Abstract
Metalloenzymes often require elaborate metallocenter assembly systems to create functional active sites. The medically important dinuclear nickel enzyme urease provides an excellent model for studying metallocenter assembly. Nickel is inserted into the urease active site in a GTP-dependent process with the assistance of UreD/UreH, UreE, UreF, and UreG. These accessory proteins orchestrate apoprotein activation by delivering the appropriate metal, facilitating protein conformational changes, and possibly providing a requisite post-translational modification. The activation mechanism and roles of each accessory protein in urease maturation are the subject of ongoing studies, with the latest findings presented in this minireview.
Keywords: Biosynthesis, Chaperone Chaperonin, GTPase, Metalloenzymes, Nickel, Urease
Introduction
Metallocenters serve essential biological functions such as transferring electrons, stabilizing biomolecules, binding substrates, and catalyzing desirable reactions. Synthesis of these sites must be tightly controlled because simple competition between metals may lead to misincorporation with loss of function and because excess cytoplasmic concentrations of free metal ions can have toxic cellular effects. In many cases, cells have evolved elaborate metallocenter assembly systems that sequester metal cofactors from the cellular milieu, thus offering protection from adventitious reactions while ensuring the fidelity of metal insertion. In addition to maintaining metal homeostasis, these assembly systems facilitate protein conformational changes and active site modifications that are required for full enzymatic activity. Several metalloproteins have been investigated as models to understand the mechanisms and dynamics of active site assembly and the complex orchestrations of their metallocenter assembly systems. In this minireview, we discuss recent findings related to maturation of the nickel-containing enzyme urease.
Introduction to Ureases
Urease is of great medical, agricultural, and historical significance. The gastric pathogen Helicobacter pylori uses urease for localized neutralization of pH, allowing it to flourish in the stomach (1), whereas the uropathogen Proteus mirabilis uses it to colonize and form stones in the urinary tract (2). In agriculture, urea is both a plant metabolite and a fertilizer degraded by plant ureases (3); however, urea is also metabolized by soil bacteria, which can lead to unproductive volatilization of ammonia and harmful soil alkylation (4). Also of interest, urease from jack bean (Canavalia ensiformis) seeds was the first enzyme to be crystallized (5) and the first protein shown to contain nickel (6). Finally, as described in subsequent sections, urease is a significant model enzyme that has advanced our understanding of the mechanisms of metallocenter assembly.
This enzyme catalyzes the hydrolysis of urea to ammonia and carbamic acid, which subsequently decomposes to another molecule of ammonia and bicarbonate (7–9): H2N-C(O)-NH2 + H2O → NH3 + H2N-COOH and H2N-COOH + H2O → NH3 + H2CO3.
Regardless of the source of the enzyme, the overall protein structures exhibit extensive similarities. Most bacterial ureases have three subunits in a (UreABC)3 configuration (Fig. 1A), as exemplified by proteins from Klebsiella aerogenes (10, 11) and Sporosarcina (formerly Bacillus) pasteurii (12). In Helicobacter species, a fusion of two genes (corresponding to K. aerogenes ureA and ureB) results in only two subunits, yielding a ((UreAB)3)4 structure (Fig. 1B) (13). In fungi and plants, all urease domains are encoded by a single gene such as JBURE-I for jack bean, with the trimeric protein forming back-to-back dimers, ((α)3)2 (Fig. 1C) (14). The metallocenter structures of these proteins are identical (Fig. 1D), with two Ni2+ ions bridged by a carbamylated Lys residue and water; one metal additionally coordinates two His residues and a terminal water molecule, and the second Ni2+ ion also coordinates two His residues, one Asp residue, and water. Aspects of the enzyme mechanism remain controversial (7, 9, 15–17), but most proposals suggest that the urea carbonyl oxygen displaces the terminal water from the Ni2+ ion shown on the left (Fig. 1D), with another Ni2+-bound water molecule acting as a nucleophile to achieve catalysis. Although much is known about the enzyme active site, the mechanism of nickel insertion into the protein is still poorly understood.
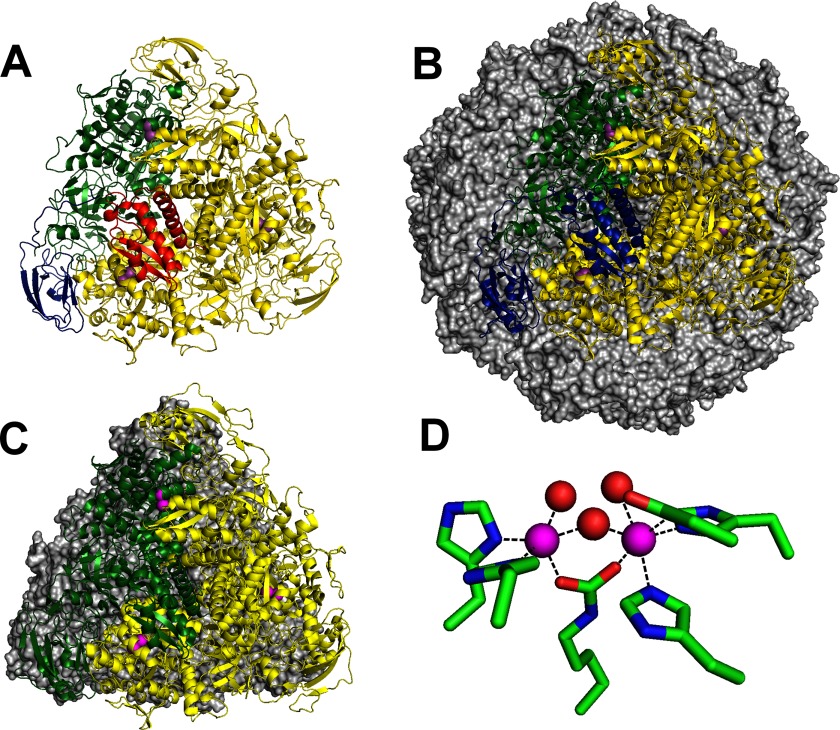
FIGURE 1.
Urease structures. A, three-subunit bacterial ureases (UreA, red; UreB, blue; UreC, green; with two more copies, yellow) assemble into a trimer of trimers (Protein Data Bank code 1FWJ). B, two-subunit Helicobacter ureases (a fusion of the two small domains, blue; large subunit, green; with two more copies, yellow) form a trimer of dimers, which interacts with three more trimers (gray surface view) to form a dodecamer of dimers (code 1E9Z). C, single-subunit urease of fungi and plants (a fusion of all three domains, green; with two more copies, yellow) forms a trimer that stacks back-to-back with a second trimer (gray surface view) (code 3LA4). D, dinuclear Ni2+ metallocenter of urease (Ni2+, magenta; solvent, red).
