Abstract
Neurofibromatosis type 1 (NF1) tumor suppressor gene product, neurofibromin, functions in part as a Ras-GAP, and though its loss is implicated in the neuronal abnormality of NF1 patients, its precise cellular function remains unclear. To study the molecular mechanism of NF1 pathogenesis, we prepared NF1 gene knockdown (KD) PC12 cells, as a NF1 disease model, and analyzed their molecular (gene and protein) expression profiles with a unique integrated proteomics approach, comprising iTRAQ, 2D-DIGE, and DNA microarrays, using an integrated protein and gene expression analysis chart (iPEACH). In NF1-KD PC12 cells showing abnormal neuronal differentiation after NGF treatment, of 3198 molecules quantitatively identified and listed in iPEACH, 97 molecules continuously up- or down-regulated over time were extracted. Pathway and network analysis further revealed overrepresentation of calcium signaling and transcriptional regulation by glucocorticoid receptor (GR) in the up-regulated protein set, whereas nerve system development was overrepresented in the down-regulated protein set. The novel up-regulated network we discovered, “dynein IC2-GR-COX-1 signaling,” was then examined in NF1-KD cells. Validation studies confirmed that NF1 knockdown induces altered splicing and phosphorylation patterns of dynein IC2 isomers, up-regulation and accumulation of nuclear GR, and increased COX-1 expression in NGF-treated cells. Moreover, the neurite retraction phenotype observed in NF1-KD cells was significantly recovered by knockdown of the dynein IC2-C isoform and COX-1. In addition, dynein IC2 siRNA significantly inhibited nuclear translocation and accumulation of GR and up-regulation of COX-1 expression. These results suggest that dynein IC2 up-regulates GR nuclear translocation and accumulation, and subsequently causes increased COX-1 expression, in this NF1 disease model. Our integrated proteomics strategy, which combines multiple approaches, demonstrates that NF1-related neural abnormalities are, in part, caused by up-regulation of dynein IC2-GR-COX-1 signaling, which may be a novel therapeutic target for NF1.
Neurofibromatosis type 1 (NF1)1 is an autosomal dominantly inherited disorder with an estimated prevalence of 1 in 3000 people (1). The hallmarks of NF1 include development of benign tumors of the peripheral nervous system and increased risk of malignancies. The phenotype of NF1 is highly variable, with several organ systems affected including the skin, bones, irises, and central and peripheral nervous systems. The effects on the nervous system are manifested in multiple neurofibroma, gliomas, and learning disabilities.
The NF1 gene is located on chromosome 17q11.2 and encodes a large protein of 2818 amino acids, neurofibromin (2). Because the great majority of NF1 gene mutations frequently found in NF1 patients disturb the expression of intact neurofibromin, functional disruption of neurofibromin is potentially relevant to the expression of some or all of the abnormalities that occur in NF1 patients (3). A region centered around 360 amino acid residues encoded by the NF1 gene shows significant homology to the known catalytic domains of mammalian Ras GTPase-activating protein (p120 GAP). This region is also similar to yeast IRA1/2 proteins, which have been shown to interact with Ras and mediate hydrolysis of Ras-bound GTP to GDP, resulting in inactivation of Ras protein function. The GAP-related domain of the NF1 gene product also stimulates Ras GTPase and consequently inactivates Ras protein (4–6). In our previous report, we demonstrated a novel role of neurofibromin in neuronal differentiation in conjunction with regulation of Ras activity via its GAP-related domain using nerve growth factor (NGF)-stimulated PC12 cells as a model for neuronal cells (6). We also found that a novel neurofibromin interacting protein CRMP-2, identified with the screening of binding proteins of neurofibromin C-terminal domain by iTRAQ, up-regulates the phosphorylation patterns in the NF1 siRNA treated PC12 cells compared with those of control cells through two-dimensional (2D) fluorescence difference gel electrophoresis (DIGE) analysis coupled with Pro-Q Diamond staining, and demonstrated that the functional association of neurofibromin and CRMP-2 is essential for neuronal cell differentiation (7). In these studies, neurofibromin expression was suppressed using small interfering RNA (siRNA) directed against NF1, and the inhibition of neurofibromin functions caused neurite retraction via the regulation of Ras-MAPK-CDK5-GSK3/ROCK activation in differentiated PC12 cells stimulated by NGF (7). These results indicated that the neurofibromin-deficient PC12 cell is a useful model for analyzing NF1-related molecular pathology in detail.
Our previous research has examined specific genes and proteins related to phenotypic changes in neural tumors using integrated proteomic techniques such as 2D-DIGE combined with Pro-Q Diamond staining to identify phosphorylated proteins, isobaric tagging for relative and absolute quantitation (iTRAQ) (7, 8), which provides information on peptide and protein quantitative expression levels from different sources in a single experiment by liquid chromatography combined with tandem mass spectrometry (LC-MS/MS), as well as DNA array technology, using the original data mining tool, MANGO (Molecular Annotation by Gene Ontology) (8). Processing the voluminous data arising from each analysis highlighted the need for a mining system for data integration. Therefore, we created the “iPEACH” (Integrated Protein Expression Analysis Chart, PCT/JP2011/58366) application to integrate information from several analysis types into a useful data file that provides comprehensive proteomic data including post-translational modification, transcriptomic data, and functional annotations from several databases, with a tool for quickly organizing, enriching, and sorting these data to reveal candidate molecules. Using iPEACH, sample data obtained from our transcriptomic and proteomic (iTRAQ and 2D-DIGE) analyses of NF1 disease model cells were integrated and stored in a database.
In this study, we constructed an iPEACH database for neurofibromin-deficient PC12 cells compared with normal cells after NGF stimulation, and used Gene Ontology (GO) and knowledge-based network analyses targeting up-regulated signals in the NF1 model cells, which lost their normal differentiation activity, to extract a novel candidate signal network for NF1-related phenotype, termed “dynein IC2-GR-COX1” signaling. Statistical analysis of the expression or modification of these proteins in NF1-knockdown (NF1-KD) cells and subsequent biological validation by sequential analyses using siRNAs and the glucocorticoid receptor (GR) antagonist led to the successful identification of protein targets of the network most likely to be involved in the abnormal PC12 differentiation caused by neurofibromin deficiency. In addition, this network involved the alternative splicing and specific phosphorylation of cytoplasmic dynein 1 intermediate chain 2 isoform B (dynein IC2-B) and isoform C (dynein IC2-C) via NGF stimulation in NF1-KD PC12 cells.
Here, we demonstrate that the function of neurofibromin for neurite outgrowth in PC12 cells may involve the regulation of cyclooxygenase-1 (COX-1) via glucocorticoid receptor (GR) and dynein IC2 splicing and phosphorylation. We also discuss the implications of a functional association between neurofibromin and COX-1 for neuronal regulation in relation to NF1 pathogenesis. This report is the first to identify a signal network related to NF1 phenotype in a neurofibromin-deficient neural model using our new integrated proteomics strategy.
EXPERIMENTAL PROCEDURES
Cell Culture, NGF Treatment, Transfection, and Preparation of Cell Lysate
PC12 cells were cultured under 5% CO2 at 37 °C in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% horse serum and 5% fetal bovine serum. Transfection of siRNA was performed with the Neon transfection system (Invitrogen) according to the manufacturer's protocol (1100 V, 20 ms, two times). PC12 cells were transfected with siRNA and then seeded onto collagen-coated culture dishes (Iwaki). At 24 h after transfection, PC12 cells were stimulated with 50 ng/ml 2.5S NGF (Wako, Richmond, VA) until harvest. Cells were washed twice in PBS and solubilized with lysis buffer containing 8 m urea, 2% (w/v) 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate (CHAPS), 1 mm dithiotreitol (DTT), 10 mm sodium fluoride, 2 mm sodium orthovanadate, 1 μm okadaic acid, and 1% (v/v) protease inhibitor mixture (Sigma). Lysates were passed through a 25-gauge syringe 20 times and centrifuged at 20,000 × g for 15 min at 4 °C, and the protein concentrations of the supernatants were determined using Protein Assay Dye Reagent Concentrate (Bio-Rad, Hercules, CA). Cellular phenotypic changes were monitored on a time-lapse microscope equipped with a CO2 incubation chamber system (IX81, Olympus) using MetaMorph software ver. 7.5.5.0 (Molecular Devices, Sunnyvale, CA).
RNA Isolation and Microarray Analysis
Total RNA was extracted and purified with the RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Isolated RNA was assessed for quantity and quality using a NanoDrop ND1000 spectrophotometer (Thermo Scientific) and a 2100 Bioanalyzer (Agilent, Santa Clara, CA). Double-stranded complementary DNA (cDNA) and labeled complementary RNA (cRNA) were synthesized from the total RNA using the 3′ IVT Express Kit and hybridized to Affymetrix Rat 230 2.0 gene chips (Affymetrix, Santa Clara, CA). The chips were scanned with a Gene Chip Scanner 3000. The probe-level raw data and CEL files were obtained using GeneChip Operating Software. The raw expression data from CEL files was normalized by MAS5 algorism and analyzed by Subio Platform software (ver. 1.12, Subio, Japan).
2D-DIGE and Image Analysis
PC12 cell lysates were desalted using a 2D Clean-up kit (GE Healthcare) and dissolved in lysis buffer containing 8 m urea, 2% (w/v) CHAPS, and 30 mm Tris-HCl (pH 8.5). Each sample (50 μg) was minimally labeled with 400 pmol of either Cy3 or Cy5 for comparison on the same gel. To facilitate image matching and cross-gel statistical comparison, a pool of all samples was prepared and labeled with Cy2 (400 pmol/50 μg) as an internal standard for all gels. The labeling mixture was incubated on ice in the dark for 30 min, and the reaction was terminated by addition of 10 nmol lysine. The differentially Cy3- and Cy5-labeled samples were mixed with Cy2-labeled internal standard and incubated with equal volumes of 2× sample buffer containing 8 m urea, 2% (w/v) CHAPS, 2.4% (v/v) Destreak Reagent (GE Healthcare), and 1% (v/v) IPG buffer (pH 3–11 or pH 4–7, GE Healthcare) for 10 min on ice. The mixed samples were supplemented with 1× sample buffer containing 8 m urea, 2% (w/v) CHAPS, 1.2% (v/v) Destreak Reagent, and 0.5% (v/v) IPG buffer to reach a final volume of 450 μl and loaded into strip holders for first-dimension isoelectric focusing (IEF). IPG strips (pH 3–11 NL or pH 4–7, 24 cm, GE Healthcare) were rehydrated with the Cy-labeled samples in the dark at room temperature overnight. IEF was performed using a Multiphor II apparatus (GE Healthcare) under the following conditions: held at 100 V for 2 h, held at 500 V for 2 h, ramped to 1000 V in 5 h, ramped to 8000 V in 3 h, and held at 800 V for 9 h for pH 3–11 NL strips; held at 100 V for 2 h, held at 500 V for 4 h, ramped to 1000 V in 5 h, ramped to 8000 V in 3 h, and held at 8000 V for 9.5 h for pH 4–7 strips. Strips were equilibrated in solution containing 2% (w/v) SDS, 50 mm Tris-HCl (pH 8.8), 6 m urea, 30% (v/v) glycerol, 0.002% (w/v) bromphenol blue, and 1 mm DTT for 20 min and then in the same buffer additionally containing 1 mm iodoacetamide for 20 min. The equilibrated strips were then transferred onto 10% gels (24 × 20 cm) for second-dimension electrophoresis using the Ettan DALTsix system (GE Healthcare). SDS-PAGE was run at 10 mA/gel for 1 h and then 12 mA/gel at 30 °C until the bromphenol dye front reached the gel end. Images of protein spots on the gels were scanned with a Typhoon 9400 variable mode imager (GE Healthcare) at 100 μm resolution using excitation/emission wavelengths of 488 nm/520 nm for Cy2, 532 nm/580 nm for Cy3, and 633 nm/670 nm for Cy5, respectively. The intensity was adjusted to ensure that the maximum volume of each image was within 60,000–90,000 pixels. Image analysis was performed using Decyder2D software version 5.2 (GE Healthcare) according to the manufacturer's instructions. The differential in-gel analysis module was used for spot detection, spot volume quantification, and volume ratio normalization of different samples in the same gel. Then, the biological variation analysis module was used to match protein spots across different gels and to identify spots demonstrating significant differences. Differentially expressed spots were determined statistically using two-way analysis of variance (condition 1: p < 0.05 for the treatment of NF1 siRNA and condition 2: p < 0.05 for the period of NGF treatment).
Staining of 2D Gels with Pro-Q Diamond Phosphoprotein Gel Stain
Two-dimensional gels were fixed in 50% methanol containing 10% acetic acid once for 1 h and again overnight. Gels were washed three times in water for 15 min, stained with Pro-Q Diamond phosphoprotein gel stain (Invitrogen) in the dark for 4 h, and then washed with destain solution (5% 1 m sodium acetate, pH 4.0, in 20% acetonitrile (ACN) three times for 60 min. Gels were washed twice with deionized water for 5 min and scanned with a Typhoon9400 variable mode imager (GE Healthcare). For SYPRO Ruby gel staining, gels were fixed in 10% methanol containing 7.5% acetic acid for 30 min, stained with SYPRO Ruby protein gel stain (Invitrogen) overnight, and washed with 10% methanol containing 7.5% acetic acid for 1 h and scanned with a Typhoon9400 imager (GE Healthcare). Fluorescent images stained with Pro-Q Diamond and SyproRuby were overlaid using Multi Gauge software (Fujifilm).
