Abstract
Shifts in bacterioplankton community composition along the salinity gradient of the Parker River estuary and Plum Island Sound, in northeastern Massachusetts, were related to residence time and bacterial community doubling time in spring, summer, and fall seasons. Bacterial community composition was characterized with denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S ribosomal DNA. Average community doubling time was calculated from bacterial production ([14C]leucine incorporation) and bacterial abundance (direct counts). Freshwater and marine populations advected into the estuary represented a large fraction of the bacterioplankton community in all seasons. However, a unique estuarine community formed at intermediate salinities in summer and fall, when average doubling time was much shorter than water residence time, but not in spring, when doubling time was similar to residence time. Sequencing of DNA in DGGE bands demonstrated that most bands represented single phylotypes and that matching bands from different samples represented identical phylotypes. Most river and coastal ocean bacterioplankton were members of common freshwater and marine phylogenetic clusters within the phyla Proteobacteria, Bacteroidetes, and Actinobacteria. Estuarine bacterioplankton also belonged to these phyla but were related to clones and isolates from several different environments, including marine water columns, freshwater sediments, and soil.
Estuarine waters contain strong biological and chemical gradients established by the mixing of freshwater and seawater and modified by autochthonous biological activity. Many of these gradients, including salinity, nutrient concentration, organic matter composition, and bacteriovore community composition, are thought to influence the composition of natural bacterioplankton communities (2, 11). Such changes in environmental conditions, when recreated in mesocosm and microcosm experiments, caused shifts in the phylogenetic composition of bacterioplankton communities (10, 19, 32, 41). It is therefore reasonable to predict that similar shifts will occur in natural freshwater and marine bacterioplankton communities when they encounter estuarine gradients, leading to the development of an estuarine community.
Several studies have described estuarine microbial diversity and some have demonstrated how freshwater and marine bacterioplankton communities mix along estuarine gradients (3, 4, 8, 14, 21, 37), but few reports have provided evidence of a unique estuarine bacterioplankton community. This is partly due to the dynamic nature of estuaries and the difficulty in distinguishing estuarine populations from those that wash in from adjacent environments. Crump et al. (6) identified putative estuarine bacteria associated with particles in the Columbia River estuarine turbidity maximum (ETM) by comparing environmental clone libraries of PCR-amplified 16S ribosomal DNA (rDNA) from the river, the estuary, and the coastal ocean. Similarly, Hollibaugh et al. (14) demonstrated the mixing of bacterial communities in the ETM of the San Joaquin River and San Francisco Bay system by characterizing communities at three sampling stations using denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S rDNA. Selje and Simon (33) used this same technique, but with greater spatial resolution (six sampling stations), in the Weser River estuary and concluded that a distinct microbial community resides in the brackish section of the system. These three studies demonstrated the presence of river and coastal ocean bacteria in estuaries and suggested that the development of unique estuarine bacterial communities may be related to the relatively long residence time of particles and particle-attached bacteria in some ETMs.
The residence time of water and free-living bacteria, however, can be too short in some estuaries, relative to bacterial growth rate, for such a shift to occur. In the Rhone River plume, where water residence time is less than 6 h, bacterioplankton appeared to be a mixture of Rhone River and Mediterranean Sea bacterioplankton (37). Similarly, in the Columbia River estuary, where water residence time averages 1 to 2 days and bacterial production is low, a mixture of freshwater and marine populations dominated the free-living bacterioplankton community (6). Compositional shifts from these advected communities to a local community should require both bacterial growth and enough time for changes in the relative growth rate (and mortality) of different populations to produce a shift in diversity. Understanding this combination of top-down (residence time) and bottom-up (growth rate) controls on bacterioplankton should predict where and when community shifts will occur.
We hypothesized that estuarine bacterioplankton include populations originating in freshwater and marine environments and that the development of a unique estuarine bacterioplankton community depends on the growth rate of the bacterioplankton community and the residence time of that community in an estuary. We tested this hypothesis in Plum Island Sound, an estuary in northern Massachusetts that exhibits large seasonal variations in residence time (39) and bacterial growth rate (46). We found that freshwater and marine populations advected into the estuary represented a large fraction of the bacterioplankton community. We also identified a unique estuarine community in summer and fall, when bacterial production was high and water residence time was long, but we found that this community was absent in spring, when bacterial production was low and residence time was short.
MATERIALS AND METHODS
Sampling sites.
Plum Island Sound (Fig. 1), located on the Gulf of Maine in northeastern Massachusetts (42°41′N, 70°46′W) is part of the Plum Island Ecosystem Long Term Ecological Research site. The estuary is 25 km long with a mean depth of 2 m at low tide (15). Tide range averages 2.6 m, salinity ranges from 0 to 32 ppt, and temperature varies seasonally from −1.0 to 28°C (44). The upper half of the estuary is a tidal river flowing through extensive salt marshes, and the lower half is a broad, shallow sound. Three watersheds drain into the estuary: Parker (155 km2), Rowley (26 km2), and Ipswich (404 km2). The Parker River, which flows over a dam and into the head of the estuary, supplies most of the freshwater that is retained in the estuary and exerts the greatest control on salinity distribution and water residence time.
FIG. 1.
Map of Plum Island Sound with subsections indicated.
The Parker River estuary and Plum Island Sound were sampled on 12-13 July 2000 (summer), 28 September 2000 (fall), and 17 April 2001 (spring). The distribution of sampling locations started above the Parker River dam and extended along the estuary to the mouth. The mouth of the estuary was sampled at high slack tide in order to collect coastal ocean water. On each sampling date, nine subsurface samples were collected with clean plastic beakers or bottles. Salinity was measured with a refractometer or a conductivity meter.
Hydrodynamic conditions.
A one-dimensional, advection-dispersion hydrodynamic model developed for the Plum Island Sound (39) provided estimates of the distribution of salinity at mean high tide for each sampling date. Drivers for this model include freshwater runoff and coastal ocean salinity. Coastal salinities for model runs were taken from samples collected at the mouth of the estuary during high slack tide. Average residence times and ages for water in individual subsections of the estuary were estimated from prior model output (39).
Measurements.
Bacterial production was measured as the rate of incorporation of l-[3H]leucine (20 to 50 nM final concentration) into the cold trichloroacetic acid (TCA; 5% final concentration)-insoluble fraction of macromolecules in four unfiltered subsamples, including one killed control, incubated for 1 h at in situ temperatures in the dark. TCA-precipitated macromolecules were collected on 0.2-μm-pore-size nitrocellulose filters (Millipore) and washed twice with ice-cold TCA, combined with 1 ml of methyl-Cellusolve and 6 ml of Scintisafe scintillation cocktail, and counted in a Beckman scintillation counter. Prokaryotic cell abundance was estimated in formaldehyde-fixed samples (2% final concentration) with direct counts using 4′,6′-diamidino-2-phenylindole nucleic acid stain (13, 27).
Bacterial community doubling times were calculated from leucine incorporation rates and cell counts. Assessments of bacterial carbon production (BP) from leucine incorporation used a ratio of cellular carbon to protein of 0.86, a fraction of leucine in protein of 0.073, and an intracellular leucine isotope dilution of 2 (18). Bacterial biomass (BB) estimated from cell concentration assumed 25 fg of carbon per cell. The equation u = ln[(BB + BP)/BB] estimated cell-specific exponential growth, and doubling time (DT) was calculated by the equation DT = ln(2)/u (1).
Community composition. (i) Sample collection.
All samples were processed without prefiltering, but portions of several samples were also gently screened with a 1-μm Nytex net and processed in parallel. Sample processing and DNA extraction procedures followed those described by Crump et al. (7).
(ii) DGGE.