Prototypical Urease Activation Pathway
The biosynthesis of the urease dinuclear nickel metallocenter generally requires the participation of several accessory proteins (7, 9). For example, the canonical urease system of K. aerogenes (involving ureDABCEFG expression in Escherichia coli) utilizes UreD, UreE, UreF, and UreG to facilitate activation of the UreABC apoprotein (Table 1) (18, 19). Many other ureolytic bacteria contain these four auxiliary genes flanking the enzyme subunit genes (7–9, 20), although the gene order often changes, and ureD is renamed ureH in Helicobacter spp. (21). Homologs of ureD/ureH, ureF, and ureG exist in urease-containing eukaryotes (3, 22, 23); however, the eukaryotic accessory genes are not adjacent to the urease structural genes, and sequences related to ureE have not been identified. Below, we describe the four prototypical urease accessory proteins, their complexes with each other and with urease, and their proposed roles in urease activation.
TABLE 1.
Selected proteins needed for urease activation
| K. aerogenesa | H. pylori | Plants | Function |
|---|---|---|---|
| UreA | Enzyme subunit | ||
| UreB | UreAb | Enzyme subunit | |
| UreC | UreBc | Ureased | Enzyme subunit |
| UreD | UreH | UreD | Scaffold protein |
| UreE | UreE | —e | Metallochaperone |
| UreF | UreF | UreF | Potential fidelity enhancer |
| UreG | UreG | UreG | GTPase |
a A similar set of proteins is present in many other bacteria, including S. pasteurii.
b Equivalent to a fusion of UreA and UreB of K. aerogenes.
c Equivalent to UreC of K. aerogenes.
d Equivalent to a fusion of UreA, UreB, and UreC of K. aerogenes.
e —, no UreE ortholog has been detected in plants.
UreD/UreH: Scaffold for Recruitment of Other Accessory Proteins and Facilitator of Activation
Characterization of UreD/UreH is most advanced for the K. aerogenes and H. pylori proteins, which exhibit only 25% identity. Heterologous expression of ureD or ureH in E. coli yields insoluble products (24, 25); however, these problems were circumvented by different approaches. For the K. aerogenes protein, a maltose-binding protein (MBP)2 fusion variant of UreD is soluble and functionally replaces the native protein (26). In the case of H. pylori ureH, solubilization is achieved by coexpression with ureF, which provides a UreH:UreF complex (25). The structure of this complex (Protein Data Bank code 3SF5) and that of a UreH:UreF:UreG complex3 reveal a novel β-helical fold for UreH, with 17 β-strands and two α-helices (Fig. 2), that resembles SufD (code 1VH4), a member of a scaffold protein complex that functions in iron-sulfur cluster biosynthesis (27). Whereas no obvious metal-binding sites are apparent in UreH, 2.7 Ni2+ ions bind per UreD protomer (26).
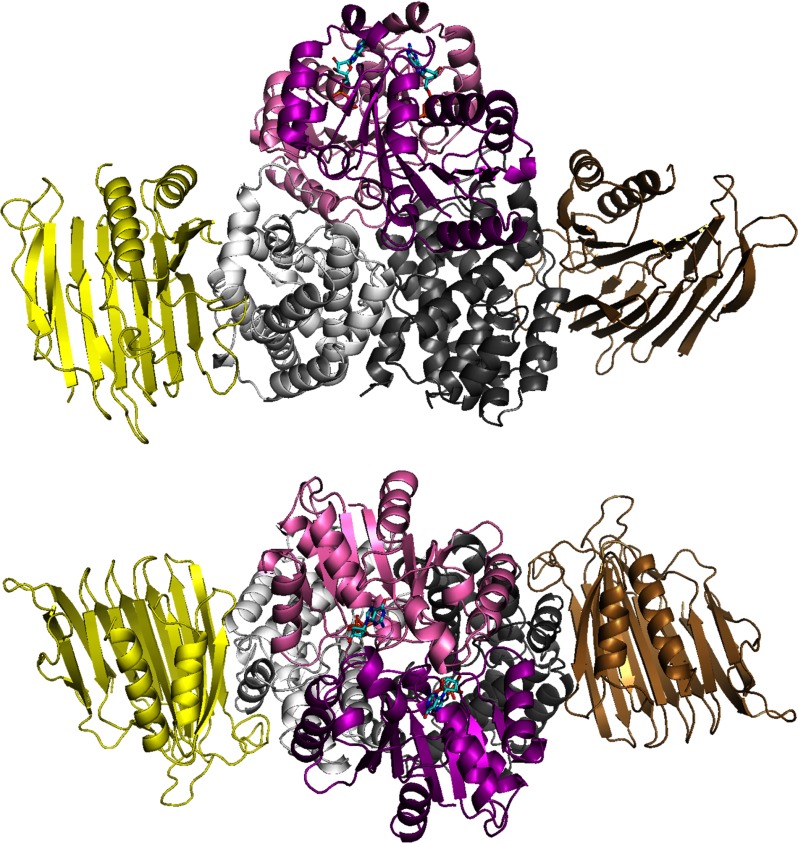
FIGURE 2.
Structure of UreH:UreF:UreG. Shown are two views of the (UreH:UreF:UreG)2 complex from H. pylori (UreH, UreF, and UreG in shades of yellow, gray, and magenta, respectively). A GDP molecule (cyan) is located in each UreG.
Several studies have demonstrated that UreD/UreH binds to urease. Enhanced expression of K. aerogenes ureD in the presence of the cognate structural proteins results in a UreABC:UreD complex that contains zero to three molecules of UreD/(UreABC)3 according to two-dimensional (native and denaturing) gel electrophoresis (24). Although a crystal structure is not available, a model of this species has been created (Fig. 3) based on several lines of evidence (28). For example, small angle x-ray scattering studies of this complex yielded data that are best modeled with UreD binding to the vertices of the triangular urease (29). Chemical cross-linking of this species confirmed that UreD binds to the UreB and UreC subunits (30). MBP-UreD associates in vivo with UreABC (26) but not with UreAC (i.e. urease missing UreB) (40). Similarly, yeast two-hybrid studies of H. pylori proteins identified interactions between UreH and UreA (31, 32). Of great functional significance, in vitro studies with purified K. aerogenes components showed that UreD enhances the extent of activation of urease apoprotein. For example, whereas ∼15% of the urease apoprotein generates functional sites when incubated with 100 μm NiCl2 and 100 mm bicarbonate (needed to carbamylate the Lys metal ligand) (33), ∼30% is made functional when using the UreABC:UreD species (34). Along with results from additional studies (see below), these findings led to the current hypothetical role of UreD/UreH as both a scaffold for recruiting other accessory proteins and a direct facilitator of nickel insertion into the active site.
