Abstract
The reactivity of the cobalt-carbon bond in cobalamins is the key to their chemical versatility, supporting both methyl transfer and isomerization reactions. During evolution of higher eukaryotes that utilize vitamin B12, the high reactivity of the cofactor coupled with its low abundance pressured development of an efficient system for uptake, assimilation, and delivery of the cofactor to client B12-dependent enzymes. Although most proteins suspected to be involved in B12 trafficking were discovered by 2009, the recent identification of a new protein reveals that the quest for elucidating the intracellular B12 highway is still far from complete. Herein, we review the biochemistry of cobalamin trafficking.
Keywords: Adenosylcobalamin, Metabolic Diseases, Metal Homeostasis, Trafficking, Vitamins, Cofactors
Introduction
Cofactors are variously deployed in nature to stabilize macromolecular structures, expand catalytic functionality, transport gases, transduce signals, and function as sensors. Due to their relative rarity and/or reactivity, cells have evolved strategies for sequestering and regulating the movement of cofactors from their point of entry into the cell to their point of docking in target proteins (1). A subset of cofactors, i.e. the vitamins, is obtained in a precursor form from the diet. Reactions catalyzing the assimilation of inactive cofactors into their active forms are integral to their trafficking pathways. Similarly, elaboration of metals into clusters often occurs on chaperones that subsequently transfer the cofactor to target proteins. The interprotein transfer of metals can occur via ligand exchange reactions that are driven by differences in metal coordination geometry and affinity between the donor and acceptor proteins (2, 3). Seclusion of cofactors in chaperones during assembly/processing into their active forms minimizes unwanted side reactions, whereas guided delivery averts dilution and promotes specificity of cofactor docking.
In contrast to our understanding of cellular strategies used for trafficking metals (4–6) and metal clusters (7), significantly less is known about strategies for shepherding organic and organometallic cofactors to target proteins. This picture has been changing, however, with the convergence of clinical genetics and biochemical approaches that are beginning to illuminate an elaborate pathway for assimilation and delivery of dietary vitamin B12 or cobalt-containing cobalamin, a complex organometallic cofactor (8–10). Much less is known about how the tetrapyrrolic cousins of B12, e.g. iron protoporphyrin (heme), nickel corphin (F430), and magnesium chlorin (chlorophyll), are guided to specific destinations.
In this minireview, we describe a model for mammalian cobalamin trafficking, which includes strategies for conversion of inactive precursors to the active cofactor forms methylcobalamin (MeCbl)3 and 5′-deoxyadenosylcobalamin (AdoCbl; coenzyme B12) and discuss the human diseases that result from impairments along the trafficking highway. We posit that the navigation strategy for B12, in which a rare, reactive, and high value cofactor is sequestered and targeted to client proteins, might represent a general archetype for the trafficking of other essential but scarce organic and organometallic cofactors.
B12 Chemistry and Catalysis
Cobalamin, discovered as the antipernicious anemia factor, was first crystallized in the cyanocobalamin (CNCbl) form (11), which is technically vitamin B12. The biologically active alkylcobalamins, MeCbl and AdoCbl, serve as cofactors for the methyltransferase and isomerase families of B12 enzymes, respectively (12). Although mammals have only two B12-dependent enzymes, methionine synthase and methylmalonyl-CoA mutase (MCM), there are many “handlers” that tailor dietary B12 and deliver it to its target enzymes (8, 9). The existence of the B12 trafficking pathway was suggested by careful clinical genetics studies spanning several decades on patients with inborn errors of cobalamin metabolism (10).
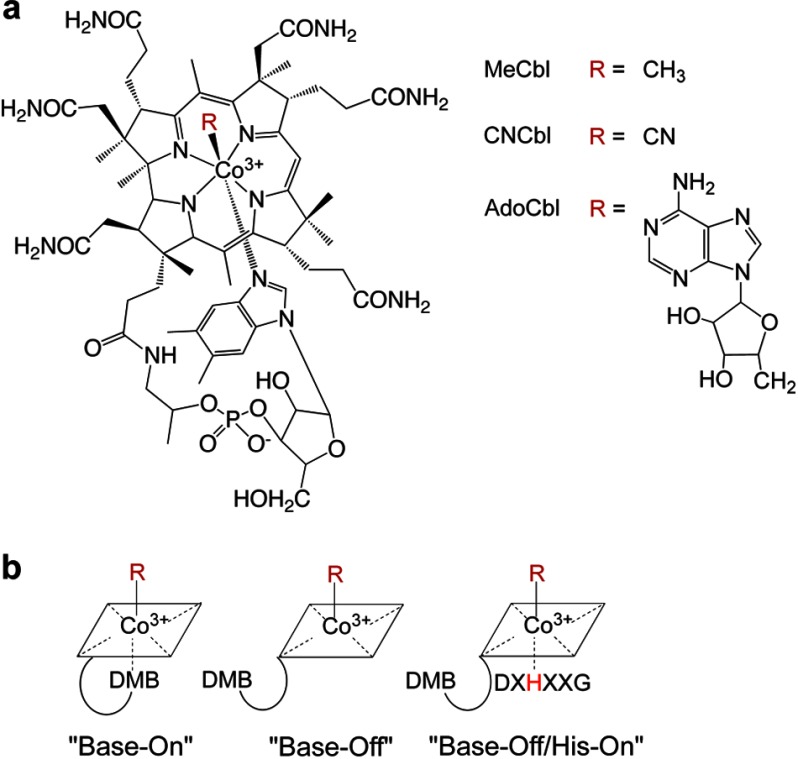
Chemically, cobalamins comprise a central cobalt atom that is coordinated by four equatorial nitrogen atoms donated by the tetrapyrrolic corrin ring (Fig. 1a). A bulky base, 5,6-dimethylbenzimidazole (DMB), extends from one edge of the corrin ring and coordinates the cobalt at the lower or α-axial position. The identity of the upper or β-axial ligand varies and includes cyano, aquo, methyl, and 5′-deoxyadenosyl groups. Ironically, 6 decades after its discovery, the origin and biological relevance of the cyano group remain unknown, although we have recently described a decyanase activity in the processing pathway (13), which allows utilization of CNCbl used in vitamin supplements. As the cobalt oxidation state decreases from 3+ → 2+ → 1+, the coordination number typically decreases from 6 → 5 → 4. In solution, alkylcobalamins preferentially exist in the six-coordinate “base-on” state, which is in contrast to the “base-off/His-on” state found in the active site of both mammalian B12 enzymes (Fig. 1b) (14, 15). Because the pKa for the base-on to base-off transition ranges from −2.13 to 3.17 depending on the identity of the upper axial ligand (16), the base-on conformation predominates at physiological pH.
FIGURE 1.
Cobalamin structure and conformations. a, cobalamin is shown in the base-on conformation, with DMB coordinating cobalt from the lower axial face of the corrin ring. The variable upper ligands are denoted as R on the right. b, alternative conformations of cobalamins differing with respect to the lower or α-axial ligation site.
The cobalt-carbon bond in alkylcobalamins is inherently weak and holds the key to the reactivity of this cofactor. The bond dissociation energies for base-on AdoCbl and MeCbl are 30 and 37 kcal/mol, respectively (17, 18). Methyltransferases utilize MeCbl, and the cobalt-carbon bond is cleaved heterolytically during nucleophilic displacement of the methyl group from MeCbl to an acceptor. In contrast, the isomerases cleave the cobalt-carbon bond homolytically generating a radical pair: 5′-deoxyadenosyl radical and cob(II)alamin. The former is the “working” radical, which abstracts a hydrogen atom from the substrate to initiate a radical-based isomerization reaction. The cobalt-carbon bond is re-formed at the end of each catalytic cycle.
