Distinct FOXO-regulated transcriptional programs stimulate cell death or enhance organism life span. Keniry et al. performed a screen for genes that modify FOXO activation of the death receptor TRAIL and identified the transcriptional repressor NFIL3. They found that NFIL3 is overexpressed in various cancers and supports tumor cell survival largely through its repression of TRAIL. Altogether, NFIL3 alters cancer cell behavior and FOXO function, suggesting that targeting NFIL3 could be of therapeutic benefit for cancer patients.
Keywords: FOXO, NFIL3, PTEN, apoptosis, chromatin
Abstract
Depending on the circumstance, FOXO (Forkhead O) (FOXO1, FOXO3, and FOXO4) transcription factors activate the expression of markedly different sets of genes to produce different phenotypic effects. For example, distinct FOXO-regulated transcriptional programs stimulate cell death or enhance organism life span. To gain insight into how FOXOs select specific genes for regulation, we performed a screen for genes that modify FOXO activation of TRAIL, a death receptor ligand capable of inducing extrinsic apoptosis. We discovered that the bZIP transcriptional repressor NFIL3 (nuclear factor interleukin 3-regulated) hindered FOXO transcription factor access to chromatin at the TRAIL promoter by binding to nearby DNA and recruiting histone deacetylase-2 (HDAC2) to reduce histone acetylation. In the same manner, NFIL3 repressed expression of certain FOXO targets—e.g., FAS, GADD45α (growth arrest and DNA damage-inducible, α), and GADD45β—but not others. NFIL3, which we found to be overexpressed in different cancers, supported tumor cell survival largely through repression of TRAIL and antagonized hydrogen peroxide-induced cell death. Moreover, its expression in cancer was associated with lower patient survival. Therefore, NFIL3 alters cancer cell behavior and FOXO function by acting on chromatin to restrict the menu of FOXO target genes. Targeting of NFIL3 could be of therapeutic benefit for cancer patients.
Upon receptor tyrosine kinase activation, class I phosphoinositide 3-kinases (PI3Ks) catalyze the formation of the lipid second messenger phosphatidylinositol (3,4,5) tris-phosphate (PIP3) (Manning and Cantley 2007). Phosphatase and tensin homolog deleted on chromosome 10 (PTEN) dephosphorylates PIP3 to antagonize PI3K. One of the major outputs of the PIP3 signal is the inactivation of FOXO (Forkhead O) transcription factors by the serine/threonine kinase AKT/PKB (v-akt murine thymoma viral oncogene homolog, protein kinase B). AKT phosphorylation of FOXO1, FOXO3, and FOXO4 (hereafter referred to as FOXOs) promotes their interaction with 14-3-3 proteins and cytoplasmic sequestration from their DNA-binding sites on chromatin (Calnan and Brunet 2008).
PI3K signaling to FOXO controls fundamental biological processes such as apoptosis, metabolism, aging, proliferation, cell cycle, and development (Manning and Cantley 2007). Paradoxically, FOXOs mediate increased life span or induce apoptosis and cell cycle arrest, depending on their regulation and environment (Calnan and Brunet 2008). Although AKT is a major FOXO regulator, a proportion of endogenous FOXO can remain active in the nucleus in the setting of PI3K pathway activation (Chen et al. 2010). In addition to phospho-regulation by AKT and other protein kinases, the acetylation and ubiquitination of FOXO control its activity (Calnan and Brunet 2008).
The PI3K/PTEN/FOXO pathway is commonly modified in cancer to promote inactivation of FOXOs. Inactivating mutations of the tumor suppressor PTEN and activating mutations of the proto-oncogene PIK3CA (encoding PI3Kα catalytic subunit) that promote AKT phosphorylation of FOXOs are found at a high frequency in an array of cancers (Li et al. 1997; Samuels et al. 2004; Saal et al. 2005). Although FOXOs are rarely directly inactivated in human cancer, mouse models indicate that FOXO genes suppress spontaneous tumor development and are partially redundant (Paik et al. 2007). On the other hand, pro-oncogenic roles for FOXO factors have been recently identified in acute myelogenous leukemia (AML) and breast cancer, highlighting that the paradoxical nature of these factors is also seen in cancer (Chen et al. 2010; Sykes et al. 2011).
One consequence of inhibition of the PI3K pathway is the induction of FOXO-mediated feedback loops that counteract inhibition of the pathway. For instance, PI3K pathway inhibitors, including exogenous PTEN, induce mRNA expression of pathway components such as INSR (insulin receptor), IRS2 (insulin receptor substrate 2) ERBB3 (v-Erb-B2 erythroblastic leukemia viral oncogene homolog 3), and IGF1R (insulin-like growth factor 1 receptor) via conserved homeostatic feedback mechanisms (Hong et al. 2000; Simpson et al. 2001; Chandarlapaty et al. 2011). We hypothesized that transcripts whose levels are altered by inhibiting the PI3K pathway via PTEN in microarray gene expression experiments could be enriched for additional as yet undetermined modifiers of FOXO transcription factor function. To explore this, expression vectors corresponding to a series of PTEN-regulated genes were constructed and screened for their ability to affect the activation of a FOXO-regulated reporter gene derived from the TRAIL promoter. In this manner, NFIL3 (nuclear factor interleukin 3-regulated) was identified as a modulator of FOXO output.
NFIL3, also known as E4BP4 (E4-binding protein 4), is a homodimer bZIP transcriptional repressor and activator that was originally identified for its ability to bind the adenoviral E4 promoter and, later, the IL3 promoter (Cowell 2002). NFIL3 inhibits apoptosis in B cells and motor neurons (Ikushima et al. 1997; Cowell 2002). NFIL3 is also required for natural killer cell development and IgE class switching (Gascoyne et al. 2009; Kashiwada et al. 2010). Several stimuli induce the expression of NFIL3, including IL3, IL4, dexamethasone, and circadian rhythm (Cowell 2002; Kashiwada et al. 2010). Here we show that NFIL3 binds to chromatin and histone deacetylase-2 (HDAC2) to favor a closed chromatin state, which prevents FOXO transactivation of the genes encoding TRAIL (tumor necrosis factor ligand superfamily, member 10A [TNFSF10A]), FAS (TNF receptor superfamily, member 6 [TNFRSF6]), GADD45α (growth arrest and DNA damage-inducible, α [GADD45A]), and GADD45β (GADD45B). We went on to find that NFIL3 regulates only a portion of FOXO target genes, maintains cancer cell survival and resistance to oxidative stress, and is often overexpressed in poor prognosis cancer.
Results
NFIL3 is a candidate regulator of the PI3K/PTEN/FOXO pathway
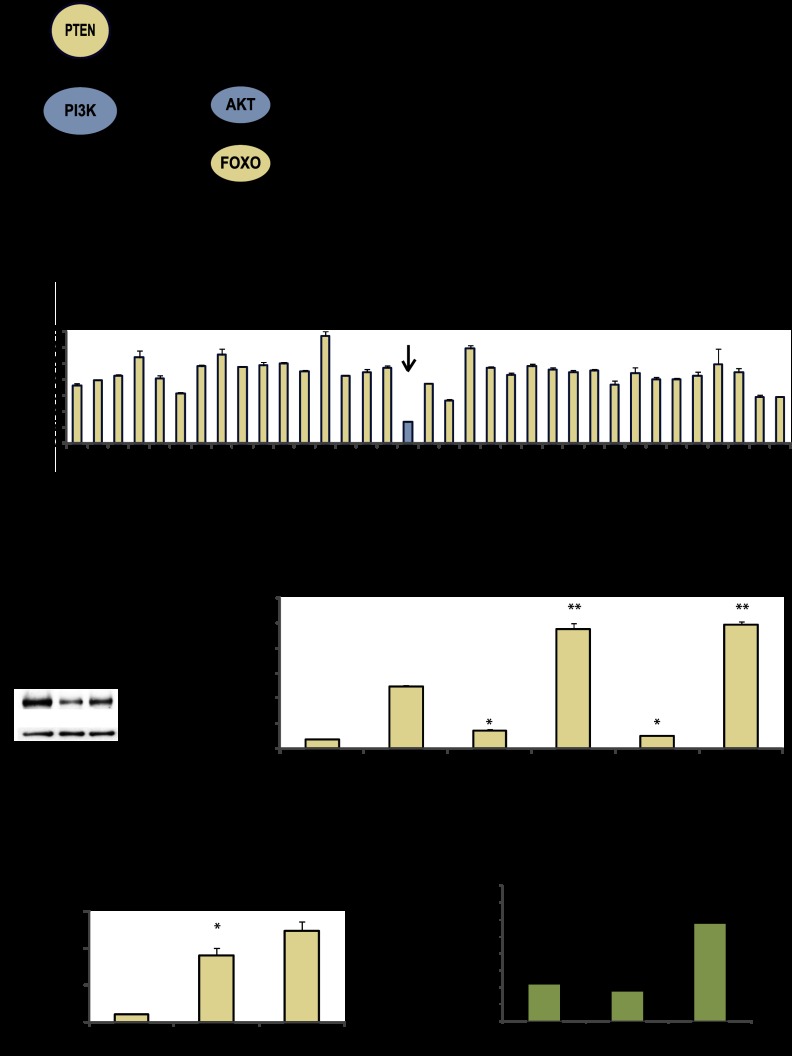
Given that PTEN induces feedback regulation of components of the PI3K pathway, we hypothesized that gene sets whose expression is altered by PTEN in microarray studies would be enriched for novel regulators of FOXO (Fig. 1A). Of the PTEN-regulated genes identified in four previously published independent genome-wide mRNA expression microarray studies, 34 were differentially expressed by at least twofold and had available cDNAs in the Human Orfeome Library 1.1 to facilitate expression vector preparation (Supplemental Fig. 1A; Hong et al. 2000; Matsushima-Nishiu et al. 2001; Simpson et al. 2001; Stolarov et al. 2001). These 34 genes were screened for their ability to affect PI3K pathway activity.
