Abstract
The iron-molybdenum cofactor (the M-cluster) serves as the active site of molybdenum nitrogenase. Arguably one of the most complex metal cofactors in biological systems, the M-cluster is assembled through the formation of an 8Fe core prior to the insertion of molybdenum and homocitrate into this core. Here, we review the recent progress in the research area of M-cluster assembly, with an emphasis on our work that provides useful insights into the mechanistic details of this process.
Keywords: Biosynthesis, Metalloproteins, Molybdenum, Nitrogenase, S-Adenosylmethionine (SAM)
Introduction
Nitrogenase catalyzes the reduction of nitrogen (N2) under ambient conditions (1–3). The molybdenum-dependent nitrogenase consists of a reductase component (NifH) and a catalytic component (NifDK). The active site of molybdenum nitrogenase is called the iron-molybdenum (FeMo) cofactor or the M-cluster (supplemental Fig. S1A). Located within NifDK (an α2β2-tetramer), the M-cluster receives electrons from NifH (a γ2-dimer) in an ATP-assisted process and subsequently serves as the site for substrate reduction upon accumulation of sufficient electrons (supplemental Fig. S1B) (4). Arguably one of the most complex metal cofactors found in nature, the M-cluster can be viewed as [Fe4S3] and [MoFe3S3] subclusters bridged by three sulfide atoms (5, 6). Additionally, it has a homocitrate moiety attached to its molybdenum end, as well as a carbide atom coordinated in the center of its structure (6–8). The M-cluster is ligated to the α-subunit of NifDK by Cysα275 at its iron end and Hisα442 at its molybdenum end, with Lysα426 providing an additional anchor for its homocitrate entity (5, 6).
The structural complexity and biological importance of the M-cluster have prompted vigorous research on the assembly mechanism of this metal cofactor, as knowledge in this regard is not only important for understanding the structure-function relationship of nitrogenase but is also instrumental in developing future synthetic strategies for nitrogenase-based biomimetic catalysts. Previous studies have established that the M-cluster is synthesized stepwise on a number of scaffold proteins before it is delivered to its destined location in NifDK and that the minimum set of factors required for this process includes the gene products of nifS, nifU, nifB, nifE, nifN, and nifH (9). Here, we briefly review the recent progress in this research area, highlighting our work on the molybdenum nitrogenase from Azotobacter vinelandii that provides meaningful insights into the biosynthetic pathway of its M-cluster. An alternative view of some aspects of M-cluster assembly can be found elsewhere (10).
Synthesis of a 4Fe Cluster Pair
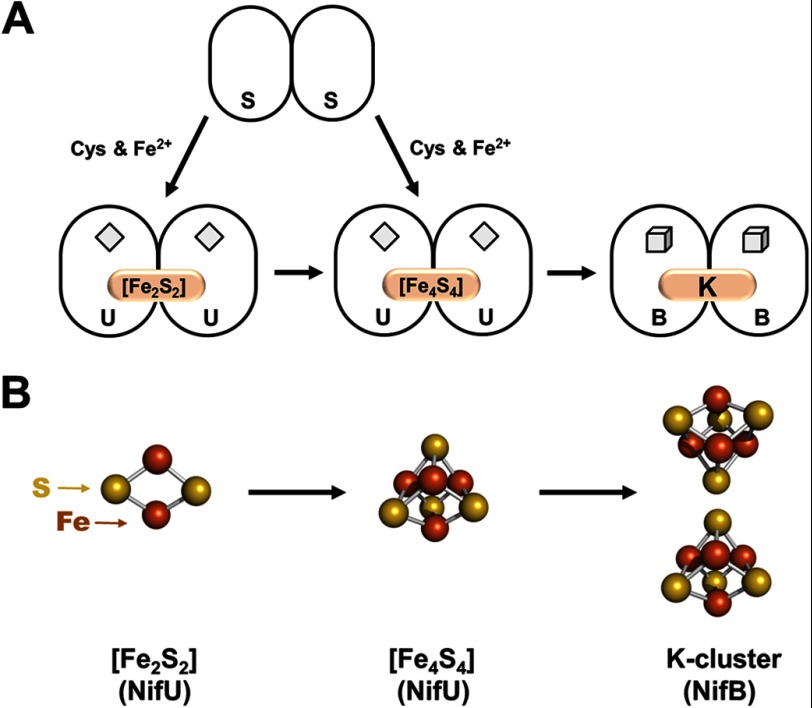