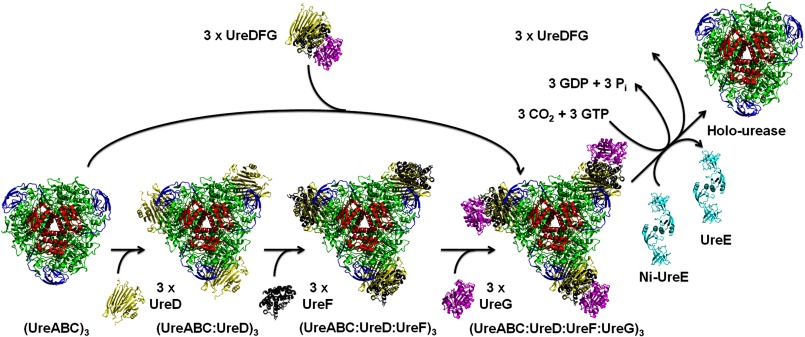
FIGURE 3.
Model of K. aerogenes urease activation. The trimer-of-trimers urease apoprotein (UreA, red; UreB, blue; UreC, green) either sequentially binds UreD (yellow), UreF (gray), and UreG (magenta) or binds the UreDFG complex (only one protomer of each protein is shown, but the isolated complex contains two protomers of each). Formation of the active enzyme requires CO2 to carbamylate Lys-217 at the native active site, GTP binding to and hydrolysis by UreG, and nickel delivery by dimeric UreE (cyan). It remains unclear whether the accessory proteins are released as a UreDFG unit or as individual proteins.
UreF: Checkpoint for Metallocenter Fidelity
UreF proteins also are best characterized for K. aerogenes and H. pylori. Heterologous expression of K. aerogenes ureF yields insoluble product (35); however, MBP-UreF (36) and UreE-UreF (37) fusion proteins are soluble, with the latter protein shown to function in cellular activation of urease. The native form of H. pylori UreF is soluble and exhibits an equilibrium between the monomeric and dimeric species (25). The protein crystallizes as an all-α-helical dimer (Protein Data Bank code 3CXN), but it lacks the C-terminal 21 residues due to proteolysis, and its N-terminal 24 residues are disordered (38). The intact structure of UreF (Fig. 2) is available from the UreH:UreF complex (code 3SF5) and the UreH:UreF:UreG complex,3 in which a UreF dimer (with bound UreG when present) bridges two UreH protomers (25). These structures confirm the proposed interactions between UreF and UreH that were based on yeast two-hybrid and tandem affinity purification studies (31, 32, 39). UreH stabilizes the N-terminal helix of UreF and exposes a conserved Tyr residue at position 48. This residue and the highly conserved C terminus make up one face of the three-dimensional UreF dimer (shown to be the UreG-binding site; see below). The interface between UreF and UreH is poorly conserved, likely due to the low similarities within the UreF and UreD/UreH sequences.
The assembly model shows UreF binding to UreABC:UreD to form UreABC:UreD:UreF (Fig. 3), a complex that can be directly isolated from cells expressing the corresponding genes (35). Alternatively, in vitro incubation of UreE-UreF with UreABC:UreD provides UreABC:UreD:UreE-UreF (37). Native gel electrophoresis of UreABC:UreD:UreF revealed multiple species, and zero to three pairs of UreD:UreF are suggested to bind per (UreABC)3. Small angle x-ray scattering experiments suggested a close proximity between UreD and UreF, with both accessory proteins binding in the vicinity of UreB (29). Chemical cross-linking results support this configuration and also provide evidence for a conformational change in urease within the UreABC:UreD:UreF complex (30); specifically, UreB is proposed to undergo a hinge-like motion that enhances access to the nascent active site (40). Following in vitro activation of UreABC:UreD:UreF, the urease-specific activity is similar to that obtained with UreABC:UreD; however, much lower concentrations of bicarbonate are required, and the process is more resistant to inhibition by Ni2+ (35). UreF serves as the binding site for the UreG GTPase within the UreABC:UreD:UreF complex (see below). A role as a GTPase-activating protein was suggested for UreF (41), but the UreH:UreF:UreG structure (which shows UreF binding UreG opposite the GTP site) (Fig. 2) and experimental evidence derived from mutagenesis and GTPase activity studies (42) argue against this proposal. For example, a urease activation complex containing a UreF variant exhibited enhanced GTPase activity compared with the complex with wild-type accessory protein. UreF thus appears to gate the GTPase activity of UreG so as to promote efficient coupling of GTP hydrolysis and metallocenter biosynthesis, thereby enhancing the fidelity of urease activation.
UreG: GTPase for Urease Activation
In contrast to UreD/UreH and K. aerogenes UreF, which are insoluble, UreG is soluble and has been characterized from several sources (43–47). The K. aerogenes protein is a monomer that binds 1 eq of Ni2+ or Zn2+ (Kd ∼ 5 μm for either metal) (48). Mycobacterium tuberculosis (45) and S. pasteurii (44) possess dimeric UreG proteins, with the latter binding two Zn2+ ions (Kd ∼ 40 μm) or, more weakly, larger numbers of Ni2+. The H. pylori protein dimerizes in the presence of Zn2+ (Kd ∼ 0.3 μm, 1/dimer) but not with Ni2+, which binds more weakly (Kd ∼ 10 μm, 1.8/monomer) (46). X-ray absorption spectroscopy of the zinc-bound protein revealed a trigonal bipyramidal site including two His and two Cys residues, likely positioned at the subunit interface (49). The soybean (Glycine max) protein also exhibits a monomer/dimer equilibrium, with the dimer stabilized by Zn2+, but the binding thermodynamics are quite complex (47). No crystal structure is available for free UreG, perhaps related to its intrinsic disorder (50); however, UreG homology models (44–47) have been created by using HypB (required for nickel insertion into [Ni-Fe] hydrogenase) (51, 52) from Methanocaldococcus jannaschii (a dimeric GTPase with a dinuclear zinc site at the subunit interface (53)) as the template, and the structure of the H. pylori UreH:UreF:UreG complex is known (Fig. 2).3 The latter complex has two protomers of each peptide, with the two UreG molecules in contact as expected for a protein able to dimerize. Although UreG is a GTPase, the free protein exhibits slow (44, 45, 47) or no (43, 46, 48) GTPase activity. When present in urease activation complexes, GTPase activity is observed (54), and substitution of a key residue in the GTP-binding P-loop motif of K. aerogenes or H. pylori UreG abolishes the cell's ability to make active urease (43, 55).