Figure 1.
Screen of PTEN-regulated genes identifies NFIL3 as a candidate modulator of FOXO. (A) Model of the PI3K/PTEN pathway. (B) NFIL3 inhibits reporter activity. PTEN-regulated genes (in pDEST40) were tested for the ability to affect PTEN-induced (pCEP4-PTEN) TRAIL luciferase reporter activity; lacZ is control. (C) NFIL3 Western blot: HEK293 samples treated with NFIL3 shRNAs (KD1 or KD2). (D) NFIL3 shRNA increased TRAIL reporter activity. (*) Significantly different from control shRNA with vector alone (P < 0.05); (**) significantly different from control shRNA with PTEN vector (P < 0.05). (E) NFIL3 shRNA-induced endogenous TRAIL gene by qRT–PCR. (*) Significantly different from control shRNA (P < 0.05). (F) ChIP analysis with FOXO1 and NFIL3 antibodies in log growth HEK293 samples. DNA was subjected to qPCR for the proximal region of the TRAIL promoter or control β-actin. Only NFIL3 associated with the TRAIL promoter. (*) Significantly different from IgG control (P < 0.05). Data are means ± SEM of three experiments.
In order to measure changes in PI3K pathway activity in response to the overexpression of the identified PTEN-regulated genes, we sought out an amenable reporter gene. A side-by-side comparison of six published FOXO-regulated luciferase reporters revealed that the TRAIL reporter gave the best induction with exogenous PTEN in log growth HEK293 (human embryonic kidney) cells (Supplemental Fig. 1B; Modur et al. 2002). The TRAIL reporter was also induced by the PI3K inhibitor wortmannin, blocked by exogenous dominant-negative FOXO1 (DN-FOXO1, containing the DNA-binding domain but not the transactivation domain), and inhibited by exogenous activated Myr-AKT (Supplemental Fig. 1C–E). These data suggest that TRAIL reporter activity reflects PI3K signaling flux through endogenous FOXOs.
Next, we tested each of the 34 PTEN-regulated genes for their ability to alter TRAIL reporter activity in HEK293 cells. Of these 34 genes, NFIL3 had the greatest effect (Fig. 1B). NFIL3 expression impeded PTEN-induced reporter activity and also antagonized wild-type FOXO1-induced TRAIL reporter activity (Fig. 1B; Supplemental Fig. 1F). Several approaches were taken to confirm whether NFIL3 was a modulator of the TRAIL promoter. We found that NFIL3 shRNA increased TRAIL reporter activity and increased endogenous TRAIL gene expression (measured by quantitative RT–PCR [qRT–PCR]) (Fig. 1C–E). We also found that exogenous PTEN cooperated with NFIL3 shRNA to induce endogenous TRAIL expression in the glioblastoma cell line U87MG (Supplemental Fig. 1G). Chromatin immunoprecipitation (ChIP) analysis showed that NFIL3 associated with the proximal region of the endogenous TRAIL promoter under uninduced conditions, whereas FOXO1 did not (Fig. 1F); as a positive control, we found that FOXO1 associated with the INSR promoter under these conditions (Supplemental Fig. 1H). NFIL3 did not, however, behave as a general modulator of FOXO transcription factor function, since it did not affect either a synthetic (IRS x 3-luciferase) or another gene-based FOXO-dependent luciferase reporter (IRS2-luciferase) (data not shown). These data are consistent with NFIL3 acting as a repressor of TRAIL gene expression and as a potential regulator of FOXO output at other genes.
FOXO1- and NFIL3-binding sites are adjacent on the TRAIL promoter
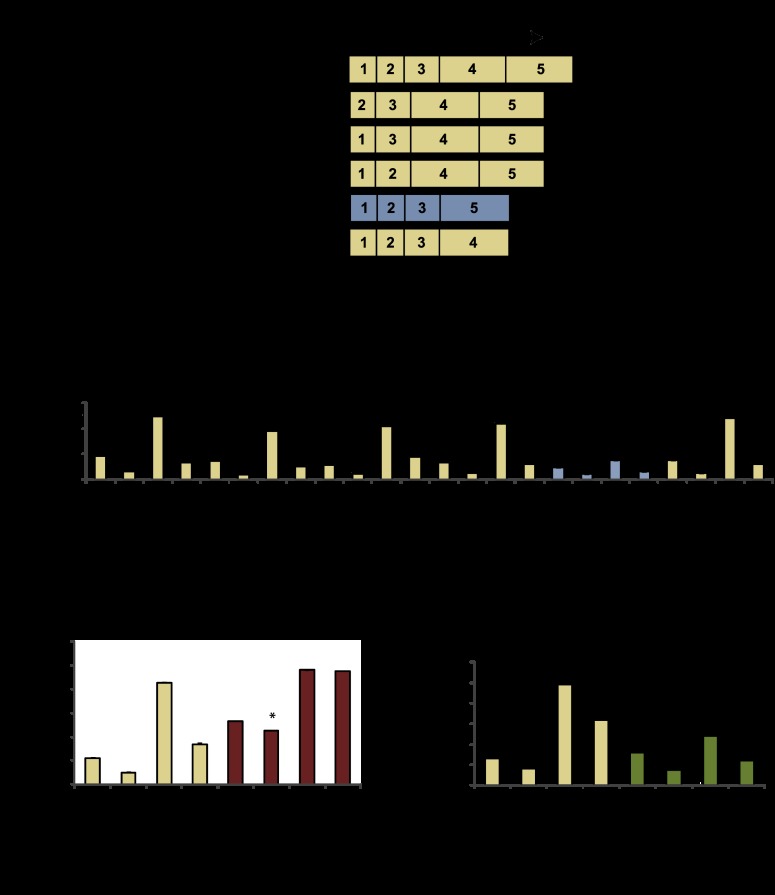
To examine the mechanism by which NFIL3 and FOXO1 regulate TRAIL, we mapped the responsible regulatory sites using mutated luciferase reporters. While reporter-based experiments have caveats such as potentially disrupting spatial relationships between factors, they still provide information that can be useful. A truncated TRAIL reporter that contains only 165 base pairs (bp) from the 1.5-kb initial sequence was also regulated by PTEN and NFIL3 (Supplemental Fig. 2A). Next, we prepared and tested a series of reporter deletions within the 165-bp region (Fig. 2A–B). Deletion-4 (D4), which removes −60 to −1 of the TRAIL promoter containing putative FOXO- and NFIL3-binding sites, was defective for regulation by PTEN and NFIL3 (Fig. 2A,B; Supplemental Fig. 2B). Despite having reduced overall activity, a reporter driven by this 60-bp sequence was modulated by PTEN and NFIL3, demonstrating that it is necessary and sufficient for PTEN/NFIL3 regulation (Supplemental Fig. 2A). Deleting the putative NFIL3 site from the TRAIL reporter led to a loss in NFIL3 repression but not PTEN (or FOXO1) induction (Fig. 2C; Supplemental Fig. 2C). Mutating the predicted FOXO site led to a loss of PTEN (and FOXO1) induction, but NFIL3 repression was retained (Fig. 2D; Supplemental Fig. 2C). Based on these results, FOXO1 and NFIL3 appear to regulate TRAIL using separable binding sites that are in close proximity to each other.
Figure 2.
NFIL3 and FOXO regulatory sequences are adjacent on the TRAIL promoter. (A) TRAIL reporter deletion mutants; a critical deletion (D4) is in blue. (B) D4 is defective for regulation by NFIL3 and PTEN. (*) Significantly less inhibited by NFIL3 compared with wild-type TRAIL reporter; (**) significantly less induced by PTEN than wild-type TRAIL reporter. (C) ΔNFIL3 site reporter was tested for PTEN and NFIL3 regulation. (*) Significantly less inhibited by NFIL3 compared with the wild-type TRAIL reporter. (D) Putative FOXO-binding site was mutated in the TRAIL reporter; this reporter was tested for PTEN and NFIL3 regulation. (*) Significantly less induced by PTEN compared with the wild-type TRAIL reporter. Data are means ± SEM of three experiments.
