Bone resorption by osteoclasts requires lysosomes to release proteases into the resorption lacuna. Ferron et al. find that the osteoclast differentiation factor RANKL promotes lysosomal biogenesis once osteoclasts are differentiated through the selective activation of TFEB. PKCβ phosphorylation of TFEB stabilizes and increases its activity. Mice lacking either TFEB or PKCβ in their osteoclasts exhibit decreased lysosomal gene expression and increased bone mass. These results uncover RANKL-dependent signaling occurring in differentiated osteoclasts to enhance lysosomal biogenesis, a necessary step for proper bone resorption.
Keywords: osteoclast, RANKL, lysosomal biogenesis, TFEB, PKCβ
Abstract
Bone resorption by osteoclasts requires a large number of lysosomes that release proteases in the resorption lacuna. Whether lysosomal biogenesis is a consequence of the action of transcriptional regulators of osteoclast differentiation or is under the control of a different and specific transcriptional pathway remains unknown. We show here, through cell-based assays and cell-specific gene deletion experiments in mice, that the osteoclast differentiation factor RANKL promotes lysosomal biogenesis once osteoclasts are differentiated through the selective activation of TFEB, a member of the MITF/TFE family of transcription factors. This occurs following PKCβ phosphorylation of TFEB on three serine residues located in its last 15 amino acids. This post-translational modification stabilizes and increases the activity of this transcription factor. Supporting these biochemical observations, mice lacking in osteoclasts—either TFEB or PKCβ—show decreased lysosomal gene expression and increased bone mass. Altogether, these results uncover a RANKL-dependent signaling pathway taking place in differentiated osteoclasts and culminating in the activation of TFEB to enhance lysosomal biogenesis—a necessary step for proper bone resorption.
Bone is constantly remodeled through the coordinated action of two cell types: the osteoblast and the osteoclast (Ducy et al. 2000; Teitelbaum 2000). While the osteoblast synthesizes and mineralizes the bone extracellular matrix (ECM), the osteoclast is responsible for resorbing this mineralized ECM. To achieve this specialized task, differentiated and multinucleated osteoclasts attach tightly to the bone surface and generate closed resorption lacunae. These resorption lacunae are characterized by an acidic pH (∼4.5) and contain numerous proteases that are exported by the large number of lysosomes present in this cell type (Coxon and Taylor 2008). Proteases and acidification of the lacunae are both necessary for efficient bone resorption. The importance of lysosomal biogenesis for osteoclast function and optimum bone resorption underscores the need to elucidate how this aspect of osteoclast biology is regulated.
Over the last two decades, our understanding of the mechanisms by which cells of the myeloid lineage differentiate into functional multinucleated osteoclasts has made considerable progress (Teitelbaum and Ross 2003; Takayanagi 2007). This led to the identification of two cytokines, RANKL and M-CSF (Yoshida et al. 1990; Lacey et al. 1998), as critical determinants of this process and the identification of numerous transcription factors acting downstream from these cytokines. Some of these factors—PU.1, c-FOS, JunB, Fra-1, NFκB, and PPARγ—act early in the differentiation process within the myeloid precursor cells (Grigoriadis et al. 1994; Iotsova et al. 1997; Tondravi et al. 1997; Matsuo et al. 2000; Kenner et al. 2004; Wan et al. 2007). In contrast, NFATc1, MITF, and TFE3 affect later aspects of osteoclast differentiation, such as the fusion of the precursor cells to form multinucleated osteoclasts (Steingrimsson et al. 2002; Takayanagi et al. 2002). It is not known however, whether these transcription factors promote lysosomal biogenesis in osteoclasts or whether this process is also under the control of a different signaling pathway and transcription factors. The fact that RANKL signaling increases the ability of already differentiated osteoclasts to resorb bone ECM through a poorly understood mechanism is another reason to ask this question (Lacey et al. 1998; Burgess et al. 1999; Hsu et al. 1999; Lomaga et al. 1999).
TFEB is a member of the MITF/TFE subfamily of basic helix–loop–helix leucine zipper (bHLH-Zip) transcription factors that comprises three other members: MITF, TFE3, and TFEC (Steingrimsson et al. 2004). Overexpression of TFEB in HeLa cells, mouse embryonic fibroblasts (MEFs), or hepatocytes can enhance lysosomal biogenesis and function through the activation of multiple genes encoding lysosomal proteins (Sardiello et al. 2009; Medina et al. 2011; Settembre et al. 2011, 2012). Interestingly, some of these genes are also implicated in mice and humans in ECM acidification and degradation by osteoclasts. Those are ACP5, ATP6V0D1, ATP6V0D2, ATP6V1C1, CTSK, CLCN7, and OSTM1 (Gelb et al. 1996; Kornak et al. 2001; Chalhoub et al. 2003; Lee et al. 2006; Feng et al. 2009). This observation raises the prospect that RANKL could increase the resorptive activity of osteoclasts in part by recruiting TFEB.
Testing this hypothesis in vivo revealed that, in a three-step pathway, RANKL signaling in osteoclasts recruits PKCβ, which phosphorylates TFEB on previously uncharacterized sites. This phosphorylation results in TFEB accumulation in osteoclasts, an increase in expression of lysosomal genes, and, ultimately, an increase in lysosomal biogenesis. Cell-specific loss-of-function experiments verified that both TFEB and PKCβ are required for lysosomal biogenesis and osteoclast function. We further show that this RANKL–PKCβ–TFEB cascade is specific to TFEB and does not affect accumulation of MITF, another member of the MITF/TFE family that is implicated in osteoclast differentiation.
Results
RANKL regulates lysosomal biogenesis in differentiated osteoclasts
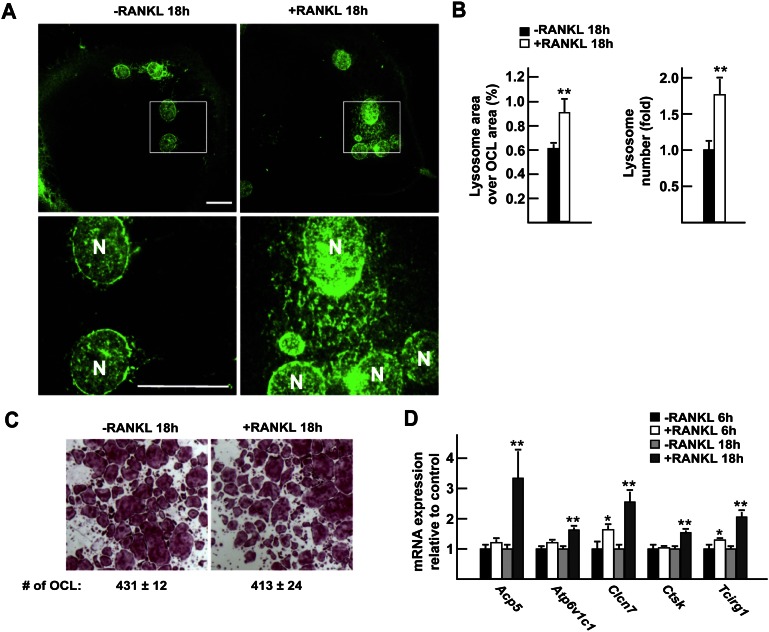
To study ex vivo how RANKL favors lysosomal biogenesis in mature osteoclasts, we generated fully differentiated multinucleated osteoclasts by culturing bone marrow-derived monocytes in the presence of M-CSF and RANKL for 6 d (Lacey et al. 1998). Thereafter, RANKL was either withdrawn from or kept in the culture medium for an additional 18 h, and lysosomal biogenesis was assessed by immunofluorescence staining for lysosomal-associated membrane protein 1 (LAMP1), a molecular marker of lysosomes. Both the area covered by lysosomes and the number of lysosomes in each osteoclast were significantly increased in RANKL-treated compared with untreated osteoclasts, whereas the total number of multinucleated tartrate-resistant acid phosphatase (TRAP)-positive osteoclasts remained the same in both groups (Fig. 1A–C). Thus, under the conditions of this cell-based assay, one can dissociate the role of RANKL in osteoclast differentiation from its regulation of lysosomal biogenesis in fully differentiated osteoclasts.
Figure 1.
Transcriptional regulation of lysosomal biogenesis by RANKL in osteoclasts. (A) LAMP1 immunofluorescence staining of differentiated osteoclasts cultured in the presence (right panels) or absence (left panels) of RANKL (30 ng/mL) for 18 h. The top panels show 63× magnification pictures, and the boxed area is further magnified in the bottom panels. Bar, 10 μm; (N) Nucleus. (B) Quantification of the percentage of the osteoclast area covered by lysosomes in individual osteoclasts and of the number of lysosomes per osteoclast relative to −RANKL, normalized by the osteoclast area and the number of nuclei per osteoclast. (C) TRAP staining of osteoclasts treated or not with RANKL for 18 h (2.5× magnification). (D) Quantitative PCR (qPCR) expression analysis of differentiated osteoclasts treated or not with RANKL for 6 h and 18 h. In all experiments, the cells were cultured in the presence of FBS (10%) and M-CSF (10 ng/mL).
This latter function of RANKL is the result of specific transcriptional events, since, when compared with untreated cells, osteoclasts treated with RANKL for 6 h or 18 h demonstrated a significant increase in the expression of Acp5, Atp6v1c1, Clcn7, Ctsk, and Tcirg1, five lysosomal genes that play a critical role during bone resorption in mice and humans (Fig. 1D; Gelb et al. 1996; Frattini et al. 2000; Kornak et al. 2001; Suter et al. 2001; Feng et al. 2009). We therefore used expression of these five genes as a readout of lysosomal biogenesis in osteoclasts in the remainder of this study.
TFEB regulates the expression of genes implicated in osteoclast function
In order to understand how RANKL signaling in differentiated osteoclasts promotes lysosomal biogenesis, we studied transcription factors that have been proposed to affect this process. We were intrigued by the fact that both overexpression in cells or mice and knockdown in cell lines of the transcription factor TFEB affect expression of Atp6v1c1 and Clcn7 genes just as RANKL treatment of differentiated osteoclasts does (Fig. 1E; Sardiello et al. 2009; Settembre et al. 2011). We therefore asked whether this transcription factor was expressed in osteoclasts, necessary for lysosomal biogenesis in vivo, and regulated transcriptionally or post-transcriptionally by RANKL.
