Abstract
Malaria infections of mammals are initiated by the transmission of Plasmodium salivary gland sporozoites during an Anopheles mosquito vector bite. Sporozoites make their way through the skin and eventually to the liver, where they infect hepatocytes. Blocking this initial stage of infection is a promising malaria vaccine strategy. Therefore, comprehensively elucidating the protein composition of sporozoites will be invaluable in identifying novel targets for blocking infection. Previous efforts to identify the proteins expressed in Plasmodium mosquito stages were hampered by the technical difficulty of separating the parasite from its vector; without effective purifications, the large majority of proteins identified were of vector origin. Here we describe the proteomic profiling of highly purified salivary gland sporozoites from two Plasmodium species: human-infective Plasmodium falciparum and rodent-infective Plasmodium yoelii. The combination of improved sample purification and high mass accuracy mass spectrometry has facilitated the most complete proteome coverage to date for a pre-erythrocytic stage of the parasite. A total of 1991 P. falciparum sporozoite proteins and 1876 P. yoelii sporozoite proteins were identified, with >86% identified with high sequence coverage. The proteomic data were used to confirm the presence of components of three features critical for sporozoite infection of the mammalian host: the sporozoite motility and invasion apparatus (glideosome), sporozoite signaling pathways, and the contents of the apical secretory organelles. Furthermore, chemical labeling and identification of proteins on live sporozoites revealed previously uncharacterized complexity of the putative sporozoite surface-exposed proteome. Taken together, the data constitute the most comprehensive analysis to date of the protein expression of salivary gland sporozoites and reveal novel potential surface-exposed proteins that might be valuable targets for antibody blockage of infection.
Malaria, a disease caused by eukaryotic parasites of the genus Plasmodium, causes hundreds of millions of clinical cases and kills approximately 1 million people annually WHO. World malaria report. 2011. http://www.who.int (last accessed 11 December 2012). Plasmodium is transmitted to humans by infected female anopheline mosquitoes seeking a blood meal, which results in the release of mosquito saliva and sporozoites into the skin (1). Sporozoites leave the bite site, enter the blood stream, and are transported to the liver where they invade hepatocytes. Sporozoites transform into liver stages that grow, mature, and release exo-erythrocytic merozoites, which enter the blood stream and invade erythrocytes (2, 3). The erythrocytic stages of the infection lead to all clinical symptoms and disease. The sporozoite and liver stages of infection are asymptomatic, constitute population bottlenecks in the life cycle, and thus are perceived as important drug and vaccine targets (4). Indeed, irradiated and genetically attenuated sporozoites (lacking genes critical for liver stage development) are powerful immunogens that confer sterile protection against malaria infection (5). Animal model data show that these attenuated sporozoites exhibit normal behavior in that they infect the liver, but then they arrest during liver stage development. Exposure to extracellular sporozoites provokes antibody responses, and the subsequent intrahepatocytic liver stages are thought to result in the presentation of intracellular antigens to the host immune system that elicit a CD8+ T cell response against infected hepatocytes (6–11). However, achieving complete sterile protection against malaria infection with sporozoite and liver-stage-based subunit vaccines has so far proven an elusive goal.
The subunit vaccine candidate RTS,S, which contains part of the major sporozoite surface protein, circumsporozoite protein (CSP),1 has been tested in phase III trials and has shown partial efficacy (12, 13). The RTS,S trial results highlight the need to identify additional antigen targets for inclusion in a next-generation malaria vaccine. Recently, a virally vectored platform of immunization has shown that recombinant chimpanzee adenovirus and modified vaccinia virus Ankara encoding thrombospondin-related adhesion protein (TRAP), a sporozoite- and early liver stage parasite-expressed antigen, can induce potent immune responses (6). In preclinical studies, this prime-boost platform induced substantial protection that depended on the induction of a potent CD8+ T cell effector memory response directed against liver stage parasites (6). The partial success of sporozoite protein subunit vaccine candidates and the sterilizing protection model of whole sporozoite vaccination can be further built upon in order to ultimately achieve complete sterilizing protection against malaria infection with a multivalent subunit vaccine. To aid this, a thorough assessment of the Plasmodium sporozoite protein arsenal is needed so that the best candidates can be identified and evaluated.
Proteomic analysis via mass spectrometry, although a well-established and routine technique, has yet to define complete proteomes in single experiments. However, given a sufficient sample and access to state-of-the-art instrumentation, upwards of 50% complete proteomes (e.g. 68% of yeast (14) and 51% of human (15–17) proteomes) can be determined with a reasonable amount of effort. A major obstacle to proteomic analysis of the Plasmodium parasite pre-erythrocytic stages is the difficulty of separating the parasites from vector or host material. The presence of excessive contaminating protein can overwhelm chromatographic column capacity and mass analyzer duty cycles, thereby limiting the number of parasite proteins that can be observed in this large dynamic range of host versus parasite protein abundance. In a study of P. falciparum mosquito stages, Lasonder et al. (18) reported that mosquito proteins made up between 65% and 89% of the samples analyzed. Despite significant effort, only 728 mosquito stage parasite proteins were identified (71.8% with multiple peptides), which constitutes ∼13% of the total protein coding capacity of P. falciparum's genome. Florens et al. (19) reported 1048 P. falciparum salivary gland sporozoite protein identifications (∼19% of P. falciparum ORFs), but only 30.0% of those were identified by multiple peptides. Florens et al. did not discuss the presence of vector material in the sample, but they did report requiring the combined protein content of five separate sporozoite preparations. A proteomic study of P. berghei by Hall et al. (20) reported only 134 salivary gland sporozoite protein identifications (78.3% identified by multiple peptides), despite finding a total of 1836 proteins across the P. berghei life cycle. Although the success of any proteomic profiling experiment is directly linked to the purification technique used and access to state-of-the-art mass analyzers, the limited success of the above-described efforts suggests that more complete proteomic characterization of Plasmodium salivary gland sporozoites will depend even more heavily upon optimized sample preparation and purification.
In this work we employed a high-resolution LTQ Orbitrap Velos mass analyzer coupled to nano–liquid chromatography (nanoLC) for the identification of proteins present in highly purified salivary gland sporozoites of both the human-infective P. falciparum and rodent-infective P. yoelii species. In addition to surveying the total salivary gland sporozoite proteome, we also identified factors and features of parasite motility and invasion processes and labeled live salivary gland sporozoites to capture and identify putative proteins present on their surface. Using our improved purification techniques for the removal of mosquito debris, we have achieved the most complete proteome coverage yet reported for a pre-erythrocytic stage of Plasmodium, and herein we identify novel, putative sporozoite surface protein candidates.
EXPERIMENTAL PROCEDURES
Experimental Animals and Parasite Production
Six- to eight-week-old female Swiss Webster mice from Harlan (Indianapolis, IN) were used for the production of wild-type P. yoelii 17XNL (Py17XNL) (a non-lethal strain) blood stage parasites. Animal handling was conducted according to protocols approved by the Institutional Animal Care and Use Committee at Seattle Biomedical Research Institute. Transgenic Py17XNL parasites with a C-terminal 4xmyc tag attached to PY01024 (PlasmoDB Accession No. PY01024) were created by means of standard single-crossover reverse genetics approaches (21). Transgenic Py17XNL parasites with a C-terminal triple HA tag attached to PY00899 (PlasmoDB Accession No. PY00899) were created via standard double-crossover reverse genetics approaches. Py17XNL wild-type and transgenic parasites were cycled between Swiss Webster mice and Anopheles stephensi mosquitoes. Infected mosquitoes were maintained on 10% w/v dextrose solution at 24 °C and 70% humidity. Salivary gland sporozoites were isolated via micro-dissection 14 days after the consumption of an infectious blood meal.
P. falciparum (NF54 strain) cultures were maintained in vitro through infections of O+ erythrocytes grown in RPMI 1640 supplemented with 50 μm hypoxanthine, 25 mm HEPES, 2 mm l-glutamine, and 10% A+ human serum in a gas mixture consisting of 5% CO2, 5% O2, and 90% N2. Gametocyte cultures were initiated at 5% hematocrit and 0.8% to 1% parasitemia and were maintained for up to 17 days with daily media changes. Adult female An. stephensi mosquitoes (3 to 7 days post-emergence) were collected into mesh-topped, wax-lined pots and were allowed to feed through Parafilm for up to 20 min upon gametocyte cultures at 40% hematocrit containing fresh A+ human serum and O+ erythrocytes. Infected mosquitoes were maintained for up to 19 days at 27 °C and 75% humidity and were provided with an 8% w/v dextrose solution in 0.05% w/v PABA water. Salivary gland sporozoites were isolated via micro-dissection 14 to 19 days after the blood meal.
Sporozoite Purification
For the identification of salivary gland sporozoite surface-exposed proteins, salivary gland sporozoites were purified on a 17% w/v Accudenz (in ddH2O) cushion via centrifugation at 2500g for 20 min (22). For determination of the total salivary gland sporozoite proteomes, salivary gland sporozoites coated with BSA on ice were purified via passage through DEAE-cellulose (for P. yoelii) or were first purified over an Accudenz cushion and then coated with BSA on ice and passed through DEAE-cellulose (for P. falciparum) (23). Purified salivary gland sporozoites were pelleted and stored at −80 °C until processed further.
Indirect Immunofluorescence Assay
Salivary gland sporozoites were processed for an indirect immunofluorescence assay essentially as described in Ref. 24. Briefly, sporozoites were air dried onto glass slides, fixed with 10% v/v formalin, permeabilized by 0.1% v/v Triton X-100, and blocked in a 3% w/v BSA solution (all diluted in 1xPBS). Samples were then stained at room temperature in blocking solution with primary antibodies (rabbit anti-c-myc (Santa Cruz Biotechnology, Santa Cruz, CA, catalog no. SC-789), 1 μg/ml final concentration; rabbit anti-HA (Santa Cruz Biotechnology, catalog no. sc-805) diluted 1:400; mouse anti-PyCSP (clone 2F6) (25); mouse anti-PyRON4 (clone 48F8) (26); Alexa Fluor–conjugated secondary antibodies specific to mouse or rabbit IgG (Alexa Fluor 488 and 594 (Invitrogen), respectively)) and 4′,6-diamidino-2-phenylindole (DAPI), which labels A-T rich regions of DNA. Samples were covered with VectaShield (Vector Laboratories, Burlingame, CA) and sealed under a coverglass slip. Fluorescent and differential interference contrast images were acquired using a DeltaVision Spectris RT microscope (Applied Precision, Issaquah, WA) using a 100× oil objective and were deconvolved using the softWoRx software package.
Biotinylation and Western Blot Detection of Putative Sporozoite Surface-exposed Proteins
Salivary gland sporozoites (4 × 106 to 6 × 106) purified over an Accudenz cushion were washed in cold 1xPBS (pH 8.0 at 4 °C) and incubated with 2 mm EZ-Link® Sulfo-NHS-LC-Biotin (Thermo Scientific) for 30 min at room temperature. The biotinylation reaction was quenched by glycine added to a final concentration of 100 mm. Sporozoites were further washed in 100 mm glycine in cold 1xPBS (pH 8.0 at 4 °C) and were lysed for 1 h on ice in a solution comprising 0.1% SDS, 4 m urea, 150 mm NaCl, 50 mm Tris-HCl (pH 8.0 at 22 °C), and protease inhibitors (Complete Mini, Roche). The lysate was centrifuged at 16,300g for 10 min at 4 °C to pellet cell debris. The supernatant was diluted 10-fold with a solution identical to that described above, but without urea. The lysate was incubated with Dynabeads® MyOne™ Streptavidin T1 (Invitrogen) for 1 h at 4 °C with end-over-end rotation. The beads were then washed in 0.1% SDS, 400 mm urea, 150 mm NaCl, and 50 mm Tris-HCl (pH 8.0 at 22 °C). Proteins were eluted from the beads by boiling them for 5 min in 1x Sample Buffer (25 mm Tris (pH 6.8 at room temperature), 2.5% w/v SDS, 2.5% v/v glycerol, 0.08% w/v bromphenol blue, and 5% beta-mercaptoethanol (added fresh immediately before use)). Proteins were assessed for biotinylation via western blotting using standard methods with streptavidin-conjugated horseradish peroxidase (HRP) or antibodies against PyCSP (monoclonal, 2F6) or PfCSP (monoclonal, 2A10) with an HRP-conjugated anti-mouse IgG secondary antibody. All samples were visualized by means of enhanced chemiluminescence (SuperSignal West Pico, Thermo Scientific). Samples were stored at −20 °C until processed further via mass spectrometry.