Biosynthesis of the M-cluster is launched by NifS and NifU, which mobilize iron and sulfur for the generation of small FeS fragments (Fig. 1). It is believed that NifS, a pyridoxal phosphate-dependent cysteine desulfurase, forms a protein-bound cysteine persulfide, which is subsequently donated to NifU for the sequential formation of [Fe2S2] and [Fe4S4] units (11). The [Fe4S4] clusters are then delivered from NifU to NifB and further processed into a large FeS core (Fig. 1). This hypothesis is supported by the identification of a pair of [Fe4S4] clusters (designated the K-cluster) on NifB, which can be easily accommodated by a sufficient amount of ligands in NifB (9). Metal analysis further reveals the presence of additional [Fe4S4] clusters on NifB, which are associated with the conserved CxxxCxxC motifs (a characteristic feature of radical S-adenosyl-l-methionine (SAM)3 enzymes) in this protein (9). Collectively, the K-cluster and the SAM motif-associated cluster (designated the SAM cluster) give rise to an S = 1/2 EPR signal at g = 2.02, 1.95, and 1.90 (12). More excitingly, this composite S = 1/2 signal disappears upon addition of SAM, suggesting that the K-cluster and the SAM cluster are located in close proximity to each other (12). Such a cluster arrangement is important, as it facilitates the subsequent coupling of the two 4Fe units of K-clusters via a radical SAM-dependent route (see below).
FIGURE 1.
Synthesis of a 4Fe cluster pair. A, NifS and NifU mobilize iron and sulfur for the sequential formation of [Fe2S2] and [Fe4S4] clusters, followed by the transfer of an [Fe4S4] cluster pair (the K-cluster) from NifU to NifB for further processing. The permanent [Fe2S2] clusters on NifU and the SAM motif-associated [Fe4S4] clusters on NifB are depicted as diamonds and cubes, respectively, whereas the transient clusters on these proteins are represented by pink ovals and are labeled accordingly. B, structural details of the [Fe2S2] cluster (left), the [Fe4S4] cluster (middle), and the [Fe4S4] cluster pair (right). The clusters are shown as ball-and-stick models, with the atoms colored as follows: iron, orange; and sulfur, yellow. PyMOL was used to create this figure (Protein Data Bank code 1N2C).
Formation of an 8Fe Core
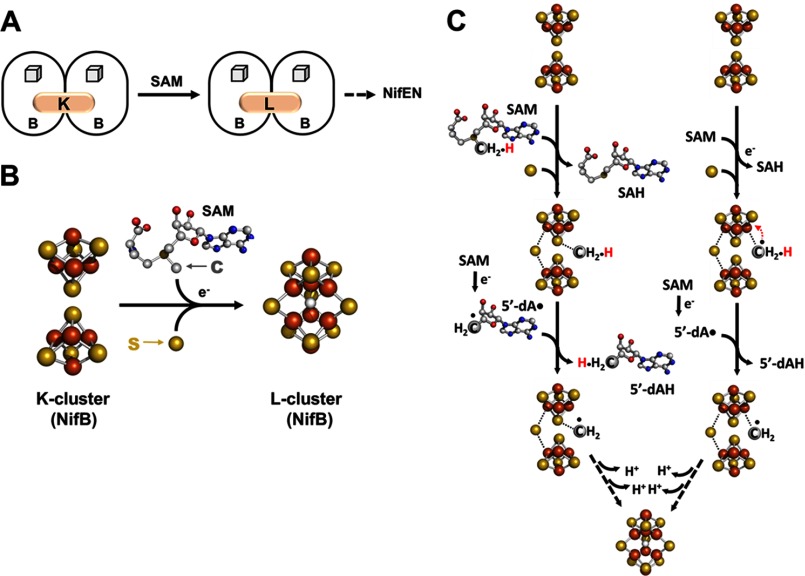
The K-cluster on NifB can be converted to an 8Fe core in the presence of SAM (Fig. 2A). The concomitant disappearance of the K-cluster- and SAM cluster-originated S = 1/2 signal upon addition of SAM is accompanied by the appearance of a unique g = 1.94 signal, which has been attributed to an [Fe8S9] cluster (designated the L-cluster) (12). Iron K-edge x-ray absorption spectroscopy (XAS)/extended x-ray absorption fine structure and crystallographic studies show that the L-cluster closely resembles the core structure of the mature M-cluster, except that the molybdenum and homocitrate components at one end of the cluster are replaced by an iron atom (13–16). Iron Kβ x-ray emission spectroscopy analysis further demonstrates the presence of a carbide atom in the center of the L-cluster (17). Together, these observations imply that the insertion of carbide may occur simultaneously with the transformation of a K-cluster into an L-cluster. A novel synthetic route to the 8Fe L-cluster can be postulated, one that inserts the carbon atom via the radical chemistry at the SAM domain of NifB while attaching an additional sulfur atom and coupling/rearranging the two 4Fe units of the K-cluster into an [Fe8S9] L-cluster at the same time (Fig. 2B).
