Abstract
MicroRNA-29b (miR-29b) expression has been shown to be reduced in non-small–cell lung cancer (NSCLC) tissues. Here, we have identified the oncogene cyclin-dependent protein kinase 6 (CDK6) as a direct target of miR-29b in lung cancer. We hypothesized that in vivo restoration of miR-29b and thus targeting of genes important to tumor initiation and progression may represent an option for lung cancer treatment. We developed a cationic lipoplexes (LPs)-based carrier that efficiently delivered miR-29b both in vitro and in vivo. LPs containing miR-29b (LP-miR-29b) efficiently delivered miR-29b to NSCLC A549 cells, reduced the expression of key targets CDK6, DNMT3B, and myeloid cell leukemia sequence 1 (MCL1), as well as cell growth and clonogenicity of A549 cells. In addition, the IC50 for cisplatin in the miR-29b–treated cells was effectively reduced. In a xenograft murine model, LPs efficiently accumulated at tumor sites. Systemic delivery of LP-miR-29b increased the tumor miR-29b expression by approximately fivefold, downregulated the tumor mRNA expression of CDK6, DNMT3B, and MCL1 by ~57.4, ~40.5, and ~52.4%, respectively, and significantly inhibited tumor growth by ~60% compared with LP-miR-NC (negative control). Our results demonstrate that cationic LPs represent an efficient delivery system that holds great potential in the development of miRNA-based therapeutics for lung cancer treatment.
Keywords: cationic lipoplexes, lung cancer, microRNA
Introduction
Lung cancer is the leading cause of cancer deaths in the United States with a disappointing 15% overall 5-year survival rate.1,2 It is estimated that in 2012, there will be approximately 226,000 new cases and 160,000 deaths from lung cancer (non-small cell and small cell combined).2 The overall 5-year mortality has changed very little since the 1970s.3 Much of this may be attributed to the lack of adequate screening, the lack of clarity in what constitutes “high risk” for the development of disease, heterogeneity of disease and the limited number of novel therapies.
MicroRNAs (miRNAs) are small (~22 nucleotide) endogenous non-coding RNAs that regulate gene expression at the posttranscriptional level through RNA interference.4,5 MiRNAs are actively involved in many biological processes and have an impact on many diseases including cancer. Differential expression of miRNAs has been demonstrated in many types of cancer. These miRNAs carry both diagnostic and prognostic information and have been shown to alter cancer cell phenotype by targeting biological pathways critical to tumorigenesis.6,7,8 For example, recent studies have suggested that members of miR-29 family may play an important role in cancer by regulating cell proliferation, differentiation, apoptosis, migration, and invasion.9,10 Downregulation of miR-29 family members has been observed in leukemia,11,12,13,14 melanoma,15 liver,16 colon,17 cervical,18 and lung cancer.19,20,21 In lung cancer, Fabbri et al. recently determined that miR-29 members functionally targeted DNA methytransferases (DNMT) 3A and 3B, two key enzymes involved in DNA methylation and often related to the survival of patients with lung cancer.20 Re-expression of miR-29 in lung cancer cells restored normal DNA methylation patterns and thus induced re-expression of methylation-silenced tumor suppressor genes, such as FHIT and WWOX, and inhibited tumorigenicity in vitro and in vivo. Rothschild et al. determined that miR-29b was involved in the Src-ID1 signaling pathway thus regulating lung cancer cell migration and invasion.21 As a result, miR-29 has become an attractive candidate for miRNA-based therapeutics.
The development of miRNA-based therapeutics represents a new strategy in cancer treatment. However, targeted in vivo delivery of miRNAs faces many challenges including limited stability in serum, rapid blood clearance, off-target effects, and poor cellular uptake.22 Chemical modifications (locked nucleic acid, 2′-O-methylation, etc.), and nanoparticle delivery systems have been developed to overcome these challenges. Alternatively, miRNAs can be delivered as a precursor shRNA encoded by a plasmid via viral vectors. Delivery by nanoparticles has the advantages of being more cost-effective, less immunogenic, less toxic and less oncogenic.23 However, only three groups have published results on the development of nanoparticle delivery systems to systemically deliver miRNAs for lung cancer treatment. Chen et al. established a liposome-polycation-hyaluronicacid–based nanoparticle to effectively deliver siRNA and miR-34a in a syngeneic model of B16F10 lung metastases.24 Wiggins et al. and Trang et al. used a neutral lipid emulsion, Max Suppressor in vivo RNA LancerII (BIOO Scientific) to deliver both miR-34a and let-7 to inhibit tumor growth in lung cancer mouse models.25,26 Our group developed a cationic lipoplexes (LPs)-based carrier system for miR-133b delivery.27 In the current study, we used a cationic LPs-based delivery system to efficiently deliver miR-29b to suppress tumorigenicity in vitro and in vivo by downregulating multiple oncogenes, including cyclin-dependent protein kinase 6 (CDK6), DNMT3B, and myeloid cell leukemia sequence 1 (MCL1).
Results
Efficient cellular uptake of LPs
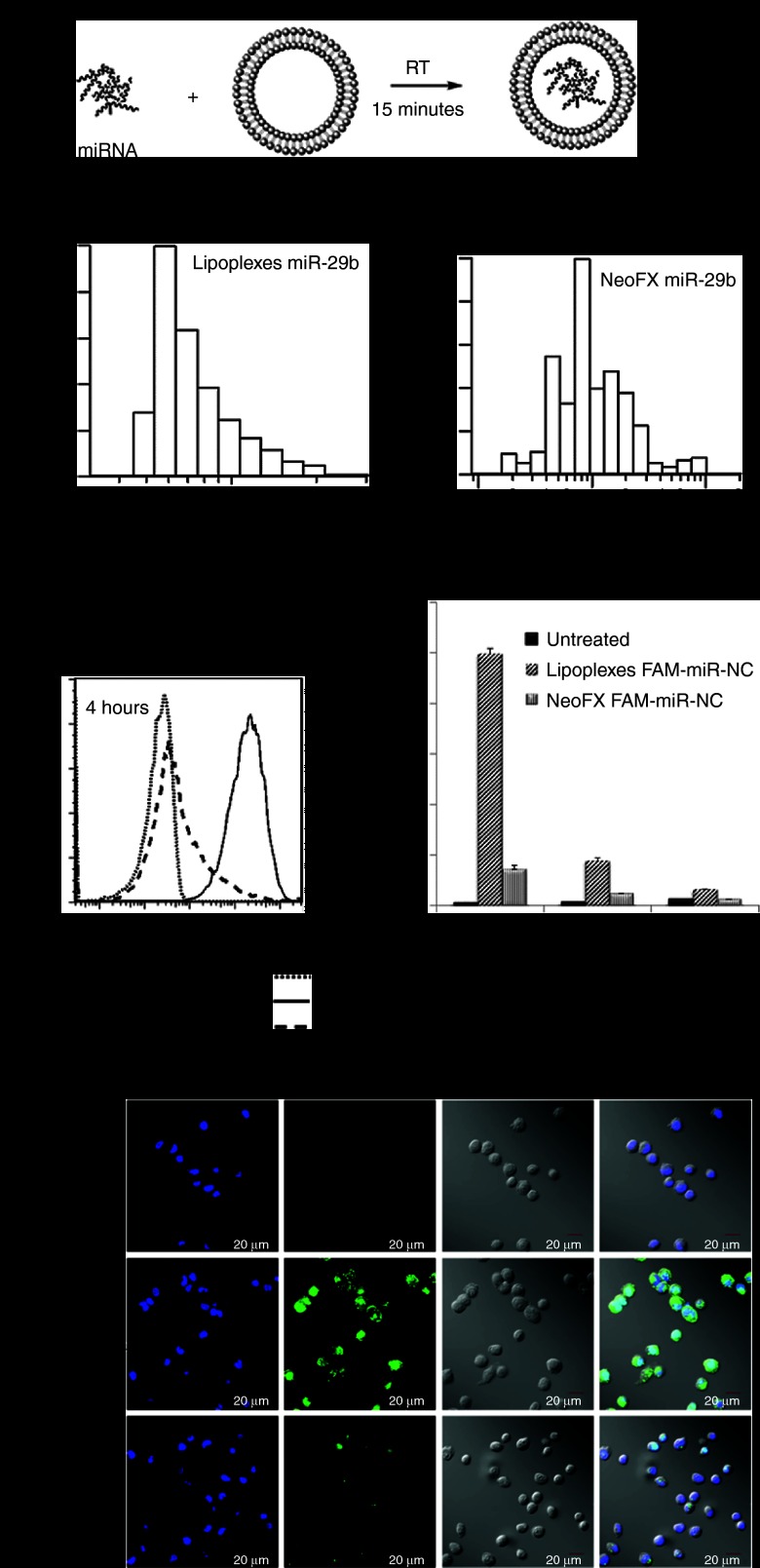
LPs containing miR-29b (LP-miR-29b) were prepared by mixing synthetic miR-29b with empty liposomes at a lipids/miR-29b mass ratio of 12.5 (Figure 1a). As a positive control, siPORT NeoFX transfection reagent was used to prepare complexes containing miR-29b (NeoFX-miR-29b) by following the protocol recommended by the manufacturer. As shown in Figure 1b, the resulting LP-miR-29b had a mean diameter weighted by volume of 84.0 ± 10.9 nm and zeta potential of 17.26 ± 3.15 mV. The resulting NeoFX-miR-29b had a mean diameter by volume of 1,276.0 ± 120.8 nm and zeta potential of −7.57 ± 2.34 mV.