Sample Fractionation via One-dimensional SDS-PAGE and In-gel Tryptic Digestion
Whole cell lysates were prepared from ∼107 P. yoelii or P. falciparum purified salivary gland sporozoites. The frozen sporozoite pellet was thawed on ice, loosened through vortexing, and lysed by the addition of an equal volume of 2x Sample Buffer and heating at 70 °C for 5 min. Purified salivary gland sporozoites or the isolated surface-exposed proteins were electrophoresed through a 4%–20% SDS-polyacrylamide gel at 180 V for 30 to 40 min at 22 °C. Gels were post-stained with Imperial Stain (Thermo Scientific) and destained in double-deionized H2O. The in-gel tryptic digestion of proteins was automated (see supplemental material) with a TECAN Freedom Evo (Männedorf, Switzerland). The whole cell lysate gels were cut into 16 or 21 fractions and analyzed via LC-MS/MS in triplicate. The surface-exposed protein gels were cut into four fractions and analyzed via LC-MS/MS in duplicate.
LC-MS
Peptide separation was performed on capillary columns packed in-house with C18 (ReproSil-Pur C18-AQ, 120 Å, 3 μm; Dr. Maisch, Ammerbuch-Entringen, Germany). Injected samples were first washed on a trap column prior to being loaded on the separation column. The sample was separated by a linear gradient changing from 95% A (0.1% v/v formic acid in water) and 5% B (0.1% v/v formic acid in acetonitrile) to 65% A and 35% B over 60 or 90 min. The mass analyzer was a Thermo Fisher Scientific LTQ Orbitrap Velos or LTQ Orbitrap Elite Velos Pro.
Peak List Generation
Raw mass spectra were converted to .mzXML format using MSConvert (27) and searched with X!Tandem (28), version 2010.10.01.1, and SEQUEST v.27 rev.0 (29). Spectra were searched against databases comprising the Plasmodium species in question, A. gambiae (to account for mosquito debris, as no comprehensive database for A. stephensi exists), common protein contaminants, and decoys. The A. gambiae database (VectorBase, version 3.6, updated October 10, 2011) contained 14,324 entries. The contaminant database was a modified version of the common Repository of Adventitious Proteins (version 2009.05.1, last updated October 18, 2011) with the Sigma Universal Standard Proteins removed and human angiotensin II and [Glu-1] fibrinopeptide B (MS calibration peptides) added, for a total of 66 entries. Decoys were generated with Mimic. To compensate for the poor sequence coverage and annotation of the P. yoelii 17XNL strain database, P. yoelii data were also searched against a P. berghei database. The P. falciparum database contained a total of 39,828 entries comprising 5524 P. falciparum entries (PlasmoDB v.8.2, updated October 27, 2011), 14,324 A. gambiae entries, 66 cRAP entries, and 19,914 decoys. The P. yoelii database contained a total of 44,746 entries comprising 7983 P. yoelii entries (based on Ref. 30 and manually curated to remove duplicate entries, correct spurious entries, and add new entries), 14,324 A. gambiae entries, 66 cRAP entries, and 22,373 decoys. The P. berghei database contained a total of 38,588 entries comprising 4904 P. berghei entries (PlasmoDB v.8.2, updated October 27, 2011), 14,324 A. gambiae entries, 66 cRAP entries, and 19,294 decoys. Similar search criteria were used for X!Tandem and SEQUEST. A wide precursor mass tolerance of ±0.1 Da was used to improve the performance of the accurate mass binning tool available in Peptide Prophet. Fragment ions were searched with a mass tolerance of ±0.4 Da in X!Tandem. Fragment ion mass tolerance is not specified in SEQUEST. Peptides were assumed to be semi-tryptic with up to two missed cleavages allowed. The search parameters included a static modification of +57.021464 Da at C for carbamidomethylation by iodoacetamide and potential modifications of +15.994915 for oxidation at M. Additionally, X!Tandem automatically searched for potential modifications of −17.026549 Da for deamidation at N-terminal Q and −18.010565 Da for loss of water at N-terminal E from the formation of pyro-Glu, as well as −17.026549 Da at N-terminal carbamidomethylated C for deamidation from the formation of S-carbamoylmethylcysteine. For surface-exposed proteins, a potential modification of +339.161662 at K was added for the biotin tag. MS/MS data were analyzed using the Trans-Proteomic Pipeline (31), version 4.5 Rev.2. Peptide spectrum matches (PSMs) generated by each search engine were analyzed separately with Peptide Prophet to assign each PSM a probability of being correct. The Peptide Prophet scores for a given analysis (e.g. all X!Tandem and SEQUEST results of all the injections of all gel fractions of one sporozoite lysate) were then combined in iProphet (32). Protein identifications were inferred with Protein Prophet (33). If multiple proteins were inferred at equal confidence by a set of peptides, the inference was counted as a single identification, and all relevant protein IDs were listed. The false positive error rate of protein identifications was estimated as the fraction of decoy protein inferences found above a given Protein Prophet probability cut-off. Only proteins identified at Protein Prophet probabilities corresponding to a false positive error rate of less than 1.0% were reported.
Spectral Counting
A normalized spectral abundance factor (NSAF) was calculated for Plasmodium proteins in order to provide an estimate of the relative abundance of proteins within a proteome and between the two Plasmodium species (34). The initial spectral count for a given protein was taken as the number of PSMs used by Protein Prophet to make the protein inference. The spectral abundance factor for each protein was calculated by dividing the spectral count by the protein's length. The NSAF for each Plasmodium protein was calculated by dividing its spectral abundance factor by the sum of all Plasmodium spectral abundance factors for that species.
RESULTS
Total Sporozoite Proteome Determination
NanoLC coupled to a high-resolution mass spectrometer was used to obtain the most comprehensive proteomic coverage to date of P. falciparum and P. yoelii salivary gland sporozoites. The numbers of PSMs and unique stripped peptides (peptides with unique sequence regardless of charge state or modifications) reported below reflect peptides with iProphet probabilities estimated to correspond to a false positive error rate of less than 1%. All protein identifications reported here were identified at Protein Prophet probabilities corresponding to a decoy-estimated false positive error rate of less than 1%. A complete list of all peptide spectrum matches and inferred protein identifications is provided in the supplemental information (supplemental Tables S1–S4). The data associated with this manuscript are available for download from PeptideAtlas (select P. falciparum and P. yoelii) (35) and have been used to create the first PeptideAtlas built for Plasmodium species in order to facilitate future malaria proteomic efforts. The data associated with this manuscript may also be downloaded from the Proteome Commons Tranche using the following hash: 7egFn9UV4wwUoNIvIF0zSs8Wwe1PcBrObDyUduwrGo83WW5+87l2ZZXuNoQhiiv9weoizb0cFXYmI1OUlbEHevk8VpYAAAAAAAKXbA==.
Approximately 107 sporozoites of P. yoelii and P. falciparum were fractionated by means of one-dimensional SDS-PAGE and analyzed via nanoLC-MS/MS in triplicate. Despite the variability in protein recovery and sporozoite purification between the P. yoelii and P. falciparum preparations (Table I), the total number of Plasmodium-specific PSMs obtained for the two species was within 26%. An NSAF based on spectral counting was calculated for each Plasmodium protein in order to provide an estimate of the relative abundance of a protein within a proteome, as well as between the P. falciparum and P. yoelii proteomes (34, 36). Analysis of P. falciparum salivary gland sporozoite whole cell lysate yielded 120,668 PSMs and 19,440 unique stripped peptides unique to Plasmodium. A total of 1991 P. falciparum proteins were identified (86.6% identified by multiple peptides), 36.0% of 5524 annotated ORFs. Of these identified proteins, 64.2% were not previously identified in proteomics efforts with P. falciparum salivary gland sporozoites (supplemental Fig. S1A) (18, 19). Additionally, 49.5% of the identified proteins were not detected in a recent asexual blood stage proteome of similar complexity (supplemental Fig. S1B) (80).
Table I. Summary of protein discovery results.
The total number of unique peptide spectrum matches (PSM), unique stripped peptides (unique peptide sequence disregarding variable modifications and charge state), and unique proteins identified are given for each of the nanoLC-MS experiments described here. The percent composition of each protein source (e.g. Plasmodium, mosquito, etc.) is given. Entries labeled as “Chimera” are peptides or protein inferences that correspond equally well to distinct proteins from different genera (e.g. both mosquito and Plasmodium). The row labeled “Contaminants” indicates the portion of the sample identified as common lab contaminants (e.g. keratin). Trypsin and BSA are listed separately from other contaminants because they are endogenous interferants rather than exogenous contaminants. The P. yoelii spectra were searched separately against a P. yoelii database and a P. berghei database. Two separate P. falciparum whole cell lysates were analyzed. The proteins identified by the second attempt constitute the P. falciparum proteome described in this paper. The results of the first attempt are summarized in this table to illustrate the negative effect of extensive contamination by mosquito debris.