FIGURE 2.
Formation of an 8Fe core. A, NifB catalyzes the SAM-dependent conversion of an [Fe4S4] cluster pair (the K-cluster) to an [Fe8S9] cluster (the L-cluster), which is subsequently delivered to NifEN. The SAM motif-associated [Fe4S4] clusters on NifB are depicted as cubes, whereas the transient clusters on this protein are represented by pink ovals and are labeled accordingly. B, structural details of the K-cluster (left) and L-cluster (right) on NifB. The clusters are shown as ball-and-stick models, with the atoms colored as follows: iron, orange; sulfur, yellow; oxygen, red; carbon, gray; and nitrogen, dark blue. PyMOL was used to create this figure (Protein Data Bank code 3PDI). C, two proposed mechanisms of carbon insertion by NifB. One involves the transfer of a methyl group via an SN2 mechanism, followed by the formation of a methylene radical upon hydrogen atom abstraction by 5′-dA• and the subsequent transfer of this radical intermediate to a sulfur atom of the K-cluster (left), whereas the other involves the formation of a methyl radical via reductive cleavage of SAM, followed by the transfer of this transient intermediate to an iron atom of the K-cluster and the subsequent processing of this intermediate into a methylene radical (right). SAH, S-adenosyl-l-homocysteine.
The cleavage pattern of SAM provides initial insights into the role of radical SAM chemistry in this process (18). In the presence of NifB, SAM is cleaved into two products: 5′-deoxyadenosine (5′-dA) and S-adenosyl-l-homocysteine. Upon substitution of [methyl-d3]SAM for unlabeled SAM, however, a mixture of deuterium-enriched and unlabeled 5′-dA can be detected along with S-adenosyl-l-homocysteine as products of SAM cleavage (18). Interestingly, two radical SAM RNA methylases, RlmN and Cfr, display the same SAM cleavage and deuterium substitution patterns as those of NifB, which have been accounted for by an SN2-type of methyl transfer from one equivalent of SAM, followed by the generation of 5′-dA• from a second equivalent of SAM and the subsequent hydrogen atom abstraction from the methyl group by 5′-dA• (19, 20). The striking parallelism between NifB and the two RNA methylases suggests that NifB utilizes a similar mechanism to mobilize the methyl group of SAM for carbon insertion. Radiolabeling experiments provide convincing evidence for this proposal, showing that the incubation of NifB with [methyl-14C]SAM leads to an accumulation of 14C label on NifB, which can be further traced to the L-cluster in this protein (18).