Tfeb expression in osteoclasts was fivefold to 10-fold lower than the one of Mitf and Tfe3, two other members of the MITF/TFE gene family implicated in osteoclasts differentiation (Supplemental Fig. S1A; Steingrimsson et al. 2002), yet Tfeb was more highly expressed in differentiated osteoclasts, as defined by their Ctsk expression, than in any other tissues tested (Fig. 2A).
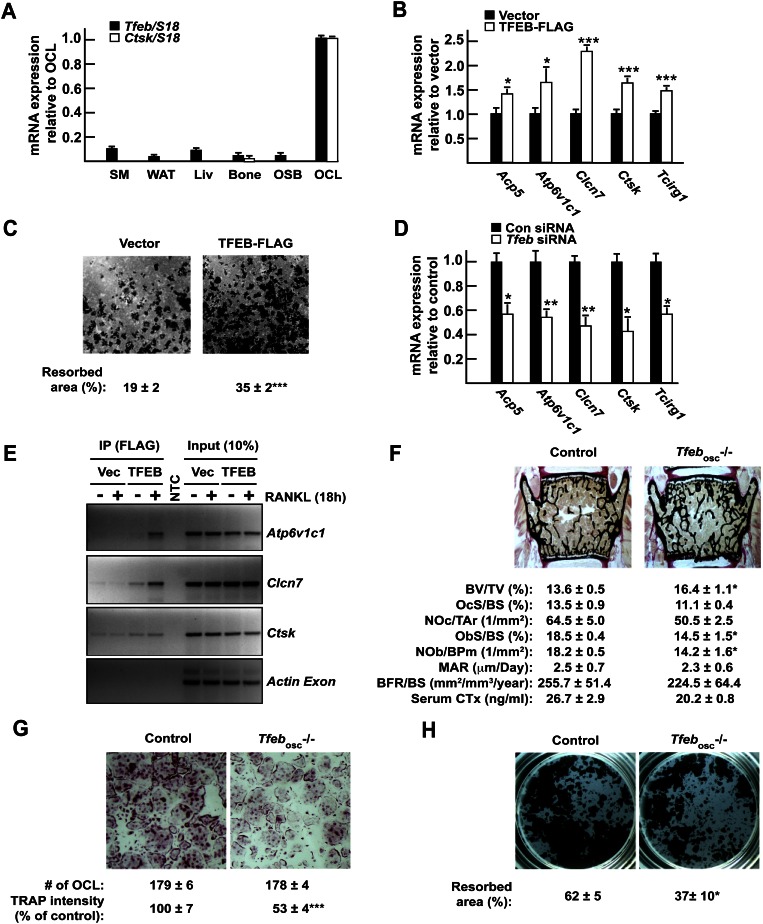
Figure 2.
TFEB is required for normal osteoclast function in vitro and in vivo. (A) Expression pattern of Tfeb and Ctsk in mouse tissues and cell types by qPCR. (SM) Skeletal muscle; (WAT) white adipose tissue; (Liv) liver; (OSB) osteoblasts; (OCL) osteoclasts. (B) qPCR expression analysis in RAW 264.7 cells stably transfected with empty vector or TFEB-Flag and treated for 72 h with RANKL (30 ng/mL). (C) Resorption assay. RAW 264.7 cells expressing TFEB-Flag or not were treated for 96 h with RANKL (30 ng/mL), and the percentage of resorbed area over total area was quantified. (D) qPCR expression analysis in RAW 264.7 cells transfected with control nontargeting siRNA (Con siRNA) or siRNA targeting Tfeb (Tfeb siRNA) and treated for 72 h with RANKL (30 ng/mL). (E) ChIP assays performed on RAW 264.7 cells transfected with vector or TFEB-Flag using Flag antibodies demonstrate binding of TFEB to indicated genes but not to the coding region of Actin (see also Supplemental Fig. S1D). (F) Bone histomorphometric analysis of lumbar vertebrae in 6-wk-old control and Tfebosc−/− female mice. (BV/TV) Bone volume over tissue volume; (OcS/BS) osteoclast surface over bone surface; (NOc/Tar) number of osteoclasts per tissue area; (ObS/BS) osteoblast surface over bone surface; (NOb/BPm) number of osteoblasts per bone perimeter; (MAR) mineral apposition rate; (BFR/BS) bone formation rate over bone surface. Fasting serum CTx levels are also included. (G) TRAP staining of control and Tfebosc−/− bone marrow-derived osteoclasts (5× magnification). The number of osteoclasts per well and the relative TRAP staining intensity are indicated. (H) Resorption assay. Control and Tfebosc−/− primary monocytes were differentiated into osteoclasts on Osteo assay for 6 d. The percentage of the resorbed area over the total area is indicated. All experiments were performed at least in quadruplicate. Four to six animals of each genotype were analyzed in F.
To determine whether TFEB was involved in lysosomal biogenesis in osteoclasts in cell culture, we performed two different types of assay. First, we generated clones of RAW 264.7 cells, which can be differentiated into osteoclast-like cells upon RANKL treatment (Hsu et al. 1999), stably overexpressing a Flag-tagged version of TFEB (Supplemental Fig. S1B). Cells were then treated with RANKL for 3 d, and gene expression was analyzed. Tfeb overexpression increased RANKL-mediated expression of Acp5, Atp6v1c1, Clcn7, Ctsk, and Tcirg1 (Fig. 2B). Moreover, increasing Tfeb expression in RAW 264.7 cells resulted in the generation of osteoclast-like cells that were more efficient in resorbing a mineralized ECM in an in vitro resorption assay (Fig. 2C). Conversely, we transfected a pool of siRNAs targeting Tfeb in RAW 264.7 cells and verified that these siRNAs efficiently decreased expression of Tfeb but not of other members of the MITF/TFE family (Supplemental Fig. S1C). Osteoclast differentiation was then induced by RANKL addition, and expression of lysosomal genes was analyzed 72 h later. Mirroring what was observed in RAW 264.7 cells overexpressing Tfeb, knockdown of Tfeb in RAW 264.7 cells resulted in a 30%–60% decrease in the ability of RANKL to induce expression of Acp5, Atp6v1c1, Clcn7, Ctsk, and Tcirg1 (Fig. 2D). Bioinformatics analysis of the promoter regions of these genes revealed the presence of at least one TFEB-binding site in each of them (Supplemental Fig. S1D). Chromatin immunoprecipitation (ChIP) assay in RAW 264.7 cells demonstrated that TFEB was binding to sites detected in Atp6v1c1, Clcn7, and Ctsk promoters and that this binding was increased in the presence of RANKL, at least for Atp6v1c1 and Clcn7 (Fig. 2E). Taken together, the results of these gain-of-function and loss-of-function experiments indicate that, in cell culture, TFEB favors lysosomal biogenesis and thereby the resorptive capacity of osteoclasts.
Tfeb is required for normal osteoclast function in vivo
Next, to determine whether TFEB affects lysosomal biogenesis in osteoclasts in vivo, we generated a mouse model lacking Tfeb specifically in osteoclasts (Tfebosc−/−) by crossing Tfebfl/+ mice with Cathepsink-Cre (Ctsk-Cre) mice that delete genes in differentiated osteoclasts specifically and do not demonstrate any bone mass phenotype (Supplemental Fig. S2D; Nakamura et al. 2007). Gene expression analysis showed that Tfeb expression was decreased at least 70% in long bones of Tfebosc−/− mice and by >90% in primary osteoclasts derived from these mice compared with control (Tfebfl/fl) osteoclasts (Supplemental Fig. S2A). Recombination at the Tfeb locus in the Tfebosc−/− mice was detected in bones but not in any of the other tissues tested (Supplemental Fig. S2B). Of note, while the expression of Tfec and Tfe3 was unchanged in Tfeb-deficient osteoclasts, the one of Mitf was increased by ∼40% (Supplemental Fig. S2C). Thus, this mouse model allowed us to study TFEB function in osteoclasts in vivo.
Bone histomorphometric analysis of lumbar vertebrae and microcomputed tomography (μCT) of tibia revealed that bone mass was significantly increased in Tfebosc−/− mice compared with Tfebfl/fl control littermates (Fig. 2F; Supplemental Fig. S2E). However, neither osteoclast number nor surface differed between control and mutant mice; likewise, bone formation remained unchanged, while osteoblast number was marginally decreased (Fig. 2F). The length and shape of Tfebosc−/− long bones were unaffected compared with control mice. However, the presence of cartilage remnants in their mineralized trabeculae and a decrease in serum CTx levels in Tfebosc−/− mice suggested a defect in osteoclast function (Fig. 2F; Supplemental Fig. S2F,G). In view of these findings and given the ability of TFEB to regulate the expression of lysosomal genes implicated in osteoclast function, we tested whether Tfeb inactivation in osteoclasts affects their resorbing capacity by decreasing expression of genes required for lysosome function. Five different lines of evidence supported this hypothesis.