In-gel Digestion of Proteins Identified in 2D-DIGE
To identify proteins detected in 2D-DIGE, 150 μg of proteins was separated onto 2D gels. The gels were fixed with 10% methanol and 7.5% acetic acid and then stained with Deep Purple total protein stain (GE Healthcare) according to the manufacturer's instructions. After scanning the gel image with a Typhoon 9400 imager (GE Healthcare), protein spots of interests were selected with DeCyder software and picked by an Ettan spot picker (GE Healthcare). The gel pieces were washed three times with 50 mm ammonium bicarbonate in 50% (v/v) ACN, dehydrated in 100% (v/v) ACN, and vacuum-dried. Sequencing grade modified trypsin (Promega, Madison, WI) was added to the gel pieces at a concentration of 50 ng/ml in 10% ACN including 50 mm ammonium bicarbonate, and the mixture was incubated at 37 °C overnight. The trypsinized peptides were sequentially extracted from the gels with 0.1% (v/v) trifluoroacetic acid (TFA) in 30% (v/v) ACN, 0.1% (v/v) TFA in 50% (v/v) ACN, and 0.1% (v/v) TFA in 80% (v/v) ACN, for 5 min each. The extracted peptides were vacuum-dried and dissolved in 20 μl of 0.1% (v/v) TFA in 2% (v/v) ACN. These samples were desalted with a ZipTip C18 pipette tip (Millipore) and subjected to MALDI-MS/MS analysis using the 4700 Proteomics analyzer (Applied Biosystems/AB SCIEX) or LC-ESI-MS/MS analysis using the API QSTAR Pulsar i time-of-flight mass spectrometer (Applied Biosystems/AB SCIEX) coupled with the UltiMate NanoLC system (Thermo Scientific Dionex).
iTRAQ Sample Preparation, Fractionation, and Desalting
Protein samples (100 μg) were precipitated using a 2-D Clean-Up kit (GE Healthcare), and the precipitates were dissolved in 10 μl of 6 m urea. iTRAQ sample labeling was performed according to the manufacturer's protocol with minimum modification (8). The digests were incubated with eight different iTRAQ reagents (113–121) for 2 h as follows: iTRAQ 113 for NGF 0 h and control siRNA, 114 for 24-h control, 115 for 48-h control, 116 for 72-h control, 117 for 0-h NF1 siRNA, 118 for 24-h NF1 siRNA, 119 for 48-h NF1 siRNA, and 121 for 72-h NF1 siRNA. The labeled samples were then mixed together and fractionated to remove excess, unbound iTRAQ reagent and to simplify the peptide mixture using a GE Healthcare AKTA system. The mixed sample was diluted in loading buffer (20% ACN and 10 mm potassium phosphate, pH 3.0) and loaded onto a Mono S column (GE Healthcare) equilibrated with loading buffer. Peptides were eluted with a gradient of solvent B (10 mm potassium phosphate, pH 3.0, and 500 mm KCl in 20% ACN) as follows: 0–2 min, 0–7% B; at 10 min, to 14% B; at 14 min, to 32% B; at 19 min, to 70% B; and at 24 min, to 100% B. Forty-four fractions that included the iTRAQ-labeled peptides were used for analysis. The fractions were dried in a vacuum centrifuge and rehydrated with solution containing 2% ACN and 0.1% TFA. The samples were desalted with ZipTip μ-C18 pipette tips (Millipore). The desalted peptides were divided into two fractions to analyze the same samples by LC-MALDI-MS/MS and LC-ESI-MS/MS analyses.
LC-MALDI-MS/MS Analysis
Samples were separated by C18 nano-LC using a DiNa MaP system (KYA Tech Corp.) equipped with a device spotting eluted fractions onto a MALDI plate. Sample was injected onto a C18 column (0.5-mm inner diameter × 1-mm length, KYA Tech Corp.) equilibrated with solvent A (2% ACN and 0.1% TFA) and resolved on a C18 nanocolumn (0.15-mm inner diameter × 50-mm length; KYA Tech Corp.) at a flow rate of 300 nl/min with a 113-min gradient of solvent B (70% ACN and 0.1% TFA) as follows: 0–22% B from 0 to 10 min, to 39% B at 53 min, to 45% B at 83 min, and to 100% B at 93 min. Column effluent was mixed with matrix (3 mg/ml alpha-cyano-4-hydroxycinnamic acid in 50% ACN and 0.1% TFA) at a flow rate of 1.4 μl/min. Fractions were spotted at 30-s intervals onto a stainless steel MALDI target plate (384 wells/plate; Applied Biosystems). Mass spectra of the peptides were acquired on an AB SCIEX TOF/TOF 5800 system (AB SCIEX) using TOF/TOF Series Explorer software (Version 4.0.0). Mass spectra from m/z 800 to 4,000 were acquired for each fraction with 1500 laser shots. To analyze the less abundant peptides, all peaks with a signal-to-noise ratio threshold from 25 to 50, from 50 to 100, and more than 100 in each MS spectrum were sequentially selected for MS/MS analysis. Fragmentation of the labeled peptides was induced by the use of atmosphere as a collision gas with a pressure of 1 × 10−6 Torr and a collision energy of 1 kV.
LC-ESI-MS/MS Analysis
Samples were analyzed by nano-LC-ESI-MS/MS using the LC Packings Ultimate instrument fitted with a 20-μl sample loop. Samples were loaded onto a 5-mm RP C18 precolumn (Thermo Scientific Dionex) at 25 μl/min and washed for 10 min before switching the precolumn in line with the separation column, which was a 75-μm internal diameter × 150-mm length PepMap RP column (Thermo Scientific Dionex) packed with 3-μm C18 beads with 100-Å pores. The flow rate used for separation on the RP column was 200 nl/min with a 120-min gradient of solvent B (85% ACN and 0.1% formic acid) as follows: 0–3% B from 0–5 min, to 15% B at 10 min, to 40% B at 95 min, to 100% B at 100 min. Sample analysis was performed using a QSTAR Elite mass spectrometer (Applied Biosystems/AB SCIEX), and Analyst QS 2.0 software (Applied Biosystems/AB SCIEX) was used for data acquisition with the scan cycles set to perform a 1-s MS scan followed by three MS/MS scans of the three most abundant peaks for 2 s each. Data acquisition was performed with an exclusion of 60 s for previous target ions. The labeled peptides were fragmented under CID conditions designed to yield iTRAQ reporter ions.
Protein Identification in 2D-DIGE and iTRAQ
Data from MALDI and ESI analysis of proteins identified in 2D-DIGE were analyzed using MASCOT software application 2.1.04 (Matrix Sciences). The UniProt database (release-2010_03) was used for the search. The following search parameters were used: taxonomy, rat; cleavage enzyme, trypsin; variable modifications, carbamidomethyl (C), oxidation (M); max missed cleavage, 1. Peptide tolerance of 0.3 Da, fragment mass tolerance of 0.3 Da, and peptide charge of +1 for MALDI or +2, +3, and +4 for ESI were considered significant. Confident identification required a statistically significant (p < 0.05) protein score based on combined MS and MS/MS spectra. Data from MALDI or ESI analysis with the iTRAQ method and from ESI analysis with 2D-DIGE were analyzed using the ParagonTM algorithm (9) of ProteinPilot Version 4.1 (AB SCIEX), and the UniProt Rat proteome database (release2012_02, 37,104 entries) was searched. Identified proteins were grouped by the Paragon algorithm to minimize redundancy. The six user-defined options included (1) cysteine alkylation, methyl methane thiosulfate in iTRAQ or iodoacetic acid in 2D-DIGE; (2) digestion, trypsin digestion; (3) special factors, none in iTRAQ or phosphorylation emphasis in 2D-DIGE; (4) species, none; (5) identification focus, biological modifications, amino acid substitutions; and (6) search effort, thorough identification search. The ProteinPilot cutoff score used was 1.3, which corresponds to a confidence limit of 95%. The protein identification confidence for the data set employing the iTRAQ method was further evaluated by false discovery rate in the concomitant search of the UniProt Rat proteome database for the reverse sequences.
Protein Quantification for iTRAQ Analysis
Quantitation of proteins was carried out based on the relative intensities of iTRAQ reporter ions released during MS/MS fragmentation of peptides according to the ParagonTM algorithm of ProteinPilot Version 4.1 (AB SCIEX). The data analysis parameters were set as follows: Sample type: iTRAQ (peptide labeled); Bias Correction: Auto; Background Correction: Yes.
Data Processing of Integrated Proteomics and Network Analysis
All the data obtained by transcriptomic and proteomic analysis were integrated into one chart by iPEACH. The iPEACH tool has following function such as, mutual conversions of probe ID, UniProt ID, and Entrez ID, calculations and outputs of iPEACH indices from DNA microarray, iTRAQ, and 2D-DIGE data, and annotations of UniProt descriptions (Organism, Amino Acid_Seqence_Length, Theoretical_MW, Keyword, GO_Biological_Process, GO_Molecular_Function, GO_Cellular_Component, SwissProt:[CC]) and original data information (Post-Translational Modification, Cleavage sites, Theoretical_pI, Observed_MW, Observed_pI) obtained from iTRAQ, and 2D-DIGE analysis. Input multi types of files for iPEACH are CEL files (DNA microarray), peptide summary and protein summary files from ProteinPilot (iTRAQ experiments using ESI and MALDI), and xml output files from Decyder 2D software (2D-DIGE experiment using pH3–11 and pH4–7 gels). First, iPEACH integrates genes and proteins identified in the different experiments using Entrez ID. Then, iPEACH calls R/Bioconductor with affy package to normalize the gene expression data with MAS5 algorithm. iTRAQ and 2D-DIGE data are already normalized by data processing software. And then fold changes (control versus treated samples at each time point: 0 h, 24 h, 48 h, 72 h) are calculated within each experiment. Finally, iPEACH integrates the calculated fold changes and outputs iPEACH_sum (sum of fold changes at same time points), iPEACH_log2 (sum of logarithmical fold changes at same time points), and iPEACH_Index (sum of absolute value of logarithmical fold changes at same time points) followed by the sorting of iPEACH_Index descending order (supplemental Table S1). iPEACH indices (z) were calculated as follows:
where x = ratio of gene or protein expression in the NF1 siRNA treatment to control siRNA treatment groups. Each suffix indicates stimulation time. UniProt release 2012_02 was used for the annotation in this study. The iPEACH column legends are listed in the supplemental information. The extraction of differentially expressed genes/proteins and cluster analysis were performed based on the protein ratio (NF1 siRNA/control treatment) using the Subio platform. Network analysis was performed using KeyMolnet (Institute of Medicinal Molecular Design). The lists of differentially expressed genes/proteins were imported into KeyMolnet, and the “start points and end points” network search algorithm was used to generate the network of all types of molecular interactions between these genes/proteins. The network included direct and indirect activation, transcriptional activation, and complex formation.
siRNA
The target sequences for rat NF1, dynein IC2-C, dynein IC2, and COX-1 siRNA were designed as follows: a 21-oligonucleotide siRNA duplex was designed as recommended elsewhere (10) and synthesized by Nippon EGT to target the rat NF1 sequences 5′-249CAAGGAGTGTCTGATCAACTT-3′ (for NF1 249 siRNA) and 5′-611GGTTACAGGAGTTGACTGTTT-3′ (for NF1 611 siRNA), the rat dynein IC2-C sequences 5′-329-GATCTAGACGAGGACCTATTT-3′ (for dynein IC2-C 329 siRNA) and 5′-331-TCTAGACGAGGACCTATTATT-3′ (for dynein IC2-C 331 siRNA), and the rat COX-1 sequences 5′-1023-CCATCGAGATTATCATCGATT-3′ (for COX-1 1023 siRNA) and 5′-2618-CAGGTGGACTCATCTACGATT-3′ (for COX-1 2618 siRNA). A 25-oligonucleotide siRNA duplex was synthesized by Nippon EGT to target the rat dynein IC2 sequence 5′-CCCTTTGCTTTGGATTGGTGTCATT-3′ (11). Silencer Negative Control siRNA 1 (Ambion) was used as a control.
Immunoblotting
Cell lysate samples were electrophoresed on SDS-polyacrylamide gels, transferred onto a PVDF membrane by electroblotting, and subjected to immunoblotting with the indicated antibody. Membranes were probed with different primary antibodies followed by horseradish peroxidase-conjugated mouse or rabbit secondary antibodies or ECL-Plex rabbit IgG-Cy2 and ECL-Plex mouse IgG-Cy5 (GE Healthcare). The blot images were developed with ECL Prime reagent and exposed to Hyperfilm ECL (GE Healthcare). Fluorescent images visualized with CyDye patterns were processed with a Typhoon 9400 imager (GE Healthcare). The following primary antibodies were used: NF1, dynein IC, TCPε1 (Santa Cruz Biotechnology), COX-1, GR, Lamin B1 (Abcam, Cambridge, MA), and beta-actin (Sigma). The intensities were measured using an ImageQuant system (GE Healthcare) with background subtraction and normalization by total spot volume. For 2D-immunoblotting, the intensities were measured using ProGenesis Work station version 2005 (PerkinElmer Life Sciences). The intensity of each spot was recorded as digital data and processed with Microsoft Office Excel. Experimental values are expressed as mean ± S.E.