DGGE procedures followed those described by Muyzer et al. (22) and Crump et al. (7). DGGE-PCR amplification (1× PCR buffer [Promega], 8 μM deoxynucleoside triphosphates [dNTPs], 1 μM primers, 0.01 U of Taq polymerase [Promega]/μl) used the bacteria-specific, GC-clamp primer 357f(g+c) (5′-CGCCCGCCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCCCCCCTACGGGAGGCAGCAG-3′) and universal primer 519r (5′-ACCGCGGCTGCTGGCAC-3′) under the following PCR conditions: an initial 5 min at 94°C followed by 20 to 25 cycles of 1 min at 94°C, 1 min at 65 to 55°C (reducing temperature by 0.5°C per cycle for 20 cycles plus further cycles at 55°C), and 1 min at 72°C followed by 5 min at 72°C. Steps involving a temperature reduction were done at 0.3°C per s. The amount of template varied with the sample and was selected to optimize PCR amplification. In general the entire volume of each 50-μl reaction mixture was used to load the DGGE gel.
PCR products were separated into bands by electrophoresis for 16 to 18 h at 70 V on acrylamide (8%) gels prepared with 30% acrylamide-bis-acrylamide (37.5:1; Bio-Rad), 0.5× TAE buffer (1× TAE is 40 mM Tris [pH 8.0], 20 mM acetic acid, 1 mM EDTA), and gradients of 30 to 50% denaturants (urea and formamide).
Magnified sections of DGGE gels (six per gel) were photographed with a ChemImager 4000 imaging system (Alpha Innotech), and complete images of each gel were reconstructed with Photoshop software (Adobe). Bands, defined as those having an intensity of at least 5% of the most intense band in the sample, were scored as present or absent at each position in the gel using the GelcomparII software package (Applied Maths). Comparison of banding profiles for different samples identified matching bands. A pairwise distance matrix (Dice), calculated from this binary data set, was analyzed using the multidimensional scaling (MDS) module of the Statistica software package (StatSoft). The graphical representation of these analyses plots the DGGE banding patterns from each sample such that samples containing many of the same bands are in close proximity to each other.
(iii) DGGE band identification.
Procedures for sequencing DNA from DGGE bands were modified from those reported by Crump et al. (7). Four samples from each season were amplified with DGGE primers and run on a DGGE gel as described above. After photographing the gel, eight bands per sample were selected for identification. DNA fragments from DGGE bands were sampled with sterile pipette tips and PCR amplified as described above. PCR products were then run on DGGE gels along with the original natural samples in order to identify the appropriate bands from the reamplifications. These bands were sampled again and PCR amplified with non-GC-clamp primers (1× PCR buffer [Promega]), 8 μM dNTPs, 1 μM primers, 0.01 U of Taq polymerase [Promega]/μl) (G. Muyzer, personal communication). PCR products were cloned with a TOPO-TA cloning kit (Invitrogen) following the manufacturer's instructions. The inserts from four clones per band were amplified with DGGE primers and run on DGGE gels along with the natural samples. Clones containing potential matches to bands in the original sample were run on DGGE gels a second time in lanes adjacent to the natural samples in order to confirm the band position match. Clones that exactly matched bands from natural samples were sequenced. In addition, some clones that did not match the original bands in natural samples, but rather aligned with bands nearby, were also sequenced.
(iv) DNA sequencing.
DNA sequences of DGGE band clones were determined with an ABI 3730 automated sequencer following the manufacturer's instructions. DGGE band sequences were determined for both complementary strands using plasmid-specific primers.
Phylogenetic analyses were accomplished with the PAUP version 4.0b10 for Macintosh program (D. L. Swofford, Sinauer Associates, Sunderland, Mass.). Substitution models were chosen using likelihood ratio tests calculated with the program Modeltest version 3.06 (28). Distance matrices were estimated using these models under maximum likelihood criteria. Minimum evolution trees were determined using three iterations of tree and parameter estimations with tree bisection-reconnection branch swapping and the last iteration including 100 random-addition replicates. Bootstrapping with both distance and parsimony estimations used the same models and parameters as heuristic searches on 100 replicates, with three random-addition replicates per bootstrap replicate, under minimum evolution criteria. Bootstrap analysis was limited to 106 rearrangements for the Bacteroidetes phylum and the plastids. DNA sequences used for the phylogenetic analysis covered helices 16, 17, and 18 of the rDNA gene and ranged from 133 to 157 bp. This portion of the gene includes one hypervariable region, helix 18 (40), that varied greatly in both sequence and length.
RESULTS
Residence time.
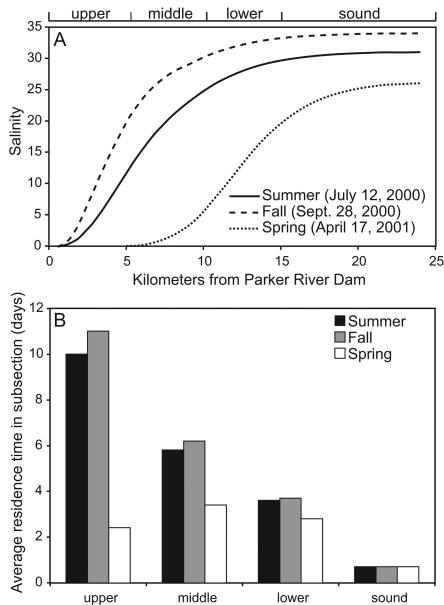
The salinity profile of the Parker River estuary and Plum Island Sound is strongly influenced by river discharge. In summer and fall the Parker River discharge was relatively low, and the majority of the salinity gradient was located in the upper and middle estuary segments (Fig. 2A). In spring, however, the Parker River discharge was 10 times greater, and the majority of the salinity gradient was displaced downstream to the lower estuary and the sound.
FIG. 2.
Salinity profile along main stem of estuary at mean high tide (A) and average residence time of water in each subsection (B) on summer, fall, and spring sampling dates.
Average residence times of water in subsections of the estuary during summer and fall were longest in the upper estuary, ranging from 10 to 11 days (Fig. 2B). During spring, residence times were highest in the middle and lower estuary but were much shorter, averaging only 3 days.
To estimate the time marine and freshwater bacterial communities resided in the estuary, the average ages of freshwater and seawater were estimated for subsections of the estuary containing the majority of the salinity gradient. In summer and fall, bacterial communities from the river and the coastal ocean were in the estuary for an average of 13 to 24 days (Table 1). In spring these ages averaged 5 to 9 days. Note that freshwater downstream of these estuarine segments and seawater upstream of these segments had longer ages but constituted much smaller fractions of the total volume of water.
TABLE 1.
Salinity range and transport time for bacteria in subsections of the estuary containing most of the salinity gradient
| Season | Subsection(s) containing salinity gradient | Salinity (ppt) | Residence time (days)
|
Age (days)
|
||
|---|---|---|---|---|---|---|
| Subsection | Estuary | Freshwater | Seawater | |||
| Summera | Upper and middle | 0-30 | 7 | 18 | 20 | 18 |
| Falla | Upper and middle | 0-25 | 8 | 17 | 18 | 17 |
| Spring | Lower | 5-20 | 3 | 5 | 9 | 5 |
Transport times for summer and fall are volume-weighted averages of times for the upper and middle estuary.
Bacterial production, cell concentration, and community doubling time.