NFIL3 regulates transcription in concert with HDACs
Transcriptional repression can be mediated via HDAC-dependent and -independent mechanisms (Fig. 3A; Minucci and Pelicci 2006). To gain insight into whether NFIL3 repressed transcription with HDACs, we investigated NFIL3- and HDAC DNA-binding properties on the TRAIL promoter by performing avidin–biotin complex DNA assays. Nuclear lysates prepared from transfected HEK293 cells were incubated with biotinylated DNA, and DNA–protein complexes were isolated. NFIL3 bound well to the minimal 60-bp wild-type TRAIL promoter fragment but poorly to a mutant fragment lacking the NFIL3-binding site. In contrast, histone H2B (control) bound the wild-type and mutant TRAIL promoter fragments equally well (Fig. 3B). We next examined the chromatin deacetylating proteins HDAC1, HDAC2, and SIRT1 (silent mating type information regulation 2 homolog 1) for TRAIL promoter binding. We found that HDAC2 DNA binding mirrored that of NFIL3 (Fig. 3B), while the other tested HDACs were not observed to bind the DNA. These data demonstrate that the NFIL3-binding site is required for in vitro association of NFIL3 and HDAC2 with the TRAIL promoter.
Figure 3.
NFIL3/HDAC2 have similar DNA-binding dynamics. (A) Schematic of possible modes of NFIL3-mediated repression. (B) Avidin–biotin complex DNA assays: Lysates from transfected HEK293 cells were incubated with biotinylated TRAIL promoter DNA to purify associated proteins. NFIL3 and HDAC2 associated with DNA that retained the mapped NFIL3-binding site. (C,D) NFIL3 and HDAC2 physically associate. Coimmunoprecipitations were performed with HEK293 cell extracts and antibodies to endogenous NFIL3 and HDAC2, which detected an interaction between these proteins; no interaction was seen between NFIL3 and HDAC1. (E) TRAIL reporter assays with or without exogenous NFIL3 and/or HDAC2. HDAC2 repressed TRAIL reporter activity. (F) Treatment of HEK293 cells with HDAC inhibitors (2 μM SAHA or 1 μM TSA) for 24 h induced endogenous TRAIL gene expression as measured by qRT–PCR.
Since both NFIL3 and HDAC2 are known transcriptional repressors (Cowell 2002; Minucci and Pelicci 2006), the above DNA-binding data for NFIL3 and HDAC2 indicated that these factors may associate with each other in a repressor complex. To address this possibility, we performed coimmunoprecipitations with transfected HEK293 nuclear extracts. V5-NFIL3 coimmunoprecipitated with FL-HDAC2 from nuclear extracts using either a monoclonal antibody to the V5 epitope tag on NFIL3 or the Flag epitope tag on HDAC2 (Supplemental Fig. 3A,B). Furthermore, endogenous HDAC2 coprecipitated with endogenous NFIL3 using antibodies that recognize NFIL3 or HDAC2, supporting the idea that these proteins reside in a repressor complex together (Fig. 3C,D); we also investigated whether NFIL3 coprecipitated with HDAC1 and failed to detect an interaction (Fig. 3C). To functionally assess HDAC2 regulation of TRAIL, we exogenously expressed HDAC2 in TRAIL reporter assays. HDAC2 inhibited TRAIL reporter activity (Fig. 3E). In addition, the HDAC inhibitors suberoylanilide hydroxamic acid (SAHA) or trichostatin A (TSA) induced endogenous TRAIL expression in HEK293 cells (Fig. 3F). Finally, HDAC inhibition with SAHA significantly reduced the ability of NFIL3 to repress TRAIL reporter activity (Supplemental Fig. 3C).
Given that NFIL3 depends on HDAC activity to inhibit the TRAIL reporter and that FOXO1 is known to be acetylated, we wanted to determine whether the acetylation state of FOXO1 is important for its antagonism by NFIL3/HDAC2. We found that the acetylation-defective mutant for FOXO1 (6KR) was still antagonized by NFIL3, suggesting that the acetylation of FOXO1 (at least on these residues) is not important for NFIL3/HDAC2 inhibition of this reporter (Supplemental Fig. 3D). The acetylation mimetic mutant of FOXO1 (6KQ) failed to induce the TRAIL reporter. In sum, HDAC activity is important for NFIL3 repression of TRAIL. This regulation is operational with the FOXO 6KR mutant, suggesting that the deacetylation of histones may be important for this regulatory mechanism.
NFIL3 shRNA targeting activates a cohort of FOXO-induced genes
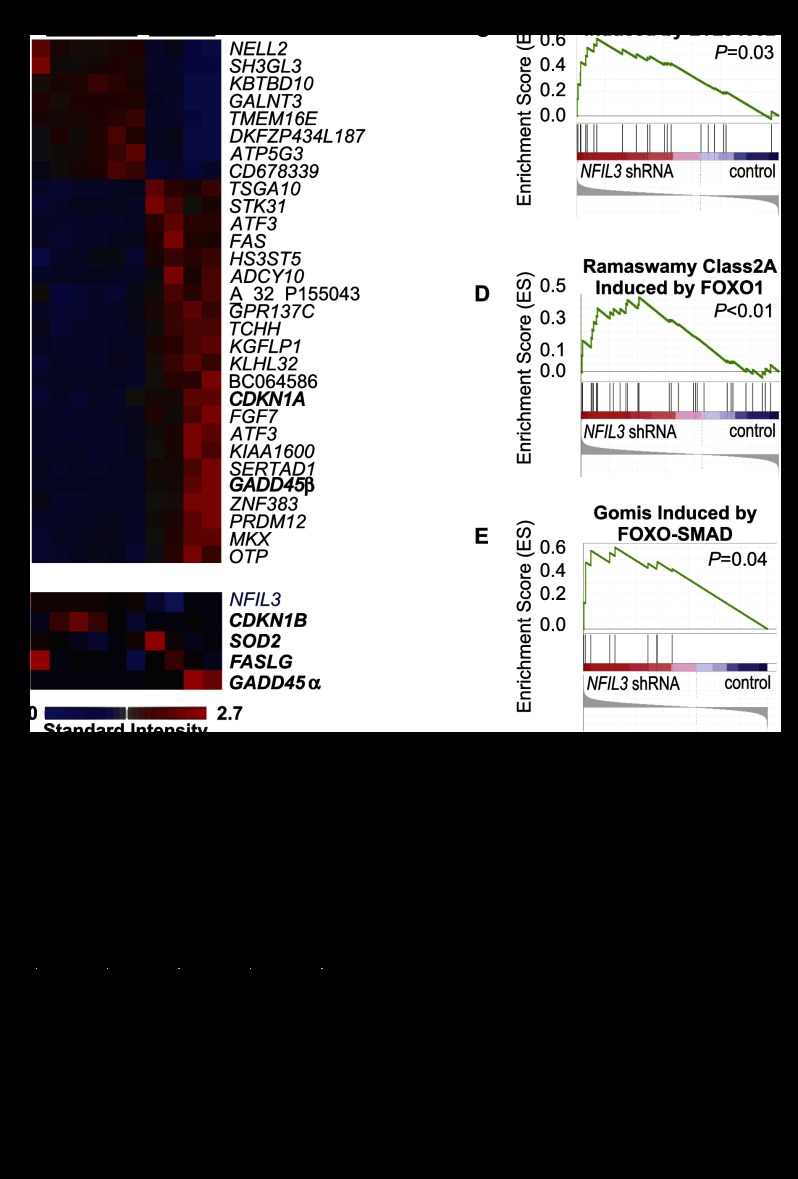
To investigate the breadth of NFIL3-regulated targets and their potential relationship to other FOXO-regulated genes, we performed gene expression profiling with total RNA from HEK293 cells treated with shRNA against NFIL3 or control shRNA. A relatively small number of genes, 289, were induced by at least twofold by NFIL3 shRNA (Supplemental Table 1). Knockdown of NFIL3 with two different shRNA hairpins strongly induced several known FOXO-activated genes, including GADD45α, GADD45β, and CDKN1A (cyclin-dependent kinase inhibitor 1A, encodes p21) (Fig. 4A,B; Supplemental Table 1; Gomis et al. 2006; Calnan and Brunet 2008), whereas other FOXO target genes were not induced (CDKN1B, SOD2, and FASLG) (Fig. 4B). A striking induction of the cell death receptor FAS was also observed. To confirm microarray findings, genes from Figure 4, A and B, and Supplemental Table 1 were examined by qRT–PCR after NFIL3 or control shRNA treatment of the cell lines HEK293 and BT549 (PTEN mutant basal-like breast cancer cell line). qRT–PCR confirmed 20 of 20 NFIL3 shRNA-induced genes in HEK293s and 16 of 24 NFIL3 shRNA-induced genes in BT549s; NFIL3 shRNA also induced FOXO target genes CDKN1A, FAS, and GADD45β in the glioblastoma cell line U87MG (Supplemental Table 2).
Figure 4.