Two-dimensional Immunoblotting Using Auto-2D
PC12 cell lysates were desalted using a 2D clean-up kit (GE Healthcare) and dissolved in lysis buffer (8 m urea, 2% (w/v) CHAPS). Samples (5 μg) were mixed with rehydration solution (8 m urea, 2% (w/v) CHAPS, 1.0% (v/v) Destreak Reagent, 0.4% (v/v) IPG buffer (pH 4–7)) and loaded into strip holders for first-dimension IEF. IEF was performed using Auto-2D (Sharp Manufacturing Systems) with the following conditions: held at 200 V for 15 min, ramped to 1000 V in 15 min, ramped to 3000 V in 15 min, ramped to 6000 V in 15 min, and held at 6000 V for 15 min. Strips were equilibrated in solution (45% (v/v) NuPAGE (Invitrogen), 50 mm DTT) for 5 min and transferred to 8% gels (5 × 7.5 cm) for second-dimension electrophoresis. After electrophoresis, gels were transferred onto a PVDF membrane by electroblotting and subjected to immunoblotting with the indicated antibody. After immunoblotting, the intensities were measured using ProGenesis Work station version 2005 (PerkinElmer Life Sciences).
Neurite Outgrowth Analysis
To quantify the neurite outgrowth of PC12 cells, cells transfected with siRNAs were cultured on collagen-coated culture dishes (Iwaki) and stimulated with 50 ng/ml 2.5S NGF (Wako) at 72 h. Total neurite length of NGF-stimulated PC12 cells was measured using MetaMorph software (Molecular Devices). The total number of tip ends was manually counted to represent the number of neurites from individual cells. For each measurement, at least 50 cells per dish were analyzed from randomly selected fields. Each experiment was repeated three times.
Immunocytochemistry
PC12 cells grown on a six-well plate were fixed with 4% paraformaldehyde in PBS for 15 min at room temperature and then permeabilized with 0.1% (w/v) Triton X-100 in PBS for 15 min. After washing with PBS, cells were incubated in primary antibodies diluted in PBS containing 0.2% (w/v) BSA, followed by a secondary antibody conjugated with a fluorescent dye for 1 h at room temperature and observed with a fluorescence microscope (Olympus IX81 at 20× magnification).
Preparation of Nuclear and Cytoplasmic Protein Extracts
PC12 cells were collected, and the nuclear and cytoplasmic proteins were obtained using the 2-D Sample Prep for Nuclear Proteins kit (Thermo Fisher scientific) according to the manufacturer's instructions.
RESULTS
Integrated Proteomics Using iPEACH to Identify Specific Molecules Related to the Phenotypic Change Caused by NF1 Knockdown in PC12 Cells
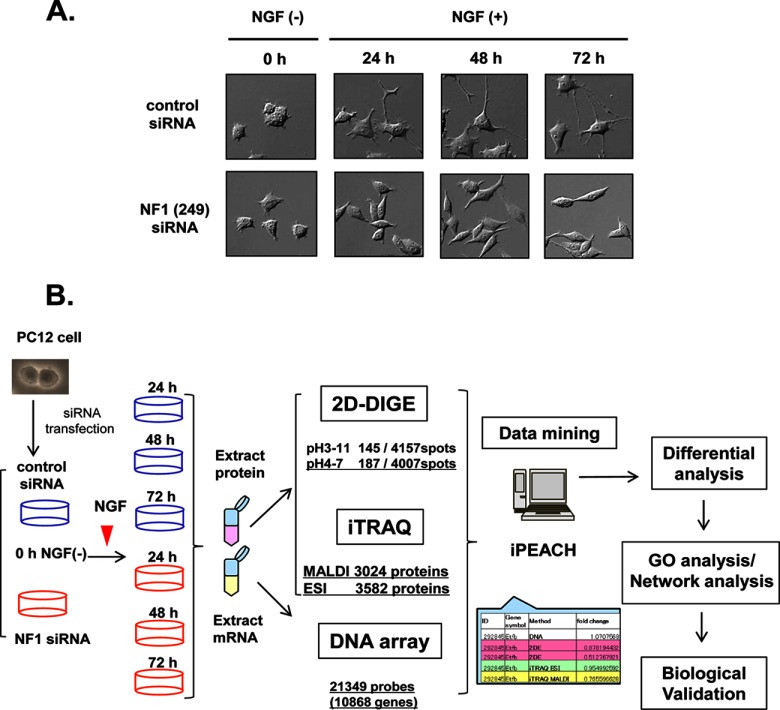
According to our previous study (7), we prepared NF1-KD PC12 cells using NF1 siRNA. PC12 cells were transfected with control siRNA or NF1 (249) siRNA and stimulated with NGF, and morphological changes were observed at the indicated time points (0 h, NGF-; 24 h, 48 h, and 72 h; NGF+). We confirmed the suppression of the NF1 gene product, neurofibromin, by immunoblot analysis and immunocytochemistry in PC12 cells (Fig. 1A, supplemental Figs. S1A- 1C). PC12 cells that express neurofibromin normally extend neurites on NGF treatment, whereas NF1-KD PC12 cells consistently adopted spindle bipolar shapes and inhibited neurite outgrowth (Fig. 1A, supplemental Fig. S1C). Cells were then subjected to integrated proteomics analyses as shown in Fig. 1B. PC12 cells were transfected with control siRNA or NF1 (249) siRNA and stimulated with NGF, and protein and mRNA samples were prepared from the cells at the indicated time points (0 h, NGF-; 24 h, 48 h, and 72 h; NGF+). The protein and mRNA samples were subjected to 2D-DIGE and iTRAQ-8plex methods and DNA microarray analysis using Rat 230 2.0 gene chips (Affymetrix). All the results were then integrated using iPEACH (Fig. 1B).
Fig. 1.
Workflow of the integrated proteomics approach to identify the abnormal network in NF1-KD PC12 cells. A, Representative images of NGF-stimulated PC12 cells treated with NF1 siRNA. PC12 cells were transfected with NF1 siRNA (lower panel) or control siRNA (upper panel), treated with NGF, and observed at the indicated time points. B, Workflow of the integrated proteomics approach. PC12 cells were transfected with control siRNA or NF1 (249) siRNA and stimulated with NGF, and protein and mRNA samples were prepared from the cells at the indicated time points (0 h, NGF-; 24 h, 48 h, and 72 h; NGF+). The protein and mRNA samples were subjected to 2D-DIGE and iTRAQ-eight-plex methods and DNA microarray Rat 230 2.0 gene chip analysis (Affymetrix), respectively. After these analyses, an integrated chart from all data was generated using iPEACH to identify differentially expressed genes and proteins. Further biological and functional interpretation of the differentially expressed genes and proteins was carried out by GO and network analyses followed by cell biological analyses.
Results of 2D-DIGE
PC12 cells were transfected with control siRNA or NF1 (249) siRNA and stimulated with NGF for 0 h, 24 h, 48 h, or 72 h. Proteins were extracted from the cells at each time point (n = 3), desalted, and subjected to 2D-DIGE. To obtain as much protein information as possible, each sample was separated into two fractions and subjected to IEF using both strip ranges of pH 3–11 and pH 4–7. An average of 4,157 ± 160 spots in pH 3–11 gels and an average of 4007 ± 176 spots in pH 4–7 gels were detected. Differentially expressed spots were statistically determined using two-way analysis of variance (condition 1: p < 0.05 for the treatment of NF1 siRNA and condition 2: p < 0.05 for the period of NGF treatment) with DeCyder 2D software. As a result, 145 spots (pH 3–11) and 187 spots (pH 4–7) were found to be significant differentially expressed, and 123 nonredundant proteins (pH 3–11: 128, pH 4–7: 124 spots) were identified by MALDI-TOF/TOF or LC-ESI-Qq-TOF MS. The proteins identified with 2D-DIGE are shown in supplemental Table S4 and integrated into the iPEACH Table (supplemental Table S1).
Results of iTRAQ (8-Plex) Analysis
iTRAQ analysis was performed with the same samples used for 2D-DIGE analysis. For 8-plex iTRAQ labeling, each protein sample obtained from the three pooled independent experiments at four time points of NGF stimulation after NF1 or control siRNA treatment was digested with trypsin after reduced alkylation and labeled with iTRAQ reagents. The labeled samples were mixed together, fractionated into 44 fractions with cation exchange column chromatography, and analyzed by nanoLC-MALD-TOF/TOF MS and nanoESI-Qq-TOF MS. Total spectral data (MALDI:141533, ESI: 286025) were analyzed with ProteinPilot 4.1 software using the UniProt Rat proteome database. In LC-MALDI-TOF/TOF analysis, 3024 proteins were identified from 46,766 peptide sequences; in LC-ESI-MS/MS analysis, 3582 proteins were identified from 68,790 peptide sequences with the criterion of unused protein score = 1.3 (95% confidence) (Fig. 1B, supplemental Fig. S2B). The iTRAQ data is shown in supplemental Table S2 and S3 and integrated into the iPEACH Table (supplemental Table S1).
DNA Array Results
Total RNAs extracted from cells prepared in the same manner described for proteome analyses at each time points were subjected to GeneChip Rat Genome 230 2.0 Array analyses. After obtaining normalized signal data, 10,868 genes (21,349 probes with present call among 31,099 total probes) were used for differential expression analysis.
Integration of All Data Obtained from Proteomics and Transcriptomics
Data obtained from four time-course samples with or without NF1 knockdown using two differential proteome analyses and a transcriptomic analysis (123 proteins from 2D-DIGE, 3239 proteins from 8-plex iTRAQ, and 10,868 genes from DNA array) were combined by iPEACH (Fig. 1B, supplemental Table S1). The iPEACH list contains all molecular information, including molecular ID (Entrez gene ID, UniProt), GO, post-translational modifications, expression ratios, and analysis methods in an integrated chart (international patent PCT/JP2011/58366). The integrated total data from the original iPEACH in this study consisted of 16,421 lines of comprehensive molecular information. After obtaining this chart, the integrated data were reprocessed for GO analysis or knowledge-based network analysis by KeyMolnet, and the specific signal networks mediated by up-regulated molecules in NF1-KD samples were extracted.
Data Mining
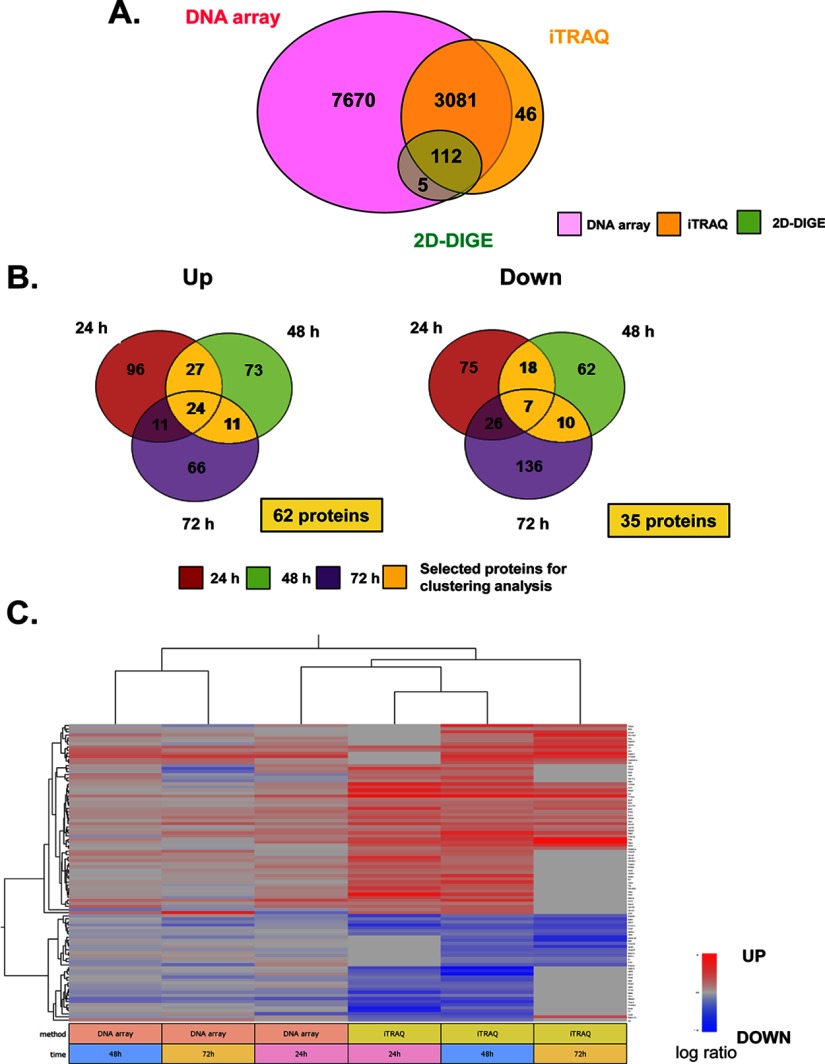
To identify the specific molecules differentially up- or down-regulated in NF1-KD cells, all data listed in iPEACH were rearranged based on the ratio of molecular expression (NF1-KD/control), and groups of up- or down-regulated molecules were extracted. In total, 3193 molecules were identified by both proteomics (2D-DIGE or iTRAQ) and transcriptomics (Fig. 2A). On NGF stimulation, proteins that were identified by iTRAQ as continuously (from 24–48 h, from 48–72 h, or from 24–72 h) up-regulated (62 proteins) or down-regulated (35 proteins) in NF1-KD cells compared with control cells were selected as the differentially expressed core proteins (Fig. 2B). The expression patterns of these 97 proteins and mRNA sequences were subjected to hierarchical clustering analysis using uncentered correlation (Fig. 2C). In the heat map, we observed that a cluster of 24 h mRNA and 24–72 h protein expression patterns were closely related, suggesting that protein expression was affected by mRNA expression at 24 h.
