Abstract
High confidence definition of protein interactions is an important objective toward the understanding of biological systems. Isotope labeling in combination with affinity-based isolation of protein complexes has increased in accuracy and reproducibility, yet, larger organisms—including humans—are hardly accessible to metabolic labeling and thus, a major limitation has been its restriction to small animals, cell lines, and yeast.
As composition as well as the stoichiometry of protein complexes can significantly differ in primary tissues, there is a great demand for methods capable to combine the selectivity of affinity-based isolation as well as the accuracy and reproducibility of isotope-based labeling with its application toward analysis of protein interactions from intact tissue.
Toward this goal, we combined isotope coded protein labeling (ICPL)1 with immunoprecipitation (IP) and quantitative mass spectrometry (MS). ICPL-IP allows sensitive and accurate analysis of protein interactions from primary tissue.
We applied ICPL-IP to immuno-isolate protein complexes from bovine retinal tissue. Protein complexes of immunoprecipitated β-tubulin, a highly abundant protein with known interactors as well as the lowly expressed small GTPase RhoA were analyzed. The results of both analyses demonstrate sensitive and selective identification of known as well as new protein interactions by our method.
Classical antibody-based strategies to determine protein interactions have long been hampered by the fact that most binders exhibit unspecific binding. Immunoprecipitations—the most widely used method—not only suffer from nonspecific binding because of compromised selectivity and specificity of the immunoglobulin, but also from nonspecific binding to the carrier beads. Because of this lack of specificity, a large proportion of reported protein interactions in the literature as well as in databases that gather interaction data are likely to be compromised by false positives. Furthermore, despite great advancements in sensitivity and accuracy of mass spectrometers and peptide separation techniques, mass spectrometry-based identifications usually fail to detect low-abundance members of protein complexes, medium affinity or transient binders. Several methods have tackled these problems. Tandem affinity purification (TAP) has resulted in an unprecedented specificity, concerning protein interaction data (1, 2). Yet this method is limited by the fact that recombinant expression of a TAP-fusion protein is required and additionally hampered by the risk that exogenous expression of the bait protein of interest may result in an artificial change of stoichiometries.
To circumvent these drawbacks, Selbach and Mann developed a quantitative immunoprecipitation, combined with RNAi (QUICK), using stable isotope labeling with amino acids in cell culture (SILAC) to gain improved selectivity (3–5). The main advantage of QUICK is that endogenous protein stoichiometries are the basis for immunoprecipitation, for the first time allowing one to accurately monitor protein interactions at endogenous protein concentrations from living cells and discriminate true positive from false positive interactions. Yet this method requires metabolic isotope labeling of whole organisms or reference cells, as described for SuperSILAC, to allow comparative analysis of two protein sets (6). Metabolic labeling, especially when applied to living organisms, requires feeding them with isotopic food (7–9). The procedure of labeling living animals or plants metabolically is time-consuming (1–2 generations for ∼93% labeled proteins) and also connected to high financial expenses (8). Because of these constraints, larger organisms like pigs, cows, as well as humans are not amenable to metabolic labeling and therefore, their tissue is not experimentally accessible in this way. The use of reference cell-line derived material bears the limitation that tissue-specific proteins that are not expressed in the reference material will not be detected at all.
To overcome these limitations and attempt quantitative analysis of protein complexes from primary tissues that cannot be metabolically labeled we combined IP, isotope coded protein labeling (ICPL) (10)—a method of chemical isotopic protein labeling—with mass spectrometry and advanced computational analysis of spectra. The major benefits of this MS-based quantitative “ICPL-IP” compared with traditional IPs are: (1) Identification of native protein complexes can be controlled via ICPL, monitoring maximal three samples in combination with comparative quantitative MS allowing highly sensitive as well as comparative detection of complex components. (2) Nonspecific binders to the bead material, the antibody as well as other contaminants, are filtered out by using a differentially isotope-labeled reference sample of the same tissue prepared as an appropriate control (e.g., beads plus unrelated immunoglobulins). Vice versa, specific binders can be clearly discriminated by their enriched abundance through quantitative MS. (3) The approach is unbiased; it does not depend on subsequent immunoblotting with antibodies against suspected bait proteins.
As a result, this workflow allows the sensitive and selective identification of protein complex components isolated by IP as well as other affinity-based methods via comparative quantitative MS. Additionally, comparative assessment of protein abundances is possible. This allows comparison of different physiological states in a given tissue or pathological changes associated with disease.
EXPERIMENTAL PROCEDURES
Cell Culture
HEK293T cells were cultured as described previously (11). For SILAC experiments, HEK293T cells were grown in SILAC DMEM (PAA Laboratories GmbH (Coelbe, Germany)) supplemented with 3 mm l-Glutamine (PAA), 10% dialyzed fetal bovine serum (PAA), 0.55 mm lysine, 0.4 mm arginine, 50 units/ml Penicillin and 0.05 mg/ml Streptomycin. Light SILAC medium was supplemented with 12C6 14N2 lysine and 12C6 14N4 arginine. Heavy SILAC medium was supplemented with 13C6 15N2 lysine and 13C6 15N4 arginine. Proline (0.5 mm) was added to all SILAC media to prevent arginine to proline conversion (12). All amino acids were purchased from Silantes (München, Germany).
Retina Preparation
Bovine eyes were obtained from a local slaughterhouse. The retinae were dissected and stored in cold isolation medium (20% (w/v) sucrose, 20 mm Hepes-HCl pH 7.2, 2 mm MgCl2, 130 mm NaCl). For dark-adapted retinae, bovine eyes were kept in CO2-independent medium (Invitrogen, Carlsbad, CA) in the dark on ice for 3 h until dissection. Subsequently retinae were transferred to clear or black 1.5 ml Eppendorf tubes, frozen in liquid nitrogen and stored at −80 °C until further use. All further preparations and experiments with dark-adapted retinae/rod outer segments (ROS) were carried out under dim red light.
Isolation of Photoreceptor Rod Outer Segments (ROS)
ROS were isolated from bovine retinae according a modified protocol from Schmitt et al. (13). Briefly, frozen retinae were thawed on ice. Three retinae were added to 8 ml of 50% sucrose in HBS (115 mm NaCl, 2 mm KCl, 2 mm MgCl2, 10 mm HEPES, pH 7), vortexed for 30 s and left on ice for 10 min. The mixture was transferred to an ultracentrifugation tube and 1 ml of HBS was added to the top. After centrifugation for 30 min at 50,000 × g the “pellet” was taken from the interphase between 50% sucrose and 1 ml HBS and transferred to a new 15 ml falcon tube, washed once with HBSS (PAA) for 5 min at 2000 × g. The pellet was collected in 1 ml of HBS and transferred on the top of a discontinuous sucrose gradient (25%, 30%, 35%, 40%, 45% in HBS). Subsequent to centrifugation (1 h, 50,000 × g) purified ROS were collected between 25 and 35% sucrose and washed once with HBSS (5 min, 2000 × g). ROS were collected in isolation medium and protein concentration was determined by Bradford (14). Three milligram aliquots were frozen in liquid nitrogen and stored at −80 °C until further use.
Immunoprecipitation
A monoclonal antibody specific for β-tubulin (Sigma Aldrich), a polyclonal anti-β-tubulin (Sigma Aldrich), and an anti-RhoA antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was used for immunoprecipitation. Mouse and rabbit IgGs (Sigma Aldrich) as well as an anti-RGS9 (Epitomics) and anti-GAPDH antibody (Millipore) were used in control experiments. Frozen retinal tissue and ROS (light- or dark-adapted), respectively, were homogenized in 0.7 ml ice-cold retina/ROS lysis buffer (20 mm Hepes pH 7.8, 150 mm NaCl, 1 mm EDTA, complete© (Roche) protease inhibitors and 1% n-dodecyl-β-d-maltoside (Sigma Aldrich)) using pellet pestles (Sigma Aldrich) and a G-20 gauge needle. Lysates were incubated for 20 min at 4 °C with overhead rotation and were subsequently subjected to centrifugation at 16,000 × g for 10 min.