| Whole cell lysate |
Affinity capture |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
P. yoelii |
P. falciparum |
P. yoelii |
P. falciparum |
|||||||||||
|
P. yoelii search |
P. berghei search |
1st attempt |
2nd attempt |
P. yoelii search |
P. berghei search |
|||||||||
| PSM | % | PSM | % | PSM | % | PSM | % | PSM | % | PSM | % | PSM | % | |
| Peptide spectrum matches | ||||||||||||||
| Plasmodium | 89,752 | 63.0 | 64,200 | 54.9 | 49,490 | 19.5 | 120,668 | 65.0 | 283 | 2.4 | 96 | 0.8 | 237 | 2.9 |
| Mosquito | 16,051 | 11.3 | 16,005 | 13.7 | 178,687 | 70.4 | 39,213 | 21.1 | 4466 | 37.5 | 4399 | 37.9 | 2517 | 31.0 |
| Chimera | 2476 | 1.7 | 2551 | 2.2 | 4642 | 1.8 | 3521 | 1.9 | 114 | 1.0 | 118 | 1.0 | 108 | 1.3 |
| Trypsin | 4548 | 3.2 | 4507 | 3.9 | 5167 | 2.0 | 7430 | 4.0 | 3565 | 29.9 | 3510 | 30.2 | 2777 | 34.2 |
| BSA | 21,730 | 15.3 | 21,767 | 18.6 | 11,861 | 4.7 | 6264 | 3.4 | 782 | 6.6 | 789 | 6.8 | 814 | 10.0 |
| Contaminants | 7869 | 5.5 | 7895 | 6.8 | 3923 | 1.5 | 8593 | 4.6 | 2709 | 22.7 | 2695 | 23.2 | 1658 | 20.4 |
| TOTAL | 142,426 | 116,926 | 253,770 | 185,689 | 11,919 | 11,607 | 8111 | |||||||
| Peptides | % | Peptides | % | Peptides | % | Peptides | % | Peptides | % | Peptides | % | Peptides | % | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unique stripped peptides | ||||||||||||||
| Plasmodium | 14,523 | 80.1 | 9631 | 73.2 | 6615 | 40.8 | 19,440 | 79.2 | 66 | 3.6 | 41 | 2.3 | 57 | 4.6 |
| Mosquito | 2973 | 16.4 | 2911 | 22.1 | 9106 | 56.2 | 4401 | 17.9 | 1427 | 77.9 | 1404 | 78.6 | 799 | 64.8 |
| Chimera | 185 | 1.0 | 183 | 1.4 | 164 | 1.0 | 264 | 1.1 | 31 | 1.7 | 33 | 1.8 | 25 | 2.0 |
| Trypsin | 39 | 0.2 | 39 | 0.3 | 30 | 0.2 | 78 | 0.3 | 64 | 3.5 | 64 | 3.6 | 62 | 5.0 |
| BSA | 170 | 0.9 | 163 | 1.2 | 149 | 0.9 | 139 | 0.6 | 54 | 2.9 | 54 | 3.0 | 57 | 4.6 |
| Contaminants | 233 | 1.3 | 226 | 1.7 | 135 | 0.8 | 217 | 0.9 | 191 | 10.4 | 191 | 10.7 | 233 | 18.9 |
| TOTAL | 18,123 | 13,153 | 16,199 | 24,539 | 1833 | 1787 | 1233 |
| Proteins | % | Proteins | % | Proteins | % | Proteins | % | Proteins | % | Proteins | % | Proteins | % | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Proteins | ||||||||||||||
| Plasmodium | 1749 | 72.6 | 1466 | 69.3 | 861 | 39.5 | 1991 | 70.5 | 27 | 6.0 | 24 | 5.4 | 22 | 8.0 |
| Mosquito | 642 | 26.7 | 628 | 29.7 | 1311 | 60.1 | 820 | 29.0 | 411 | 90.7 | 403 | 91.2 | 227 | 82.8 |
| Chimera | 1 | 0.04 | 2 | 0.09 | 1 | 0.05 | 0 | 0.00 | 1 | 0.2 | 1 | 0.2 | 0 | 0.0 |
| Other | 17 | 0.7 | 18 | 0.9 | 9 | 0.4 | 15 | 0.5 | 14 | 3.1 | 14 | 3.2 | 25 | 9.1 |
| TOTAL | 2409 | 2114 | 2182 | 2826 | 453 | 442 | 274 |
Mass spectra obtained from triplicate nanoLC-MS/MS analysis of a one-dimensional SDS-PAGE-fractionated P. yoelii salivary gland sporozoite whole cell lysate were searched separately against a P. yoelii database and a database for P. berghei, a rodent-infective Plasmodium species that is closely related to P. yoelii. The P. yoelii database suffers from suboptimal genome sequence coverage and gene annotation (30); therefore the more complete P. berghei database was searched for possible orthologs to augment the data set. The P. yoelii search yielded 89,752 PSMs, 14,523 unique stripped peptides, and 1749 parasite-specific protein identifications (89.8% identified by multiple peptides). The P. berghei search yielded 64,200 PSMs, 9631 unique stripped peptides, and 1466 parasite-specific protein identifications (86.0% identified by multiple peptides). From this search of the P. berghei database, 127 proteins were confidently identified whose orthologs were not identified from the search of the P. yoelii database (supplemental Table S3). Combining the P. yoelii and P. berghei search results yielded a total of 1876 protein identifications (88.2% identified by multiple peptides), 23.5% of 7983 manually curated ORFs.
Several noteworthy findings arose from the P. falciparum and P. yoelii proteomic datasets. First, a high overlap of orthologous genes was detected from both species (71.9% of P. falciparum proteins overlap with P. yoelii proteins; 78.9% of P. yoelii proteins overlap with P. falciparum proteins). Also, many well-characterized sporozoite proteins were detected, including those involved in gliding motility (e.g. TRAP, myosin A, GAP50, inner membrane complex (IMC) proteins), cell traversal (e.g. SPECT, SPECT2, CelTOS), and hepatocyte invasion (e.g. CSP, TRAP, SIAP-1, P52, P36), which are described in further detail below. Proteins that were expected to be highly expressed in sporozoites (e.g. CSP, TRAP, SIAP, AMA1, and SPECT2) (37) were found to be among the proteins with the highest NSAF values (supplementary Table S1). Interestingly, when compared to a list of genes found by means of targeted subtraction hybridization and microarray assays to be up-regulated in infectious sporozoites (UIS) (38, 39), our analysis of sporozoites identified the corresponding proteins for less than half of the UIS genes: 58/124 (46.8%) for P. falciparum and 54/124 (43.5%) for P. yoelii (44 UIS proteins were detected in both species). Several of the UIS proteins that were not detected in our data set are critical for the liver stage of infection that follows the sporozoite stage, which could indicate that a significant proportion of these UIS transcripts are translated only after sporozoite transmission (40). Finally, an unexpectedly large number of uncharacterized, conserved proteins without predictable functional domains were also detected in high abundance (supplemental Table S1).
Sporozoite Purity is a Major Determinant of Proteome Detection
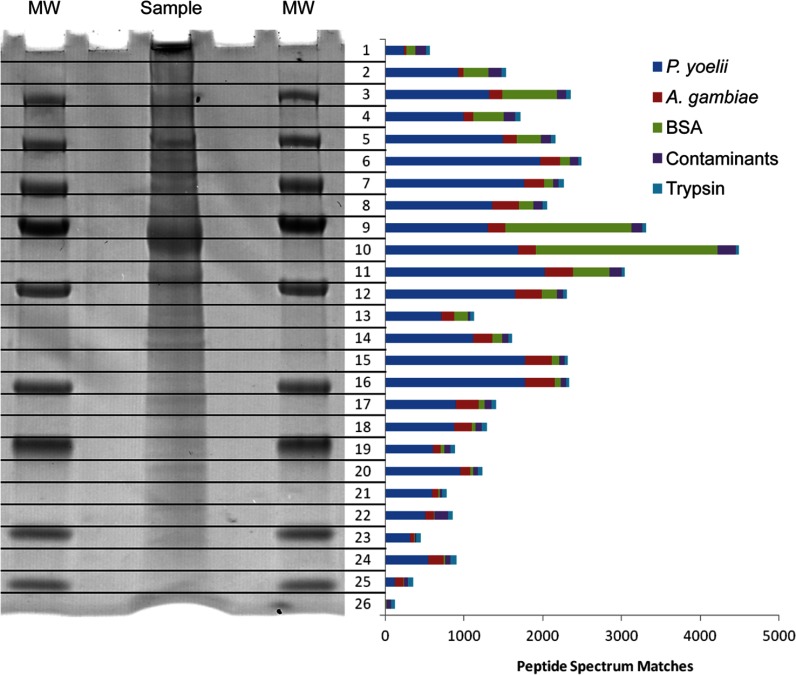
The purification protocols described here significantly reduced the presence of mosquito debris in the sample (Table I). For instance, 63% of the PSMs from the P. yoelii salivary gland sporozoite whole cell lysate were for Plasmodium proteins, whereas only 11% were for Anopheles proteins. Because of the technical requirement of coating the parasites in BSA for the chromatographic purification approach, BSA was a significant additional source of contamination, accounting for 15% of PSMs. However, our use of one-dimensional SDS-PAGE fractionation for these samples restrained the majority of BSA-specific peptide spectra to two gel fractions (Fig. 1). Illustrating the importance of sample purity for protein discovery, our initial attempt to analyze P. falciparum salivary gland sporozoite whole cell lysate yielded only 861 Plasmodium-specific proteins, despite the fact that it was analyzed in a fashion essentially identical to that used for P. yoelii (data not shown). Only 20% of the peptide spectrum matches were for Plasmodium proteins, and 70% were for mosquito proteins, despite the fact that the total number of peptide spectrum matches for the experiment was significantly greater than for the comparable P. yoelii experiment. The P. falciparum whole cell lysate data presented here were generated from a subsequent attempt in which greater care was taken to minimize the amount of mosquito debris by purifying the parasites first on a 17% w/v Accudenz cushion (22) and then via DEAE-cellulose chromatography. The Plasmodium portion of peptide spectrum matches was increased to 65%, and the mosquito portion was decreased to 20%. Only 7 of the 861 Plasmodium proteins identified in the first attempt were not among the 1991 proteins identified in the second attempt, and each of these 7 proteins was identified from only a single peptide.
Fig. 1.
One-dimensional SDS-PAGE fractionation of sporozoite proteins. The one-dimensional SDS-PAGE separation of a P. yoelii salivary gland sporozoite whole cell lysate (flanked on both sides by a molecular weight (MW) marker) is shown with a bar graph representing the total number of high-quality (false positive error rate < 1.0%) peptide spectrum matches found from a single injection of each fraction after in-gel tryptic digest. Bovine serum albumin (BSA) from the purification protocol can be seen as the large band in fractions 9 and 10. In every other fraction, P. yoelii proteins were the major component.
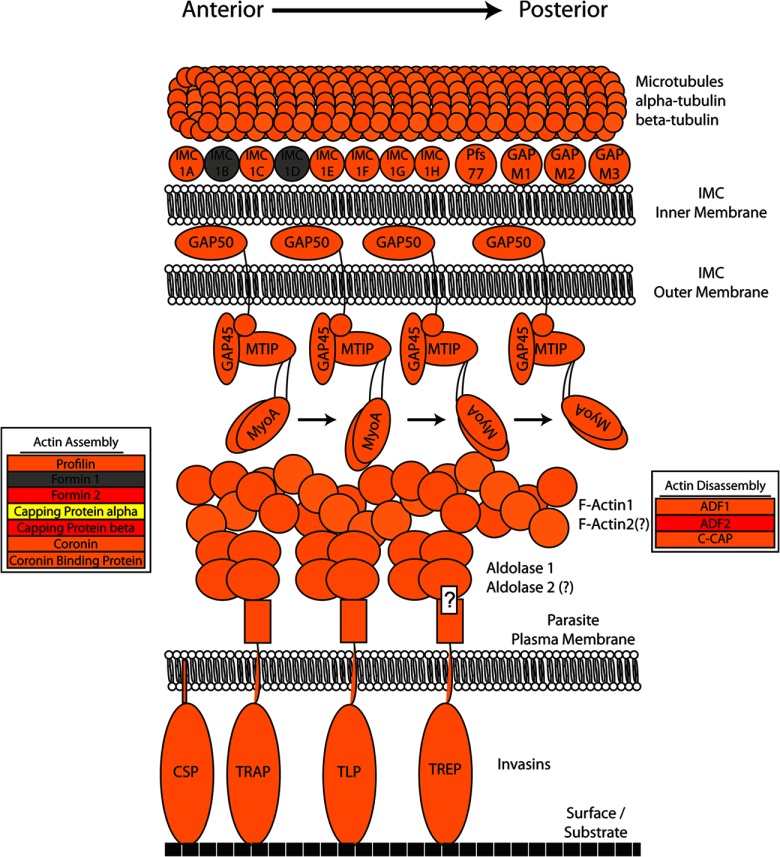
The Sporozoite Glideosome
Invasive forms of apicomplexan parasites use a complex molecular motor (termed the glideosome) to move via gliding motility, traverse cells, and invade host cells for subsequent life cycle progression (41). Salivary gland sporozoites actively seek the host vasculature upon deposition in the skin and traverse multiple hepatocytes prior to the productive infection of a single hepatocyte (reviewed in Ref. 42). However, delineation of the sporozoite glideosome has not been attempted, and few components of the glideosome have been experimentally verified in sporozoites (43). Making use of the high number of glideosome-associated proteins known from blood stage parasites that were also identified in P. falciparum and P. yoelii salivary gland sporozoite total proteomes (capping protein alpha and beta (44), profilin (45), etc.), we compiled the available structural and molecular data from all Plasmodium life stages, as well as from the related apicomplexan parasite Toxoplasma gondii, into a single preliminary model and determined which proteins were absent (gray in Fig. 2) or present in salivary gland sporozoites in P. falciparum (red in Fig. 2), P. yoelii (yellow in Fig. 2), or both (orange in Fig. 2) (supplemental Table S5). The relative abundances of the major components of the sporozoite glideosomes of both P. falciparum and P. yoelii sporozoites (as estimated by the NSAF) are consistent with current models of motility functions. For instance, the major coat protein CSP is robustly shed during gliding motility and is therefore one of the most abundant sporozoite proteins (18). CSP was indeed among the most abundant Plasmodium proteins we identified in both P. falciparum and P. yoelii (fifth and twenty-second most abundant, respectively, as estimated by the NSAF).
Fig. 2.