A UreABC:UreD:UreF:UreG complex (Fig. 3) forms in K. aerogenes cultures grown without Ni2+ (56). The complex can also be accessed by mixing UreG with UreABC:UreD:UreF (54). Furthermore, UreABC:UreD:UreE-UreF:UreG is made in cells producing the UreE-UreF fusion protein (38). Mutagenesis studies identified Asp-80 as a key UreG residue involved in this interaction (48) and defined several residues along one face of UreF as the UreG-docking site (42). Using standard activation conditions, ∼60% of the nascent active sites in UreABC:UreD:UreF:UreG become active (54). Significantly, when using more physiological levels of bicarbonate and Ni2+, the resulting activity is decreased, but activation is greatly facilitated by GTP. UreG is active as a GTPase when present in this complex.
UreD/UreH:UreF:UreG: Molecular Chaperone Complex for Urease Activation
As an alternative to sequentially adding each accessory protein to the urease apoenzyme, the heterotrimer may bind as a unit to urease (Fig. 3). A UreD:UreF:UreG complex forms in vivo when the corresponding K. aerogenes genes are expressed independently of the structural components (43); however, this species is poorly soluble and not well characterized. This solubility problem is overcome in the MBP-UreD:UreF:UreG complex, and this species binds to urease but not to urease lacking UreB (40). MBP-UreD:UreF:UreG contains two copies of each protomer according to gel filtration and mass spectrometric studies.4 The structure of the analogous UreH:UreF:UreG complex from H. pylori (Fig. 2)3 reveals two UreG protomers binding to one face of the UreF dimer, with each UreG interacting with both UreF protomers and with each UreH interacting with a single UreF. A GDP is bound opposite of UreF within each UreG, confirming that the former protein is not a GTPase-activating protein. A potential metal-binding site is deeply buried and bridges the two UreG molecules, with each protomer providing His and Cys residues.
UreE: A Nickel Metallochaperone
A hint that UreE might be involved in nickel delivery to urease is given in the sequence of the K. aerogenes protein, which reveals 10 His residues in the C-terminal 15 residues, thus resembling a His-tagged protein (18). Indeed, the purified protein binds ∼6 eq of Ni2+/dimer (57). Not all UreE proteins contain this His-rich extension (58), and a truncation variant of the K. aerogenes protein lacking this region (H144* UreE) retains its ability to facilitate urease activation (59). The crystal structure of copper-bound K. aerogenes H144* UreE (Protein Data Bank code 1GMW) reveals three metal-binding sites, including an interfacial site with His-96 from each subunit and peripheral sites in each protomer involving His-110 and His-112 (60). Equilibrium dialysis measurements confirm the binding of Ni2+ to multiple distinct sites in the truncated protein (61). Mutagenesis studies demonstrated that only the interfacial site is required for UreE function (62). The full-length zinc-bound S. pasteurii UreE dimeric structure (code 1EAR) exhibits close similarity to the K. aerogenes protein, lacks the C-terminal His-rich region and the two peripheral sites, and binds a single Zn2+ ion (presumably substituting for Ni2+) at the interfacial site (63). Structures of several forms of H. pylori UreE are known, including the nickel-bound species (codes 3L9Z and 3TJ8) (64, 65), in which the Ni2+ is coordinated at the interfacial site with an additional His residue provided from the C terminus. These highly soluble proteins are proposed to bind metal ions in the cytoplasm and specifically deliver nickel to urease within the complex of other accessory proteins.
UreE forms UreG:UreE and UreABC:UreD:UreF:UreG:UreE complexes, with the latter species likely to serve as the ultimate urease activation machinery (Fig. 3). For the H. pylori components, two UreG protomers bind the UreE dimer, with the interaction stabilized by Zn2+ but not Ni2+ (66). In contrast, one UreG monomer from K. aerogenes binds to its cognate UreE dimer, with the interaction stabilized by either Zn2+ or Ni2+ (48). Ligand identities in the metal-stabilized UreE:UreG complexes have not been reported. The transient formation of a UreABC:UreD:UreF:UreG:UreE complex is suggested by the generation of fully active urease when UreABC:UreD:UreF:UreG is incubated with UreE, bicarbonate, Ni2+, and GTP (67). In addition, UreABC:UreD:UreF:UreG:UreE can be directly isolated from cells that synthesize a G11P UreB variant (29) or a Strep tag II variant of UreG when the culture contains Ni2+ (48).
The current working model for urease activation with the prototypical accessory proteins (Fig. 3) involves the binding of UreD, UreF, and UreG to the urease apoprotein, either sequentially or as a molecular chaperone unit, followed by interaction with the metallochaperone UreE. This activation complex carries out metallocenter assembly by steps that include Lys carbamylation, nickel incorporation, and GTP hydrolysis. No evidence indicates that accessory proteins facilitate the interaction of Lys with carbon dioxide at the nascent active site, but this possibility is not excluded, and it is reasonable to suspect that the nearby His residues assist in this reaction. The structure of UreABC bound to UreD/UreH:UreF:UreG is not defined, but two distinct models have been proposed. Fig. 3 shows a computational model (28) derived from studies using the K. aerogenes components. In this case, each vertex of the urease trimer binds a single molecule of UreD, UreF, and UreG, requiring dissociation of (UreD:UreF:UreG)2. By contrast, (UreH:UreF:UreG)2 of H. pylori (Fig. 2) has been proposed to bind at 2-fold symmetry sites of its cognate urease dodecamer so that the accessory protein complex remains intact (25). Such an interaction is precluded for the three-subunit bacterial ureases and the single-subunit eukaryotic ureases (Fig. 1), suggesting possible species-specific differences in the properties of this accessory protein complex. It remains unclear how UreE binds to the urease activation complex and how nickel is transferred from UreE to the active site; one proposal suggests intermediate binding sites on UreG and UreD (26). The function of GTP hydrolysis by UreG in this process remains poorly understood. The net outcome from the activation machinery is to transform urease apoprotein into the holoprotein, with dissociation of the accessory proteins for possible reuse.
Variations in Urease Activation Systems
Additional Accessory Proteins
Additional genes have been shown to facilitate urease activation in some microorganisms. For example, located just 5′ of the typical urease genes in Yersinia pseudotuberculosis is yntABCDE, which encodes an ATP-binding cassette-type metal transporter; deletion of these genes eliminates urease activity and reduces the Ni2+ uptake rate (68). Evidence for a similar Ni2+ transporter dedicated to urease in Actinobacillus pleuropneumoniae comes from recombinant expression of the urease gene cluster with or without its adjacent cbiKLMQ genes in E. coli (69). In the same manner, heterologous expression of the Bacillus sp. TB-90 urease gene cluster and its deletion mutants indicates a nickel-dependent role for ureH (unrelated to ureH of H. pylori) in urease activation; Bacillus ureH is suspected to encode a Ni2+ permease (70). Many other microorganisms contain Ni2+ transporters and Ni2+ permeases (with their levels often controlled by nickel-dependent transcriptional regulators) that enhance urease activity by providing the essential metal ion (52, 71, 72), but generally the corresponding genes are distant from the urease genes. For example, H. pylori uses NixA, AbcABCD, and the outer membrane transporter HP1512 to take up Ni2+, and deletion of these genes leads to reductions in urease activity (73–76). Of additional interest, hypA and hypB of this microorganism are required for urease activity, but the corresponding gene products are not involved in Ni2+ uptake (77). A direct competition is observed between HypA and UreG for binding UreE (78). HypA and HypB are generally associated with metallocenter biosynthesis of [Ni-Fe] hydrogenases, but they appear to serve a dual role here of still undefined function.