Figure 1.
Efficient cellular uptake of lipoplexes. (a) Preparation of lipoplexes containing miRNAs. (b) Typical size distributions of LP-miR-29b (left) and NeoFX-miR-29b (right). (c) Flow cytometry analysis of A549 cellular uptake of LP-FAM-miR-NC and NeoFX-FAM-miR-NC at FAM-miR-NC concentration of 100 nmol/l. A typical flow cytometry dataset at 4 hours after transfection (left). Mean fluorescence intensity of FAM averaged on three replicates 4, 24, and 48 hours after transfection (right, n = 3). (d) Confocal microscopy analysis of cellular uptake of LP-FAM-miR-NC and NeoFX-FAM-miR-NC 4 hours after transfection at FAM-miR-NC concentration of 100 nmol/l.
To demonstrate more efficient cellular uptake of LPs, A549 cells were transfected with LPs containing FAM-labeled miR-Negative Control (LP-FAM-miR-NC) and NeoFX complexes containing FAM-labeled miR-NC (NeoFX-FAM-miR-NC) at FAM-miR-NC concentration of 100 nmol/l. Cellular uptake was evaluated at 4, 24, and 48 hours after transfection by flow cytometry (Figure 1c). At 4 hours after transfection the mean fluorescence intensity of FAM in A549 cells treated by LP-FAM-miR-NC was much higher than that of NeoFX-FAM-miR-NC. In addition, LP-FAM-miR-NC transfected cells demonstrated a narrower FAM fluorescence distribution than NeoFX-FAM-miR-NC. Confocal microscopy images also showed stronger and more uniform cellular uptake of LP-FAM-miR-NC than NeoFX-FAM-miR-NC (Figure 1d). These results demonstrate that LPs are a more efficient delivery system in vitro than NeoFX complexes. At 24 and 48 hours after transfection, the mean fluorescence intensity of FAM was progressively decreased in A549 cells transfected by both LPs and NeoFX complexes, which was due to the degradation of FAM-labeled miR-NC and the “dilution” effect by the cell growth. On the basis of the observation, delivery of miR-29b every 48 hours might represent a reasonable frequency if repeated delivery is desired. Therefore, LP-miR-29b was delivered every other day in the subsequent in vivo studies.
MiR-29b expression is reduced in human non-small–cell lung cancer
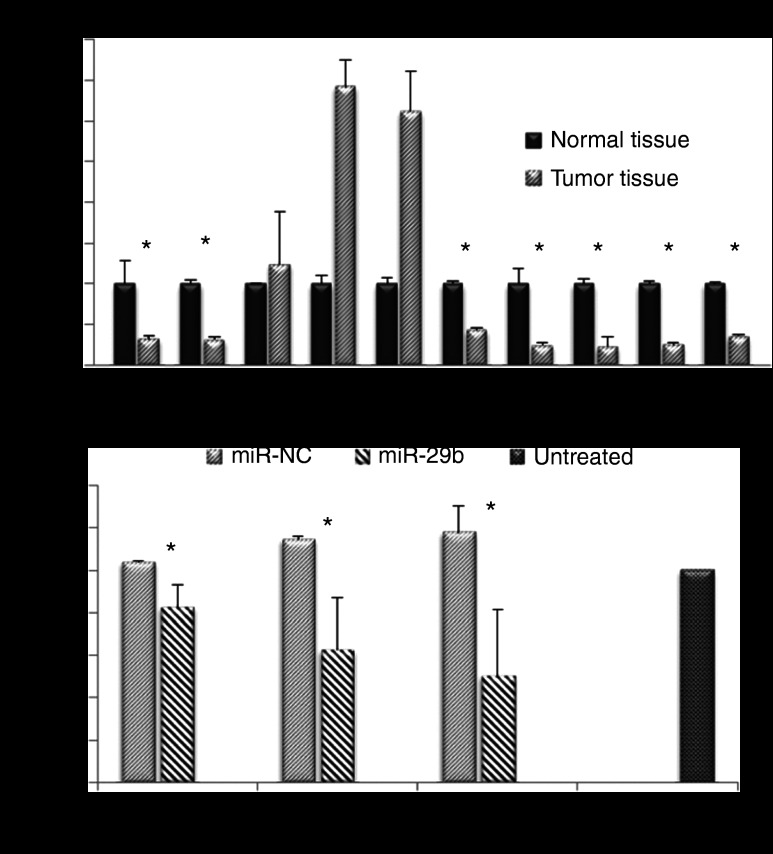
We analyzed the expression of miR-29b and CDK6 in 10 pairs of human non-small cell lung cancer (NSCLC) tumor tissue samples and adjacent normal tissue samples by quantitative real-time PCR (qRT-PCR). As shown in Figure 2a, the expression of miR-29b was significantly downregulated in 7 out of 10 NSCLC tumor tissue samples compared with normal tissue samples (P < 0.05).
Figure 2.
MiR-29b dysregulation and target expression. (a) Expression of miR-29b was significantly downregulated in 7 out of 10 NSCLC tumor tissue samples compared with normal tissue samples. (b) CDK6 was a direct target of miR-29b. Three CDK6 complementary sites were cloned into the 3′-UTR of the firefly luciferase gene and transfected with miR-29b in CHO cells. MiR-29b significantly reduced the luciferase activity with respect to the scrambled miR-NC sequences (n = 3, *P < 0.05).
MiR-29b directly targets CDK6
Next, we sought to determine whether CDK6 was a direct target of miR-29b in lung cancer. Three CDK6 complementary sites were cloned into the 3′-UTR (untranslated region) of the firefly luciferase gene and transfected with miR-29b in CHO cells. As shown in Figure 2b, miR-29b significantly reduced the luciferase activity compared with the scrambled sequences (P < 0.05).
