metab 12: 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryo A, Wulf G, Lee TH, Lu KP 2009. Pinning down HER2–ER crosstalk in SMRT regulation. Trends Biochem Sci 34: 162–165 [DOI] [PubMed] [Google Scholar]
- Saltiel AR 2011. Derepressing nuclear receptors for metabolic adaptation. Cell 147: 717–718 [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Laplante M, Oh S, Sabatini DM 2010. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 468: 1100–1104 [DOI] [PubMed] [Google Scholar]
- Seol W, Mahon MJ, Lee YK, Moore DD 1996. Two receptor interacting domains in the nuclear hormone receptor corepressor RIP13/N-CoR. Mol Endocrinol 10: 1646–1655 [DOI] [PubMed] [Google Scholar]
- Shi Y, Downes M, Xie W, Kao HY, Ordentlich P, Tsai CC, Hon M, Evans RM 2001. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev 15: 1140–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha RA, You SH, Zhou J, Siddique MM, Bay BH, Zhu X, Privalsky ML, Cheng SY, Stevens RD, Summers SA, et al. 2012. Thyroid hormone stimulates hepatic lipid catabolism via activation of autophagy. J Clin Invest 122: 2428–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R, et al. 2012. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 485: 62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Yao X, Ying H 2011. Thyroid hormone action in metabolic regulation. Protein Cell 2: 358–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanya KJ, Liu Y, Means AR, Kao HY 2008. Cdk2 and Pin1 negatively regulate the transcriptional corepressor SMRT. J Cell Biol 183: 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, et al. 2006. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell 127: 397–408 [DOI] [PubMed] [Google Scholar]
- Sun Z, Singh N, Mullican SE, Everett LJ, Li L, Yuan L, Liu X, Epstein JA, Lazar MA 2011. Diet-induced lethality due to deletion of the Hdac3 gene in heart and skeletal muscle. J Biol Chem 286: 33301–33309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Miller RA, Patel RT, Chen J, Dhir R, Wang H, Zhang D, Graham MJ, Unterman TG, Shulman GI, et al. 2012. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat Med 18: 934–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutanto MM, Ferguson KK, Sakuma H, Ye H, Brady MJ, Cohen RN 2010. The silencing mediator of retinoid and thyroid hormone receptors (SMRT) regulates adipose tissue accumulation and adipocyte insulin sensitivity in vivo. J Biol Chem 285: 18485–18495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata Y, Liu J, Yin F, Collins AR, Lyon CJ, Lee CH, Atkins AR, Downes M, Barish GD, Evans RM, et al. 2008. PPARδ-mediated antiinflammatory mechanisms inhibit angiotensin II-accelerated atherosclerosis. Proc Natl Acad Sci 105: 4277–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiefenbach J, Novac N, Ducasse M, Eck M, Melchior F, Heinzel T 2006. SUMOylation of the corepressor N-CoR modulates its capacity to repress transcription. Mol Biol Cell 17: 1643–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CC, Kao HY, Yao TP, McKeown M, Evans RM 1999. SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol Cell 4: 175–186 [DOI] [PubMed] [Google Scholar]
- Tsuda L, Kaido M, Lim YM, Kato K, Aigaki T, Hayashi S 2006. An NRSF/REST-like repressor downstream of Ebi/SMRTER/Su(H) regulates eye development in Drosophila. EMBO J 25: 3191–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlakhanova N, Hahm JB, Privalsky ML 2011. Regulation of SMRT corepressor dimerization and composition by MAP kinase phosphorylation. Mol Cell Endocrinol 332: 180–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes SA, Ji H, Wong WH, McMahon AP 2008. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev 22: 2651–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PJ, Fairall L, Santos GM, Schwabe JW 2012a. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature 481: 335–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PJ, Fairall L, Schwabe JW 2012b. Nuclear hormone receptor co-repressors: Structure and function. Mol Cell Endocrinol 348: 440–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb P, Anderson CM, Valentine C, Nguyen P, Marimuthu A, West BL, Baxter JD, Kushner PJ 2000. The nuclear receptor corepressor (N-CoR) contains three isoleucine motifs (I/LXXII) that serve as receptor interaction domains (IDs). Mol Endocrinol 14: 1976–1985 [DOI] [PubMed] [Google Scholar]
- Wong CW, Privalsky ML 1998. Transcriptional repression by the SMRT-mSin3 corepressor: Multiple interactions, multiple mechanisms, and a potential role for TFIIB. Mol Cell Biol 18: 5500–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Li K, Tian M, Hu P, Song W, Chen J, Gao X, Zhao Q 2009. N-CoR is required for patterning the anterior–posterior axis of zebrafish hindbrain by actively repressing retinoid signaling. Mech Dev 126: 771–780 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Williams EG, Mouchiroud L, Canto C, Fan W, Downes M, Heligon C, Barish GD, Desvergne B, Evans RM, et al. 2011. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell 147: 827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Lazar MA 2005. The orphan nuclear receptor Rev-erbα recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol 19: 1452–1459 [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, et al. 2007. Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science 318: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Lazar MA 2010. Nuclear receptor Rev-erbα: A heme receptor that coordinates circadian rhythm and metabolism. Nucl Recept Signal 8: e001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HG, Wong J 2006. The corepressors silencing mediator of retinoid and thyroid hormone receptor and nuclear receptor corepressor are involved in agonist- and antagonist-regulated transcription by androgen receptor. Mol Endocrinol 20: 1048–1060 [DOI] [PubMed] [Google Scholar]
- You SH, Lim HW, Sun Z, Broache M, Won KJ, Lazar MA 2013. Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat Struct Mol Biol 20: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Li Y, Ishizuka T, Guenther MG, Lazar MA 2003. A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. EMBO J 22: 3403–3410 [DOI] [PMC free article] [PubMed] [Google Scholar]