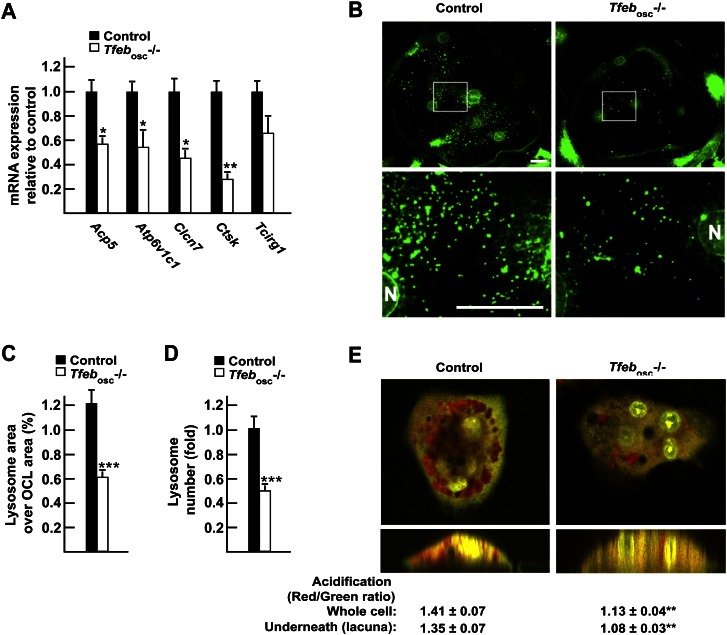
First, while bone marrow cells originating from Tfebosc−/− or control mice differentiated equally well in TRAP-positive osteoclasts in culture, the TRAP staining was consistently weaker in Tfeb−/− than in control osteoclasts (Fig. 2G). This observation suggested that TFEB might be required for the full induction of lysosomal genes in osteoclasts. Second, Tfeb−/− osteoclasts were resorbing bone or a mineralized substrate less efficiently than control osteoclasts, even though the number of TRAP-positive osteoclasts was not decreased in the culture (Fig. 2G,H; Supplemental Fig. S3A). Third, gene expression analysis of Tfebosc−/− and control long bones demonstrated that the expression of all five lysosomal genes implicated in osteoclastic activity—including Acp5, the gene encoding TRAP—was decreased between 40% and 75% when Tfeb was deleted in osteoclasts (Fig. 3A). Fourth, LAMP1 immunofluorescence staining of primary osteoclasts revealed that the number of LAMP1-positive lysosomes was significantly reduced in Tfeb−/− compared with control osteoclasts grown on glass (Fig. 3B). Quantitative analysis of osteoclasts of each genotype demonstrated that the total area covered by lysosomes and the total number of lysosomes in each osteoclast were reduced by half in Tfeb−/− osteoclasts (Fig. 3C,D). Of note, when grown on bone, the LAMP1-positive area within the actin ring was decreased in Tfeb−/− osteoclasts compared with control (Supplemental Fig. S3B). Fifth, acridine orange staining of osteoclasts grown on bone reveals that the intracellular pH and the one underneath the cell were less acidic in Tfeb−/− osteoclasts compared with control (Fig. 3E). Together, these results support the notion that, in vivo, TFEB is needed to maintain osteoclast function by promoting expression of several lysosomal genes.
Figure 3.
TFEB regulates lysosomal biogenesis in osteoclasts. (A) qPCR expression analysis in control and Tfebosc−/− flushed long bones. (B) LAMP1 immunofluorescence staining of control (left panels) and Tfebosc−/− (right panels) primary osteoclasts. The top panels show 40× magnification pictures, and the boxed area is further magnified in the bottom panels. Bar, 10 μm. (N) Nucleus. (C) Quantification of the percentage of the osteoclast area covered by lysosomes in individual osteoclasts. (D) Quantification of the number of lysosomes per osteoclast relative to control, normalized by the osteoclast area and the number of nuclei per osteoclast. (E) Acridine orange staining of live osteoclasts differentiated on bone slices imaged in a Z-stack using a confocal microscope. The ratio of red (acidic) over green (neutral) signal was quantified in several osteoclasts (n = 10–12) within and underneath the cells. The bottom panels display an orthogonal projection of the top panels. All experiments were performed at least in quadruplicate, and four to six animals of each genotype were analyzed in A.
RANKL induces TFEB protein accumulation in cell culture
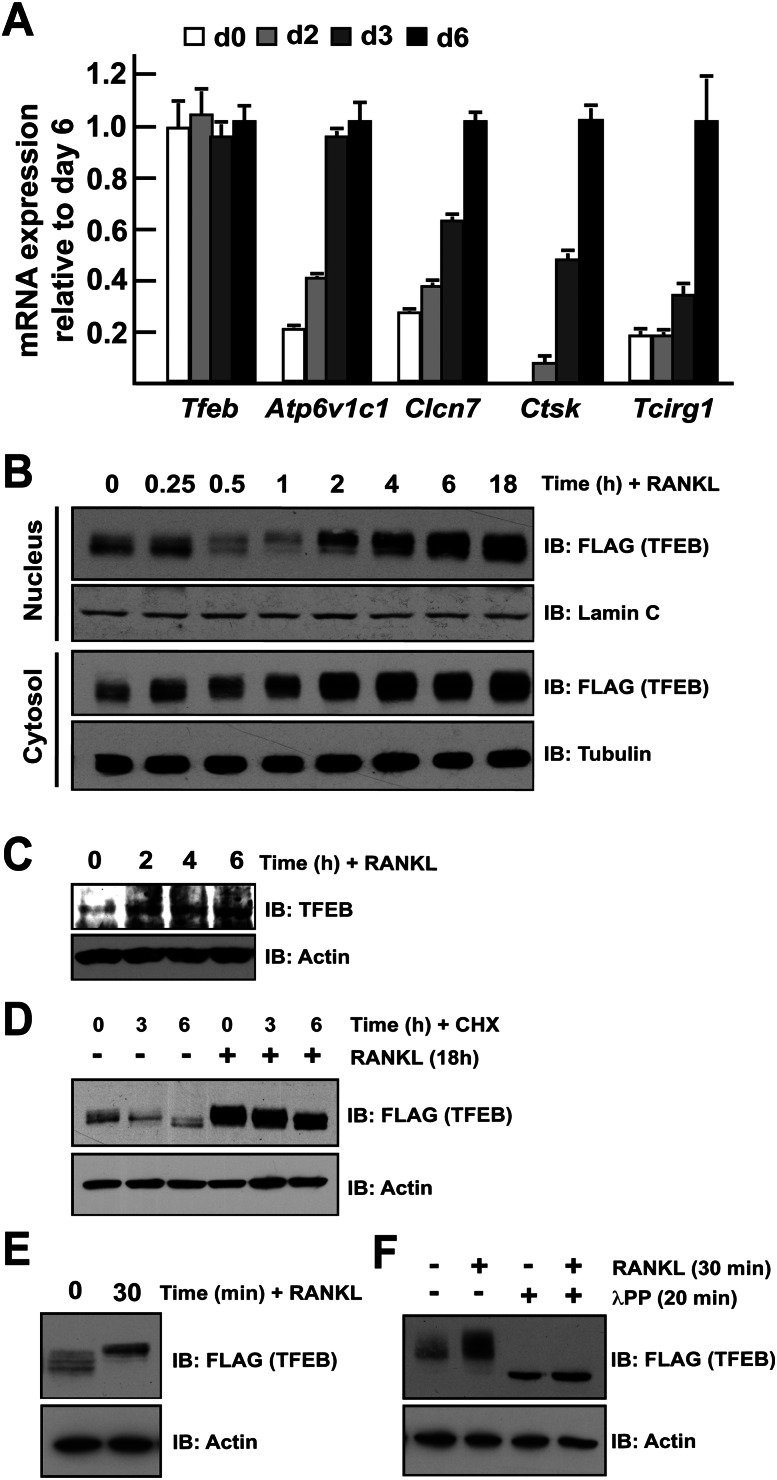
The above-described functions of TFEB prompted us to ask whether it was a transcriptional mediator of RANKL regulation of lysosomal biogenesis. While Tfeb expression in primary osteoclasts was unaffected by RANKL treatment, expression of its target genes—Atp6v1c1, Clcn7, Ctsk, and Tcirg1—was markedly increased over the course of osteoclast differentiation (Fig. 4A). Furthermore, cell fractionation studies demonstrated that TFEB protein accumulates in the nucleus of RAW 264.7 cells as early as 4 h after addition of RANKL in the medium (Fig. 4B, top panels). Immunofluorescence staining revealed that the TFEB-Flag protein was localized in the nucleus of osteoclast-like cells following RANKL treatment (Supplemental Fig. S4). Importantly, the cytosolic levels of TFEB were also increased within 2 h of RANKL treatment (Fig. 4B, bottom panels), suggesting that the increase in cellular TFEB levels could be secondary to the stabilization of TFEB. Stabilization of endogenous TFEB protein was also observed following RANKL treatment of primary osteoclast precursors (Fig. 4C). Treatment with cycloheximide, an inhibitor of protein synthesis, decreased TFEB levels in RAW 264.7 cells in the absence of RANKL but not in RANKL-treated cells (Fig. 4D). These results suggest that one mechanism by which RANKL induces TFEB accumulation is by increasing its stability.
Figure 4.
TFEB is regulated post-translationally by RANKL. (A) qPCR expression analysis of the indicated genes in bone marrow-derived osteoclasts differentiated in the presence of M-CSF and RANKL for 0–6 d. The expression level of each gene is normalized to the expression at day 6. (B) RAW 264.7 cells stably transfected with TFEB-Flag were treated for the indicated times with RANKL (50 ng/mL). Nucleus and cytosol extracts were prepared as described in the Materials and Methods, and TFEB protein was revealed by Western blotting using Flag antibodies. Lamin C and tubulin were used as nuclear- and cytosolic-specific markers, respectively. (C) Bone marrow-derived monocytes were treated with RANKL (50 ng/mL) for the indicated times, and endogeous TFEB protein was detected by Western blotting. (D) RAW 264.7 cells stably transfected with TFEB-Flag were treated with cycloheximide (CHX at 100 μg/mL) and/or RANKL (50 ng/mL) for the indicated times, and TFEB protein in total cell extracts was revealed by Western blotting using Flag antibodies. (E) RAW 264.7 cells stably transfected with TFEB-Flag were treated with RANKL (50 ng/mL) for the indicated times, and TFEB protein in total cell extracts was revealed by Western blotting using Flag antibodies. (F) RAW 264.7 cells stably transfected with TFEB-Flag were treated with RANKL (50 ng/mL) for 30 min. The total cell extracts were next subjected to λPPase treatment as described in the Materials and Methods. In D–G, actin was used as a loading control. In all experiments, the cells were cultured in the presence of FBS (10%).
PKCβ mediates RANKL-induced lysosomal biogenesis by phosphorylating TFEB on novel sites
In trying to understand how RANKL may increase TFEB stability, we noticed that treatment of RAW 264.7 cells with RANKL consistently caused an electrophoretic shift of TFEB migration whether one looks at nuclear, cytosolic, or total cell extracts (Fig. 4B,E). This suggested that RANKL signaling in osteoclasts triggers a post-translational modification of TFEB. Since TFEB activity is regulated by phosphorylation on at least two serine residues (Settembre et al. 2011), we tested whether RANKL induces phosphorylation of TFEB. For that purpose, RAW 264.7 cells expressing TFEB-Flag were treated with or without RANKL, and cell extracts were incubated in the presence or absence of λ protein phosphatase (λPPase). This treatment resulted in a downward shift of TFEB in electrophoresis and abrogated the electrophoretic shift of TFEB induced by RANKL treatment, indicating that RANKL signaling is responsible of at least one phosphorylation event in TFEB (Fig. 4F).
