Background: AIM2 binds dsDNA and associates with ASC through their PYDs to form an inflammasome.
Results: The AIM2 PYD structure illustrates distinct charge distribution and a unique hydrophobic patch.
Conclusion: The AIM2 PYD may bind the ASC PYD and the AIM2 HIN domain through overlapping surface.
Significance: These findings provide insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly.
Keywords: Crystal Structure, Death Domain, Immunology, Infectious Diseases, Innate Immunity, AIM2, PYD
Abstract
Absent in melanoma 2 (AIM2) is a cytosolic double-stranded (dsDNA) sensor essential for innate immune responses against DNA viruses and bacteria such as Francisella and Listeria. Upon dsDNA engagement, the AIM2 amino-terminal pyrin domain (PYD) is responsible for downstream signaling to the adapter protein apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC) through homotypic PYD-PYD interactions and the assembly of an inflammasome. Toward a better understanding of the AIM2 signaling mechanism, we determined the crystal structure of the human AIM2 PYD. The structure reveals a death domain fold with a short α3 helix that is buttressed by a highly conserved lysine residue at the α2 helix, which may stabilize the α3 helix for potential interaction with partner domains. The surface of the AIM2 PYD exhibits distinct charge distribution with highly acidic α1-α2 helices and highly basic α5-α6 helices. A prominent solvent-exposed hydrophobic patch formed by residues Phe-27 and Phe-28 at the α2 helix resembles a similar surface involved in the death effector domain homotypic interactions. Docking studies suggest that the AIM2 PYD may bind the AIM2 hematopoietic interferon-inducible nuclear (HIN) domain or ASC PYD using overlapping surface near the α2 helix. This may ensure that AIM2 interacts with the downstream adapter ASC only upon release of the autoinhibition by the dsDNA ligand. Our work thus unveils novel structural features of the AIM2 PYD and provides insights into the potential mechanisms of the PYD-HIN and PYD-PYD interactions important for AIM2 autoinhibition and inflammasome assembly.
Introduction
AIM22 is a major cytosolic dsDNA sensor that forms an inflammasome with the adapter ASC and procaspase-1 to activate proinflammatory cytokine processing (1–4). The AIM2 inflammasome is essential for immune responses against DNA viruses such as vaccinia virus and mouse cytomegalovirus and bacteria such as Francisella tularensis and Listeria monocytogenes (5–9). AIM2 was also reported to play a role in autoimmune disorders such as psoriasis through recognition of host DNA (10). It belongs to the pyrin and HIN protein (PYHIN) family of proteins that includes interferon γ-inducible protein 16 (11), IFN-inducible protein X/PYHIN1, and myeloid cell nuclear differentiation antigen in the human genome (12, 13). AIM2 contains a carboxyl-terminal HIN domain that binds dsDNA and an amino-terminal PYD that is responsible for downstream signaling to the adapter protein ASC. Previous phylogenetic analysis of the PYD sequences revealed two separate clades among the mammalian PYHIN proteins, one containing the AIM2 orthologs and the other containing all other PYHIN proteins (14). In agreement with its distinct sequence, the AIM2 PYD was shown to be the only PYHIN PYD that associates with ASC to form an inflammasome (1, 2). However, it remains unclear how the distinct sequence of the AIM2 PYD contributes to this unique function.
The pyrin domains were found in both PYHIN proteins and Nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) as signal transduction modules that adopt the six-helix bundle fold typical of the death domain superfamily (15–19). Besides PYD, the death domain superfamily includes the death domain (DD), death effector domain (DED), and caspase recruitment domain (CARD). Many of them are involved in the assembly of oligomeric multiprotein signaling complexes such as the PIDDosome (DD) (20), MyDDosome (DD) (21), apoptosomes (CARD) (22, 23), and inflammasomes (PYD) (24). Despite progress in the structural characterization of the DD, DED, and CARD signaling complexes, the molecular mechanisms of PYD-mediated signaling events remain poorly understood largely because of the lack of structural information on PYD-PYD complexes. To date, the structures of eight human PYDs and two mouse PYDs have been experimentally characterized, largely through nuclear magnetic resonance (NMR) spectroscopy. These include the PYDs from human ASC (Protein Data Bank codes 1UCP (25) and 2KN6 (26)), NLRP1 (Protein Data Bank code 1PN5) (27), NLRP3 (Protein Data Bank code 3QF2) (28), NLRP4 (Protein Data Bank code 4EWI) (29), NLRP7 (Protein Data Bank code 2KM6) (30), NLRP12 (Protein Data Bank code 2L6A) (31), and POP1/ASC2 (Protein Data Bank code 2HM2) (32) and mouse NLRP10 (Protein Data Bank code 2DO9) as well as the PYHIN family members myeloid cell nuclear differentiation antigen (Protein Data Bank code 2DBG; human) and p205b (Protein Data Bank code 2YU0; mouse). Interestingly, all known PYD structures have a uniquely short α3 helix compared with other DD superfamily members (33).