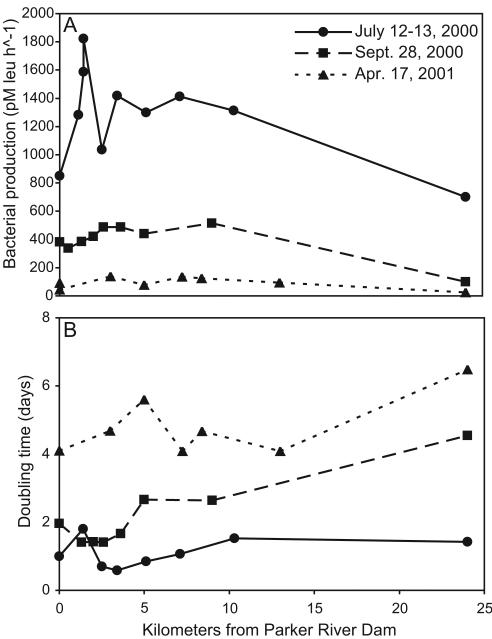
Bacterial production was always higher in the estuary than in the river and coastal ocean and was constant along the salinity gradient, except in the summer in the upper estuary, where high bacterial production was associated with the seasonal phytoplankton bloom (Fig. 3A). Production was highest in summer, lowest in spring, and intermediate in fall. Community doubling time at intermediate salinities was between 1 and 3 days in summer and fall and greater than 4 days in spring (Fig. 3B).
FIG. 3.
Bacterial production rate (A) and average community doubling time (B) along main stem of estuary on spring, summer, and fall sampling dates.
DGGE.
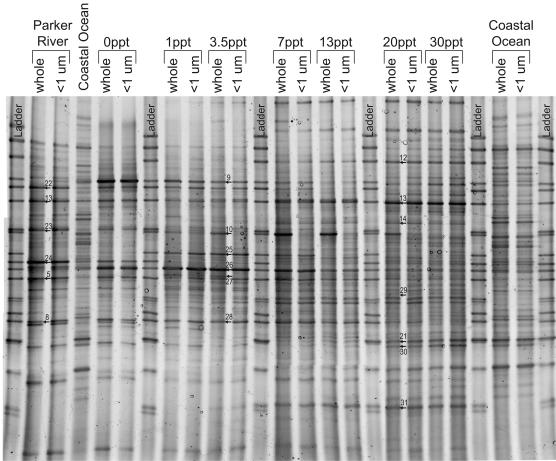
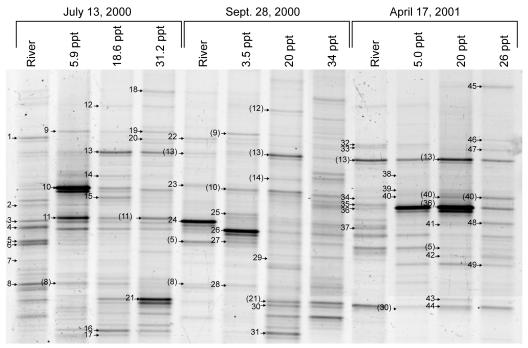
In each season, DGGE banding patterns of samples collected from the salinity gradient shared many of the same bands as those from just up or down estuary, but patterns from river and coastal ocean samples shared very few bands (Fig. 4). Among seasons, DGGE banding patterns in summer and fall had many more bands per sample and nearly twice the total number of band positions (all samples combined) than DGGE patterns in spring (Table 2). Close inspection of these patterns revealed that individual bands originated from the river, the estuary, or the coastal ocean (Table 2). Most of the bands from the river and coastal ocean also appeared in the banding patterns of samples from the estuary, but they became gradually dimmer relative to other bands in banding patterns of samples collected farther along the salinity gradient. This gave the impression that these phylotypes were gradually diluted by other bacterial phylotypes along the estuarine mixing gradient. For this to be true, PCR must have provided consistent amplification of each phylotype in proportion to the total number of target sequences in each sample. Bands of estuarine origin were absent from the river and the coastal ocean, or they appeared only faintly in the coastal ocean while appearing much darker in the estuary.
FIG. 4.
DGGE gel of PCR-amplified 16S rDNA genes from samples collected along the salinity gradient on 28 September 2000. Bands from which DNA was sequenced are marked and numbered, corresponding to band numbers in Table 3.
TABLE 2.
Seasonal comparisons of DGGE bands
| Season | No. of DGGE band positions
|
% Estuarine | Avg no. of bands per sample | No. of bands that differed in unfiltered vs filtered | ||||
|---|---|---|---|---|---|---|---|---|
| River | Estuary | Coastal ocean | Othera | Total | ||||
| Summer | 41 | 29 | 35 | 4 | 109 | 27 | 42 | 1-4 |
| Fall | 43 | 46 | 31 | 8 | 128 | 36 | 48 | 0-9 |
| Spring | 24 | 6 | 23 | 3 | 56 | 11 | 28 | 0-6 |
No clear origin.
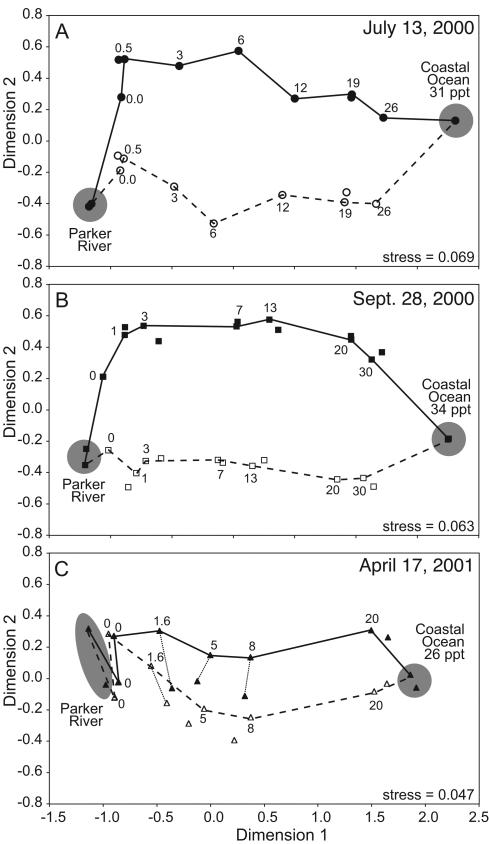
MDS analyses of pairwise similarities between DGGE banding patterns revealed shifts in bacterial community composition along the salinity gradient between the freshwater and marine environments (Fig. 5). In summer and fall the largest shift occurred at the head of the estuary between the river and about 1-ppt salinity. No such shift appeared in the spring, when changes in bacterial community composition occurred more gradually and tended to track salinity.
FIG. 5.
Multidimensional scaling diagrams (with stress values) of Dice distance matrices calculated from DGGE banding patterns of samples collected in spring (A), summer (B), and fall (C). Solid symbols represent real DGGE banding patterns. Open symbols represent artificial banding patterns from which estuarine bands were removed. Symbols representing unfiltered water samples are connected with lines according to the salinity gradient. Symbols representing 1-μm- screened samples are either the closest symbols to the unfiltered samples or are connected to the unfiltered samples with a line. Salinity of samples is indicated.
Artificial banding patterns were created by altering the presence-absence data set so that bands categorized as being of estuarine origin (Table 2) were removed. These artificial banding patterns were then included in MDS analyses to show how the estuarine bacterial community influenced our picture of community structure along the salinity gradient. Differences between the original patterns and these artificial patterns of community composition along the salinity gradient were due to the estuarine bacterial community (Fig. 5). In summer and fall, the estuarine community contributed greatly to compositional shifts along the salinity gradient and was responsible for the rapid shift in community composition at low salinities. In spring, the estuarine community represented a very small fraction of the total community in the estuary and therefore did not greatly influence our picture of community composition.
DGGE banding patterns of prefiltered samples appeared very similar to those of unfiltered samples in both number and density of bands, but there were some differences (Table 2). In summer and fall these differences were small relative to the total number of bands in each sample and relative to the differences between samples (Fig. 5A and B). In spring these differences were more significant (Fig. 5C) because they represented a larger fraction of the total number of bands in the samples (Table 2).
DGGE band sequencing.