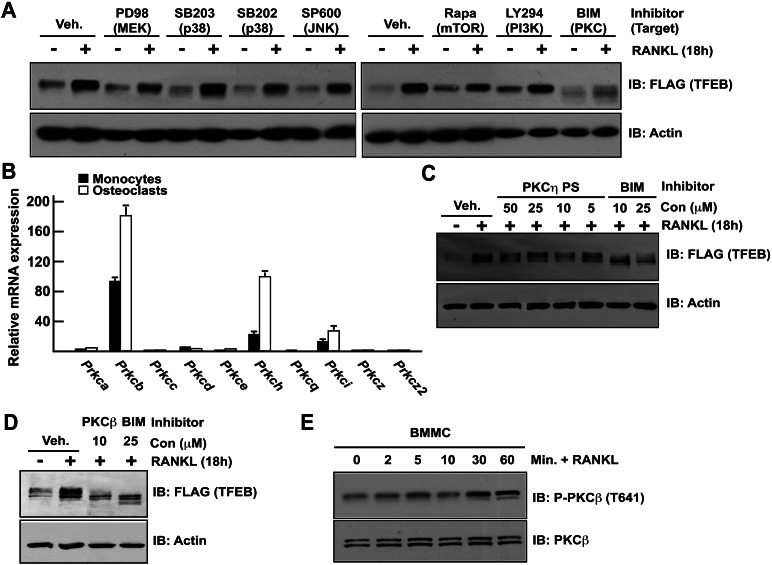
To identify the kinase recruited by RANKL and phosphorylating TFEB, pharmacological inhibitors of the major kinases acting downstream from RANK, the receptor for RANKL, were applied on RAW 264.7 cells expressing TFEB-Flag for 2 h. Cells were then either untreated or stimulated with RANKL for 18 h, and TFEB accumulation was assessed by Western blot. Among the different kinase inhibitors that were tested, only bisindolylmaleimide I (BIM), a pan-protein kinase C (PKC) inhibitor (Williams et al. 2003), suppressed RANKL-induced TFEB accumulation. This class of kinases has not been previously shown to regulate TFEB.
To identify which members of the PKC family of kinases regulate TFEB accumulation in osteoclasts, we first analyzed the expression of the genes encoding all members of the PKC family in osteoclasts. Prkcb and Prkch, the genes encoding for PKCβ and PKCη, respectively, were by far the most highly expressed in osteoclasts, and their expression increased during differentiation of monocytes into osteoclasts (Fig. 5B). Of note, expression of Prkcb was 600-fold higher in osteoclasts than in osteoblasts (Supplemental Fig. S5A). To determine whether PKCβ and/or PKCη was/were required for RANKL-induced TFEB accumulation, RAW 264.7 cells expressing TFEB-Flag were treated with either a PKCη pseudosubstrate inhibitor (PKCη PS) or a PKCβ-specific inhibitor. While the PKCη PS did not affect RANKL-induced accumulation of TFEB (Fig. 5C), the PKCβ inhibitor was as efficient as BIM in preventing RANKL-induced TFEB accumulation in RAW 264.7 cells (Fig. 5D) and primary osteoclast precursors (Supplemental Fig. S5B). Moreover, that PKCβ was phosphorylated in response to RANKL in primary osteoclast precursors (Fig. 5E; Supplemental Fig. S5C) suggested that RANKL signaling in these cells might activate this kinase. Collectively, these results suggest that PKCβ is required for RANKL-induced TFEB protein accumulation in RAW 264.7 cells. Consistent with this idea, examination of the TFEB amino acid sequence revealed the presence in its C-terminal sequence of three serine residues predicted to be potential PKC phosphorylation sites (Fig. 6A).
Figure 5.
RANKL-induced TFEB stabilization is PKCβ-dependent. (A) RAW 264.7 cells stably transfected with TFEB-Flag were treated with vehicle (DMSO) or the indicated inhibitors as described in the Materials and Methods and with or without RANKL (50 ng/mL) for 16 h. TFEB protein in total cell extracts was determined by Western blotting using Flag antibody. (B) Relative qPCR expression analysis of PKCs encoding genes in primary monocytes and osteoclasts. Values are expressed as fold of Prcka expression levels in monocytes. (C,D) RAW 264.7 cells stably transfected with TFEB-Flag were treated with vehicle (DMSO) or the indicated inhibitors and with or without RANKL (50 ng/mL) for 16 h. TFEB accumulation in cell extracts was measured by Western blotting using Flag antibody. In all experiments, the cells were cultured in the presence of FBS (10%). (E) Bone marrow-derived monocytes were serum-starved for 3 h and treated for the indicated times with RANKL (50 ng/mL), and PKCβ phosphorylation was assessed by Western blotting using an antibody previously validated using Prkcb−/− cell extracts (see Supplemental Fig. S5C).
Figure 6.
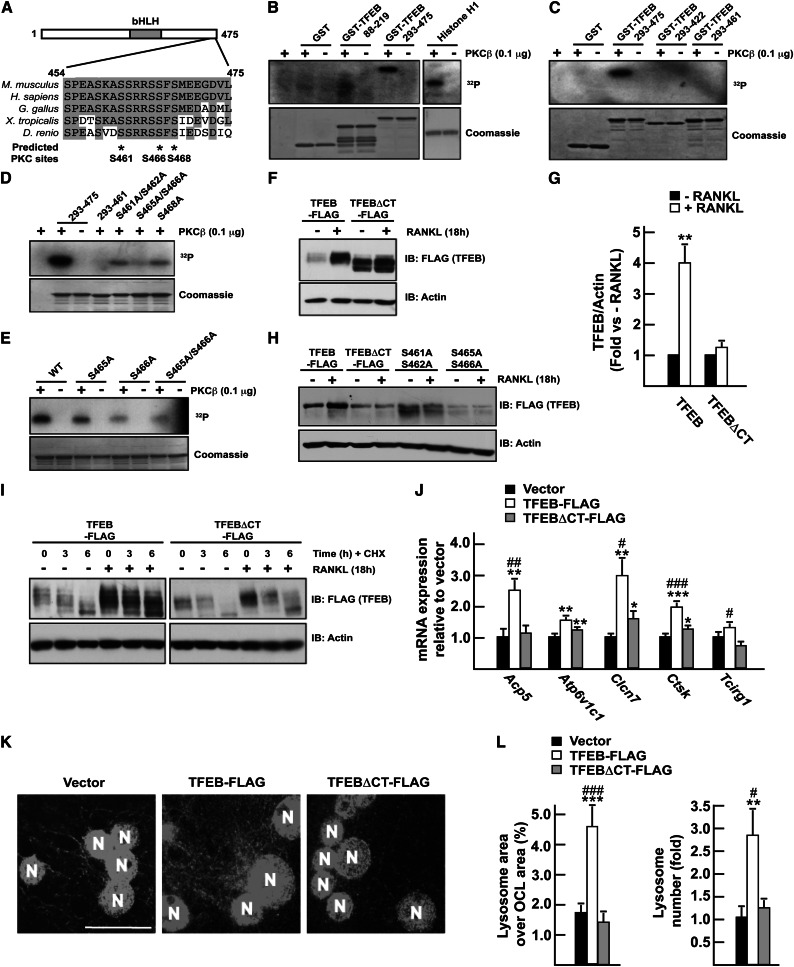
PKCβ phosphorylates TFEB in a C-terminal motif. (A) Sequence alignment of TFEB protein sequence from different vertebrates. The positions of three putative PKCβ phosphorylation sites (http://scansite.mit.edu) are indicated by asterisks. In B–E, kinase assays were performed on recombinant GST fusion proteins as described in the Materials and Methods. In D, 293–475 and 293–460 indicate GST-TFEB293–475 and GST-TFEB293–460, respectively. S461A/S462A, S465A/S466A, and S468A mutations were all introduced into the GST-TFEB293–475 protein. In E, wild type (WT) indicates GST-TFEB293–475. S465A, S466A, and S465A/S466A mutations were introduced into the GST-TFEB293–475 protein. (F) RAW 264.7 cells stably transfected with TFEB-Flag or with a TFEB construct lacking the last 15 amino acids (TFEBΔCT-Flag) were treated or not with RANKL (50 ng/mL) for 16 h. TFEB protein in total cell extracts was determined by Western blotting using Flag antibody. (G) Quantification of the results presented in F. The levels of the Flag proteins were quantified, normalized to actin levels, and expressed as a fold induction normalized to each protein level in the absence of RANKL. Three independent experiments were analyzed. (H) RAW 264.7 cells transiently transfected with TFEB-Flag, TFEBΔCT-Flag, or TFEB-Flag containing the indicated mutation were treated or not with RANKL (50 ng/mL). (I) RAW 264.7 cells stably transfected with TFEB-Flag or TFEBΔCT-Flag were treated with RANKL (50 ng/mL) and/or cycloheximide (CHX at 100 μg/mL) for the indicated times. (J) qPCR expression analysis in RAW 264.7 cells stably transfected with the empty vector or TFEB-Flag or TFEBΔCT-Flag construct and treated for 72 h with RANKL (30 ng/mL). (K) LAMP1 immunofluorescence staining on RAW 264.7 osteoclast-like cells following 96 h of treatment with RANKL (30 ng/mL). Magnification, 63×. Bar, 10 μm. (N) Nucleus. (L) Quantification of the percentage of the osteoclast area covered by lysosomes in individual osteoclasts and of the number of lysosomes per osteoclast relative to vector transfected cells, normalized by the osteoclast area and the number of nuclei per osteoclast. (#) P < 0.05; (##) P < 0.01; (###) P < 0.001 when comparing TFEB-Flag and TFEBΔCT-Flag. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 when compared with vector.
To test whether TFEB was a direct target of PKCβ, we performed in vitro kinase assays and incubated GST-TFEB in the presence of purified PKCβ and γ-ATP32. PKCβ phosphorylated a segment of TFEB containing its conserved C-terminal motif (amino acids 293–475) but not a more N-terminal one (amino acids 88–219). Histone H1, used here as a positive control, was efficiently phosphorylated by PKCβ, while GST was not (Fig. 6B). We excluded from these experiments amino acids 1–88 and 220–292 of TFEB, since these two regions do not contain a putative PKC phosphorylation site and, when included, considerably hampered expression of TFEB in bacteria (data not shown). Further deletion analysis revealed that PKCβ phosphorylated TFEB in its last 15 amino acids (Fig. 6C), a region containing five serine residues, all conserved in vertebrate TFEB homologs (Fig. 6A), and site-specific mutagenesis followed by in vitro kinase assays identified S461 and/or S462, S466, and S468 as being phosphorylated by PKCβ in vitro (Fig. 6D,E).