Fig. 2.
Cluster analysis and pathway-based characterization of differentially expressed genes and proteins from integrated proteomics. A, Venn diagrams of the number of total mRNAs/proteins identified by 2D-DIGE, iTRAQ, and DNA array. B, Venn diagrams of the number of up/down-regulated proteins identified by iTRAQ. In total, 3239 proteins were identified with a confidence limit of 95% by MALDI and ESI. The fold change of each protein expression ratio (NF1 siRNA/control siRNA) was calculated by average iTRAQ ratio in MALDI and ESI analyses at each time point. Proteins quantified with a fold change more than 20% (average iTRAQ ratio >1.20 or <0.83) were identified as differentially expressed proteins. For cluster analysis, continuously up/down-regulated proteins were selected by Venn diagrams (yellow area, 62 up-regulated proteins; 35 down-regulated proteins). C, Hierarchical clustering of the 97 continuously up/down-regulated proteins. Each vertical column represents a sample map, whereas each horizontal row represents an individual protein, with relative expression values displayed as an expression matrix (heat map) using a standardized log abundance scale ranging from −0.5 (blue) to +0.5 (red). Blue and red colors indicate decreased and increased expression, respectively: the brighter the color, the stronger the change.
GO Analysis
GO analysis was performed by an analysis with “pathway based on molecule” (based on the GO criterion) in KeyMolnet. Up-regulated molecules comprised 62 continuously up-regulated proteins identified in iTRAQ, 32 up-regulated proteins identified in 2D-DIGE, 185 genes selected based on fold-change analysis of DNA array data (24 h > 1.2 and 48 h > 1.5) supplemental Table S5. Down-regulated molecules comprised 35 proteins continuously down-regulated in iTRAQ, 20 proteins down-regulated in 2D-DIGE, and 186 genes selected based on fold-change analysis of DNA arrays (24 h < 0.83 and 48 h < 0.67) supplemental Table S. GO analysis identified “Calcium signaling” (p = 1.380 × 10−5), “Transcriptional regulation by GR” (p = 4.963 × 10−5), “Granzyme signaling pathway” (p = 8.112 × 10−5), “MMP (matrix metalloproteinase) signaling pathway” (p = 1.118 × 10−4), and “Intermediate filament signaling pathway” (p = 1.789 × 10−4) as up-regulated in NF1-KD cells (Table IA), and “Serotonin signaling pathway” (p = 6.580 × 10−6), “CaSR (calcium-sensing receptor) signaling pathway” (p = 3.507 × 10−5), ”AMPAR (alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor) signaling pathway” (p = 5.266 × 10−5), “Calcium signaling pathway” (p = 6.266 × 10−5), and “GABA (gamma-aminobutyric acid) signaling pathway” (p = 9.489 × 10−5) as down-regulated in NF1-KD cells (Table 1B). In addition, cell communication, epidermis development, and apoptotic process were noted as significant biological events in the up-regulated group, whereas those of the down-regulated group were localization, transport, and response to wounding (data not shown). These gene ontologies identified in NF1-KD cells may relate to the tumorigenesis and abnormal neuronal differentiation in NF1 pathology.
Table I. Pathway-based characterization of up- and down-regulated genes and proteins in PC12 cells identified with 2D-DIGE, iTRAQ, and DNA array analysis. KeyMolnet software was used to determine statistically overrepresented pathway categories among the up- or down-regulated molecules in NF1-KD PC12 cells. The list contained 263 up-regulated genes and proteins (iTRAQ, 62 proteins; 2D-DIGE, 32 proteins; DNA array, 185 genes) and 239 down-regulated genes and proteins (iTRAQ, 35 proteins; 2D-DIGE, 20 proteins; DNA array, 186 genes) (Fig. 2A, 2B, supplemental Table S6). The top 10 KeyMolnet pathways showing a significant association with up- or down-regulated genes or proteins are listed with rank, KeyMolnet pathway, score, and p value of the score. Bold molecules were identified by proteome analysis.
| A. Upregulated pathways. | ||||
|---|---|---|---|---|
| Rank | Keymolnet pathway | Score | Score (p value) | Protein |
| 1 | Calcium signaling pathway | 16.145 | 1.38 × 10−5 | 14-3-3, Annexin, cadherin, Crm1, DGK, Importin, Kinesin, nAChR, S100, VILIP1, vinculin |
| 2 | Transcriptional regulation by GR | 14.298 | 4.96 × 10−5 | 14-3-3h, ANXA1, AP-1, CGA, collagenase, CRF, GR, GRb, HSP70, IBAT, RGS2 |
| 3 | Wnt signaling pathway | 13.59 | 8.11 × 10−5 | b-TRCP, cadherin, CRMP, MAP, MAP1B, tub, Wnt |
| 3 | Granzyme signaling pathway | 13.59 | 8.11 × 10−5 | a2M, COLIV, HSP70, lamin, Laminin, PI-9, tub |
| 5 | MMP signaling pathway | 13.127 | 1.12 × 10−4 | a2M, COLIV, Laminin, laminin5, LRP, MMP, MMP-10, MMP-13, MMP-19, MMP-3, PAI-2 |
| 6 | Intermediate filament signaling pathway | 12.449 | 1.79 × 10−4 | 14-3-3, 14-3-3z, AP3-C, dynein, IF-II, K8, Kinesin, lamin, laminA, laminC, tub |
| 7 | Inflammasome signaling pathway | 11.866 | 2.68 × 10−4 | AIM2, caspase-1, inflammasome, IPAF, NALP1, NALP3, PI-9 |
| 8 | CD44 signaling pathway | 11.135 | 4.45 × 10−4 | COLIV, collagen, ERMP, ezrin, Laminin, MMP, tub |
| 9 | CYP family | 10.729 | 5.89 × 10−4 | CYP3A, CYP3A4, CYP3A43, CYP3A5, CYP3A7, CYP4, CYP4B1 |
| 10 | LHR signaling pathway | 10.609 | 6.40 × 10−4 | CGA, hCG, LH, LHR |
| B. Downregulated pathways. | ||||
|---|---|---|---|---|
| Rank | Keymolnet pathway | Score | Score (p value) | Protein |
| 1 | Serotonin signaling pathway | 17.213 | 6.58 × 10−6 | 5HT3R, 5HT5AR, 5HT5R, EPAC, Ga(q/11), Ga(q/11)bg, nNOS, PKC, PLA2 |
| 2 | CaSR signaling pathway | 14.799 | 3.51 × 10−5 | Ga(q/11), Ga(q/11)bg, PI4K, PKC, PMCA |
| 3 | AMPAR signaling pathway | 14.213 | 5.27 × 10−5 | Actin, AKAP5, Liprin-a, PKC, Protein 4.1G, tub |
| 4 | Calcium signaling pathway | 13.962 | 6.27 × 10−5 | Actin, CRT, Ga(q/11), Ga(q/11)bg, Kinesin, P2X, PKCg, PMCA4, troponin |
| 5 | GABA signaling pathway | 13.363 | 9.49 × 10−5 | Actin, AKAP5, GABAAR, GABAARg, GABAARg3, GABAARq, GABAR, PKC |
| 6 | Calpain signaling pathway | 13.188 | 1.07 × 10−4 | Actin, calpain, calpain8, integrin b, NF-M, PKC, PKCg, PMCA, TN-T, tub |
| 7 | HDAC signaling pathway | 13.1 | 1.14 × 10−4 | Ets, HDAC, HDAC11, HSP90, IV-HDAC |
| 8 | Melanopsin signaling pathway | 11.955 | 2.52 × 10−4 | Ga(q/11), Ga(q/11)bg, melanopsin |
| 9 | JAM family signaling pathway | 11.769 | 2.87 × 10−4 | Actin, int-aL/b2, int-aM/b2, int-aX/b2, int-b2 |
| 10 | Guanylate cyclase(receptor type) signaling pathway | 10.916 | 5.18 × 10−4 | CNGC, HSP70, HSP90, phospholamban, PMCA, Tyr3MOX |
Network Analysis
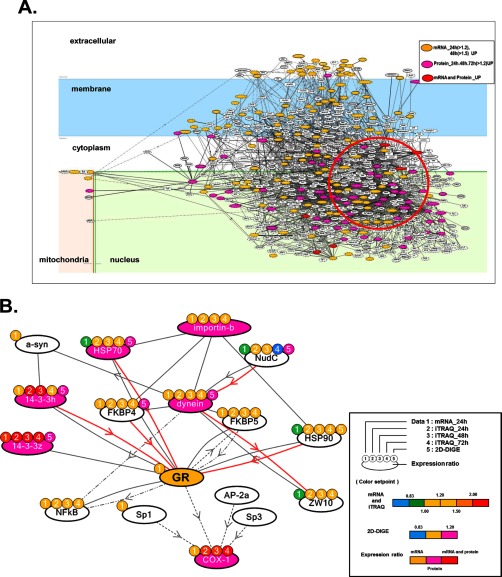
To extract abnormal signaling patterns in NF1-KD PC12 cells, we conducted a precise network analysis focused on up-regulated proteins. We performed the “start points and end points” network search algorithm using the start points of mRNA at 24 h > 1.2 and at 48 h > 1.5 and end points of up-regulated proteins identified by iTRAQ and 2D-DIGE. Fig. 3A shows the up-regulated total network in NF1-KD cells illustrated by KeyMolnet. We focused on a highly clustered area consisting of up-regulated proteins in this network (Fig. 3A, red circle), and particularly a specific network consisting of COX-1, which was continuously up-regulated at all time points in iTRAQ analysis, dynein IC2, which was highly differentially expressed by 2D-DIGE (listed as a top molecule in the iPEACH index), and GR, which was detected as an up-regulated gene in DNA array and a significant key molecule in the GO-based pathway analysis. We named this signaling pathway “dynein IC2-GR-COX-1” (Fig. 3B).
Fig. 3.
Molecular network analysis of up-regulated genes and proteins in NF1-KD PC12 cells. A, The list of genes/proteins up-regulated by the NF1 siRNA treatment in PC12 cells (supplemental Table S7) was imported into KeyMolnet. Using the “start points and end points” network search algorithm, KeyMolnet illustrated a highly complex network of targets with the most statistically significant relationships. Orange nodes represent up-regulated genes, pink nodes represent up-regulated proteins, and red nodes represent up-regulated genes and proteins. White nodes indicate additional nodes extracted automatically from the core contents of KeyMolnet to establish molecular connections. B, Extraction of the abnormal molecular network including dynein IC2, GR, and COX-1 from the molecular network shown in A. Color marks represent numerical data for each method as follows: color mark 1: 24 h mRNA, 2: 24 h iTRAQ, 3: 48 h iTRAQ, 4: 72 h iTRAQ, 5: 2D-DIGE. Color set point is described in right panel of (B). The cluster of up-regulated proteins including dynein IC2, GR, and COX-1 is highlighted by a red circle. The molecular relationships are indicated by a solid line with arrow (direct binding or activation), solid line with arrow and stop (direct inactivation), solid line without arrow (complex formation), dashed line with arrow (transcriptional activation), and dashed line with arrow and stop (transcriptional repression).
This in silico network analysis by KeyMolnet indicated that dynein is associated with GR via cargo proteins such as heat shock protein 90 and FK506-binding protein, and that the dynein complex can transport GR into the nucleus, where this GR signal may function as a transcriptional factor to up-regulate COX-1 expression. This network was strongly suggested to be up-regulated and related to the abnormal neuronal differentiation of NF1-KD cells.