Metabolically labeled (SILAC light and heavy) and normal HEK293T cells were washed with PBS (PAA), harvested in ice-cold cell lysis buffer (50 mm Hepes pH 7.2, 150 mm NaCl, 0.5% Nonidet P-40, complete© (Roche) protease inhibitors) homogenized with a G-20 gauge needle, lyzed on ice for 30 min and cleared by 10 min centrifugation at 16,000 × g. The cleared supernatants of either retinal origin or HEK cells were transferred to fresh microcentrifuge tubes and protein concentrations were determined by Bradford (14). For ICPL-triplex IPs light- and dark-adapted ROS equal to 3 mg protein were used and for SILAC-IPs or ICPL-duplex IPs lysates (HEK or light-adapted retina) equal to 10 mg protein were transferred to microspin columns (Sartorius Stedim Biotech). Subsequently samples were incubated with 20 μg antibodies or 20 μg control immunoglobulins/antibodies (IgGs, anti-RGS9, anti-GAPDH) for 2 h at 4 °C with overhead rotation. 90 μl protein G plus agarose (Santa Cruz Biotechnology) was added to each tube before overnight incubation (4 °C) with overhead rotation. The precipitates were washed three times with 0.5 ml retina/ROS or cell lysis buffer. Bound proteins were eluted two times by incubation with 200 μl 6 m guanidine HCl pH 8.5. Samples were concentrated using Vivaspin 500 columns (10 kDa cutoff) to a volume of 20 μl and washed once with 0.5 ml 6 m guanidine HCl pH 8.5. After transfer to new microcentrifuge tubes, ICPL-labeling was applied for the control and IP as described below.
ICPL-Labeling (SERVA) and In-solution Cleavage
Labeling was performed according to the manufacturer's instructions. Briefly 0.5 μl reduction solution was added to each sample and the mixtures were incubated for 30 min at 60 °C. The samples were cooled down to room temperature and 0.5 μl of freshly prepared alkylation reagent was added to each sample before the samples were incubated for 30 min at room temperature in the dark. To stop the reaction, 0.5 μl stop solution 1 was added to each sample and the samples were incubated for 15 min at 25 °C. For duplex ICPL labeling, 3 μl of ICPL0-Nic-reagent solution was added to the control sample and 3 μl of ICPL6-Nic-reagent solution to the IP sample. Triplex ICPL-labeling conditions were as follows: ICPL0 for control sample, ICPL6 for IP-light-sample and ICPL10 for IP-dark-sample. All samples were overlaid with argon to exclude oxidation, vortexed (10 s), sonicated for 1 min and incubated for 2 h at 25 °C. Two microliters of stop solution 2 was added to each sample before the samples were further incubated for 20 min at 25 °C to destroy excess reagent. Both ICPL-light and -heavy labeled samples were combined and vortex thoroughly. The pH of the mixture was adjusted to 11.9 ± 0.1 by adding 2 N NaOH to destroy possible esterification products. After 20 min the same amount of 2 N HCl was added to neutralize the sample. SILAC samples were reduced and alkylated correspondingly. For duplex ICPL- and SILAC-labeled samples 650 μl of 25 mm Hepes pH 8 and for triplex ICPL-labeled samples 950 μl of 25 mm Hepes pH 8 was added, to dilute salt concentration and samples were cleaved by sequence grade modified trypsin (Sigma Aldrich) overnight at 37 °C.
Peptide Purification and Mass Spectrometry
In-solution cleaved samples were desalted and preconcentrated using 200 μl StageTips (Thermo Fisher Scientific) before LC-MS analysis according to the manufacturer's instructions with modifications. In brief, StageTips were initialized with 20 μl 80% acetonitrile (ACN) and 5% trifluoroacetic acid (TFA). Re-equilibration was performed with 20 μl 5% TFA. Seven hundred microliters of sample containing of 5% TFA were applied to the StageTips in 200 μl steps. Washing was performed with 20 μl 5% TFA and the samples were eluted once with 20 μl 50% ACN, 5% TFA and twice with 20 μl 80% ACN, 5% TFA. Samples were vacuum-concentrated to almost complete dryness and re-suspended in 20 μl 0.5% TFA (15, 16).
LC-MS/MS analysis was performed on an Ultimate3000 nano rapid separation LC system (Dionex) coupled to a LTQ Velos mass spectrometer (Thermo Fisher Scientific) by a nano spray ion source basically as described earlier (17). Tryptic peptide mixtures were automatically injected and loaded at flow rate of 6 μl/min in 0.5% TFA in HPLC grade water onto a Acclaim® Pepmap100 (75 μm × 2 cm, C18, 3 μm, 100 Å, Dionex) column. After 5 min, peptides were eluted and separated on a Acclaim® Pepmap RSLC (75 μm × 25 cm, C18, 2 μm, 100 Å, Dionex) column by a linear gradient from 2% to 35% of buffer B (80% acetonitrile, 0.08% formic acid in HPLC grade water) in buffer A (2% acetonitrile, 0.1% formic acid in HPLC grade water) at a flow rate of 300 nl/min over 150 min. Remaining peptides were eluted by a short gradient from 35% to 100% buffer B in 5 min. The eluted peptides were analyzed by the LTQ Velos mass spectrometer. From the high resolution MS prescan with a mass range of 300 to 1500, the 10 most intense peptide ions were automatically selected for fragment analysis in the linear ion trap if they exceeded an intensity of at least 200 counts and if they were at least doubly charged. The normalized collision energy for CID was set to a value of 35 and the resulting fragments were detected with normal resolution in the linear ion trap. The lock mass option was activated; the background signal with a mass of 445.12002 was used as lock mass (18). Every ion selected for fragmentation was excluded for 20 s by dynamic exclusion.
Data Analysis
For ICPL and SILAC experiments all acquired spectra were processed and analyzed using the MaxQuant software (19, 20) (version 1.1.1.19 and 1.3.0.5) and the IPI (www.ebi.ac.uk) bovine database (version 3.73, 30,403 entries) for bovine retinal tissue/ROS or the human specific IPI database (version 3.80, 86,719 entries) for HEK293T cells.
Endoproteinase Arg-C was set as cleaving enzyme. Cysteine carbamidomethylation was selected as fixed modification, methionine oxidation and protein acetylation were allowed as variable modifications. Peptide minimum length 6 and two missed cleavages were allowed. As labels ICPL0 (105.02146 Da) and ICPL6 (111.04159 Da) for duplex and additional ICPL10 (115.06669 Da) for triplex were chosen. For SILAC analysis Lys8/Arg10 was adjusted. The peptide and protein false discovery rates were set to 1%. The initial mass tolerance for precursor ions was set to 6 ppm. The mass tolerance for fragment ions was set to 0.5 Da. Contaminants like keratins or immunoglobulins were removed. For the β-tubulin experiments only proteins identified and quantified by at least two peptides and two heavy/light counts per experiment in at least two of three (polyclonal anti β-tubulin antibody) or four of five (monoclonal anti β-tubulin antibody) independent biological experiments were considered for further analysis. For statistical analysis the Perseus software (version 1.2.0.17 and 1.3.0.4) was used (20). The normalized ratios were log2 transformed and the significance A was calculated. The threshold for significant enrichment was set to p value<0.01. For the detection of the RhoA protein complex, only proteins identified and quantified by at least one unique peptide and one ratio count that were enriched in at least three of six biological replicates for light- or dark-adapted samples were considered for further analysis. Only proteins that were detected as significantly enriched compared with the corresponding control (significance A, p < 0.05) were considered as potential components of the RhoA-complex. The detection of light-induced alterations was achieved by direct comparison of RhoA-complexes, purified from light- and dark-adapted ROS, only considering proteins identified to be components of the RhoA-complex. Proteins identified and quantified in at least six of 12 biological replicates by at least one unique peptide and one ratio count were considered for further analysis. A significance A p value below 0.05 was set as threshold for light-induced alterations. Visualization of the data was done with the R software (http://www.r-project.org). Full data analysis (e.g. peptide and protein identification, post-translational modifications, quantification data) and protein lists are supplied as supplementary material.