To determine whether the last 15 amino acids of TFEB are required for RANKL-mediated accumulation of TFEB, RAW 264.7 cells stably transfected with either full-length TFEB-Flag or a TFEB construct lacking only these last 15 amino acids (TFEBΔCT) were treated with RANKL or vehicle. RANKL markedly increased (about fourfold) the amount of full-length TFEB but not of TFEBΔCT mutant protein (<1.2-fold) (Fig. 6F,G). Mutation of S461 and S462 or of S465 and S466 to alanine residues also resulted in an inhibition of RANKL-induced TFEB accumulation (Fig. 6H), verifying that these serine residues are required for the stabilization of TFEB in response to RANKL. Moreover, while RANKL treatment prevented the decrease in TFEB accumulation induced by a cycloheximide treatment, it had no effect on the reduction of the TFEBΔCT mutant protein (Fig. 6I). These three experiments indicate that RANKL-induced TFEB stabilization is dependent on the presence of these serine residues. Consistent with this notion, overexpressing full-length TFEB increased expression of the lysosomal genes implicated in bone resorption following RANKL treatment, while overexpression of the TFEBΔCT mutant did not (Fig. 6J). Moreover, while the number and the area of LAMP1-positive lysosomes were increased in osteoclast-like cells when full-length TFEB was overexpressed, they remained unchanged in the cells overexpressing the TFEBΔCT mutant (Fig. 6K,L). Together, these results establish that PKCβ directly phosphorylates TFEB in its last 15 amino acids. Deletion of this sequence abolishes RANKL-induced TFEB stabilization and reduces TFEB's ability to induce lysosomal genes.
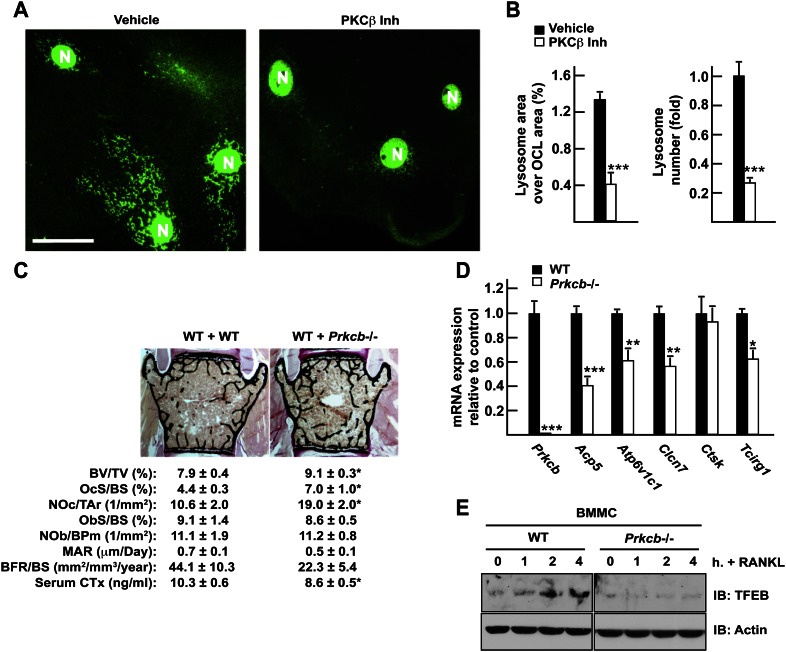
PKCβ is required for normal osteoclast function in vivo
An implication of the results presented above is that PKCβ should be required for proper osteoclast function. This was tested in two ways. First, we assessed the effect of a pharmacological inhibition of PKCβ on lysosomal biogenesis in mature osteoclasts in culture. Fully differentiated bone marrow-derived osteoclasts treated for 18 h with the PKCβ inhibitor displayed a significant reduction in the number and the area of lysosomes when compared with vehicle-treated osteoclasts (Fig. 7A,B). To obtain an in vivo confirmation of this function of PKCβ and in the absence of a floxed allele of Prkcb, we transplanted wild-type or Prkcb−/− bone marrow hematopoietic stem cells (HSCs) into wild-type irradiated adult mice. Since the osteoclasts derived from a hematopoietic progenitor cell, the osteoclasts of the transplanted mice will derive from the donor cells, while the nonhematopoietic cell types such as osteoblasts will originate from the wild-type host. Genotyping PCR on long bone DNA confirmed the proper engraftment of the Prkcb−/− cells in the recipient animals (Supplemental Fig. S5D). Three months post-transplantation, the bones of these animals were analyzed.
Figure 7.
PKCβ is required for normal osteoclast function in vivo. (A) LAMP1 immunofluorescence staining of differentiated osteoclasts cultured in the presence of 10 μM PKCβ inhibitor (right panels) or vehicle (DMSO) (left panels) for 18 h. Magnification, 63×. Bar, 10 μm. (N) Nucleus. (B) Quantification of the percentage of the osteoclast area covered by lysosomes in individual osteoclasts and of the number of lysosomes per osteoclast relative to vehicle, normalized by the osteoclast area and the number of nuclei per osteoclast. (C) Bone histomorphometric analysis of lumbar vertebrae in mice transplanted with wild-type (WT + WT) or Prkcb−/− (WT + Prkcb−/−) bone marrow cells (n = 6–13). (BV/TV) Bone volume over tissue volume; (OcS/BS) osteoclast surface over bone surface; (NOc/Tar) number of osteoclasts per tissue area; (ObS/BS) osteoblast surface over bone surface; (NOb/BPm) number of osteoblasts per bone perimeter; (MAR) mineral apposition rate; (BFR/BS) bone formation rate over bone surface. Fasting serum CTx levels are also included. (D) qPCR expression analysis in wild-type and Prkcb−/− flushed long bones. (E) Wild-type or Prkcb−/− bone marrow-derived monocytes were cultured in the presence of M-CSF and with or without RANKL (50 ng/mL) for the indicated times. Accumulation of endogenous TFEB assessed by Western blotting.
Bone histomorphometric analysis revealed that mice transplanted with PKCβ-deficient bone marrow cells display a significant increase in their bone mass compared with mice transplanted with wild-type bone marrow cells (Fig. 7C). This increase in bone mass was not due to either a defect in osteoclast differentiation, since the number of osteoclasts was not decreased, or to an increase in bone formation, since bone formation markers were slightly reduced in the animals transplanted with PKCβ-deficient bone marrow (Fig. 7C). Thus, we hypothesized that, just as it was the case in Tfebosc−/− mice, the increased bone mass observed in mice transplanted with PKCβ-deficient cells was secondary to a decrease in osteoclast function. That serum CTx levels and expression of several lysosomal genes needed for optimal bone resorption and previously identified as being regulated by RANKL and TFEB in osteoclasts were decreased in Prkcb−/− bones and mice (Fig. 7C,D) is consistent with the hypothesis that PKCβ is required for lysosomal biogenesis in osteoclasts and therefore for bone resorption. In addition, TFEB protein accumulates in wild-type but not in Prkcb−/− primary osteoclast precursors in response to RANKL, while Tfeb mRNA levels remained unchanged (Fig. 7E; Supplemental Fig. S5E).
RANKL favors accumulation of TFEB but not MITF
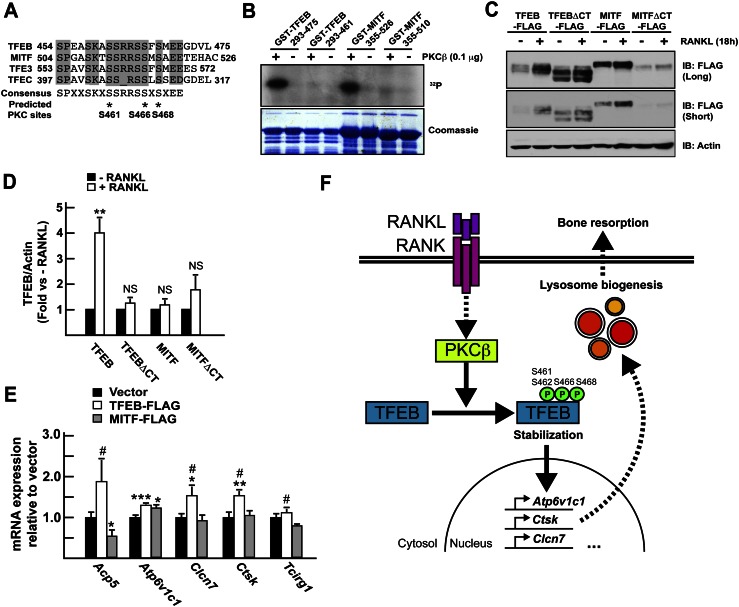
The motif phosphorylated by PKCβ in the TFEB C-terminal region is conserved among all four members of the MITF/TFE transcription factor family (Fig. 8A), thus raising the question of the specificity of this function of RANKL. Given the fact that MITF is implicated in osteoclast differentiation, we asked whether the RANKL/PKCβ cascade identified here was specific to TFEB.
Figure 8.
RANKL induces accumulation of TFEB but not MITF. (A) Sequence alignment of MITF/TFE mouse proteins. The positions of three putative conserved PKCβ phosphorylation sites are indicated by asterisks. (B) Kinase assay was performed on recombinant GST fusion proteins as described in the Materials and Methods. (C) RAW 264.7 cells stably transfected with TFEB-Flag, TFEBΔCT-Flag, MITF, or a MITF construct lacking the last 16 amino acids (MITFΔCT-Flag) were treated or not with RANKL (50 ng/mL) for 16 h. TFEB and MITF proteins in total cell extracts were revealed by Western blotting using Flag antibodies. Two exposure times of the same blot are included. (D) Quantification of the results presented in C. The levels of the Flag proteins were quantified, normalized to actin levels, and expressed as a fold induction normalized to the protein level in the absence of RANKL. Three independent experiments were analyzed. (E) qPCR expression analysis in RAW 264.7 cells stably transfected with the empty vector or TFEB-Flag or MITF-Flag construct and treated for 72 h with RANKL (30 ng/mL). (#) P < 0.05 when comparing TFEB-Flag and MITF-Flag. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 when compared with vector. (F) Model. To promote lysosomal biogenesis, RANKL signaling uses PKCβ that phosphorylates TFEB on at least three serine residues located in its C terminus. This phosphorylation stabilizes TFEB, leading to its accumulation and the activation of target genes implicated in lysosomal biogenesis and bone resorption.