The shRNA-mediated knockdown of NFIL3 activates FOXO-induced genes. Total RNA was analyzed from HEK293 cells treated with NFIL3 or control shRNA. (A) Heat map of differentially expressed genes (false discovery rate [FDR], 0.05 and ≥10-fold expression difference); known FOXO targets are in bold. (B) Heat map of additional FOXO target genes as well as NFIL3. (C–E) Enrichment plots from GSEA with experimentally identified PTEN pathway gene sets and microarray data from HEK293 cells. Gene sets induced by LY294002 and FOXO1 (Ramaswamy class 2A and Gomis FOXO–SMAD) were significantly enriched in NFIL3 shRNA samples. (F) GSEA results (normalized enrichment score [NES]) from analysis with PTEN/PI3K pathway gene sets. (G) GSEA with >1400 curated gene sets from Broad Institute Molecular Signatures Database and HEK293 (NFIL3 shRNA) microarray data was performed; none of these sets was FOXO-regulated. Eight of the top 50 most enriched gene sets in NFIL3 shRNA samples were up-regulated by HDAC inhibition. See Supplemental Table 5 for a complete list of the 50 most enriched curated gene sets in NFIL3 shRNA samples.
We further investigated the regulation of NFIL3 target genes and found that HDAC2 shRNA induced a high fraction of these genes (Supplemental Tables 2, 3). To examine whether the NFIL3 targets could be FOXO-regulated as well, exogenous activated FOXO1-AAA (mutated on AKT phosphorylation sites) was transfected into HEK293 and BT549 cells, and gene expression was measured. FOXO1-AAA induced many of the NFIL3 shRNA-induced genes (Supplemental Tables 2, 3), demonstrating a partial overlap between NFIL3 and FOXO1-regulated genes. To ensure that gene expression changes induced by NFIL3 shRNA were dependent on the loss of NFIL3 expression, we treated BT549 cells that expressed lentivirally delivered mouse Nfil3 (which is insensitive to the human NFIL3 shRNA) or a GFP control with human NFIL3 shRNA and examined gene expression. We found that the induction of GADD45α and TRAIL was substantially rescued in samples that expressed mouse Nfil3 (Supplemental Fig. 4A–C). Overall, these results show that a reduction in NFIL3 or HDAC2 induced a number of FOXO target genes, many of which are known to be involved in stress response and apoptosis.
To further investigate the ability of NFIL3 to regulate FOXO targets, we performed gene set enrichment analysis (GSEA) with experimentally determined PI3K/PTEN/FOXO pathway gene sets (Matsushima-Nishiu et al. 2001; Ramaswamy et al. 2002; Subramanian et al. 2005; Gomis et al. 2006; Terragni et al. 2008). GSEA with HEK293- and BT549-derived microarray data revealed that genes induced by FOXO1, the PI3K inhibitor LY294002, and exogenous PTEN were enriched in NFIL3 knockdown samples (Fig. 4C–F; Supplemental Fig. 4D). However, even in the case of the Ramaswamy class 2A FOXO1 gene set, which had a normalized enrichment score (NES) of 1.63, many FOXO1 targets were not induced by NFIL3 shRNA. GSEA was also performed with >1400 curated gene sets (Broad Institute Molecular Signatures Database, none of which were PI3K/PTEN/FOXO-regulated) in combination with experimentally determined PI3K/PTEN/FOXO pathway gene sets to compare enrichment levels. Three of the experimentally derived PI3K/PTEN/FOXO pathway-induced sets were in the top 10% of enriched gene sets in NFIL3 shRNA samples, grouping them at the leading edge of enrichment (Supplemental Table 4). The partial enrichment of FOXO target genes after NFIL3 shRNA supports the idea that NFIL3 regulates only a subset of FOXO target genes.
We next examined the GSEA results with NFIL3 shRNA microarray data and >1400 curated sets (Molecular Signatures Database). Gene sets were ranked by NES. Strikingly, eight of the 50 most enriched gene sets (comprising the top third percentile of NFIL3 shRNA-enriched gene sets) were induced by HDAC inhibition (Fig. 4G; Supplemental Table 5), suggesting that HDACs act in concert with NFIL3 to regulate these genes. In a converse manner, GSEA was performed using microarray data from the cancer cell line RJ225 (Burkitt's lymphoma) treated with TSA (Gialitakis et al. 2006). We found that TSA-treated samples had significant enrichment for gene sets induced by FOXO1, LY294002, and PTEN (Supplemental Fig. 4E). These results indicate that NFIL3 and HDACs repress a cohort of FOXO target genes. However, the scope of NFIL3 gene regulation (only 289 genes were induced by twofold or more by NFIL3 shRNA) is much narrower than the scope of genes globally regulated by HDACs. NFIL3 likely hones HDACs to a small portion of their targets, including a cohort of FOXO-regulated genes. Moreover, NFIL3 and FOXO target genes only partially overlap, suggesting that NFIL3 hinders only a portion of FOXO activity.
Residual nuclear FOXO is present in PTEN-null settings
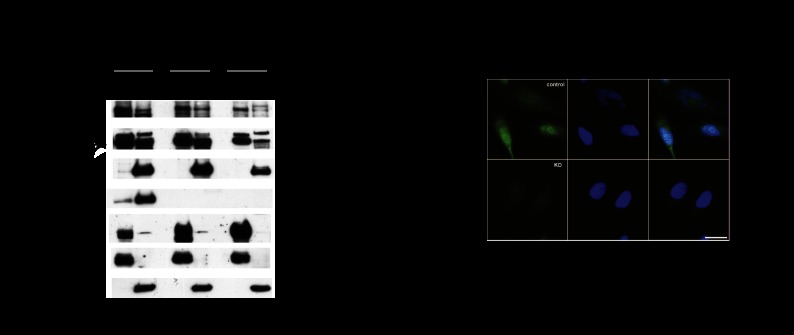
We were surprised that NFIL3 shRNA induced FOXO target genes in PTEN mutant BT549 cells, since strong AKT activation would be expected to phosphorylate FOXO and signal its retention in the cytoplasm. To examine whether FOXO is present in the nuclei, we performed subcellular fractionation on cell extracts from PTEN wild-type HEK293 cells, mouse embryonic fibroblasts (MEFs; PTEN wild-type and deleted), and PTEN mutant BT549 and MDA-MB-468 cancer cells (Fig. 5A; Supplemental Fig. 5A–E). FOXO was detected in the nucleus and cytoplasm of both PTEN wild-type and PTEN mutant cells, with less nuclear FOXO in the PTEN mutant cells (Fig. 5A; Supplemental Fig. 5A). Antibodies were validated with siRNA against FOXO1 and FOXO3 and Cre treatment of MEFs in which the Foxo genes are flanked by lox sites (Supplemental Fig. 5B,D,E). In addition, indirect immunofluorescence of FOXO3 in BT549 cells revealed that 16.9% of the 225 cells checked had nuclear FOXO3 (Fig. 5B). Once we established that FOXO was present in the nucleus, we next sought to determine whether NFIL3 had an impact on FOXO localization. Subcellular fractionation experiments with BT549 cells treated with NFIL3 shRNA revealed that reduction of NFIL3 had no effect on FOXO localization (Supplemental Fig. 5F). In sum, these data indicate that FOXO is present in the nucleus even when AKT is highly activated and therefore susceptible to regulation by NFIL3 on chromatin.
Figure 5.
Residual nuclear FOXO is observed in PTEN-null cancer cell lines. (A) Subcellular fractionation of HEK293, MDA-MB-468 (labeled as 468), and BT549 cell lines; MAX and β-tubulin mark nuclear (N) and cytoplasmic (C) fractions, respectively, with the percentage of fraction loaded shown. PTEN genotype is indicated as wild-type (WT) and mutant (mut.). FOXO1 and FOXO3 are in the nucleus and cytoplasm; “9462” and “Upstate” indicate the particular FOXO antibodies used. The FOXO3 band (indicated by and asterisk) is reduced by siRNA. NFIL3 is nuclear, whereas PTEN and AKT are mostly cytoplasmic. (B) Immunofluorescence with BT549 cells using anti-FOXO3 antibody.