The DNA fragments contained in DGGE bands were sequenced from the river, estuary, and coastal ocean in all three seasons (Fig. 6). Most DGGE bands identified through DNA sequencing (41 out of 49) represented one unique DNA sequence (Table 3). However, four bands contained pairs of DNA fragments that differed by 1 bp (bands 12, 24, 44, and 47), and one band contained three DNA fragments that differed by 1 or 2 bp (band 11). Three other bands contained pairs of DNA fragments that were very different from each other (bands 5, 13, and 48). Several pairs of sequences with only 1-bp difference formed DGGE bands at different positions in the gel (5a and 7, 12a and 20, 26 and 27, and 22 and 32). We found no consistent pattern to distinguish these nearly identical sequences with disparate mobilities from nearly identical sequences with identical mobilities (e.g., transition versus transversion, location of differences on DNA strands). We conclude that DGGE does not always provide single base pair resolution of this segment of the 16S rDNA gene.
FIG. 6.
DGGE gel of select samples from three sampling series indicating bands from which DNA was sequenced. Parentheses indicate bands containing DNA with sequence identical to a previously sequenced band. Numbers correspond to the band numbers in Table 3.
TABLE 3.
DGGE band DNA sequences
| Band no. | Accession no. | Origin | Date of sequence collection(s) | Phylum or class | Category | Closest match | Source | Accession no. | % Simi- larity |
|---|---|---|---|---|---|---|---|---|---|
| 21 | AY308684 | Marine | July, Sept. | Alpha | SAR11a | C. P. ubiqueb | Coastal Pacific Ocean | AF510192 | 100 |
| 30 | AY308694 | Marine | Sept., Apr. | Alpha | Roseobactera | NAC11-3 | Atlantic Ocean | AF245632 | 100 |
| 42 | AY308706 | Marine | Apr. | Alpha | Roseobacter | Shippagan | Larval haddock culture | AF100168 | 100 |
| 49 | AY308716 | Marine | Apr. | Alpha | Roseobacter | Isolate ARK10207 | Arctic sea ice/water | AF468373 | 100 |
| 44a | AY308708 | Marine | Apr. | Alpha | Roseobacter | ZD0207 | North Sea | AJ400341 | 100 |
| 44b | AY308709 | Marine | Apr. | Alpha | Roseobacter | ZD0207 | North Sea | AJ400341 | 99 |
| 18 | AY308681 | Marine | July | Gamma | SAR86a | EBAC31A08 | Coastal Pacific Ocean | AF279106 | 99 |
| 19 | AY308682 | Marine | July | Bacteroidetesc | Unknown | Isolate HOS19 | Halichondria panicea | Z88577 | 100 |
| 20 | AY308683 | Marine | July | Bacteroidetes | Unknown | FL-16 | Coastal Pacific Ocean | AY028182 | 100 |
| 45 | AY308710 | Marine | Apr. | Bacteroidetes | Unknown | SS1 | Japan Sea | AB035018 | 97 |
| 46 | AY308711 | Marine | Apr. | Bacteroidetes | Unknown | CRE-FL57 | Columbia River estuary | AF141474 | 100 |
| 47a | AY308712 | Marine | Apr. | Bacteroidetes | Flavobacteriales | GOBB3-CL102 | Baltic Sea | AF388899 | 100 |
| 47b | AY308713 | Marine | Apr. | Bacteroidetes | Flavobacteriales | GOBB3-CL102 | Baltic Sea | AF388899 | 99 |
| 48a | AY308714 | Marine | Apr. | Bacteroidetes | Unknown | OTU_A | North Sea | AF207850 | 100 |
| 48b | AY308715 | Marine | Apr. | Bacteroidetes | Unknown | MoDE-8 | Coastal Pacific Ocean | AF419358 | 100 |
| 38 | AY308702 | Marine | Apr. | Plastid | Coscinodiscophyceae | CRE-PA60 | Columbia River estuary | AF141534 | 99 |
| 40 | AY308704 | Marine | Apr. | Plastid | Haptophyceae | Plastid | Ochrosphaera neapolitan | X80390 | 100 |
| 17 | AY308680 | Estuarine | July | Actinobacteria | Unknown | CL-S77-289 | Salt marsh | AF547402 | 98 |
| 28 | AY308692 | Estuarine | Sept. | Alpha | Roseobacter | R. gallaeciensisd | Marine | Y13244 | 98 |
| 29 | AY308693 | Estuarine | Sept. | Alpha | Rickettsiales | CRO-FL16 | Coastal Pacific Ocean | AF141594 | 99 |
| 25 | AY308689 | Estuarine | Sept. | Beta | Unknown | Mul01-10 | Freshwater sediments | AJ518107 | 100 |
| 16 | AY308679 | Estuarine | July | Gamma | Unknown | Isolate ODIII6 | Marine hydrothermal vent | AF170422 | 96 |
| 31 | AY308695 | Estuarine | Sept. | Gamma | Unknown | Isolate ODIII6 | Marine hydrothermal vent | AF170422 | 99 |
| 43 | AY308707 | Estuarine | Apr. | Gamma | Unknown | OM182 | Coastal Atlantic Ocean | U70699 | 96 |
| 14 | AY308677 | Estuarine | July, Sept. | Bacteroidetes | Unknown | OM271 | Coastal Atlantic Ocean | U70708 | 100 |
| 12a | AY308673 | Estuarine | July, Sept. | Bacteroidetes | Unknown | FL-16 | Coastal Pacific Ocean | AY028182 | 99 |
| 12b | AY308674 | Estuarine | July, Sept. | Bacteroidetes | Unknown | FL-16 | Coastal Pacific Ocean | AY028182 | 98 |
| 11a | AY308670 | Estuarine | July | Cyanobacteria | Synechococcus | NAC1-5 | Atlantic Ocean | AF245618 | 100 |
| 11b | AY308671 | Estuarine | July | Cyanobacteria | Synechococcus | NAC1-5 | Atlantic Ocean | AF245618 | 99 |
| 11c | AY308672 | Estuarine | July | Cyanobacteria | Synechococcus | NAC1-5 | Atlantic Ocean | AF245618 | 99 |
| 9 | AY308668 | Estuarine | July, Sept. | Plastid | Unknown | Plastid | Chilomonas paramecium | AF545624 | 96 |
| 10 | AY308669 | Estuarine | July, Sept. | Plastid | Coscinodiscophyceae | CRE-PA60 | Columbia River estuary | AF141534 | 99 |
| 15 | AY308678 | Estuarine | July | Plastid | Prasinophyceae | Plastid EBAC38 | Coastal Pacific Ocean | AF268228 | 99 |
| 26 | AY308690 | Estuarine | Sept. | Plastid | Coscinodiscophyceae | Plastid MoDE-3 | Coastal Pacific Ocean | AF419353 | 99 |
| 27 | AY308691 | Estuarine | Sept. | Plastid | Coscinodiscophyceae | Plastid MoDE-3 | Coastal Pacific Ocean | AF419353 | 98 |
