Abstract
The cyanobacterium Synechocystis sp. PCC 6803 possesses two leader peptidases, LepB1 (Sll0716) and LepB2 (Slr1377), responsible for the processing of signal peptide-containing proteins. Deletion of the gene for LepB1 results in an inability to grow photoautotrophically and an extreme light sensitivity. Here we show, using a combination of Blue Native/SDS-PAGE, Western blotting and iTRAQ analysis, that lack of LepB1 strongly affects the cell's ability to accumulate wild-type levels of both photosystem I (PSI) and cytochrome (Cyt) b6f complexes. The impaired assembly of PSI and Cyt b6f is considered to be caused by the no or slow processing of the integral subunits PsaF and Cyt f respectively. In particular, PsaF, one of the PSI subunits, was found incorporated into PSI in its unprocessed form, which could influence the assembly and/or stability of PSI. In contrast to these results, we found the amount of assembled photosystem II (PSII) unchanged, despite a slower processing of PsbO. Thus, imbalance in the ratios of PSI and Cyt b6f to photosystem II leads to an imbalanced photosynthetic electron flow up- and down-stream of the plastoquinone pool, resulting in the observed light sensitivity of the mutant. We conclude that LepB1 is the natural leader peptidase for PsaF, PsbO, and Cyt f. The maturation of PsbO and Cyt f can be partially performed by LepB2, whereas PsaF processing is completely dependent on LepB1. iTRAQ analysis also revealed a number of indirect effects accompanying the mutation, primarily a strong induction of the CydAB oxidase as well as a significant decrease in phycobiliproteins and chlorophyll/heme biosynthesis enzymes.
Cyanobacteria comprise a diverse group of photoautotrophic prokaryotes with an oxygen evolving photosynthetic apparatus very similar to that of higher plants. Similarly to plant chloroplasts, they contain three different types of membranes, an outer membrane, a plasma membrane (PM), and a thylakoid membrane (TM). The thylakoid membrane is the site not only for photosynthesis but also the main site for respiration. The ultrastructure and organization of the membranes is however still under debate and mainly two different opinions prevail concerning the organization of plasma and thylakoid membranes. One is that the two membranes are continuous, making the periplasm and lumen a common compartment, the second that the membranes are completely separated (1–9).
The complex membrane organization within the cyanobacterium implies the existence of a sophisticated system for the sorting and transport of extracytoplasmic proteins to the different membranes and compartments. Targeting of these proteins in eubacteria and archaea occurs through the well-characterized Sec and Tat translocon pathways (10). Protein substrates for the Sec and Tat systems were shown to be distinguished by specific N-terminal signal sequences (11). However, most thylakoid and plasma membrane integral proteins do not have N-terminal signal peptides. For insertion into the membrane all TM and PM integral proteins require, in addition to the Sec translocon, a protein insertase of the Alb3/Oxa1 family, YidC (12–14), as shown for the core photosystem II (PSII) protein D1 (PsbA) (15, 16). The Synechocystis sp. PCC 6803 (hereafter referred to as Synechocystis) genome, as well as all other completely sequenced cyanobacterial genomes, contains only a single set of genes encoding the subunits of the Sec and Tat translocons (17–19). This suggests that the same translocation machineries are working in plasma and thylakoid membranes and this in turn indicates that information additional to the signal sequence is needed for correct targeting (20, 21). The question of where in the intricate Synechocystis membrane system translocation and protein sorting takes place is still subject for intense research.
Sec and Tat signal peptides are both cleaved by Type I signal peptidases (also called leader peptidase, Lep) (11). In contrast to other Gram-negative bacteria, almost all the cyanobacterial genomes analyzed so far contain two lep genes. Although the overall sequence identity of leader peptidases from various species is relatively low, five conserved regions were identified in the catalytic domain, called boxes B, C, D, and E, as well as the membrane anchor domain A (22, 23). Both LepB1 and LepB2 in Synechocystis have the conserved A-E boxes (21, 23), including the invariant amino acid residues demonstrated to be essential for catalytic activity. No other proteins with these conserved regions are found in the Synechocystis genome.
In subproteomic analyses LepB2 was found in the plasma membrane, whereas LepB1 was found in the thylakoid membrane (7, 24, 25). Both LepB1 and LepB2 are integral membrane proteins with one transmembrane helix, present in very low amounts in the membrane. The above observations may therefore not implicate completely unique membrane localizations for the two leader peptidases. Knockout mutations of the two genes, however, give significantly different results: whereas LepB2 appears to be totally essential for cell viability, cells with a deletion in LepB1 are still able to grow under heterotrophic conditions in dim light (26). The light sensitivity disappeared when 3-(3,4-dichlorophenyl)-1,1-dimetylurea (DCMU)1 was present. DCMU inhibits the reduction of the plastoquinone (PQ) pool by photosystem II (PSII) and this indicates that the primary cause for the photosensitivity observed is the impairment in electron flow downstream of PSII. The reported decrease in the amount of cytochrome f (Cyt f), a component of the cytochrome b6f (Cyt b6f) complex, further strengthens this hypothesis (26).
In the present study, we have analyzed the LepB1 mutant strain by using two-dimensional Blue Native/SDS-PAGE (2D BN/SDS-PAGE), Western blotting with specific antibodies and isobaric tags for relative and absolute quantitation (iTRAQ), with special emphasis on the integrity of the photosynthetic apparatus, to reach a better understanding of the role of LepB1 in protein maturation in Synechocystis. Our studies show that LepB1 has irreplaceable substrate specificity, as in the case of PsaF, an integral membrane subunit of the photosystem I (PSI). The processing of other photosynthetic proteins (PsbO and Cyt f) is also affected although LepB2 is able to partly compensate the loss of LepB1. This defect in processing severely disrupts the ability to maintain proper organization of the thylakoid membrane complexes and also affects the cellular proteome in a far-reaching manner.
EXPERIMENTAL PROCEDURES
Cell Culture and Growth Conditions
Synechocystis sp. PCC 6803 glucose-sensitive strain (Wild type, WT GS) and the sll0716 gene inactivation mutant strain (ΔLepB1) were grown under heterotrophic conditions in BG-11 medium in the presence of 5 mm glucose (an amount well tolerated by the WT GS strain under the conditions used in this study) (27). Independent cultures, 300 ml for each strain, were maintained at 32 °C, under dim light (<1 μE m−2 s−1 of warm white light) and vigorous bubbling with filtered air (0.45 μm, Millipore Millex). ΔLepB1 was routinely grown in the presence of 25 μg/ml kanamycin (26). Cell growth was monitored by measuring OD730 with a spectrophotometer (BioSpec-Mini, Shimadzu).
Isolation of the Total Membrane Fraction
The membrane fractions were prepared as described elsewhere (28), with slight modifications. Three independent cultures for each strain were harvested in mid-logarithmic phase (OD730 = 1) by centrifugation at 6000 × g, 4 °C, 10 min. Cultures from each strain were resuspended in washing buffer (50 mm HEPES-NaOH, pH 7.5, and 30 mm CaCl2), mixed in equal proportion and recentrifuged in the same conditions to have a pooled sample for each condition, containing cells from three independent cultures. Sample pooling was previously shown to be an effective mean to reduce the impact of biological variation even though it may hinder detection of differential expression when biological variability is small (29). Cells were broken by vigorous vortexing for 4 × 2 min with glass beads (0.15–0.212 mm in diameter) in isolation buffer (50 mm HEPES-NaOH, pH 7.5, 30 mm CaCl2, 800 mm sorbitol, and 1 mm ε-amino-n-caproic acid), supplemented with 1 mm phenylmethylsulfonyl fluoride (30). The glass beads were washed five times with isolation buffer. Whole cell lysates obtained from each wash were collected together and centrifuged at 4500 × g, 4 °C, 10 min, to remove unbroken cells and cell debris. Total membrane and soluble fractions were separated by centrifugation at 230,000 × g, 4 °C for 1 h. The pellet (total membrane fraction) was washed three times with washing buffer, and UV-visible spectra of each supernatant wash were recorded (using a Varian CARY 300 Bio; Agilent, Santa Clara, CA) to monitor the content of phycobiliproteins until no obvious phycocyanin peak could be observed (31). The washed membrane samples were resuspended in storage buffer (50 mm HEPES- NaOH, pH 7.5, 600 mm sucrose, 30 mm CaCl2, and 1 m glycinebetaine, 1 mm PMSF) and equal protein amounts (20 μg) were used to record UV-Vis spectra, to check the pigment content prior to freezing at −80 °C.
Sample Preparation for 2D BN/SDS-PAGE
2D BN/SDS-PAGE was performed as previously described (28), with some modifications. Isolated total membranes corresponding to 150 μg protein (quantified using the Peterson method (32)) were washed once with 10 volumes of washing buffer (300 mm sorbitol, 50 mm Bis-Tris, pH 7.0, and 250 μg/ml of Pefabloc [Roche Applied Science]) and resuspended in 7 μl resuspension buffer, consisting of 20% glycerol (w/v), 25 mm Bis-Tris (pH 7.0), 10 mm MgCl2, and 0.1 unit/μl DNaseI (Sigma-Aldrich). The samples were then gently mixed with an equal volume of resuspension buffer supplemented with 4% n-dodecyl-β-d-maltoside (DDM, Sigma-Aldrich). Solubilization was performed for 50 min on ice, following by incubation at room temperature for 10 min. Insoluble material was removed by centrifugation at 18,000 × g for 15 min (4 °C) and the solubilized membrane protein complexes were mixed with one-tenth volume of Coomassie blue solution, consisting of 100 mm Bis Tris pH 7.0, 30% (w/v) sucrose, 500 mm ε-amino-n-caproic acid, 10 mm EDTA pH 7.0, 5% (w/v) Serva Blue G. The samples were loaded onto a 0.75 mm thick 5–12.5% acrylamide gradient gel (Hoefer SE250, GE Healthcare) and electrophoresis was performed at 4 °C, on ice, by increasing voltage gradually from 50 V up to 200V during the run.