RESULTS
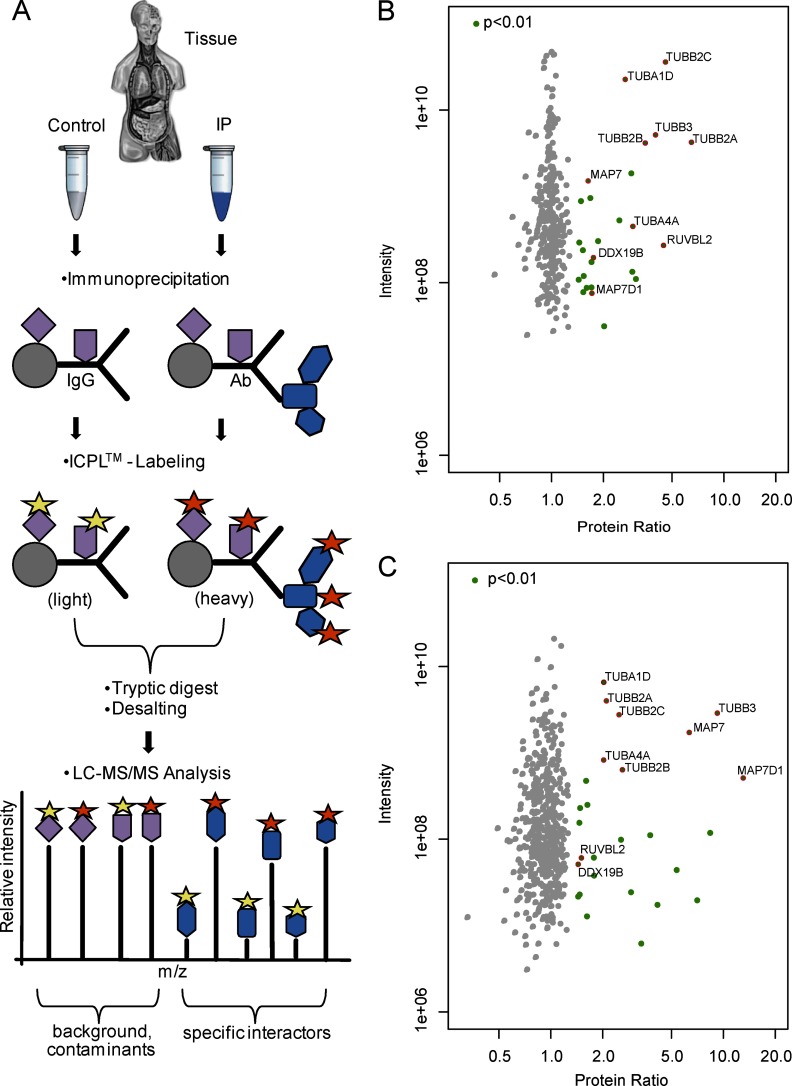
Native bovine retinal tissue was analyzed with β-tubulin as first bait. Bovine retinae were lysed and equal amounts of the protein lysate were used either as a control or for the IP. To purify the β-tubulin protein complex, a monoclonal β-tubulin antibody and a second polyclonal antibody directed against β-tubulin was used, whereas controls were incubated with species-specific IgGs or an anti-RGS9 antibody as a nonspecific control antibody. Precipitated protein complexes were washed, eluted, and—after reduction and alkylation—labeled either with the light ICPL0 (control) or with the heavy ICPL6 (IP). Following labeling, both samples were combined. Combining the samples early on minimizes the experimental error and appears as an advantage over sample combination on the peptide level as necessary for e.g., iTRAQ labeling (21) as well as over separated mass spectrometric analysis of the samples in label-free approaches. As a result, ICPL increases the likelihood of identifying protein complex components by decreasing the quantitative inaccuracy especially once lowly abundant peptides are compared. In our workflow, combined samples were tryptically cleaved in solution. Peptides were desalted by the use of StageTips (15, 16) and analyzed by online liquid chromatography-tandem mass spectrometry (LC-MS/MS) on a LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). MaxQuant with the integrated Andromeda Search Engine was used for both, protein identification, and quantification (19, 20). Subsequently, specific protein complex components were reliably distinguished from nonspecific binders and contaminants by significant enrichment in the IP, compared with the control sample (Fig. 1A).
Fig. 1.
ICPL-IP: A novel approach for the quantitative protein complex analysis from native tissue. A, Experimental scheme of the ICPL-IP by using a combination of immunoprecipitation, isotope coded protein labeling, and mass spectrometry. Equal amounts of lyzed tissue are split into a control and IP. The IP contains the specific antibody, whereas the control only contains species-specific IgGs. Following IP, the samples are differently labeled (yellow and orange stars), mixed and analyzed by LC-MS/MS. After software-based analysis, nonspecific binders (purple) can be easily determined by ratios of 1:1. Specific binders to the protein of interest (blue) are detected by significant enrichment in the IP. B–C, Detection of β-tubulin protein complex components by ICPL-IP in retinal tissue. Proteins were immunoprecipitated using a monoclonal (B) or a polyclonal (C) β-tubulin antibody, respectively. Plotted are log10 ratios (x-axis) and log10 intensities (y-axis) for each quantified protein. Significantly enriched proteins in one of the two ICPL-IPs (green, p < 0.01), nonspecific binders (gray), and proteins significantly enriched in both cases (red, p < 0.01) are indicated (for details see Experimental Procedures and Table I).
As expected, we found different β-tubulin subunits as the major components of the precipitated protein complex. Overall we identified and quantified 327 proteins (monoclonal β-tubulin antibody) and 524 proteins (polyclonal β-tubulin antibody), respectively, with a minimum of two identified peptides and two heavy/light counts (supplemental Tables S1 and S2). Most of the proteins were not significantly enriched and were equally abundant in both, the control and the IP sample. Therefore, these were considered as nonspecific contaminants and background (Fig. 1B and 1C). We found a total of 10 proteins with significantly increased abundance ratios (p < 0.01) in both experiments. Six of those are either β-tubulin or α-tubulin subunits, two are microtubule-associated proteins (22, 23). The two remaining proteins DDX19B and RUVBL2 were not considered so far as tubulin interacting proteins (Table I).
Table I. Proteins found significantly (p < 0.01) enriched in two different β-tubulin ICPL-IPs [(β-tubulin(m), β-tubulin(p)] from bovine retinal tissue. Monoclonal β-tubulin antibody [β-tubulin(m)], polyclonal β-tubulin antibody [β-tubulin(p)].
| Ratio H/L normalized β-tubulin(m) | Ratio H/L normalized β-tubulin(m) Significance B | Ratio H/L normalized β-tubulin(p) | Ratio H/L normalized β-tubulin(p) Significance B | Uniprot ID | Gene Name | Protein Names | Tubulin/Interactor |
|---|---|---|---|---|---|---|---|
| 6.26 | 3.47E-36 | 2.58 | 1.08E-09 | Q6B856 | TUBB2A | Tubulin beta-2B chain | yes |
| 4.46 | 7.09E-25 | 2.46 | 5.46E-09 | Q3MHM5 | TUBB2C | Tubulin beta-2C chain | yes |
| 4.31 | 8.07E-24 | 1.43 | 7.29E-03 | Q2TBU9 | RUVBL2 | RuvB2-like protein | unknown |
| 3.87 | 8.69E-21 | 9.15 | 2.80E-42 | Q2T9S0 | TUBB3 | Tubulin beta-3 chain | yes |
| 3.38 | 3.28E-17 | 2.46 | 3.28E-17 | Q2KJD0 | TUBB2B | Tubulin beta-5 chain | yes |
| 2.80 | 6.14E-13 | 2.00 | 3.73E-06 | P81948 | TUBA4A | Tubulin alpha-4A chain | yes |
| 2.58 | 2.76E-11 | 2.01 | 3.59E-06 | Q2HJ86 | TUBA1D | Tubulin alpha-1D chain | yes |
| 1.69 | 1.08E-04 | 1.49 | 3.45E-03 | Q2YDF3 | DDX19B | (Asp-Glu-Ala-As) box polypeptide 19B | unknown |
| 1.57 | 6.36E-04 | 6.29 | 4.68E-30 | A6QNZ5 | MAP7 | Microtubule-associated protein 7 | yes |
| 1.48 | 2.66E-03 | 12.93 | 1.77E-55 | F1N1I9 | MAP7D1 | MAP7 domain-containing protein 1 | yes |
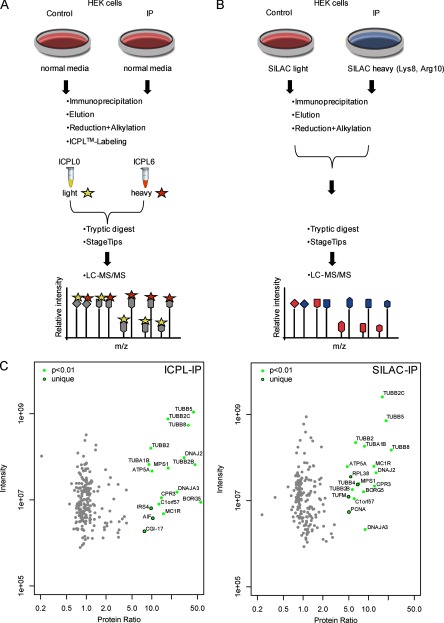
As a proof of the accuracy and reliability of the ICPL-IP approach, we benchmarked our method to SILAC (4–8). To this end, we used cultured HEK cells instead of tissue and again enriched the β-tubulin complex. HEK293T cells were metabolically labeled via SILAC (Fig. 2B); simultaneously HEK293T cells were grown in normal media for subsequent ICPL labeling (Fig. 2A). After lysis of the cells, equal amounts of lysates were used for SILAC and ICPL-IP experiments. The IP samples were incubated with the monoclonal β-tubulin antibody; controls were incubated with species-specific IgGs instead. Precipitated protein complexes were washed, eluted, and combined (SILAC light, heavy) or labeled with light ICPL0 (control) and heavy ICPL6 (IP) and combined following the chemical labeling. All proteins were cleaved in solution with trypsin, resulting peptides were desalted using StageTips (15, 16) and analyzed by online liquid chromatography-tandem mass spectrometry (LC-MS/MS) on a LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). For protein identification and quantification the MaxQuant Software with the integrated Andromeda Search Engine was used (19, 20).