Missing Accessory Proteins
Plants appear to lack homologs to ureE (3), and a large number of ureolytic microorganisms lack one or more of the standard set of urease genes (9, 79). A dramatic example of this situation exists in Bacillus subtilis, where the genome reveals the presence of only the structural urease genes; nevertheless, the cells synthesize an active nickel urease, although with poor efficiency (80). In many other cases, however, the sequenced microorganisms were not examined for urease activity.
Iron Urease
Helicobacter mustelae, a gastric pathogen of ferrets, contains two urease gene clusters: ureABIEFGH and ureA2B2 (81). The former cluster, closely related to that found in H. pylori, is induced by Ni2+ and encodes two structural genes, a proton-gated urea channel (82), and the four standard maturation proteins. The latter cluster is inversely regulated by Ni2+ and encodes only the two structural genes (83). Urease activity is retained in ureB and ureB/ureG mutants, indicating that ureA2B2 encodes an active urease, and its activation does not require the standard urease-specific GTPase. This finding was confirmed and extended by results showing that heterologous expression of ureA2B2 in E. coli generates active enzyme (84). Purified UreAB is a conventional nickel urease, whereas isolated UreA2B2 is an oxygen-labile iron-containing enzyme. The structure of oxidized UreA2B2, a dodecamer like that shown in Fig. 1B, reveals a dinuclear active site that is remarkably similar to the metallocenter of conventional ureases (84). This finding is consistent with the high degree of similarity in their sequences, e.g. UreA is 57% identical to UreA2, and UreB is 70% identical to UreB2. Two other strains of Helicobacter, Helicobacter felis and Helicobacter acinonychis, have similar arrangements of urease genes. The hosts of these pathogens, both in the Felidae (cat) family, are carnivores like the ferret, leading to speculation that these bacteria have evolved an iron urease because of their association with meat diets that are rich in iron and depleted in nickel (83). When the three UreA2B2 sequences are aligned and compared with the sequences of nickel ureases, a prominent cluster of distinct residues are seen to encircle the channel into the active site (84). These results are compatible with an interaction between UreA2B2 apoprotein and an iron delivery protein. One possibility is that activation of iron urease makes use of a general iron delivery system that is used for maturation of the many iron proteins in the cell. The oxidized state of UreA2B2, suggested to be a μ-oxo-bridged Fe(III)-O-Fe(III) species, probably forms naturally within the microaerophilic microorganism, and the cell likely has a mechanism to regenerate the active diferrous species (85). The ability to form active iron urease in E. coli using only ureA2B2 implies that urease-specific accessory proteins are not required for Lys carbamylation.
Future Directions
As should be clear from the preceding discussion, many questions remain to be answered about how the urease metallocenter is synthesized. Significantly, these questions also often apply to the biosynthesis of other types of metallocenters such as that found in [Ni-Fe] hydrogenases, which utilize the HypB GTPase along with SlyD and HypA metallochaperones for their activation (51, 52). For example, it is unknown whether metallochaperones such as UreE interact with membrane-associated transport proteins to couple metal binding to metal transport. It is also unclear how UreE, a protein that binds several metal ions, is able to specifically deliver Ni2+ to urease and to function in an in vitro activation system even when a Ni2+ chelator with greater affinity is present (67). The known interactions between UreE and UreG, along with the nickel-binding capabilities of some UreG proteins, suggest that nickel may be delivered to UreG before subsequently making its way to the nascent active site. Further effort is needed to ascertain the function of the UreG GTPase activity; it may be associated with a nickel transfer step, a conformational change of a protein, a protein dissociation step, or some other process. The mechanism by which UreF enhances urease activation fidelity (42) is unknown. UreD appears to serve as a scaffold for binding other proteins but also exhibits the direct effect of increasing activation efficiency by an unknown mechanism. Following activation, it is unclear whether UreDFG is released as a unit or as the individual proteins. The mechanisms used for activation of nickel urease in organisms lacking one or more accessory proteins demand further clarification. Finally, the discovery of iron urease in H. mustelae raises questions such as which other organisms contain this type of enzyme, how the iron is delivered, what dictates the metal specificity, and could other metals be used in selected cases.
Acknowledgments
We thank Prof. Kam-Bo Wong for coordinates of the structure in Fig. 2 and Prof. Célia Carlini for coordinates of the computational model in Fig. 3.
This work was supported, in whole or in part, by National Institutes of Health Grant DK045686 (to R. P. H.). This is the third article in the Thematic Minireview Series on Metals in Biology 2013.
K.-B. Wong, personal communication.
M. A. Farrugia, L. Han, Y. Zhong, J. L. Boer, B. J. Ruotolo, and R. P. Hausinger, unpublished data.
- MBP
- maltose-binding protein.