Biological Validation of a Unique Pathway of Dynein IC2 Regulation, COX-1 Expression, and GR Nuclear Translocation in NF1-KD PC12 Cells
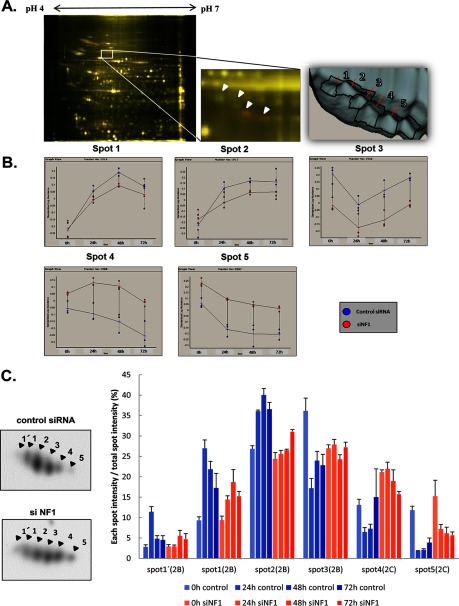
Cytoplasmic dynein 1 intermediate chain 2 (dynein IC2) was the most significantly altered protein in 2D-DIGE analysis. Dynein IC2 was identified in 5 spots, and each spot was differentially expressed on NF1 knockdown (Fig. 4A). Each spot was defined as spots 1, 2, 3, 4, and 5 from the lower pI on the 2D-DIGE, and the alteration of sequential expression levels in each spot is shown in Fig. 4B. Interestingly, spots 1–3 were continuously down-regulated, whereas spots 4 and 5 were up-regulated in NF1-KD cells. After NGF stimulation in control cells, the temporal expression of spots 1 and 2 increased, and that of spots 3 and 5 decreased. Spot 4 was also decreased in the control condition but increased in NF1-KD cells. Rat dynein IC2 is known to have two alternative splicing sites, and three isoforms were identified as IC2-A, IC2-B, and IC2-C in rat hippocampal neurons (12). The combined information of the observed molecular weight and pI in 2D-DIGE and the MS/MS analysis of spots 1–5 (supplemental Table S5, supplemental Fig. S3B) identified the detected spots as IC2-B or IC2-C.
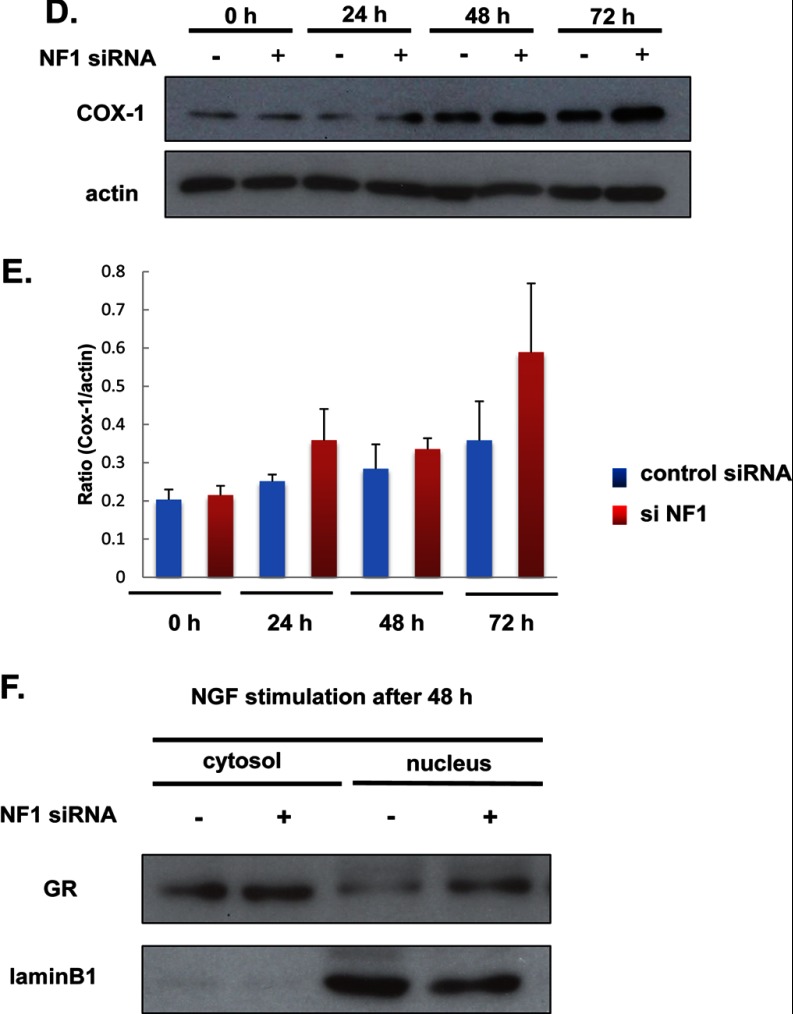
Fig. 4.
Validation of dynein IC2, GR, and COX-1 from the results of integrated proteomics. A and B, Cytoplasmic dynein 1 intermediate chain 2 (dynein IC2) in 2D-DIGE. Cellular proteins prepared from PC12 cells transfected with NF1 (249) siRNA and control siRNA were labeled with Cy3 (red) and Cy5 (green), respectively. A, Representative images of the dynein IC2 spot pattern in 2D-DIGE analysis. Dynein IC2 was identified as differentially expressed protein spots 1–5 between control and NF1 siRNA-treated PC12 cells. Arrows indicate the dynein IC2 spots (right panel). B, Time-course expression analysis of dynein IC2 by DeCyder 2D software (2-way ANOVA analysis). Suppression of NF1 expression by siRNA leads to alteration of the 2D-DIGE pattern in PC12 cells. The average normalized intensities of spots 1, 2, 3, 4, and 5 (n = 3) are shown in the graphs. Blue circle: control siRNA, red circle: NF1 siRNA, x axis: period of NGF stimulation, y axis: normalized spot intensity. C, Identification and quantification of dynein IC2 protein spots by 2D-immunoblot analysis. Left panel shows the representative dynein IC2 protein spot pattern in 2D-immunoblotting with anti-dynein IC antibodies (lower: NF1 siRNA-treated cells, upper: control siRNA-treated cells, after 48-h NGF treatment). Protein spots corresponding to spots 1′, 1, 2, 3, 4, and 5. PC12 cells were transfected with NF1 (249) siRNA or control siRNA, stimulated with NGF, and harvested after 24 h, 48 h, or 72 h. Each cell lysate sample was subjected to 2D-immunoblot analysis (7 × 5 cm) using anti-dynein IC antibodies. Percentage ratios of the intensity of each spot to total spot intensity are indicated in each histogram. The data were obtained from three separate identical experiments; error bars represent S.E. D and E, Immunoblot images and quantification of COX-1 in PC12 cells transfected with NF1 siRNA or control siRNA. D, Representative image of COX-1 immunoblot analysis. The expression of COX-1 was analyzed by immunoblotting using anti-COX-1 antibody. E, The intensity of the COX-1 band was quantified with ImageQuant software and normalized to that of actin to calculate the ratio. Data were obtained from three separate identical experiments. Error bars represent S.E. F, Analysis of GR nuclear localization by immunoblotting using anti-GR antibody. PC12 cells were transfected with NF1 (249) siRNA or control siRNA, stimulated with NGF, and harvested after 48-h NGF treatment. The cytoplasmic and nuclear proteins enriched in the cells were subjected to immunoblot analysis using anti-GR antibody. Lamin B1 was used as a marker of the nuclear fraction. Representative images of three reproducible experiments are shown.
To confirm each spot identity, we next designed specific dynein IC2-C siRNAs (termed 329 and 331) able to distinguish IC2-C from IC2-B. Knockdown of IC2-C by treatment with both 329 and 331 siRNAs significantly diminished both spots 4 and 5, which were thus concluded to be IC2-C (supplemental Fig. S3D, S3E). These results further indicated that spots 1, 2, and 3 are IC-2B. The pIs of these dynein IC2 spots were shifted in 2D-PAGE, suggesting that dynein IC2 was post-translationally modified. We analyzed their phosphorylation patterns, which may reflect dynein IC2-related cellular signaling, using a phospho-specific protein stain, Pro-Q Diamond (supplemental Fig. S3A). Dynein IC2 spots 1, 2, and 4 were responsive to the Pro-Q Diamond staining (yellow), indicating that these spots were phosphorylated. The phosphorylation of spots 2 and 4 was demonstrated on Ser 87/81/81 (dynein IC2-A/-B/-C) by MS/MS analyses (supplemental Table 5, supplemental Fig. S3A and S3C).
To observe the sequential alternation of dynein IC2 isoform expression patterns, we next analyzed them by 2D-immunoblotting using a specific dynein IC antibody. Cell lysates were prepared identically to the method used for 2D-DIGE analyses with NF1 siRNA-treated or control siRNA-treated PC12 cells and subjected to 2D-immunoblotting analysis. Six spots were identified by the anti-dynein IC antibody (Fig. 4C, left upper and lower panel), and the ratios of the intensity of each spot relative to total spot intensity were calculated (Fig. 4C, right panel). Similar results to those of 2D-DIGE, except for an additional spot 1′, were obtained. Spots 1′, 1, 2, and 3 were down-regulated, and spots 4 and 5 were up-regulated in NF1-KD cells compared with control cells. In response to NGF stimulation, phospho-staining-positive spots 1′, 1, and 2 were increased, and phospho-staining-negative spots 3, 4, and 5 were decreased in control siRNA-treated cells (Fig. 4C, right panel blue column), whereas, in NF1 siRNA-treated cells, spots 3, 4, and 5 were increased and spots 1′, 1, and 2 were decreased. Notably, the expression ratio of spot 4 (phosphorylated IC-2C) was increased in NF1-KD PC12 cells. These results suggest that suppression of NF1 causes dynamic changes of the regulation pattern of dynein IC2 alternative splicing and phosphorylation in NGF-treated PC12 cells, which may affect the abnormal neuronal differentiation related to the NF1 disease phenotype.
COX-1 and GR Protein Up-regulation in NF1-KD Cells
Next, we analyzed the functional regulation of COX-1, which was detected as a significantly up-regulated protein in NF1-KD cells by iTRAQ analysis. To observe the expression pattern of COX-1, PC12 cells were transfected with control siRNA or NF1 (249) siRNA and stimulated with NGF. Proteins were harvested from the cells at each time point (n = 3) and subjected to immunoblotting using a COX-1-specific antibody. Compared with control siRNA-treated cells, COX-1 expression was up-regulated up to 2-fold in NF1-KD cells, and after 72 h of NGF stimulation, it reached three times that of nonstimulated NF1-KD cells (Fig. 4D, 4E, n = 3). These results are consistent with the result of quantitative analysis by iTRAQ. In the network extracted by KeyMolnet, GR was assumed as an expression regulator of COX-1, and its mRNA expression was up-regulated 2-fold in NF1-KD cells compared with that of control cells after 48-h NGF stimulation, suggesting that GR activation causes the elevation of COX-1 expression.
Thus, we next analyzed the GR activation pattern in NF1-KD cells. Usually, GR exists in cytoplasm in an inactive state, and on activation, it is transported to the nucleus (13, 14). We prepared both cytoplasmic and nuclear protein fractions from NGF-stimulated NF1-KD or control cells and subjected these fractions to immunoblot analysis using an anti-GR antibody. No difference in GR expression between NF1-KD cells and control cells was observed in the cytoplasmic fraction, whereas in the nuclear fraction, GR expression was significantly higher in NF1-KD cells compared with control cells (Fig. 4F), suggesting that elevated nuclear translocation of GR caused the increase of transcriptional activity.
These results demonstrate that suppression of NF1 causes the down-regulation of dynein IC2-B expression and its phosphorylation, up-regulation of alternative splicing and phosphorylation of dynein IC2-C, and also up-regulates the nuclear translocation and activation of GR followed by up-regulation of COX-1, supporting the involvement of dynein IC2-GR-COX-1 network activation in NF1-KD cells.
Regulation of Dynein IC2 Splicing and Phosphorylation by NF1 is Related to NGF-Induced Neurite Outgrowth in PC12 Cells
This study demonstrated that the up-regulation of dynein IC2-C expression and phosphorylation is a significant event in NF1-KD cells. To investigate the function of dynein IC2-C in NF1-KD cells, the effects of the dynein IC2-C siRNA knockdown on cellular phenotype were analyzed. PC12 cells were cotransfected with NF1 and dynein IC2-C siRNAs for 24 h. Then, after 48-h NGF treatment, dynein expression in cells was analyzed by 2D-immunoblotting using anti-dynein IC antibody, confirming that expression of spots 4 and 5 (corresponding to dynein IC2-C) were reproducibly suppressed (Fig. 5A). We observed PC12 phenotypes using time-lapse microscopy and found that knockdown of cytoplasmic dynein IC2-C effectively recovered the inhibition of neurite outgrowth observed in NF1-KD PC12 cells (Fig. 5B). We measured the total length of neurites in siRNA-treated cells and found that neurite extension in cotransfected cells significantly recovered to the same level as that of control siRNA-treated cells (Fig. 5C). The number of neurites did not differ between NF1 siRNA-treated cells and both NF1 and dynein IC2-C siRNA-treated cells (data not shown). These results strongly suggest that dynein IC2-C is an important effector for the inhibition of neurite outgrowth in NF1-KD PC12 cells.