Lanes from the BN gel were excised and equilibrated in 2 × SDS sample buffer containing 10% β-mercaptoethanol and 8 m Urea for 45 min at room temperature. The gel pieces were covered by a small volume of SDS sample buffer and fresh buffer was added during the incubation period to avoid gel drying. The lanes were rinsed in SDS-PAGE running buffer and laid onto in-house-cast 1 mm thick 14% SDS-PAGE gels with 6 m urea (Hoefer SE260, GE Healthcare) (33). After electrophoresis proteins were visualized by Coomassie Brilliant Blue R250 staining solution (Bio-Rad).
Immunoblotting
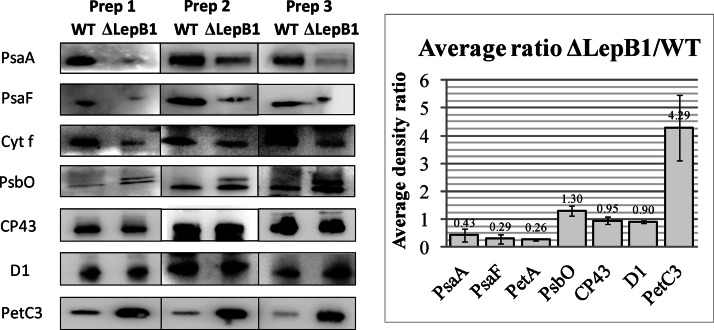
WT and ΔLepB1 samples containing 10 μg protein in SDS-PAGE loading buffer with 4 m urea were applied on precast 12% acrylamide gels (Mini-Protean TGX gels; Bio-Rad, Hercules, CA) or in-house-cast 1 mm thick 12% SDS-PAGE gel (Hoefer SE250, GE Healthcare) with 6 m urea (33). For immunodetection the proteins were electrotransferred with a semidry blotter (Trans-Blot S.D., Bio-Rad) to PVDF membranes (Hybond-P, GE Healthcare). Membranes were blocked in 5% fat-free milk and incubated with primary antibodies as indicated in Fig. 3. Advanced ECL Prime (GE Healthcare) was used to develop the membrane after incubation with horseradish peroxidase-conjugated secondary antibodies (GE Healthcare). All the blots were done at least three times with different preparations from independent cultures. The images were captured using an ImageQuant LAS 4000 system equipped with a Fujifilm Super CCD camera and were quantified using the MultiGauge software (v 3.0, Fuji). Antibodies against PsaA, CP43 and D1 were purchased from Agrisera AB, PsbO antibodies from AbCam. Cyt f and PetC3 antibodies were kind gifts from M. Rogner, PsaF antibodies a gift from J. D. Rochaix.
Fig. 3.
Western blot analyses comparing expression of proteins in WT and ΔLepB1. Samples containing 10 μg protein were loaded in each lane. Dilutions of primary antibodies (in TBS-T) were as follows: anti-PsaA and anti-PsaF 1:1000; anti-Cyt f, 1:500; anti-PsbO and anti-CP43, 1:3000; anti-D1, 1:5000; anti-PetC3, 1:2000. All individual blots were carried out three times using samples from independent cultures. Band intensities were quantified using MultiGauge and used to calculate the average ΔLepB1/WT ratios of the different proteins (histogram). Error bars refer to standard deviation in the averaging calculations.
iTRAQ Proteome Analysis
The iTRAQ proteome analysis was performed as previously described (34) with some modifications. Three independent cultures were pooled together to make the total membrane fractions, as described above. Two hundred and fifty micrograms of total membrane proteins from the WT and LepB1 mutant strains were separated on 12% precast SDS-PAGE gels. The gel was run for one fourth of its total length for sample concentration and pigment removal. Lanes of each sample were sliced and diced into small pieces. Gel pieces were extensively washed with 50 mm triethylammonium bicarbonate (TEAB) buffer and then dehydrated by acetonitrile (ACN). Proteins were reduced by 5 mm Tris-(2-carboxyethyl)-phosphine (TCEP) in TEAB buffer for 1 h at 60 °C and alkylated by 10 mm methyl methanethiosulfate (MMTS) in TEAB for 30 min at RT in the dark. In-gel digestion was performed at 37 °C overnight. The tryptic peptides were extracted by 5% acetic acid in 50% ACN and vacuum dried by SpeedVac.
iTRAQ Labeling and ERLIC Fractionation
The tryptic peptides were labeled with four-plex iTRAQ reagents (Applied Biosystems, Foster City, CA) according to the instructions from the manufacturer. Two different labeling schemes were applied in our experiments to minimize both systematic and experimental errors- Experiment 1: WT, 114; ΔLepB1, 115/116/117 and Experiment 2: WT, 114/116, ΔLepB1, 115/117. These labeled peptides were pooled and desalted using Sep-Pak C18 cartridges (Waters) and vacuum dried. The mixture of iTRAQ-labeled peptides was fractionated using a PolyWAX LP weak anion-exchange column (4.6 × 200 mm; 5 μm, 300 Å; PolyLC) on the same UFLC system, with UV detection monitored at a wavelength of 280 nm. Mobile Phase A (10 mm CH3COONH4 in 85% ACN/1% formic acid [FA]) and mobile phase B (30% ACN/0.1% FA) were used to establish a 50 min gradient of 0–28% buffer B for 40 min, 28–100% buffer B for 5 min, and 100% buffer B for 5 min at a flow rate of 1 ml/min, with 30 fractions collected. The peptides were dissolved in 200 μl mobile phase A for complete solution before being loaded to the column. Mobile phase A was prepared by dissolving CH3COONH4directly in 85% ACN/1% FA without any adjustment of its pH. The collected fractions were then dried by vacuum centrifuge at 45 °C, combined into 20 fractions and reconstituted in 3% ACN/0.1% FA for LC-MS/MS analysis.
LC-MS/MS Analysis
The peptides were separated and analyzed on a home-packed nanobore C18 column (15 cm x 75 μm; 5 μm particles) with a Picofrit nanospray tip (New Objectives, Woburn, MA) on a TempoTM nano-MDLC system coupled with a QSTAR® Elite Hybrid LC-MS/MS system (Applied Biosystems). Peptides from each fraction were analyzed in triplicate by LC-MS/MS over a gradient of 90 min. The flow rate of the LC system was set to a constant 300 nl/min. Data acquisition in QSTAR Elite was set to positive ion mode using Analyst® QS 2.0 software (Applied Biosystems). MS data was acquired in positive ion mode with a mass range of 300–1600 m/z. Peptides with +2 to +4 charge states were selected for MS/MS. For each MS spectrum, the three most abundant peptides above a five-count threshold were selected for MS/MS and dynamically excluded for 30 s with a mass tolerance of 0.03 Da. Smart information-dependent acquisition was activated with automatic collision energy and automatic MS/MS accumulation. The fragment intensity multiplier was set to 20 and maximum accumulation time was 2 s.
Data Analysis
Protein identification and quantification were performed using ProteinPilot Software Version 2.01 (Applied Biosystems). The Paragon algorithm in the ProteinPilot software was used for the peptide identification, further processed by the Pro Group algorithm, where isoform-specific quantification was adopted to trace the differences between expressions of various isoforms. User defined parameters were as follows: (1) Sample Type, iTRAQ 4-plex (Peptide Labeled); (2) Cysteine alkylation, MMTS; (3) Digestion, Trypsin; (4) Instrument, QSTAR ESI; (5) Special factors, None; (6) Species, None; (7) Specify Processing, Quantitate; (8) ID Focus, biological modifications, amino acid substitutions; (9) Database, the Synechocystis PCC 6803 protein sequence database (from 2004) was downloaded from Cyanobase (http://genome.kazusa.or.jp/cyanobase/Synechocystis/genes.faa), which includes 3672 sequences and 1143081 residues; (10) Search effort, thorough; (11) MS and MS/MS mass tolerance, 0.2 Da. Protein confidence was set at 95% (equivalent to Unused ProtScore of 1.3). The peptide for iTRAQ quantification was automatically selected by the Pro Group algorithm to calculate the reporter peak area, error factor (EF) and p value. The resulting data was auto bias-corrected by the built-in ProteinPilot algorithm to eliminate any variations imparted because of the unequal mixing during combination of differentially labeled samples. During bias correction, the software identifies the median average protein ratio, corrects it to unity and then applies this factor to all results for quantification (35).
Respiration Measurements
Cells from three independent cultures for each strain were collected at OD730 = 1 by low speed centrifugation (1500 × g) for 10 min at room temperature (RT). The cell pellet was gently washed once with BG-11 and resuspended to a final OD730 = 5 in fresh BG-11medium supplemented with 5 mm glucose. All cell culture concentration steps were performed under dim light. The concentrated cell suspensions were stirred with a magnetic stirrer to prevent cell sinking. Respiratory electron transport was measured at RT by using a Clark-type electrode (Hansatech), as described below. One milliliter of the cell suspension was added in the electrode chamber which was immediately covered with a thick black cloth. After 5 min of dark incubation, different electron transport inhibitors (KCN, PCP, or DBMIB) were added from freshly prepared stock solutions to the final concentrations, as indicated in the Results section. Following a further 5 min incubation, respiratory rates were read and calculated using the Oxygraph Plus software (Hansatech) using data up to 2 min of the start of the measurement. All individual experiments were repeated three times under the same conditions.