Absorption, Transport, and Storage of Cobalamin
The recommended dietary allowance for cobalamin is 1–5 μg/day. A multistep process delivers the cofactor from the mouth into circulation and thereon to cells (Fig. 2) (19). Cobalamin released from food is first bound by haptocorrin, a salivary glycoprotein with broad specificity and high affinity for B12 at both neutral and acidic pH (20). In the duodenum, pancreatic proteases release cobalamin from the haptocorrin-B12 complex and from other proteins containing bound B12 that have been ingested. Subsequent binding of cobalamin to a second glycoprotein, intrinsic factor, facilitates its uptake by intestinal cells via cubilin/AMN receptor-mediated endocytosis (21). Intrinsic factor is highly selective for the physiologically relevant precursors of cobalamin containing an intact lower axial DMB ligand (22). Following internalization into enterocytes, intrinsic factor is degraded in the lysosome, and cobalamin is released into the blood stream. The ATP-dependent transporter ABCC1 (also known as MRP1 (multidrug resistance protein 1)), present in the basolateral membrane of intestinal epithelial and other cells, exports cobalamin bound to transcobalamin out of the cell (23). MRP1 knock-out mice accumulate cobalamin in the distal part of the intestine and exhibit low plasma, liver, and kidney cobalamin levels (23).
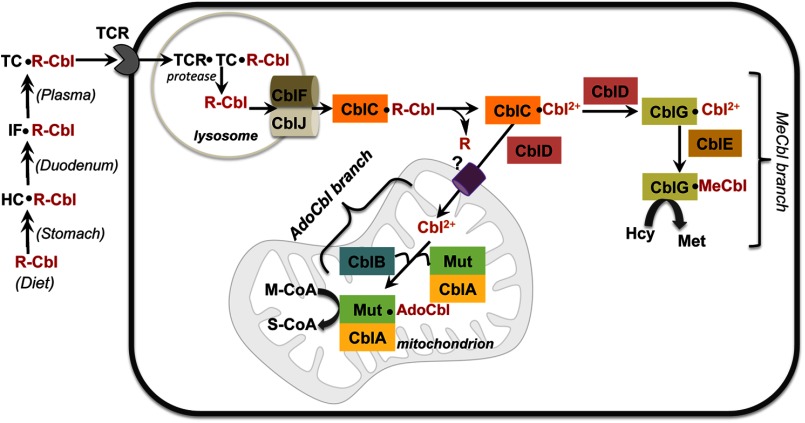
FIGURE 2.
Model for cobalamin utilization and intracellular trafficking in mammals. Dietary cobalamin (R-Cbl) is bound by a series of proteins found in the saliva and stomach (haptocorrin (HC)), in the duodenum (intrinsic factor (IF)), and in blood (transcobalamin (TC)). The details of these early trafficking steps are not shown in this figure. Transcobalamin is recognized by the transcobalamin receptor (TCR) found on cell surfaces and endocytosed into the lysosome, where R-Cbl is released following proteolytic digestion of transcobalamin. R-Cbl exits the lysosome in a process that requires the products of the cblJ and cblF loci. In the cytoplasm, CblC converts R-Cbl (and CNCbl) to cob(II)alamin (Cbl2+), which is partitioned to (i) the MeCbl pathway in the cytoplasm, which involves CblG (methionine synthase), CblE (methionine synthase reductase), and CblD, and (ii) the AdoCbl pathway in the mitochondrion, which requires CblD, CblB (ATR), and CblA (G-protein or MeaB in bacteria) in addition to MCM (Mut). The question mark by the mitochondrial cobalamin transporter denotes that it is unidentified. M-CoA, methylmalonyl-CoA; S-CoA, succinyl-CoA.
In the bloodstream, cobalamin is associated with two carriers, transcobalamin and haptocorrin. It is estimated that ∼20% of the circulating cobalamin is bound to transcobalamin, whereas the remainder, including incomplete B12 derivatives, is bound to haptocorrin (24, 25). Transcobalamin preferentially binds the intact cobalamin cofactor, representing a second level of molecular sieving out of degraded derivatives that could potentially compete with and inhibit B12-dependent enzymes (20, 26). Transcobalamin binds B12 avidly and mediates its transport across cells following complexation with the transcobalamin receptor, which is internalized in the lysosome (27). Lysosomal degradation of transcobalamin by resident hydrolases releases cobalamin, which is retained and further processed intracellularly.
Despite their low sequence identity (∼25%), the human cobalamin-binding proteins share a common evolutionary origin, with transcobalamin being the oldest, followed by intrinsic factor and haptocorrin (28). Biochemical studies with native or recombinant proteins indicate that all three proteins bind a single equivalent of B12 with high affinity (Kd < 1 pm), albeit with different specificities (20). Both transcobalamin and intrinsic factor bind cobalamin in a base-on conformation, whereas the binding mode for haptocorrin depends on the lower axial ligand. In addition to cobalamins, haptocorrin can bind cobinamides, B12 analogs that lack the DMB moiety, cannot be converted to the active cofactor forms by mammals, and account for ∼40% of the total plasma corrins (29). Although a definitive role for haptocorrin remains to be established, its suggested functions include a role in B12 storage and in removal of inhibitory corrinoid derivatives.
Lysosomal Egress of Cobalamin
Transport of cobalamin across the lysosomal membrane requires two membrane proteins with apparently distinct functions: LMBD1 and ABCD4 (Fig. 2) (30, 31). Defects in the genes encoding these two transporters result in accumulation of B12 in the lysosome and are classified as cblF (LMBD1) and the recently discovered cblJ (ABCD4) complementation groups (31, 32). Subcellular localization studies indicate that both LMBD1 and ABCD4 co-localize with other lysosomal proteins such as LAMP1; however, the precise role of each protein in the lysosomal export of cobalamin is unclear. Transport of free cobalamin into prokaryotes and the export of cobalamin from mammalian cells are fuelled by ATP hydrolysis (23, 33). ABCD4 is an ATP-binding cassette transporter, which might be the true lysosomal cobalamin transporter that is assisted by LMBD1. Alternatively, LMBD1, a putative transmembrane protein, might facilitate passive transport of cobalamin across the lysosomal membrane. In this model, ABCD4 could be involved in an ATPase-driven loading of B12 from the lysosome onto LMBD1 and/or in releasing the cobalamin cargo from LMBD1. Functional studies using fibroblasts from patients with cblF and cblJ defects indicate that the two transporters can only partially complement each other, and therefore, it is likely that they act synergistically. Clearly, biochemical studies are needed to decipher their function in the lysosomal egress of cobalamin. We have previously suggested that the release of cobalamin in the acidic environment of the lysosome favors formation of the base-off conformation of the cofactor and that once cobalamin exits this compartment, it retains this conformation by being protein-bound (9).
Early Steps in the Cytosolic Processing of Cobalamin
Upon arrival in the cytosol, cobalamins with various upper axial ligands must be processed to a common intermediate that can be allocated to the MeCbl and AdoCbl synthesis pathways to meet cellular needs. The protein that is postulated to accept the cobalamin cargo exiting the lysosomal compartment is MMACHC (methylmalonic aciduria type C and homocystinuria), the product of the cblC locus and hereafter referred to as CblC (Fig. 2) (9). Mutations in CblC impair both AdoCbl and MeCbl synthesis and are the most common cause of inherited cobalamin disorders (34).