| 36 | AY308700 | Estuarine | Apr. | Plastid | Coscinodiscophyceae | HstpL29 | H. stipulacea leavese | AF159634 | 100 |
| 8 | AY308667 | Freshwater | July, Sept. | Actinobacteria | ACK-MIf | TLM6 | Toolik Lake | AF534430 | 100 |
| 1 | AY308659 | Freshwater | July | Beta | Isolate BAL47f | TLM4 | Toolik Lake | AF534428 | 96 |
| 22 | AY308685 | Freshwater | Sept. | Beta | Isolate BAL47 | TLM4 | Toolik Lake | AF534428 | 99 |
| 32 | AY308696 | Freshwater | Apr. | Beta | Isolate BAL47 | TLM4 | Toolik Lake | AF534428 | 100 |
| 5b | AY308664 | Freshwater | Apr. | Beta | Unknown | OM180 | Coastal Atlantic Ocean | U70707 | 100 |
| 3 | AY308661 | Freshwater | July | Delta | Bdellovibrio | HC-20 | Arsenite-oxidizing biofilm | AY168736 | 99 |
| 4 | AY308662 | Freshwater | July | Bacteroidetes | Unknown | CL0-55 | Crater Lake | AF316796 | 94 |
| 23 | AY308686 | Freshwater | Sept. | Bacteroidetes | Unknown | Clone M09 | Wasterwater sludge | AF495416 | 97 |
| 33 | AY308697 | Freshwater | Apr. | Bacteroidetes | Flavobacteriales | F. gelidilacusg | Ace Lake microbial mat | AJ440996 | 98 |
| 34 | AY308698 | Freshwater | Apr. | Bacteroidetes | Unknown | WL5-12 | Freshwater Weser estuary | AF497890 | 95 |
| 35 | AY308699 | Freshwater | Apr. | Bacteroidetes | Flavobacteriales | DGGE band7-3 | ChangJiang River | AY071888 | 99 |
| 37 | AY308701 | Freshwater | Apr. | Bacteroidetes | Flavobacteriales | DGGE band7-3 | ChangJiang River | AY071888 | 98 |
| 39 | AY308703 | Freshwater | Apr. | Bacteroidetes | Unknown | AEGEAN_179 | North Aegean Sea | AF406541 | 98 |
| 41 | AY308705 | Freshwater | Apr. | Bacteroidetes | Unknnown | COL-28 | Marine mesocosm | AY028189 | 93 |
| 5a | AY308663 | Freshwater | July, Sept., Apr. | Bacteroidetes | PRD01a001Bf | CRE-FL39 | Columbia River estuary | AF141460 | 100 |
| 7 | AY308666 | Freshwater | July | Bacteroidetes | PRD01a001B | CRE-FL39 | Columbia River estuary | AF141460 | 99 |
| 2 | AY308660 | Freshwater | July | Plastid | Cryptophyta | LCK-26 | Lake Cadagno | AF107323 | 100 |
| 6 | AY308665 | Freshwater | July | Plastid | Chrysophyceae | TLMdgge17 | Toolik Lake | AF534453 | 99 |
| 24a | AY308687 | Freshwater | Sept. | Plastid | Cryptophyta | Plastid | Hemiselmis virescens | AB073112 | 100 |
| 24b | AY308688 | Freshwater | Sept. | Plastid | Cryptophyta | Plastid | Hemiselmis virescens | AB073112 | 99 |
| 13a | AY308675 | Unknown | Sept., Apr. | Beta | Isolate BAL47 | CRE-FL19 | Columbia River estuary | AF141446 | 100 |
| 13b | AY308676 | Unknown | July, Sept., Apr. | Bacteroidetes | Flavobacteriales | OTU_C | North Sea | AF207852 | 100 |
In several cases the same DGGE band was identified in more than one sample. For example, the DNA contained in band 8, which came from a freshwater Actinobacteria, was sequenced from the summer and fall river samples and also from a summer estuarine sample (Fig. 6). The DNA contained in band 21, which came from a marine α-Proteobacteria, was sequenced from a summer coastal ocean sample and from a fall estuarine sample.
Phylotypes identified through sequencing of DNA from DGGE bands were related to organisms within the α-, β-, γ-, and δ-Proteobacteria, Actinobacteria, cyanobacteria, and Bacteroidetes (otherwise known as Cytophaga-Flavobacteria-Bacteroides) phyla (Table 3). Several bands contained DNA sequences matching chloroplast 16S rDNA genes from several clades of phytoplankton, including centric diatoms, cryptophytes, chrysophytes, and chlorophytes.
Phylotypes originating from the coastal ocean were, in most cases, 100% identical in DNA sequence to environmental clones and isolates from other coastal ocean environments (Table 3; Fig. 7). Similarly, most phylotypes originating from the Parker River were nearly identical to environmental clones and isolates from other freshwater systems. Estuarine phylotypes and their closest relatives, however, could not be classified by ecosystem type in the same way. Those related to the α-Proteobacteria, Bacteroidetes, and cyanobacteria phyla were most closely related to clones and isolates from coastal ocean environments, but the phylotypes related to β-Proteobacteria were identical to environmental clones from groundwater, freshwater sediments, and soils. Furthermore, two estuarine γ-Proteobacteria phylotypes were most closely related to a sulfur-oxidizing isolate from a shallow marine hydrothermal vent, and the estuarine Actinobacteria phylotype, though somewhat related to globally distributed freshwater cluster Urk0-14 (47), was most closely related to an environmental clone collected from a salt marsh.
FIG. 7.
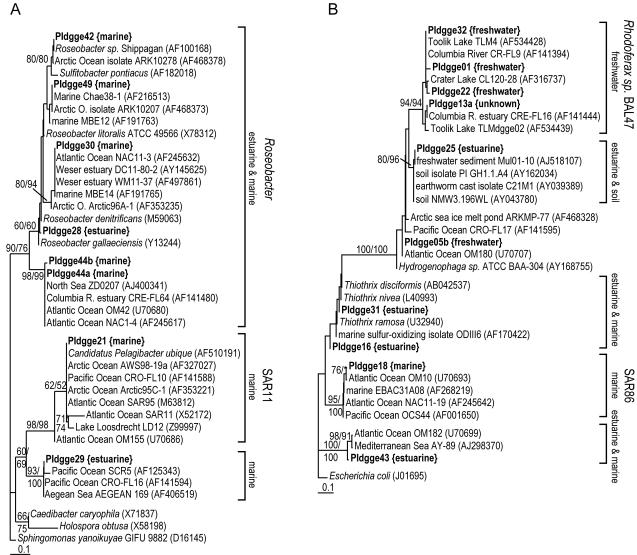
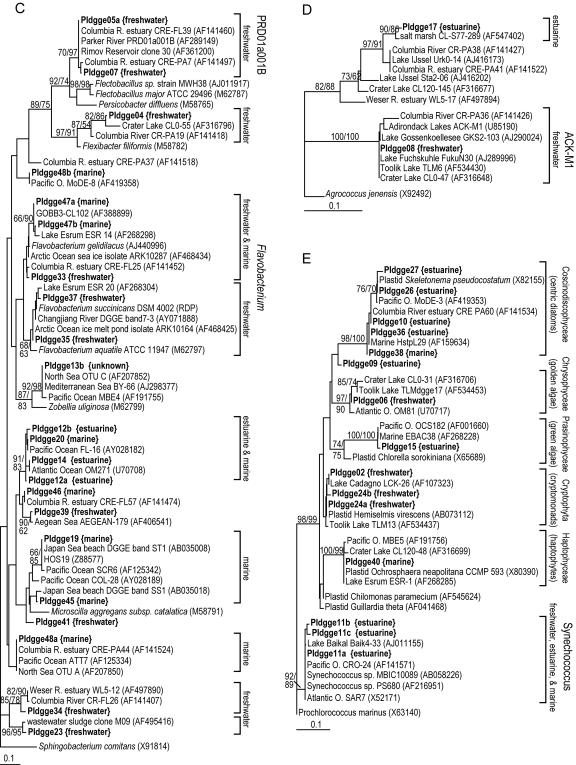
Minimum evolution trees showing phylogenetic positions of organisms within α-Proteobacteria (A), β- and γ-Proteobacteria (B), Bacteroidetes (C), Actinobacteria (D), and cyanobacteria and chloroplasts (E). Sequences from this study are in boldface type, with origin indicated. Labels on square brackets indicate the phylogenetic cluster (11, 47) and environmental sources of clones and isolates. Each set of identical sequences (100% similarity) was treated as a single sequence during analyses. Bootstrap values for distance and parsimony estimations are shown when both values are greater than 50. Analyses were based on alignments of 133 to 157 bp, including the hypervariable region in helix 18 (40).