Components of the Plasmodium glideosome identified in sporozoite stages. The currently known components of Plasmodium's gliding motility and invasion machinery (the “glideosome”) mainly identified from asexual blood stages and orthologs from Toxoplasma gondii are illustrated in this model and are depicted based on their presence in the sporozoite proteome. Sporozoite proteins are colored to indicated whether they were detected in P. falciparum sporozoites only (red), P. yoelii sporozoites only (yellow), both species (orange), or neither species (gray). Two shades of orange were used to provide contrast between the intertwined tubulin and F-actin molecules. The anchoring of CSP in the sporozoite plasma membrane occurs via a glycosylphosphatidylinositol anchor, whereas invasins do so via a transmembrane domain and a cytoplasmic C-terminal tail that interacts with aldolase through a conserved penultimate tryptophan (this interaction for Thrombospondin Related Anonymous Protein (TRAP)-related protein has not been experimentally validated and is indicated by a question mark). Aldolase 2 and Actin 2 were previously thought to be present only in blood stage parasites but were confidently identified in these experiments. They are marked with question marks, as their involvement in gliding motility has not been experimentally validated.
Unexpectedly, two proteins thought to be restricted to sexual or asexual erythrocytic stage expression (Actin 2 (46) and Aldolase 2 (47), respectively) were detected in addition to the forms expected to be found in salivary gland sporozoites (Actin 1, Aldolase 1; see supplemental Table S5). Despite some sequence homology among various isoforms of actin found in both Plasmodium and Anopheles, Actin 2 (P. falciparum gene PF14 0124, P. yoelii gene PY01545) was confidently identified here by multiple non-degenerate peptides (peptides with sequences specific to the protein in question; see supplemental Table S1). The NSAF values suggest that Actin 2 is significantly less abundant than Actin 1 in sporozoites (10-fold in P. falciparum and 7-fold in P. yoelii). Aldolase 2 (P. falciparum gene PF14 0425, P. yoelii gene PY03709) is thought to be predominantly expressed in blood stage parasites. However, we saw strong evidence that it is present in sporozoites; the protein was observed with very good sequence coverage (79% for P. falciparum, 61% for P. yoelii), and most of the peptides used to make the protein inference for Aldolase 2 were non-degenerate. Interestingly, we observed Aldolase 2 to be significantly more abundant than Aldolase 1 (4-fold in P. falciparum and 9-fold in P. yoelii). Because both Aldolase 1 and Actin 1 play a role in gliding motility in blood stage parasites, Aldolase 2 and Actin 2 have been included in our preliminary model for sporozoites as well.
Only three proteins implicated in glideosome function were not detected in sporozoites of either species: IMC1b, IMC1d, and Formin 1. Each of these proteins can theoretically generate several tryptic peptides that would be observable by means of mass spectrometry according to predicted charge and hydrophobicity (data not shown), yet not a single peptide matching the sequence of any of these three proteins was detected. Only the P. falciparum form of IMC1d and the P. yoelii form of Formin 1 have a pI greater than 8.5; otherwise there is no indication that the proteins should not be detectable via the methods described here. The absence of these proteins from our dataset might indicate that these proteins were present at levels below the detection limit of the assay, or that they were not actually present in the sample.
Calcium Signaling Pathways in Sporozoites
Several recent reports have demonstrated the importance of calcium signaling and related pathways for the proper response of Plasmodium blood stage parasites to external stimuli (48–55). As strong evidence exists that calcium signaling plays a role in microneme secretion and sporozoite dedifferentiation (55–57), we sought to determine which members of these pathways were present in sporozoites, implicating their involvement in pre-erythrocytic stages as well. Again, we have reconstructed the available data into a model with proteins shaded as before (Fig. 3; absent, gray; P. falciparum only, red; P. yoelii only, yellow; both species, orange) (supplemental Table S6). Of particular note, we detected G-protein-coupled receptors, adenylyl and guanylyl cyclases, and a carbonic anhydrase, which might allow the sporozoite to detect external stimuli and trigger calcium-dependent signaling cascades. Internal calcium levels are likely affected by both SERCA and non-SERCA calcium pumps, along with two receptors (the inositol trisphosphate receptor and the ryanodine receptor). Although neither inositol trisphosphate receptors nor ryanodine receptors have predictable ORFs in P. falciparum or P. yoelii, their associated actions could nonetheless be inhibited chemically (denoted by green shading) (58). In turn, changes in calcium concentrations by these pumps and receptors can affect downstream effectors, such as calmodulin and the subset of calcium-dependent protein kinases (CDPK1, -4, -6, and -7) that are detected in sporozoites.
Fig. 3.
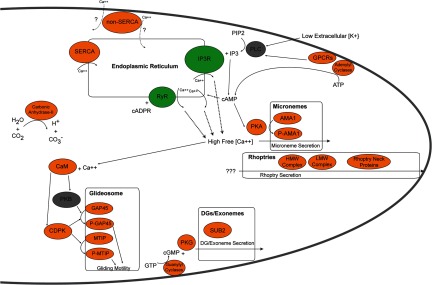
Calcium signaling pathways identified in the sporozoite proteome. A composite model of known calcium-dependent signaling molecules and their related pathways mainly identified from asexual blood stages are illustrated in this model and depicted based on their presence in the sporozoite proteome. Sporozoite proteins are colored according to their presence in both P. falciparum and P. yoelii sporozoites (orange) or their absence from both species (gray). Two endoplasmic-reticulum-associated receptors (inositol trisphosphate receptor (IP3R) and ryanodine receptor (RyR), in green) have no identified orthologous genes but have been detected based on the chemical inhibition of their functions. The triggering stimuli for rhoptry release and the directionality of the Non-SERCA calcium pump are not known and are denoted by question marks.
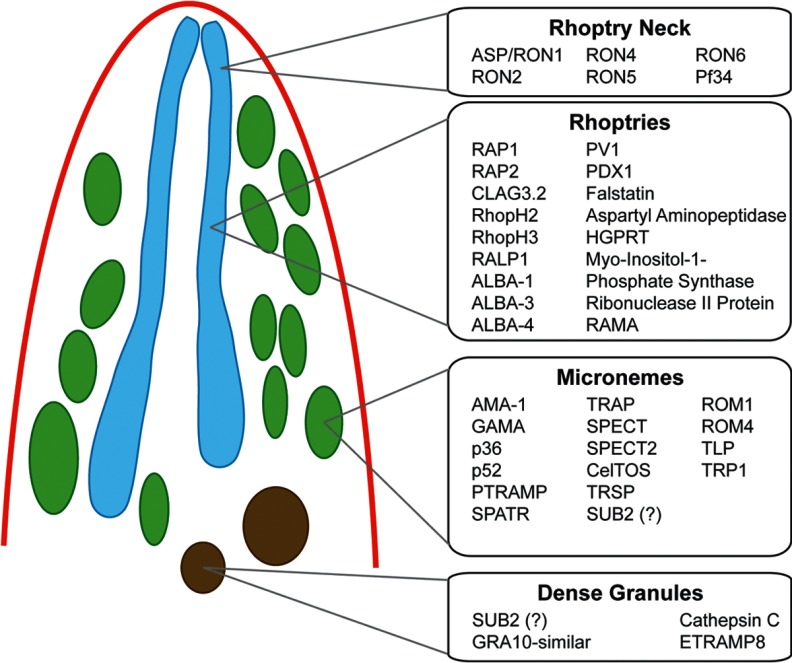
Putative Sporozoite Secretory Organelle Proteins
Many proteins thought to function during or shortly after parasite invasion typically localize to three types of apical secretory organelles: micronemes, rhoptries, and dense granules. Several key proteins and their associated mechanisms of invasion have been well characterized in the blood stage merozoite form of the parasite (59–61). Building upon these findings and what is known from sporozoite invasion studies, we sought to determine whether these proteins are also present in the highly infectious salivary gland sporozoite. To this end, we searched our total sporozoite proteomes for characterized Plasmodium apical organelle proteins described in the literature. In addition, we also extended our search to include proteins bearing significant amino acid similarity/identity to invasion proteins of a related apicomplexan parasite, Toxoplasma gondii. We detected proteins implicated at all points of the characterized invasion process, including those that localize to the micronemes (20 proteins detected), the rhoptry neck (6 proteins detected), the rhoptry body (20 proteins detected), and the dense granules (4 proteins detected) and which are typically released in this sequence before, during, and after host cell invasion (Fig. 4, supplemental Table S7). The appropriate signaling responses to specific external stimuli result in the ordered secretion of proteins to first promote gliding motility and traversal activities, thus enabling the sporozoite to migrate through the host's skin and enter the vasculature. When the sporozoite senses that is has arrived in the liver, additional proteins are released that are important for the traversal of the endothelial lining, invasion, and the productive infection of hepatocytes.
Fig. 4.
Proteins associated with the invasion organelles of Plasmodium sporozoites. A composite model of known proteins identified in the apical invasive organelles mainly identified from asexual Plasmodium blood stages is illustrated in this model. Proteins are listed if they were detected in sporozoites. Orthologs of invasion-related proteins from Toxoplasma gondii were also detected (supplemental Table S7).
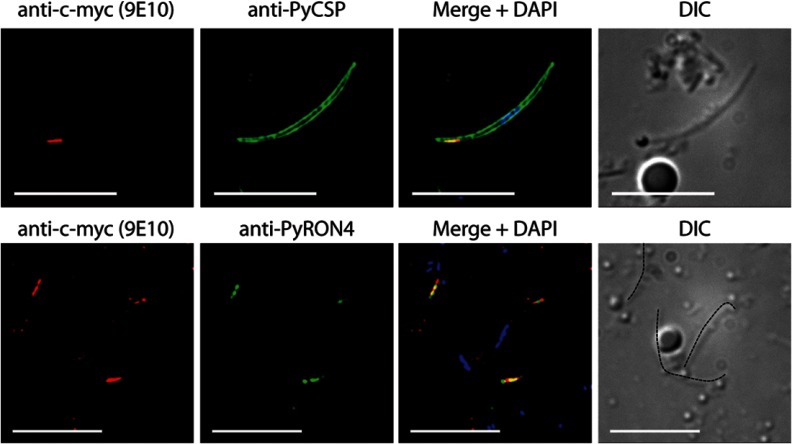
Validation of the Expression and Localization of a Low Abundance Sporozoite Protein
We next sought to validate our detection of low abundance proteins that could be constituents of the apical secretory organelles by attempting to detect one such representative protein from our P. yoelii data set. To this end, we created a transgenic P. yoelii parasite that bore a quadruple Myc tag on the C terminus of the PY01024 protein (supplemental Table S1: one unique peptide, four peptide spectrum matches; lower 15th percentile of abundance as estimated by NSAF) via standard reverse genetics approaches. This protein was of particular interest, as it bore both a predicted signal peptide and sequence homology to a known transcriptional modulator family (CCAAT-box DNA binding proteins), and such a protein might take part in modulating the infected hepatocyte. The expression of PY01024myc in transgenic salivary gland sporozoites was observed in the anterior apical end of the sporozoite (Fig. 5). Partial co-localization with PyRON4, a known rhoptry neck protein involved in parasite invasion, indicated that PY01024 also was localized to the neck of the rhoptries (26, 62). Interestingly, this protein has to date not been implicated in sporozoite host cell infection, and thus further characterization of its role is certainly warranted.
Fig. 5.
Expression and localization of a representative low abundance sporozoite protein as detected via indirect immunofluorescence assay. The protein PY01024 (CCAAT-box DNA binding protein subunit B) was confidently identified in P. yoelii salivary gland sporozoites by means of mass spectrometry, but with poor sequence coverage (one unique peptide, four peptide spectrum matches) and low abundance (lower 15th percentile as estimated based on the normalized spectral abundance factor). To confirm the presence of the protein, a transgenic P. yoelii salivary gland sporozoite bearing a C-terminal 4xMyc tag on PY01024 was subjected to an indirect immunofluorescence assay with anti-c-myc (red) that confirmed the presence of the protein. Anti-PyCSP (circumsporozoite protein, a major surface protein; top, green) and the DNA label 4′,6-diamidino-2-phenylindole (DAPI) (blue) showed PY01024 to be located at the anterior apical end of the sporozoite. Anti-PyRON4 (Rhoptry Neck Protein 4, known to localize to the rhoptry neck; bottom, green) and DAPI indicated localization of PY01024 to the rhoptry organelles. Sporozoites are marked by dotted lines layered onto the differential interference contrast images on the lower panel for clarity. Scale bar = 10 μm.