Fig. 2.
Comparison of ICPL and SILAC for quantitative immunoprecipitation. A, Experimental scheme of the quantitative β-tubulin ICPL-IP using cultured HEK293T cells. Equal amounts of cell lysate are taken either for the control or for the IP. After IP, samples are ICPL-labeled and analyzed by LC-MS/MS. Specific interaction partners are determined by significant enrichment in the IP, whereas nonspecific binders are identified by ratios of 1:1. B, Experimental scheme of the quantitative β-tubulin IP with SILAC-labeled HEK293T cells. The workflow is equivalent to the ICPL-IP procedure except that proteins in the cells are already metabolically SILAC-labeled. C, Detection of specific protein complex components and nonspecific background of the β-tubulin ICPL-IP and SILAC-IP in HEK293T cells. Plotted are log10 ratios (x axis) and log10 intensities (y axis) for each protein quantified. Significantly enriched proteins (p < 0.01) found in common are plotted in green. Uniquely detected β-tubulin complex components (p < 0.01) for each approach are shown with black circles, nonspecific binders in gray (for details see Experimental Procedures and Table II).
As for the tissue-based approach, we could identify different subunits of β-tubulin as major components of the precipitated protein complexes for both, the ICPL and the SILAC-labeled proteins. Altogether 17 of 228 proteins were found to be significantly enriched (p < 0.01) with the ICPL-IP and 18 (p < 0.01) of 205 with the SILAC-IP (Fig. 2C, Table II, supplemental Tables S3 to S7). All specifically enriched proteins have been previously described as either tubulin/microtubule-associated proteins or associated with its binary interactors. A comparison shows that 14 of the 17/18 significantly enriched proteins were identical for both labeling approaches (Table II, supplemental Table S7).
Table II. Comparison of the enriched (p < 0.01) proteins of the β-tubulin ICPL-IP versus SILAC-IP from HEK293T cells. Ratios are the mean of three biological replicates.
| ICPL-IP |
SILAC-IP |
Uniprot ID | Gene Name | Protein Names | Tubulin/Interactor | ||
|---|---|---|---|---|---|---|---|
| Ratio β-tubulin/control Normalized | Ratio β-tubulin/control Normalized Significance A | Ratio β-tubulin/control Normalized | Ratio β-tubulin/control Normalized Significance A | ||||
| 56.51 | 2.14E-06 | 8.55 | 5.93E-04 | Q00587-1 | BORG5 | Binder of Rho GTPases 5 | yes |
| 46.29 | 6.22E-06 | 5.76 | 4.39E-03 | Q9BVA1 | TUBB2B | Tubulin beta-2B chain | yes |
| 43.83 | 8.26E-06 | 18.86 | 3.57E-06 | P07437 | TUBB | Tubulin beta chain;Tubulin beta-5 chain | yes |
| 36.51 | 2.07E-05 | 22.72 | 8.64E-07 | Q3ZCM7 | TUBB8 | Tubulin beta-8 chain | yes |
| 31.40 | 4.29E-05 | 13.29 | 4.13E-05 | P31689 | DNAJ2 | DnaJ homolog subfamily A member 1 | yes |
| 24.53 | 1.33E-04 | 9.04 | 4.33E-04 | Q96EY1-1 | DNAJA3 | DnaJ homolog subfamily A member 3 | yes |
| 17.94 | 5.02E-04 | 7.25 | 1.43E-03 | P42677 | MPS1 | 40S ribosomal protein S27 | yes |
| 17.81 | 5.17E-04 | 16.67 | 8.74E-06 | P68371 | TUBB2C | Tubulin beta-2 chain;Tubulin beta-2C chain | yes |
| 15.20 | 9.65E-04 | 12.33 | 6.69E-05 | Q01726 | MC1R | Melanocortin receptor 1 | yes |
| 14.18 | 1.26E-03 | 12.58 | 5.88E-05 | O60884 | CPR3 | Cell cycle progression restoration gene 3 protein | yes |
| 12.97 | 1.75E-03 | 6.16 | 3.21E-03 | Q9BSD7 | C1orf57 | Nucleoside triphosphate phosphohydrolase | yes |
| 10.38 | 3.84E-03 | - | - | O95831-1 | AIF | Apoptosis-inducing factor 1 | yes |
| 10.17 | 4.11E-03 | 4.83 | 9.60E-03 | P25705 | ATP5A | ATP synthase subunit alpha | yes |
| 9.81 | 4.64E-03 | - | - | O14654 | IRS4 | 160 kDa phosphotyrosine protein; Insulin receptor substrate 4 | yes |
| 9.76 | 4.72E-03 | 6.47 | 2.53E-03 | Q13885 | TUBB2 | Tubulin beta-2A chain | yes |
| 9.15 | 5.83E-03 | 8.84 | 4.91E-04 | P68363 | TUBA1B | Alpha-tubulin ubiquitous;Tubulin alpha-1B chain | yes |
| 7.74 | 9.87E-03 | - | - | Q9BRX2 | CGI-17 | Protein pelota homolog | yes |
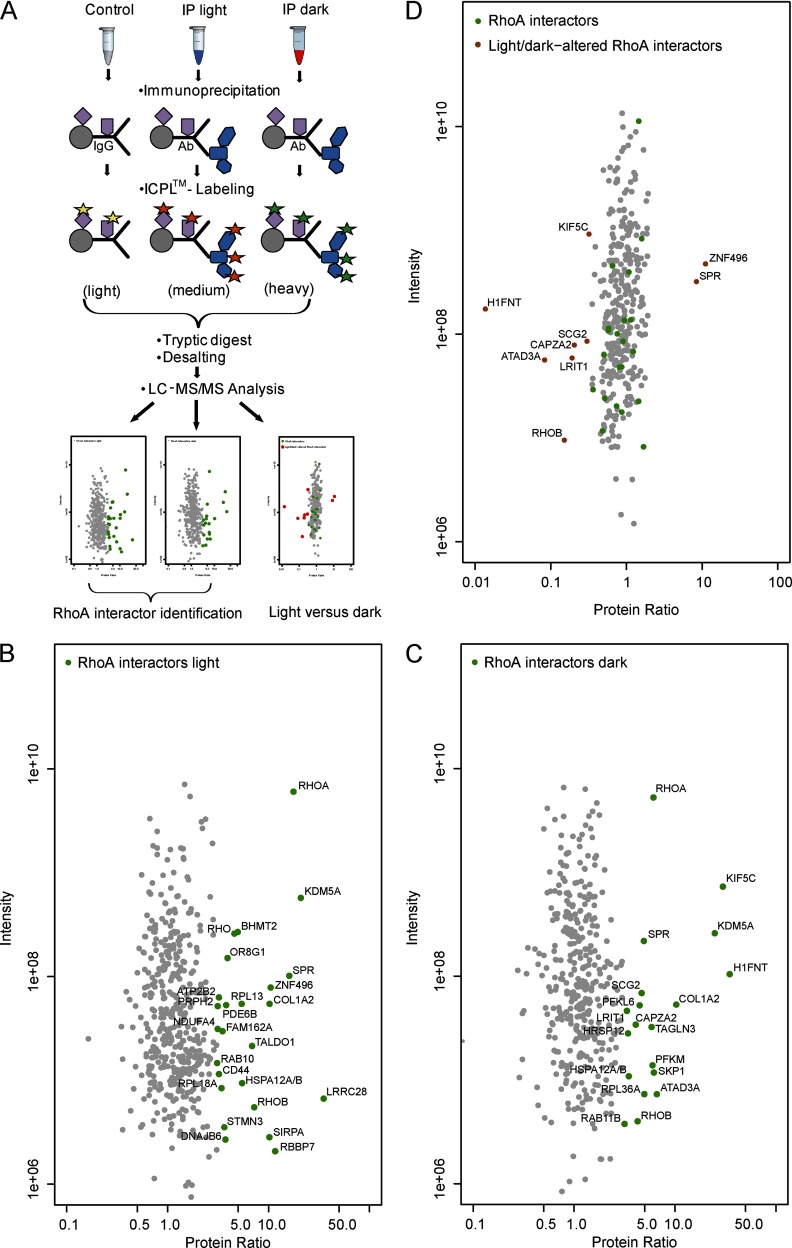
To challenge our ICPL-IP approach and to demonstrate the possibility also to compare the protein complex composition of two physiological states, enabled by applying triplex ICPL-labeling, we selected the low abundantly expressed small GTPase RhoA. This GTPase plays a key role in the regulation of actomyosin contractility. RhoA was shown to be expressed 12-times lower than β-tubulins in bovine rod photoreceptor outer segments (ROS) (24). To determine and compare RhoA-associated complexes, ROS were isolated from light- or dark-adapted bovine retinae, lyzed and equal amounts of protein lysate were used for either controls or IPs. For one half of the ICPL-IP triplex approach we compared an IP of light-adapted ROS (IPlight) with the corresponding light-adapted control (Clight). For the second half, we performed the corresponding dark-adapted ROS approach (IPdark, Cdark). To detect light-induced alterations, we directly compared IPlight and IPdark in each experiment (Fig. 3A). The use of the different controls (Clight or Cdark) was necessary, because of the light-induced translocation of several proteins from the photoreceptor outer to the inner segment as well as the other way around, leading to concentration changes during light- and dark-adaptation (25).