NFIL3 blocks FOXO promoter recruitment on chromatin
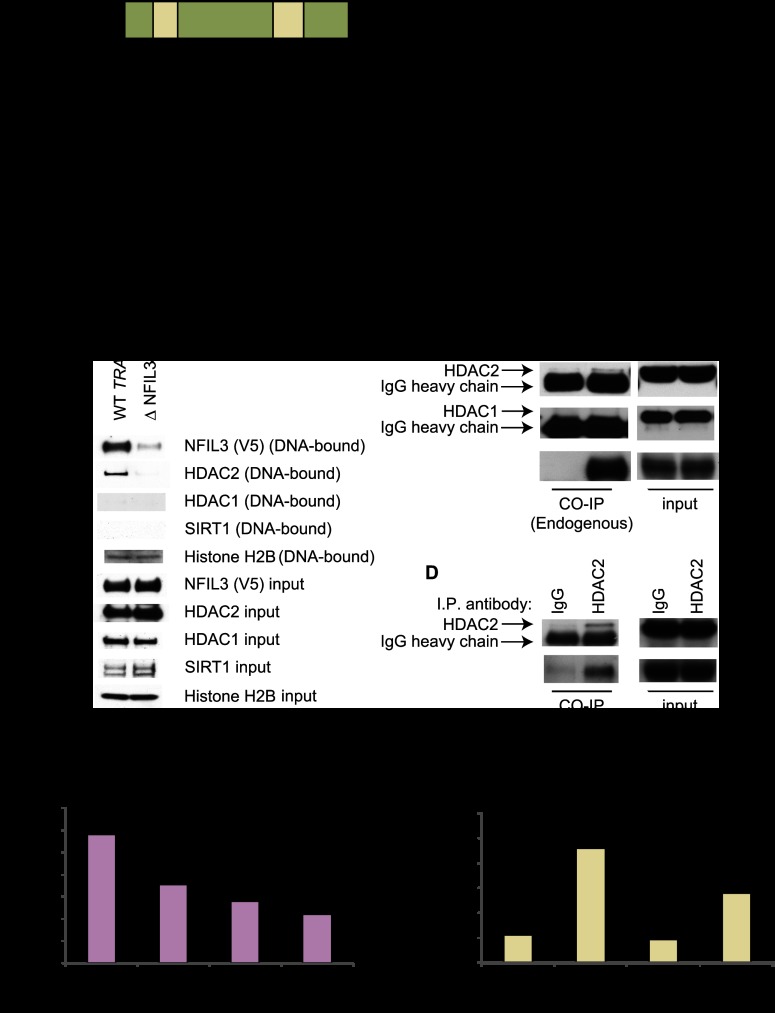
We next set out to determine whether NFIL3 interferes with FOXO's ability to bind to the promoters of endogenous FOXO target genes. Reporter assays had revealed that the NFIL3-binding site was required for TRAIL repression. In addition, predicted NFIL3-binding sites were present in the promoters of FOXO target genes induced by NFIL3 shRNA. To shed further light on the mechanism employed by NFIL3 to regulate FOXO1 output, we performed ChIPs using antibodies to endogenous proteins with extracts from HEK293 or BT549 cells in which NFIL3 had or had not been diminished with shRNA. We found that NFIL3 shRNA-treated samples had significantly lower NFIL3 binding and higher FOXO1 binding to promoters for genes such as TRAIL, GADD45α, GADD45β, and FAS (Fig. 6A,B; Supplemental Table 6). Furthermore, these FOXO target genes had higher histone H3 and H4 acetylation in NFIL3 shRNA-treated samples as measured by ChIP, whereas total histone H3 binding remained the same or decreased on these promoters (Fig. 6A,B; Supplemental Table 6).
Figure 6.
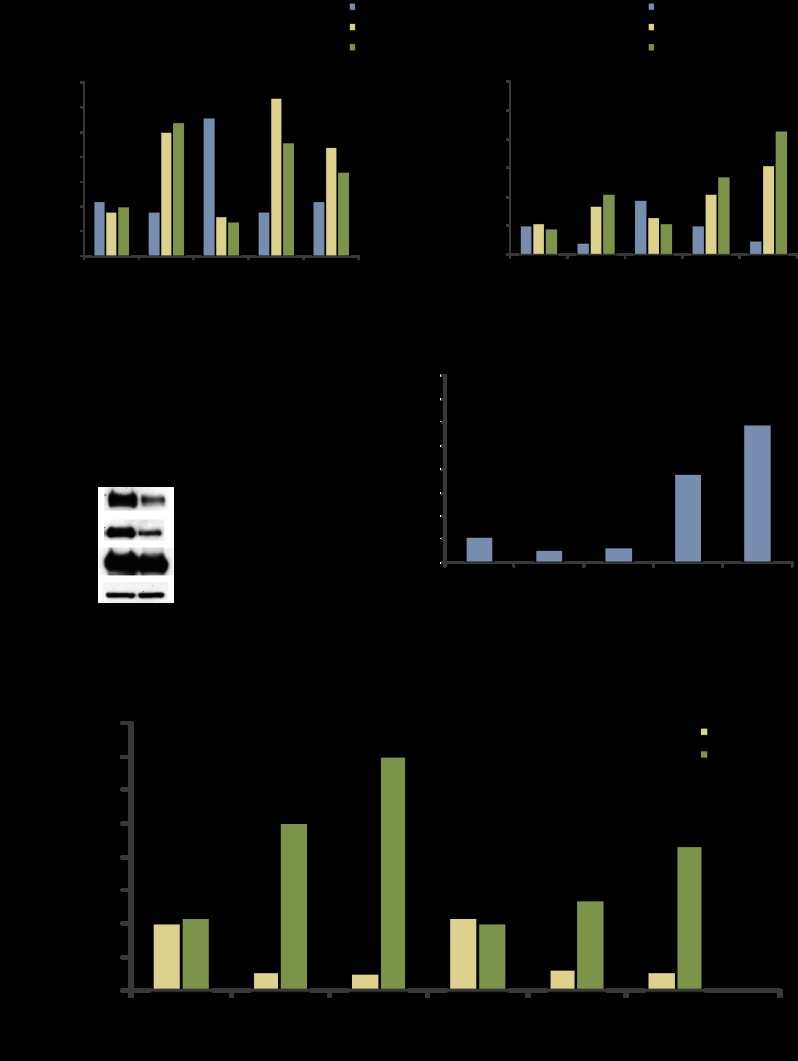
NFIL3 hinders FOXO recruitment to target genes. (A,B) ChIPs with BT549 samples treated with control shRNA or NFIL3 shRNA (KD1 or KD2). Promoter binding was quantified by qPCR using β-actin as a control. NFIL3 shRNA increased FOXO1 TRAIL promoter binding, decreased NFIL3 TRAIL promoter binding, and increased acetyl histone H3 and H4 on the TRAIL promoter. Similar results were obtained for the GADD45α promoter. (C) Western blot for BT549 samples treated with water or 250 μM H2O2 for 18 h. (D) ChIPs with lysates from H2O2-treated (250 μM, 18 h) BT549s. NFIL3 and HDAC2 binding decreased, and FOXO1 binding and acetyl histone H4 increased on the GADD45α promoter. (*) Significantly different from control IgG (P < 0.05). (E) Nfil3 shRNA-induced endogenous Gadd45α gene in 1° MEFs (Rosa26CreERT2, floxed/floxed FoxO1, FoxO3, and FoxO4) that were infected with Ad-GFP or Ad-Cre; Gadd45α induction was reduced in MEFs that were treated with Ad-Cre to remove FoxO genes. (**) Samples have significantly lower Nfil3 expression than control replicates. Data are means ± SEM.
We also examined whether an environmental cue could recapitulate the FOXO1/NFIL3 chromatin-binding dynamics that were observed with NFIL3 shRNA treatment. To do this, we examined DNA binding after cells were treated with 250 μM H2O2 (conditions that are known to activate FOXO). We found that H2O2 treatment markedly reduced total NFIL3 protein, decreased NFIL3 and HDAC2 recruitment to the GADD45α promoter, and increased FOXO1 binding as well as histone H3 and H4 acetylation on the GADD45α promoter (Fig. 6C,D). Interestingly, the reduced NFIL3 protein after H2O2 treatment was associated with induction of NFIL3 mRNA, which may be indicative of feedback regulation of NFIL3 protein levels (Supplemental Fig. 6A). Thus, depletion of NFIL3 protein after treatment with H202 induced FOXO1 promoter recruitment and increased histone acetylation of particular FOXO target genes.
The recruitment of FOXO1 to target promoters following NFIL3 knockdown suggests that these genes were induced in a FOXO-dependent manner. To test this hypothesis, we measured the induction of FOXO target genes in FoxO1, FoxO3, and FoxO4 triple-knockout MEFs. To do this, 1° MEFs with flox/flox FoxO1, FoxO3, and FoxO4 were treated with either Ad-GFP or Ad-Cre to obtain triple-knockout cells and controls. The Cre-mediated loss in FoxO expression was confirmed by qRT–PCR for recombinant deletion products (Supplemental Fig. 6B). The resultant control and triple-knockout MEFs were treated with Nfil3 targeting shRNA, and gene expression was measured by qRT–PCR. We found that triple-knockout MEFs had substantially diminished inductions of the FoxO target genes Gadd45α, Gadd45β, and Fas in Nfil3 shRNA-treated samples (Fig. 6E; Supplemental Fig. 6C). The partial rescue found with triple-knockout cells could be due to incomplete recombination, which we detected in parallel isolates of these cells, and/or the participation of other factors in this regulation. Hence, the Nfil3 regulation of Gadd45α, Gadd45β, and Fas in MEFs is substantially dependent on FoxO.