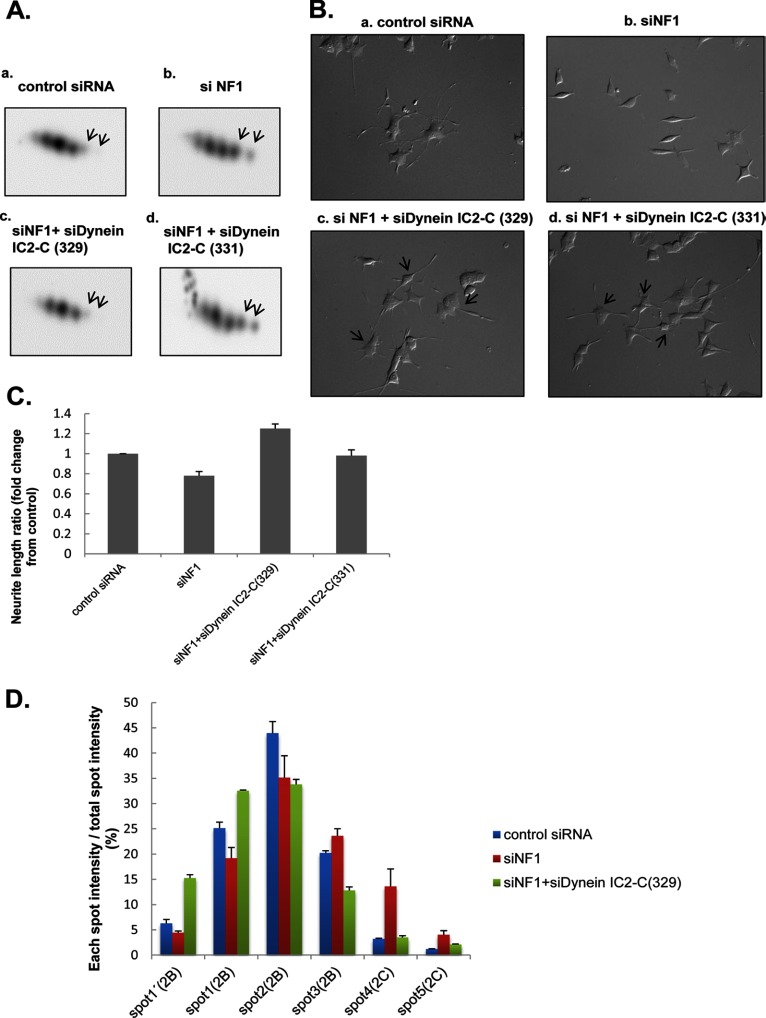
Fig. 5.
siRNA knockdown of dynein IC2-C recovered the inhibition of neurite outgrowth in NF1-KD PC12 cells. PC12 cells were transfected for 24 h with control siRNA, NF1 (249) siRNA, both NF1 (249) and dynein IC2-C (329) siRNAs, or both NF1 (249) and dynein IC2-C (331) siRNAs before treatment with NGF. After 48-h NGF treatment, dynein expression in the cells was analyzed by 2-D immunoblotting using anti-dynein IC antibody, and the neurite length of the cells was measured. A, Representative spot patterns of dynein IC2 in 2D immunoblotting of PC12 cells treated with control siRNA [a], NF1 (249) siRNA [b], NF1 (249) siRNA, and dynein IC2-C (329) siRNA [c], and NF1 (249) siRNA and dynein IC2-C (331) siRNA [d]. B, Differential interference contrast images of PC12 cells treated with control siRNA [a], NF1 (249) siRNA [b], NF1 (249) siRNA and dynein IC2-C (329) siRNA [c], and NF1 (249) siRNA and dynein IC2-C (331) siRNA [d]. C, Measurement of neurite length of PC12 cells treated with the siRNAs. The average of the total length of PC12 neurites are shown on the y axis. Data are expressed as means and S.E. of three independent experiments. For each experiment, more than 50 cells were counted. D, Comparison of the intensity of each dynein IC2 spot after transfection with NF1 siRNA and dynein IC2-C (329) siRNA. The histogram shows the percentage ratio of the intensity of each spot to the total spot intensity of dynein IC2 in individual samples. The intensity of each sample was obtained by 2D-immunoblotting using anti-dynein IC antibody followed by Cy5-labeled secondary antibody. The data were obtained from the average values of three separate identical experiments. Error bars represent S.E. of each set of three experiments.
To study the alternative splicing and phosphorylation patterns of dynein IC2 after the knockdown both of NF1 and dynein IC2-C, we analyzed changes in the intensity of six dynein IC2 spots with 2D-immunoblotting. We confirmed that inhibition of dynein IC2-C decreased the intensities of spots 4 and 5 (corresponding to dynein IC2-C), and this inhibition was subsequently related to the increase in dynein IC2-B spots. In particular, spots 1′ and 1 (representing the phosphorylated form of dynein IC2-B) were increased with concomitant decrease of spot 3 (nonphosphorylated dynein IC2-B) and spot 2 compared with those of NF1-KD cells (Fig. 5D). These findings suggest that phosphorylation of dynein IC2-B is promoted by suppression of dynein IC2-C in NF1-KD cells and is an important process underlying neurite extension in PC12 cells. This observation was also validated by the use of pan-dynein IC siRNA, which inhibits both dynein IC2-B and -C. Because neurite retraction by NF1 siRNA was not recovered by pan-dynein siRNA (supplemental Fig. S4A), increased expression and phosphorylation of dynein IC2-B may be important for the recovery of neurite outgrowth.
Validation of the Dynein IC2-GR-COX-1 Network
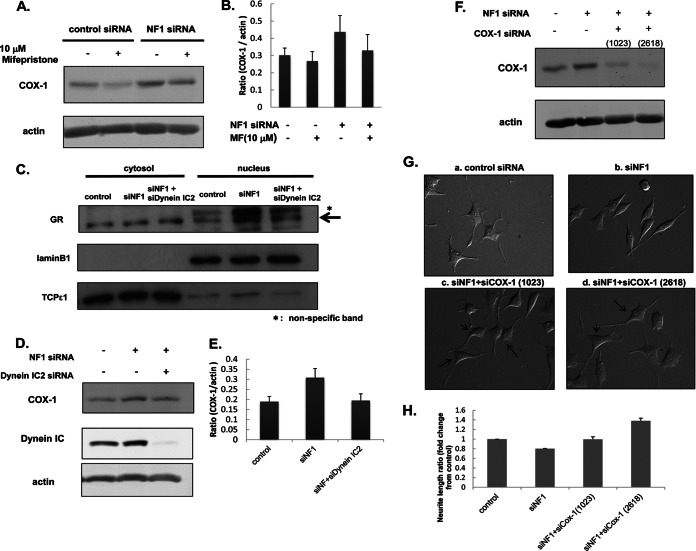
Having demonstrated the potential for GR up-regulation in NF1-KD cells to directly affect COX-1 up-regulation (Figs. 3B and 4F), we next analyzed the effect of treatment with a GR antagonist, mifepristone, on COX1 expression to investigate the interaction between GR and COX-1 in more detail. After 24-h NF1 or control siRNA transfection, PC12 cells were treated with 10 μm mifepristone for 30 min before treatment with NGF. After 48-h NGF treatment, cells were harvested for measurement of COX-1 expression by immunoblotting analysis. Up-regulation of COX-1 by NF1 siRNA was significantly decreased by mifepristone treatment to the basal level of control siRNA-treated cells (Fig. 6A, 6B), suggesting that the expression of COX-1 is regulated by GR activity in NF1-KD cells. As shown in Fig. 4F, in NF1-KD cells, GR was transported to the nucleus from cytoplasm, and this transportation could be supported by other binding proteins such as chaperones and motor proteins to form a cargo complex (15–17). Notably, dynein intermediate chain is known to play an important role in this transportation as a cargo protein (18). Thus, we speculated that dynein IC2 may be associated with this transportation of GR to induce COX-1 expression in NF1-KD cells.
Fig. 6.
Biological validation of the abnormal dynein IC2-GR-COX1 network in NF1-KD PC12 cells identified by integrated proteomics. A and B, Up-regulation of COX-1 is suppressed by GR antagonist mifepristone. After 24-h NF1 or control siRNA transfection, PC12 cells were treated with 10 μm mifepristone for 30 min before treatment with NGF. After 48-h NGF treatment, cells were harvested for measurements of COX1 by immunoblot analysis. A, representative immunoblot image of COX1. Actin was used as the internal loading control. B, Quantification of COX1 expression. The normalized intensities of COX1 obtained from three separate identical experiments are shown in the histogram. Error bars represent S.E. of each sets of three experiments. C, Nuclear translocation of GR was decreased by the knockdown of dynein IC2. PC12 cells were transfected for 24 h with control siRNA, NF1 (249) siRNA, both NF1 (249) and dynein IC2 siRNAs before treatment with NGF. After 48-h NGF treatment, cells were harvested, and the cytoplasmic and nuclear proteins were extracted. Both cytoplasmic and nuclear fractions were subjected to immunoblot analysis using anti-GR antibody. TCPε1 and Lamin B1 were used as markers of the cytoplasmic and nuclear fraction, respectively. Representative images of three reproducible experiments are shown. D and E, COX-1 expression is down-regulated by dynein IC2 siRNA treatment. PC12 cells were transfected for 24 h with control siRNA, NF1 (249) siRNA, or both NF1 (249) and dynein IC siRNAs before treatment with NGF. After 48-h NGF treatment, COX-1 expression in the cells was analyzed by immunoblotting using anti-COX-1 antibody. D, Representative immunoblot images of COX1. Actin was used as the internal loading control. Representative images of three reproducible experiments are shown. E, Quantification of COX-1 expression. The normalized COX-1 intensities obtained from three separate identical experiments are shown in the histogram. Error bars represent S.E. of each set of three experiments. F, G, and H, COX-1 knockdown recovered the inhibition of neurite outgrowth in NF1-KD PC12 cells. PC12 cells were transfected for 24 h with control siRNA, NF1 (249) siRNA, both NF1 (249) and COX-1 (1023) siRNAs, or both NF1 (249) and COX-1 (2618) siRNAs before treatment with NGF. After 48-h NGF treatment, COX-1 expression in cells was analyzed by immunoblotting with anti-COX-1 antibody, and the neurite length were measured. F, Representative immunoblot images of COX-1. Actin was used as the internal loading control. G, Differential interference contrast images of PC12 cells treated with control siRNA [a], NF1 (249) siRNA [b], NF1 (249) and COX-1 (1023) siRNAs [c], and NF1 (249) and COX-1 (2618) siRNAs [d]. H, Measurement of neurite length of PC12 cells treated with the siRNAs. The average of the total length of PC12 neurites are shown on the y axis. The data are expressed as means and S.E. of three independent experiments. For each experiment, more than 50 cells were counted.
To validate this hypothesis, we analyzed the effect of dynein IC2 knockdown on the activation of GR and COX-1 expression in NF1-KD cells. First, to examine whether dynein IC2 is involved in the increase of GR nuclear translocation triggered by NF1 knockdown, PC12 cells were cotransfected with dynein IC2 siRNA and NF1 siRNA and fractionated into cytoplasmic and nuclear proteins, which were subjected to immunoblot analysis using anti-GR antibody. Nuclear translocation of GR by NF1 knockdown was greatly decreased by cotransfection of both NF1 and dynein IC2 siRNAs (Fig. 6C).
Next, we analyzed the effects of double-knockdown of NF1 and dynein IC2 on COX-1 expression in NGF-stimulated PC12 cells. Up-regulation of COX-1 by NF1 knockdown was significantly decreased by cotransfection with NF1 and dynein IC2 siRNAs to the level of control siRNA-treated cells (Figs. 6D, 6E), whereas no effects on COX-1 expression in cells cotransfected with NF1 siRNA and dynein IC2-C siRNA were observed (supplemental Fig. S4C). From these experimental results, we speculate that dynein IC2-B function or the mutual interaction of both dynein IC2-B and -C may play an important role in the transportation of GR that affects the expression of COX-1.
Finally, we analyzed the effect of COX-1 knockdown on the cellular phenotypes of NF1-KD cells. COX-1 up-regulation in NF1-KD cells was suppressed by the cotransfection of COX-1 siRNA (Fig. 6F), and, interestingly, the neurite retraction and spindle-like alteration observed in NF1-KD cells were recovered by treatment with COX-1 siRNA (Fig. 6G). We calculated the total length of neurites in the double-knockdown PC12 cells and observed significant extension up to 1.8-times longer than those of NF1-KD cells (Fig. 6H, n = 3). The number of neurites demonstrated no difference between NF1 siRNA-treated cells and NF1 siRNA- and COX-1 siRNA-treated cells (data not shown). These results suggest that the inhibition of COX-1 up-regulation in NF1-KD cells recovers the cellular phenotypes that are abnormally altered in NF1-KD PC12 cells.
DISCUSSION
In this study, we clarified global alterations of the molecular network in NF1-disease model PC12 cells, which demonstrate abnormal phenotypes caused by NF1 knockdown. Using an integrated proteomics approach followed by network analysis in silico, we extracted key molecules that were validated after sequential cellular functional analysis using siRNA and found that a novel dynein IC2-GR-COX-1 signaling pathway is specifically activated in NF1-KD cells and related to the abnormal neuronal phenotype observed in the NF1 disease model.