Two mechanisms can be proposed for the formation of the L-cluster on NifB (18). One involves the transfer of the methyl group of SAM to a sulfide atom of the K-cluster via an SN2 mechanism, followed by the formation of a methylene radical through the abstraction of a hydrogen atom from this methyl group (Fig. 2C, left); the other involves the formation of a methyl radical via the reductive cleavage of SAM, followed by the transfer of this radical to an iron atom of the K-cluster and the subsequent rearrangement of this radical into a methylene radical (Fig. 2C, right). In either scenario, the transfer of the carbon intermediate to the K-cluster may play a pivotal role in coupling the two 4Fe units of the K-cluster into an 8Fe L-cluster, initiating radical chemistry for the initial bond rearrangement that is required for the subsequent restructuring of cluster units. Regardless of the type of the initial carbon intermediate, it must undergo additional deprotonation and/or dehydrogenation steps until a carbide atom is generated in the center of the L-cluster (Fig. 2C).
Maturation of the 8Fe Core
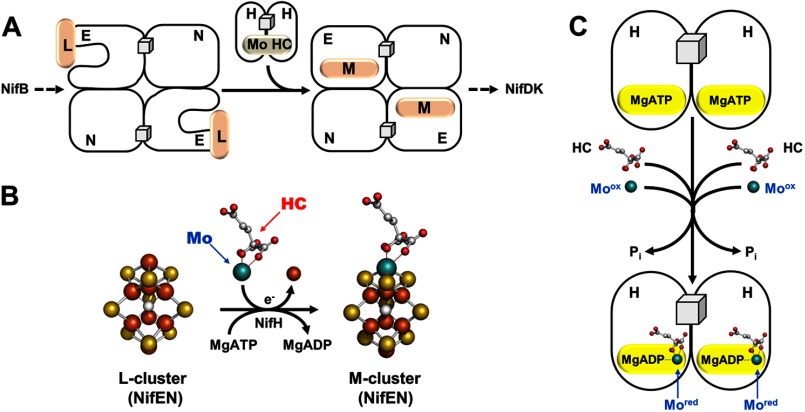
Once formed, the L-cluster is transferred from NifB to NifEN, where it is matured into an M-cluster upon the insertion of molybdenum and homocitrate (Fig. 3A). Such a conversion can be achieved in vitro by incubating L-cluster-bound NifEN with NifH, MgATP, dithionite, molybdate (MoO42−) and homocitrate, generating an NifEN-bound M-cluster (Fig. 3B) that can be used for the subsequent reconstitution and activation of apo-NifDK (21, 22). Maturation of the cluster species on NifEN is reflected by the disappearance of the L-cluster-specific g = 1.94 signal and the concurrent appearance of a small M-cluster-like signal at g = 4.45, 3.96, 3.60, and 2.03 (21, 22). Iron and molybdenum K-edge XAS/extended x-ray absorption fine structure analyses further confirm that the structure of the M-cluster on NifEN (Fig. 3B, right) is nearly identical to that of the M-cluster on NifDK (supplemental Fig. S1A), except for a somewhat asymmetric coordination of molybdenum that arises from a different ligand environment in NifEN (21, 22).
FIGURE 3.
Maturation of the 8Fe core. A, NifEN houses the conversion of the L-cluster to the M-cluster upon NifH-mediated insertion of molybdenum and homocitrate (HC). The maturation of the L-cluster is likely accompanied by transfer of the cluster from the surface of NifEN to the M-cluster-binding site within the protein. The permanent [Fe4S4] clusters in NifEN are represented by cubes, whereas the transient clusters on this protein are represented by pink ovals and are labeled accordingly. B, structural details of the L-cluster (left) and M-cluster (right) on NifEN, with one terminal iron atom of the L-cluster replaced by molybdenum and homocitrate in the M-cluster through an ATP-dependent process. C, NifH serves as an ATP-dependent molybdenum/homocitrate insertase and possibly mobilizes these two missing elements in the form of NifH/ADP-molybdenum-homocitrate for the maturation of the L-cluster. The clusters are shown as ball-and-stick models, with the atoms colored as follows: iron, orange; sulfur, yellow; molybdenum, cyan; oxygen, red; and carbon, gray. PyMOL was used to create this figure (Protein Data Bank codes 3PDI and 1M1N).