Downregulation of CDK6, DNMT3B, and MCL1 expression by LP-miR-29b
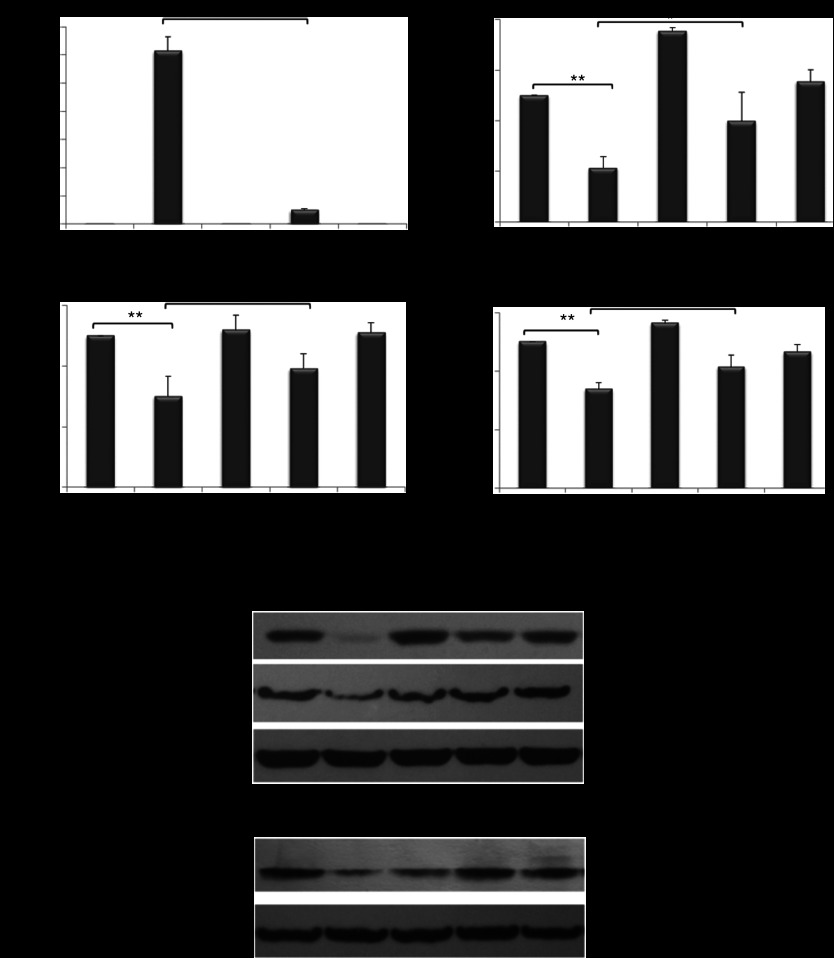
A549 cells were transfected with LP-miR-29b, NeoFX-miR-29b, LP-miR-NC, or NeoFX-miR-NC at miRNA concentration of 100 nmol/l. At 48 hours after transfection, compared with untreated control, mature miR-29b expression was increased ~1,230-folds in cells transfected with LP-miR-29b, and ~98-fold in those transfected by NeoFX-miR-29b, demonstrating that LPs were a more efficient carrier system than NeoFX complexes (Figure 3a). The expression of target genes, CDK6, DNMT3B, and MCL1, was more efficiently downregulated at both mRNA and protein levels in cells transfected with LP-miR-29b compared with NeoFX-miR-29b (Figure 3b–e). LP-miR-NC and NeoFX-miR-NC did not cause a nonspecific effect on the expression of mature miR-29b or three target genes.
Figure 3.
LP-miR-29b downregulates target gene expression in vitro. (a) Expression of mature miR-29b and downregulation of CDK6, DNMT3B, and MCL1 expression at (b–d) mRNA level and (e) protein level 48 hours after A549 cells were transfected with LP-miR-29b, NeoFX-miR-29b, LP-miR-NC, and NeoFX-miR-NC at microRNA concentration of 100 nmol/l (*P < 0.05 and **P < 0.01) (n = 3).
Inhibition of A549 tumorigenicity by LP-miR-29b in vitro
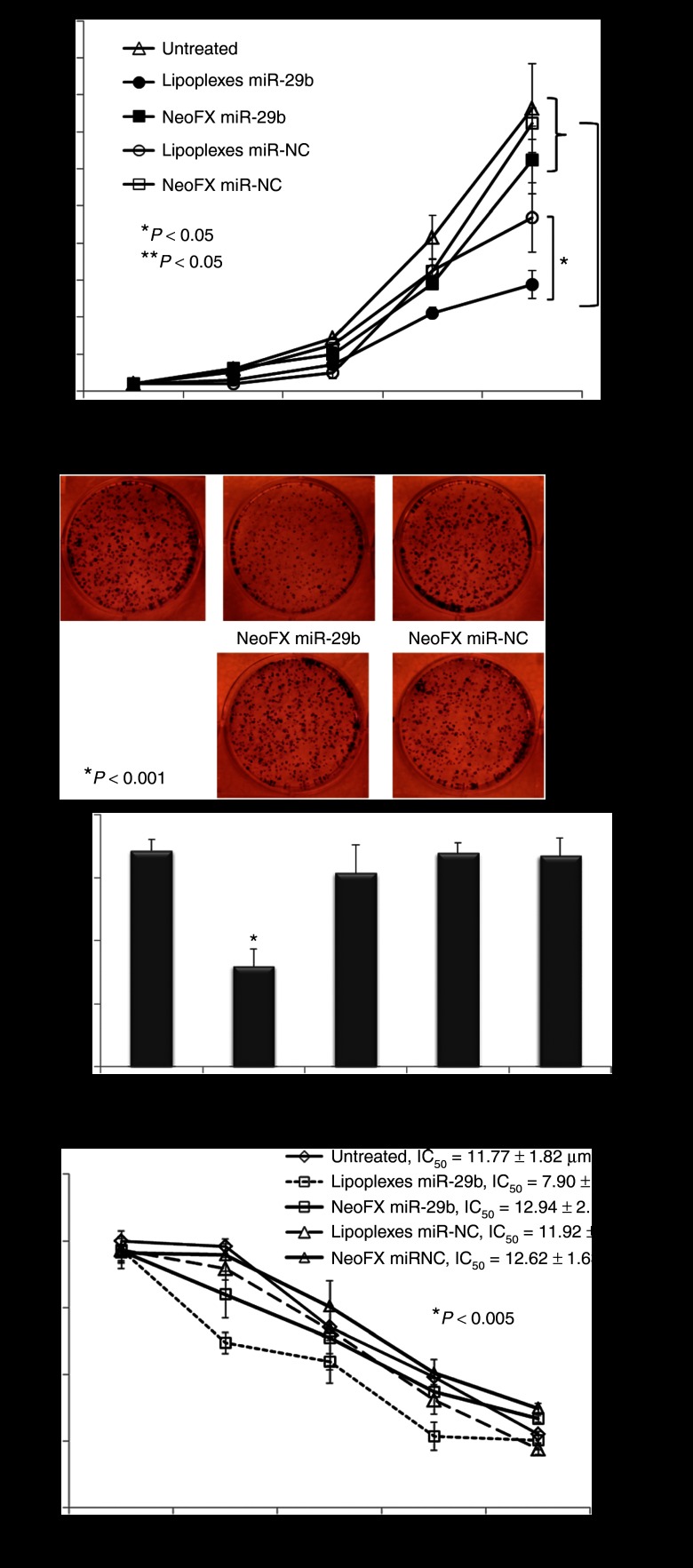
A549 cells were transfected with LP-miR-29b, NeoFX-miR-29b, LP-miR-NC, or NeoFX-miR-NC at microRNA concentration of 100 nmol/l. After transfection, the tumorigenicity of A549 cells was evaluated by measuring cell growth, clonogenic survival and IC50 following treatment with cisplatin. As shown in Figure 4a, LP-miR-29b significantly inhibited the growth of A549 cells compared with NeoFX-miR-29b, LP-miR-NC, NeoFX-miR-NC, and untreated control. In the clonogenic survival assay, the number of colonies was reduced by ~50% in the cells transfected with LP-miR-29b compared with untreated control (Figure 4b). No significant difference was found between untreated cells and cells transfected by NeoFX-miR-29b, LP-miR-NC, and NeoFX-miR-NC. LP-based transfection also increased the sensitivity of A549 cells to chemotherapeutic agent cisplatin. The IC50 of cisplatin was reduced from ~11.77 µmol/l in untreated cells to ~7.90 µmol/l in cells transfected by LP-miR-29b (Figure 4c). NeoFX-miR-29b, LP-miR-NC, and NeoFX-miR-NC did not affect the sensitivity of A549 cells to cisplatin treatment.