Human CblC is a soluble protein in which the C-terminal ∼40 amino acid residues appear to be a recent evolutionary addition and are predicted to be highly disordered (35). Two CblC variants, a full-length (32 kDa) and a truncated (26 kDa) form, have been reported in human fibroblasts and in murine tissue (35, 36). The formation of the truncated CblC form is attributed to a predicted splicing variant (35). Although the prevalence of the truncated CblC form is not known, the presence of the C-terminal domain diminishes the stability of human CblC and is not required for its function (35). The crystal structures of the truncated variant of human CblC do not reveal the presence of a C-terminally located bacterial TonB-like domain, as predicted previously (35, 37).
CblC binds B12 stoichiometrically and exhibits a broad specificity for the β-axial ligand accommodating C1–C6 alkyl, adenosyl, cyano, and hydroxyl groups. Cobalamin is bound to CblC in a five-coordinate base-off conformation, where the DMB tail is dissociated from the cobalt. In the structure of MeCbl-bound CblC, the cobalamin tail is secured away from the corrin ring in a pocket dominated by hydrophobic interactions (Fig. 3a) (35). CblC exhibits remarkable chemical versatility by its ability to cleave cobalt-carbon bonds via both homolytic and heterolytic mechanisms, depending on the nature of the upper axial ligand (Fig. 3b). Thus, when an alkylcobalamin is bound to the active site, CblC catalyzes a nucleophilic displacement reaction in the presence of glutathione (Fig. 3b) (38). Mechanistically, the dealkylation reaction resembles the first half-reaction catalyzed by methionine synthase, in which the thiolate of homocysteine displaces the methyl group in MeCbl to form the thioether methionine and cob(I)alamin (Fig. 4a). Similarly, in the CblC-catalyzed dealkylation reaction, the glutathione thioether forms upon transfer of the alkyl group in addition to cob(I)alamin. When CNCbl is in the active site, electrons provided by free or protein-bound reduced flavin promote reductive homolytic cleavage, leading to cyanide elimination (13, 35). Based on UV-visible and EPR spectroscopy, base-off cob(II)alamin has been identified as the other product of the reductive elimination, consistent with a homolytic cleavage reaction mechanism. The modest decyanation and dealkylation rates exhibited by CblC are apparently sufficient to handle the flux through the cobalamin processing pathway to meet cellular needs. Fibroblasts derived from cblC patients confirm that CblC is required for processing alkyl- and cyanocobalamins for apportioning dietary B12 into MeCbl and AdoCbl, respectively (39).
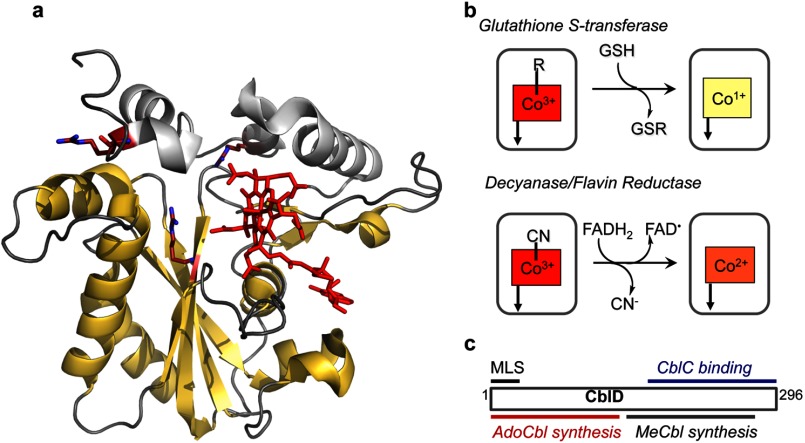
FIGURE 3.
Biochemical functions of CblC and CblD. a, the structure of human CblC with MeCbl (Protein Data Bank code 3SC0). MeCbl (red) is bound in a base-off conformation, with the DMB tail located in a side pocket. The flavin reductase domain is shown in yellow. The arginine residues that are mutated in patients are shown in stick representation. b, reactions catalyzed by CblC. c, localization of mutations in CblD that lead to impaired AdoCbl or MeCbl synthesis and the minimal length required for binding to CblC. MLS, mitochondrial leader sequence.
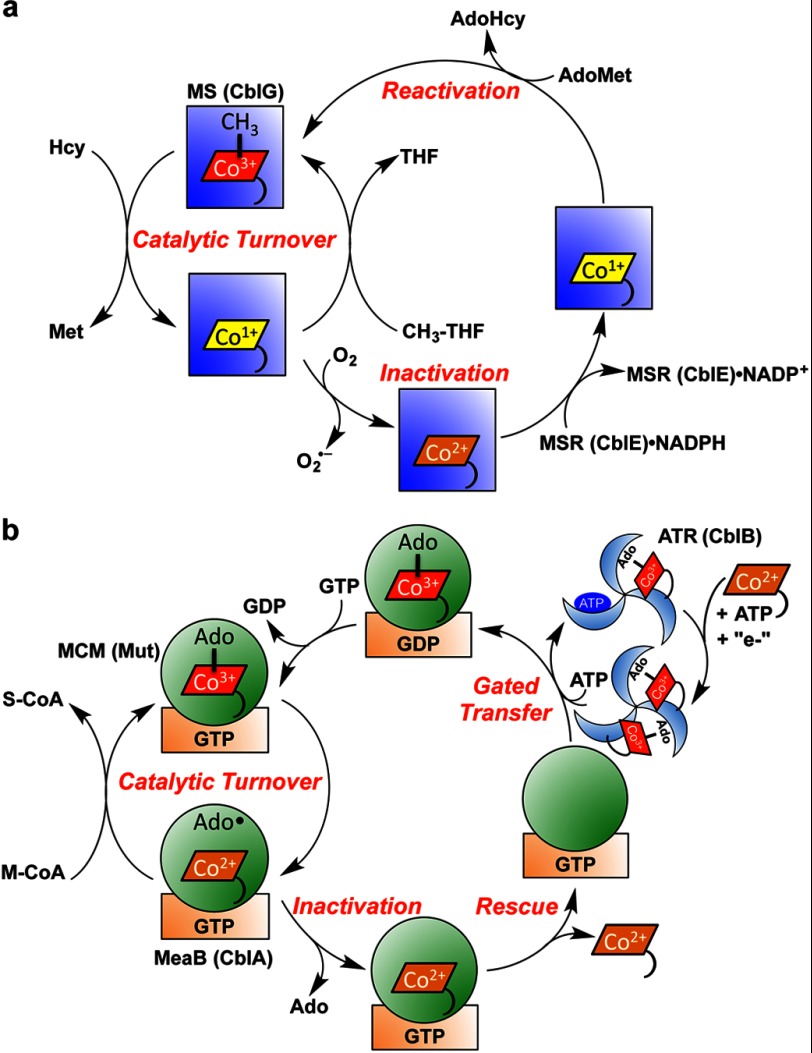
FIGURE 4.