Sporozoite Surface-exposed Proteome Determination
Putative surface-exposed proteins from P. falciparum and P. yoelii salivary gland sporozoites were identified by specific labeling of live sporozoites with a biotin-conjugated, amine-reactive cross-linker and subsequent pull-down by streptavidin-coated Dynabeads (Table II, supplemental Table S1). General biotin addition to proteins was confirmed via western blot (supplemental Fig. S2A). Specific biotinylation of the major sporozoite surface protein CSP was also detected via western blot with monoclonal antibodies specific to P. yoelii CSP (supplemental Fig. S2B), with similar patterns observed with monoclonal antibodies against P. falciparum CSP (data not shown). The affinity-captured proteins were fractionated via one-dimensional SDS-PAGE and analyzed via nanoLC-MS/MS in duplicate. P. falciparum proteins captured by means of surface-exposed labeling yielded 237 peptide spectrum matches, 57 unique stripped peptides, and 22 parasite-specific proteins (68% identified by multiple peptides; supplemental Table S1). A search of the results of duplicate nanoLC-MS/MS analyses of one-dimensional SDS-PAGE fractionated, surface-exposed labeled P. yoelii proteins against a P. yoelii database yielded 283 peptide spectrum matches, 66 unique stripped peptides, and 27 parasite-specific proteins (59% identified by multiple peptides), and searching the data against a P. berghei database yielded 96 peptide spectrum matches, 41 unique stripped peptides, and 24 parasite-specific proteins (50% identified by multiple peptides). Combining the P. yoelii and P. berghei search results yielded a total of 35 unique protein identifications. Comparison across the P. falciparum and P. yoelii/P. berghei data sets revealed 13 shared proteins. This biologically relevant subset of parasite proteins was identified despite the presence of a significant amount of soluble mosquito proteins also present in the solution (Table I). Among all of the surface-exposed proteins identified were several known surface and secreted proteins: CSP, TRAP, thrombospondin-related sporozoite protein, apical membrane antigen-1, and a hexose transporter (63). As was observed by Wass and colleagues (64), several proteins associated with the glideosome that is located just below the plasma membrane (IMC1c; aldolase; actin; myosin; and glideosome-associated proteins with multiple membrane spans 1, 2, and 3) were also identified (discussed below). Additionally, many types of transporters (sugar, nucleotide, metabolite/drug), heat shock proteins, and several other uncharacterized proteins were detected. Lastly, the qualitative abundances of this proteome data set are consistent with previous work describing the relative expression of these proteins, with CSP being by far the most abundant for both P. falciparum and P. yoelii data sets (42).
Table II. A selected group of putative surface-exposed sporozoite proteins identified via biotin-conjugated cross-linker addition and capture with live sporozoites.
The presence of predicted signal peptide sequences, transmembrane (TM) domains, and glycosylphosphatidylinositol (GPI) anchor sequences is indicated. Peptide spectra numbers (total and unique) and the presence of signal peptides, transmembrane domains, and GPI anchor sequences are listed for a subset of putative sporozoite surface proteins. The complete listing is available in supplemental Table S1.
| Protein | P. falciparum gene ID | Unique peptides | Peptide spectrum matches | P. yoelii gene ID | Unique peptides | Peptide spectrum matches | Signal peptide | TM domains | GPI anchor |
|---|---|---|---|---|---|---|---|---|---|
| Hexose transporter | PFB0210c | 2 | 3 | PY00899 | 2 | 3 | No | Yes | No |
| CSP | PFC0210c | 17 | 140 | SBRIPY_PFC0210c, SBRIPY_PY07368 | 13 | 166 | Yes | Yes | Yes |
| TRSP | PFA0200w | 2 | 7 | PY07092 | 4 | 14 | Yes | Yes | No |
| TRAP | PF13_0201 | 1 | 1 | PY03052 | 8 | 16 | Yes | Yes | No |
| Sporozoite conserved orthologous transcript | PF11_0545 | 2 | 6 | PY02432 | 2 | 3 | No | No | No |
| Sugar transporter | PFI0955w | 1 | 2 | PY05332 | 2 | 7 | Yes | Yes | No |
| Sporozoite conserved orthologous transcript | PF10_0112 | 2 | 4 | PY06766 | 1 | 2 | No | Yes | No |
| AMA1 | PF11_0344 | 2 | 5 | PY01581 | - | - | Yes | Yes | No |
| Sporozoite conserved orthologous transcript | PFL0650c | 2 | 5 | PY05921 | - | - | No | No | No |
| Sporozoite conserved orthologous transcript | PF08_0088 | 1 | 1 | PY01796 | 1 | 3 | Yes | Yes | No |
| GEST | PF14_0467 | - | - | PY05966 | 1 | 4 | Yes | No | No |
| Conserved Plasmodium membrane protein, unknown function | PFL1825w | 1 | 3 | PY03591 | - | - | No | Yes | Yes |
| Conserved Plasmodium protein, unknown function | MAL7P1.67 | - | - | SBRIPY_MAL7P1.67 | 1 | 2 | Yes | Yes | No |
| Conserved Plasmodium protein, unknown function | PFL0370w | 1 | 1 | PY04162 | - | - | No | No | No |
To provide preliminary experimental validation of the surface localization of these identified proteins, a triple HA epitope tag was appended to the C terminus of one representative candidate (PY00899, a hexose transporter) that bears 12 predicted transmembrane domains. The resulting transgenic sporozoites were subjected to an indirect immunofluorescence assay (supplemental Fig. S3), and strong circumferential staining of the sporozoite was obtained with antibodies to the HA tag that co-localized well with CSP. These data support our observation that putative surface-exposed antigens were in fact captured through this cross-linking strategy.
DISCUSSION
Determining the presence of gene products at the various stages of the Plasmodium life cycle is critical to understanding the parasite's mechanisms of infection (for the ookinete, sporozoite, and merozoite) and replication (the midgut oocyst, the liver stage schizont, and the blood stage schizont). To this end, we present here key findings for the most complete proteome coverage to date of the invasive salivary gland sporozoite stage for two species of the malaria parasite, P. falciparum and P. yoelii. Although a lack of mass spectrometric evidence for a protein does not necessarily imply that protein's absence from a sample, positive identification of a protein confers strong evidence of its presence, especially if the identification is corroborated by multiple unique PSMs, as was possible with >86% of the identified proteins. The high-confidence identification of proteins via mass spectrometry has enabled us to assess various features of the salivary gland sporozoite and reliably detect components of pathways that have been characterized only in other forms of the malaria parasite, which are more easily accessible for biochemical characterization. In the absence of any internal standards for absolute or relative quantification, we have used normalized spectral abundance factors to estimate relative protein abundance for our characterizations of these data. Proteins that were expected to be highly expressed and secreted in sporozoites were found to be among the proteins identified with the largest NSAFs. Furthermore, we have experimentally validated this approach through our successful detection of a low abundance protein of interest (PY01024) in the apical rhoptry organelles of P. yoelii salivary gland sporozoites.
Careful sample preparation and improved purification protocols combined with mass spectrometric analysis using an LTQ Orbitrap Velos enabled the most complete proteome coverage to date of Plasmodium salivary gland sporozoites. Employing one-dimensional SDS-PAGE for whole cell fractionation, we were able to obtain 1991 P. falciparum salivary gland sporozoite protein identifications (86.6% identified by multiple peptides) from a single preparation of 107 purified sporozoites. Florens et al. (19) employed the MudPIT fractionation strategy (65) to identify 1048 proteins from P. falciparum salivary gland sporozoites, of which only 30.0% were identified by multiple peptides. The authors of that study did not discuss the extent of contamination by mosquito vector proteins, but it is notable that five separate preparations of 107 sporozoites were required in order to obtain the reported protein identifications. In their analysis of P. falciparum salivary gland sporozoites, Lasonder et al. (18) divided lysates of 107 cells into soluble and insoluble fractions, each of which was then fractionated via one-dimensinoal SDS-PAGE. Despite running up to four LC-MS/MS technical replicates, they were able to identify only 477 Plasmodium proteins (72.5% identified by multiple peptides) from the sporozoite sample. Their work clearly illustrates how host material contamination impedes the discovery of parasite proteins; only 31% of the proteins identified from their salivary gland sporozoite samples were of parasite origin, with the remainder being primarily contaminating mosquito proteins.
As illustrated here, the fractionation of complex samples prior to analysis, combined with mass analyzers with rapid duty cycle and high mass accuracy, enables increased protein identification. However, the success of protein discovery experiments is equally, if not predominantly, determined by the quality of the sample preparation. Sample preparation is a unique factor of the work presented here, as parasites were first manually isolated from infected mosquitoes, which presented opportunities for extensive contamination. With large variations in the dynamic range of host versus parasite proteins, the presence of contaminating proteins can obscure parasite analytes of interest. Furthermore, as the mass analyzer has a defined duty cycle, peptide spectra generated for contaminating protein tryptic peptides represent wasted instrument time and possible lost parasite protein identifications. In proteomic analyses of salivary gland sporozoites by others, as well as in our own initial attempt to analyze P. falciparum, a large proportion of mosquito protein in the sample resulted in significantly reduced parasite protein discovery relative to what was achieved with highly purified samples. The purification protocols used here (combined with a careful micro-dissection technique) facilitated a much more effective investigation of mosquito-stage parasites than has previously been reported.
Many components of the Plasmodium glideosome have been described only with respect to the asexual blood stages of infection. We have discovered that the vast majority of these glideosome proteins are also present in salivary gland sporozoites and are likely utilized in similar ways to generate a directional locomotive force toward the posterior of the sporozoite (Fig. 2). Absent from our proteome datasets are only two proteins associated with the inner membrane complex (IMCb and IMCd) and another involved in actin dynamics (Formin 1). The amino acid sequences of these proteins contain domains that should result in detectable tryptic peptides, suggesting that these proteins are either of very low abundance or actually absent from the sporozoite glideosome. Interestingly, two proteins previously characterized as being stage-restricted to sexual and asexual parasites (Actin 2 and Aldolase 2, respectively) were confidently detected in relatively high abundance in sporozoites, along with the expected homologs. All together, the major components of the locomotive machinery and its supporting molecular infrastructure (microtubules, filamentous actin, and assembly/disassembly proteins) are present in our datasets.
As the sporozoite must properly detect and respond to various extracellular stimuli, it expresses a variety of plasma-membrane-associated sensor proteins. In the current version of PlasmoDB (Version 9.2), there currently are only one G-protein-coupled receptor, one rhodopsin-like receptor, and four serpentine-receptor-like proteins (SR1, SR10, SR12, and SR25) annotated for Plasmodium species (66). We have detected a subset of these receptors in salivary gland sporozoites (SR1, SR10, rhodopsin-like receptor, G-protein-coupled receptor), which the parasite might use to sense key attributes about its ambient environment, such as specific cues to indicate whether it remains in the mosquito vector or has been transmitted to the mammalian host. In conjunction with these receptors, both adenylyl and guanylyl cyclases are expressed in Plasmodium to produce cAMP and cGMP secondary messengers for downstream signaling events, which we have also detected in our sporozoite proteomes (67). These putative sensors can then feed into intracellular signaling cascades to stimulate the parasite to respond appropriately to its environment (e.g. maintain preparations for vector/host transmission, initiate motility/traversal/invasion programs). Interestingly, phospholipase C, a common enzyme in this signaling pathway, was not detected in either species. It is also noteworthy that tyrosine kinases, which have also been implicated with phospholipase C in producing inositol 1,4,5-trisphosphate in other eukaryotes, are apparently absent from Plasmodium genomes (68). Finally, we detected a non-SERCA type calcium pump (also called ATPase 4) that also likely affects this signaling pathway and has been reported to traffic to punctate foci near the parasite periphery in infected RBCs (69). However, little is known about its functions, including into what intracellular space it pumps calcium.