Interestingly, biochemical experiments suggest that NifEN undergoes a conformational change upon the conversion of the L-cluster to the M-cluster, as only the M-cluster-bound form of NifEN is capable of complex formation with NifDK (23). Consistent with this suggestion, crystallographic analysis of the L-cluster-bound form of NifEN reveals an unusual, nearly surface-exposed location of the L-cluster (16). This observation is unexpected, as the highly homologous NifEN and NifDK proteins should have homologous cluster-binding sites that are buried within their respective polypeptides. It is conceivable that the surface location of L-cluster permits an easier access of molybdenum and homocitrate for cluster maturation, whereas a concurrent conformational change of NifEN enables subsequent relocation of the matured cluster from the surface to the binding site within the protein (Fig. 3A).
The conformational change of NifEN is likely induced by its interaction with NifH upon cluster maturation, which parallels the interaction between NifDK and NifH upon substrate turnover. Indeed, NifH exhibits a strict dependence on ATP hydrolysis and redox potential to carry out its function both in catalysis (1–3) and in assembly (21, 22). However, the role of this ATPase in assembly appears more complicated, as NifH can be “loaded” with molybdenum and homocitrate and subsequently used as a donor of these two missing components to the NifEN-bound L-cluster (24). Biochemical analysis shows a mandatory co-mobilization of molybdenum and homocitrate by NifH, whereas molybdenum K-edge XAS analysis reveals that the molybdenum species is “processed” upon binding to NifH, displaying a change in its formal oxidation state and/or the ligation pattern (24). More interestingly, EPR analysis suggests that NifH binds nucleotide along with molybdenum and homocitrate, showing that the loaded NifH protein gives rise to a signal that has an intermediary line shape between those of the ADP- and ATP-bound forms of NifH (24). This observation coincides with the outcome of the initial crystallographic analysis of an ADP-bound form of NifH, which places molybdenum at a position that corresponds to the γ-phosphate of ATP (25). A binding pattern of NifH/ADP-molybdenum-homocitrate can be proposed based on these results, which could represent the initial step of molybdenum mobilization by NifH (Fig. 3C). Such a binding pattern parallels the formation of a possible adenylylated molybdate intermediate during the biosynthesis of molybdopterin cofactors (9). Moreover, another ATPase, CooC, has been implicated in the insertion of nickel into the C-cluster of the carbon monoxide dehydrogenase from Rhodospirillum rubrum (26). Thus, the mobilization of molybdenum by NifH may exemplify a general scheme for metal trafficking in biological systems.
Delivery of the M-cluster to Its Target Location
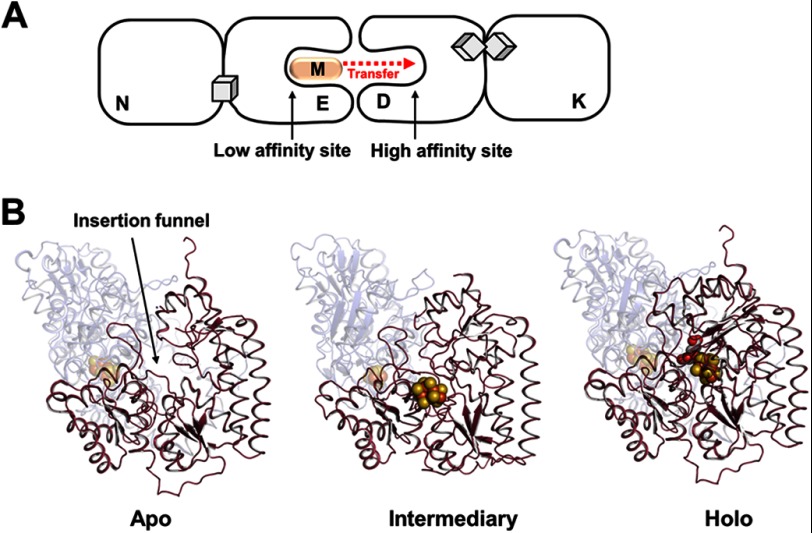
The final “touch-up” of the structure of the M-cluster is achieved upon delivery of this cluster from NifEN to NifDK. The completion of M-cluster assembly on NifEN signals the cluster transfer from NifEN to apo-NifDK, which begins with the complex formation between these two homologous proteins (23). Sequence analysis suggests the presence of homologous M-cluster-binding sites in NifEN and NifDK; however, certain residues that either provide a covalent ligand to the M-cluster or tightly pack the M-cluster within the polypeptide matrix of NifDK cannot be found in the sequence of NifEN (9). Thus, it can be postulated that the M-cluster-binding sites in NifEN and NifDK are “lined up” upon