Cofactor loading, activity, and repair of mammalian cobalamin-dependent enzymes. a, methionine synthase (MS; CblG; blue square) catalyzes the overall transfer of a methyl group from N5-methyltetrahydrofolate to homocysteine to give methionine and tetrahydrofolate (THF). Occasional oxidative escape of the cob(I)alamin intermediate during the catalytic cycle leads to the inactive cob(II)alamin species. The latter is rescued to MeCbl in a reductive methylation reaction needing NADPH, methionine synthase reductase (MSR; CblE), and AdoMet. This repair reaction is also likely to represent the route for formation of MeCbl following transfer of cob(II)alamin to apomethionine synthase. The mechanism for cob(II)alamin transfer during methionine synthase reconstitution is not known. AdoHyc, S-adenosylhomocysteine. b, ATR (CblB; blue wheel) converts ATP and cob(II)alamin in the presence of a reductant to AdoCbl. Two equivalents of AdoCbl are bound at one time, and binding of ATP to the vacant site triggers transfer of one AdoCbl to the MCM (Mut; green circle) active site in a reaction that is gated by GTP hydrolysis in the G-protein chaperone (MeaB or CblA; orange rectangle). The GTP-bound state of MeaB blocks transfer of cob(II)alamin from ATR to the complex between MCM and G-protein. During catalysis, the cobalt-carbon bond is cleaved homolytically to initiate a radical-based mechanism for the conversion of methylmalonyl-CoA (M-CoA) to succinyl-CoA (S-CoA). Occasional loss of 5′-deoxyadenosine (Ado) from the active site precludes re-formation of AdoCbl at the end of the catalytic cycle and leads to inactive mutase. In this situation, the GTP-containing chaperone promotes dissociation of cob(II)alamin, permitting reconstitution of the mutase with active cofactor.
Unexpectedly, the crystal structure of CblC revealed a flavin reductase fold (Fig. 3a) (35). Flavin binds at the dimer interface in two structurally related flavin reductases, BluB and iodotyrosine deiodinase. However, solution studies indicate that CblC exists predominantly as a monomer (13, 37). Unlike the structure of apo- and MeCbl-bound CblC, a dimeric crystal structure has been reported for AdoCbl-bound CblC, where the dimer interface is stabilized by interactions between residues belonging to a highly conserved “PNRRP” loop (37). The increase in the proportion of dimeric CblC in the presence of AdoCbl and FMN to ∼50% of the total protein in solution but to a lesser extent in the presence of MeCbl raises questions about the physiological relevance of the dimeric structure. Further studies are clearly needed to address this issue. An arginine-rich pocket located in the vicinity of the cobalamin-binding site (Fig. 3a) is suggested to be important for glutathione binding because mutations of highly conserved arginine residues impair binding (37).
The cblD complementation group represents the most complex and intriguing class of inherited cobalamin defects. Patients classified in this group exhibit isolated or combined methylmalonic aciduria and homocystinuria (40, 41). Mutations located in the N and C termini of the encoded CblD protein (also known as MMADHC (methylmalonic aciduria type D and homocystinuria)) are associated with methylmalonic aciduria and homocystinuria, respectively (Fig. 3c) (40, 41). Only the full-length CblD protein is seen in normal cell lines (42). However, translation initiation at two alternative sites, Met-62 and Met-116, has been invoked to explain how patient mutations predicted to lead to premature termination are bypassed in a subset of cblD patients. These shorter CblD variants are competent in the MeCbl (but not AdoCbl) synthesis pathway.
Studies on the ΔN11 (lacking the mitochondrial leader sequence) and ΔN61 (starting at the second methionine initiation codon) CblD variants identified disordered regions in the N terminus of the protein that decrease its stability (36). Studies on fibroblasts derived from cblD patients that express shorter CblD variants revealed that the N-terminal 115 amino acids are not required for MeCbl synthesis (42). They are also not required for binding to CblC (43). The structural determinants needed for AdoCbl synthesis are harbored within a stretch of residues extending from positions 62 to 116 (42).
Despite the presence of a predicted B12-binding sequence, CblD does not bind B12, suggesting that its involvement in intracellular B12 delivery might be exerted indirectly (36). The ability of CblC and CblD to form a complex that is stabilized particularly in the presence of alkylcobalamin and glutathione (43) indicates that CblD might assist in the delivery of cobalamin from CblC to downstream targets.
MeCbl Synthesis
Methionine synthase, encoded by the cblG locus in humans (44–46), catalyzes the methyl transfer from N5-methyltetrahydrofolate to homocysteine in two half-reactions (Fig. 4a) (47). First, the methyl group is transferred from MeCbl to homocysteine to give methionine and cob(I)alamin. Second, the supernucleophilic cob(I)alamin removes the methyl group from N5-methyltetrahydrofolate to give tetrahydrofolate and re-forms MeCbl. Occasional oxidation of the bound cob(I)alamin to the inactive cob(II)alamin state necessitates repair via a reductive methylation reaction, in which the methyl group of S-adenosylmethionine (AdoMet) is transferred to cobalamin in the presence of the electron donor NADPH and the diflavin oxidoreductase methionine synthase reductase, encoded by the cblE locus (Fig. 2).
Mutations in cblG and cblE loci result in isolated hyperhomocysteinemia, i.e. without methylmalonic aciduria (48). Human methionine synthase is an ∼140-kDa monomeric protein that is predicted to be modular, like its better studied bacterial counterpart. The four modules in bacterial methionine synthase bind homocysteine, N5-methyltetrahydrofolate, cobalamin, and AdoMet, respectively (49). Methionine synthase reductase shuttles electrons derived from NADPH to methionine synthase-bound cobalamin (50, 51). The reduction of cob(II)alamin to cob(I)alamin by methionine synthase reductase is thermodynamically unfavorable (52, 53) and driven by kinetic coupling to the exothermic methyl group transfer reaction (54). The coupled reactions convert cob(II)alamin to MeCbl. Hence, in addition to rescuing inactive enzyme generated during the catalytic turnover cycle, it also represents a mechanism for the in situ synthesis of MeCbl from cob(II)alamin, loaded into the active site of apomethionine synthase. It is unclear how cob(II)alamin bound to CblC is transferred to the methionine synthase and what role CblD plays in this process. Methionine synthase reductase has been postulated to assist in the cofactor docking process (55). However, although the interaction between these proteins appears to stimulate cofactor docking to methionine synthase in vitro, a compulsory role for methionine synthase reductase in vivo is unlikely, as fibroblasts derived from patients with cblE defects have 84–100% holomethionine synthase (56).
Tailoring of AdoCbl in the Mitochondrion
ATP-dependent cob(I)alamin adenosyltransferase (ATR) catalyzes the synthesis of AdoCbl and is encoded by the cblB locus (57, 58). ATR is a bifunctional mitochondrial enzyme that catalyzes the formation of AdoCbl and subsequently transfers the cofactor to the lone AdoCbl-utilizing enzyme, MCM (Fig. 4b) (59). UV-visible, magnetic circular dichroism, and EPR spectroscopy studies have established that cob(II)alamin is bound to ATR in a four-coordinate base-off state (60–62). The base-off state makes reduction of cob(II)alamin to cob(I)alamin, which precedes the adenosyl transfer step, more facile. In solution, the redox potential of cob(I)alamin/cob(II)alamin is −610 mV versus approximately −500 mV for the base-on versus base-off states (63). It is not known if a dedicated reductase couples to the ATR. In vitro studies have demonstrated that flavoprotein oxidoreductases such methionine synthase reductase, ferredoxin, and flavodoxin can couple to ATRs (64, 65).