REFERENCES
- 1. Scott D. R., Marcus E. A., Weeks D. L., Sachs G. (2002) Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology 123, 187–195 [DOI] [PubMed] [Google Scholar]
- 2. Nielubowicz G. R., Mobley H. L. T. (2010) Host-pathogen interactions in the urinary tract interaction. Nat. Rev. Urol. 7, 430–441 [DOI] [PubMed] [Google Scholar]
- 3. Witte C.-P. (2011) Urea metabolism in plants. Plant Sci. 180, 431–438 [DOI] [PubMed] [Google Scholar]
- 4. Bremner J. M. (1995) Recent research on problems in the use of urea as a nitrogen fertilizer. Fertilizer Res. 42, 321–329 [Google Scholar]
- 5. Sumner J. B. (1926) The isolation and crystallization of the enzyme urease. J. Biol. Chem. 69, 435–441 [Google Scholar]
- 6. Dixon N. E., Gazzola C., Blakeley R. L., Zerner B. (1975) Jack bean urease (EC 3.5.1.5). A metalloenzyme. A simple biological role for nickel? J. Am. Chem. Soc. 97, 4131–4133 [DOI] [PubMed] [Google Scholar]
- 7. Carter E. L., Flugga N., Boer J. L., Mulrooney S. B., Hausinger R. P. (2009) Interplay of metal ions and urease. Metallomics 1, 207–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krajewska B. (2009) Ureases I. Functional, catalytic and kinetic properties: a review. J. Mol. Catal. B Enzym. 59, 9–21 [Google Scholar]
- 9. Zambelli B., Musiani F., Benini S., Ciurli S. (2011) Chemistry of Ni2+ in urease: sensing, trafficking, and catalysis. Acc. Chem. Res. 44, 520–530 [DOI] [PubMed] [Google Scholar]
- 10. Jabri E., Carr M. B., Hausinger R. P., Karplus P. A. (1995) The crystal structure of urease from Klebsiella aerogenes. Science 268, 998–1004 [PubMed] [Google Scholar]
- 11. Pearson M. A., Michel L. O., Hausinger R. P., Karplus P. A. (1997) Structure of Cys319 variants and acetohydroxamate-inhibited Klebsiella aerogenes urease. Biochemistry 36, 8164–8172 [DOI] [PubMed] [Google Scholar]
- 12. Benini S., Rypniewski W. R., Wilson K. S., Miletti S., Ciurli S., Mangani S. (1999) A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels. Structure 7, 205–216 [DOI] [PubMed] [Google Scholar]
- 13. Ha N.-C., Oh S.-T., Sung J. Y., Cha K. A., Lee M. H., Oh B.-H. (2001) Supramolecular assembly and acid resistance of Helicobacter pylori urease. Nat. Struct. Biol. 8, 505–509 [DOI] [PubMed] [Google Scholar]
- 14. Balasubramanian A., Ponnuraj K. (2010) Crystal structure of the first plant urease from jack bean: 83 years of journey from its first crystal to molecular structure. J. Mol. Biol. 400, 274–283 [DOI] [PubMed] [Google Scholar]
- 15. Estiu G., Merz K. M., Jr. (2006) Catalyzed decomposition of urea. Molecular dynamics simulations of the binding of urea to urease. Biochemistry 45, 4429–4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Estiu G., Merz K. M., Jr. (2007) Competitive hydrolytic and elimination mechanisms in the urease catalyzed decomposition of urea. J. Phys. Chem. B 111, 10263–10274 [DOI] [PubMed] [Google Scholar]
- 17. Carlsson H., Nordlander E. (2010) Computational modeling of the mechanism of urease. Bioinorg. Chem. Appl. 2010, 364891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mulrooney S. B., Hausinger R. P. (1990) Sequence of the Klebsiella aerogenes urease genes and evidence for accessory proteins facilitating nickel incorporation. J. Bacteriol. 172, 5837–5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee M. H., Mulrooney S. B., Renner M. J., Markowicz Y., Hausinger R. P. (1992) Klebsiella aerogenes urease gene cluster: sequence of ureD and demonstration that four accessory genes (ureD, ureE, ureF, and ureG) are involved in nickel metallocenter biosynthesis. J. Bacteriol. 174, 4324–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mobley H. L. T., Island M. D., Hausinger R. P. (1995) Molecular biology of microbial ureases. Microbiol. Rev. 59, 451–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cussac V., Ferrero R. L., Labigne A. (1992) Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J. Bacteriol. 174, 2466–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bacanamwo M., Witte C.-P., Lubbers M. W., Polacco J. C. (2002) Activation of the urease of Schizosaccharomyces pombe by the UreF accessory protein from soybean. Mol. Genet. Genomics 268, 525–534 [DOI] [PubMed] [Google Scholar]
- 23. Polacco J. C., Mazzafera P., Tezotto T. (2013) Opinion–nickel and urease in plants: still many knowledge gaps. Plant Sci. 199–200, 79–90 [DOI] [PubMed] [Google Scholar]
- 24. Park I.-S., Carr M. B., Hausinger R. P. (1994) In vitro activation of urease apoprotein and role of UreD as a chaperone required for nickel metallocenter assembly. Proc. Natl. Acad. Sci. U.S.A. 91, 3233–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fong Y. H., Wong H. C., Chuck C. P., Chen Y. W., Sun H., Wong K.-B. (2011) Assembly of the preactivation complex for urease maturation in Helicobacter pylori. Crystal structure of the UreF-UreH protein complex. J. Biol. Chem. 286, 43241–43249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carter E. L., Hausinger R. P. (2010) Characterization of Klebsiella aerogenes urease accessory protein UreD in fusion with the maltose binding protein. J. Bacteriol. 192, 2294–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wollers S., Layer G., Garcia-Serres R., Signor L., Clemancey M., Latour J.-M., Fontecave M., Ollagnier de Choudens S. (2010) Iron-sulfur (Fe-S) cluster assembly. The SufBCD complex is a new type of Fe-S scaffold with a flavin redox cofactor. J. Biol. Chem. 285, 23331–23341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ligabue-Braun R., Real-Guerra R., Carlini C. R., Verli H. (2012) Evidence-based docking of the urease activation complex. J. Biomol. Struct. Dyn., DOI: 10.1080/07391102.2012.713782 [DOI] [PubMed] [Google Scholar]
- 29. Quiroz-Valenzuela S., Sukuru S. C. K., Hausinger R. P., Kuhn L. A., Heller W. T. (2008) The structure of urease activation complexes examined by flexibility analysis, mutagenesis, and small-angle X-ray scattering. Arch. Biochem. Biophys. 480, 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chang Z., Kuchar J., Hausinger R. P. (2004) Chemical cross-linking and mass spectrometric identification of sites of interaction for UreD, UreF, and urease. J. Biol. Chem. 279, 15305–15313 [DOI] [PubMed] [Google Scholar]
- 31. Rain J.-C., Selig L., de Reuse H., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schächter V., Chemama Y., Labigne A., Legrain P. (2001) The protein-protein interaction map of Helicobacter pylori. Nature 409, 211–215 [DOI] [PubMed] [Google Scholar]