complex formation, which enables the “diffusion” of the M-cluster from its transient binding site in NifEN (“low affinity site”) to its final binding site in NifDK (“high affinity site”) (Fig. 4A). Given the structural homology between NifEN and NifDK, the process of M-cluster insertion into apo-NifDK could be depicted by three available “snapshots” of NifDK and/or NifEN: (i) an apo conformation, which contains a positively charged cluster insertion funnel; (ii) an intermediate conformation, which is generated upon docking of the cluster at the entrance of the insertion funnel; and (iii) a holo conformation, which has the cluster secured at its final binding site (Fig. 4B). The negative charge of homocitrate is crucial for the insertion of the M-cluster along the positively charged funnel (23, 27), and the M-cluster interacts with a number of NifDK residues en route to its target binding site (28–30), where the final structure of the M-cluster is achieved through proper ligand coordination.
FIGURE 4.
Delivery of the M-cluster to its target location. A, NifEN docks on apo-NifDK upon the completion of M-cluster assembly, allowing the cluster to diffuse from the low affinity site in NifEN to the high affinity site in NifDK. The permanent P-cluster ([Fe8S7]) in NifDK is represented by a pair of cubes. B, the conformations of NifDK before (apo; left), during (intermediate; middle), and after (holo; right) the insertion of the M-cluster. The apo and holo conformations were taken from the crystal structures of apo-NifDK and holo-NifDK, respectively, whereas the intermediate conformation was adapted from the crystal structure of the L-cluster-bound form of NifEN. The intermediate conformation has the M-cluster attached at the surface of NifDK in a manner analogous to the attachment of the L-cluster at the surface of NifEN. Apo-NifDK contains an open cluster insertion funnel (left), which is believed to be partially closed upon the docking of the M-cluster at its entrance (middle) and fully closed upon the incorporation of the M-cluster and the accompanying structural rearrangement of NifDK (right). The proteins are shown as ribbon diagrams, with the α-subunits colored red and presented in the foreground and the β-subunits colored blue and rendered transparent in the background. The clusters are shown as ball-and-stick models, with the atoms colored as follows: iron, orange; sulfur, yellow; molybdenum, cyan; oxygen, red; and carbon, gray. PyMOL was used to create this figure (Protein Data Bank codes 1L5H, 3PDI, and 1M1N).
Conclusion
The M-cluster is assembled on a number of scaffold proteins prior to its delivery to its target location in NifDK. This process involves two key steps: the stepwise formation of an 8Fe core and the subsequent maturation of this core upon incorporation of molybdenum and homocitrate. The former utilizes radical SAM chemistry for the insertion of interstitial carbide, which likely occurs concomitantly with the coupling and restructuring of two 4Fe units, whereas the latter employs a nucleotide-binding protein for the mobilization of molybdenum and homocitrate, which may represent a common strategy for metal trafficking. Future studies will focus on the mechanistic investigation of radical SAM-dependent carbon insertion, as well as the structural details of the NifH/ATPase-based molybdenum mobilization.
This work was supported, in whole or in part, by National Institutes of Health Grant GM67626 (to M. W. R.). This is the second article in the Thematic Minireview Series on Metals in Biology 2013.

This article contains supplemental Fig. S1.