The structures of mammalian and bacterial ATRs reveal a homotrimeric organization in which the active sites are located between adjacent subunits (66). For the ATR from Methylobacterium extorquens, which is the best studied, both AdoCbl and ATP bind with negative cooperativity, and only two of the three available active sites are used at any given time (59, 67). The non-equivalence of the active sites appears to be an allosteric strategy for controlling the delivery of AdoCbl from ATR (67). Binding of ATP triggers the ejection of a single equivalent of AdoCbl, presumably from the low affinity site of ATR to MCM, resulting in direct transfer of the cofactor (Fig. 4b). This strategy of chaperoned delivery averts loss of the cofactor by dilution in the cellular milieu and its conversion to the unwanted base-on state. The pathogenic C-terminal truncation mutation compromises the ability of ATR to sequester AdoCbl and instead promotes its release into solution (68).
Although the base-off conformation of AdoCbl in ATR is mirrored in the active site of MCM, the coordination environments are distinct, an important geometric consideration for the interprotein cofactor transfer process. Thus, in ATR, the cobalamin is four-coordinate, and the funnel-shaped B12-binding site leaves the DMB tail exposed to solvent. In MCM, the cobalamin is five-coordinate by virtue of a histidine ligand donated by the protein serving as a lower axial ligand. The histidine residue appears to be crucial for the translocation of AdoCbl from the active site of ATR to the mutase, and its substitution with alanine or asparagine impairs the transfer process (59). In contrast, the histidine mutations have little impact on the KD for AdoCbl binding from solution. These results suggest a model for cofactor transfer in which the histidine residue in the mutase transiently coordinates the cobalt in ATR, facilitating the relocation of AdoCbl to the mutase. Interestingly, mutation of conserved residues in a hinged lid motif that enforces the base-off conformation in ATR compromises mutase activity in vivo (66).
A G-protein Editor of MCM
In the reaction catalyzed by MCM, the only isomerase found in mammals, AdoCbl serves as a radical reservoir, generating cob(II)alamin and the reactive 5′-deoxyadenosyl radical that initiates the radical-based 1,2-rearrangement of the substrate (Fig. 4b). The cofactor-derived radicals recombine at the end of each catalytic cycle. Occasional escape of the 5′-deoxyadenosine intermediate during catalytic turnover leads to inactive enzyme and is rescued by a G-protein chaperone, which uses the binding energy of GTP to power the expulsion of inactive cob(II)alamin from the active site (69). MCM and the G-protein chaperone are encoded by the mut and cblA loci, respectively (Fig. 2).
CblA or MMAA (methylmalonic aciduria type A) is a P-loop GTPase (69, 70). Mutations in the cblA gene are associated with methylmalonic aciduria, reduced AdoCbl levels, and low mutase activity in patient fibroblasts (71, 72). A bacterial ortholog of CblA from M. extorquens, MeaB, is the best studied member of this class of G-proteins and has been used as a model for the human CblA protein (69, 73–76).
MeaB exhibits nanomolar affinity for MCM, which is modulated by the ligands and substrates bound to each protein (75). The activities of MeaB and the mutase are each influenced by the other. MeaB has low intrinsic GTPase activity, which is enhanced by ∼100-fold in the presence of the mutase. Hence, the mutase exhibits GTPase-activating protein activity. In turn, MeaB enhances the kcat of the mutase reaction by ∼2-fold. Additionally, MeaB influences other mutase functions. It (i) allows the mutase to discriminate between inactive cob(II)alamin and active AdoCbl forms of the cofactor during ATR-dependent docking, (ii) protects the mutase against inactivation during turnover, and (iii) promotes the release of cob(II)alamin from inactive mutase.
MeaB exhibits almost equal affinity for GTP and GDP and is expected to be predominantly GTP-loaded in cells due to the higher concentration of this nucleotide form. The GTPase activity of MeaB gates transfer of AdoCbl to the mutase active site (69). Thus, MeaB functions as a molecular screen, preventing assembly of MCM with incomplete cofactor precursors. The susceptibility of the mutase to inactivation during turnover would lead to its gradual accumulation in an inactive form. A two-pronged strategy is used by MeaB to avert this situation. First, MeaB diminishes the inactivation rate of the mutase by ∼3- and 15-fold in the presence of GDP and GTP, respectively (74). CblA has a similar effect on the human mutase (69). Second, when inactive MCM is generated by the escape of 5′-deoxyadenosine from the active site, MeaB utilizes the binding energy of GTP to power the ejection of cob(II)alamin from MCM (69). CblA appears to utilize a similar mechanism as MeaB (70). The remarkable sensing of the 5′-deoxyadenosine moiety by MeaB avoids the inappropriate ejection of cob(II)alamin formed during turnover. The molecular basis of communication between MeaB and MCM awaits elucidation.
Similarly, the mechanistic basis for the intricate bidirectional signaling between MeaB and MCM remains to be elucidated. Interestingly, a pathogenic mutation of a conserved arginine to cysteine at the surface of the B12 domain in the mutase nearly abolishes its GTPase-activating protein activity and destabilizes the MCM-MeaB complex (69). Strikingly, this mutation also corrupts the ability of MeaB to block cob(II)alamin binding to MCM and to promote release of cob(II)alamin from the inactive mutase. Most G-proteins use structural motifs known as switch I and II loops that exhibit conformational sensitivity to nucleotide binding and hydrolysis for communicating with client proteins (76, 77). Missense mutations in the switch I and II loops are pathogenic and are located at the surface of MeaB and CblA, which might be important for interactions with the mutase (71, 78). Interestingly, a conserved region adjacent to the switch I and II regions in both MeaB and CblA also displays nucleotide-dependent conformational plasticity, and mutations in this region are also pathogenic (79). We speculate that this region might play a critical role in bidirectional communication between MCM and its G-protein partner.
Summary
The exciting discoveries over the past decade of genes that are culpable for cobalamin disorders have opened doors to biochemical investigations of their functions. Although homology has served to clue us into function in some cases (e.g. ATR), the lack of relatedness at a sequence level to any known protein (e.g. CblC and CblD) has challenged efforts in others. The recent discovery of a duo of membrane proteins that lead to trapping of the cofactor in the lysosome when mutated raises questions about their individual function and whether one serves as a bona fide transporter and the other as an assistant. The multifunctionality of the CblC protein, which keeps busy as a decyanase, a dealkylase, and a flavin reductase, raises many question about how this monomeric protein functions as a proverbial “jack-of-all trades” and how it transfers the tightly bound cob(II)alamin product to client proteins. The role of CblD in this transfer process is a complete mystery. Similarly, the identities of the mitochondrial membrane importers for cobalamin are unknown, as is the reductase on which the ATR function depends. The mechanism of busy bidirectional signaling between MCM and the G-protein chaperone, which orchestrates gating, guiding, and repair functions, awaits elucidation. The combination of structural and functional studies on cobalamin trafficking proteins promises to illuminate this pathway and possibly general strategies for how rare cofactors are handled within cells.
This work was supported, in whole or in part, by National Institutes of Health Grant DK45776. This is the fourth article in the Thematic Minireview Series on Metals in Biology 2013.
- MeCbl
- methylcobalamin
- AdoCbl
- 5′-deoxyadenosylcobalamin
- CNCbl
- cyanocobalamin
- MCM
- methylmalonyl-CoA mutase
- DMB
- 5,6-dimethylbenzimidazole
- AdoMet
- S-adenosylmethionine
- ATR
- adenosyltransferase.