Fig. 3.
Light-induced alterations within the RhoA-complex. Applying an ICPL triple labeling approach, RhoA interactors (p < 0.05) were identified by affinity-purifying the complexes from light- and dark-adapted ROS using a RhoA-specific antibody. A, Schematic representation of the ICPL-IP triplex workflow. IP of light-adapted (IP light) and dark-adapted ROS (IP dark) are compared with appropriate controls. Controls are labeled with ICPL0 (light), IP light with ICPL6 (medium) and IP dark with ICPL10 (heavy). RhoA interactors in light and dark are determined as well as RhoA-complex alterations in IP light versus IP dark. Specifically bound proteins are detected by their significant enrichment compared with the control sample. B, Significantly enriched (p < 0.05) RhoA interactors in the IP light. Potential RhoA interactors are highlighted in green. C, Significantly enriched (p < 0.05) RhoA interactors in the IP dark. Potential RhoA interactors are highlighted in green. D, Light-induced alterations (p < 0.05) are detected by direct comparison of RhoA-complexes purified from light- and dark-adapted ROS. Only potential RhoA interactors (highlighted in green) were considered for this comparative study of different physiological states. Proteins showing a light-induced alteration in association to the RhoA-complex are additionally labeled by red circles. (B–D) Plotted are the log10 ratios on the x-axes and the log10 intensities on the y-axes. (for details see Experimental Procedures, Table III and supplemental Table S8).
Precipitation of RhoA was achieved by using a monoclonal antibody against RhoA, whereas controls were incubated with species-specific IgGs or an anti-GAPDH antibody as a nonspecific control antibody. Samples were treated the same as described for the β-tubulin duplex ICPL-IP assay before, besides the fact that dark- or light-adapted retinae were used as protein source and that labeling was performed in triplex settings. The light ICPL0 label was used either for Clight or Cdark, whereas ICPL6 (medium) was used for the IPlight and ICPL10 (heavy) for the IPdark. With MS-derived peptide identification and quantification based on MaxQuant (19), specific protein complex components were reliably distinguished from nonspecific binders as well as contaminants discriminated by significant enrichment in the IP compared with the corresponding control sample. IPlight and IPdark were directly compared and differences in the protein complex composition in the light- versus dark-adapted state were analyzed. Overall 439 (IPlight) and 392 (IPdark) proteins, respectively, could be identified and quantified (supplemental Table S8). Again most proteins were not significantly enriched, therefore these were considered as nonspecific contaminants and background. Twenty-five proteins were found to be significantly enriched in the IPlight (p < 0.05; Fig. 3B, supplemental Table S8) whereas 19 showed significant enrichment in the IPdark (p < 0.05, Fig. 3C, supplemental Table S8). One important interaction of RhoA with the visual G protein-coupled receptor rhodopsin, already described by Kiel et al. (26), was found as significantly enriched in light-adapted ROS. Also enriched under light conditions are rod cGMP-specific PDE6B, PRPH2 as well as two other small GTPases, RhoB and Rab10. Proteins exclusively enriched in the dark-adapted RhoA-complex are the small GTPase Rab11B and LRIT1.
The direct comparison of light- and dark-adapted RhoA-complexes revealed that the majority of its protein interactions in the retina are not affected by these two different physiological states. Nine proteins, however, showed light-induced alteration in binding to RhoA. Two of those showed increased binding (SPR, ZNF496), whereas the remaining (ATAD3A, CAPZA2, H1FNT, KIF5C, LRIT1, RhoB, and SCG2) were weaker associated to the RhoA-complex in light (Fig. 3D, Table III, supplemental Table S8).
Table III. Light-induced alterations in the RhoA-complex. Ratios are the mean of 12 biological replicates. Proteins highlighted by “+” show a significant light-induced alteration in association to the RhoA-complex.