Eleven of the 49 sequenced bands represented chloroplasts from several clades of phytoplankton, indicating that chloroplasts influenced our picture of bacterioplankton community composition (Fig. 7E). We expected bands representing chloroplasts to be absent or not as dense in DGGE patterns from 1-μm-filtered water samples when compared to unfiltered water, but in all but one case (band 36 [Table 1]) they were not. Most of these phytoplankton cells were too large to pass through a 1-μm Nytex net, so it is likely that the chloroplasts of these organisms were released from the cells and subsequently passed through the screen.
DISCUSSION
A native estuarine bacterioplankton community exists at intermediate salinity in the Parker River estuary and Plum Island Sound that is phylogenetically distinct from allochthonous communities advected into the estuary by tidal action and river flow. This estuarine community occurs only in the summer and fall, when the average doubling time of bacteria is much shorter than the residence time of water in the estuary. In spring, bacterial doubling time was about the same as residence time, and consequently no estuarine community developed. Allochthonous communities from the river and the coastal ocean were phylogenetically distinct and were composed of typical freshwater and marine phylotypes. These communities were always present in the estuary, but because they are continuously advected into the system, we cannot determine whether they remain active at intermediate salinities and grow alongside native estuarine communities.
Bacteria growth rate and residence time.
Residence time is a fundamental concept for understanding the hydrodynamic structure of aquatic systems and is perhaps the most important physical control on ecological processes in estuaries (17). This is especially true for plankton, which are always subject to the movement of water through an estuary. For example, residence time can control the location and magnitude of estuarine phytoplankton blooms (9, 45) and can influence the distribution of bacteria (25). Monsen et al. (20) described three such transport time scales commonly used by ecologists to relate hydrodynamics to biological and chemical processes: residence time, age, and flushing time. Two of these measures are particularly useful when thinking about bacterioplankton communities in estuaries. Residence time, or the average time it takes for a parcel of water in a section of an estuary to leave that section, is most applicable when discussing the time available for the development of native bacterial populations. The complement of residence time, age, or the average time a parcel of water in a section of an estuary has been in that section, is also a useful concept, because it describes the time that allochthonous bacterial communities have been exposed to estuarine conditions. Since these two values are calculated for discrete parcels of water, they can also be applied to bacteria and other neutrally buoyant particles contained in those parcels.
Transport time scales such as these are often calculated for whole estuaries and can vary over a huge range (e.g., residence times of 1 to 2 days in the Columbia River estuary [23], 4 to 8 weeks in the York River estuary [35], and 7.6 months in the Chesapeake Bay [24]). We found whole-estuary estimates for the combined Parker River estuary and Plum Island Sound to be deceptively short because of the very short residence time of water in the marine “sound” subsection of the system (Fig. 1) (see reference 39). In reality, transport time varies along the length of the estuary, being influenced by river discharge at the freshwater end and tidal mixing at the marine end. Because we were interested in what happens at intermediate salinities, we focused our estimates of transport time on the estuarine subsections that contained the majority of the salinity gradient.
In summer and fall, most of the salinity gradient was located in the upper and middle estuary, where allochthonous freshwater and marine bacteria have an average age of 17 to 20 days. Residence time of bacteria averaged 7 to 8 days within these subsections and 17 to 18 days in the estuary as a whole. These residence times are much longer than the average doubling time of the bacterial community in summer (average, 1.1 days) and fall (average, 1.9 days). DGGE analyses for these seasons identified unique estuarine bacterial communities, indicating that residence time was long enough to allow new bacterial populations to become adequately abundant for detection. In contrast, during spring most of the salinity gradient centered in the lower estuary, where allochthonous bacteria had much shorter average ages in the estuary (5 to 9 days). Residence time of bacteria in this subsection was also much shorter, averaging 3 days within the subsection and 5 days in the estuary as a whole. These residence times were approximately the same as the average doubling time of the bacterioplankton community (4.6 days) and, based on results from DGGE analyses, were not long enough for new bacterioplankton populations to develop.
Just as residence time describes the time available for new bacterial populations to develop in the estuary, age describes the time that freshwater and marine bacteria are exposed to what might be less than ideal growing conditions. In summer and fall, the average age of allochthonous freshwater and marine bacteria was 17 to 20 days. Our DGGE patterns clearly showed that these organisms are present at intermediate salinities, confirming the results of several other studies (6, 14, 21), but the development of unique estuarine bacterial populations calls into question the growth and survival of freshwater and marine populations. Only a few studies have addressed this question for natural bacterial communities, and all have focused on the survival of freshwater bacteria. Three studies of cultivatable bacteria suggested that most freshwater bacteria cannot survive increased concentrations of salt (16, 29, 38). Similar results were found when freshwater bacteria were captured in diffusion chambers and incubated at marine salinities (38, 43). However, one study exposed freshwater bacterioplankton to a range of estuarine salinities in the St. Lawrence estuary and found that they grew equally well when exposed to 5-ppt salinity but grew at reduced rates at 10 ppt and above (26). We detected a change in the composition of bacterioplankton communities at salinities as low as 1 ppt in summer and fall, but this change did not represent a wholesale replacement of the river populations. In fact, most DGGE bands at salinities below 5 represented river populations, and a large fraction of the river community remained detectable even at very high salinity in the estuary. It is likely that freshwater bacteria remain active in the estuary, but at a reduced rate due to a combination of increased salinity and other environmental conditions. We do not yet know how natural marine bacterioplankton respond to reduced salinity.
The connections we draw between bacterial doubling time and shifts in bacterial community composition assume an overlap between the bacterial community that assimilates leucine and the community detected with DGGE. Leucine is assimilated by most known heterotrophic bacteria and is commonly used to estimate the growth of natural bacterioplankton communities. Most of the organisms we detected by DGGE were typical bacterioplankton closely related to organisms found in many other systems, with the exception of a small number of sequences from chloroplast genes. One recent study labeled estuarine bacterioplankton with [3H]leucine and, using a combination of microautoradiography and rRNA probe hybridization, demonstrated that leucine was assimilated by all major phylogenetic groups, including those of organisms identified in our study (5). However, it is important to recognize one feature of our techniques. We only measured the leucine assimilated by bacteria that were actively producing new protein (i.e., growing), whereas our measurements of cell abundance and DGGE-based community composition did not differentiate between growing and nongrowing bacteria. This was not a problem for our community composition measurements, because we wanted to capture both active and inactive populations. However, our calculations of doubling time were averaged over all organisms, whether they were growing or not, and were therefore probably overestimates of the doubling time of the actively growing fraction of the community.
The above discussion focuses on growth as the driving force behind changes in community composition, but grazing and other forms of mortality are equally important. In planktonic systems, the grazing and mortality rate is thought to be nearly equivalent to the growth rate, thus accounting for the fairly constant abundance of bacterioplankton cells (36). Also, grazing has been shown to influence bacterial community composition in laboratory and field experiments (34, 42). The abundance of bacterial cells in our samples was fairly constant, suggesting that new cell production was balanced by grazing and mortality and indicating that shifts in community composition are most likely the result of both variations in growth rate and grazing and mortality of different bacterial populations
Bacterioplankton community composition.
Results from the comparison of bacterial community composition between the Parker River and the coastal Gulf of Maine agree with those of several other studies indicating that freshwater and marine bacterioplankton communities are phylogenetically distinct (6, 12, 31, 47). DNA sequences from DGGE bands showed that coastal communities were composed of typical marine populations, most of which were 99 to 100% identical in DNA sequence to clones or isolates from other coastal marine systems, including the North Sea, the Japan Sea, and the Columbia River estuary (Table 2). Proteobacteria phylotypes included members of the common marine phylogenetic clades SAR11 (α), Roseobacter (α), and SAR86 (γ) (11), and two of these phylotypes were 100% identical to the recently cultivated SAR11 organism Pelagibacter ubique (30) and the Roseobacter isolate ARK10207 from Arctic Sea ice (Table 2). Several of the Bacteroidetes phylotypes also belonged to phylogenetic clusters composed entirely of marine clones and isolates (clusters including PIdgge48b, PIdgge19 and PIdgge45, and PIdgge48a), but others belonged to clusters containing clones from different environments (Fig. 7C). The Flavobacterium cluster around marine phylotypes PIdgge47a and PIdgge47b included a clone from freshwater Lake Esrum and freshwater clone PIdgge33. In addition, the cluster around PIdgge20 included several estuarine-specific clones from this study. Although most marine phylotypes are related to other marine organisms, several of the Bacteroidetes belong to phylogenetic clades with no clear environmental origin.