Consistent with their high degree of sequence conservation, both TFEB and MITF were phosphorylated by PKCβ within their C terminus in vitro (Fig. 8B). MITF protein levels were also marginally increased (20% compared with 300% for TFEB) after treatment of cells with RANKL. However, deletion of the C-terminal motif of MITF did not abrogate the regulation by RANKL as it did on TFEB (Fig. 8C,D). These results suggest that although PKCβ can phosphorylate a conserved C-terminal region in TFEB and MITF in vitro, only TFEB requires this region to accumulate in response to RANKL. Consistent with this notion, overexpression of TFEB in RAW 264.7 cells increased expression of the lysosomal genes implicated in bone resorption following RANKL treatment, while overexpression of the MITF did not (Fig. 8E).
Discussion
Beside its osteoclast differentiation ability, RANKL promotes the resorptive function of osteoclasts once differentiated (Lacey et al. 1998; Burgess et al. 1999; Hsu et al. 1999; Lomaga et al. 1999). Here we show that this occurs in part through a PKCβ–TFEB-dependent pathway and independently of a change in the function of MITF, another member of the same family of transcription factor that is involved in osteoclast differentiation. These findings distinguish between RANKL regulation of osteoclast differentiation and osteoclast functions and provide a molecular basis for this latter action. They do not exclude, however, that transcription factors involved in osteoclast differentiation may also favor lysosomal biogenesis.
Lysosomal biogenesis and bone resorption
In order to resorb the bone ECM, osteoclasts need to generate a resorption lacuna, which is characterized by an acidic pH and the presence of specific proteases, including cathepsin K, a lysosomal cysteine protease responsible for collagen degradation by osteoclasts (Gelb et al. 1996). The generation of the resorption lacuna is achieved through exocytosis and fusion of numerous lysosomes, illustrating how critical the generation and function of lysosomes are for osteoclast function. In support of this notion, the most frequently mutated genes in autosomal recessive human osteopetrosis (i.e., CLCN7, OSTM1, and TCIRG1) are also implicated in the acidification of the lysosomal compartment in osteoclasts (Kornak et al. 2001; Chalhoub et al. 2003; Schinke et al. 2009), and CTSK, the gene encoding for cathepsin K, is mutated in pycnodysostosis, a lysosomal storage disease with defective bone resorption (Gelb et al. 1996).
Role of TFEB in osteoclast function
That mice harboring a cell-specific deletion of TFEB in osteoclasts demonstrated decreased expression of lysosomal genes, a reduced number of lysosomes, and a decreased ability to resorb the bone matrix indicates that TFEB is needed for lysosomal biogenesis in osteoclasts. Moreover, the fact that an osteoclast-specific ablation of Tfeb decreased lysosomal biogenesis in these cells, while mice lacking Mitf or Mitf and one allele of Tfeb did not show any osteoclast dysfunction (Steingrimsson et al. 2002), suggests that MITF and TFEB fulfill distinct roles in osteoclasts. Moreover, forced expression of MITF does not induce lysosomal biogenesis in osteoclast-like cells (Fig. 8E) or HeLa cells (Sardiello et al. 2009). However, expression of Mitf was increased in Tfeb-deficient osteoclasts, suggesting that overexpression of this gene might be partially compensating for the absence of Tfeb in these cells. This possibility, supported by the observation that MITF can directly bind and activate the promoter of Acp5, Ctsk, and Clcn7 in osteoclasts (Motyckova et al. 2001; Meadows et al. 2007), can only be addressed in vivo through the generation of mice lacking both Mitf and Tfeb in osteoclasts only. Unfortunately, no floxed allele of Mitf is currently available to test this hypothesis.
It was recently shown that osteoclast-specific ablation of Ctsk resulted in increased bone formation by osteoblasts (Lotinun et al. 2013), a phenotype that we did not observe. This difference may be due to the fact that Lotinun et al. (2013) used Cd11b-Cre and Mx1-Cre, while we used Ctsk-Cre. In addition, in their case, the reduction in Ctsk expression in osteoclasts approached 95%, while in our case, it did not go below 60%.
Regulation of TFEB by PKCβ
Multiple assays indicated that RANKL induction of lysosomal biogenesis in osteoclasts depends on their expression of Tfeb. For this purpose, RANKL recruits PKCβ, which phosphorylates TFEB in its C-terminal sequence on at least three serine residues. This sequence is conserved in all four vertebrate members of the MITF/TFE family (Fig. 8A) but is absent from its Caenorhabditis elegans and Drosophila melanogaster homologs, an observation suggesting that this motif is implicated in vertebrate-specific functions of these factors. Remarkably, this C-terminal motif has not been associated with any TFEB function previously.
Although both TFEB and MITF were phosphorylated in vitro by PKCβ, only TFEB was induced at the protein level by RANKL. This result suggests that the RANKL–PKCβ–TFEB cascade is specific to TFEB function. Of note, phosphorylation of MITF by p90 Rsk-1 in the same C-terminal region reduced its stability in response to c-Kit signaling in melanoma cells (Wu et al. 2000), suggesting that this conserved motif may have multiple functions in the MITF/TFE factors and that these functions may be either cell type- or signal-specific. Consistent with this idea, several unbiased mass spectrometry studies performed in various cell types and tissues have found that TFEB, MITF, and TFE3 can be phosphorylated on one or several serine residues contained in the C-terminal motif (Beausoleil et al. 2004; Huttlin et al. 2010; Rigbolt et al. 2011; Yu et al. 2011; Goswami et al. 2012). Finally, our findings do not exclude, as shown previously, that MITF and TFE3 are regulated by RANKL and/or M-CSF during osteoclast differentiation but through PKCβ-independent pathways (Weilbaecher et al. 2001; Meadows et al. 2007).
Although members of the PKC family have been proposed to be involved in bone resorption by osteoclasts ex vivo (Lee et al. 2003; Williams et al. 2003; Sorensen et al. 2010), the specific involvement of PKCβ in this process has never been tested in vivo. Our data showing that PKCβ regulation of osteoclast function occurs in part by phosphorylating TFEB and promoting its accumulation (Fig. 8F) are also consistent with the fact that treatment of mice with enzastaurin, a PKCβ inhibitor, can reduce osteoclastic bone destruction caused by metastatic breast cancer cells (Dudek et al. 2008). Interestingly, Cremasco et al. (2012) have shown that another PKC, PKCδ, was also required for osteoclast function in vivo because it promotes cathepsin K exocytosis. Together, these and our findings suggest that multiple members of the PKC family might contribute nonredundantly to the resorptive function of osteoclasts.
Post-translational modification of TFEB and its regulation by RANKL
TFEB is known to be phosphorylated by many kinases. For instance, it is phosphorylated by ERK or mammalian target of rapamycin (mTOR) on S142 and/or S211 in the presence of nutrients, and these phosphorylation events prevent TFEB from entering the nucleus (Settembre et al. 2011, 2012; Martina et al. 2012; Roczniak-Ferguson et al. 2012). Likewise, mTOR is also implicated in TFEB cytosolic–nuclear transport through a different mechanism (Pena-Llopis et al. 2011). In contrast, our results point toward a novel post-translational regulation of TFEB by RANKL in osteoclasts. Indeed, RANKL treatment in RAW 264.7 cells does not specifically promote TFEB nuclear accumulation but instead globally increases TFEB protein levels in the cells. Moreover, pharmacological inhibition of ERK or mTOR has no significant effect on RANKL-dependent TFEB accumulation in osteoclasts (Fig. 5A).
To be fully activated, classical PKCs, such as PKCβ, require diacylglycerol (DAG), a second messenger that is generated through the hydrolysis of PI(4,5)P2 by phospholipase C (PLC). It was shown that PLCγ2, acting downstream from RANK, regulates both osteoclast differentiation in an NFATc1-dependent manner (Mao et al. 2006) and osteoclast function independently of NFATc1 (Epple et al. 2008). Thus, one can hypothesize that generation of DAG by PLCγ2 could be a signal triggering PKCβ activation downstream from RANK. In conclusion, our results support the existence of a distinct pathway in mature osteoclasts regulating lysosomal biogenesis downstream from RANK and implicating the stabilization of TFEB in a PKCβ-dependent manner (Fig. 8F).
Materials and methods
Mouse models
Generation of Tfebfl/fl (C57BL/6) mice was reported previously (Settembre et al. 2012). To generate Tfeb osteoclast-specific knockout mice, Tfebfl/fl mice were crossed with Ctsk-Cre mice (C57BL/6), which express the Cre recombinase under the control of the osteoclast-specific Cathepsin K locus (Nakamura et al. 2007). The obtained progeny, Tfebfl/+;Ctsk-Cre and Tfebfl/+, were intercrossed to obtain controls (Tfebfl/fl) and Tfeb osteoclast knockout (Tfebfl/fl;Ctsk-Cre) animals. To generate mice lacking PKCβ in osteoclasts, wild-type or Prkcb−/− (BALB/c) bone marrow cells (Su et al. 2002) were transplanted (2 × 106 cells per mouse) via tail vein injection into 5-wk-old irradiated wild-type (BALB/c) recipient mice. All of the transplanted mice survived, and bone histological analysis was performed 12 wk later.
Reagents
PD98059, SB203580, SB202190, and SP600125 were obtained from Tocris Bioscience; cyclohexamide, rapamycin, and LY294002 were obtained from Sigma Aldrich; and BIM, PKCη pseudosubstrate, and PKCβ inhibitor were obtained from EMD Millipore. TFEB monoclonal antibody was from MyBiosource. Flag and β-actin antibodies were from Sigma Aldrich, and PKCβ antibodies were from Santa Cruz Biotechnology. Tubulin and Lamin A/C antibodies were obtained from BD Bioscience. λPPase and P-PKCβ (T641) antibody were obtained from New England Biolabs. Recombinant active PKCβ and LAMP1 antibody were obtained from Abcam.