| Ratio light/dark Normalized | Ratio light/dark Normalized A significant | Ratio light/dark Normalized Significance A | Uniprot ID | Gene Name | Protein Names |
|---|---|---|---|---|---|
| 0.01 | + | 3.38E-20 | E1BIJ7 | H1FNT | Testis-specific H1 histone |
| 10.74 | + | 5.57E-09 | E1B7Z6 | ZNF496 | ZNF496 Zinc finger protein 496 |
| 8.16 | + | 1.83E-07 | Q17QK8 | SPR | Sepiapterin reductase |
| 0.08 | + | 2.74E-07 | A7YWC4 | ATAD3A | ATPase family AAA domain-containing protein 3 |
| 0.15 | + | 1.50E-04 | Q3ZBW5 | RHOB | Rho-related GTP-binding protein RhoB |
| 0.19 | + | 1.13E-03 | E1BKZ2 | LRIT1 | LRIT1 Leucine-rich repeat-containing protein 21 |
| 0.20 | + | 1.91E-03 | Q5E997 | CAPZA2 | CAPZA2 F-actin-capping protein subunit alpha-2 |
| 0.29 | + | 0.03 | P20616 | SCG2 | SCG2 Secretogranin-2 |
| 0.31 | + | 0.04 | Q0II58 | KIF5C | Kinesin family member 5C |
| 0.35 | 0.07 | Q0IIG5 | PFKM | 6-phosphofructokinase, muscle type | |
| 1.63 | 0.10 | F1MFX1 | SIRPA | Tyrosine-protein phosphatase non-receptor type substrate 1 | |
| 1.54 | 0.13 | F1MQ59 | KDM5A | Lysine-specific demethylase 5A | |
| 1.41 | 0.19 | P61585 | RHOA | Transforming protein RhoA | |
| 1.41 | 0.19 | Q3T003 | RPL18A | 60S ribosomal protein L18a | |
| 0.47 | 0.24 | Q3MHP2 | RAB11B | Ras-related protein Rab-11B | |
| 0.49 | 0.28 | A1A4J1 | PFKL | 6-phosphofructokinase, liver type | |
| 0.50 | 0.31 | Q3ZCF3 | SKP1 | S-phase kinase-associated protein 1 | |
| 1.19 | 0.35 | Q2NKR7 | FAM162A | Protein FAM162A | |
| 0.56 | 0.44 | P02465 | COL1A2 | Collagen alpha-2(I) chain | |
| 1.11 | 0.45 | P23439 | PDE6B | Rod cGMP-specific 3,5-cyclic phosphodiesterase subunit beta | |
| 0.57 | 0.46 | P17810 | PRPH2 | Peripherin-2 | |
| 1.05 | 0.52 | F1N4C6 | BHMT2 | S-methylmethionine–homocysteine S-methyltransferase | |
| 0.63 | 0.63 | P02699 | RHO | Rhodopsin | |
| 0.91 | 0.74 | F1MN60 | ATP2B2 | Plasma membrane calcium-transporting ATPase 2 | |
| 0.87 | 0.81 | Q56JZ1 | RPL13 | 60S ribosomal protein L13 | |
| 0.72 | 0.84 | F1MJ70 | HSPA12A | Heat shock 70 kDa protein 12A | |
| 0.73 | 0.88 | Q2TBL6 | TALDO1 | Transaldolase 1 | |
| 0.84 | 0.88 | Q29423 | CD44 | CD44 antigen | |
| 0.84 | 0.88 | A6QLS9 | RAB10 | Ras-related protein Rab-10 | |
| 0.78 | 1.00 | Q01321 | NDUFA4 | NADH dehydrogenase 1 alpha | |
| n.def. | 1.00 | G3MZZ2 | OR8G1 | Olfactory receptor 8G1 | |
| n.def. | 1.00 | A4IFK9 | STMN3 | Stathmin-3 | |
| n.def. | 1.00 | Q32KX5 | LRRC28 | Leucine-rich repeat-containing protein 28 | |
| n.def. | 1.00 | Q3SWX8 | RBBP7 | Histone-binding protein RBBP7 | |
| n.def. | 1.00 | Q0III6 | DNAJB6 | HSP40 homolog | |
| n.def. | 1.00 | Q3ZBY2 | TAGLN3 | Transgelin-3 | |
| n.def. | 1.00 | Q3SZ59 | RPL36A | 60S ribosomal protein L36a | |
| n.def. | 1.00 | Q3T114 | HRSP12 | Ribonuclease UK114 |
DISCUSSION
Here we describe a new method to accurately analyze protein-protein interactions from primary tissues using isotope coded protein labeling (ICPL), immunoprecipitation, and quantitative mass spectrometry.
Via dissection of two interactomes, the β-tubulin and the RhoA interactomes, immunoprecipitated from retinal tissue and from ROS, respectively, we can show that this method can determine protein interactions with high confidence. Because of its high content of disk membranes ROS are a challenging tissue. The results of the β-tubulin IP demonstrate that the differential enrichment of specific binders over the control allows discrimination between binders and nonspecific background, as virtually all significantly enriched proteins are either tubulin subunits or tubulin-associated proteins. No obvious false positive complex components were detected. Among the significantly enriched interactors one likely new β-tubulin interacting protein was identified (DDX19B) in both IPs, using different antibodies and appropriate controls. Because this is a RNA helicase, there is no obvious functional link to tubulin yet. Its presence in both experiments, however, strongly suggests that there is a physical link between tubulin and DDX19B. To further validate this and to identify the functional consequences of this interaction, further studies, beyond the scope of this manuscript would be necessary. The second candidate which was not described yet to be a β-tubulin interacting protein, RUVBL2, is closely related and shares high sequence homology to RUVBL1 that is reported to be a tubulin interactor (27).
The benchmark of the ICPL-IP to the SILAC-IP shows that it is at least comparable to SILAC as only in three (ICPL), as opposed to four (SILAC) significantly enriched proteins differ between the two experimental setups, all of which were described to be tubulin/microtubule interactors as well (Fig. 2C, Table II, supplemental Table S7). Two (AIF, IRS4) of the three proteins, significantly enriched in the ICPL-IP only, were also detected in the SILAC-IP, although below the stringent significance threshold. The remaining protein (CGI-17) could not be identified and quantified with the SILAC approach. From the four proteins only significantly (p < 0.01) enriched with the SILAC-IP, three proteins (TUBB4, PCNA, TUFM) could not be quantified with ICPL, because the identified peptides are devoid of lysine and therefore not labeled. The fourth protein (RPL38) was found only in one out of three ICPL-IP experiments and was therefore filtered out by our stringent filter criteria. We identified the IRS4 as a specific β-tubulin interactor by the ICPL-IP (p < 0.01), whereas a previous study described β-tubulin to be a contaminant in a nonquantitative IRS4-pulldown (28). Because we could not detect IRS4 as a specific tubulin interactor in our SILAC approach (p value 0.052), the conclusiveness of the interaction still remains ambiguous.
Tubulins are highly abundant proteins and therefore not the most challenging targets. We therefore selected the low abundant small GTPase RhoA and immunoprecipitated its complexes from bovine photoreceptor outer segments. Preparation of immunoprecipitates from this specialized part of the photoreceptor cell is highly challenging as ROS are tightly packed with membranous discs and RhoA has to be solubilized out of the intermembranous space. Nevertheless, we were able to identify components of the RhoA-complex in ROS. The low abundance of RhoA resulted in an increased variability, given that we were working close to the limit of detection.
Generally, the family of Rho GTPases seems to be involved in the regulation of the cytoskeleton. The activation of Rho leads to the assembly of contractile actin-myosin filaments and of associated focal adhesion complexes. The current hypothesis is that Rho family GTPases act as molecular switches to control a signal transduction pathway that links membrane receptors to the cytoskeleton (29–31).
In addition to the known RhoA interactor rhodopsin (26), structurally a membrane integral G-protein coupled receptor, we were able to identify novel components of the RhoA-complex (Figs. 3B and 3C, supplemental Table 8). These are for example RhoB and TAGLN3, which both are involved in actin organization and vesicle transport (32–35). As rhodopsin was previously shown to be interconnected with RhoA and involved in actin cytoskeleton assembly and dynamics, both interactors strengthen the hypothesis that rhodopsin, besides its role of initiating the signal transduction of light that enables us to see, is involved in regulating outer segment structure via interaction with proteins regulating cytoskeletal dynamics.
STMN3, another interactor identified by the ICPL-IP of RhoA, further points to its role in regulating microtubular dynamics, as STMN3 was described, when activated, to sequester microtubular filaments (36, 37). Rab10 and Rab11, both previously identified as proteins expressed in photoreceptor outer segments, are likely to participate in vesicle trafficking along cytoskeletal routes (38), as is KIF5C (39, 40). These interactors indicate a potential role of RhoA in cytoskeletal reorganization in ROS.
Given, that RhoA was found to act downstream of the light receptor rhodopsin and to test, whether complex composition may be altered by light, we employed a triple ICPL-labeling strategy to detect and quantify eventual light-induced alterations within the RhoA-complex. As expected, the majority of the RhoA protein interactions are not affected by light (Fig. 3D, Table III, supplemental Table S8). However, the abundance of nine proteins (H1FNT, ZNF496, SPR, ATAD3A, RHOB, LRIT1, CAPZA2, SCG2, KIF5C) is significantly altered within the RhoA-complex in reaction to light. Seven out of these nine proteins are involved in endocytic trafficking.