Figure 4.
LP-miR-29b inhibit tumorigenicity of A549 cells in vitro. (a) Growth curve, (b) colonogenic survival, and (c) IC50 of cisplatin for A549 cells after transfection with LP-miR-29b, NeoFX-miR-29b, LP-miR-NC, and NeoFX-miR-NC at microRNA concentration of 100 nmol/l (*P < 0.05 and **P < 0.005) (n = 3).
Inhibition of A549 tumorigenicity by LP-miR-29b in vivo
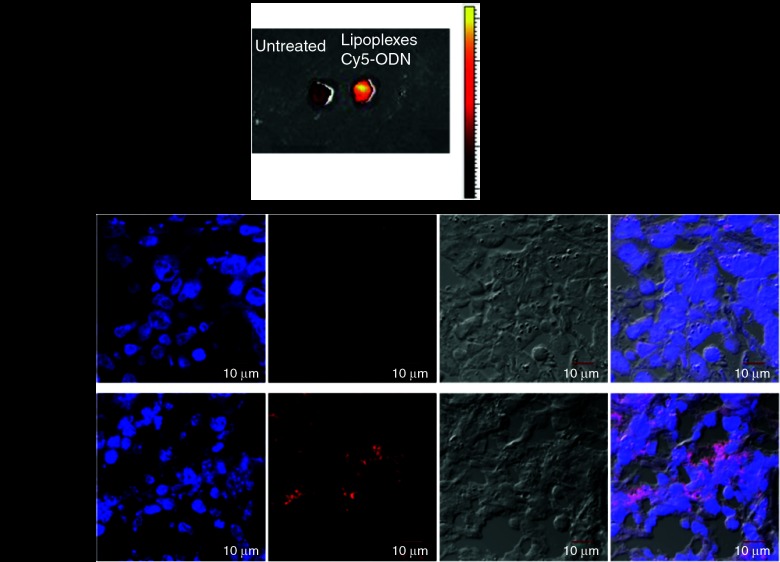
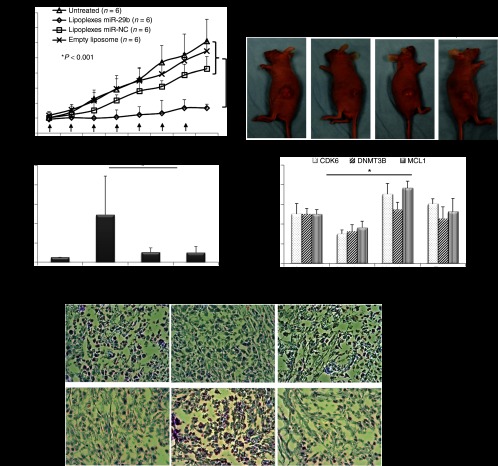
The antitumor effects of LP-miR-29b were evaluated in a xenograft murine model using A549 cells. A549 cells were implanted into the flanks of 6-week-old, female athymic mice. Tumors were allowed to develop to ~100 mm3 before treatment. To demonstrate the delivery of LPs to the tumor site, LPs containing Cy5-labeled oligodeoxynucleotide (LP-Cy5-ODN) were injected into mice through tail vein at a dosage of 1.5 mg Cy5-ODN/kg mouse weight. Four hours later, the mice were killed and tumor tissues were collected for IVIS imaging (Figure 5a) and confocal microscopy analysis (Figure 5b). Results showed that the LPs successfully delivered Cy5-ODN to the flank tumor sites demonstrating that they were an effective carrier system to systemically deliver miRNAs in vivo. Next, at 22 days following implantation, mice with established tumors (tumor volume ~100 mm3) were randomly assigned to four groups (n = 6). LP-miR-29b, LP-miR-NC, at dosages of 1.5 mg microRNA/kg mouse weight, or matching lipid dose of empty liposomes were given via intravenous injection every other day for a total of seven injections (Q2Dx7). Mice were killed 2 days after the last injection. As shown in Figure 6a,b, at the conclusion of the experiment, the mice treated with LP-miR-29b had significantly smaller tumors compared with LP-miR-NC (60.8% inhibition, n = 6, P < 0.001), empty liposomes (69.3% inhibition, n = 6, P < 0.001), and untreated control group (72.5% inhibition, n = 6, P < 0.001). Tumor tissues were collected and evaluated for the expression of mature miR-29b (Figure 6c) and three target genes (Figure 6d) using qRT-PCR. In the tumors of mice treated with LP-miR-29b, the expression of mature miR-29b was increased ~4.7-fold and the mRNA expression of CDK6, DNMT3B, and MCL1 was downregulated by ~57.4, ~40.5, and ~52.4%, respectively compared with mice treated with LP-miR-NC. No significant difference in the expression of mature miR-29b and three target genes were found between untreated control group and the two groups treated with LP-miR-NC or empty liposomes (P < 0.05). Tumor tissues from LP-miR-29b–treated mice showed reduced expression of Ki-67 and increased number of apoptotic cells (TUNEL) (Figure 6e), indicating LP-miR-29b actively inhibit cell proliferation and stimulated the apoptosis process.
Figure 5.
Accumulation of LPs at tumor sites. A549 cells were implanted into the flanks of 6-week-old, nude mice and tumors were allowed to develop to ~100 mm3 without treatment. LP-Cy5-ODN was injected through tail vein at 1.5 mg Cy5-ODN/kg mouse weight. After 4 hours, mice were killed and tumor tissues were analyzed using (a) IVIS imaging system and (b) confocal microscopy. The Cy5 fluorescence from the tumor tissues indicated the accumulation of LPs in the tumor sites (n = 6).
Figure 6.
LP-miR-29b inhibit tumorigenicity of A549 cells in vivo. (a,b) Tumor growth. A549 cells were implanted into the flanks of 6-week-old, nude mice and tumors were allowed to develop ~100 mm3 without treatment. At 22 days after implantation, mice were treated with LP-miR-29b, LP-miR-NC, and empty liposomes by intravenous injection at 1.5 mg miR/kg mouse weight every other day for seven injections. Tumors were measured using a digital caliper and tumor volumes were calculated (n = 6,*P < 0.001). (c) Expression of mature miR29b in tumors (n = 6, *P < 0.005). (d) Expression of CDK6, DNMT3B, and MCL1 was significantly downregulated in the tumors of mice treated with LP-miR-29b than those treated with LP-miR-NC (n = 6, *P < 0.01). (e) Ki-67 staining and TUNEL staining of tumor tissues of untreated mice and mice treated with LP-miR-29b and LP-miR-NC.