Plasmodium parasites are able to store calcium at least in part because of a SERCA-type calcium pump (also called ATPase 6 or ATP6, detected in both species) that drives free calcium ions from the cytoplasm to the endoplasmic reticulum. Several reports have implicated ATP6 as a target of the front-line artemisinin family of antimalarials (70), but recent efforts have demonstrated that this is not the case (71, 72). However, these calcium stores can be released from the endoplasmic reticulum to the cytosol by increased levels of inositol trisphosphate, likely via a yet-to-be-identified inositol trisphosphate receptor or ryanodine receptor associated with the endoplasmic reticulum (51).
Upon receiving external stimuli (such as highly sulfated heparan sulfate proteoglycans, albumin, or bicarbonate) that indicate that the sporozoite has arrived in the mammalian host (reviewed in Ref. 3), the parasite engages its gliding motility machinery to make its way through the skin, into the vasculature, and finally into the liver. Therein it undergoes a rapid dedifferentiation process that is associated with a rapid increase of intracellular calcium levels (56). This free calcium not only induces secretion by the micronemes during motility and invasion, but also can be utilized by calmodulin in remodeling itself structurally in order to act upon downstream effector molecules, such as calcium-dependent protein kinases (CDPKs). Several recent studies have shown that CDPKs are important for parasite infectivity and transmission, and not surprisingly, at least two substrates of CDPK1 (GAP45 and MTIP) are members of the glideosome apparatus required for both processes (52, 73). Interestingly, only CDPKs 1, 4, and 7 were detected in both sporozoite datasets, with CDPK6 being solely identified in P. falciparum as well. The estimated abundances of these proteins were sufficiently high to make the absence of CDPKs 2, 3, and 5 especially noticeable (supplemental Table S6). Lastly, another sporozoite sensor of host arrival is the detection of increased external bicarbonate levels (74), and this has been implicated as a key stimulus triggering the sporozoite to become invasive. We have detected that both P. falciparum and P. yoelii express a cytosolic carbonic anhydrase (type II) that might function in this process, as it converts CO2 and H2O to bicarbonate and hydrogen ions. The balance of these molecules might play a role in responding to external stimuli.
The end result of responses to the host environment is the ordered activation and secretion of components of the invasion organelles (micronemes, rhoptry necks, rhoptries, and then dense granules/exonemes) in order to productively infect the host hepatocyte (reviewed in Ref. 75). The protein composition of these organelles has been scantly studied in sporozoites but has been better characterized in the blood stages. Using this information in aggregate, we have determined which apical organelle proteins are detected in the sporozoite (supplemental Table S7). In addition to the canonical proteins known to be abundant in and secreted from sporozoites (e.g. CSP, TRAP, AMA1, SPECT, SPECT2, CelTOS), we have also identified proteins not previously identified in Plasmodium sporozoites (e.g. SUB2, Pf34, RON6, RAP1, CLAG3.2, RhopH2, RhopH3, RAMA, RALP1, and ASP/RON1). The precise subcellular localization of the latter proteins in sporozoites awaits further validation, but our data set will serve as a good starting point for further characterizing apical secreted proteins. For instance, whereas sporozoite rhoptries are easily identified based on morphology, micronemes and dense granules have so far not been clearly distinguished, and published claims have placed some proteins (e.g. SUB2) in one or the other organelle (76, 77). Thus, our data set will also enhance the clarification of sporozoite cell biology. Moreover, we uncovered a novel putative rhoptry-neck-localized protein (PY01024) through our efforts to experimentally validate our approach of using spectral counting as a general proxy for protein abundance in these analyses. Also, we have identified several secreted proteins that are present in our analyses of P. yoelii sporozoites but absent in those from P. falciparum sporozoites. Most noteworthy of these is UIS3, which was also absent in a previous proteomic effort to characterize the P. falciparum sporozoite (18). Understanding the similarity in the roles these secreted proteins play during infection across malaria species, or whether they have been adapted for additional sporozoite-specific roles, will improve our knowledge of how the parasite navigates its transmission to the mammalian host. Moreover, it can guide our efforts to strategically drive vaccination efforts.
The combination of our improved sporozoite purification methods with careful mass-spectrometer-based identification and informatics analysis have now allowed the biochemical labeling and proteomic assessment of targeted protein groups. For instance, the identification of additional surface-exposed proteins could prove exceptionally useful in aiding the design of antibody-based vaccines to prevent infection by sporozoites, and thus malaria. Although CSP induces an immune response against sporozoites, recent studies have demonstrated that antibodies to other sporozoite proteins play a large role in limiting infection (78, 79). Because of the medical relevance of such a finding, we chose to biotinylate putative surface-exposed sporozoite proteins on live sporozoites, enrich for them from whole parasite lysates by virtue of the tight interaction of biotin with streptavidin-conjugated Dynabeads, and then detect and identify even very low abundance proteins. Because we were able to observe the biotinylation of CSP and other proteins via our initial metric (western blotting), we subjected these samples to mass spectrometric analysis. In addition to the “gold standard” proteins that have been shown to traffic to the sporozoite surface prior to (e.g. CSP, hexose transporter (supplemental Fig. S3)) or following sporozoite activation (e.g. TRAP, thrombospondin-related sporozoite protein, apical membrane antigen-1), we have identified several novel surface-exposed proteins as well. Many of these proteins exhibit signal peptides and either transmembrane domains or a glycosylphosphatidylinositol anchor sequence and additionally are predicted to have functions consistent with a surface localization, such as the active transport of small molecules into and out of the sporozoite (Table II, supplemental Table S1). Moreover, several other putative surface-exposed proteins that we have also identified bear no predictable functional domains and have not been previously characterized. Altogether, these proteins warrant further investigation to determine their roles in sporozoite biology, to provide additional experimental validation of their localization in sporozoites, and ultimately to determine whether targeting them with antibodies would inhibit sporozoite motility, cell traversal, and invasion capabilities and thus block sporozoite infection.
It is important to point out that within the putative sporozoite surface proteomes we also observed proteins associated with the glideosome, which is a subsurface complex. These proteins were also detected by Wass and colleagues in their efforts to determine the ookinete surface proteome using similar approaches (64). In their studies, the detection of these glideosome proteins was experimentally explained by the partial permeability of the parasite plasma membrane to the cross-linking agents. This hypothesis was confirmed by the increasing accessibility of propidium iodide to the parasite under the progressive conditions used in the cross-linking process. We believe a similar phenomenon is occurring in our hands with sporozoites as well. Because of this, we have placed stronger confidence in the designation of proteins as surface localized if they are predicted to have a signal peptide and either a transmembrane domain(s) or a predicted glycosylphosphatidylinositol anchor (Table II), as proteins detected from the glideosome do not fit these criteria. However, as this is not a strict requirement for surface localization, it will be critical to further characterize and validate the localization of these proteins.
In conclusion, the sample preparation and LC-MS/MS techniques described here for uncovering the total and putative surface proteome of Plasmodium sporozoites have demonstrated that careful sample preparation and purification techniques are required in order to produce high-quality proteomic data, especially when sample amounts are limiting. In anticipation of the great effect that further proteomics efforts will have on our understanding of the malaria parasite, we have made our high-mass-accuracy peptide detection data freely available in PeptideAtlas, which we hope will expedite high-confidence peptide identification in future efforts. Not only have these data sets yielded a valuable resource for ongoing experimentation with pre-erythrocytic stages of the parasite and for constructing more comprehensive models of their functions, but they also have produced potentially novel targets for producing humoral immunity. Currently the most advanced malaria vaccine (RTS,S, GlaxoSmithKline) induces an immune response to a single recombinant fusion protein containing a repeat domain from the major surface-exposed sporozoite coat protein (CSP) by fusing it to a highly immunogenic hepatitis B antigen. The initial results of an ongoing Phase 3 trial of this vaccine indicate that this approach provides only moderate levels of protection (12, 13). We propose that antibody-based vaccine efficacy could perhaps be significantly improved by producing a multivalent malaria subunit vaccine. Should one or more of these newly characterized surface-exposed antigens prove to be an effective antigen, it could be combined with CSP to induce a broader immune response to Plasmodium parasites and provide greater levels of protection. Taken together, the salivary gland sporozoite proteomes of P. falciparum and P. yoelii have provided further insights into critical aspects of basic sporozoite biology, and have also identified potentially targetable components of large multi-protein complexes necessary in order for the sporozoite to infect within a new host.
Supplementary Material
Acknowledgments
We thank the members of the Kappe and Moritz Laboratories for both technical assistance and critical discussion of this work. We also thank Mark Kennedy (Seattle BioMed) for advanced access to a density gradient sporozoite purification method. S.E.L., K.E.S., A.H., and A.M.V. performed research; P.S. contributed critical reagents; S.E.L., K.E.S., R.L.M., and S.H.I.K. designed research, analyzed and interpreted data, and wrote the manuscript.
Footnotes
* This investigation was supported by the National Institutes of Health under a Ruth L. Kirschstein National Research Service Award (F32GM083438) to S.E.L., by the NIGMS (Center for Systems Biology Grant No. 2P50 GM076547) to R.L.M., by the NIGMS (Grant No. GM087221 to R.L.M.), and by the NIAID (Grant No. R01 AI053709-07A2 to S.H.I.K.). This investigation was also supported by the National Science Foundation MRI (Grant No. 0923536 to R.L.M.) and by the Bill and Melinda Gates Foundation (Grant No. OPP1067687 to S.E.L., R.L.M., P.S., and S.H.I.K.).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- CDPK
- calcium-dependent protein kinase
- CSP
- circumsporozoite protein
- IMC
- inner membrane complex
- PSM
- peptide spectrum match
- NSAF
- normalized spectral abundance factor
- TRAP
- thrombospondin-related adhesion protein
- UIS
- up-regulated in infectious sporozoites.