- SAM
- S-adenosyl-l-methionine
- XAS
- x-ray absorption spectroscopy
- 5′-dA
- 5′-deoxyadenosine.
REFERENCES
- 1. Burgess B. K., Lowe D. J. (1996) Mechanism of molybdenum nitrogenase. Chem. Rev. 96, 2983–3012 [DOI] [PubMed] [Google Scholar]
- 2. Howard J. B., Rees D. C. (1996) Structural basis of biological nitrogen fixation. Chem. Rev. 96, 2965–2982 [DOI] [PubMed] [Google Scholar]
- 3. Howard J. B., Rees D. C. (2006) How many metals does it take to fix N2? A mechanistic overview of biological nitrogen fixation. Proc. Natl. Acad. Sci. U.S.A. 103, 17088–17093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schindelin H., Kisker C., Schlessman J. L., Howard J. B., Rees D. C. (1997) Structure of ADP·AIF4−-stabilized nitrogenase complex and its implications for signal transduction. Nature 387, 370–376 [DOI] [PubMed] [Google Scholar]
- 5. Kirn J., Rees D. C. (1992) Crystallographic structure and functional implications of the nitrogenase molybdenum iron protein from Azotobacter vinelandii. Nature 360, 553–560 [DOI] [PubMed] [Google Scholar]
- 6. Einsle O., Tezcan F. A., Andrade S. L., Schmid B., Yoshida M., Howard J. B., Rees D. C. (2002) Nitrogenase MoFe-protein at 1.16 Å resolution: a central ligand in the FeMo-cofactor. Science 297, 1696–1700 [DOI] [PubMed] [Google Scholar]
- 7. Lancaster K. M., Roemelt M., Ettenhuber P., Hu Y., Ribbe M. W., Neese F., Bergmann U., DeBeer S. (2011) X-ray emission spectroscopy evidences a central carbon in the nitrogenase iron-molybdenum cofactor. Science 334, 974–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spatzal T., Aksoyoglu M., Zhang L., Andrade S. L., Schleicher E., Weber S., Rees D. C., Einsle O. (2011) Evidence for interstitial carbon in nitrogenase FeMo cofactor. Science 334, 940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schwarz G., Mendel R. R., Ribbe M. W. (2009) Molybdenum cofactors, enzymes and pathways. Nature 460, 839–847 [DOI] [PubMed] [Google Scholar]
- 10. Rubio L. M., Ludden P. W. (2008) Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu. Rev. Microbiol. 62, 93–111 [DOI] [PubMed] [Google Scholar]
- 11. Smith A. D., Jameson G. N. L., Dos Santos P. C., Agar J. N., Naik S., Krebs C., Frazzon J., Dean D. R., Huynh B. H., Johnson M. K. (2005) NifS-mediated assembly of [4Fe-4S] clusters in the N- and C-terminal domains of the NifU scaffold protein. Biochemistry 44, 12955–12969 [DOI] [PubMed] [Google Scholar]
- 12. Wiig J. A., Hu Y., Ribbe M. W. (2011) NifEN-B complex of Azotobacter vinelandii is fully functional in nitrogenase FeMo cofactor assembly. Proc. Natl. Acad. Sci. U.S.A. 108, 8623–8627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Corbett M. C., Hu Y., Fay A. W., Ribbe M. W., Hedman B., Hodgson K. O. (2006) Structural insights into a protein-bound iron-molybdenum cofactor precursor. Proc. Natl. Acad. Sci. U.S.A. 103, 1238–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fay A. W., Blank M. A., Lee C. C., Hu Y., Hodgson K. O., Hedman B., Ribbe M. W. (2011) Spectroscopic characterization of the isolated iron-molybdenum cofactor (FeMoco) precursor from the protein NifEN. Angew. Chem. Int. Ed. Engl. 50, 7787–7790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu Y., Fay A. W., Ribbe M. W. (2005) Identification of a nitrogenase iron-molybdenum cofactor precursor on NifEN complex. Proc. Natl. Acad. Sci. U.S.A. 102, 3236–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaiser J. T., Hu Y., Wiig J. A., Rees