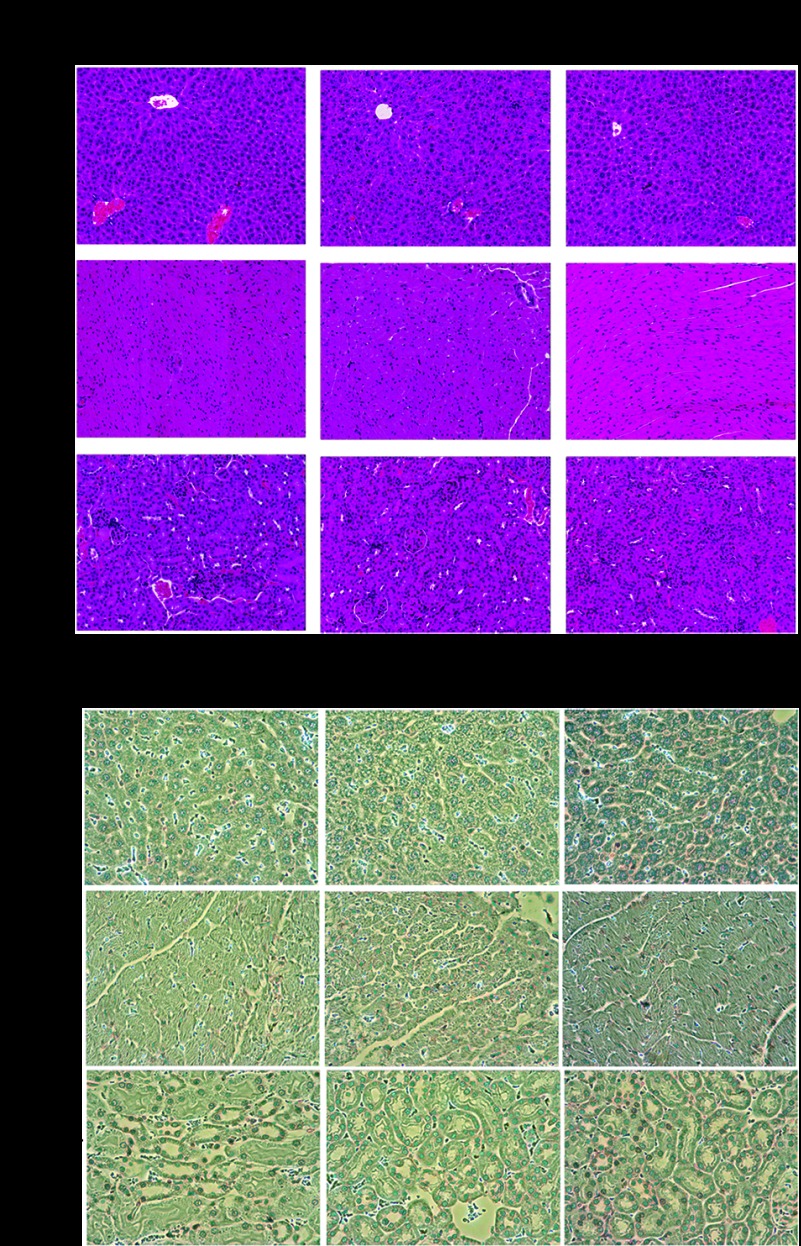
Cytotoxicity evaluation of LP-miR-29b in vivo
The cytotoxicity effect of LP-miR-29b in the major mouse organs including heart, liver, and kidney was evaluated by H&E and TUNEL staining and compared with those of untreated control mice and LP-miR-NC–treated mice. We have previously demonstrated that our nanoparticles did not induce toxicity in the lung tissues.27 As shown in Figure 7a,b, no significant pathology was observed among untreated control, LP-miR-29b–treated mice and LP-miR-NC–treated mice, suggesting that LP-miR-29b did not induce significant cytotoxicity to major organs.
Figure 7.
Cytotoxicity evaluation of LP-miR-29b in major organs. (a) H&E staining of liver, heart, and kidney. (b) TUNEL staining for liver, heart, and kidney.
Discussion
MiRNAs can serve as tumor suppressors or oncogenes by regulating processes essential to cancer development including proliferation, survival, apoptosis, and metastasis.6,7,8 Through the simultaneous regulation of multiple biological pathways, miRNA mimics may represent a powerful therapeutic strategy in cancer treatment.7,8 However, there are several obstacles to reaching the in vivo application of miRNAs as therapy. Here, we present a cationic LPs-based carrier system that efficiently delivered miR-29b to A549 NSCLC cells demonstrating an in vitro and in vivo reduction in tumorigenicity by targeting multiple oncogenes including CDK6, DNMT3B, and MCL1.
In our LPs formulation, 1,2-di-O-octadecenyl-3-trimethylammonium propane (DOTMA, chloride salt), a cationic lipid previously used in nonviral vectors of gene therapy, was selected as the building block of our miRNA delivery system. DOTMA has shown high transfection activities in vitro and in vivo.28 DOTMA also confers a positive surface charge to the LPs which in turn facilitates the interaction between LPs and the negatively charged cell membrane providing more efficient and uniform cellular uptake. Cholesterol, a neutral lipid, was chosen as a helper lipid because it can improve transfection efficiency in vivo, protecting oligonucleotides from degradation, and facilitating the interaction between LPs and the cell membranes.29 D-α-Tocopheryl polyethyleneglycol 1,000 succinate (vitamin E TPGS), a short poly(ethylene glycol) (PEG) molecule linked to vitamin E, was added to increase the stability and circulation time of LPs in vivo. In addition, confocal microscopy images showed that the LPs were able to deliver miR-29b to the nucleus of A549 cells, which is important because endogenous miR-29b is significantly enriched in the cell nucleus.9
MiR-29b, a potential prognostic marker, targets oncogenes and antiapoptotic genes, and is usually silenced or downregulated in many cancers. In acute myeloid leukemia and mantle cell lymphoma, CDK6 has been validated as a direct target of miR-29b.13,30 In HPV-mediated cervical cancer, restoration of miR-29b expression was shown to prevent malignant transformation of cervical cells by targeting CDK6.18 In NSCLC, miR-29b directly targets DNMT3A and 3B. The expression of miR-29b is inversely correlated to DNMT3A and 3B expression.20 In acute myeloid leukemia, cholangiocarcinoma, and hepatocellular carcinoma, miR-29b has been shown to target the expression of MCL1 protein, which affects malignant cell survival.13,16,31 To the best of our best knowledge, no studies have shown the relationship between miR-29b expression and CDK6 expression in NSCLC tumor tissue samples; therefore, we evaluated the expression of miR-29b in 10 pairs of human NSCLC tumor tissue samples and adjacent normal tissue samples. We found that the expression of miR-29b was downregulated in the majority of NSCLC tissue samples compared with normal tissue samples (Figure 2a). We then demonstrated that CDK6 was a direct target of miR-29b (Figure 2b), which was further validated by the downregulation of CDK6 gene expression by LP-miR-29b in vitro (Figure 3) and in vivo (Figure 6).
The antitumorigenic properties of LP-miR-29b were first evaluated in A549 cells. Because the delivery efficiency of LPs was superior to that of NeoFX complexes, mature miR-29b expression in A549 cells transfected by LPs was ~12.5 times higher, and all three targets genes (CDK6, DNMT3B, and MCL1) were successfully downregulated at both mRNA and protein levels (Figure 3). LP-miR-29b also showed greater biological activity than NeoFX-miR-29b in A549 cells. Compared with untreated control, ~62 and ~18% inhibition in A549 cell growth was observed in cells treated by LP-miR-29b and NeoFX-miR-29b respectively. The clonogenic survival of A549 cells was reduced by ~53%, and the IC50 of cisplatin was decreased by ~33% after treatment with LP-miR-29b compared with untreated control (Figure 4). No significant effects were observed in the cells transfected by NeoFX-miR-29b. In the xenograft mouse model, we observed that LPs successfully accumulated at the tumor sites (Figure 5). Following seven intravenous injections of LP-miR-29b at a dosage of 1.5 mg miR-29b/kg mouse weight, tumor growth was significantly inhibited (Figure 6a,b). Systemic miR-29b delivery by LPs increased tumor miR-29b expression by ~4.7-fold compared with mice treated with LP-miR-NC (Figure 6c). The tumor expression of CDK6, DNMT3B, and MCL1 was significantly downregulated compared with the other three control groups (Figure 6d). Results of Ki-67 and TUNEL staining showed that tumor tissues from LP-miR-29b–treated mice had lower expression of Ki-67 and more apoptotic cells which was in agreement with the observed downregulation of CDK6 and MCL1 expression (Figure 6e). In addition, we did not observe significant cytotoxicity effects of LP-miR-29b on the vital organs of mice, suggesting the off-target effects of miR-29b were minimal (Figure 7a,b).