RESULTS
Deletion of LepB1 Specifically Decreases the Amount of PSI and Cyt b6f Complexes but not PSII
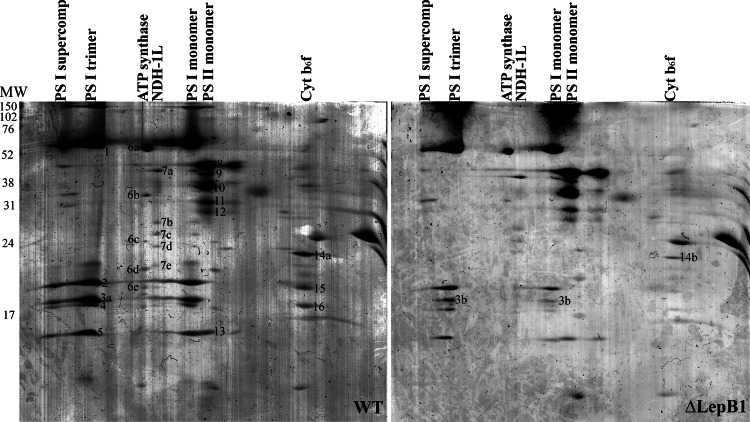
To gain insight into the consequences of the LepB1 deletion for the organization of the photosynthetic apparatus, total membranes from WT and ΔLepB1 strains were analyzed by BN-PAGE (Fig. 1), which has been shown to be a powerful tool in the analysis of membrane protein complexes in cyanobacteria (36). A remarkable decrease in the amount of PSI complexes, both in the trimer and the monomer form in ΔLepB1 (Figs. 1 and 2) was observed on the gel. In contrast, the amount of PSII was approximately the same. BN gel lanes for the WT and ΔLepB1 were further separated in the second dimension SDS-PAGE gel (Fig. 2), to resolve the protein composition of each complex. Spots from different complexes were identified by LC-MS/MS, confirming their respective identities (supplemental Data File S1). The 2D gel of ΔLepB1 showed a general decrease of all observable PSI subunits in comparison to WT (Fig. 2). The Cyt b6f complex could not be identified in the first dimension BN-PAGE gel. However, after the second dimension SDS gel run three subunits of Cyt b6f could be identified on the WT gel, corresponding to Cyt f (spot 14a), Cyt b6 (spot 15), as well as subunit IV (spot 16). In the ΔLepB1 gel, three weak spots could be observed in the same positions but, of these, only Cyt f (spot 14b) could be identified by LC-MS/MS (Fig. 2 and supplemetal Data File S1).
Fig. 1.
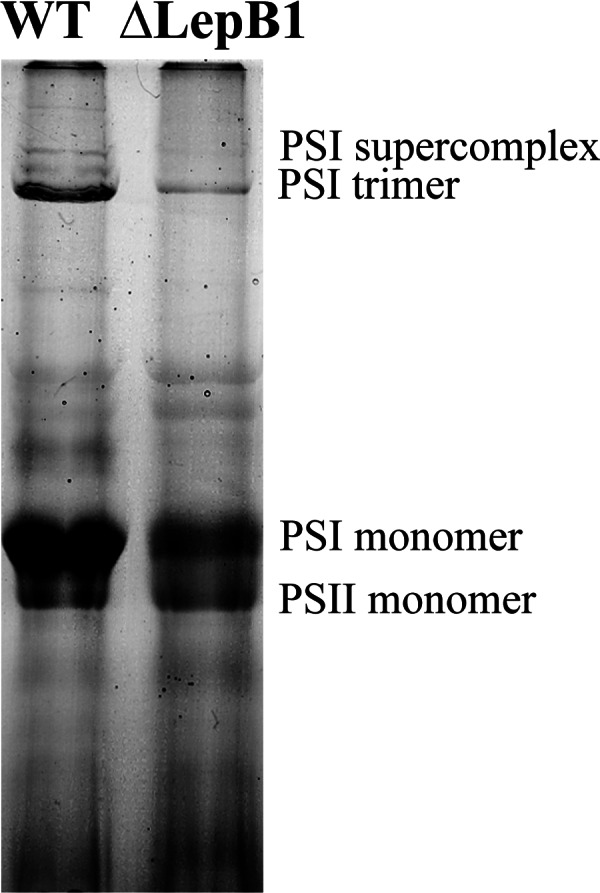
First dimension BN/PAGE separation of membrane protein complexes of WT and ΔLepB1. Membrane proteins (150 μg) were solubilized in 2% DDM, as described in “Experimental Procedures” and ran in a 5–12.5% acrylamide gradient gel.
Fig. 2.
Two-dimensional BN/SDS-PAGE separation of membrane protein complexes of WT and ΔLepB1. Protein complexes in WT and ΔLepB1 lanes from the first dimension were separated into their subunits by denaturating 14% SDS-PAGE containing 6 m urea. The numbered protein spots were excised from the Coomassie blue stained gel and identified by LC-MS/MS (See supplemental Data File S1).
PSII is mainly detectable in its monomer form in our membrane samples, both for WT and ΔLepB1, with similar intensity as shown in Fig. 1. In the second dimension SDS-PAGE, none of the four core subunits of PSII - CP47 (spot 8), CP43 (spot 10), D2 (spot 11), and D1 (spot 12), identified by LC-MS/MS - appeared to change in abundance in the mutant, in comparison to the WT (Fig. 2 and supplemental Data File S1).
To complement this analysis, several antibodies were used to compare the amount of specific proteins in total membrane samples of WT and ΔLepB1. Blot signal intensities from three independent preparations were quantified (using MultiGauge - Fig. 3) and presented in the form of a histogram, with results expressed as abundance ratios between ΔLepB1 and WT. In agreement with the results from 2D/BN-PAGE, the PSII core subunits CP43 and D1 remained largely unchanged, whereas the PSI core subunit PsaA and the Cyt b6f complex subunit Cyt f decreased to less than half the WT amounts in ΔLepB1 (Fig. 3).
Influence of the LepB1 Deletion on Preprotein Maturation
Out of 300–350 Synechocystis proteins with predicted (37) or verified signal sequences only limited numbers are part of the photosynthetic apparatus or important for its proper function and maintenance. PsaF in PSI and Cyt f in Cyt b6f complex, both integral membrane subunits, are synthesized with presequences and integrated into the respective complexes (38–40). PSII contains at least three proteins with Sec-signal peptides, PsbO, PsbU, and PsbV, which constitute the membrane associated oxygen evolving complex (OEC), situated on the lumenal side of the PSII complex (41). As potential substrates of LepB1, PsaF and Cyt f, as well as PsbO, were investigated by either Western blots or LC-MS/MS.
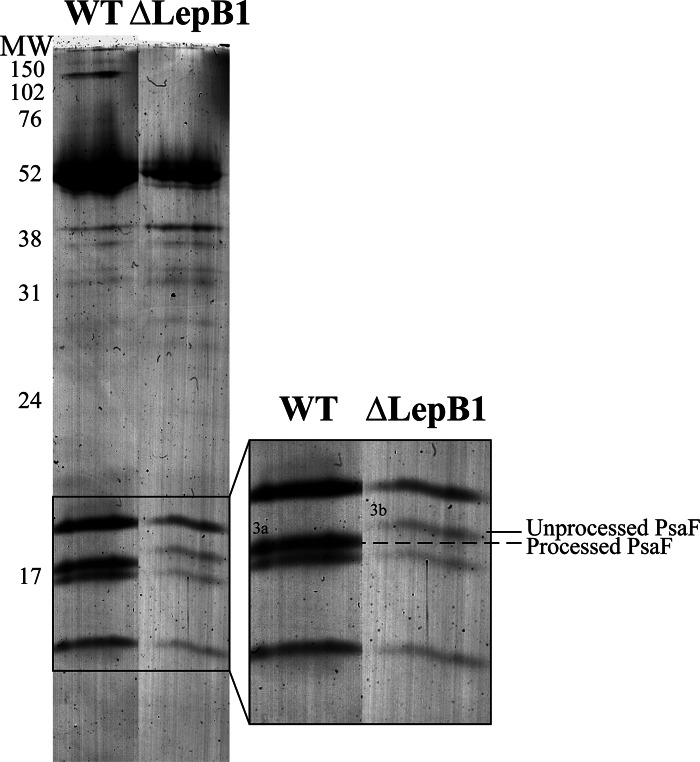
PSI monomer bands from the WT and the ΔLepB1 were cut out from the BN gel and separated on an SDS gel side by side (Fig. 4), more differences could be seen on the gel apart from the remarkable decrease of the amount in the mutant. Both cell types have four bands in the low molecular area, but it is clearly seen that the second band from the top migrates at a position 2–3 kDa higher in the mutant (marked with 3b in Fig. 4) than in the WT (marked with 3a in Fig. 4). The identity of these bands (3a and 3b) as PsaF was confirmed by LC-MS/MS as well as Western blot (Figs. 2, 3, and 4 and supplemental Data File S1). Synechocystis PsaF was previously shown to be an integral membrane protein with a cleavable signal peptide in the first 23 amino acids (38) and the 2–3 kDa increase in band 3b corresponds well with what is expected for pre-PsaF (38). All our attempts to identify PsaF presequence peptides by LC-MS/MS failed, most probably because of their highly hydrophobic nature. No processed PsaF was observed, either in 2D BN/SDS-PAGE gels (Fig. 4) or in Western blots (Fig. 3) of ΔLepB1 total membranes. It can be concluded that the majority of PSI complexes in the mutant contain PsaF in the unprocessed form (Fig. 2).
Fig. 4.
PSI monomer band from WT and ΔLepB1 separated into subunits by SDS-PAGE. PSI monomer bands were solubilized as described for the whole BN gel lane and ran side by side on 14% SDS-PAGE with 6 m urea. The area of the low molecular subunits was enlarged in the right part, showing the difference in migration between PsaF in WT and ΔLepB1. The band in WT (3a) and ΔLepB1 (3b) was excised for LC-MS/MS analysis.