Despite its role in regulating the cytoskeleton, there is increasing evidence describing the Rho GTPase subfamily in several aspects of endocytic trafficking (41). The endocytic system carries out specialized tasks in receptor recycling, degradation, cargo sorting, and transporting. This is depending on a network of interacting proteins like the Rho GTPase family, actin, and also on microtubules (42, 43) The light-induced alterations that we found within the RhoA-complex suggest that RhoA could be involved in light-induced endocytic-like processes, as most of the interactors that showed a light-induced alteration in binding to the RhoA-complex have been described to be involved in endocytic processes or in the regulation of both, the actin and microtubulin cytoskeleton.
RhoB, for example, has been suggested to participate in regulating endosomal trafficking (34, 44). Specifically, RhoB seems to recruit proteins to endosomes and apparently activates them (33). Because of the high sequence homology between RhoA and RhoB (82% sequence homology) and as RhoA is reported to bind with itself (45), an interaction between both proteins, as we could detect it, is likely to be true. CAPZA2 and SCG2, both play an active and essential role in assisting and even driving certain stages of the endocytic process, like the formation and movement of endocytic vesicles or participation in the vesicle sorting and packaging (46, 47). SPR might also be involved in endocytic processes; however, its involvement is rather indirect by regulating the levels of nitric oxide (48), which in turn has an impact on RhoA activity (49). The Kinesin family member KIF5C, a microtubule-based motor protein, is an example for the functional interplay between the actin and tubulin filament systems, as it is necessary for endocytic processes. While the C-terminal tail domain interacts with actin filaments, the motor domain of KIF5C binds to microtubules (39, 40). Furthermore, we found a light-dependent binding of two proteins (H1FNT, ATAD3A) to the RhoA-complex, whose family members are known to be involved in microtubule dynamics and vesicle-mediated protein transport (50–52).
The two remaining proteins (LRIT1, ZNF496) do not have any obvious connection to the above mentioned processes, although LRIT1 was discussed to be involved in phototransduction or photoreceptor morphogenesis and maintenance (53). The outer segments of photoreceptors are continuously renewed and maintain a constant length through disc formation at their base and disc shedding at their tip. Because of the high rate of disc turnover in ROS (10% per day) (54), there is a need for constant renewal by the delivery of membrane components at the base of ROS. This is achieved by the transfer of membrane material and proteins to and along the connecting cilium and by an endocytic-like process that leads to the invagination and secession of membrane structures (54, 55). RhoA seems to participate in these processes and the light-induced alterations we detected on the protein complex level could reflect the changes in the necessity for outer segment renewal in light and dark conditions on the molecular level. Further functional studies will be necessary to affirm the physiological relevance of these alterations.
The results of these experiments demonstrate that the ICPL-IP allows sensitive detection of quantitative changes that are due to altered physiological states. Taken together, the ICPL-IP proves to be a highly selective and confident method to determine interactions of proteins at their endogenous cellular levels in primary tissue, devoid of any limitation of species or tissue type. ICPL-IP also allows the analysis of human biopsy material and opens the door to correlate and validate work performed in human cell lines with primary biopsy material, generating new opportunities especially for medical research.
Supplementary Material
Acknowledgments
We thank N. Horn, J. Ott for their excellent technical support and Dr. D. Rathbun for critical reading of the manuscript.
Footnotes
* This work was supported by the German Federal Ministry of Education and Research (BMBF: DYNAMO, FKZ: 0315513A; to M.U.) and the Kerstan Foundation (to M.U.) and the European Community's Seventh Framework Program FP7/2009 under grant agreement no: 241955, SYSCILIA (to M.U.), no. 278568, PRIMES (to M.U. and K.B.), and 241481, AFFINOMICS (to M.U.).
 This article contains supplemental Tables S1 to S8.
This article contains supplemental Tables S1 to S8.
Author Contributions: AV and MU designed the research. AV, KB, NK and MU wrote the paper. AV, KB, BF performed experiments. AV, KB, NK, and MU analyzed the data and/or provided data analyses expertise.
1 The abbreviations used are:
- SILAC
- stable isotopic labeling with amino acids in cell culture
- ICPL
- isotope coded protein labeling
- IP
- immunoprecipitation
- TAP
- tandem affinity purification
- MS
- mass spectrometry
- DMEM
- Dulbecco′s modified eagle medium
- PBS
- phosphate buffered saline
- HCl
- hydrochloric acid
- LC
- liquid chromatography
- ROS
- rod outer segments
- TFA
- trifluoroacetic acid
- iTRAQ
- isobaric tag for relative and absolute quantification.
REFERENCES
- 1. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 2. Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 3. Selbach M., Mann M. (2006) Protein interaction screening by quantitative immunoprecipitation combined with knockdown (QUICK). Nat. Methods 3, 981–983 [DOI] [PubMed] [Google Scholar]
- 4. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 5. Ong S. E., Mann M. (2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 [DOI] [PubMed] [Google Scholar]
- 6. Geiger T., Cox J., Ostasiewicz P., Wisniewski J. R., Mann M. (2010) Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods 7, 383–385 [DOI] [PubMed] [Google Scholar]
- 7. Krijgsveld J., Ketting R. F., Mahmoudi T., Johansen J., Artal-Sanz M., Verrijzer C. P., Plasterk R. H., Heck A. J. (2003) Metabolic labeling of C. elegans and D. melanogaster for quantitative proteomics. Nat. Biotechnol. 21, 927–931 [DOI] [PubMed] [Google Scholar]
- 8. Krüger M., Moser M., Ussar S., Thievessen I., Luber C. A., Forner F., Schmidt S., Zanivan S., Fässler R., Mann M. (2008) SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 134, 353–364 [DOI] [PubMed] [Google Scholar]
- 9. McClatchy D. B., Liao L., Park S. K., Venable J. D., Yates J. R. (2007) Quantification of the synaptosomal proteome of the rat cerebellum during post-natal development. Genome Res. 17, 1378–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmidt A., Kellermann J., Lottspeich F. (2005) A novel strategy for quantitative proteomics using isotope-coded protein labels. Proteomics 5, 4–15 [DOI] [PubMed] [Google Scholar]
- 11. Gloeckner C. J., Boldt K., Schumacher A., Roepman R., Ueffing M. (2007) A novel tandem affinity purification strategy for the efficient isolation and characterisation of native protein complexes. Proteomics 7, 4228–4234 [DOI] [PubMed] [Google Scholar]
- 12. Bendall S. C., Hughes C., Stewart M. H., Doble B., Bhatia M., Lajoie G. A. (2008) Prevention of amino acid conversion in SILAC experiments with embryonic stem cells. Mol. Cell. Proteomics 7, 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmitt A., Wolfrum U. (2001) Identification of novel molecular components of the photoreceptor connecting cilium by immunoscreens. Exp. Eye Res. 73, 837–849 [DOI] [PubMed] [Google Scholar]
- 14. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 15. Ishihama Y., Rappsilber J., Mann M. (2006) Modular stop and go extraction tips with stacked disks for parallel and multidimensional Peptide fractionation in proteomics. J. Proteome Res. 5, 988–994 [DOI] [PubMed] [Google Scholar]