In our previous study, molecules related to NGF-inducible neurite outgrowth in PC12 cells were analyzed using software called MANGO (8), which was reported as a novel, unique, and useful strategy in the global study of neural cells (8, 19). We identified 39 up-regulated-proteins and 33 down-regulated-proteins in 48 h of NGF-treated PC12 cells in that study, and unique anti-apoptotic proteins were extracted as NGF induced proteins in the PC12 cells. Among those differentiated proteins, 21 (13 up-, 8 down-regulated) proteins and 12 (7 up-, 5 down-regulated) proteins were identified as up- or down-regulated in NF1-KD cells during NGF stimulation (0, 24, 48, and/or 72 h) in this proteomics study, respectively. Interestingly, among up-regulated proteins induced by the NGF stimulation in the normal PC12 cells, the specific down-regulated proteins in NF1-KD cells were found to be related to the neuronal differentiation and the stress responses. On the other hand, the up-regulated proteins in NF1-KD cells were related to cell morphology, motility, and cell survival. It was suggested that these specific proteins found commonly in the previous study and the present study regulate cellular differentiation, motility, and apoptosis. Here, in addition to MANGO, we performed a unique comprehensive assessment using integrated proteomics with iPEACH software, a sophisticated new strategy that combines the technology of proteomic techniques, such as 2D-DIGE and iTRAQ, and transcriptomic methods, such as DNA array, and tried to identify more specific signaling in NF1-KD cells. This study is the first to report the global molecular alteration in NF1 disease model PC12 cells, and is the largest study to comprehensively identify both proteins and mRNAs in neuronal disease model cells.
Neurofibromin, the gene product of NF1, has a region homologous to the Ras-GAP domain, a negative regular of Ras, whose precise cellular signals related to NF1 disease phenotypes have yet to be clarified. We previously reported NF1-KD PC12 cells as useful NF1 disease model cells (7) to elucidate the function of clinical therapies and drug development for NF1 patients. In that study, we identified CRMP-2, which was also identified as a differentially expressed protein in this study (in the 2D-DIGE result; pH-4–7, spot no. 2389, supplemental Table S4), as a functionally related protein to the neural differentiation in NF1 model cells, and demonstrated that several inhibitors for the CRMP-2 phosphorylation responsible kinases could be useful for improving NF1 disease phenotypes (7). Using this model system, we performed integrated proteomics, which enabled us to obtain global differential information on proteins and mRNAs. The advantage of this integrated proteomics approach is that a global alteration of molecules, linked with information concerning both protein and mRNA expression levels as well as post-translational modification, can be obtained using the same sets of sample sources, such as model cells compared with normal cells. In this study, using this strategy, we precisely analyzed the molecules with respect to expression levels, alternative splicing, and modification as they continuously changed during the process of neurite outgrowth mediated by NGF treatment, and observed the specific up-regulation of dynein IC2-GR-COX-1 signaling in NF1-KD PC12 cells.
This signal consisted of dynein IC2 uniquely found in 2D-DIGE, GR detected by DNA array, and COX-1 identified by iTRAQ analysis; therefore, this finding could be the first successful example of novel identifications after the integration and extraction of all information obtained from those three methods. Based on in silico analysis of protein-protein interaction, we speculate that the dynein complex transports GR to the nucleus, where it acts as a transcriptional regulator of COX-1. Dynein is a motor protein that forms a cargo complex to transport various proteins and organelles to appropriate cellular positions (20, 21), and, especially in neuronal cells, the complex associates with kinesin to transport and import neurotransmitters (22). The dynein subunit is constituted of heavy and light chains, as well as intermediate chains, which associate with cargo proteins (23) and mediate their cellular transport function (18). Three splicing isoforms of dynein IC2, dynein IC2-A, -B, and -C, have been reported (12), with dynein IC2-B and -C predominantly expressed in PC12 cells (24).
Our study clearly demonstrated that NF1 knockdown caused the up-regulation of the alternative splicing of dynein IC2-C isoform even before and after NGF stimulation as shown in Fig. 4C. Decreased dynein IC2-C isoform and increased IC2-B isoform were observed in NGF stimulated PC12 cells (as suggested in the previous report (24)), however, in NF1-KD cells, dynein IC2-C isoform was increased and further up-regulates its phosphorylation in response to NGF stimulation. Simultaneously, the phosphorylation of dynein IC2-B splice form was suppressed in the NF1-KD cells, suggesting that neurofibromin deficiency caused the up-regulation of alternative splicing/phosphorylation of dynein IC2-C, and down-regulation of that of IC2-B. We further confirmed that dynein IC2-C knockdown using siRNA significantly recovered the inhibition of neurite outgrowth in NF1-KD PC12 cells as well as the phosphorylation of dynein IC2-B. This abnormal regulation of splicing and phosphorylation of dynein IC2 found in NF1-KD cells, may be caused by the functionally associated proteins of neurofibromin and/or other factors altered in NF1-KD cells which are under our investigation.
The enzyme responsible for phosphorylation in this system has not been identified; however, polo-like kinase (25) and casein kinase (26, 27) have been reported to phosphorylate dynein IC in vitro and in Xenopus melanophores (27), and also activations of MAPK, CDK5, GSK3, and ROCK in NF1-KD PC12 cells were found in our previous study (7). Thus, these enzymes may be involved in the regulation of neurite outgrowth in NF1-KD cells.
GR up-regulation in NF1-KD cells was detected by DNA array and extracted by the network analysis as an important factor related to the NF1-KD phenotypes. GR belongs to the nucleus receptor superfamily, members of which are normally activated in the nucleus after transportation from cytoplasm, and functions as a regulatory factor in physiological and metabolic processes (13, 14). GR up-regulation was observed during involution of hippocampus and related to depression (28); thus, GR may be a factor related to neuronal cell abnormalities. In our study, NF1 knockdown mediated the up-regulation of GR protein nuclear translocation/accumulation. An interacting protein with GR in the nucleus, 14–3-3 eta, which is activated by hormonal stimulation (29), was also identified as an up-regulated molecule in NF1-KD cells using both transcriptomics and proteomics. Binding of 14–3-3 eta inhibits the ubiquitination and degradation of GR protein (28); therefore, up-regulation of 14–3-3 eta mediated by NF1 knockdown can be speculated to contribute to GR accumulation and activation as a transcription factor, in addition to the association of the dynein complex transporting GR into the nucleus.
COX-1, a prostaglandin synthase that synthesizes prostaglandin G/H from arachidonic acid (30, 31), was one of the most highly up-regulated proteins identified among the downstream targets of GR (32) using our integrated proteomics approach. The COX family has two isoforms, COX-1 and COX-2, which are independently regulated in different manners. COX-2 expression is mediated by inflammatory response, whereas COX-1 is expressed constitutively (33). Up-regulation of COX-2 expression has been reported in cancer cells including malignant neural cancer and malignant peripheral nerve sheath tumor, and in response to inflammation and cell proliferation, it up-regulates prostaglandin synthases (34–36). Thus, many studies on COX-2 as a target of drug development have been conducted (37, 38). However, limited information concerning the function of COX-1, such as for the mucosal protection of the stomach and blood flow maintenance, has been available (39). Recently, COX-1, but not COX-2, was reported to show involvement in brain inflammation (40, 41), and, importantly, COX-1 expression is regulated in PC12 cells in response to NGF stimulation (42, 43), suggesting that COX-1 is associated with neuronal differentiation. In our study, down-regulation of COX-1 using COX-1 siRNA rescued the abnormal phenotypes observed in NF1-KD cells; thus, COX-1 may be a candidate clinical target for NF1-related disease pathogenesis. The precise mechanism of COX-1 expression and function in neuronal differentiation related to NF1 pathogenesis remains to be clarified in future investigations.
In this study, we found that COX-1 expression was regulated by GR, and dynein IC2-GR-COX-1 signaling was up-regulated in NF1-KD cells. In NF1-KD cells, suppression of dynein IC2 inhibits nuclear translocation of GR and decreases COX-1 expression. However, which dynein isoform is associated with GR transport and what mechanisms of dynein phosphorylation are involved remain unclear. Because no change in COX-1 expression was induced by dynein IC2-C siRNA treatment, dynein IC2-B rather than dynein IC2-C may be associated with GR translocation. Interestingly, COX-1 siRNA effectively recovered the neurite retraction of NF1-KD cells. We speculate that prostaglandins up-regulated by COX-1 may be associated with neurite outgrowth. Four prostaglandin E2 (PGE2) receptors, EP1, EP2, EP3, and EP4, have been identified (44), and EP2 and EP3 expression was up-regulated at 48 h and 72 h, respectively, in our DNA array analysis. EP3 has been reported to couple with G protein receptors to activate RhoA (45, 46), which was also identified as up-regulated in NF1-KD HeLa cells in our previous report (10). Several studies show that activation of RhoA mediates neurite retraction in PC12 cells (45, 46). Therefore, up-regulation of COX-1 in NF1-KD cells is speculated to promote PGE2 synthesis and the activation of RhoA via EP3, followed by the inhibition of neurite outgrowth in PC12 cells. COX-1 knockdown may also block this signaling activation and improve the abnormal phenotype observed in NF1-KD cells.
In NF1 patients, a cognitive deficit has frequently been found as one of the typical NF1-related phenotypes and suggested that neurofibromin plays an essential role in neuronal cells. NF1 heterozygous (Nf1+/−) mice indicated spatial learning disability by the Morris water maze test (3, 47) and other reports also showed retraction of neurite and growth corn in NF1 heterozygous (Nf1+/−) hippocampal neurons (48). Therefore abnormal neuronal assembly such as neurite retraction is related to learning disability in NF1 patients. In our study, we used PC12 cells as model cells of neuronal differentiation to evaluate the function of neurofibromin. We firstly demonstrated that neurite retraction observed in NF1-KD cells is recovered by cotransfected dynein IC2-C siRNA (Figs 5B, 5C) or COX-1 siRNA (Figs. 6G, 6H). These experimental evidences may explain the possible functional relevance of the abnormal regulation of dynein IC2-GR-COX1 signaling to the NF1 pathology. Further studies based on our findings will elucidate the mechanism of NF1-related neuronal pathogenesis, such as learning disability.
This study is the first demonstration using an integrated proteomics approach that the specific signaling of dynein IC2-GR-COX-1 extracted from NF1-disease model neuronal cells may be an important mechanism of NF1-related pathogenesis. Drugs inhibiting this signaling such as specific COX-1 inhibitors may be useful candidates for the treatment of NF1 patients with neuronal abnormalities.
Acknowledgments
We thank the entire staff of the Department of Tumor Genetics and Biology at Kumamoto University for helpful support, especially M. Nagai for experimental assistances, and C. Midorikawa, K. Tanoue, and M. Nakamura for secretarial support.
Footnotes
* This study was supported by grants from the Ministry of Health, Labor and Welfare of Japan (NA), Kiban B Research (NA), Houga Research (NA), Exploratory Research for Young Scientists (B) (DK) from the Ministry of Education, Science, Sports, and Culture of Japan; and from the COE project B of Kumamoto University for proteomic research and education (NA).
 This article contains supplemental Figs. S1 to S4 and Tables S1 to S7.
This article contains supplemental Figs. S1 to S4 and Tables S1 to S7.
Authors' present addresses: S Mizuguchi, Dept. Neurosurgery, Faculty of Medicine, University of Miyazaki, 5200 Kihara Kiyotake Miyazaki, 889-1692, Japan; T Morikawa, Mitsubishi Chemical Medience Corporation, 14, Sunayama, Kamisu, Ibaraki 314-0255, Japan.; U. Midorikawa, Healthcare Systems Laboratories, Sharp Corp. 1-9-2, Nakase, Mihama-ku, Chiba, 261-8520, Japan; A.C. Yoshizawa, Koichi Tanaka Laboratory of Advanced Science and Technology, Shimadzu Corporation, Nishinokyo-Kuwabara-cho, Nakagyo-ku, Kyoto 604-8511, Japan.
1 The abbreviations used are:
- NF1-KD
- NF1 knockdown
- 2D-DIGE
- two-dimensional fluorescence difference gel electrophoresis
- iTRAQ
- isobaric tagging for relative and absolute quantitation
- iPEACH
- Integrated Protein Expression Analysis Chart
- NGF
- nerve growth factor
- GO
- gene ontology
- MANGO
- Molecular Annotation by Gene Ontology
- Qq-TOF
- quadrupole/quadrupole/time-of-flight mass spectrometers
- dynein IC
- dynein intermediate chain
- GR
- glucocorticoid receptor
- COX-1
- cyclooxygenase-1
- siRNA
- short interfering RNA
- MPNST
- malignant peripheral nerve sheath tumor
- PGE2
- prostaglandin E2.