Cyt f and PsbO, were both affected by the LepB1 mutation, as shown in Fig. 3. Cyt f decreased in the mutant, similarly to PsaF. However, in contrast to PsaF, cyt f is only found in the processed form, which is in agreement with the prior observations of Zhbanko et al. (26). The signal peptide cleavage site of PsbO was previously verified and the mature protein has a MW that is 3 kDa lower than the preprotein (41). Using a PsbO specific antibody, we observed an accumulation of small amounts of pre-PsbO protein, despite the lack of decrease in the amount of mature PsbO. Consequently, 30% more of total PsbO (processed plus unprocessed) was found in the mutant (Fig. 3).
Protein Abundance as Revealed by iTRAQ
Our analysis using 2D BN/SDS-PAGE and specific antibodies revealed differences in the amount of some membrane complexes in the mutant. To obtain a more complete analysis of the changes induced by the mutation we subjected the membranes to iTRAQ. iTRAQ has previously revealed useful information in understanding the changes in the Synechocystis proteomic response to CO2 limitation (42), increased salt concentration (43), and temperature shifts (44). The total membranes of WT and ΔLepB1 were analyzed by combining iTRAQ and LC-MS/MS with SDS-PAGE gel-assisted digestion. Approximately 1350 proteins from Experiment 1 (Exp. 1) and 1200 proteins from Experiment 2 (Exp. 2) were identified with over 95% confidence (Unused ProtScore > 1.3), of which more than 90% of the proteins were overlapping in the two experiments. About 470 and 440 proteins were confidently quantified having p value < 0.05, respectively. All three data sets from the Exp. 1, together with the four data sets from the Exp. 2 were combined for statistical analysis. The geometric mean of the iTRAQ ratios and standard deviation (S.D. log ratios) of each identified protein from the seven sets of iTRAQ ratios of the two independent experiments were calculated, as shown in supplemental Data File S2. Only the proteins identified in both of the iTRAQ experiments with p value < 0.05 and ΔLepB1/WT <0.7 and >1.5 fold difference in expression were selected as significantly down and up-regulated proteins (42, 44). There were 46 down-regulated and 11 up-regulated proteins in the mutant (supplemental Data File S2), in comparison with the WT. Of the down-regulated eight proteins have a predicted/proven signal peptide and of the up-regulated three. Most of the quantified proteins were not significantly changed in the lepB1 mutant under the growth conditions used in this work. Table I shows the selected proteins present in the membrane fraction relevant to photosynthesis and respiration, which will be further described and discussed.
Table I. The ratio of expression LepB1 mutant/WT for protein complexes/proteins involved/related to photosynthesis and respiration. In the first iTRAQ analyses WT was labeled with iTRAQ reagent 114 and LepB1 mutant with 115, 116, and 117. In the second iTRAQ analyses WT was labeled with 114 and 116 and lepB1 mutant with 115 and 117.
| ORF | Name | TMHMMa, SPb | 1st iTRAQ |
2nd iTRAQ |
Average ratio (geometric) | S.D. (log scale) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 115:114 | 116:114 | 117:114 | 115:114 | 117:114 | 115:116 | 117:116 | |||||
| PSI, plastocyanin | |||||||||||
| Slr1834 | PsaA | 11 | 0.42 | 0.42 | 0.43 | 0.58 | 0.49 | 0.60 | 0.51 | 0.49 | 0.14 |
| Slr1835 | PsaB | 11 | 0.44 | 0.47 | 0.44 | 0.61 | 0.54 | 0.62 | 0.55 | 0.52 | 0.13 |
| Sll0819 | PsaF | 2, sec | 0.36 | 0.36 | 0.40 | 0.55 | 0.47 | 0.57 | 0.48 | 0.45 | 0.18 |
| Slr1655 | PsaL | 3 | 0.36 | 0.43 | 0.46 | 0.55 | 0.48 | 0.54 | 0.47 | 0.46 | 0.14 |
| Ssl0563 | PsaC | 0.48 | 0.49 | 0.27 | 0.59 | 0.45 | 0.63 | 0.47 | 0.47 | 0.25 | |
| Slr0737 | PsaD | 0.27 | 0.27 | 0.33 | 0.54 | 0.46 | 0.57 | 0.48 | 0.40 | 0.30 | |
| Ssr2831 | PsaE | 0.35 | 0.33 | 0.36 | 0.51 | 0.45 | 0.52 | 0.46 | 0.42 | 0.18 | |
| Sll0199 | Plastocyanin | sec | 1.36 | 1.27 | 1.20 | 1.44 | 1.65 | 1.25 | 1.43 | 1.37 | 0.10 |
| Cytb6f | |||||||||||
| Sll1316 | Rieske protein PetC1 | 1 | 0.66 | 0.65 | 0.69 | 0.66 | 0.61 | 0.70 | 0.65 | 0.66 | 0.04 |
| Sll1317 | Cyt f PetA | 1, sec | 0.59 | 0.53 | 0.55 | 0.68 | 0.60 | 0.71 | 0.62 | 0.61 | 0.10 |
| Slr0342 | Cyt b6 PetB | 4 | 0.43 | 0.43 | 0.54 | 0.71 | 0.71 | 0.72 | 0.72 | 0.59 | 0.23 |
| Slr0343 | Subunit IV PetD | 3 | 0.55 | 0.54 | 0.68 | 0.67 | 0.60 | 0.65 | 0.58 | 0.61 | 0.16 |
| PSII | |||||||||||
| Slr1311/Sll1867 | D1 | 5 | 1.03 | 1.06 | 1.28 | 1.04 | 1.07 | 1.09 | 1.09 | 1.09 | 0.07 |
| Slr0927/Sll0849 | D2 | 6 | 1.33 | 1.33 | 1.18 | 1.06 | 1.01 | 1.07 | 1.02 | 1.14 | 0.11 |
| Slr0906 | CP47 | 6 | 1.13 | 1.08 | 1.12 | 1.08 | 1.09 | 1.07 | 1.07 | 1.09 | 0.02 |
| Sll0851 | CP43 | 7 | 1.08 | 1.08 | 1.08 | 1.07 | 1.03 | 1.09 | 1.04 | 1.07 | 0.02 |
| Ssr3451 | Cyt b559 alpha subunit | 1 | 1.56 | 1.32 | 1.06 | 1.04 | 1.02 | 0.98 | 0.96 | 1.12 | 0.17 |
| Ssl2598 | PsbH | 1 | 2.22 | 1.63 | 0.82 | 1.09 | 0.94 | 1.19 | 1.02 | 1.21 | 0.32 |
| Sll0427 | PsbO | sec | 1.30 | 1.22 | 1.18 | 1.22 | 1.16 | 1.24 | 1.17 | 1.21 | 0.03 |
| Sll1194 | PsbU | sec | 1.63 | 1.56 | 1.60 | 1.31 | 1.38 | 1.35 | 1.42 | 1.46 | 0.08 |
| Sll0258 | PsbV, cyt c550 | sec | 1.28 | 1.28 | 1.32 | 1.18 | 1.08 | 1.22 | 1.11 | 1.21 | 0.07 |
| Sll1638 | PsbQ | LP | 1.18 | 1.07 | 0.79 | 1.11 | 1.11 | 1.10 | 1.10 | 1.06 | 0.12 |
| Sll1398 | Psb28 | 1.21 | 1.06 | 0.73 | 0.98 | 1.05 | 0.92 | 0.99 | 0.98 | 0.15 | |
| Slr1645 | Psb27 | LP | 1.61 | 1.45 | 0.88 | 1.11 | 1.07 | 1.05 | 1.00 | 1.14 | 0.20 |
| Phycobiliproteins | |||||||||||
| Slr2067 | Allophycocyanin alpha subunit | 0.46 | 0.44 | 0.47 | 0.66 | 0.59 | 0.65 | 0.58 | 0.54 | 0.15 | |
| Ssl3093 | Phycobilisome small rod linker, CpcD | 0.58 | 0.57 | 0.51 | 0.65 | 0.63 | 0.67 | 0.66 | 0.61 | 0.09 | |
| Sll1577 | Phycocyanin b subunit | 0.60 | 0.56 | 0.61 | 0.74 | 0.64 | 0.75 | 0.64 | 0.65 | 0.10 | |
| Sll1578 | Phycocyanin alpha subunit | 0.62 | 0.63 | 0.64 | 0.67 | 0.62 | 0.68 | 0.63 | 0.64 | 0.03 | |
| Slr2051 | Phycobilisome rod-core linker, CpcG | 0.65 | 0.62 | 0.67 | 0.75 | 0.70 | 0.77 | 0.72 | 0.70 | 0.07 | |
| Sll1580 | Phycobilisome Linker polypeptide | 0.68 | 0.68 | 0.74 | 0.84 | 0.84 | 0.88 | 0.88 | 0.79 | 0.11 | |
| Sll0928 | Allophycocyanin B | 0.70 | 0.67 | 0.67 | 0.73 | 0.68 | 0.71 | 0.67 | 0.69 | 0.03 | |
| Ssr3383 | Phycobilisome linker domain protein CpcD | 0.77 | 0.65 | 0.46 | 0.79 | 0.79 | 0.80 | 0.80 | 0.71 | 0.19 | |
| Sll1579 | Phycobilisome linker polypeptide | 0.72 | 0.73 | 0.72 | 0.87 | 0.84 | 0.87 | 0.84 | 0.80 | 0.08 | |
| Slr1986 | Allophycocyanin beta subunit | 0.82 | 0.72 | 0.65 | 0.72 | 0.69 | 0.73 | 0.70 | 0.72 | 0.07 | |
| Sll1471 | Phycobilisome rod-core linker, CpcG | 0.72 | 0.74 | 0.83 | 0.88 | 0.90 | 0.89 | 0.91 | 0.84 | 0.09 | |
| Slr1459 | Allophycocyanin, beta subunit | 0.76 | 0.76 | 0.71 | 0.74 | 0.68 | 0.75 | 0.69 | 0.73 | 0.04 | |
| Slr0335 | Phycobilisome core membrane linker, ApcE | 0.89 | 0.86 | 0.89 | 0.93 | 0.92 | 0.91 | 0.90 | 0.90 | 0.02 | |
| Pigment biosynthesis | |||||||||||
| Slr0536 | Uroporphyrinogen decarboxylase | 0.63 | 0.60 | 0.69 | 0.68 | 0.69 | 0.73 | 0.74 | 0.68 | 0.07 | |
| Sll1214 | Magnesium-protoporphyrin IX monomethyl ester cyclase | 0.33 | 0.34 | 0.35 | 0.56 | 0.48 | 0.58 | 0.50 | 0.44 | 0.23 | |
| Sll1091 | Geranylgeranyl reductase, ChlP | 0.45 | 0.45 | 0.46 | 0.68 | 0.61 | 0.67 | 0.60 | 0.55 | 0.18 | |
| Slr0144 | hypothetical protein 4-vinyl reductase 4VR | 0.49 | 0.53 | 0.56 | 0.63 | 0.65 | 0.61 | 0.62 | 0.58 | 0.10 | |
| Redox proteins | |||||||||||
| Slr5088 | short-chain dehydrogenase/ reductase SDR | 2.94 | 2.70 | 3.24 | 1.97 | 2.23 | 1.80 | 2.05 | 2.37 | 0.21 | |
| Slr1379 | Cyt bd ubiquinol oxidase, CydA | 9 | 2.01 | 1.93 | 2.23 | 1.73 | 1.93 | 1.70 | 1.90 | 1.91 | 0.08 |
| Sll1182 | Rieske iron-sulfur protein, PetC3 | Tat/LP | 2.20 | 2.39 | 2.36 | 1.27 | 1.17 | 1.42 | 1.31 | 1.66 | 0.30 |
| Slr1239 | Pyridine nucleotide transhydrogenase, alpha subunit | 5 | 0.50 | 0.52 | 0.56 | 0.69 | 0.63 | 0.74 | 0.66 | 0.61 | 0.14 |
a Number of transmembrane helices known or predicted by the TMHMM program (http://www.cbs.dtu.dk/services/TMHMM-2.0/).