- 32. Voland P., Weeks D. L., Marcus E. A., Prinz C., Sachs G., Scott D. (2003) Interactions among the seven Helicobacter pylori proteins encoded by the urease gene cluster. Am. J. Physiol. Gastrointest. Liver Physiol. 284, G96–G106 [DOI] [PubMed] [Google Scholar]
- 33. Park I.-S., Hausinger R. P. (1995) Requirement of carbon dioxide for in vitro assembly of the urease nickel metallocenter. Science 267, 1156–1158 [DOI] [PubMed] [Google Scholar]
- 34. Park I.-S., Hausinger R. P. (1996) Metal ion interactions with urease and UreD-urease apoproteins. Biochemistry 35, 5345–5352 [DOI] [PubMed] [Google Scholar]
- 35. Moncrief M. B. C., Hausinger R. P. (1996) Purification and activation properties of UreD-UreF-urease apoprotein complexes. J. Bacteriol. 178, 5417–5421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim K. Y., Yang C. H., Lee M. H. (1999) Expression of the recombinant Klebsiella aerogenes UreF protein as a MalE fusion. Arch. Pharm. Res. 22, 274–278 [DOI] [PubMed] [Google Scholar]
- 37. Kim J. K., Mulrooney S. B., Hausinger R. P. (2006) The UreEF fusion protein provides a soluble and functional form of the UreF urease accessory protein. J. Bacteriol. 188, 8413–8420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lam R., Romanov V., Johns K., Battaile K. P., Wu-Brown J., Guthrie J. L., Hausinger R. P., Pai E. F., Chirgadze N. Y. (2010) Crystal structure of a truncated urease accessory protein UreF from Helicobacter pylori. Proteins 78, 2839–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stingl K., Schauer K., Ecobichon C., Labigne A., Lenormand P., Rousselle J.-C., Namane A., de Reuse H. (2008) In vivo interactome of Helicobacter pylori urease revealed by tandem affinity purification. Mol. Cell. Proteomics 7, 2429–2441 [DOI] [PubMed] [Google Scholar]
- 40. Carter E. L., Boer J. L., Farrugia M. A., Flugga N., Towns C. L., Hausinger R. P. (2011) Function of UreB in Klebsiella aerogenes urease. Biochemistry 50, 9296–9308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Salomone-Stagni M., Zambelli B., Musiani F., Ciurli S. (2007) A model-based proposal for the role of UreF as a GTPase-activating protein in the urease active site biosynthesis. Proteins 68, 749–761 [DOI] [PubMed] [Google Scholar]
- 42. Boer J. L., Hausinger R. P. (2012) Klebsiella aerogenes UreF: identification of the UreG binding site and role in enhancing the fidelity of urease activation. Biochemistry 51, 2298–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moncrief M. B. C., Hausinger R. P. (1997) Characterization of UreG, identification of a UreD-UreF-UreG complex, and evidence suggesting that a nucleotide-binding site in UreG is required for in vivo metallocenter assembly of Klebsiella aerogenes urease. J. Bacteriol. 179, 4081–4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zambelli B., Stola M., Musiani F., De Vriendt K., Samyn B., Devreese B., Van Beeumen J., Turano P., Dikiy A., Bryant D. A., Ciurli S. (2005) UreG, a chaperone in the urease assembly process, is an intrinsically unstructured GTPase that specifically binds Zn2+. J. Biol. Chem. 280, 4684–4695 [DOI] [PubMed] [Google Scholar]
- 45. Zambelli B., Musiani F., Savini M., Tucker P., Ciurli S. (2007) Biochemical studies on Mycobacterium tuberculosis UreG and comparative modeling reveal structural and functional conservation among the bacterial UreG family. Biochemistry 46, 3171–3182 [DOI] [PubMed] [Google Scholar]
- 46. Zambelli B., Turano P., Musiani F., Neyroz P., Ciurli S. (2009) Zn2+-linked dimerization of UreG from Helicobacter pylori, a chaperone involved in nickel trafficking and urease activation. Proteins 74, 222–239 [DOI] [PubMed] [Google Scholar]
- 47. Real-Guerra R., Staniscuaski F., Zambelli B., Musiani F., Ciurli S., Carlini C. R. (2012) Biochemical and structural studies on native and recombinant Glycine max UreG: a detailed characterization of a plant urease accessory gene. Plant Mol. Biol. 78, 461–475 [DOI] [PubMed] [Google Scholar]
- 48. Boer J. L., Quiroz-Valenzuela S., Anderson K. L., Hausinger R. P. (2010) Mutagenesis of Klebsiella aerogenes UreG to probe nickel binding and interactions with other urease-related proteins. Biochemistry 49, 5859–5869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martin-Diaconescu V., Bellucci M., Musiani F., Ciurli S., Maroney M. J. (2012) Unraveling the Helicobacter pylori UreG zinc binding site using x-ray absorption spectroscopy (XAS) and structural modeling. J. Biol. Inorg. Chem. 17, 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zambelli B., Cremades N., Neyroz P., Turano P., Uversky V. N., Ciurli S. (2012) Insights in the (un)structural organization of Bacillus pasteurii UreG, an intrinsically disordered GTPase enzyme. Mol. Biosyst. 8, 220–228 [DOI] [PubMed] [Google Scholar]
- 51. Kaluarachchi H., Chan Chung K. C., Zamble D. B. (2010) Microbial nickel proteins. Nat. Prod. Rep. 27, 681–694 [DOI] [PubMed] [Google Scholar]
- 52. Higgins K. A., Carr C. E., Maroney M. J. (2012) Specific metal recognition in nickel trafficking. Biochemistry 51, 7816–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gasper R., Scrima A., Wittinghofer A. (2006) Structural insights into HypB, a GTP-binding protein that regulates metal binding. J. Biol. Chem. 281, 27492–27502 [DOI] [PubMed] [Google Scholar]
- 54. Soriano A., Hausinger R. P. (1999) GTP-dependent activation of urease apoprotein in complex with the UreD, UreF, and UreG accessory proteins. Proc. Natl. Acad. Sci. U.S.A. 96, 11140–11144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mehta N., Benoit S., Maier R. J. (2003) Roles of conserved nucleotide-binding domains in accessory proteins, HypB and UreG, in the maturation of nickel-enzymes required for efficient Helicobacter pylori colonization. Microb. Pathog. 35, 229–234 [DOI] [PubMed] [Google Scholar]
- 56. Park I.-S., Hausinger R. P. (1995) Evidence for the presence of urease apoprotein complexes containing UreD, UreF, and UreG in cells that are competent for in vivo enzyme activation. J. Bacteriol. 177, 1947–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee M. H., Pankratz H. S., Wang S., Scott R. A., Finnegan M. G., Johnson M. K., Ippolito J. A., Christianson D. W., Hausinger R. P. (1993) Purification and characterization of Klebsiella aerogenes UreE protein: a nickel-binding protein that functions in urease metallocenter assembly. Protein Sci. 2, 1042–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Musiani F., Zambelli B., Stola M., Ciurli S. (2004) Nickel trafficking: insights into the fold and function of UreE, a urease metallochaperone. J. Inorg. Biochem. 98, 803–813 [DOI] [PubMed] [Google Scholar]