Gene expression analysis
Real-time quantitative PCR (qPCR) was performed on DNaseI-treated cell or bone total RNA converted to cDNA by MMLV reverse transcriptase using Taq SYBR Green Supermix (Bio-Rad) with ROX on an MX3000 instrument; ribosomal S18 and Hprt were used as internal references. ChIP assays were performed on nuclear extracts of RAW 264.7 cells expressing an empty vector or TFEB-Flag protein using anti-Flag antibodies according to previously described methods (Lacombe et al. 2013). Sequences of the primers used for each gene and ChIP assays are provided in Supplemental Table 1.
Bone histomorphometry
Bone histomorphometry analysis on lumbar vertebrae embedded in methyl methacrylate resin was performed as previously described (Chappard et al. 1987) using the Osteomeasure analysis system (Osteometrics). Von Kossa/Van Gieson staining was performed for bone volume over tissue volume (BV/TV) measurement. Bone formation rate parameters were analyzed by the calcein double-labeling method. For the analysis of osteoblast and osteoclast, sections were stained with toluidine blue and TRAP, respectively.
Plasmid constructs
GST-TFEB and GST-MITF deletion constructs were generated by PCR amplification and cloning into the EcoRI and SalI site of pGEX4T3 vector. TFEB-3XFlag construct (TFEB-Flag) was described previously (Sardiello et al. 2009). MITF-Flag, MITFΔCT-Flag, and TFEBΔCT-Flag were cloned in the same mammalian expression vector (p3XFlag-CMV-14; Sigma).
Cell culture, transfection, and immunofluorescence
RAW 264.7 cells were cultured according to the conditions provided by the supplier (American Type Culture Collection). siRNA knockdown in RAW 264.7 cells was achieved by transfection with siRNA pools (On-target, Dharmacon) using Lipofectamin LTX as previously described (Carralot et al. 2009). Stable transfectants were generated by electroporating linearized vectors into RAW 264.7 cells followed by selection with G418 (Invitrogen) (Ferron et al. 2011). Osteoclast-like cells were obtained by differentiating RAW 264.7 stable transfectants in the presence of RANKL (30 ng/mL). Primary osteoclast precursors (monocytes) were obtained by culturing bone marrow cells with M-CSF containing L-929 cell supernatant (10%) for 6 d. Primary osteoclasts were obtained by treating osteoclast precursors with RANKL (30 ng/mL) and M-CSF (10 ng/mL) for the indicated times. Multinucleated osteoclasts were identified by TRAP staining, and pit resorption assay was performed using Osteo Assay Stripwell Plate (Corning) or bovine bone slices stained with WGA-lectin-HRP. Mouse primary osteoblasts were isolated as previously described (Ducy et al. 1997). For immunofluorescence experiments, stable transfectants or primary osteoclasts were plated on glass coverslips or on bovine bone slices fixed in 4% paraformaldehyde and labeled with a 1:100 dilution of anti-LAMP1 antibody (Abcam) before being stained with Alexa Fluor 488- or 637-conjugated donkey anti-rabbit secondary antibody (Molecular Probes). Nuclei were stained with DAPI. ImageJ software was used to quantify the number of lysosomes normalized to the number of nuclei and the area covered by each osteoclast. Actin was stained with Phalloidin-Alexa Fluor 555. Cells were imaged using a fluorescent or confocal microscope. To assess intracellular pH, osteoclasts differentiated on bone slices were incubated in MEM buffered at pH 7.4 with 20 mM HEPES and containing 5 μg/mL acridine orange (Invitrogen) for 15 min at 37°C, washed, and imaged using a confocal microscope (excitation, 488 nm; emission, 500–550 nm [green form] and 570–620 nm [red/orange acidic form]).
Kinase inhibitor treatment
RAW 264.7 cells stably transfected with TFEB-Flag construct were pretreated for 1 h with vehicle (DMSO) or the following inhibitors: 10 μM PD98059, 5 μM SB203580, 2 μM SB202190, 2 μM SP600125, 20 nM rapamycin, 20 μM LY294002, 10–25 μM BIM, 5–50 μM PKCη pseudosubstrate, or 10 μM PKCβ inhibitor.
Cell fractionation and λPPase assay
To obtain cytosolic and nuclear extracts, cells were suspended in hypotonic buffer (10 mM Tris at pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 20 mM NaF, 1 mM sodium pyrophosphate, 20 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM PMSF, 1× protease inhibitor cocktail [Roche]). After 15-min incubations on ice, cells were lysed by adding NP-40 (0.625% final). Nuclei were collected by centrifugation, and the cytosolic fraction (i.e., supernatant) was saved. The pellet was resuspended in hypertonic buffer (20 mM Tris at pH 7.9, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 20 mM NaF, 1 mM sodium pyrophosphate, 20 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM PMSF, 1× protease inhibitor cocktail), and nuclear proteins were extracted by vortexing for 30 min at 4°C followed by centrifugation. In some experiments, total cell extracts were obtained by lysing the cells with immunoprecipitation BF (20 mM Tris at pH 7.4, 150 mM NaCl, 1% NP-40, 2 mM EDTA, 1 mM NaF, 1 mM sodium pyrophosphate, 2.5 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM PMSF 1× protease inhibitor cocktail) and sonicating the extract three times for 5 sec at 25% intensity. For the λPPase treatment, total cell extracts were prepared using the same method, except that NaF, sodium pyrophosphate, β-glycerophosphate, and sodium orthovanadate were not included in the lysis buffer. Ten milligrams of extract were diluted in 1× phosphatase buffer (50 mM HEPES at pH 7.5, 100 mM NaCl, 2 mM DTT, 0.01% Brij 35, 1 mM MnCl2) and treated or not with 800 U of λPPase for 20 min at 30°C. The reaction was stopped by adding 3× SDS loading buffer and Flag protein detected by standard Western blotting method.
Kinase assay
Recombinant proteins (10 μg each), prepared as previously described (Ferron et al. 2010), were incubated for 30 min at 30°C with recombinant PKCβ kinase (0.1 μg), 10 μM ATP, and 5 μCi of [γ32P]ATP in 1× kinase buffer (20 mM HEPES at pH 7.5, 10 mM MgCl2, 2 mM CaCl2). The reaction was stopped by the addition of Laemmli buffer, and proteins were resolved on SDS-PAGE gel. Phosphorylation of proteins was detected by exposing the gel to a radiographic film.
Statistics
Results are given as means ± standard error of the mean (SEM). Statistical analyses were performed using unpaired, two-tailed Student's t-test. In all figures, single asterisk or hashmark (* or #) indicates P < 0.05, double asterisks or hashmarks (** or ##) indicate P < 0.01, triple asterisk or hashmarks (*** or ###) indicate P < 0.001, and NS indicates not significant.
Acknowledgments
We are grateful to S. Khim for collecting the PKCβ−/− bones; B. Zhou, Z. Zhang, and Dr. X.E. Guo for the μCT analysis; Dr. W. Zou for providing bovine bone slices; Dr. R. De Cegli for bioinformatic analyses; Dr. T. Swayne for microscopy support; and M. Pata and Dr. J. Vacher for technical advices on immunofluorescence. We thank Dr. S. Teitelbaum and Dr. P. Ducy for their critical reading of the manuscript. This work was supported by funding from the U.S. National Institutes of Health (M.F., A.B., and G.K.), an American and Canadian Diabetes Associations post-doctoral fellowship (M.F.), the Italian Telethon Foundation (C.S. and A.B), the Beyond Batten Disease Foundation (A.B.), the European Research Council (A.B.), and the March of Dimes (A.B).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.213827.113.