b Known or putative sec or lipoprotein signals using SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP-3.0/) and LipoP (http://www.cbs.dtu.dk/services/LipoP/), respectively.
The decrease in expression in ΔLepB1 of subunits of the PSI and Cyt b6f complexes, seen with 2D/BN-PAGE and specific antibodies, was confirmed by the iTRAQ analyses, with lower abundance ratios for all subunits quantified. Four integral and three peripheral proteins in the PSI complex decreased to 40–52% of the WT levels and the Cyt b6f complex subunits to 59–66% of WT amounts. Plastocyanin, which contains a Sec signal peptide, is a water soluble electron carrier found in the thylakoid lumen, carrying electrons from Cyt b6f to PSI. This protein was shown by our iTRAQ analyses to remain unchanged in the mutant, with a ratio of 1.37 fold, under the established limit for significance. Whether plastocyanin has an impaired processing like PsbO still needs to be confirmed.
All the PSII integral membrane subunits identified and quantified in this work (D1, D2, CP43, CP47, PsbH, and Cyt b559α) had abundance ratios of roughly 1 between LepB1 mutant and WT (Table I). The lumenal subunits of the PSII complex, PsbO, PsbU, and PsbV, forming the OEC in a ratio of 1:1:1, contain Sec signal peptides, were also shown to be unchanged in the iTRAQ analyses. As described above, PsbO blots showed an increase in the mutant of roughly 30%. The average ratio from the iTRAQ data for PsbO is 1.21 (±0.03), a 21% increase in ΔLepB1, below our established significance threshold. The abundance ratios for the other two proteins in the OEC, PsbU, and PsbV, are 1.46 and 1.21 respectively and they must all be defined as proteins with unchanged abundance. The expression of the other PSII associated proteins identified (Psb27, PsbQ, PsbP, and Psb28) also remains at same level in mutant and WT and the consistency in these results clearly indicates that all PSII subunits are expressed in similar amounts in WT and LepB1 depletion mutant.
Both the mutant and WT strains were grown in low light (<1μE m−2 s−1) with glucose, and therefore both strains are totally dependent on respiration as a source for both ATP and reducing equivalents. In our iTRAQ data, we found no changes in abundance between WT and the mutant for the subunits belonging to NADH dehydrogenase type 1 or ATP synthase (supplemental Data File S2). Synechocystis contain three distinct respiratory terminal oxidases, CtaI, CtaII and CydAB (45, 46). We were only able to identify and quantify CydA (Slr1379), which increased almost twofold (Table I) in the LepB1 mutant. PetC3, a Rieske protein with unknown function in respiration was also increased in the mutant (Table I), as also seen in the Western blot (Fig. 3).
All 13 subunits of the phycobilisome complex were quantitatively identified and, of these, six had ratios < 0.7, indicating a significant decrease, whereas seven others had ratios between 0.7 and 0.9 (Table I). The decrease of the phycobiliproteins was also reflected by the difference observed in the absorbance spectra of total membranes. The absorbance at 618 nm, which is the phycocyanin absorption peak (31), was only 70% of the WT intensity in the mutant (supplemental Data File S3). Three enzymes involved in the heme/chlorophyll biosynthesis pathway were found to decrease significantly in ΔLepB1 (Table I), uroporphyrinogen decarboxylase (Slr0536), magnesium-protoporphyrin IX monomethyl ester cyclase (Sll1214) and geranylgeranyl reductase or ChlP (Sll1091). Synthesis of chlorophyll is correlated to the amount of chlorophyll binding proteins (47) and it was previously shown that deletion of PSI in Synechocystis leads to a reduced chlorophyll content to about 15% (31). In the LepB1 mutant the chlorophyll content is decreased, in comparison to WT (supplemental Data File S3), because of a lower amount of PSI. Thus it seems reasonable that the amount of chlorophyll synthesizing enzymes is decreased in the mutant.
We have also identified several other proteins with changed abundance ratios in the mutant (supplemental Data File S2), including some periplasmic and cytoplasmic proteins. These changes most likely reflect indirect effects of the mutation and work is currently ongoing in our laboratory to further investigate and clarify their importance for the overall adaptations of the LepB1 deficient strain.
Respiratory Electron Pathway Mainly Proceeds Through CydAB
As pointed out earlier, the mutant cells can only grow under conditions of low light (<1μE m−2 s−1) in the presence of glucose. Under these conditions, the cells rely on respiration for continuous growth and three terminal oxidases, CtaI, CtaII, and CydAB, have been identified which could participate in this respiration (45, 46).
Respiratory activity measured in the dark was quite similar in WT and ΔLepB1, about 0.09 mmol O2 h−1 (ml at OD730 = 1)−1 (Table II). Respiration in both strains was inhibited 90% by KCN (not shown), which inhibits all three terminal oxidases. PCP, a specific inhibitor for the quinol oxidase CydAB (46), caused 80 and 60% inhibition in WT and ΔLepB1, respectively, indicating that this might be the major route for the electrons in dark grown cells. DBMIB is a potent inhibitor of the Cyt b6f complex (48), inhibiting the reduction of Cyt b6, with an inhibition constant of 80 nm (49). In our experiments, no inhibitory effect on the respiration in either strain was observed at any of the concentrations tested (from 40 nm to 40 μm, not shown). Interestingly, DBMIB caused a concentration-dependent increase in the rate of O2-consumption. This was particularly noteworthy in the mutant, with an increase in respiratory activity of more than threefold at 40 μm DBMIB (Table II). The addition of 5 mm KCN caused about 90% inhibition of oxygen consumption both in the absence and presence of DBMIB, for both strains, demonstrating that the DBMIB stimulated rate is dependent on a CN− sensitive oxidase (not shown).
Table II. Respiratory O2 uptake in darkness by Synechocystis WT and ΔLepB1 strains - effect of PCP and DBMIB. All rates are given as μmol O2 h−1 (ml at OD 730 = 1)−1 ± standard deviation, measured in the presence of 5 mm glucose. Values in parentheses are expressed as percentage of control rate.
| Addition | Rate of respiration |
|
|---|---|---|
| WT | ΔLepB1 | |
| Control | 0.090 ± 0.002 (100) | 0.087 ± 0.002 (100) |
| 3 mm PCP | 0.018 ± 0.003 (20) | 0.035 ± 0.001 (40) |
| 10 μm DBMIB | 0.122 ± 0.001 (136) | 0.205 ± 0.002 (236) |
| 40 μm DBMIB | 0.130 ± 0.003 (144) | 0.291 ± 0.004 (334) |
DISCUSSION
In the previous study of the knockout mutants of the two leader peptidases in Synechocystis it was shown that LepB2 is essential for cell viability (26). The LepB1 mutant is highly prone to photoinhibition unless DCMU, an inhibitor of PSII is present indicating that the electron flow from PSII is the cause for the observed photodamage. In the present study we show a dramatic decrease in Cyt b6f and PSI, in combination with an unchanged amount of PSII in ΔLepB1. This could produce an imbalance in electron flow upstream and downstream the PQ pool and be the direct reason for the light sensitivity. The LepB1 mutant could be propagated under heterotrophic conditions, although only at very low light intensity. It was previously shown that a PSI-less mutant, could only survive in low light, though keeping normal amounts of PSII (50), a direct parallel to the LepB1 mutant.