REFERENCES
- 1. Waldron K. J., Rutherford J. C., Ford D., Robinson N. J. (2009) Metalloproteins and metal sensing. Nature 460, 823–830 [DOI] [PubMed] [Google Scholar]
- 2. Boal A. K., Rosenzweig A. C. (2009) Structural biology of copper trafficking. Chem. Rev. 109, 4760–4779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hu Y., Ribbe M. W. (2012) Nitrogenase assembly. Biochim. Biophys. Acta, DOI 10.1016/j.bbabio.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reddi A. R., Jensen L. T., Culotta V. C. (2009) Manganese homeostasis in Saccharomyces cerevisiae. Chem. Rev. 109, 4722–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lutsenko S. (2010) Human copper homeostasis: a network of interconnected pathways. Curr. Opin. Chem. Biol. 14, 211–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fukada T., Yamasaki S., Nishida K., Murakami M., Hirano T. (2011) Zinc homeostasis and signaling in health and diseases: zinc signaling. J. Biol. Inorg. Chem. 16, 1123–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lill R. (2009) Function and biogenesis of iron-sulphur proteins. Nature 460, 831–838 [DOI] [PubMed] [Google Scholar]
- 8. Banerjee R., Gherasim C., Padovani D. (2009) The tinker, tailor, soldier in intracellular B12 trafficking. Curr. Opin. Chem. Biol. 13, 484–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Banerjee R. (2006) B12 trafficking in mammals: A for coenzyme escort service. ACS Chem. Biol. 1, 149–159 [DOI] [PubMed] [Google Scholar]
- 10. Shevell M. I., Rosenblatt D. S. (1992) The neurology of cobalamin. Can. J. Neurol. Sci. 19, 472–486 [PubMed] [Google Scholar]
- 11. Hodgkin D. C., Kamper J., Mackay M., Pickworth J., Trueblood K. N., White J. G. (1956) Structure of vitamin B12. Nature 178, 64–66 [DOI] [PubMed] [Google Scholar]
- 12. Banerjee R., Ragsdale S. W. (2003) The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu. Rev. Biochem. 72, 209–247 [DOI] [PubMed] [Google Scholar]
- 13. Kim J., Gherasim C., Banerjee R. (2008) Decyanation of vitamin B12 by a trafficking chaperone. Proc. Natl. Acad. Sci. U.S.A. 105, 14551–14554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Drennan C. L., Huang S., Drummond J. T., Matthews R. G., Lidwig M. L. (1994) How a protein binds B12: a 3.0 Å x-ray structure of B12-binding domains of methionine synthase. Science 266, 1669–1674 [DOI] [PubMed] [Google Scholar]
- 15. Mancia F., Keep N. H., Nakagawa A., Leadlay P. F., McSweeney S., Rasmussen B., Bösecke P., Diat O., Evans P. R. (1996) How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2 Å resolution. Structure 4, 339–350 [DOI] [PubMed] [Google Scholar]
- 16. Brown K. L., Hakimi J. M. (1984) Heteronuclear NMR studies of cobalamins. 3. Phosphorus-31 NMR of aquocobalamin and various organocobalamins. J. Am. Chem. Soc. 106, 7894–7899 [Google Scholar]
- 17. Brown K. L., Zou X. (1999) Thermolysis of coenzymes B12 at physiological temperatures: activation parameters for cobalt-carbon bond homolysis and a quantitative analysis of the perturbation of the homolysis equilibrium by the ribonucleoside triphosphate reductase from Lactobacillus leichmannii. J. Inorg. Biochem. 77, 185–195 [DOI] [PubMed] [Google Scholar]
- 18. Waddington M. D., Finke R. G. (1993) Neopentylcobalamin (neopentyl-B12) cobalt-carbon bond thermolysis products, kinetics, activation parameters, and bond dissociation energy: a chemical model exhibiting 106 of the 1012 enzymic activation of coenzyme B12's cobalt-carbon bond. J. Am. Chem. Soc. 115, 4629–4640 [Google Scholar]
- 19. Stupperich E., Nexø E. (1991) Effect of the cobalt-N coordination on the cobamide recognition by the human vitamin B12-binding proteins intrinsic factor, transcobalamin and haptocorrin. FEBS J. 199, 299–303 [DOI] [PubMed] [Google Scholar]
- 20. Fedosov S. N., Berglund L., Fedosova N. U., Nexø E., Petersen T. E. (2002) Comparative analysis of cobalamin binding kinetics and ligand protection for intrinsic factor, transcobalamin, and haptocorrin. J. Biol. Chem. 277, 9989–9996 [DOI] [PubMed] [Google Scholar]
- 21. Aminoff M., Carter J. E., Chadwick R. B., Johnson C., Gräsbeck R., Abdelaal M. A., Broch H., Jenner L. B., Verroust P. J., Moestrup S. K., de la Chapelle A., Krahe R. (1999) Mutations in CUBN, encoding the intrinsic factor-vitamin B12 receptor, cubilin, cause hereditary megaloblastic anaemia 1. Nat. Genet. 21, 309–313 [DOI] [PubMed] [Google Scholar]
- 22. Quadros E. V. (2010) Advances in the understanding of cobalamin assimilation and metabolism. Br. J. Haematol. 148, 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Beedholm-Ebsen R., van de Wetering K., Hardlei T., Nexø E., Borst P., Moestrup S. K. (2010) Identification of multidrug resistance protein 1 (MRP1/ABCC1) as a molecular gate for cellular export of cobalamin. Blood 115, 1632–1639 [DOI] [PubMed] [Google Scholar]
- 24. Hall C. A. (1977) The carriers of native vitamin B12 in normal human serum. Clin. Sci. Mol. Med. 53, 453–457 [DOI] [PubMed] [Google Scholar]
- 25. Seetharam B., Yammani R. R. (2003) Cobalamin transport proteins and their cell-surface receptors. Expert Rev. Mol. Med. 5, 1–18 [DOI] [PubMed] [Google Scholar]
- 26. Wuerges J., Garau G., Geremia S., Fedosov S. N., Petersen T. E., Randaccio L. (2006) Structural basis for mammalian vitamin B12 transport by transcobalamin. Proc. Natl. Acad. Sci. U.S.A. 103, 4386–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Quadros E. V., Nakayama Y., Sequeira J. M. (2009) The protein and the gene encoding the receptor for the cellular uptake of transcobalamin-bound cobalamin. Blood 113, 186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li N., Seetharam S., Seetharam B. (1995) Genomic structure of human transcobalamin II: comparison to human intrinsic factor and transcobalamin I. Biochem. Biophys. Res. Commun. 208, 756–764 [DOI] [PubMed] [Google Scholar]
- 29. Kolhouse J. F., Kondo H., Allen N. C., Podell E., Allen R. H. (1978) Cobalamin analogues are present in human plasma and can mask cobalamin deficiency because current radioisotope dilution assays are not specific for true cobalamin. N. Engl. J. Med. 299, 785–792 [DOI] [PubMed] [Google Scholar]
- 30. Rutsch F., Gailus S., Miousse I. R., Suormala T., Sagné C., Toliat M. R., Nürnberg G., Wittkampf T., Buers I., Sharifi A., Stucki M., Becker C., Baumgartner M., Robenek H., Marquardt T., Höhne W., Gasnier B., Rosenblatt D. S., Fowler B., Nürnberg P. (2009) Identification of a putative lysosomal cobalamin exporter altered in the cblF defect of vitamin B12 metabolism. Nat. Genet. 41, 234–239 [DOI] [PubMed] [Google Scholar]