In summary, we have developed a novel cationic LPs-based delivery system that efficiently delivered miR-29b and thus inhibited the tumorigenicity of NSCLC in vitro and in vivo. Our results demonstrate a potential for LPs in the development of miRNA-based therapeutics for lung cancer treatment. In the future, chemotherapeutic drugs, such as cisplatin, might be combined with miRNA-based therapeutic strategies to enhance therapeutic efficacy.
Materials and Methods
Materials. 1,2-Di-O-octadecenyl-3-trimethylammonium propane (DOTMA, chloride salt) was purchased from Avanti Polar Lipids (Alabaster, AL). Cholesterol was purchased from Sigma-Aldrich (St Louis, MO). D-α-Tocopheryl polyethyleneglycol 1,000 succinate (vitamin E TPGS) was purchased from Eastman (Kingsport, TN). MiRIDIAN mimic hsa-miR-29b (C-300521-05, mature miR sequence: 5′-UAGCACCAUUUGAAAUCAGUGUU-3′) and miRIDIAN mimic NC#1 (CN-001000-01, mature miR sequence: 5′-UCAC AACCUCCUAGAAAGAGUAGA-3′) were purchased from Dharmacon RNAi Technologies (Lafayette, CO). FAM dye–labeled pre-miR negative control #1 (FAM-miR-NC, Ambion, AM17121) was purchased from Ambion (Austin, TX). Cy5 dye–labeled oligodeoxynucleotides (Cy5-ODN, 5′-Cy5-TCT CCC AGC GTG CGC CAT-3′) was custom synthesized by Alpha DNA (Montreal, Quebec, Canada).
Experimental animals. Female athymic nude mice of age 4–6 weeks (weight 18–20 g) were purchased from Charles River Laboratories International (strain code 490; Wilmington, MA). All work performed on animals was in accordance with a protocol approved by the IACUC at the Ohio State University.
Human tissues. Ten pairs of human non-small cell lung adenocarcinomas tissue samples and adjacent uninvolved tissue samples were obtained the Cooperative Human Tissue Network through an IRB approved protocol. qRT-PCR was then performed for miR-29b (Applied Biosystems, Foster City, CA; assay ID 002247) and presented as 2−ΔCt. Statistical significance was based on a Student's t-test.
Cell culture. A549 cells and CHO cells were obtained from the American Type Culture Collection (Manassas, VA). A549 cells were routinely cultured in a 75 cm2 T flask containing 15 ml of RPMI 1640 media supplemented with 10% heat-inactivated fetal bovine serum (16,000; Invitrogen, Carlsbad, CA). CHO cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA), 1× antibiotic-antimycotic (Invitrogen), and 0.0175 mg/ml L-proline (Sigma-Aldrich). The cells were seeded into T flasks at a concentration of 1 × 105 viable cells/ml and incubated at 37 °C in a humidified atmosphere containing 5% CO2. The cells were subcultured every 2 days.
Preparation of LP-miR-29b. Empty liposomes were prepared first by injecting a lipid mixture in ethanol (DOTMA:cholesterol:TPGS at 49.5:49.5:1 molar ratio) into 20 mmol/l HEPES buffer (pH = 7.4) to achieve 10% ethanol and 90% aqueous in the final mixture. LP-miR-29b was prepared by adding miR-29b to empty liposomes at the lipids to miR-29b mass ratio of 12.5. The mixture was incubated at room temperature for 15 minutes and used immediately.
Particle size and surface charge measurement. The size distributions of LP-miR-29b were measured by dynamic light scattering (Brookhaven Instruments, Holtsville, NY; BI 200SM). The wavelength of the laser was 632.8 nm, and the detection angle was 90°. The size distributions of three batches of LP-miR-29b prepared independently were measured at 20 °C, and the mean diameter by volume ± SD was reported. The surface charges of LP-miR-29b were measured using ZetaPALS zeta potential analyzer (Brookhaven Instruments, Holtsville, NY). Three batches of independently prepared LPs were diluted in 20 mmol/l HEPES buffer. Three measurements, each consisting of five runs, were performed at 20 °C. The Smoluchowski model was used to calculate the zeta potential, and the mean ± SD was reported.
Luciferase reporter constructs. A 1,271-bp fragment of the human CDK6 3′-UTR (NM_001259, transcript variant 1, mRNA) that encompassed three putative miR-29 binding sites (http://www.targetscan.org, TargetScan 6.2, release date June, 2012) was PCR-amplified using the following sense (5′-ATACTCGAGTTCCCTGCTACCATCCTGGCTTGTCC-3′) and antisense (5′-ATAGCGGCCGCCATCATCTGACAATAAATACCTAC-3′) primers using standard procedures, a proofreading polymerase (Platinum Pfu; Invitrogen) and human genomic DNA as template. The XhoI and NotI restriction endonuclease sites incorporated into the sense and antisense primers above are shown in boldface. After PCR amplification, the amplicon was double digested with XhoI/NotI for 2 hours following the manufacturer's protocol. The PCR product was subsequently subcloned into the XhoI/NotI sites downstream of the Renilla luciferase (r-luc) reporter gene (psiCHECK-2; Promega). The authenticity and orientation of the inserts relative to the Renilla luciferase gene were confirmed by dideoxy sequencing. The resulting recombinant plasmids were designated psiCHECK/CDK6-UTR. Finally, transformed bacterial cultures were grown and each reporter construct was purified using the PureLink Hipure Plasmid Maxiprep kit (Invitrogen).
Transfection and luciferase assay. MiR-29b and negative control mimics (partially double-stranded RNAs that mimic the Dicer cleavage product and are subsequently processed into their respective mature miRNAs) were obtained from Dharmacon (Lafayette, CO). Transfection of CHO cells were transfected with psiCHECK/CDK6-UTR construct (10 ng per well) using Lipofectamine 2000 (Invitrogen) and the appropriate miRNA precursor as indicated. After 24 hours, CHO cells were washed and lysed with Passive Lysis Buffer (Promega), and firefly and Renilla luciferase activities were determined using the Dual-Luciferase Reporter Assay System (Promega) and a luminometer. Renilla luciferase expression in the psiCHECK vector is generated via an SV40 promoter. In addition, the psiCHECK-2 vector possesses a secondary firefly reporter expression cassette which is under the control of the HSV-TK promoter. This firefly reporter cassette has been specifically designed to be an intraplasmid transfection normalization reporter; thus when using the psiCHECK-2 vector, the Renilla luciferase signal is normalized to the firefly luciferase signal.
Transfection of A549 cells with LP-miR-29b. A549 cells were seeded at 2 × 105 viable cells/well in six-well plates containing 2 ml of culture medium supplemented with 10% fetal bovine serum. The cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2 overnight. The culture medium was then replaced with medium containing no fetal bovine serum. LP-miR-29b was then added to the cells at miR-29b concentration of 100 nmol/l. The cells were incubated at 37 °C for 4 hours, and then transferred into 2 ml fresh culture medium supplemented with 10% fetal bovine serum. All transfection experiments were performed in triplicate.