PsaF was exclusively observed in the unprocessed form in the mutant, strongly suggesting that when LepB1 is deleted, the remaining leader peptidase, LepB2, is not able to cleave the presequence of PsaF. The structure of Thermosynechococcus elongatus PSI has been determined at 2.5 Å resolution (39) and it was shown that PsaA and PsaB form the core reaction center located in the center of the monomer, surrounded by seven small integral membrane proteins. PsaF is situated in the periphery of the PSI complex according to the crystal structure. Studies on the PSI assembly process in Synechocystis concluded that PsaL is the last subunit to be added to the monomer, and that subsequent addition of PsaK leads to trimerization of PSI monomers (51). PsaF should consequently be inserted in an earlier step of the monomer assembly at least before the PsaL subunit is inserted, although the dynamics leading to early PSI complex formation are still unknown (51, 52). Furthermore, PsaF has been proposed to be important for the stabilization of both surfaces, lumenal and cytoplasmic, of PSI (53). The lower amounts of PSI that we observe could be explained by a slower assembly rate or a lower stability of the PSI monomer resulting from the presence of the signal peptide in PsaF. PsaF function has been well studied in plants and green algae, as well as in cyanobacteria. Unlike the eukaryotic PsaF, the cyanobacterial PsaF is not essential for docking of plastocyanin to PSI (54), because of absence of the basic N-terminal region present in plant and green algae, which is important for plastocyanin binding. The cyanobacterial PsaF-null mutant showed normal photoautotrophic growth when compared with the WT, suggesting that PsaF has dispensable accessory roles in the function and organization of PSI complex (38, 55). A chimeric PsaF, containing the N-terminal domain of PsaF from C. reinhardtii and the C-terminal sequence of the cyanobacterial PsaF was found to assemble into PSI complexes in Synechococcus with plastocyanin docking as well as PSI activity (56). Similarly to what we observe in the Synechocystis LepB1 mutant, the incorporation of this chimeric PsaF into PSI decreased the amount of PSI to one third of WT. The presence of the signal peptide introduces an extra N-terminal domain on the lumenal side of PSI which could result in a slower assembly rate or a lower stability of PSI in the mutant.
In contrast to PsaF, Cyt f in ΔlepB1 was only found in its processed form, albeit in significantly lower amounts. The Cyt b6f complex is shared by both the photosynthetic and the respiratory chains, catalyzing electron transfer between the plastoquinone pool and either plastocyanin or Cyt c6, The crystal structures of the Cyt b6f complexes from both the thermophilic cyanobacterium Mastigocladus laminosus (40) and the filamentous cyanobacterium Nostoc sp. PCC 7120 (57) contain four large subunits (Cyt f, Cyt b6, PetC1, and subunit IV) and four small subunits (PetG, -L, -M, and -N). Of all these, Cyt f is the largest subunit, containing a prosthetic c-type heme group. Mutants of CcsB (Sll1513), a protein responsible for heme insertion in c-type cytochromes, showed large accumulation of free pre-apo Cyt f in the thylakoid membrane lacking the covalently bound heme group. This indicates that heme binding and processing of Cyt f are closely coordinated in Synechocystis (58). In the iTRAQ analysis CcsB amounts did not change in the mutant (supplemental Data File S2). The normal level of CcsB and the decreased level of Cyt f in the LepB1 mutant may indicate that the processing by the remaining LepB2 is too slow and that the newly synthesized but unprocessed pre-Cyt f is degraded because of the presence of a degradation system that is able to remove nonfunctional proteins (58, 59).
A third scenario is observed for the OEC subunit PsbO, which is found both in processed and unprocessed forms in the mutant. The presence of pre-PsbO in the mutant despite the WT levels of the mature form indicates a LepB1 substrate preference and low activity of LepB2 on the processing of pre-PsbO. In contrast to PsaF and Cyt f, PsbO is a soluble lumenal protein. As such it could be grouped together with the PsbU, PsbV and plastocyanin, three other soluble lumen located proteins containing a sec signal peptide. Neither of these decreased in the mutant, even if, because of the lack of proper antibodies and an inability to detect their respective presequence peptides in our iTRAQ data, we were unable to confirm whether or not these proteins have deficient processing as well.
The different behavior observed for individual LepB1 substrate proteins leads to the conclusion that the deletion has a more profound effect on integral membrane proteins than on lumenal or periplasmic proteins. This could, in the case of integral membrane proteins, reflect a need for correlation between processing and integration into the complex and also a lower stability of nonassembled proteins.
As previously mentioned, LepB1 and LepB2 were identified in TM and PM, respectively (24, 25). However, the results in this work seem to implicate substrate overlapping and a dual location for LepB2. In E. coli the single leader peptidase always works together with the Sec translocon. When LepB1 is mutated, LepB2 becomes the only leader peptidase in Synechocystis and, even though the mutation caused a severe decrease of the PSI and Cyt b6f and concomitant light sensitivity, the cells can still grow in dim light. Most of the proteins we identified and quantified in the iTRAQ analysis were kept at the same expression level as in WT cells, including the PSII, NADH dehydrogenase 1 and ATP synthase complexes. The limited effect of the LepB1 mutation indicates that LepB2 is the major leader peptidase and probably has a dual membrane location in Synechocystis. Should LepB2 only exist in the plasma membrane of Synechocystis, this would implicate a very complicated movement of proteins between the periplasm and the lumen, as well as between PM and TM. YidC, which is required together with the Sec translocon for insertion of integral membrane proteins was previously shown to be present exclusively in TM (7). Therefore LepB1 might also exist only in TM to process photosynthetic signal peptide containing integral membrane proteins, inserted by the Sec translocon-YidC complex, which is indicated from the lack of PsaF processing.
Fig. 5 shows the proposed scheme for respiratory and photosynthetic electron transport in TM of Synechocystis (46, 48). In dark respiration, different dehydrogenases reduce PQ, which is then oxidized by CtaI (via Cyt b6f) or CydAB. It was previously concluded that in WT cells grown at normal light intensities (50 μE m−2 s−1) in the presence of glucose the Cyt b6f complex provides the major route for PQH2 oxidation in the respiration electron flow (48, 60). On the other hand, in cells grown in low light (<1 μE m−2 s−1 light and glucose) the CydAB oxidase seems to be the major route for oxidation of PQH2, because addition of PCP caused 80 and 60% inhibition in WT and mutant, respectively (Table II). WT and ΔLepB1 grown at low light intensities showed similar respiratory rates despite the large decrease in Cyt b6f content in the mutant cells. This could be explained by a compensation by the almost twofold increase in CydAB in the mutant (Table I) DBMIB was shown to inhibit Cyt b6f with high affinity in cultures grown at normal light intensities (48, 49). Surprisingly, in our case there was no inhibition of oxygen consumption but rather a dose-dependent stimulation. DBMIB is also a redox active molecule and is able to act as an electron shuttle (48, 61). Therefore, the stimulatory effect of DBMIB could caused by electron shuttling from the PQ pool, or dehydrogenases, to the quinol oxidase CydAB. In the absence of DBMIB the rate-limiting step in respiration is before CydAB, because the same rate of respiration is obtained for WT and mutant. On the other hand in the presence of DBMIB CydAB becomes rate-limiting and the more than doubled respiration rate observed in ΔLepB1 (Table II) corresponds well with the increase in content of CydAB, as shown by the iTRAQ analysis (Table I). Increased CydAB activity has earlier been associated with impairments in Cyt b6f (62) and a ΔPetC1 mutant was shown to have increased levels of CydA (63).
Fig. 5.
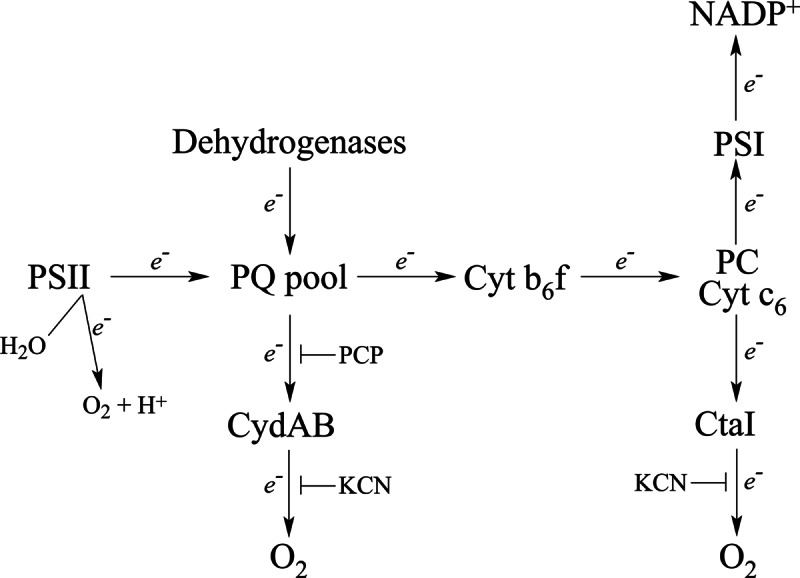
A proposed scheme for photosynthetic and respiratory electron transport in the TM of Synechocystis (adapted from (46, 48)).
The lower extent of inhibiton by PCP (60% versus 80% in the WT) could indicate additional unidentified changes in the respiratory electron transport in the mutant. Though relatively little is still known about respiration in PM (64), CtaII was found only in this subfraction (7, 24, 46, 48, 65) and CydAB in both TM and PM (45, 48). The Rieske protein PetC3, which was found to increase in the mutant as seen in Western blot and iTRAQ analyses, was shown to be present only in PM (64) and, similarly, a GFP-PetC3 construct was targeted to PM (66). Because of its unusually low redox potential (+135 mV), PetC3 cannot be a component of the “classical” Cyt b6f complex and oxidize plastoquinone (60). There is little information regarding the electron transport chain in very low light conditions and the discrepancy seen between protein expression levels and oxygen consumption could perhaps be caused by the presence of uncharacterized respiratory chain components in the plasma membrane (64).