REFERENCES
- 1. Stephens K., Riccardi V. M., Rising M., Ng S., Green P., Collins F. S., Rediker K. S., Powers J. A., Parker C., Donis-Keller H. (1987) Linkage studies with chromosome 17 DNA markers in 45 neurofibromatosis 1 families. Genomics 1, 353–357 [DOI] [PubMed] [Google Scholar]
- 2. Cawthon R. M., Weiss R., Xu G. F., Viskochil D., Culver M., Stevens J., Robertson M., Dunn D., Gesteland R., O'Connell P., et al. (1990) A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell 62, 193–201 [DOI] [PubMed] [Google Scholar]
- 3. Costa R. M., Federov N. B., Kogan J. H., Murphy G. G., Stern J., Ohno M., Kucherlapati R., Jacks T., Silva A. J. (2002) Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature 415, 526–530 [DOI] [PubMed] [Google Scholar]
- 4. Guo H. F., Tong J., Hannan F., Luo L., Zhong Y. (2000) A neurofibromatosis-1-regulated pathway is required for learning in Drosophila. Nature 403, 895–898 [DOI] [PubMed] [Google Scholar]
- 5. Gregory P. E., Gutmann D. H., Mitchell A., Park S., Boguski M., Jacks T., Wood D. L., Jove R., Collins F. S. (1993) Neurofibromatosis type 1 gene product (neurofibromin) associates with microtubules. Somat. Cell Mol. Genet. 19, 265–274 [DOI] [PubMed] [Google Scholar]
- 6. Yunoue S., Tokuo H., Fukunaga K., Feng L., Ozawa T., Nishi T., Kikuchi A., Hattori S., Kuratsu J., Saya H., Araki N. (2003) Neurofibromatosis type I tumor suppressor neurofibromin regulates neuronal differentiation via its GTPase-activating protein function toward Ras. J. Biol. Chem. 278, 26958–26969 [DOI] [PubMed] [Google Scholar]
- 7. Patrakitkomjorn S., Kobayashi D., Morikawa T., Wilson M. M., Tsubota N., Irie A., Ozawa T., Aoki M., Arimura N., Kaibuchi K., Saya H., Araki N. (2008) Neurofibromatosis type 1 (NF1) tumor suppressor, neurofibromin, regulates the neuronal differentiation of PC12 cells via its associating protein, CRMP-2. J. Biol. Chem. 283, 9399–9413 [DOI] [PubMed] [Google Scholar]
- 8. Kobayashi D., Kumagai J., Morikawa T., Wilson-Morifuji M., Wilson A., Irie A., Araki N. (2009) An integrated approach of differential mass spectrometry and gene ontology analysis identified novel proteins regulating neuronal differentiation and survival. Mol. Cell. Proteomics 8, 2350–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., Schaeffer D. A. (2007) The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 10. Ozawa T., Araki N., Yunoue S., Tokuo H., Feng L., Patrakitkomjorn S., Hara T., Ichikawa Y., Matsumoto K., Fujii K., Saya H. (2005) The neurofibromatosis type 1 gene product neurofibromin enhances cell motility by regulating actin filament dynamics via the Rho-ROCK-LIMK2-cofilin pathway. J. Biol. Chem. 280, 39524–39533 [DOI] [PubMed] [Google Scholar]
- 11. Ha J., Lo K. W., Myers K. R., Carr T. M., Humsi M. K., Rasoul B. A., Segal R. A., Pfister K. K. (2008) A neuron-specific cytoplasmic dynein isoform preferentially transports TrkB signaling endosomes. J. Cell Biol. 181, 1027–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pfister K. K., Salata M. W., Dillman J. F., 3rd, Vaughan K. T., Vallee R. B., Torre E., Lye R. J. (1996) Differential expression and phosphorylation of the 74-kDa intermediate chains of cytoplasmic dynein in cultured neurons and glia. J. Biol. Chem. 271, 1687–1694 [DOI] [PubMed] [Google Scholar]
- 13. Picard D., Yamamoto K. R. (1987) Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J. 6, 3333–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Madan A. P., DeFranco D. B. (1993) Bidirectional transport of glucocorticoid receptors across the nuclear envelope. Proc. Natl. Acad. Sci. U.S.A. 90, 3588–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Galigniana M. D., Radanyi C., Renoir J. M., Housley P. R., Pratt W. B. (2001) Evidence that the peptidylprolyl isomerase domain of the hsp90-binding immunophilin FKBP52 is involved in both dynein interaction and glucocorticoid receptor movement to the nucleus. J. Biol. Chem. 276, 14884–14889 [DOI] [PubMed] [Google Scholar]
- 16. Harrell J. M., Murphy P. J., Morishima Y., Chen H., Mansfield J. F., Galigniana M. D., Pratt W. B. (2004) Evidence for glucocorticoid receptor transport on microtubules by dynein. J. Biol. Chem. 279, 54647–54654 [DOI] [PubMed] [Google Scholar]
- 17. Wochnik G. M., Ruegg J., Abel G. A., Schmidt U., Holsboer F., Rein T. (2005) FK506-binding proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. J. Biol. Chem. 280, 4609–4616 [DOI] [PubMed] [Google Scholar]
- 18. Vaughan P. S., Leszyk J. D., Vaughan K. T. (2001) Cytoplasmic dynein intermediate chain phosphorylation regulates binding to dynactin. J. Biol. Chem. 276, 26171–26179 [DOI] [PubMed] [Google Scholar]
- 19. BioBit, ASBMB Journal Science (2009) A Mango Smoothies. ASBMB Today August 2009, 31 [Google Scholar]
- 20. Schnapp B. J., Reese T. S. (1989) Dynein is the motor for retrograde axonal transport of organelles. Proc. Natl. Acad. Sci. U.S.A. 86, 1548–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schroer T. A., Steuer E. R., Sheetz M. P. (1989) Cytoplasmic dynein is a minus end-directed motor for membranous organelles. Cell 56, 937–946 [DOI] [PubMed] [Google Scholar]
- 22. Hirokawa N., Niwa S., Tanaka Y. (2010) Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68, 610–638 [DOI] [PubMed] [Google Scholar]
- 23. Pfister K. K., Shah P. R., Hummerich H., Russ A., Cotton J., Annuar A. A., King S. M., Fisher E. M. (2006) Genetic analysis of the cytoplasmic dynein subunit families. PLoS Genet 2, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Salata M. W., Dillman J. F., 3rd, Lye R. J., Pfister K. K. (2001) Growth factor regulation of cytoplasmic dynein intermediate chain subunit expression preceding neurite extension. J. Neurosci. Res. 65, 408–416 [DOI] [PubMed] [Google Scholar]
- 25. Bader J. R., Kasuboski J. M., Winding M., Vaughan P. S., Hinchcliffe E. H., Vaughan K. T. (2011) Polo-like kinase1 is required for recruitment of dynein to kinetochores during mitosis. J. Biol. Chem. 286, 20769–20777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Karki S., Tokito M. K., Holzbaur E. L. (1997) Casein kinase II binds to and phosphorylates cytoplasmic dynein. J. Biol. Chem. 272, 5887–5891 [DOI] [PubMed] [Google Scholar]
- 27. Ikeda K., Zhapparova O., Brodsky I., Semenova I., Tirnauer J. S., Zaliapin I., Rodionov V. (2011) CK1 activates minus-end-directed transport of membrane organelles along microtubules. Mol. Biol. Cell 22, 1321–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Anacker C., Zunszain P. A., Cattaneo A., Carvalho L. A., Garabedian M. J., Thuret S., Price J., Pariante C. M. (2011) Antidepressants increase human hippocampal neurogenesis by activating the glucocorticoid receptor. Mol. Psychiatry 16, 738–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim Y. S., Jang S. W., Sung H. J., Lee H. J., Kim I. S., Na D. S., Ko J. (2005) Role of 14-3-3 eta as a positive regulator of the glucocorticoid receptor transcriptional activation. Endocrinology 146, 3133–3140 [DOI] [PubMed] [Google Scholar]
- 30. Marnett L. J., Rowlinson S. W., Goodwin D. C., Kalgutkar A. S., Lanzo C. A. (1999) Arachidonic acid oxygenation by COX-1 and COX-2. Mechanisms of catalysis and inhibition. J. Biol. Chem. 274, 22903–22906 [DOI] [PubMed] [Google Scholar]
- 31. Smith W. L., Garavito R. M., DeWitt D. L. (1996) Prostaglandin endoperoxide H synthases (cyclooxygenases)-1 and -2. J. Biol. Chem. 271, 33157–33160 [DOI] [PubMed] [Google Scholar]
- 32. Sun H., Sheveleva E., Chen Q. M. (2008) Corticosteroids induce cyclooxygenase 1 expression in cardiomyocytes: role of glucocorticoid receptor and Sp3 transcription factor. Mol. Endocrinol. 22, 2076–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smith W. L., DeWitt D. L., Garavito R. M. (2000) Cyclooxygenases: structural, cellular, and molecular biology. Annu. Rev. Biochem. 69, 145–182 [DOI] [PubMed] [Google Scholar]
- 34. Dang I., De Vries G. H. (2011) Aberrant cAMP metabolism in NF1 malignant peripheral nerve sheath tumor cells. Neurochem. Res. 36, 1697–1705 [DOI] [PubMed] [Google Scholar]
- 35. Park S. W., Kim H. S., Choi M. S., Jeong W. J., Heo D. S., Kim K. H., Sung M. W. (2011) The effects of the stromal cell-derived cyclooxygenase-2 metabolite prostaglandin E2 on the proliferation of colon cancer cells. J. Pharmacol. Exp. Ther. 336, 516–523 [DOI] [PubMed] [Google Scholar]
- 36. Rundhaug J. E., Pavone A., Kim E., Fischer S. M. (2007) The effect of cyclooxygenase-2 overexpression on skin carcinogenesis is context dependent. Mol. Carcinog. 46, 981–992 [DOI] [PubMed] [Google Scholar]
- 37. Masferrer J. L., Zweifel B. S., Manning P. T., Hauser S. D., Leahy K. M., Smith W. G., Isakson P. C., Seibert K. (1994) Selective inhibition of inducible cyclooxygenase 2 in vivo is antiinflammatory and nonulcerogenic. Proc. Natl. Acad. Sci. U.S.A. 91, 3228–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Muraki C., Ohga N., Hida Y., Nishihara H., Kato Y., Tsuchiya K., Matsuda K., Totsuka Y., Shindoh M., Hida K. (2012) Cyclooxygenase-2 inhibition causes antiangiogenic effects on tumor endothelial and vascular progenitor cells. Int. J. Cancer 130, 59–70 [DOI] [PubMed] [Google Scholar]
- 39. Peskar B. M. (2001) Role of cyclooxygenase isoforms in gastric mucosal defence. J. Physiol. Paris 95, 3–9 [DOI] [PubMed] [Google Scholar]
- 40. Anrather J., Gallo E. F., Kawano T., Orio M., Abe T., Gooden C., Zhou P., Iadecola C. (2011) Purinergic signaling induces cyclooxygenase-1-dependent prostanoid synthesis in microglia: roles in the outcome of excitotoxic brain injury. PLoS One 6, e25916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Choi S. H., Aid S., Bosetti F. (2009) The distinct roles of cyclooxygenase-1 and -2 in neuroinflammation: implications for translational research. Trends Pharmacol. Sci. 30, 174–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kaplan M. D., Olschowka J. A., O'Banion M. K. (1997) Cyclooxygenase-1 behaves as a delayed response gene in PC12 cells differentiated by nerve growth factor. J. Biol. Chem. 272, 18534–18537 [DOI] [PubMed] [Google Scholar]
- 43. Yung H. S., Chow K. B., Lai K. H., Wise H. (2009) Gi-coupled prostanoid receptors are the likely targets for COX-1-generated prostanoids in rat pheochromocytoma (PC12) cells. Prostaglandins Leukot. Essent. Fatty Acids 81, 65–71 [DOI] [PubMed] [Google Scholar]
- 44. Sugimoto Y., Narumiya S. (2007) Prostaglandin E receptors. J. Biol. Chem. 282, 11613–11617 [DOI] [PubMed] [Google Scholar]
- 45. Tamiji J., Crawford D. A. (2010) Prostaglandin E(2) and misoprostol induce neurite retraction in Neuro-2a cells. Biochem. Biophys. Res. Commun. 398, 450–456 [DOI] [PubMed] [Google Scholar]
- 46. Katoh H., Negishi M., Ichikawa A. (1996) Prostaglandin E receptor EP3 subtype induces neurite retraction via small GTPase Rho. J. Biol. Chem. 271, 29780–29784 [DOI] [PubMed] [Google Scholar]
- 47. Silva A. J., Frankland P. W., Marowitz Z., Friedman E., Laszlo G. S., Cioffi D., Jacks T., Bourtchuladze R. (1997) A mouse model for the learning and memory deficits associated with neurofibromatosis type I. Nat. Genet. 15, 281–284 [DOI] [PubMed] [Google Scholar]
- 48. Brown J. A., Gianino S. M., Gutmann D. H. (2010) Defective cAMP generation underlies the sensitivity of CNS neurons to neurofibromatosis-1 heterozygosity. J. Neurosci. 30, 5579–5589 [DOI] [PMC free article] [PubMed] [Google Scholar]