NeoFX-miR-29b was used as the positive control. The transfection of miR-29b via siPORT NeoFX transfection reagent (Invitrogen; AM4511) was performed by following the manufacturer's protocol. Briefly, 5 μl of NeoFX was mixed with 95 μl OptiMEM (Invitrogen; 11058021) and the mixture was incubated at room temperature for 10 minutes. MiR-29b was diluted in OptiMEM and then added into the NeoFX transfection agent. The final mixture was incubated at room temperature for another 10 minutes and used for transfection by following the same transfection procedure as for transfection of LPs. Untreated cells and cells transfected by scrambled microRNA mimic with no target, miR-NC, which were delivered by both LPs and NeoFX, were negative controls in our work.
Cellular uptake of LP-FAM-miR-NC. A549 cellular uptake of LP-FAM-miR-NC was studied by flow cytometry (BD LSR II, San Jose, CA, USA) and laser scanning confocal microscopy (Olympus FV1000, Center Valley, PA). A549 cells were transfected with LP-FAM-miR-NC or NeoFX-FAM-miR-NC. The cells were harvested 0, 24, and 48 hours after transfection. Briefly, the cells were first detached from culture plates using 0.25% trypsin, washed with PBS twice and fixed using 4% paraformaldehyde. In the flow cytometry experiments, the fluorescence signals of FAM were observed in the FITC channel. For each sample, 10,000 events were collected, and the average results of three replicates were reported. In confocal microscopy experiments, A549 cells were counterstained with DAPI and mounted on glass slides. The fluorescence signals of DAPI and FAM were observed in the DAPI (dichroic mirror 430–470 nm) and FAM (dichroic mirror 505–605 nm) channels respectively.
Expression of mature miR-29b, CDK6, DNMT3B, and MCL1 mRNAs in A549 cells by qRT-PCR. Total RNA was extracted using TRIzol reagent (Invitrogen; 15596-018) and chloroform, further purified by isopropanol precipitation and washed by 70% ethanol. To measure mature miR-29b expression, the total RNA was first reverse transcribed into cDNA using the TaqMan MicroRNA reverse transcription kit (Applied Biosystems; 4366596). The qRT-PCR amplification of cDNA was then performed using TaqMan MicroRNA assay (Applied Biosystems; assay ID 002247). The mature miR-29b expression was determined by the ΔΔCt method and normalized to RNU48 (Applied Biosystems; assay ID 001006), which was the endogenous control in the corresponding samples, and relative to the untreated control cells.
To measure the expression of CDK6, DNMT3B, and MCL1 mRNAs, the total RNA was transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems; 4368814). The resulting cDNA was amplified by qRT-PCR (Applied Biosystems; CDK6 assay ID: Hs01026371_m1, DNMT3B assay ID: Hs00171876_m1, and MCL1 assay IDHs03043899_m1). Relative gene expression values were determined by the ΔΔCt method. The expression of CDK6, DNMT3B, and MCL1 mRNAs was normalized to GAPDH (Applied Biosystems; assay ID: Hs02758991_g1), which was the endogenous reference in the corresponding samples, and relative to the untreated control cells.
Expression of CDK6, DNMT3B, and MCL1 protein in A549 cells by western blotting. Protein from A549 cells was extracted using RIPA lysis buffer (Sigma-Aldrich; R0278) containing a protease inhibitor cocktail (Sigma-Aldrich; P8340) on ice for 15 minutes. The cell lysate was centrifuged for 15 minutes at 12,000g at 4 °C. The protein concentration of the supernatant was measured by bicinchoninic acid (BCA) assay (Biorad, Hercules, CA; 500-0006), and 50μg of protein from each sample was loaded in a 4–15% Ready Gel Tris–HCl polyacrylamide gel (BioRad), and then transferred to a PVDF membrane (GE). After blocking with 5% nonfat milk in Tris-buffered saline/Tween-20 for 1 hour, the membranes were incubated with mouse antihuman CDK6 antibody (1:2,000; Cell Signaling Technology, Danvers, MA; 3136S), rabbit antihuman DNMT3B antibody (1:1,000; Cell Signaling Technology; 2161S), rabbit antihuman MCL1 antibody (1:500; Cell Signaling Technology; 4572S), or mouse antihuman β-tubulin antibody (1:10,000; Sigma; T8328) at 4 °C overnight, followed by incubation with horseradish peroxidase-conjugated goat antirabbit IgG (1:2,000; Santa Cruz Biotechnology; SC2004) and horseradish peroxidase-conjugated goat antimouse IgG (1:2,000; Santa Cruz Biotechnology, Dallas, TX; SC2005) for 1 hour at room temperature. The membrane was then developed with Pierce SuperSignal West Pico or Dura Extended DurationSubstrate (Pierce, Rockford, IL) and imaged with KodakX-OMAT film (Kodak, Rochester, NY).
Growth curve of A549 cells. At 1, 2, 3, and 4 days after transfection, the A549 cells were stained with Trypan Blue (0.4%) (Invitrogen; 15250-061) and viable cells were counted using Hemocytometer. Each sample had three replicates and the mean ± SD was reported.
Clonogenic assay. At 24 hours after transfection, A549 cells were harvested and resuspended in the culture medium. In six-well plates, 2,000 cells/well were seeded and allowed to grow until visible colonies formed (6 days). Cell colonies were fixed using 4% paraformaldehyde and stained with 0.5% crystal violet in water at room temperature for 2 hours. Cell colonies were then washed three times with tap water and air-dried. Colonies with >50 cells were counted under microscope. Each sample had three replicates and the mean ± SD was reported.
LP-Cy5-ODN accumulation at tumor sites. Three million A549 cells were resuspended in PBS and implanted into the flanks of 6-week-old, female athymic nude mice. Tumors were allowed to develop to ~100 mm3 without any treatment. LP-Cy5-ODN was injected through tail vein at dosage of 1.5 mg Cy5-ODN/kg mouse weight. After 4 hours, mice were euthanized and tumor tissues were collected and fixed in 10% formalin for 24 hours. The tumor tissues were then soaked in 30% sucrose solution for another 24 hours. The fluorescence signals of Cy5 emitted by the whole tumor tissues were measured using Xenogen IVIS-200 Optical In Vivo Imaging System (Caliper Life Sciences, Hopkinton, MA). All tumor tissues were then cryopreserved in optimal cutting temperature (OCT) compound. The cross sections of tumor tissues were counterstained with Hoechst and mounted on glass slides for confocal microscopy analysis. The fluorescence signals of Hoechst and Cy5 were observed in the DAPI (dichroic mirror 430–470 nm) and Cy5 (dichroic mirror 655–755 nm) channels, respectively.
In vivo delivery of LP-miR-29b. Three million A549 cells were resuspended in PBS and implanted into the flanks of 6-week-old, nude mice. Tumors were allowed to develop to ~100 mm3 without any treatment till day 22. LP-miR-29b, LP-miR-NC and empty liposomes were administered to nude mice by intravenous injection at a dosage level of 1.5 mg miR/kg mouse weight every other day for total seven injections (n = 6). Tumors were measured using a digital caliper. Tumor volumes were calculated using the equation V (in mm3) = A × B2/2, where A is the largest diameter and B is the perpendicular diameter. At 48 hours after the last administration, mice were euthanized and tumor tissues were collected and quickly frozen in liquid nitrogen. The frozen tumor tissues were grounded to powders. The total RNA was extracted using TRIzol and chloroform further purified by isopropanol precipitation and washed by 70% ethanol. The total RNA was then reverse transcribed into cDNA using the TaqMan MicroRNA reverse transcription kit. The qRT-PCR amplification of cDNA was then performed using TaqMan MicroRNA assay (Applied Biosystems; assay ID 002247). The mature miR-29b expression was determined by the ΔΔCt method and normalized to sno135 (Applied Biosystems; assay ID 001230), which was the endogenous control in the corresponding samples, and relative to the untreated control tissue samples. To measure the expression of CDK6, DNMT3B, and MCL1 mRNAs in the tumors, the total RNA was transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems; 4368814). The resulting cDNA was amplified by qRT-PCR (Applied Biosystems; CDK6 assay ID: Mm01311342_m1, DNMT3B assay ID: Mm01240113_m1, and MCL1 assay ID Mm0725832_s1). Relative gene expression values were determined by the ΔΔCt method. The expression of CDK6, DNMT3B, and MCL1 mRNAs was normalized to GAPDH (Applied Biosystems; assay ID: Mm99999915_g1), which was the endogenous reference in the corresponding samples, and relative to the untreated control cells.