- 31. Coelho D., Kim J. C., Miousse I. R., Fung S., du Moulin M., Buers I., Suormala T., Burda P., Frapolli M., Stucki M., Nürnberg P., Thiele H., Robenek H., Höhne W., Longo N., Pasquali M., Mengel E., Watkins D., Shoubridge E. A., Majewski J., Rosenblatt D. S., Fowler B., Rutsch F., Baumgartner M. R. (2012) Mutations in ABCD4 cause a new inborn error of vitamin B12 metabolism. Nat. Genet. 44, 1152–1155 [DOI] [PubMed] [Google Scholar]
- 32. Rosenblatt D. S., Hosack A., Matiaszuk N. V., Cooper B. A., Laframboise R. (1985) Defect in vitamin B12 release from lysosomes: newly described inborn error of vitamin B12 metabolism. Science 228, 1319–1321 [DOI] [PubMed] [Google Scholar]
- 33. Borths E. L., Poolman B., Hvorup R. N., Locher K. P., Rees D. C. (2005) In vitro functional characterization of BtuCD-F, the Escherichia coli ABC transporter for vitamin B12 uptake. Biochemistry 44, 16301–16309 [DOI] [PubMed] [Google Scholar]
- 34. Lerner-Ellis J. P., Tirone J. C., Pawelek P. D., Doré C., Atkinson J. L., Watkins D., Morel C. F., Fujiwara T. M., Moras E., Hosack A. R., Dunbar G. V., Antonicka H., Forgetta V., Dobson C. M., Leclerc D., Gravel R. A., Shoubridge E. A., Coulton J. W., Lepage P., Rommens J. M., Morgan K., Rosenblatt D. S. (2006) Identification of the gene responsible for methylmalonic aciduria and homocystinuria, cblC type. Nat. Genet. 38, 93–100 [DOI] [PubMed] [Google Scholar]
- 35. Koutmos M., Gherasim C., Smith J. L., Banerjee R. (2011) Structural basis of multifunctionality in a vitamin B12-processing enzyme. J. Biol. Chem. 286, 29780–29787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Deme J. C., Miousse I. R., Plesa M., Kim J. C., Hancock M. A., Mah W., Rosenblatt D. S., Coulton J. W. (2012) Structural features of recombinant MMADHC isoforms and their interactions with MMACHC, proteins of mammalian vitamin B12 metabolism. Mol. Genet. Metab. 107, 352–362 [DOI] [PubMed] [Google Scholar]
- 37. Froese D. S., Krojer T., Wu X., Shrestha R., Kiyani W., von Delft F., Gravel R. A., Oppermann U., Yue W. W. (2012) Structure of MMACHC reveals an arginine-rich pocket and a domain-swapped dimer for its B12 processing function. Biochemistry 51, 5083–5090 [DOI] [PubMed] [Google Scholar]
- 38. Kim J., Hannibal L., Gherasim C., Jacobsen D. W., Banerjee R. (2009) A human vitamin B12 trafficking protein uses glutathione transferase activity for processing alkylcobalamins. J. Biol. Chem. 284, 33418–33424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hannibal L., Kim J., Brasch N. E., Wang S., Rosenblatt D. S., Banerjee R., Jacobsen D. W. (2009) Processing of alkylcobalamins in mammalian cells: a role for the MMACHC (cblC) gene product. Mol. Genet. Metab. 97, 260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Coelho D., Suormala T., Stucki M., Lerner-Ellis J. P., Rosenblatt D. S., Newbold R. F., Baumgartner M. R., Fowler B. (2008) Gene identification for the cblD defect of vitamin B12 metabolism. N. Engl. J. Med. 358, 1454–1464 [DOI] [PubMed] [Google Scholar]
- 41. Suormala T., Baumgartner M. R., Coelho D., Zavadakova P., Kozich V., Koch H. G., Berghaüser M., Wraith J. E., Burlina A., Sewell A., Herwig J., Fowler B. (2004) The cblD defect causes either isolated or combined deficiency of methylcobalamin and adenosylcobalamin synthesis. J. Biol. Chem. 279, 42742–42749 [DOI] [PubMed] [Google Scholar]
- 42. Stucki M., Coelho D., Suormala T., Burda P., Fowler B., Baumgartner M. R. (2012) Molecular mechanisms leading to three different phenotypes in the cblD defect of intracellular cobalamin metabolism. Hum. Mol. Genet. 21, 1410–1418 [DOI] [PubMed] [Google Scholar]
- 43. Gherasim C., Hannibal L., Rajagopalan D., Jacobsen D. W., Banerjee R. (2013) The C-terminal domain of CblD interacts with CblC and influences intracellular cobalamin partitioning. Biochemie, 95, 1023–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gulati S., Baker P., Li Y. N., Fowler B., Kruger W., Brody L. C., Banerjee R. (1996) Mutations in human methionine synthase in cblG patients. Hum. Mol. Genet. 5, 1859–1865 [DOI] [PubMed] [Google Scholar]
- 45. Leclerc D., Campeau E., Goyette P., Adjalla C. E., Christensen B., Ross M., Eydoux P., Rosenblatt D. S., Rozen R., Gravel R. A. (1996) Human methionine synthase: cDNA cloning and identification of mutations in patients of the cblG complementation group of folate/cobalamin disorders. Hum. Mol. Genet. 5, 1867–1874 [DOI] [PubMed] [Google Scholar]
- 46. Chen L. H., Liu M.-L., Hwang H.-Y., Chen L.-S., Korenberg J., Shane B. (1997) Human methionine synthase. cDNA cloning, gene localization, and expression. J. Biol. Chem. 272, 3628–3634 [PubMed] [Google Scholar]
- 47. Matthews R. G., Koutmos M., Datta S. (2008) Cobalamin-dependent and cobamide-dependent methyltransferases. Curr. Opin. Struct. Biol. 18, 658–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Watkins D., Rosenblatt D. S. (1989) Functional methionine synthase deficiency (cblE and cblG): clinical and biochemical heterogeneity. Am. J. Med. Genet. 34, 427–434 [DOI] [PubMed] [Google Scholar]
- 49. Ludwig M. L., Matthews R. G. (1997) Structure-based perspectives on B12-dependent enzymes. Annu. Rev. Biochem. 66, 269–313 [DOI] [PubMed] [Google Scholar]
- 50. Olteanu H., Banerjee R. (2001) Human methionine synthase reductase, a soluble P-450 reductase-like dual flavoprotein, is sufficient for NADPH-dependent methionine synthase activation. J. Biol. Chem. 276, 35558–35563 [DOI] [PubMed] [Google Scholar]
- 51. Olteanu H., Munson T., Banerjee R. (2002) Differences in the efficiency of reductive activation of methionine synthase and exogenous electron acceptors between the common polymorphic variants of human methionine synthase reductase. Biochemistry 41, 13378–13385 [DOI] [PubMed] [Google Scholar]
- 52. Olteanu H., Wolthers K. R., Munro A. W., Scrutton N. S., Banerjee R. (2004) Kinetic and thermodynamic characterization of the common polymorphic variants of human methionine synthase reductase. Biochemistry 43, 1988–1997 [DOI] [PubMed] [Google Scholar]
- 53. Wolthers K. R., Basran J., Munro A. W., Scrutton N. S. (2003) Molecular dissection of human methionine synthase reductase: determination of the flavin redox potentials in full-length enzyme and isolated flavin-binding domains. Biochemistry 42, 3911–3920 [DOI] [PubMed] [Google Scholar]