NFIL3 expression in cancer is associated with poor prognosis
Given that NFIL3 modulates FOXO activity, which is important for tumor development, we investigated NFIL3 expression in cancer. Query of the Oncomine Database revealed that NFIL3 was highly expressed in many different cancers, including basal-like breast cancer and glioblastoma multiforme (Supplemental Fig. 7A; Rhodes et al. 2007). NFIL3 expression in basal-like breast cancer was confirmed by qRT–PCR (Fig. 7A). NFIL3 levels were higher in the more aggressive luminal B breast cancer subtype relative to the less aggressive luminal A (Fig. 7B,C; van de Vijver et al. 2002).
Figure 7.
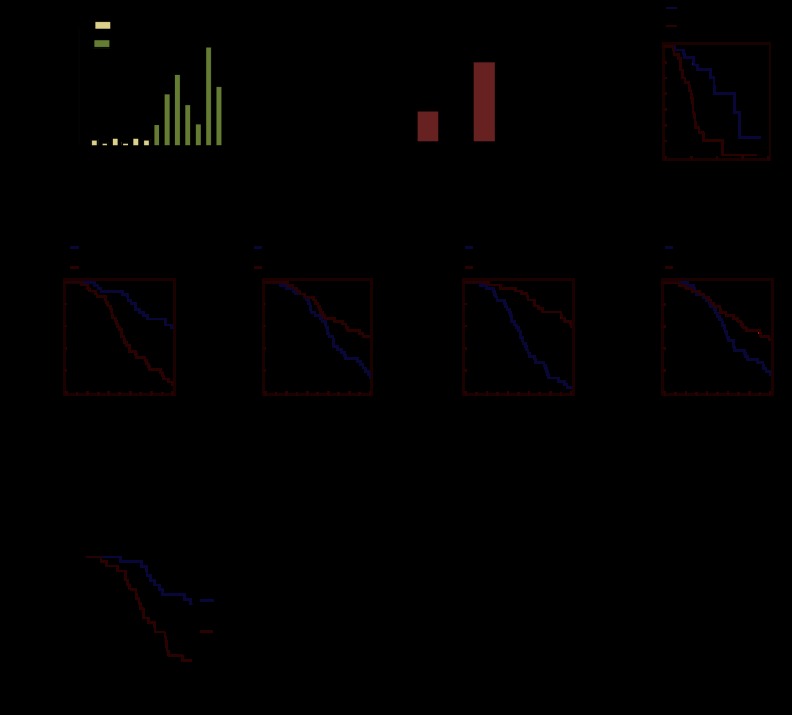
High NFIL3 expression and repression of FOXO targets are associated with poor prognosis in breast cancer. (A) qRT–PCR analysis with tumor samples shows NFIL3 is elevated in basal-like breast cancer. (B) Differential NFIL3 expression in luminal A and luminal B breast tumors (295 from van de Vijver set). (C) Kaplan-Meier analysis of same luminal samples; luminal B had a worse prognosis. (D–G) Kaplan-Meier analysis with 295 samples from the van de Vijver breast tumor data set. Samples were divided into two equal groups based on gene expression levels; these genes were repressed by NFIL3 and induced by FOXO1. (H) Kaplan Meier analysis with the 249 non-basal-like breast cancer samples from NKI (Netherlands Cancer Institute) data set; NFIL3 expression is associated with poor prognosis.
We examined the relationship between NFIL3 expression and cancer survival by performing Kaplan-Meier analysis with a data set of 295 breast tumors that included corresponding patient survival data (van de Vijver et al. 2002). Samples were divided into two equal groups based on NFIL3 levels. High NFIL3 expression was strongly correlated with poor survival (P = 0.003) (Fig. 7D). Because basal-like breast cancer confers a poor prognosis and NFIL3 is highly expressed in basal-like breast cancer, we wanted to determine whether NFIL3 correlated with prognosis in other subtypes of breast cancer. The association between NFIL3 and survival was also found in 249 non-basal-like breast cancer samples (P = 0.0186) (Fig. 7H), indicating that NFIL3 expression is strongly correlated with poor prognosis in breast cancer regardless of the subtype. Furthermore, expression levels of FOXO1 target genes that were repressed by NFIL3 were found to be associated with good prognosis in breast cancer (Fig. 7E–G; Supplemental Fig. 7B–D), suggesting that the repression of these genes by NFIL3 facilitates poor prognosis. In contrast, a number of previously published FOXO1 target genes that were not repressed by NFIL3 in our microarray analysis were strongly associated with poor prognosis in breast cancer: CD72, NFAT5, CD14, and SOD2 (Supplemental Fig. 7E–H; Fan et al. 2010; Ochiai et al. 2012). For these FOXO target genes that were not repressed by NFIL3, we observed the constitutive association of FOXO1 to the promoters by ChIP (Supplemental Fig. 7I). Thus, in poor prognosis tumors, NFIL3 appears to hinder FOXO1 transactivation of target genes involved in triggering cell death and cell cycle arrest but does not hinder the expression of other sets of FOXO1 targets, which are often well expressed in these tumors.
NFIL3 inhibits apoptosis in solid tumor cell lines
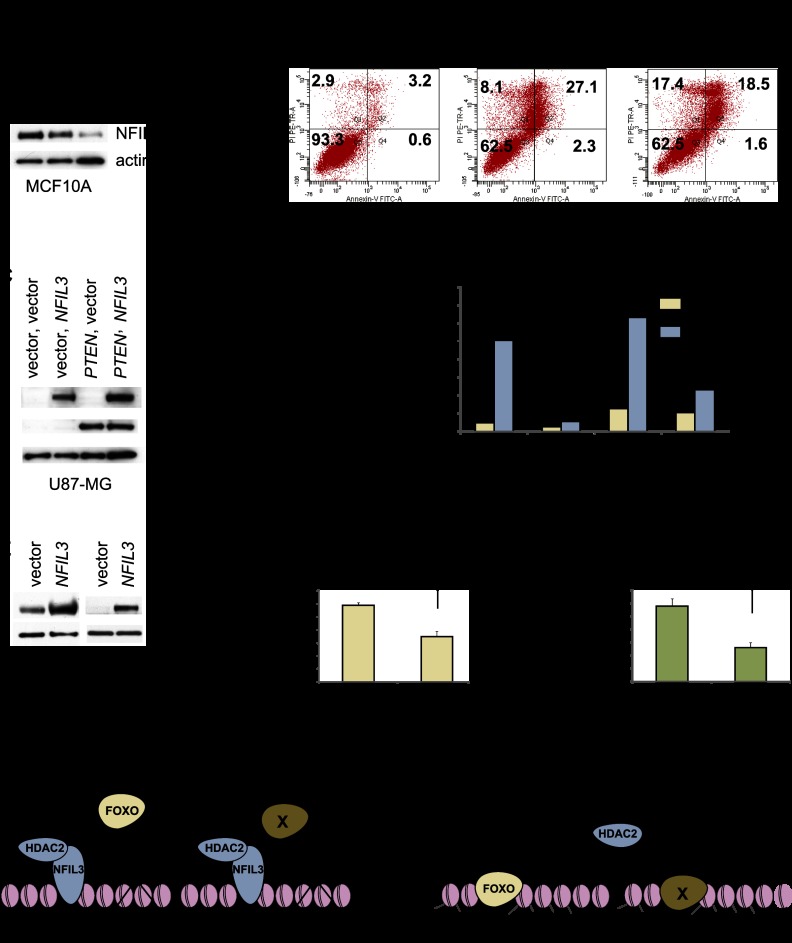
NFIL3 is a survival factor in B lymphocytes and neurons (Ikushima et al. 1997; Cowell 2002). To investigate the role of NFIL3 in cell survival in epithelial cell lines, we reduced its expression in MCF10A, BT549, and MDA-MB-468 cells by shRNA and found that NFIL3 reduction induced apoptosis and, depending on the cell line, some combination of FOXO target genes (TRAIL, GADD45α, GADD45β, and/or FAS) (Fig. 8A,B; Supplemental Fig. 8C,D; Supplemental Table 7). HDAC2 shRNA similarly induced apoptosis in MCF10A cells (Supplemental Fig. 8A,B). To determine the specificity of human NFIL3 shRNA in triggering cell death, experiments were performed in the presence of lentivirally delivered mouse Nfil3, which is resistant to human NFIL3 shRNA. Cell death was substantially rescued by mouse Nfil3 (Supplemental Fig. 8E–G). The cell death was also significantly rescued by shRNA targeting of TRAIL (Supplemental Fig. 8H). In a converse manner, NFIL3 overexpression inhibited H2O2-induced apoptosis in both the presence and absence of exogenous PTEN in the glioblastoma cell line U87MG (Fig. 8C,D) and also suppressed H2O2-induced inhibition of viable cell growth in PTEN mutant BT549 and MDA-MB-468 cancer cells (Fig. 8E–G). Consistent with these results, H2O2 induced FOXO target genes (a combination of GADD45α, GADD45β, and/or FAS, depending on the cell line) (Supplemental Tables 3, 7). These results suggest a role for NFIL3 in cancer cell survival and highlight the ability of NFIL3 to antagonize the proapoptotic effect of PTEN and FOXO.
Figure 8.