REFERENCES
- 1. Sinnis P., Zavala F. (2008) The skin stage of malaria infection: biology and relevance to the malaria vaccine effort. Future Microbiol. 3, 275–278 [DOI] [PubMed] [Google Scholar]
- 2. Baer K., Klotz C., Kappe S. H., Schnieder T., Frevert U. (2007) Release of hepatic Plasmodium yoelii merozoites into the pulmonary microvasculature. PLoS Pathog. 3, e171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lindner S. E., Miller J. L., Kappe S. H. (2012) Malaria parasite pre-erythrocytic infection: preparation meets opportunity. Cell. Microbiol. 14, 316–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kappe S. H., Vaughan A. M., Boddey J. A., Cowman A. F. (2010) That was then but this is now: malaria research in the time of an eradication agenda. Science 328, 862–866 [DOI] [PubMed] [Google Scholar]
- 5. Vaughan A. M., Wang R., Kappe S. H. (2010) Genetically engineered, attenuated whole-cell vaccine approaches for malaria. Hum. Vaccin. 6, 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reyes-Sandoval A., Wyllie D. H., Bauza K., Milicic A., Forbes E. K., Rollier C. S., Hill A. V. (2011) CD8+ T effector memory cells protect against liver-stage malaria. J. Immunol. 187, 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Epstein J. E., Tewari K., Lyke K. E., Sim B. K., Billingsley P. F., Laurens M. B., Gunasekera A., Chakravarty S., James E. R., Sedegah M., Richman A., Velmurugan S., Reyes S., Li M., Tucker K., Ahumada A., Ruben A. J., Li T., Stafford R., Eappen A. G., Tamminga C., Bennett J. W., Ockenhouse C. F., Murphy J. R., Komisar J., Thomas N., Loyevsky M., Birkett A., Plowe C. V., Loucq C., Edelman R., Richie T. L., Seder R. A., Hoffman S. L. (2011) Live attenuated malaria vaccine designed to protect through hepatic CD8 T cell immunity. Science 334, 475–480 [DOI] [PubMed] [Google Scholar]
- 8. Butler N. S., Schmidt N. W., Vaughan A. M., Aly A. S., Kappe S. H., Harty J. T. (2011) Superior antimalarial immunity after vaccination with late liver stage-arresting genetically attenuated parasites. Cell Host Microbe 9, 451–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schmidt N. W., Podyminogin R. L., Butler N. S., Badovinac V. P., Tucker B. J., Bahjat K. S., Lauer P., Reyes-Sandoval A., Hutchings C. L., Moore A. C., Gilbert S. C., Hill A. V., Bartholomay L. C., Harty J. T. (2008) Memory CD8 T cell responses exceeding a large but definable threshold provide long-term immunity to malaria. Proc. Natl. Acad. Sci. U.S.A. 105, 14017–14022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hafalla J. C., Rai U., Bernal-Rubio D., Rodriguez A., Zavala F. (2007) Efficient development of plasmodium liver stage-specific memory CD8+ T cells during the course of blood-stage malarial infection. J. Infect. Dis. 196, 1827–1835 [DOI] [PubMed] [Google Scholar]
- 11. Hafalla J. C., Cockburn I. A., Zavala F. (2006) Protective and pathogenic roles of CD8+ T cells during malaria infection. Parasite Immunol. 28, 15–24 [DOI] [PubMed] [Google Scholar]
- 12. Agnandji S. T., Lell B., Soulanoudjingar S. S., Fernandes J. F., Abossolo B. P., Conzelmann C., Methogo B. G., Doucka Y., Flamen A., Mordmuller B., Issifou S., Kremsner P. G., Sacarlal J., Aide P., Lanaspa M., Aponte J. J., Nhamuave A., Quelhas D., Bassat Q., Mandjate S., Macete E., Alonso P., Abdulla S., Salim N., Juma O., Shomari M., Shubis K., Machera F., Hamad A. S., Minja R., Mtoro A., Sykes A., Ahmed S., Urassa A. M., Ali A. M., Mwangoka G., Tanner M., Tinto H., D'Alessandro U., Sorgho H., Valea I., Tahita M. C., Kabore W., Ouedraogo S., Sandrine Y., Guiguemde R. T., Ouedraogo J. B., Hamel M. J., Kariuki S., Odero C., Oneko M., Otieno K., Awino N., Omoto J., Williamson J., Muturi-Kioi V., Laserson K. F., Slutsker L., Otieno W., Otieno L., Nekoye O., Gondi S., Otieno A., Ogutu B., Wasuna R., Owira V., Jones D., Onyango A. A., Njuguna P., Chilengi R., Akoo P., Kerubo C., Gitaka J., Maingi C., Lang T., Olotu A., Tsofa B., Bejon P., Peshu N., Marsh K., Owusu-Agyei S., Asante K. P., Osei-Kwakye K., Boahen O., Ayamba S., Kayan K., Owusu-Ofori R., Dosoo D., Asante I., Adjei G., Chandramohan D., Greenwood B., Lusingu J., Gesase S., Malabeja A., Abdul O., Kilavo H., Mahende C., Liheluka E., Lemnge M., Theander T., Drakeley C., Ansong D., Agbenyega T., Adjei S., Boateng H. O., Rettig T., Bawa J., Sylverken J., Sambian D., Agyekum A., Owusu L., Martinson F., Hoffman I., Mvalo T., Kamthunzi P., Nkomo R., Msika A., Jumbe A., Chome N., Nyakuipa D., Chintedza J., Ballou W. R., Bruls M., Cohen J., Guerra Y., Jongert E., Lapierre D., Leach A., Lievens M., Ofori-Anyinam O., Vekemans J., Carter T., Leboulleux D., Loucq C., Radford A., Savarese B., Schellenberg D., Sillman M., Vansadia P. (2011) First results of phase 3 trial of RTS,S/AS01 malaria vaccine in African children. N. Engl. J. Med. 365, 1863–1875 [DOI] [PubMed] [Google Scholar]
- 13. RTS,S Clinical Trials Partnership, Agnandji ST, Lell B, Fernandes JF, Abossolo BP, Methogo BG, Kabwende AL, Adegnika AA, Mordmüller B, Issifou S, Kremsner PG, Sacarlal J, Aide P, Lanaspa M, Aponte JJ, Machevo S, Acacio S, Bulo H, Sigauque B, Macete E, Alonso P, Abdulla S, Salim N, Minja R, Mpina M, Ahmed S, Ali AM, Mtoro AT, Hamad AS, Mutani P, Tanner M, Tinto H, D'Alessandro U, Sorgho H, Valea I, Bihoun B, Guiraud I, Kaboré B, Sombié O, Guiguemdé RT, Ouédraogo JB, Hamel MJ, Kariuki S, Oneko M, Odero C, Otieno K, Awino N, McMorrow M, Muturi-Kioi V, Laserson KF, Slutsker L, Otieno W, Otieno L, Otsyula N, Gondi S, Otieno A, Owira V, Oguk E, Odongo G, Woods JB, Ogutu B, Njuguna P, Chilengi R, Akoo P, Kerubo C, Maingi C, Lang T, Olotu A, Bejon P, Marsh K, Mwambingu G, Owusu-Agyei S, Asante KP, Osei-Kwakye K, Boahen O, Dosoo D, Asante I, Adjei G, Kwara E, Chandramohan D, Greenwood B, Lusingu J, Gesase S, Malabeja A, Abdul O, Mahende C, Liheluka E, Malle L, Lemnge M, Theander TG, Drakeley C, Ansong D, Agbenyega T, Adjei S, Boateng HO, Rettig T, Bawa J, Sylverken J, Sambian D, Sarfo A, Agyekum A, Martinson F, Hoffman I, Mvalo T, Kamthunzi P, Nkomo R, Tembo T, Tegha G, Tsidya M, Kilembe J, Chawinga C, Ballou WR, Cohen J, Guerra Y, Jongert E, Lapierre D, Leach A, Lievens M, Ofori-Anyinam O, Olivier A, Vekemans J, Carter T, Kaslow D, Leboulleux D, Loucq C, Radford A, Savarese B, Schellenberg D, Sillman M, Vansadia P. (2012) A Phase 3 trial of RTS,S/AS01 malaria vaccine in African infants. N. Engl. J. Med. 367, 2284–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thakur S. S., Geiger T., Chatterjee B., Bandilla P., Frohlich F., Cox J., Mann M. (2011) Deep and highly sensitive proteome coverage by LC-MS/MS without prefractionation. Mol. Cell. Proteomics 10, M110.003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beck M., Schmidt A., Malmstroem J., Claassen M., Ori A., Szymborska A., Herzog F., Rinner O., Ellenberg J., Aebersold R. (2011) The quantitative proteome of a human cell line. Mol. Syst. Biol. 7, 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Geiger T., Wehner A., Schaab C., Cox J., Mann M. (2012) Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics 11, M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nagaraj N., Wisniewski J. R., Geiger T., Cox J., Kircher M., Kelso J., Paabo S., Mann M. (2011) Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lasonder E., Janse C. J., van Gemert G. J., Mair G. R., Vermunt A. M., Douradinha B. G., van Noort V., Huynen M. A., Luty A. J., Kroeze H., Khan S. M., Sauerwein R. W., Waters A. P., Mann M., Stunnenberg H. G. (2008) Proteomic profiling of Plasmodium sporozoite maturation identifies new proteins essential for parasite development and infectivity. PLoS Pathog. 4, e1000195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Florens L., Washburn M. P., Raine J. D., Anthony R. M., Grainger M., Haynes J. D., Moch J. K., Muster N., Sacci J. B., Tabb D. L., Witney A. A., Wolters D., Wu Y., Gardner M. J., Holder A. A., Sinden R. E., Yates J. R., Carucci D. J. (2002) A proteomic view of the Plasmodium falciparum life cycle. Nature 419, 520–526 [DOI] [PubMed] [Google Scholar]
- 20. Hall N., Karras M., Raine J. D., Carlton J. M., Kooij T. W., Berriman M., Florens L., Janssen C. S., Pain A., Christophides G. K., James K., Rutherford K., Harris B., Harris D., Churcher C., Quail M. A., Ormond D., Doggett J., Trueman H. E., Mendoza J., Bidwell S. L., Rajandream M. A., Carucci D. J., Yates J. R., 3rd, Kafatos F. C., Janse C. J., Barrell B., Turner C. M., Waters A. P., Sinden R. E. (2005) A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 307, 82–86 [DOI] [PubMed] [Google Scholar]
- 21. Jongco A. M., Ting L. M., Thathy V., Mota M. M., Kim K. (2006) Improved transfection and new selectable markers for the rodent malaria parasite Plasmodium yoelii. Mol. Biochem. Parasitol. 146, 242–250 [DOI] [PubMed] [Google Scholar]
- 22. Kennedy M., Fishbauger M. E., Vaughan A. V., Patrapuvich R., Boonhok R., Yimamnuaychok N., Rezakhani N., Metzger P., Ponpuak M., Sattabongkot J., Kappe S. H., Hume J. C. C., Lindner S. E. (2012) A rapid and scalable density gradient purification method for plasmodium sporozoites. Malar. J. 2012 Dec 17;11:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mack S. R., Vanderberg J. P., Nawrot R. (1978) Column separation of Plasmodium berghei sporozoites. J. Parasitol. 64, 166–168 [PubMed] [Google Scholar]
- 24. Aly A. S., Lindner S. E., MacKellar D. C., Peng X., Kappe S. H. (2011) SAP1 is a critical post-transcriptional regulator of infectivity in malaria parasite sporozoite stages. Mol. Microbiol. 79, 929–939 [DOI] [PubMed] [Google Scholar]
- 25. Vera I. M., Beatty W. L., Sinnis P., Kim K. (2011) Plasmodium protease ROM1 is important for proper formation of the parasitophorous vacuole. PLoS Pathog. 7, e1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Narum D. L., Nguyen V., Zhang Y., Glen J., Shimp R. L., Lambert L., Ling I. T., Reiter K., Ogun S. A., Long C., Holder A. A., Herrera R. (2008) Identification and characterization of the Plasmodium yoelii PyP140/RON4 protein, an orthologue of Toxoplasma gondii RON4, whose cysteine-rich domain does not protect against lethal parasite challenge infection. Infect. Immun. 76, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kessner D., Chambers M., Burke R., Agus D., Mallick P. (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 29. Yates J. R., 3rd, Eng J. K., McCormack A. L., Schieltz D. (1995) Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67, 1426–1436 [DOI] [PubMed] [Google Scholar]
- 30. Vaughan A., Chiu S. Y., Ramasamy G., Li L., Gardner M. J., Tarun A. S., Kappe S. H., Peng X. (2008) Assessment and improvement of the Plasmodium yoelii yoelii genome annotation through comparative analysis. Bioinformatics 24, i383-i389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Deutsch E. W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B., Eng J. K., Martin D. B., Nesvizhskii A. I., Aebersold R. (2010) A guided tour of the Trans-Proteomic Pipeline. Proteomics 10, 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shteynberg D., Deutsch E. W., Lam H., Eng J. K., Sun Z., Tasman N., Mendoza L., Moritz R. L., Aebersold R., Nesvizhskii A. I. (2011) iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell. Proteomics 10, M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 34. Gokce E., Andrews G. L., Dean R. A., Muddiman D. C. (2011) Increasing proteome coverage with offline RP HPLC coupled to online RP nanoLC-MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 879, 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deutsch E. W. (2010) The PeptideAtlas Project. Methods Mol. Biol. 604, 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lundgren D. H., Hwang S. I., Wu L., Han D. K. (2010) Role of spectral counting in quantitative proteomics. Expert Rev. Proteomics 7, 39–53 [DOI] [PubMed] [Google Scholar]