As with the coastal communities, the Parker River communities included typical freshwater populations, most of which were closely related in DNA sequence to clones and isolates from other freshwater systems, including Toolik Lake in Alaska, Crater Lake in Oregon, and the ChangJiang (Yangtze) River in China (Table 2). Several phylotypes were members of the common freshwater phylogenetic clades ACK-M1 (Actinobacteria), Rhodoferax sp. strain BAL47 (β-Proteobacteria), and PRD01a001B (Bacteroidetes) (47). Four Bacteroidetes phylotypes were also part of four freshwater-specific phylogenetic clusters (PIdgge04, PIdgge37, PIdgge34, and PIdgge23) (Fig. 7C). However, two Bacteroidetes phylotypes and one β-Proteobacteria phylotype were most closely related to clones and isolates from marine systems. PIdgge35 was 100% identical to the 16S rDNA sequence of an isolate from an Arctic Ocean sea ice melt pond, but these melt ponds are typically very low in salinity and thus may be characterized as freshwater environments. The other two phylotypes were related to clones from coastal marine environments: PIdgge39 is very similar to a clone from the Aegean Sea (Fig. 7C), and PIdgge05b is identical to a clone from the coastal Atlantic Ocean (Fig. 7B). These clones from the Aegean Sea and the Atlantic Ocean may be part of phylogenetic clades with no clear environmental origin, or they may simply be freshwater organisms that washed in from land and were detected in PCR-amplified clone libraries.
Unlike the river and coastal ocean communities, the estuarine community was a mixture of populations related in DNA sequence to clones and isolates from many different environments. Many of these estuarine phylotypes belong to marine bacterioplankton clades, suggesting that they are marine populations capable of adapting to estuarine conditions, including reduced salinity. Among the Proteobacteria, PIdgge28 appears to be a species of Roseobacter (Fig. 7A) and PIdgge29 and PIdgge43 are closely related to several coastal ocean clones (Fig. 7A and B). Also, three estuarine Bacteroidetes phylotypes (PIdgge12a, PIdgge12b, and PIdgge14) fall into a cluster of marine clones that includes the marine phylotype PIdgge20 from this study (Fig. 7C). In contrast, PIdgge25 is identical to a clone from freshwater sediment and is part of a cluster of clones and isolates from sediments and soils (Fig. 7B). Also, three estuarine phylotypes may have their origins in the salt marsh that flanks the estuary. PIdgge17 is very similar to a salt marsh clone from the Georgia coast of the southeastern United States (Fig. 7D), and PIdgge16 and PIdgge31 are related to a sulfur-oxidizing isolate from a shallow hydrothermal vent (Fig. 7B) and to several species of Thiothrix, a group of chemolithotrophic sulfide oxidizers. Three other phylotypes, PIdgge11a, PIdgge11b, and PIdgge11c, are clearly Synechococcus species (Fig. 7E), but this genus cannot be separated into freshwater and marine clades using 16S rDNA gene sequences. Absent from this collection of estuarine phylotypes are any organisms that are clearly related to freshwater bacterioplankton. Thus, estuarine bacterioplankton communities appear to be composed of marine bacterioplankton that are tolerant of reduced salinity and sediment and soil bacteria that can also exist free living in an estuarine water column.
PCR primers targeting 16S rDNA from bacteria also amplify those genes from chloroplasts. Chloroplast sequences from DGGE bands show the diversity of the eukaryotic phytoplankton community in Plum Island Sound and can be categorized into marine, estuarine, and freshwater populations, much like the bacterioplankton (Fig. 7E). Chrysophytes and cryptomonads appear to wash into the estuary from the river, while haptophytes wash in from the coastal ocean. Estuarine phylotypes include centric diatoms, green algae, and one unknown phylotype. Most of these plastid sequences are 99 to 100% similar to plastids of cultivated phytoplankton or clones from other planktonic systems (Table 2).
Conclusions.
Physical and chemical properties of estuarine water columns can be very different than adjacent freshwater and marine environments. Strong gradients in salinity, temperature, nutrient concentration, and organic matter composition can result from the mixing of freshwater and seawater. Moreover, the concentration and composition of organic and inorganic nutrients can be greatly altered by autochthonous production and consumption. Our study shows how bacterioplankton community composition changes along estuarine gradients and demonstrates that shifts from a mixture of allochthonous communities to a native estuarine community require adequate bacterial growth rates and a relatively long residence time. However, we do not yet know the chemical and biological conditions that drive microbial community shifts, because many of them covary. Identifying these drivers will require higher-resolution quantitative studies of bacterial communities and a better understanding of the natural history and optimal growth conditions of individual bacterial populations.
Acknowledgments
This work was supported by two grants from the National Science Foundation (LTER grant OCE-9726921 and Microbial Observatory grant MCB-9977897) and the NASA Astrobiology Institute (cooperative agreement NCC2-1054 to M.L.S.).
We thank J. J. Vallino for calculating salinity profiles with the advection-dispersion hydrodynamic model, M. Bahr for cell count data, P. R. Raymond for assistance in the field, B. Zuniga for the map of Plum Island Sound, and the Bay Paul Center for Comparative Molecular Biology and Evolution for laboratory facilities and DNA sequencing.
REFERENCES
- 1.Barbosa, A. B., H. M. Galvao, P. A. Mendes, X. A. Alvarez-Salgado, F. G. Figueiras, and I. Joint. 2001. Short-term variability of heterotrophic bacterioplankton during upwelling off the NW Iberian margin. Prog. Oceanogr. 51:339-359. [Google Scholar]
- 2.Barcina, I., P. Lebaron, and J. Vives-Rego. 1997. Survival of allochthonous bacteria in aquatic systems: a biological approach. FEMS Microbiol. Ecol. 23:1-9. [Google Scholar]
- 3.Bidle, K. D., and M. Fletcher. 1995. Comparison of free-living and particle-associated bacterial communities in the Chesapeake Bay by stable low-molecular-weight RNA analysis. Appl. Environ. Microbiol. 61:944-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouvier, T. C., and P. A. del Giorgio. 2002. Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol. Oceanogr. 47:453-470. [Google Scholar]
- 5.Cottrell, M. T., and D. L. Kirchman. 2003. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol. Oceanogr. 48:168-178. [Google Scholar]
- 6.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crump, B. C., G. W. Kling, M. Bahr, and J. E. Hobbie. 2003. Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Appl. Environ. Microbiol. 69:2253-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Bie, M. J. M., A. Speksnijder, G. A. Kowalchuk, T. Schuurman, G. Zwart, J. R. Stephen, O. E. Diekmann, and H. J. Laanbroek. 2001. Shifts in the dominant populations of ammonia-oxidizing beta-subclass Proteobacteria along the eutrophic Schelde estuary. Aquat. Microb. Ecol. 23:225-236. [Google Scholar]
- 9.Doering, P. H., C. A. Oviatt, J. H. McKenna, and L. W. Reed. 1994. Mixing behavior of dissolved organic-carbon and its potential biological significance in the Pawcatuck River estuary. Estuaries 17:521-536. [Google Scholar]
- 10.Gasol, J. M., M. Comerma, J. C. Garcia, J. Armengol, E. O. Casamayor, P. Kojecka, and K. Simek. 2002. A transplant experiment to identify the factors controlling bacterial abundance, activity, production, and community composition in a eutrophic canyon-shaped reservoir. Limnol. Oceanogr. 47:62-77. [Google Scholar]
- 11.Giovannoni, S. J., and M. S. Rappe. 2000. Evolution, diversity, and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, Inc., New York, N.Y.