References
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP 2004. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci 101: 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess TL, Qian Y, Kaufman S, Ring BD, Van G, Capparelli C, Kelley M, Hsu H, Boyle WJ, Dunstan CR, et al. 1999. The ligand for osteoprotegerin (OPGL) directly activates mature osteoclasts. J Cell Biol 145: 527–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carralot JP, Kim TK, Lenseigne B, Boese AS, Sommer P, Genovesio A, Brodin P 2009. Automated high-throughput siRNA transfection in raw 264.7 macrophages: A case study for optimization procedure. J Biomol Screen 14: 151–160 [DOI] [PubMed] [Google Scholar]
- Chalhoub N, Benachenhou N, Rajapurohitam V, Pata M, Ferron M, Frattini A, Villa A, Vacher J 2003. Grey-lethal mutation induces severe malignant autosomal recessive osteopetrosis in mouse and human. Nat Med 9: 399–406 [DOI] [PubMed] [Google Scholar]
- Chappard D, Palle S, Alexandre C, Vico L, Riffat G 1987. Bone embedding in pure methyl methacrylate at low temperature preserves enzyme activities. Acta Histochem 81: 183–190 [DOI] [PubMed] [Google Scholar]
- Coxon FP, Taylor A 2008. Vesicular trafficking in osteoclasts. Semin Cell Dev Biol 19: 424–433 [DOI] [PubMed] [Google Scholar]
- Cremasco V, Decker CE, Stumpo D, Blackshear PJ, Nakayama KI, Nakayama K, Lupu TS, Graham DB, Novack DV, Faccio R 2012. Protein kinase C-δ deficiency perturbs bone homeostasis by selective uncoupling of cathepsin K secretion and ruffled border formation in osteoclasts. J Bone Miner Res 27: 2452–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G 1997. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell 89: 747–754 [DOI] [PubMed] [Google Scholar]
- Ducy P, Schinke T, Karsenty G 2000. The osteoblast: A sophisticated fibroblast under central surveillance. Science 289: 1501–1504 [DOI] [PubMed] [Google Scholar]
- Dudek AZ, Zwolak P, Jasinski P, Terai K, Gallus NJ, Ericson ME, Farassati F 2008. Protein kinase C-β inhibitor enzastaurin (LY317615.HCI) enhances radiation control of murine breast cancer in an orthotopic model of bone metastasis. Invest New Drugs 26: 13–24 [DOI] [PubMed] [Google Scholar]
- Epple H, Cremasco V, Zhang K, Mao D, Longmore GD, Faccio R 2008. Phospholipase Cγ2 modulates integrin signaling in the osteoclast by affecting the localization and activation of Src kinase. Mol Cell Biol 28: 3610–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Deng L, Chen W, Shao J, Xu G, Li YP 2009. Atp6v1c1 is an essential component of the osteoclast proton pump and in F-actin ring formation in osteoclasts. Biochem J 417: 195–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron M, Wei J, Yoshizawa T, Del Fattore A, DePinho RA, Teti A, Ducy P, Karsenty G 2010. Insulin signaling in osteoblasts integrates bone remodeling and energy metabolism. Cell 142: 296–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron M, Boudiffa M, Arsenault M, Rached M, Pata M, Giroux S, Elfassihi L, Kisseleva M, Majerus PW, Rousseau F, et al. 2011. Inositol polyphosphate 4-phosphatase B as a regulator of bone mass in mice and humans. Cell Metab 14: 466–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frattini A, Orchard PJ, Sobacchi C, Giliani S, Abinun M, Mattsson JP, Keeling DJ, Andersson AK, Wallbrandt P, Zecca L, et al. 2000. Defects in TCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis. Nat Genet 25: 343–346 [DOI] [PubMed] [Google Scholar]
- Gelb BD, Shi GP, Chapman HA, Desnick RJ 1996. Pycnodysostosis, a lysosomal disease caused by cathepsin K deficiency. Science 273: 1236–1238 [DOI] [PubMed] [Google Scholar]
- Goswami T, Li X, Smith AM, Luderowski EM, Vincent JJ, Rush J, Ballif BA 2012. Comparative phosphoproteomic analysis of neonatal and adult murine brain. Proteomics 12: 2185–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriadis AE, Wang ZQ, Cecchini MG, Hofstetter W, Felix R, Fleisch HA, Wagner EF 1994. c-Fos: A key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science 266: 443–448 [DOI] [PubMed] [Google Scholar]
- Hsu H, Lacey DL, Dunstan CR, Solovyev I, Colombero A, Timms E, Tan HL, Elliott G, Kelley MJ, Sarosi I, et al. 1999. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc Natl Acad Sci 96: 3540–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP 2010. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143: 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iotsova V, Caamano J, Loy J, Yang Y, Lewin A, Bravo R 1997. Osteopetrosis in mice lacking NF-κB1 and NF-κB2. Nat Med 3: 1285–1289 [DOI] [PubMed] [Google Scholar]
- Kenner L, Hoebertz A, Beil FT, Keon N, Karreth F, Eferl R, Scheuch H, Szremska A, Amling M, Schorpp-Kistner M, et al. 2004. Mice lacking JunB are osteopenic due to cell-autonomous osteoblast and osteoclast defects. J Cell Biol 164: 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornak U, Kasper D, Bosl MR, Kaiser E, Schweizer M, Schulz A, Friedrich W, Delling G, Jentsch TJ 2001. Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man. Cell 104: 205–215 [DOI] [PubMed] [Google Scholar]
- Lacey DL, Timms E, Tan HL, Kelley MJ, Dunstan CR, Burgess T, Elliott R, Colombero A, Elliott G, Scully S, et al. 1998. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93: 165–176 [DOI] [PubMed] [Google Scholar]
- Lacombe J, Hanley O, Jung H, Philippidou P, Surmeli G, Grinstein J, Dasen JS 2013. Genetic and functional modularity of Hox activities in the specification of limb-innervating motor neurons. PLoS Genet 9: e1003184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SW, Kwak HB, Chung WJ, Cheong H, Kim HH, Lee ZH 2003. Participation of protein kinase C β in osteoclast differentiation and function. Bone 32: 217–227 [DOI] [PubMed] [Google Scholar]
- Lee SH, Rho J, Jeong D, Sul JY, Kim T, Kim N, Kang JS, Miyamoto T, Suda T, Lee SK, et al. 2006. v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat Med 12: 1403–1409 [DOI] [PubMed] [Google Scholar]
- Lomaga MA, Yeh WC, Sarosi I, Duncan GS, Furlonger C, Ho A, Morony S, Capparelli C, Van G, Kaufman S, et al. 1999. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev 13: 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotinun S, Kiviranta R, Matsubara T, Alzate JA, Neff L, Lüth A, Koskivirta I, Kleuser B, Vacher J, Vuorio E, et al. 2013. Osteoclast-specific cathepsin K deletion stimulates S1P-dependent bone formation. J Clin Invest 123: 666–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao D, Epple H, Uthgenannt B, Novack DV, Faccio R 2006. PLCγ2 regulates osteoclastogenesis via its interaction with ITAM proteins and GAB2. J Clin Invest 116: 2869–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Chen Y, Gucek M, Puertollano R 2012. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8: 903–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K, Owens JM, Tonko M, Elliott C, Chambers TJ, Wagner EF 2000. Fosl1 is a transcriptional target of c-Fos during osteoclast differentiation. Nat Genet 24: 184–187 [DOI] [PubMed] [Google Scholar]
- Meadows NA, Sharma SM, Faulkner GJ, Ostrowski MC, Hume DA, Cassady AI 2007. The expression of Clcn7 and Ostm1 in osteoclasts is coregulated by microphthalmia transcription factor. J Biol Chem 282: 1891–1904 [DOI] [PubMed] [Google Scholar]
- Medina DL, Fraldi A, Bouche V, Annunziata F, Mansueto G, Spampanato C, Puri C, Pignata A, Martina JA, Sardiello M, et al. 2011. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell 21: 421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motyckova G, Weilbaecher KN, Horstmann M, Rieman DJ, Fisher DZ, Fisher DE 2001. Linking osteopetrosis and pycnodysostosis: Regulation of cathepsin K expression by the microphthalmia transcription factor family. Proc Natl Acad Sci 98: 5798–5803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Imai Y, Matsumoto T, Sato S, Takeuchi K, Igarashi K, Harada Y, Azuma Y, Krust A, Yamamoto Y, et al. 2007. Estrogen prevents bone loss via estrogen receptor α and induction of Fas ligand in osteoclasts. Cell 130: 811–823 [DOI] [PubMed] [Google Scholar]
- Pena-Llopis S, Vega-Rubin-de-Celis S, Schwartz JC, Wolff NC, Tran TA, Zou L, Xie XJ, Corey DR, Brugarolas J 2011. Regulation of TFEB and V-ATPases by mTORC1. EMBO J 30: 3242–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, Blagoev B 2011. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal 4: rs3. [DOI] [PubMed] [Google Scholar]
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM 2012. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 5: ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. 2009. A gene network regulating lysosomal biogenesis and function. Science 325: 473–477 [DOI] [PubMed] [Google Scholar]
- Schinke T, Schilling AF, Baranowsky A, Seitz S, Marshall RP, Linn T, Blaeker M, Huebner AK, Schulz A, Simon R, et al. 2009. Impaired gastric acidification negatively affects calcium homeostasis and bone mass. Nat Med 15: 674–681 [DOI] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. 2011. TFEB links autophagy to lysosomal biogenesis. Science 332: 1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, et al. 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 31: 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen MG, Karsdal MA, Dziegiel MH, Boutin JA, Nosjean O, Henriksen K 2010. Screening of protein kinase inhibitors identifies PKC inhibitors as inhibitors of osteoclastic acid secretion and bone resorption. BMC Musculoskelet Disord 11: 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E, Tessarollo L, Pathak B, Hou L, Arnheiter H, Copeland NG, Jenkins NA 2002. Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc Natl Acad Sci 99: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E, Copeland NG, Jenkins NA 2004. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet 38: 365–411 [DOI] [PubMed] [Google Scholar]
- Su TT, Guo B, Kawakami Y, Sommer K, Chae K, Humphries LA, Kato RM, Kang S, Patrone L, Wall R, et al. 2002. PKC-β controls I κ B kinase lipid raft recruitment and activation in response to BCR signaling. Nat Immunol 3: 780–786 [DOI] [PubMed] [Google Scholar]
- Suter A, Everts V, Boyde A, Jones SJ, Lullmann-Rauch R, Hartmann D, Hayman AR, Cox TM, Evans MJ, Meister T, et al. 2001. Overlapping functions of lysosomal acid phosphatase (LAP) and tartrate-resistant acid phosphatase (Acp5) revealed by doubly deficient mice. Development 128: 4899–4910 [DOI] [PubMed] [Google Scholar]
- Takayanagi H 2007. Osteoimmunology: Shared mechanisms and crosstalk between the immune and bone systems. Nat Rev Immunol 7: 292–304 [DOI] [PubMed] [Google Scholar]
- Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, et al. 2002. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell 3: 889–901 [DOI] [PubMed] [Google Scholar]
- Teitelbaum SL 2000. Bone resorption by osteoclasts. Science 289: 1504–1508 [DOI] [PubMed] [Google Scholar]
- Teitelbaum SL, Ross FP 2003. Genetic regulation of osteoclast development and function. Nat Rev Genet 4: 638–649 [DOI] [PubMed] [Google Scholar]
- Tondravi MM, McKercher SR, Anderson K, Erdmann JM, Quiroz M, Maki R, Teitelbaum SL 1997. Osteopetrosis in mice lacking haematopoietic transcription factor PU.1. Nature 386: 81–84 [DOI] [PubMed] [Google Scholar]
- Wan Y, Chong LW, Evans RM 2007. PPAR-γ regulates osteoclastogenesis in mice. Nat Med 13: 1496–1503 [DOI] [PubMed] [Google Scholar]
- Weilbaecher KN, Motyckova G, Huber WE, Takemoto CM, Hemesath TJ, Xu Y, Hershey CL, Dowland NR, Wells AG, Fisher DE 2001. Linkage of M-CSF signaling to Mitf, TFE3, and the osteoclast defect in Mitf(mi/mi) mice. Mol Cell 8: 749–758 [DOI] [PubMed] [Google Scholar]
- Williams JP, Thames AM III, McKenna MA, McDonald JM 2003. Differential effects of calmodulin and protein kinase C antagonists on bone resorption and acid transport activity. Calcif Tissue Int 73: 290–296 [DOI] [PubMed] [Google Scholar]
- Wu M, Hemesath TJ, Takemoto CM, Horstmann MA, Wells AG, Price ER, Fisher DZ, Fisher DE 2000. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev 14: 301–312 [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Hayashi S, Kunisada T, Ogawa M, Nishikawa S, Okamura H, Sudo T, Shultz LD 1990. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature 345: 442–444 [DOI] [PubMed] [Google Scholar]
- Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al. 2011. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332: 1322–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]