- 37. Santos J. M., Lebrun M., Daher W., Soldati D., Dubremetz J. F. (2009) Apicomplexan cytoskeleton and motors: key regulators in morphogenesis, cell division, transport and motility. Int. J. Parasitol. 39, 153–162 [DOI] [PubMed] [Google Scholar]
- 38. Matuschewski K., Ross J., Brown S. M., Kaiser K., Nussenzweig V., Kappe S. H. (2002) Infectivity-associated changes in the transcriptional repertoire of the malaria parasite sporozoite stage. J. Biol. Chem. 277, 41948–41953 [DOI] [PubMed] [Google Scholar]
- 39. Mikolajczak S. A., Silva-Rivera H., Peng X., Tarun A. S., Camargo N., Jacobs-Lorena V., Daly T. M., Bergman L. W., de la Vega P., Williams J., Aly A. S., Kappe S. H. (2008) Distinct malaria parasite sporozoites reveal transcriptional changes that cause differential tissue infection competence in the mosquito vector and mammalian host. Mol. Cell. Biol. 28, 6196–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mueller A. K., Labaied M., Kappe S. H., Matuschewski K. (2005) Genetically modified Plasmodium parasites as a protective experimental malaria vaccine. Nature 433, 164–167 [DOI] [PubMed] [Google Scholar]
- 41. Opitz C., Soldati D. (2002) ‘The glideosome’: a dynamic complex powering gliding motion and host cell invasion by Toxoplasma gondii. Mol. Microbiol. 45, 597–604 [DOI] [PubMed] [Google Scholar]
- 42. Kappe S. H., Buscaglia C. A., Nussenzweig V. (2004) Plasmodium sporozoite molecular cell biology. Annu. Rev. Cell Dev. Biol. 20, 29–59 [DOI] [PubMed] [Google Scholar]
- 43. Baum J., Richard D., Healer J., Rug M., Krnajski Z., Gilberger T. W., Green J. L., Holder A. A., Cowman A. F. (2006) A conserved molecular motor drives cell invasion and gliding motility across malaria life cycle stages and other apicomplexan parasites. J. Biol. Chem. 281, 5197–5208 [DOI] [PubMed] [Google Scholar]
- 44. Ganter M., Schuler H., Matuschewski K. (2009) Vital role for the Plasmodium actin capping protein (CP) beta-subunit in motility of malaria sporozoites. Mol. Microbiol. 74, 1356–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kursula I., Kursula P., Ganter M., Panjikar S., Matuschewski K., Schuler H. (2008) Structural basis for parasite-specific functions of the divergent profilin of Plasmodium falciparum. Structure 16, 1638–1648 [DOI] [PubMed] [Google Scholar]
- 46. Wesseling J. G., Snijders P. J., van Someren P., Jansen J., Smits M. A., Schoenmakers J. G. (1989) Stage-specific expression and genomic organization of the actin genes of the malaria parasite Plasmodium falciparum. Mol. Biochem. Parasitol. 35, 167–176 [DOI] [PubMed] [Google Scholar]
- 47. Meier B., Dobeli H., Certa U. (1992) Stage-specific expression of aldolase isoenzymes in the rodent malaria parasite Plasmodium berghei. Mol. Biochem. Parasitol. 52, 15–27 [DOI] [PubMed] [Google Scholar]
- 48. Billker O., Dechamps S., Tewari R., Wenig G., Franke-Fayard B., Brinkmann V. (2004) Calcium and a calcium-dependent protein kinase regulate gamete formation and mosquito transmission in a malaria parasite. Cell 117, 503–514 [DOI] [PubMed] [Google Scholar]
- 49. Garcia C. R., de Azevedo M. F., Wunderlich G., Budu A., Young J. A., Bannister L. (2008) Plasmodium in the postgenomic era: new insights into the molecular cell biology of malaria parasites. Int. Rev. Cell Mol. Biol. 266, 85–156 [DOI] [PubMed] [Google Scholar]
- 50. Khan S. M., Franke-Fayard B., Mair G. R., Lasonder E., Janse C. J., Mann M., Waters A. P. (2005) Proteome analysis of separated male and female gametocytes reveals novel sex-specific Plasmodium biology. Cell 121, 675–687 [DOI] [PubMed] [Google Scholar]
- 51. Moreno S. N., Ayong L., Pace D. A. (2011) Calcium storage and function in apicomplexan parasites. Essays Biochem. 51, 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ojo K. K., Pfander C., Mueller N. R., Burstroem C., Larson E. T., Bryan C. M., Fox A. M., Reid M. C., Johnson S. M., Murphy R. C., Kennedy M., Mann H., Leibly D. J., Hewitt S. N., Verlinde C. L., Kappe S., Merritt E. A., Maly D. J., Billker O., Van Voorhis W. C. (2012) Transmission of malaria to mosquitoes blocked by bumped kinase inhibitors. J. Clin. Invest. 122, 2301–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Singh S., Chitnis C. E. (2012) Signalling mechanisms involved in apical organelle discharge during host cell invasion by apicomplexan parasites. Microbes Infect. 14, 820–824 [DOI] [PubMed] [Google Scholar]
- 54. Sebastian S., Brochet M., Collins M. O., Schwach F., Jones M. L., Goulding D., Rayner J. C., Choudhary J. S., Billker O. (2012) A Plasmodium calcium-dependent protein kinase controls zygote development and transmission by translationally activating repressed mRNAs. Cell Host Microbe 12, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kebaier C., Vanderberg J. P. (2010) Initiation of Plasmodium sporozoite motility by albumin is associated with induction of intracellular signalling. Int. J. Parasitol. 40, 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Doi Y., Shinzawa N., Fukumoto S., Okano H., Kanuka H. (2011) Calcium signal regulates temperature-dependent transformation of sporozoites in malaria parasite development. Exp. Parasitol. 128, 176–180 [DOI] [PubMed] [Google Scholar]
- 57. Singh S., Alam M. M., Pal-Bhowmick I., Brzostowski J. A., Chitnis C. E. (2010) Distinct external signals trigger sequential release of apical organelles during erythrocyte invasion by malaria parasites. PLoS Pathog. 6, e1000746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Raabe A. C., Wengelnik K., Billker O., Vial H. J. (2011) Multiple roles for Plasmodium berghei phosphoinositide-specific phospholipase C in regulating gametocyte activation and differentiation. Cell. Microbiol. 13, 955–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Harvey K. L., Gilson P. R., Crabb B. S. (2012) A model for the progression of receptor-ligand interactions during erythrocyte invasion by Plasmodium falciparum. Int. J. Parasitol. 42, 567–573 [DOI] [PubMed] [Google Scholar]
- 60. Chen Z., Harb O. S., Roos D. S. (2008) In silico identification of specialized secretory-organelle proteins in apicomplexan parasites and in vivo validation in Toxoplasma gondii. PloS One 3, e3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sam-Yellowe T. Y., Banks T. L., Fujioka H., Drazba J. A., Yadav S. P. (2008) Plasmodium yoelii: novel rhoptry proteins identified within the body of merozoite rhoptries in rodent Plasmodium malaria. Exp. Parasitol. 120, 113–117 [DOI] [PubMed] [Google Scholar]
- 62. Giovannini D., Spath S., Lacroix C., Perazzi A., Bargieri D., Lagal V., Lebugle C., Combe A., Thiberge S., Baldacci P., Tardieux I., Menard R. (2011) Independent roles of apical membrane antigen 1 and rhoptry neck proteins during host cell invasion by apicomplexa. Cell Host Microbe 10, 591–602 [DOI] [PubMed] [Google Scholar]
- 63. Blume M., Hliscs M., Rodriguez-Contreras D., Sanchez M., Landfear S., Lucius R., Matuschewski K., Gupta N. (2011) A constitutive pan-hexose permease for the Plasmodium life cycle and transgenic models for screening of antimalarial sugar analogs. FASEB J. 25, 1218–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wass M. N., Stanway R., Blagborough A. M., Lal K., Prieto J. H., Raine D., Sternberg M. J., Talman A. M., Tomley F., Yates J., Sinden R. E. (2012) Proteomic analysis of Plasmodium in the mosquito: progress and pitfalls. Parasitology 2012 Aug;139(9):1131–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Washburn M. P., Wolters D., Yates J. R., 3rd (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 66. Madeira L., Galante P. A., Budu A., Azevedo M. F., Malnic B., Garcia C. R. (2008) Genome-wide detection of serpentine receptor-like proteins in malaria parasites. PloS One 3, e1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Carucci D. J., Witney A. A., Muhia D. K., Warhurst D. C., Schaap P., Meima M., Li J. L., Taylor M. C., Kelly J. M., Baker D. A. (2000) Guanylyl cyclase activity associated with putative bifunctional integral membrane proteins in Plasmodium falciparum. J. Biol. Chem. 275, 22147–22156 [DOI] [PubMed] [Google Scholar]
- 68. Ward P., Equinet L., Packer J., Doerig C. (2004) Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics 5, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dyer M., Jackson M., McWhinney C., Zhao G., Mikkelsen R. (1996) Analysis of a cation-transporting ATPase of Plasmodium falciparum. Mol. Biochem. Parasitol. 78, 1–12 [DOI] [PubMed] [Google Scholar]
- 70. Eckstein-Ludwig U., Webb R. J., Van Goethem I. D., East J. M., Lee A. G., Kimura M., O'Neill P. M., Bray P. G., Ward S. A., Krishna S. (2003) Artemisinins target the SERCA of Plasmodium falciparum. Nature 424, 957–961 [DOI] [PubMed] [Google Scholar]
- 71. Cardi D., Pozza A., Arnou B., Marchal E., Clausen J. D., Andersen J. P., Krishna S., Moller J. V., le Maire M., Jaxel C. (2010) Purified E255L mutant SERCA1a and purified PfATP6 are sensitive to SERCA-type inhibitors but insensitive to artemisinins. J. Biol. Chem. 285, 26406–26416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cui L., Wang Z., Jiang H., Parker D., Wang H., Su X. Z. (2012) Lack of association of the S769N mutation in Plasmodium falciparum SERCA (PfATP6) with resistance to artemisinins. Antimicrob. Agents Chemother. 56, 2546–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Green J. L., Rees-Channer R. R., Howell S. A., Martin S. R., Knuepfer E., Taylor H. M., Grainger M., Holder A. A. (2008) The motor complex of Plasmodium falciparum: phosphorylation by a calcium-dependent protein kinase. J. Biol. Chem. 283, 30980–30989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hegge S., Kudryashev M., Barniol L., Frischknecht F. (2010) Key factors regulating Plasmodium berghei sporozoite survival and transformation revealed by an automated visual assay. FASEB J. 24, 5003–5012 [DOI] [PubMed] [Google Scholar]
- 75. Baum J., Gilberger T. W., Frischknecht F., Meissner M. (2008) Host-cell invasion by malaria parasites: insights from Plasmodium and Toxoplasma. Trends Parasitol. 24, 557–563 [DOI] [PubMed] [Google Scholar]
- 76. Barale J. C., Blisnick T., Fujioka H., Alzari P. M., Aikawa M., Braun-Breton C., Langsley G. (1999) Plasmodium falciparum subtilisin-like protease 2, a merozoite candidate for the merozoite surface protein 1–42 maturase. Proc. Natl. Acad. Sci. U.S.A. 96, 6445–6450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hackett F., Sajid M., Withers-Martinez C., Grainger M., Blackman M. J. (1999) PfSUB-2: a second subtilisin-like protein in Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 103, 183–195 [DOI] [PubMed] [Google Scholar]
- 78. Mauduit M., Gruner A. C., Tewari R., Depinay N., Kayibanda M., Chavatte J. M., Franetich J. F., Crisanti A., Mazier D., Snounou G., Renia L. (2009) A role for immune responses against non-CS components in the cross-species protection induced by immunization with irradiated malaria sporozoites. PloS One 4, e7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mauduit M., Tewari R., Depinay N., Kayibanda M., Lallemand E., Chavatte J. M., Snounou G., Renia L., Gruner A. C. (2010) Minimal role for the circumsporozoite protein in the induction of sterile immunity by vaccination with live rodent malaria sporozoites. Infect. Immun. 78, 2182–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Oehring S. C., Woodcroft B. J., Moes S., Wetzel J., Dietz O., Pulfer A., Dekiwadia C., Maeser P., Flueck C., Witmer K., Brancucci N. M., Niederwieser I., Jenoe P., Ralph S. A., Voss T. S. (2012) Organellar proteomics reveals hundreds of novel nuclear proteins in the malaria parasite Plasmodium falciparum. Genome Biol. 2012 Nov 26;13(11):R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.