- 12.Glockner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollibaugh, J. T., P. S. Wong, and M. C. Murrell. 2000. Similarity of particle-associated and free-living bacterial communities in northern San Francisco Bay, California. Aquat. Microb. Ecol. 21:103-114. [Google Scholar]
- 15.Hopkinson, C. S., A. E. Giblin, J. Tucker, and R. H. Garritt. 1999. Benthic metabolism and nutrient cycling along an estuarine salinity gradient. Estuaries 22:863-881. [Google Scholar]
- 16.Hyun, J. H., J. K. Choi, K. H. Chung, E. J. Yang, and M. K. Kim. 1999. Tidally induced changes in bacterial growth and viability in the macrotidal Han River estuary, Yellow Sea. Estuar. Coast. Shelf Sci. 48:143-153. [Google Scholar]
- 17.Jay, D. A., W. R. Geyer, and D. R. Montgomery. 2000. An ecological perspective on estuarine classification, p. 149-176. In J. E. Hobbie (ed.), Estuarine science, a synthetic approach to research and practice. Island Press, Washington, D.C.
- 18.Kirchman, D. L. 1993. Leucine incorporation as a measure of biomass production by heterotrophic bacteria, p. 513-517. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, Fla.
- 19.Lebaron, P., P. Servais, M. Troussellier, C. Courties, G. Muyzer, L. Bernard, H. Schafer, R. Pukall, E. Stackebrandt, T. Guindulain, and J. Vives-Rego. 2001. Microbial community dynamics in Mediterranean nutrient-enriched seawater mesocosms: changes in abundances, activity and composition. FEMS Microbiol. Ecol. 34:255-266. [DOI] [PubMed] [Google Scholar]
- 20.Monsen, N. E., J. E. Cloern, L. V. Lucas, and S. G. Monismith. 2002. A comment on the use of flushing time, residence time, and age as transport time scales. Limnol. Oceanogr. 47:1545-1553. [Google Scholar]
- 21.Murray, A. E., J. T. Hollibaugh, and C. Orrego. 1996. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl. Environ. Microbiol. 62:2676-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S ribosomal RNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neal, V. T. 1972. Physical aspects of the Columbia River and its estuary, p. 19-40. In A. T. Pruter and D. L. Alverson (ed.), The Columbia River estuary and adjacent coastal waters. University of Washington Press, Seattle.
- 24.Nixon, S. W., J. W. Ammerman, L. P. Atkinson, V. M. Berounsky, G. Billen, W. C. Boicourt, W. R. Boynton, T. M. Church, D. M. Ditoro, R. Elmgren, J. H. Garber, A. E. Giblin, R. A. Jahnke, N. J. P. Owens, M. E. Q. Pilson, and S. P. Seitzinger. 1996. The fate of nitrogen and phosphorus at the land sea margin of the North Atlantic Ocean. Biogeochemistry 35:141-180. [Google Scholar]
- 25.Painchaud, J., D. Lefaivre, J. C. Therriault, and L. Legendre. 1996. Bacterial dynamics in the upper St. Lawrence estuary. Limnol. Oceanogr. 41:1610-1618. [Google Scholar]
- 26.Painchaud, J., D. Lefaivre, J. C. Therriault, and L. Legendre. 1995. Physical processes controlling bacterial distribution and variability in the upper St. Lawrence estuary. Estuaries 18:433-444. [Google Scholar]
- 27.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 28.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 29.Prieur, D., M. Troussellier, A. Romana, S. Chamroux, G. Mevel, and B. Baleux. 1987. Evolution of bacterial communities in the Gironde estuary (France) according to a salinity gradient. Estuar. Coast. Shelf Sci. 24:95-108. [Google Scholar]
- 30.Rappe, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 31.Rappe, M. S., K. Vergin, and S. J. Giovannoni. 2000. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol. Ecol. 33:219-232. [DOI] [PubMed] [Google Scholar]
- 32.Schafer, H., L. Bernard, C. Courties, P. Lebaron, P. Servais, R. Pukall, E. Stackebrandt, M. Troussellier, T. Guindulain, J. Vives-Rego, and G. Muyzer. 2001. Microbial community dynamics in Mediterranean nutrient-enriched seawater mesocosms: changes in the genetic diversity of bacterial populations. FEMS Microbiol. Ecol. 34:243-253. [DOI] [PubMed] [Google Scholar]
- 33.Selje, N., and M. Simon. 2003. Composition and dynamics of particle-associated and free-living bacterial communities in the Weser estuary, Germany. Aquat. Microb. Ecol. 30:221-237. [Google Scholar]
- 34.Simek, K., J. Nedoma, J. Pernthaler, T. Posch, and J. R. Dolan. 2002. Altering the balance between bacterial production and protistan bacterivory triggers shifts in freshwater bacterial community composition. Antonie Leeuwenhoek 81:453-463. [DOI] [PubMed] [Google Scholar]
- 35.Sin, Y., R. L. Wetzel, and I. C. Anderson. 2000. Seasonal variations of size-fractionated phytoplankton along the salinity gradient in the York River estuary, Virginia (USA). J. Plankton Res. 22:1945-1960. [Google Scholar]
- 36.Strom, S. L. 2000. Bacterivory: interactions between bacteria and their grazers, p. 351-386. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, Inc., New York, N.Y.
- 37.Troussellier, M., H. Schafer, N. Batailler, L. Bernard, C. Courties, P. Lebaron, G. Muyzer, P. Servais, and J. Vives-Rego. 2002. Bacterial activity and genetic richness along an estuarine gradient (Rhone River plume, France). Aquat. Microb. Ecol. 28:13-24. [Google Scholar]
- 38.Valdés, M., and L. J. Albright. 1981. Survival and heterotrophic activities of Fraser River and Strait of Georgia bacterioplankton within the Fraser River plume. Mar. Biol. 64:231-241. [Google Scholar]
- 39.Vallino, J. J., and C. S. Hopkinson. 1998. Estimation of dispersion and characteristic mixing times in Plum Island Sound estuary. Estuar. Coast. Shelf Sci. 46:333-350. [Google Scholar]
- 40.Van de Peer, Y., S. Chapelle, and R. De Wachter. 1996. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 24:3381-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Hannen, E. J., W. Mooij, M. P. van Agterveld, H. J. Gons, and H. J. Laanbroek. 1999. Detritus-dependent development of the microbial community in an experimental system: qualitative analysis by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2478-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Hannen, E. J., M. Veninga, J. Bloem, H. J. Gons, and H. J. Laanbroek. 1999. Genetic changes in the bacterial community structure associated with protistan grazers. Arch. Hydrobiol. 145:25-38. [Google Scholar]
- 43.Vasconcelos, G. J., and R. G. Swartz. 1976. Survival of bacteria in seawater using a diffusion chamber apparatus in situ. Appl. Environ. Microbiol. 31:913-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vorosmarty, C. J., and T. C. Loder. 1994. Spring neap tidal contrasts and nutrient dynamics in a marsh-dominated estuary. Estuaries 17:537-551. [Google Scholar]
- 45.Welch, E. B., R. M. Emery, R. I. Matsuda, and W. A. Dawson. 1972. The relation of periphytic and planktonic algal growth in an estuary to hydrographic factors. Limnol. Oceanogr. 17:731-737. [Google Scholar]
- 46.Wright, R. T., R. B. Coffin, and M. E. Lebo. 1987. Dynamics of planktonic bacteria and heterotrophic microflagellates in the Parker estuary, northern Massachusetts. Cont. Shelf Res. 7:1383-1397. [Google Scholar]
- 47.Zwart, G., B. C. Crump, M. Agterveld, F. Hagen, and S. K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]