- 54. Banerjee R. V., Harder S. R., Ragsdale S. W., Matthews R. G. (1990) Mechanism of reductive activation of cobalamin-dependent methionine synthase: an electron paramagnetic resonance spectroelectrochemical study. Biochemistry 29, 1129–1135 [DOI] [PubMed] [Google Scholar]
- 55. Yamada K., Gravel R. A., Toraya T., Matthews R. G. (2006) Human methionine synthase reductase is a molecular chaperone for human methionine synthase. Proc. Natl. Acad. Sci. U.S.A. 103, 9476–9481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gulati S., Chen Z., Brody L. C., Rosenblatt D. S., Banerjee R. (1997) Defects in auxiliary redox proteins lead to functional methionine synthase deficiency. J. Biol. Chem. 272, 19171–19175 [DOI] [PubMed] [Google Scholar]
- 57. Dobson C. M., Wai T., Leclerc D., Kadir H., Narang M., Lerner-Ellis J. P., Hudson T. J., Rosenblatt D. S., Gravel R. A. (2002) Identification of the gene responsible for the cblB complementation group of vitamin B12-dependent methylmalonic aciduria. Hum. Mol. Genet. 11, 3361–3369 [DOI] [PubMed] [Google Scholar]
- 58. Leal N. A., Park S. D., Kima P. E., Bobik T. A. (2003) Identification of the human and bovine ATP:cob(I)alamin adenosyltransferase cDNAs based on complementation of a bacterial mutant. J. Biol. Chem. 278, 9227–9234 [DOI] [PubMed] [Google Scholar]
- 59. Padovani D., Labunska T., Palfey B. A., Ballou D. P., Banerjee R. (2008) Adenosyltransferase tailors and delivers coenzyme B12. Nat. Chem. Biol. 4, 194–196 [DOI] [PubMed] [Google Scholar]
- 60. Stich T. A., Yamanishi M., Banerjee R., Brunold T. C. (2005) Spectroscopic evidence for the formation of a four-coordinate Co2+ cobalamin species upon binding to the human ATP:cobalamin adenosyltransferase. J. Am. Chem. Soc. 127, 7660–7661 [DOI] [PubMed] [Google Scholar]
- 61. Yamanishi M., Labunska T., Banerjee R. (2005) Mirror “base-off” conformation of coenzyme B12 in human adenosyltransferase and its downstream target, methylmalonyl-CoA mutase. J. Am. Chem. Soc. 127, 526–527 [DOI] [PubMed] [Google Scholar]
- 62. Yamanishi M., Vlasie M., Banerjee R. (2005) Adenosyltransferase: an enzyme and an escort for coenzyme B12? Trends Biochem. Sci. 30, 304–308 [DOI] [PubMed] [Google Scholar]
- 63. Lexa D., Saveant J.-M. (1983) The electrochemistry of vitamin B12. Acc. Chem. Res. 16, 235–243 [Google Scholar]
- 64. Mera P. E., Escalante-Semerena J. C. (2010) Dihydroflavin-driven adenosylation of 4-coordinate Co(II) corrinoids: are cobalamin reductases enzymes or electron transfer proteins? J. Biol. Chem. 285, 2911–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Leal N. A., Olteanu H., Banerjee R., Bobik T. A. (2004) Human ATP:cob(I)alamin adenosyltransferase and its interaction with methionine synthase reductase. J. Biol. Chem. 279, 47536–47542 [DOI] [PubMed] [Google Scholar]
- 66. Mera P. E., Escalante-Semerena J. C. (2010) Multiple roles of ATP:cob(I)alamin adenosyltransferases in the conversion of B12 to coenzyme B12. Appl. Microbiol. Biotechnol. 88, 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Padovani D., Banerjee R. (2009) A rotary mechanism for coenzyme B12 synthesis by adenosyltransferase. Biochemistry 48, 5350–5357 [DOI] [PubMed] [Google Scholar]
- 68. Lofgren M., Banerjee R. (2011) Loss of allostery and coenzyme B12 delivery by a pathogenic mutation in adenosyltransferase. Biochemistry 50, 5790–5798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Padovani D., Banerjee R. (2009) A G-protein editor gates coenzyme B12 loading and is corrupted in methylmalonic aciduria. Proc. Natl. Acad. Sci. U.S.A. 106, 21567–21572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Takahashi-Íñiguez T., García-Arellano H., Trujillo-Roldán M. A., Flores M. E. (2011) Protection and reactivation of human methylmalonyl-CoA mutase by MMAA protein. Biochem. Biophys. Res. Commun. 404, 443–447 [DOI] [PubMed] [Google Scholar]
- 71. Lerner-Ellis J. P., Dobson C. M., Wai T., Watkins D., Tirone J. C., Leclerc D., Doré C., Lepage P., Gravel R. A., Rosenblatt D. S. (2004) Mutations in the MMAA gene in patients with the cblA disorder of vitamin B12 metabolism. Hum. Mutat. 24, 509–516 [DOI] [PubMed] [Google Scholar]
- 72. Dobson C. M., Wai T., Leclerc D., Wilson A., Wu X., Doré C., Hudson T., Rosenblatt D. S., Gravel R. A. (2002) Identification of the gene responsible for the cblA complementation group of vitamin B12-responsive methylmalonic acidemia based on analysis of prokaryotic gene arrangements. Proc. Natl. Acad. Sci. U.S.A. 99, 15554–15559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Korotkova N., Lidstrom M. E. (2004) MeaB is a component of the methylmalonyl-CoA mutase complex required for protection of the enzyme from inactivation. J. Biol. Chem. 279, 13652–13658 [DOI] [PubMed] [Google Scholar]
- 74. Padovani D., Banerjee R. (2006) Assembly and protection of the radical enzyme, methylmalonyl-CoA mutase, by its chaperone. Biochemistry 45, 9300–9306 [DOI] [PubMed] [Google Scholar]
- 75. Padovani D., Labunska T., Banerjee R. (2006) Energetics of interaction between the G-protein chaperone, MeaB, and B12-dependent methylmalonyl-CoA mutase. J. Biol. Chem. 281, 17838–17844 [DOI] [PubMed] [Google Scholar]
- 76. Hubbard P. A., Padovani D., Labunska T., Mahlstedt S. A., Banerjee R., Drennan C. L. (2007) Crystal structure and mutagenesis of the metallochaperone MeaB: insight into the causes of methylmalonic aciduria. J. Biol. Chem. 282, 31308–31316 [DOI] [PubMed] [Google Scholar]
- 77. Froese D. S., Kochan G., Muniz J. R., Wu X., Gileadi C., Ugochukwu E., Krysztofinska E., Gravel R. A., Oppermann U., Yue W. W. (2010) Structures of the human GTPase MMAA and vitamin B12-dependent methylmalonyl-CoA mutase and insight into their complex formation. J. Biol. Chem. 285, 38204–38213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pérez B., Angaroni C., Sánchez-Alcudia R., Merinero B., Pérez-Cerdá C., Specola N., Rodríguez-Pombo P., Wajner M., de Kremer R. D., Cornejo V., Desviat L. R., Ugarte M. (2010) The molecular landscape of propionic acidemia and methylmalonic aciduria in Latin America. J. Inherit. Metab. Dis. 33, S307–S314 [DOI] [PubMed] [Google Scholar]
- 79. Dempsey-Nunez L., Illson M. L., Kent J., Huang Q., Brebner A., Watkins D., Gilfix B. M., Wittwer C. T., Rosenblatt D. S. (2012) High resolution melting analysis of the MMAA gene in patients with cblA and in those with undiagnosed methylmalonic aciduria. Mol. Genet. Metab. 107, 363–367 [DOI] [PubMed] [Google Scholar]