- 59. Brayman T. G., Hausinger R. P. (1996) Purification, characterization, and functional analysis of a truncated Klebsiella aerogenes UreE urease accessory protein lacking the histidine-rich carboxyl terminus. J. Bacteriol. 178, 5410–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Song H. K., Mulrooney S. B., Huber R., Hausinger R. P. (2001) Crystal structure of Klebsiella aerogenes UreE, a nickel-binding metallochaperone for urease activation. J. Biol. Chem. 276, 49359–49364 [DOI] [PubMed] [Google Scholar]
- 61. Grossoehme N. E., Mulrooney S. B., Hausinger R. P., Wilcox D. E. (2007) Thermodynamics of Ni2+, Cu2+, and Zn2+ binding to urease metallochaperone UreE. Biochemistry 46, 10506–10516 [DOI] [PubMed] [Google Scholar]
- 62. Colpas G. J., Brayman T. G., Ming L.-J., Hausinger R. P. (1999) Identification of metal-binding residues in the Klebsiella aerogenes urease nickel metallochaperone, UreE. Biochemistry 38, 4078–4088 [DOI] [PubMed] [Google Scholar]
- 63. Remaut H., Safarov N., Ciurli S., Van Beeumen J. (2001) Structural basis for Ni2+ transport and assembly of the urease active site by the metallochaperone UreE from Bacillus pasteurii. J. Biol. Chem. 276, 49365–49370 [DOI] [PubMed] [Google Scholar]
- 64. Banaszak K., Martin-Diaconescu V., Bellucci M., Zambelli B., Rypniewski W., Maroney M. J., Ciurli S. (2012) Crystallographic and x-ray absorption spectroscopic characterization of Helicobacter pylori UreE bound to Ni2+ and Zn2+ reveals a role for the disordered C-terminal arm in metal trafficking. Biochem. J. 441, 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shi R., Munger C., Asinas A., Benoit S. L., Miller E., Matte A., Maier R. J., Cygler M. (2010) Crystal structures of apo and metal-bound forms of the UreE protein from Helicobacter pylori: role of multiple metal binding sites. Biochemistry 49, 7080–7088 [DOI] [PubMed] [Google Scholar]
- 66. Bellucci M., Zambelli B., Musiani F., Turano P., Ciurli S. (2009) Helicobacter pylori UreE, a urease accessory protein: specific Ni2+- and Zn2+ -binding properties and interaction with its cognate UreG. Biochem. J. 422, 91–100 [DOI] [PubMed] [Google Scholar]
- 67. Soriano A., Colpas G. J., Hausinger R. P. (2000) UreE stimulation of GTP-dependent urease activation in the UreD-UreF-UreG-urease apoprotein complex. Biochemistry 39, 12435–12440 [DOI] [PubMed] [Google Scholar]
- 68. Sebbane F., Mandrand-Berthelot M.-A., Simonet M. (2002) Genes encoding specific nickel transport systems flank the chromosomal urease locus of pathogenic Yersiniae. J. Bacteriol. 184, 5706–5713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bossé J. T., Gilmour H. D., MacInnes J. I. (2001) Novel genes affecting urease activity in Actinobacillus pleuropneumoniae. J. Bacteriol. 183, 1242–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Maeda M., Hidaka M., Nakamura A., Masaki H., Uozumi T. (1994) Cloning, sequencing, and expression of thermophilic Bacillus sp. strain TB-90 urease gene complex in Escherichia coli. J. Bacteriol. 176, 432–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rodionov D. A., Hebbeln P., Gelfand M. S., Eitinger T. (2006) Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 188, 317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mulrooney S. B., Hausinger R. P. (2003) Nickel uptake and utilization by microorganisms. FEMS Microbiol. Rev. 27, 239–261 [DOI] [PubMed] [Google Scholar]
- 73. Bauerfeind P., Garner R. M., Mobley H. L. T. (1996) Allelic exchange mutagenesis of nixA in Helicobacter pylori results in reduced nickel transport and urease activity. Infect. Immun. 64, 2877–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hendricks J. K., Mobley H. L. T. (1997) Helicobacter pylori ABC transporter: effect of allelic exchange mutagenesis on urease activity. J. Bacteriol. 179, 5892–5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Davis G. S., Flannery E. L., Mobley H. L. T. (2006) Helicobacter pylori HP1512 is a nickel-responsive NikR-regulated outer membrane protein. Infect. Immun. 74, 6811–6820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Schauer K., Gouget B., Carrière M., Labigne A., de Reuse H. (2007) Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Mol. Microbiol. 63, 1054–1068 [DOI] [PubMed] [Google Scholar]
- 77. Olson J. W., Mehta N. S., Maier R. J. (2001) Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol. Microbiol. 39, 176–182 [DOI] [PubMed] [Google Scholar]
- 78. Benoit S. L., McMurry J. L., Hill S. A., Maier R. J. (2012) Helicobacter pylori hydrogenase accessory protein HypA and urease accessory protein UreG compete with each other for UreE recognition. Biochim. Biophys. Acta 1820, 1519–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mizuki T., Kamekura M., DasSarma S., Fukushima T., Usami R., Yoshida Y., Horikoshi K. (2004) Ureases of extreme halophiles of the genus Haloarcula with a unique structure of gene cluster. Biosci. Biotechnol. Biochem. 68, 397–406 [DOI] [PubMed] [Google Scholar]
- 80. Kim J. K., Mulrooney S. B., Hausinger R. P. (2005) Biosynthesis of active Bacillus subtilis urease in the absence of known urease accessory proteins. J. Bacteriol. 187, 7150–7154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. O'Toole P. W., Snelling W. J., Canchaya C., Forde B. M., Hardie K. R., Josenhans C., Graham R. L. J., McMullan G., Parkhill J., Belda E., Bentley S. D. (2010) Comparative genomics and proteomics of Helicobacter mustelae, an ulcerogenic and carcinogenic gastric pathogen. BMC Genomics 11, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Strugatsky D., McNulty R., Munson K., Chen C.-K., Soltis S. M., Sachs G., Luecke H. (2013) Structure of the proton-gated urea channel from the gastric pathogen Helicobacter pylori. Nature 493, 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Stoof J., Breijer S., Pot R. G. J., van der Neut D., Kuipers E. J., Kusters J. G., van Vliet A. H. M. (2008) Inverse nickel-responsive regulation of two urease enzymes in the gastric pathogen Helicobacter mustelae. Environ. Microbiol. 10, 2586–2597 [DOI] [PubMed] [Google Scholar]
- 84. Carter E. L., Tronrud D. E., Taber S. R., Karplus P. A., Hausinger R. P. (2011) Iron-containing urease in a pathogenic bacterium. Proc. Natl. Acad. Sci. U.S.A. 108, 13095–13099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Carter E. L., Proshlyakov D. A., Hausinger R. P. (2012) Apoprotein isolation and activation, and vibrational structure of the Helicobacter mustelae iron urease. J. Inorg. Biochem. 111, 195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]