In conclusion, deletion of LepB1 strongly affects the assembly and integration of both PSI and Cyt b6f in the thylakoid membrane, resulting in considerably reduced amounts of these complexes. This leads to cells that are prone to photoinhibiton and unable to grow photoautotrophically because of an imbalance in photosynthetic electron transport. The obvious targets of the mutation are PsaF and Cyt f, integral membrane proteins with a signal peptide. The assembly of PSII is not affected by the mutation, even though PsbO processing is slower than in the WT. The different effects on PSI and Cyt b6f relative to PSII most likely reflects the fact that PsaF and Cyt f are integral membrane proteins inserted into the core complexes in early steps of assembly, whereas PsbO is a soluble extrinsic PSII subunit and assembled late. Our results also indicate a difference in specificity for the two leader peptidases, as well as an overlapping function for LepB2. LepB1 seems to be the natural peptidase for processing of PsaF, Cyt f, and PsbO. LepB2 is able to partially replace LepB1 in the processing of Cyt f and PsbO—although at lower rates—but not of PsaF. When cells were grown in the dim light with glucose added a number of adaptive changes for the LepB1 mutant to overcome the loss of Cyt b6f, such as twofold increase of CydAB oxidase, as well as decreased expression of chlorophyll biosynthesis pathway enzymes and phycobiliproteins were also established.
Supplementary Material
Acknowledgments
We thank Bertil Andersson (NTU, Singapore) for valuable discussions and Sierin Lim (School of Chemical and Biomedical Engineering, NTU, Singapore) for the use of her O2 electrode. We thank Ralf Klösgen (Martin Luther University Halle-Wittenberg, Germany) for the ΔLepB1 strain, Matthias Rögner (Ruhr-Universität Bochum, Germany) for the kind gift of PetC3 and Cyt f antibodies and Jean-David Rochaix (University of Geneva, Switzerland) for PsaF antibodies.
Footnotes
* This work was supported by grants (M4080306 to BN and RG 51/10 to SSK) from Nanyang Technological University (NTU, Singapore).
 This article contains supplemental Data Files S1 to S3.
This article contains supplemental Data Files S1 to S3.
1 The abbreviations used are:
- DBMIB
- 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone
- DCMU
- 3-(3,4-dichlorophenyl)-1,1-dimetylurea
- ERLIC
- electrostatic repulsion-hydrophilic interaction chromatography
- iTRAQ
- isobaric tags for relative and absolute quantitation
- MDLC
- multidimensional liquid chromatography
- MS/MS
- tandem mass spectrometry
- PCP
- pentachlorophenol.
REFERENCES
- 1. Nierzwicki-Bauer S. A., Balkwill D. L., Stevens S. E., Jr. (1983) Three-dimensional ultrastructure of a unicellular cyanobacterium. J. Cell Biol. 97, 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gantt E. (1994) Supramolecular membrane organization. In: Bryant D. A., ed. The molecular biology of cyanobacteria, pp. 119–138, Kluwer, Dordrecht [Google Scholar]
- 3. Liberton M., Howard Berg R., Heuser J., Roth R., Pakrasi H. B. (2006) Ultrastructure of the membrane systems in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. Protoplasma 227, 129–138 [DOI] [PubMed] [Google Scholar]
- 4. van de Meene A. M., Hohmann-Marriott M. F., Vermaas W. F., Roberson R. W. (2006) The three-dimensional structure of the cyanobacterium Synechocystis sp. PCC 6803. Arch. Microbiol. 184, 259–270 [DOI] [PubMed] [Google Scholar]
- 5. Nevo R., Charuvi D., Shimoni E., Schwarz R., Kaplan A., Ohad I., Reich Z. (2007) Thylakoid membrane perforations and connectivity enable intracellular traffic in cyanobacteria. EMBO J. 26, 1467–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schneider D., Fuhrmann E., Scholz I., Hess W. R., Graumann P. L. (2007) Fluorescence staining of live cyanobacterial cells suggest non-stringent chromosome segregation and absence of a connection between cytoplasmic and thylakoid membranes. BMC Cell Biol. 8, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pisareva T., Kwon J., Oh J., Kim S., Ge C., Wieslander A., Choi J. S., Norling B. (2011) Model for membrane organization and protein sorting in the cyanobacterium Synechocystis sp. PCC 6803 inferred from proteomics and multivariate sequence analyses. J. Proteome Res. 10, 3617–3631 [DOI] [PubMed] [Google Scholar]
- 8. Nickelsen J., Rengstl B., Stengel A., Schottkowski M., Soll J., Ankele E. (2011) Biogenesis of the cyanobacterial thylakoid membrane system-an update. FEMS Microbiol. Lett. 315, 1–5 [DOI] [PubMed] [Google Scholar]
- 9. van de Meene A. M., Sharp W. P., McDaniel J. H., Friedrich H., Vermaas W. F., Roberson R. W. (2012) Gross morphological changes in thylakoid membrane structure are associated with photosystem I deletion in Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 1818, 1427–1434 [DOI] [PubMed] [Google Scholar]
- 10. Yuan J., Zweers J. C., van Dijl J. M., Dalbey R. E. (2010) Protein transport across and into cell membranes in bacteria and archaea. Cell. Mol. Life Sci. 67, 179–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dalbey R. E., Von Heijne G. (1992) Signal peptidases in prokaryotes and eukaryotes-a new protease family. Trends Biochem. Sci. 17, 474–478 [DOI] [PubMed] [Google Scholar]
- 12. Yi L., Jiang F., Chen M., Cain B., Bolhuis A., Dalbey R. E. (2003) YidC is strictly required for membrane insertion of subunits a and c of the F(1)F(0)ATP synthase and SecE of the SecYEG translocase. Biochemistry 42, 10537–10544 [DOI] [PubMed] [Google Scholar]
- 13. Wang P., Dalbey R. E. (2011) Inserting membrane proteins: the YidC/Oxa1/Alb3 machinery in bacteria, mitochondria, and chloroplasts. Biochim. Biophys. Acta 1808, 866–875 [DOI] [PubMed] [Google Scholar]
- 14. Wickstrom D., Wagner S., Simonsson P., Pop O., Baars L., Ytterberg A. J., van Wijk K. J., Luirink J., de Gier J. W. (2011) Characterization of the consequences of YidC depletion on the inner membrane proteome of E. coli using 2D blue native/SDS-PAGE. J. Mol. Biol. 409, 124–135 [DOI] [PubMed] [Google Scholar]
- 15. Spence E., Bailey S., Nenninger A., Møller S. G., Robinson C. (2004) A homolog of Albino3/OxaI is essential for thylakoid biogenesis in the cyanobacterium Synechocystis sp. PCC6803. J. Biol. Chem. 279, 55792–55800 [DOI] [PubMed] [Google Scholar]
- 16. Ossenbühl F., Inaba-Sulpice M., Meurer J., Soll J., Eichacker L. A. (2006) The Synechocystis sp PCC 6803 oxa1 homolog is essential for membrane integration of reaction center precursor protein pD1. Plant Cell 18, 2236–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fulda S., Huang F., Nilsson F., Hagemann M., Norling B. (2000) Proteomics of Synechocystis sp. strain PCC 6803. Identification of periplasmic proteins in cells grown at low and high salt concentrations. Eur. J. Biochem. 267, 5900–5907 [DOI] [PubMed] [Google Scholar]
- 18. Cao T. B., Saier M. H., Jr. (2003) The general protein secretory pathway: phylogenetic analyses leading to evolutionary conclusions. Biochim. Biophys. Acta 1609, 115–125 [DOI] [PubMed] [Google Scholar]
- 19. Spence E., Sarcina M., Ray N., Møller S. G., Mullineaux C. W., Robinson C. (2003) Membrane-specific targeting of green fluorescent protein by the Tat pathway in the cyanobacterium Synechocystis PCC6803. Mol. Microbiol. 48, 1481–1489 [DOI] [PubMed] [Google Scholar]
- 20. Hegde R. S., Bernstein H. D. (2006) The surprising complexity of signal sequences. Trends Biochem. Sci. 31, 563–571 [DOI] [PubMed] [Google Scholar]
- 21. Rajalahti T., Huang F., Klement M. R., Pisareva T., Edman M., Sjöstrom M., Wieslander A., Norling B. (2007) Proteins in different Synechocystis compartments have distinguishing N-terminal features: a combined proteomics and multivariate sequence analysis. J. Proteome Res. 6, 2420–2434 [DOI] [PubMed] [Google Scholar]
- 22. Dalbey R. E., Lively M. O., Bron S., van Dijl J. M. (1997) The chemistry and enzymology of the type I signal peptidases. Protein Sci. 6, 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Auclair S. M., Bhanu M. K., Kendall D. A. (2012) Signal peptidase I: cleaving the way to mature proteins. Protein Sci. 21, 13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang F., Parmryd I., Nilsson F., Persson A. L., Pakrasi H. B., Andersson B., Norling B. (2002) Proteomics of Synechocystis sp. strain PCC 6803: identification of plasma membrane proteins. Mol. Cell. Proteomics 1, 956–966 [DOI] [PubMed] [Google Scholar]
- 25. Srivastava R., Pisareva T., Norling B. (2005) Proteomic studies of the thylakoid membrane of Synechocystis sp. PCC 6803. Proteomics 5, 4905–4916 [DOI] [PubMed] [Google Scholar]
- 26. Zhbanko M., Zinchenko V., Gutensohn M., Schierhorn A., Klösgen R. B. (2005) Inactivation of a predicted leader peptidase prevents photoautotrophic growth of Synechocystis sp. strain PCC 6803. J. Bacteriol.187, 3071–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Allen M. M. (1968) Simple conditions for growth of unicellular blue-green algae on plates. J. Phycol. 4, 1–4 [DOI] [PubMed] [Google Scholar]