D. C., Ribbe M. W. (2011) Structure of precursor-bound NifEN: a nitrogenase FeMo cofactor maturase/insertase. Science 331, 91–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lancaster K. M., Hu Y., Bergmann U., Ribbe M. W., DeBeer S. (2013) X-ray spectroscopic observation of an interstitial carbide in NifEN-bound FeMoco precursor. J. Am. Chem. Soc. 135, 610–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wiig J. A., Hu Y., Lee C. C., Ribbe M. W. (2012) Radical SAM-dependent carbon insertion into nitrogenase M-cluster. Science 337, 1672–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grove T. L., Benner J. S., Radle M. I., Ahlum J. H., Landgraf B. J., Krebs C., Booker S. J. (2011) A radically different mechanism for S-adenosylmethionine-dependent methyltransferases. Science 332, 604–607 [DOI] [PubMed] [Google Scholar]
- 20. Boal A. K., Grove T. L., McLaughlin M. I., Yennawar N. H., Booker S. J., Rosenzweig A. C. (2011) Structural basis for methyl transfer by a radical SAM enzyme. Science 332, 1089–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hu Y., Corbett M. C., Fay A. W., Webber J. A., Hodgson K. O., Hedman B., Ribbe M. W. (2006) FeMo cofactor maturation on NifEN. Proc. Natl. Acad. Sci. U.S.A. 103, 17119–17124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yoshizawa J. M., Blank M. A., Fay A. W., Lee C. C., Wiig J. A., Hu Y., Hodgson K. O., Hedman B., Ribbe M. W. (2009) Optimization of FeMoco maturation on NifEN. J. Am. Chem. Soc. 131, 9321–9325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fay A. W., Blank M. A., Yoshizawa J. M., Lee C. C., Wiig J. A., Hu Y., Hodgson K. O., Hedman B., Ribbe M. W. (2010) Formation of a homocitrate-free iron-molybdenum cluster on NifEN: implications for the role of homocitrate in nitrogenase assembly. Dalton Trans. 39, 3124–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu Y., Corbett M. C., Fay A. W., Webber J. A., Hodgson K. O., Hedman B., Ribbe M. W. (2006) Nitrogenase Fe protein: A molybdate/homocitrate insertase. Proc. Natl. Acad. Sci. U.S.A. 103, 17125–17130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Georgiadis M. M., Komiya H., Chakrabarti P., Woo D., Kornuc J. J., Rees D. C. (1992) Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science 257, 1653–1659 [DOI] [PubMed] [Google Scholar]
- 26. Jeoung J. H., Giese T., Grünwald M., Dobbek H. (2009) CooC1 from Carboxydothermus hydrogenoformans is a nickel-binding ATPase. Biochemistry 48, 11505–11513 [DOI] [PubMed] [Google Scholar]
- 27. Schmid B., Ribbe M. W., Einsle O., Yoshida M., Thomas L. M., Dean D. R., Rees D. C., Burgess B. K. (2002) Structure of a cofactor-deficient nitrogenase MoFe protein. Science 296, 352–356 [DOI] [PubMed] [Google Scholar]
- 28. Fay A. W., Hu Y., Schmid B., Ribbe M. W. (2007) Molecular insights into nitrogenase FeMoco insertion–the role of His 274 and His 451 of MoFe protein α subunit. J. Inorg. Biochem. 101, 1630–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu Y., Fay A. W., Ribbe M. W. (2007) Molecular insights into nitrogenase FeMoco insertion–the role of His362 of MoFe protein α subunit in FeMoco incorporation. J. Biol. Inorg. Chem. 12, 449–460 [DOI] [PubMed] [Google Scholar]
- 30. Hu Y., Fay A. W., Schmid B., Makar B., Ribbe M. W. (2006) Molecular insights into nitrogenase FeMoco insertion. Trp-444 of MoFe protein α subunit locks FeMoco in its binding site. J. Biol. Chem. 281, 30534–30541 [DOI] [PubMed] [Google Scholar]