- 16. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 17. Boldt K., Mans D. A., Won J., van Reeuwijk J., Vogt A., Kinkl N., Letteboer S. J., Hicks W. L., Hurd R. E., Naggert J. K., Texier Y., den Hollander A. I., Koenekoop R. K., Bennett J., Cremers F. P., Gloeckner C. J., Nishina P. M., Roepman R., Ueffing M. Disruption of intraflagellar protein transport in photoreceptor cilia causes Leber congenital amaurosis in humans and mice. J. Clin. Invest. 121, 2169–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 19. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 20. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 21. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 22. Hutchins J. R., Toyoda Y., Hegemann B., Poser I., Heriche J. K., Sykora M. M., Augsburg M., Hudecz O., Buschhorn B. A., Bulkescher J., Conrad C., Comartin D., Schleiffer A., Sarov M., Pozniakovsky A., Slabicki M. M., Schloissnig S., Steinmacher I., Leuschner M., Ssykor A., Lawo S., Pelletier L., Stark H., Nasmyth K., Ellenberg J., Durbin R., Buchholz F., Mechtler K., Hyman A. A., Peters J. M. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328, 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bulinski J. C., Bossler A. (1994) Purification and characterization of ensconsin, a novel microtubule stabilizing protein. J. Cell Sci. 107 (Pt 10), 2839–2849 [DOI] [PubMed] [Google Scholar]
- 24. Kwok M. C., Holopainen J. M., Molday L. L., Foster L. J., Molday R. S. (2008) Proteomics of photoreceptor outer segments identifies a subset of SNARE and Rab proteins implicated in membrane vesicle trafficking and fusion. Mol. Cell. Proteomics 7, 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Calvert P. D., Strissel K. J., Schiesser W. E., Pugh E. N., Jr., Arshavsky V. Y. (2006) Light-driven translocation of signaling proteins in vertebrate photoreceptors. Trends Cell Biol. 16, 560–568 [DOI] [PubMed] [Google Scholar]
- 26. Kiel C., Vogt A., Campagna A., Chatr-aryamontri A., Swiatek-de Lange M., Beer M., Bolz S., Mack A. F., Kinkl N., Cesareni G., Serrano L., Ueffing M. Structural and functional protein network analyses predict novel signaling functions for rhodopsin. Mol. Syst. Biol. 7, 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dobreva I., Fielding A., Foster L. J., Dedhar S. (2008) Mapping the integrin-linked kinase interactome using SILAC. J. Proteome Res. 7, 1740–1749 [DOI] [PubMed] [Google Scholar]
- 28. Krebs D. L., Uren R. T., Metcalf D., Rakar S., Zhang J. G., Starr R., De Souza D. P., Hanzinikolas K., Eyles J., Connolly L. M., Simpson R. J., Nicola N. A., Nicholson S. E., Baca M., Hilton D. J., Alexander W. S. (2002) SOCS-6 binds to insulin receptor substrate 4, and mice lacking the SOCS-6 gene exhibit mild growth retardation. Mol. Cell. Biol. 22, 4567–4578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hall A. (1998) Rho GTPases and the actin cytoskeleton. Science 279, 509–514 [DOI] [PubMed] [Google Scholar]
- 30. Ridley A. J. (1996) Rho: theme and variations. Current Biol. 6, 1256–1264 [DOI] [PubMed] [Google Scholar]
- 31. Ridley A. J., Hall A. (1992) The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70, 389–399 [DOI] [PubMed] [Google Scholar]
- 32. Ishida H., Zhang X., Erickson K., Ray P. (2004) Botulinum toxin type A targets RhoB to inhibit lysophosphatidic acid-stimulated actin reorganization and acetylcholine release in nerve growth factor-treated PC12 cells. J. Pharmacol. Exp. Ther. 310, 881–889 [DOI] [PubMed] [Google Scholar]
- 33. Mellor H., Flynn P., Nobes C. D., Hall A., Parker P. J. (1998) PRK1 is targeted to endosomes by the small GTPase, RhoB. J. Biol. Chem. 273, 4811–4814 [DOI] [PubMed] [Google Scholar]
- 34. Robertson D., Paterson H. F., Adamson P., Hall A., Monaghan P. (1995) Ultrastructural localization of ras-related proteins using epitope-tagged plasmids. J. Histochem. Cytochem. 43, 471–480 [DOI] [PubMed] [Google Scholar]
- 35. Assinder S. J., Stanton J. A., Prasad P. D. (2009) Transgelin: an actin-binding protein and tumour suppressor. Int. J. Biochem. Cell Biol. 41, 482–486 [DOI] [PubMed] [Google Scholar]
- 36. Cassimeris L. (2002) The oncoprotein 18/stathmin family of microtubule destabilizers. Curr. Opin. Cell Biol. 14, 18–24 [DOI] [PubMed] [Google Scholar]
- 37. Grenningloh G., Soehrman S., Bondallaz P., Ruchti E., Cadas H. (2004) Role of the microtubule destabilizing proteins SCG10 and stathmin in neuronal growth. J. Neurobiol. 58, 60–69 [DOI] [PubMed] [Google Scholar]
- 38. Zhu A. X., Zhao Y., Flier J. S. (1994) Molecular cloning of two small GTP-binding proteins from human skeletal muscle. Biochem. Biophys. Res. Commun. 205, 1875–1882 [DOI] [PubMed] [Google Scholar]
- 39. Iwai S., Ishiji A., Mabuchi I., Sutoh K. (2004) A novel actin-bundling kinesin-related protein from Dictyostelium discoideum. J. Biol. Chem. 279, 4696–4704 [DOI] [PubMed] [Google Scholar]
- 40. Eckmiller M. S., Toman A. (1998) Association of kinesin with microtubules in diverse cytoskeletal systems in the outer segments of rods and cones. Acta Anatomica 162, 133–141 [DOI] [PubMed] [Google Scholar]
- 41. Ellis S., Mellor H. (2000) Regulation of endocytic traffic by rho family GTPases. Trends Cell Biol. 10, 85–88 [DOI] [PubMed] [Google Scholar]
- 42. Kirkham M., Parton R. G. (2005) Clathrin-independent endocytosis: new insights into caveolae and non-caveolar lipid raft carriers. Biochim. Biophys. Acta 1746, 349–363 [DOI] [PubMed] [Google Scholar]
- 43. Parton R. G., Richards A. A. (2003) Lipid rafts and caveolae as portals for endocytosis: new insights and common mechanisms. Traffic 4, 724–738 [DOI] [PubMed] [Google Scholar]
- 44. Adamson P., Paterson H. F., Hall A. (1992) Intracellular localization of the P21rho proteins. J. Cell Biol. 119, 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rual J. F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G. F., Gibbons F. D., Dreze M., Ayivi-Guedehoussou N., Klitgord N., Simon C., Boxem M., Milstein S., Rosenberg J., Goldberg D. S., Zhang L. V., Wong S. L., Franklin G., Li S., Albala J. S., Lim J., Fraughton C., Llamosas E., Cevik S., Bex C., Lamesch P., Sikorski R. S., Vandenhaute J., Zoghbi H. Y., Smolyar A., Bosak S., Sequerra R., Doucette-Stamm L., Cusick M. E., Hill D. E., Roth F. P., Vidal M. (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 [DOI] [PubMed] [Google Scholar]
- 46. Miyazaki T., Yamasaki M., Uchigashima M., Matsushima A., Watanabe M. (2011) Cellular expression and subcellular localization of secretogranin II in the mouse hippocampus and cerebellum. Eur. J. Neurosci. 33, 82–94 [DOI] [PubMed] [Google Scholar]
- 47. Robertson A. S., Smythe E., Ayscough K. R. (2009) Functions of actin in endocytosis. Cell. Mol. Life Sci. 66, 2049–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Werner E. R., Blau N., Thöny B. (2011) Tetrahydrobiopterin: biochemistry and pathophysiology. Biochem. J. 438, 397–414 [DOI] [PubMed] [Google Scholar]
- 49. Zuckerbraun B. S., Stoyanovsky D. A., Sengupta R., Shapiro R. A., Ozanich B. A., Rao J., Barbato J. E., Tzeng E. (2007) Nitric oxide-induced inhibition of smooth muscle cell proliferation involves S-nitrosation and inactivation of RhoA. Am. J. Physiol. 292, C824–831 [DOI] [PubMed] [Google Scholar]
- 50. Mithieux G., Alquier C., Roux B., Rousset B. (1984) Interaction of tubulin with chromatin proteins. H1 and core histones. J. Biol. Chem. 259, 15523–15531 [PubMed] [Google Scholar]
- 51. Multigner L., Gagnon J., Van Dorsselaer A., Job D. (1992) Stabilization of sea urchin flagellar microtubules by histone H1. Nature 360, 33–39 [DOI] [PubMed] [Google Scholar]
- 52. Snider J., Thibault G., Houry W. A. (2008) The AAA+ superfamily of functionally diverse proteins. Genome Biol. 9, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gomi F., Imaizumi K., Yoneda T., Taniguchi M., Mori Y., Miyoshi K., Hitomi J., Fujikado T., Tano Y., Tohyama M. (2000) Molecular cloning of a novel membrane glycoprotein, pal, specifically expressed in photoreceptor cells of the retina and containing leucine-rich repeat. J. Neurosci. 20, 3206–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Young R. W. (1967) The renewal of photoreceptor cell outer segments. J. Cell Biol. 33, 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Obata S., Usukura J. (1992) Morphogenesis of the photoreceptor outer segment during postnatal development in the mouse (BALB/c) retina. Cell Tissue Res. 269, 39–48 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.