- 28. Zhang P., Battchikova N., Jansen T., Appel J., Ogawa T., Aro E. M. (2004) Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp PCC 6803. Plant Cell 16, 3326–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gan C. S., Chong P. K., Pham T. K., Wright P. C. (2007) Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ). J. Proteome Res. 6, 821–827 [DOI] [PubMed] [Google Scholar]
- 30. Norling B., Zak E., Andersson B., Pakrasi H. (1998) 2D-isolation of pure plasma and thylakoid membranes from the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 436, 189–192 [DOI] [PubMed] [Google Scholar]
- 31. Shen G., Boussiba S., Vermaas W. F. (1993) Synechocystis sp PCC 6803 strains lacking photosystem I and phycobilisome function. Plant Cell 5, 1853–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peterson G. L. (1977) A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 83, 346–356 [DOI] [PubMed] [Google Scholar]
- 33. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 34. Hao P., Qian J., Ren Y., Sze S. K. (2011) Electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) versus strong cation exchange (SCX) for fractionation of iTRAQ-labeled peptides. J. Proteome Res. 10, 5568–5574 [DOI] [PubMed] [Google Scholar]
- 35. Kuss C., Gan C. S., Gunalan K., Bozdech Z., Sze S. K., Preiser P. R. (2012) Quantitative proteomics reveals new insights into erythrocyte invasion by Plasmodium falciparum. Mol. Cell. Proteomics 11, M111.010645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Herranen M., Battchikova N., Zhang P., Graf A., Sirpiö S., Paakkarinen V., Aro E. M. (2004) Towards functional proteomics of membrane protein complexes in Synechocystis sp. PCC 6803. Plant Physiol. 134, 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6 [DOI] [PubMed] [Google Scholar]
- 38. Chitnis P. R., Purvis D., Nelson N. (1991) Molecular cloning and targeted mutagenesis of the gene psaF encoding subunit III of photosystem I from the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 266, 20146–20151 [PubMed] [Google Scholar]
- 39. Jordan P., Fromme P., Witt H. T., Klukas O., Saenger W., Krauss N. (2001) Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 411, 909–917 [DOI] [PubMed] [Google Scholar]
- 40. Kurisu G., Zhang H., Smith J. L., Cramer W. A. (2003) Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science 302, 1009–1014 [DOI] [PubMed] [Google Scholar]
- 41. Kashino Y., Lauber W. M., Carroll J. A., Wang Q., Whitmarsh J., Satoh K., Pakrasi H. B. (2002) Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41, 8004–8012 [DOI] [PubMed] [Google Scholar]
- 42. Battchikova N., Vainonen J. P., Vorontsova N., Keranen M., Carmel D., Aro E. M. (2010) Dynamic changes in the proteome of Synechocystis 6803 in response to CO2 limitation revealed by quantitative proteomics. J. Proteome Res. 9, 5896–5912 [DOI] [PubMed] [Google Scholar]
- 43. Pandhal J., Ow S. Y., Wright P. C., Biggs C. A. (2009) Comparative proteomics study of salt tolerance between a nonsequenced extremely halotolerant cyanobacterium and its mildly halotolerant relative using in vivo metabolic labeling and in vitro isobaric labeling. J. Proteome Res. 8, 818–828 [DOI] [PubMed] [Google Scholar]
- 44. Rowland J. G., Simon W. J., Nishiyama Y., Slabas A. R. (2010) Differential proteomic analysis using iTRAQ reveals changes in thylakoids associated with Photosystem II-acquired thermotolerance in Synechocystis sp. PCC 6803. Proteomics 10, 1917–1929 [DOI] [PubMed] [Google Scholar]
- 45. Howitt C. A., Vermaas W. F. (1998) Quinol and cytochrome oxidases in the cyanobacterium Synechocystis sp. PCC 6803. Biochemistry 37, 17944–17951 [DOI] [PubMed] [Google Scholar]
- 46. Pils D., Schmetterer G. (2001) Characterization of three bioenergetically active respiratory terminal oxidases in the cyanobacterium Synechocystis sp. strain PCC 6803. FEMS Microbiol. Lett. 203, 217–222 [DOI] [PubMed] [Google Scholar]
- 47. Vavilin D. V., Vermaas W. F. (2002) Regulation of the tetrapyrrole biosynthetic pathway leading to heme and chlorophyll in plants and cyanobacteria. Physiol Plant 115, 9–24 [DOI] [PubMed] [Google Scholar]
- 48. Berry S., Schneider D., Vermaas W. F., Rögner M. (2002) Electron transport routes in whole cells of Synechocystis sp. strain PCC 6803: the role of the cytochrome bd-type oxidase. Biochemistry 41, 3422–3429 [DOI] [PubMed] [Google Scholar]
- 49. Jones R. W., Whitmarsh J. (1988) Inhibition of electron transfer and the electrogenic reaction in the cytochrome b/f complex by 2-n-nonyl-4-hydroxyquinoline N-oxide (NQNO) and 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB). Biochim. Biophys. Acta 933, 258–268 [Google Scholar]
- 50. Smart L. B., McIntosh L. (1993) Genetic inactivation of the psaB gene in Synechocystis sp. PCC 6803 disrupts assembly of photosystem I. Plant Mol. Biol. 21, 177–180 [DOI] [PubMed] [Google Scholar]
- 51. Duhring U., Ossenbuhl F., Wilde A. (2007) Late assembly steps and dynamics of the cyanobacterial photosystem I. J. Biol. Chem. 282, 10915–10921 [DOI] [PubMed] [Google Scholar]
- 52. Ozawa S., Nield J., Terao A., Stauber E. J., Hippler M., Koike H., Rochaix J. D., Takahashi Y. (2009) Biochemical and structural studies of the large Ycf4-photosystem I assembly complex of the green alga Chlamydomonas reinhardtii. Plant Cell 21, 2424–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Grotjohann I., Fromme P. (2005) Structure of cyanobacterial photosystem I. Photosynth. Res. 85, 51–72 [DOI] [PubMed] [Google Scholar]
- 54. Hippler M., Reichert J., Sutter M., Zak E., Altschmied L., Schröer U., Herrmann R. G., Haehnel W. (1996) The plastocyanin binding domain of photosystem I. EMBO J. 15, 6374–6384 [PMC free article] [PubMed] [Google Scholar]
- 55. Xu Q., Yu L., Chitnis V. P., Chitnis P. R. (1994) Function and organization of photosystem I in a cyanobacterial mutant strain that lacks PsaF and PsaJ subunits. J. Biol. Chem. 269, 3205–3211 [PubMed] [Google Scholar]
- 56. Hippler M., Drepper F., Rochaix J. D., Mühlenhoff U. (1999) Insertion of the N-terminal part of PsaF from Chlamydomonas reinhardtii into photosystem I from Synechococcus elongatus enables efficient binding of algal plastocyanin and cytochrome c(6). J. Biol. Chem. 274, 4180–4188 [DOI] [PubMed] [Google Scholar]
- 57. Baniulis D., Yamashita E., Whitelegge J. P., Zatsman A. I., Hendrich M. P., Hasan S. S., Ryan C. M., Cramer W. A. (2009) Structure-Function, Stability, and Chemical Modification of the Cyanobacterial Cytochrome b(6)f Complex from Nostoc sp PCC 7120. J. Biol. Chem. 284, 9861–9869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tichy M., Vermaas W. (1999) Accumulation of pre-apocytochrome f in a Synechocystis sp. PCC 6803 mutant impaired in cytochrome c maturation. J. Biol. Chem. 274, 32396–32401 [DOI] [PubMed] [Google Scholar]
- 59. Howe G., Mets L., Merchant S. (1995) Biosynthesis of cytochrome f in Chlamydomonas reinhardtii: analysis of the pathway in gabaculine-treated cells and in the heme attachment mutant B6. Mol. Gen. Genet. 246, 156–165 [DOI] [PubMed] [Google Scholar]
- 60. Schneider D., Skrzypczak S., Anemuller S., Schmidt C. L., Seidler A., Rogner M. (2002) Heterogeneous Rieske proteins in the cytochrome b6f complex of Synechocystis PCC6803? J. Biol. Chem. 277, 10949–10954 [DOI] [PubMed] [Google Scholar]
- 61. Pisciotta J. M., Zou Y., Baskakov I. V. (2011) Role of the photosynthetic electron transfer chain in electrogenic activity of cyanobacteria. Appl. Microbiol. Biotechnol. 91, 377–385 [DOI] [PubMed] [Google Scholar]
- 62. Schneider D., Berry S., Volkmer T., Seidler A., Rögner M. (2004) PetC1 is the major Rieske iron-sulfur protein in the cytochrome b6f complex of Synechocystis sp. PCC 6803. J. Biol. Chem. 279, 39383–39388 [DOI] [PubMed] [Google Scholar]
- 63. Tsunoyama Y., Bernat G., Dyczmons N. G., Schneider D., Rögner M. (2009) Multiple Rieske proteins enable short- and long-term light adaptation of Synechocystis sp. PCC 6803. J. Biol. Chem. 284, 27875–27883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schultze M., Forberich B., Rexroth S., Dyczmons N. G., Roegner M., Appel J. (2009) Localization of cytochrome b6f complexes implies an incomplete respiratory chain in cytoplasmic membranes of the cyanobacterium Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 1787, 1479–1485 [DOI] [PubMed] [Google Scholar]
- 65. Huang F., Fulda S., Hagemann M., Norling B. (2006) Proteomic screening of salt-stress-induced changes in plasma membranes of Synechocystis sp. strain PCC 6803. Proteomics 6, 910–920 [DOI] [PubMed] [Google Scholar]
- 66. Aldridge C., Spence E., Kirkilionis M. A., Frigerio L., Robinson C. (2008) Tat-dependent targeting of Rieske iron-sulphur proteins to both the plasma and thylakoid membranes in the cyanobacterium Synechocystis PCC6803. Mol. Microbiol. 70, 140–150 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.