Tumor histology. Tissues were fixed in formalin and embedded in paraffin. Sections (4 μm) were cut and stained with H&E. Representative sections of liver, heart, and kidney were scanned with a Scanscope CS slide scanner (Aperio Technologies, Vista, CA), visualized with ImageScope software (Aperio Technologies), and composed in Adobe Photoshop (San Jose, CA). Images were adjusted in brightness, contrast, and color balance for a more uniform appearance of the H&E stain. These adjustments do not obscure, eliminate, or misrepresent any information presented in the original slides. Original objective lens magnification was 20×. All images were reviewed by a veterinary pathologist.
Immunohistochemistry. Paraffin embedded tissues were cut at 4 μm and placed on positively charged slides. Slides were then placed in a 60 °C oven for 1 hour, cooled, and deparaffinized and rehydrated through xylenes and graded ethanol solutions to water. For Ki-67 staining, all slides were quenched for 5 minutes in a 3% hydrogen peroxide solution in water to block for endogenous peroxidase. The slides were antigen retrieved in Dako's Target Retrieval Solution (pH6.0) in a vegetable steamer. The slides were incubated with the rat antimouse Ki-67 primary antibody (DakoM7249, clone TEC-3) at the dilution of 1:500 for 1 hour at room temperature. Endogenous biotin was blocked using the Avidin/Biotin Blocking Kit (Dako, X0590; 20 minutes for the avidin block, 20 minutes for the biotin block). The slides were then incubated with the rabbit antirat secondary antibody (Vector Labs, BA-4001) at the dilution of 1:200 and incubated for 20 minutes. The detection system used was Vectastain Elite (Vector Labs, PK-6100) for 30 minutes. The substrate chromogen used was DAB+ (Dako K3468). The slides were then counterstained with Richard Allen hematoxylin, dehydrated through graded ethanol solutions and sealed with cover slips.
For TUNEL staining, the slides were pretreated with Dako RTU Proteinase K (S3020) for 5 minutes at room temperature, and then quenched in 3% H2O2 for 5 minutes. Next, the slides were incubated with RTU equibibration buffer for 10 seconds at room temperature, and then incubated with Tdt enzyme in hybridizer at 37 °C. The reaction was stopped by rinsing with stop/wash buffer and PBS. The slides were incubated with RTU anti-Digoxigenin-Peroxidase conjugate for 30 minutes and then rinsed with PBS. The slides were incubated with peroxidase substrate (DAB) for 5 minutes and rinsed with diH2O. The slides were counterstained with Richard Allen hematoxylin, dehydrate in alcohols, clear in xylene and sealed with cover slips.
Statistical analysis. Data are presented as the mean ± SD. The statistical significance was determined using JMP Pro 9 software.
Acknowledgments
This work was supported in part by the National Science Foundation grant EC-425625 to LJ LEE and National Cancer Institute grant CA150297 to S.P. Nana-Sinkam. The authors declare no conflict of interest.
References
- Howlader N, Noone AM, Krapcho M, Neyman N, Aminou R, Altekruse SF.et al. Seer Cancer Statistics Review 1975-2009 (Vintage 2009 Populations). National Cancer Institute: Bethesda; < http://seer.cancer.gov/csr/1975_2009_pops09 >. [Google Scholar]
- Jemal A, Siegel R, Ward E, Murray T, Xu J., and, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- Marshall E. Cancer research and the $90 billion metaphor. Science. 2011;331:1540–1541. doi: 10.1126/science.331.6024.1540-a. [DOI] [PubMed] [Google Scholar]
- He L., and, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Chen K., and, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- Garzon R, Marcucci G., and, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D.et al. (2005MicroRNA expression profiles classify human cancers Nature 435834–838. [DOI] [PubMed] [Google Scholar]
- Kriegel AJ, Liu Y, Fang Y, Ding X., and, Liang M. The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol Genomics. 2012;44:237–244. doi: 10.1152/physiolgenomics.00141.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt MJ, Margue C, Behrmann I., and, Kreis S.2012miRNA-29: A microRNA Family with Tumor-Suppressing and Immune-Modulating Properties Curr Mol Medin press). [DOI] [PubMed]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE.et al. (2005A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia N Engl J Med 3531793–1801. [DOI] [PubMed] [Google Scholar]
- Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C.et al. (2008Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin Proc Natl Acad Sci USA 1053945–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Heaphy CE, Havelange V, Fabbri M, Volinia S, Tsao T.et al. (2009MicroRNA 29b functions in acute myeloid leukemia Blood 1145331–5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V.et al. (2006Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181 Cancer Res 6611590–11593. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Kuo C, Nicholl MB, Sim MS, Turner RR, Morton DL.et al. (2011Downregulation of microRNA-29c is associated with hypermethylation of tumor-related genes and disease outcome in cutaneous melanoma Epigenetics 6388–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH.et al. (2010Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma Hepatology 51836–845. [DOI] [PubMed] [Google Scholar]
- Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T.et al. (2006The colorectal microRNAome Proc Natl Acad Sci USA 1033687–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang F, Xu J, Ye F, Shen Y, Zhou J.et al. (2011Progressive miRNA expression profiles in cervical carcinogenesis and identification of HPV-related target genes for miR-29 J Pathol 224484–495. [DOI] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M.et al. (2006Unique microRNA molecular profiles in lung cancer diagnosis and prognosis Cancer Cell 9189–198. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E.et al. (2007MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B Proc Natl Acad Sci USA 10415805–15810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild SI, Tschan MP, Federzoni EA, Jaggi R, Fey MF, Gugger M.et al. (2012MicroRNA-29b is involved in the Src-ID1 signaling pathway and is dysregulated in human lung adenocarcinoma Oncogene 314221–4232. [DOI] [PubMed] [Google Scholar]
- Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G., and, Sood AK. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen J., and, Szoka FC. Nucleic acid delivery: the missing pieces of the puzzle. Acc Chem Res. 2012;45:1153–1162. doi: 10.1021/ar3000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhu X, Zhang X, Liu B., and, Huang L. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol Ther. 2010;18:1650–1656. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D.et al. (2010Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34 Cancer Res 705923–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trang P, Wiggins JF, Daige CL, Cho C, Omotola M, Brown D.et al. (2011Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice Mol Ther 191116–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Crawford M, Yu B, Mao Y, Nana-Sinkam SP., and, Lee LJ. MicroRNA delivery by cationic lipoplexes for lung cancer therapy. Mol Pharm. 2011;8:1381–1389. doi: 10.1021/mp2002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T, Song YK, Zhang G., and, Liu D. Structural basis of DOTMA for its high intravenous transfection activity in mouse. Gene Ther. 2000;7:764–768. doi: 10.1038/sj.gt.3301153. [DOI] [PubMed] [Google Scholar]
- Schroeder A, Levins CG, Cortez C, Langer R., and, Anderson DG. Lipid-based nanotherapeutics for siRNA delivery. J Intern Med. 2010;267:9–21. doi: 10.1111/j.1365-2796.2009.02189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JJ, Lin J, Lwin T, Yang H, Guo J, Kong W.et al. (2010microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma Blood 1152630–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott JL, Kobayashi S, Bronk SF., and, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]