NFIL3 shRNA induces cell death, whereas NFIL3 overexpression inhibits cell death, in immortalized and cancer cell lines. (A) NFIL3 was diminished in MCF10A cells with shRNA (KD1 or KD2); Western blot is shown. (B) MCF10A shRNA-treated samples were subjected to cell viability analysis as described in the Supplemental Material. Loss of NFIL3 induced cell death. (C) Western analysis of U87MG cells that overexpress NFIL3 and/or PTEN. (D) Cell viability analysis was performed on transduced U87MG cells that were treated with water or 250 μM H2O2 for 18 h and analyzed for apoptosis as described in the Supplemental Material. The percentage of apoptotic cells equals the propidium iodide and Annexin V single- and double-positive cells divided by the total number of cells; NFIL3 attenuated apoptosis. (E–G) Cell viability assays were performed with BT549 and MDA-MB-468 cells that overexpress NFIL3 (MSCV-NFIL3). Subconfluent cells were treated with 250 μM H2O2 for 18 h. Cells remaining after treatment were stained with crystal violet, and OD 565 nm was measured to quantify growth inhibition. (H) Model for NFIL3 action: The NFIL3/HDAC2 complex alters histone acetylation and blocks transcription factor association (FOXO and, presumably, other transcription factors, denoted as X) with target genes on chromatin. Data are means ± SEM; n = 3.
Discussion
Reporter screen identifies NFIL3 as a modulator of FOXO transcription factor output
PI3K pathway inhibition induces the transcription of a number of its components, including INSR, IRS2, IGF1R, and ERBB3. We exploited this aspect of known PI3K/PTEN homeostatic transcriptional circuitry to identify a novel pathway modulator, NFIL3. We screened PTEN-regulated genes from microarray studies for the ability to affect a reporter of FOXO transcription factor output derived from the TRAIL promoter. As a result, we found that NFIL3 in concert with HDAC2 repressed certain FOXO targets involved in tumor suppression (TRAIL, GADD45α, GADD45β, and FAS) by interfering with nuclear FOXO1 promoter binding and histone acetylation (Figs. 1–6). Our findings support a model in which NFIL3 influences cancer patient survival by working with HDAC2 to interfere with the ability of FOXOs to activate target promoters involved in apoptosis and other responses to stress (Figs. 7, 8). This mechanism appears to be distinct from the regulation of FOXO by AKT or acetylation and is specific to only a subset of FOXO target genes. In addition, NFIL3/HDAC2 also regulate non-FOXO targets, which may also impact cancer patient and cell survival (Fig. 8H).
NFIL3 blocks FOXO1 regulation of a cohort of tumor-suppressive target genes
FOXO transcription factors are controlled by numerous mechanisms such as phosphorylation, acetylation, methylation, and ubiquitination, which direct changes in stability, localization, DNA binding, and protein–protein interactions (Calnan and Brunet 2008). Here, we present a model in which NFIL3 binds to particular FOXO target promoters, thereby blocking FOXO recruitment (Fig. 8H). Gene expression profiling and GSEA indicated that NFIL3 regulates a subset of FOXO target genes (Fig. 4A–F; Supplemental Tables 1–4). Specifically, NFIL3 shRNA treatment induced a subset of FOXO targets that regulate the cell cycle and stress response. This regulation was seen in many contexts, including HEK293s, BT549s, U87MGs, MDA-MB-468s, and a series of immortalized and primary MEF samples (Supplemental Fig. 6; Supplemental Tables 2, 3, 7). Interestingly, NFIL3 blocks the association of another bZIP factor, CEBPβ, to a portion of its target promoters; our GSEA results concur with this finding, as CEBPβ targets were highly induced by NFIL3 shRNA (the CEBP gene set ranked second out of >1400 examined sets in Supplemental Table 5 with an NES of 2.0; Macgillavry et al. 2011). Based on this, a model emerges whereby NFIL3 promoter binding diverts transcription factors away from subsets of target genes.
Mechanistically, we found that NFIL3 required its binding site to repress TRAIL reporter activity (Fig. 2C) and acted at least in part with HDAC2 (Figs. 3B–E, 6D; Supplemental Fig. 3A,B). Interestingly, NFIL3 antagonized FOXOs even in the absence of PTEN (Fig. 4A). This led to the realization that at select target genes, NFIL3 represses the function of FOXOs that persist in the nucleus in PTEN mutant cells (Fig. 5A; Supplemental Fig. 5A). To further scrutinize how NFIL3 regulates FOXO output, we examined endogenous promoter recruitment of these factors and found that reduced promoter binding of NFIL3 caused by shRNA led to increased FOXO1 promoter association as well as increased acetylation of histones H3 and H4 on promoters such as TRAIL, GADD45α, and GADD45β (Fig. 6A,B; Supplemental Table 6). Therefore, we identified a direct mechanism whereby NFIL3 promoter binding blocks FOXO recruitment to certain genes. We further showed that the induction of FoxO target genes by Nfil3 shRNA was diminished in MEFs in which FoxOs were largely deleted (Fig. 6E; Supplemental Fig. 6B,C), showing that NFIL3 regulates FOXO targets in a FOXO-dependent manner.
NFIL3 only repressed a subset of FOXO targets (Fig. 4C–F, Supplemental Figs. 4D, 6A; Supplemental Table 1), allowing nuclear FOXO to activate transcriptional programs (Supplemental Fig. 7E–I) that potentially promote cancer. In line with this idea, many of the FOXO1 targets that we found to be repressed by NFIL3 were more highly expressed in good prognosis breast cancer, whereas other FOXO1-regulated genes, not found to be repressed by NFIL3 in our work, were associated with poor prognosis breast cancer (Fig. 7E–G; Supplemental Fig. 7E–H). FOXO factors indeed have key positive roles in the oncogenesis of AML and breast cancer (Chen et al. 2010; Sykes et al. 2011). Interestingly, FOXO appears to be resistant to high AKT activity in these settings, as evidenced by surprisingly high levels of nuclear FOXO observed in settings with high AKT activity, suggesting that independent FOXO regulatory mechanisms escape AKT-negative regulation (Fig. 5; Supplemental Fig. 5). FOXO1 also induces OCT4 and SOX2 expression in human embryonic stem cells in a setting of high AKT activity (Zhang et al. 2011). Our data would support the notion that elevated NFIL3 expression in cancer could modulate FOXO factors to mitigate their tumor-suppressive outputs and enhance their pro-oncogenic ones. Identifying the full spectrum of nuclear FOXO action and regulation in cancer will shed light on this emerging issue.
NFIL3 as a cell survival factor in cancer
NFIL3 was previously described as a survival factor in B lymphocytes and neurons, but the mechanism has remained elusive (Ikushima et al. 1997; Cowell 2002). Here we show that NFIL3 shRNA up-regulated cell death genes such as TRAIL (Fig. 4; Supplemental Tables 2, 3) and induced apoptosis that could be substantially rescued by either nontargetable Nfil3 or the diminishment of TRAIL by shRNA (Fig. 8A,B; Supplemental Figs. 4A–C, 8C–H). Moreover, in cell survival assays, PTEN, likely through activation of FOXO transcription factors, suppressed the ability of NFIL3 to hinder apoptosis (Fig. 8D). Finally, H202-induced apoptosis was inhibited by NFIL3, and, interestingly, H202 treatment of cells led to the reduction of total and chromatin-bound NFIL3 and reciprocal induction of histone acelylation and activation of FOXO binding (Fig. 6C,D). Induction of apoptosis following NFIL3 knockdown may be a feature of cancers that are adapted to high levels of NFIL3, as the knockout mouse for Nfil3 is viable (Gascoyne et al. 2009). It will be interesting to determine whether NFIL3 or perhaps other transcriptional repressors contribute to the selection of available FOXO target genes during normal development.
NFIL3 is highly expressed in poor prognosis cancers such as basal-like breast cancer and glioblastoma (Fig. 7A–C; Supplemental Fig. 7A,B), and its expression is associated with poor prognosis in breast cancer (Fig. 7D,H). The mechanisms that drive elevated NFIL3 expression in cancer remain to be elucidated. Multiple mechanisms are likely to cause high NFIL3, which could include factors known to induce (interleukins, circadian rhythm, and glucocorticoids) or repress (TGF-β) its expression (Cowell 2002; Gascoyne et al. 2009). Interestingly, GSEA with >1400 curated genes sets from the Molecular Signatues Database showed that genes associated with good prognosis in breast cancer were strongly enriched in NFIL3 knockdown samples (Supplemental Table 5). In addition, many of the FOXO target genes that we found to be repressed by NFIL3 were also associated with good prognosis in breast cancer (Fig. 7E–G; Supplemental Fig. 7B–D). Thus, NFIL3, when highly expressed in certain cancers, alters the epigenetic landscape of chromatin to functionally regulate prognosis genes.
The ability of NFIL3/HDAC2 to inhibit cell death in poor prognosis cancer has important therapeutic implications. NFIL3 shRNA only induced 289 genes by twofold in our microarray analysis (Supplemental Table 1), suggesting a specific transcriptional response that could conceivably be blocked by inhibitors that abrogate the NFIL3/HDAC2 interaction. An inhibitor of HDACs restricted to the HDAC2/NFIL3 transcriptional repressor complex may prove to be of great therapeutic benefit for patients with poor prognosis cancer.
Materials and methods
Reporter assays
HEK293 cells were transfected with 250 ng of luciferase reporter, 100 ng of the TK Renilla control reporter, and additional plasmids as noted using FuGENE 6 (Roche). Five-hundred nanograms of expression vectors (cDNAs in pDEST40, pCEP4 PTEN, DN-FOXO1, and Myr-AKT) was used, whereas only 100 ng of the FOXO1 vector was used. Reporter assays were done 48 h post-transfection using the Dual Luciferase assay kit (Promega) and LUMAT LB9501 luminometer (Berthold). Firefly luciferase activities were normalized using renilla luciferase.
Western blotting
Protein lysates were prepared in 2× sample buffer (125 mM Tris-HCl at pH 6.8, 10% βME, 2% SDS, 20% glycerol, 0.05% Bromophenol Blue, 8 M urea); protein was resolved on 4%–20% Tris-glycine gels (Invitrogen), transferred to PVDF, and probed with indicated antibodies. The following antibodies were used: NFIL3 (H-300), Myc-9E10 (SC-40), HDAC2 (H-54), p300 (N15), and FOXO4 (N-19) from Santa Cruz Biotechnology; V5 from from Invitrogen; α-Flag (F1804), β-actin (AC-74), and HDAC3 (H3034) from Sigma; PTEN (6H2.1) from Cascade Bioscience; β-tubulin (TU-27) from Covance; FOXO3 (06-951), acetyl histone H3 (06-0599), and acetyl histone H4 (06-866) from Upstate Biotechnolog/Millipore; total histone H3 (ab1791) from Abcam; and HDAC1 (CC-2062), FOXO1 (9462), FOXO1 (C29H4), FOXO3 (75D8), AKT (9272), P-AKT-S473 (9271), P-AKT-T308 (2965), P-FOXO T24/32 (9464), and cleaved caspase 3 D175 (9661) from Cell Signaling. The SIRT1 antibody (rabbit polyclonal) was from W. Gu.
Survival analysis
Kaplan-Meier analysis was performed using microarray and survival data (van de Vijver et al. 2002) and MedCalc software. A classification of the van de Vijver 295 tumor set was used to identify non-basal-like breast cancer samples (Chang et al. 2005).
Details of plasmid construction, subcellular fractionation, ChIPs, and qRT–PCR are in the Supplemental Material. See also Supplemental Table 8 for primer sequences.
Acknowledgments
We thank members of the Parsons, Gu, Gautier, Ferrando, Califano, Accili, Abeliovich, Maurer, Plevy, and Dalla-Favera laboratories for reagents and helpful discussions. We thank Adolfo Ferrando and Anna Lasorella for critical reading of the manuscript. We also thank Tao Su and Hanina Hibshoosh from the Herbert Irving Comprehensive Cancer Center (HICCC) Shared Resource for Molecular Pathology, and Kristie Gordon from the HICCC FACS Facility. This work was supported by grants CA082783 and CA97403 from the U.S. National Institutes of Health (to R.P.) and grants BC075515 and PC050068 from the Department of Defense (to M.K.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.214049.113.
References
- Calnan DR, Brunet A 2008. The FoxO code. Oncogene 27: 2276–2288 [DOI] [PubMed] [Google Scholar]
- Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, Majumder PK, Baselga J, Rosen N 2011. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell 19: 58–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sorlie T, Dai H, He YD, van't Veer LJ, Bartelink H, et al. 2005. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci 102: 3738–3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Gomes AR, Monteiro LJ, Wong SY, Wu LH, Ng TT, Karadedou CT, Millour J, Ip YC, Cheung YN, et al. 2010. Constitutively nuclear FOXO3a localization predicts poor survival and promotes Akt phosphorylation in breast cancer. PLoS ONE 5: e12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell IG 2002. E4BP4/NFIL3, a PAR-related bZIP factor with many roles. Bioessays 24: 1023–1029 [DOI] [PubMed] [Google Scholar]
- Fan W, Morinaga H, Kim JJ, Bae E, Spann NJ, Heinz S, Glass CK, Olefsky JM 2010. FoxO1 regulates Tlr4 inflammatory pathway signalling in macrophages. EMBO J 29: 4223–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, Brady HJ 2009. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol 10: 1118–1124 [DOI] [PubMed] [Google Scholar]
- Gialitakis M, Kretsovali A, Spilianakis C, Kravariti L, Mages J, Hoffmann R, Hatzopoulos AK, Papamatheakis J 2006. Coordinated changes of histone modifications and HDAC mobilization regulate the induction of MHC class II genes by trichostatin A. Nucleic Acids Res 34: 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis RR, Alarcon C, He W, Wang Q, Seoane J, Lash A, Massagué J 2006. A FoxO-Smad synexpression group in human keratinocytes. Proc Natl Acad Sci 103: 12747–12752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong TM, Yang PC, Peck K, Chen JJ, Yang SC, Chen YC, Wu CW 2000. Profiling the downstream genes of tumor suppressor PTEN in lung cancer cells by complementary DNA microarray. Am J Respir Cell Mol Biol 23: 355–363 [DOI] [PubMed] [Google Scholar]
- Ikushima S, Inukai T, Inaba T, Nimer SD, Cleveland JL, Look AT 1997. Pivotal role for the NFIL3/E4BP4 transcription factor in interleukin 3-mediated survival of pro-B lymphocytes. Proc Natl Acad Sci 94: 2609–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada M, Levy DM, McKeag L, Murray K, Schroder AJ, Canfield SM, Traver G, Rothman PB 2010. IL-4-induced transcription factor NFIL3/E4BP4 controls IgE class switching. Proc Natl Acad Sci 107: 821–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. 1997. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275: 1943–1947 [DOI] [PubMed] [Google Scholar]
- Macgillavry HD, Cornelis J, van der Kallen LR, Sassen MM, Verhaagen J, Smit AB, van Kesteren RE 2011. Genome-wide gene expression and promoter binding analysis identifies NFIL3 as a repressor of C/EBP target genes in neuronal outgrowth. Mol Cell Neurosci 46: 460–468 [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC 2007. AKT/PKB signaling: Navigating downstream. Cell 129: 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima-Nishiu M, Unoki M, Ono K, Tsunoda T, Minaguchi T, Kuramoto H, Nishida M, Satoh T, Tanaka T, Nakamura Y 2001. Growth and gene expression profile analyses of endometrial cancer cells expressing exogenous PTEN. Cancer Res 61: 3741–3749 [PubMed] [Google Scholar]
- Minucci S, Pelicci PG 2006. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer 6: 38–51 [DOI] [PubMed] [Google Scholar]
- Modur V, Nagarajan R, Evers BM, Milbrandt J 2002. FOXO proteins regulate tumor necrosis factor-related apoptosis inducing ligand expression. Implications for PTEN mutation in prostate cancer. J Biol Chem 277: 47928–47937 [DOI] [PubMed] [Google Scholar]
- Ochiai K, Maienschein-Cline M, Mandal M, Triggs JR, Bertolino E, Sciammas R, Dinner AR, Clark MR, Singh H 2012. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat Immunol 13: 300–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, et al. 2007. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128: 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR 2002. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell 2: 81–91 [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. 2007. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9: 166–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmstrom PO, Mansukhani M, Enoksson J, et al. 2005. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res 65: 2554–2559 [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. 2004. High frequency of mutations of the PIK3CA gene in human cancers. Science 304: 554. [DOI] [PubMed] [Google Scholar]
- Simpson L, Li J, Liaw D, Hennessy I, Oliner J, Christians F, Parsons R 2001. PTEN expression causes feedback upregulation of insulin receptor substrate 2. Mol Cell Biol 21: 3947–3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolarov J, Chang K, Reiner A, Rodgers L, Hannon GJ, Wigler MH, Mittal V 2001. Design of a retroviral-mediated ecdysone-inducible system and its application to the expression profiling of the PTEN tumor suppressor. Proc Natl Acad Sci 98: 13043–13048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes SM, Lane SW, Bullinger L, Kalaitzidis D, Yusuf R, Saez B, Ferraro F, Mercier F, Singh H, Brumme KM, et al. 2011. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell 146: 697–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terragni J, Graham JR, Adams KW, Schaffer ME, Tullai JW, Cooper GM 2008. Phosphatidylinositol 3-kinase signaling in proliferating cells maintains an anti-apoptotic transcriptional program mediated by inhibition of FOXO and non-canonical activation of NFκB transcription factors. BMC Cell Biol 9: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. 2002. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347: 1999–2009 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yalcin S, Lee DF, Yeh TY, Lee SM, Su J, Mungamuri SK, Rimmele P, Kennedy M, Sellers R, et al. 2011. FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nat Cell Biol 13: 1092–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]