Previously, we reported the crystal structure of the AIM2 HIN domain in complex with dsDNA and found that the HIN domain binds dsDNA through electrostatic attraction, and the AIM2 PYD and HIN domain interaction maintains the receptor in an autoinhibited state in the absence of dsDNA (34). To examine the structure and function of the AIM2 PYD, we carried out crystallographic studies of the AIM2 PYD. The AIM2 PYD was crystallized as a fusion with maltose-binding protein (MBP). The structure reveals a typical death domain fold for the PYD with distinct surface charges and hydrophobic patches. We identify a highly conserved lysine residue at the α2 helix that stabilizes the short α3 helix, a common feature for all known PYD structures that has not been described previously. Our docking and binding studies suggest potential modes of the PYD-PYD and PYD-HIN associations through overlapping surface at the AIM2 PYD such that the AIM2 receptor signal transduction only occurs upon ligand engagement.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The pyrin domain of human AIM2 (NCBI accession number NP_004824; residues 1–107) was cloned into a pET30a-derived vector with a non-cleavable amino-terminal MBP tag and a carboxyl-terminal hexahistidine tag. The MBP tag harbors mutations (D82A/K83A/E172A/N173A/K239A) designed to enhance its crystallization propensity (35, 36). Transformed BL21(DE3) Codon Plus RIPL cells (Stratagene, Santa Clara, CA) were grown at 37 °C, induced with 0.3 mm isopropyl 1-thio-β-d-galactopyranoside at 18 °C for 4 h, harvested, and lysed in buffer A (20 mm Tris-HCl, pH 8.0, 100 mm NaCl) plus 5 mm imidazole, DNase (Biomatik, Wilmington, DE), and protease inhibitors (Roche Applied Science). Soluble protein from the cell lysate was purified by Hisprep IMAC column (GE Healthcare) and further purified using a XK26/60 Superdex 200 size exclusion column in buffer A supplemented with 5 mm maltose (Research Products International Corp., Mount Prospect, IL). The mutant AIM2 PYD constructs were produced using the Phusion site-directed mutagenesis kit (Thermo Scientific, Waltham, MA), and the expression and purification were carried essentially the same as for the wild type protein. The human ASC PYD (residues 1–91) coding sequence was cloned into a pET30a-derived vector with a tobacco etch virus protease cleavable amino-terminal GB1 tag. Expression and purification were carried out similarly to the AIM2 PYD except that the GB1 tag was removed by tobacco etch virus cleavage.
Crystallization
Purified MBP-PYD protein was concentrated to 20 mg/ml before setting up hanging drops for vapor diffusion crystallization. Multiple commercial crystallization screens were tested using the Mosquito crystallization robot (TTP Labtech, UK). Rod-shaped crystals grew at room temperature in 1 week using a well solution containing 20% polyethylene glycol (PEG) 3350 and 0.1 m potassium acetate at pH 4.0. 20% ethylene glycol (v/v) was added to the reservoir solution as the cryoprotectant to freeze the crystals in liquid nitrogen for x-ray diffraction data collection.
X-ray Diffraction, Structure Determination, and Refinement
X-ray diffraction data were collected at the General Medical Sciences and Cancer Institutes Collaborative Access Team (GM/CA-CAT) of the Advanced Photon Source, Argonne National Laboratory. Data were processed with the HKL2000 program suite (37). The solvent content was 53.2% for one molecule of the fusion protein per asymmetric unit. The structure was determined by molecular replacement with Phaser (38) from the CCP4 program suite (39). An MBP structure from the Protein Data Bank (code 3DM0) was used as the search model (35). Electron density maps calculated with phases from the MBP search model clearly showed positive densities for the AIM2 PYD. The PYD model was manually built with Coot (40) and refined with Phenix.refine (41). The final structure contains 467 residues of which residues Met-371 to Lys-467 correspond to the first 97 residues of the AIM2 receptor (NCBI accession number NP_004824). Validation by the MolProbity server (42) and Research Collaboratory for Structural Bioinformatics ADIT validation server (43) showed that 97.5% of all protein residues were in the favored regions of the Ramachandran plot with no outliers. Electrostatic surfaces were calculated with program Delphi (v4) (44) and displayed with PyMOL (Schrödinger, LLC). Calculation of the solvent-accessible area was performed with the program Areaimol from the CCP4 program suite (39, 45).
Sequence Alignment and Calculation of Conservation Scores
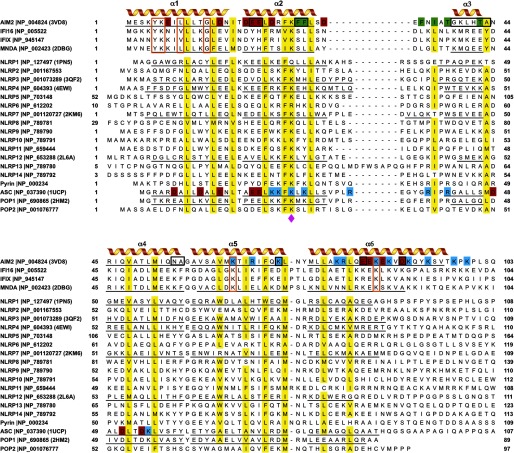
Sequence alignment of the PYDs was performed with the program MEGA5 (46) with minor adjustments. The phylogenic tree was calculated using the maximum likelihood method in MEGA5, and the reliability of the tree was tested with bootstrapping for 1000 replications. The sequence conservation scores were calculated by the ConSurf server (47) using the sequence alignment in Fig. 1.
FIGURE 1.
Sequence alignment of the human PYDs. The six α helices of the PYDs with known structures are underlined and marked. The highly conserved residues (>80%) are shaded in yellow. The conserved lysine residue at the α2 helix is indicated with a magenta diamond. The acidic residues at the α1, α2, and α6 helices of the AIM2 PYD are shaded in red, the basic residues at the α5 and α6 helices are shaded in blue, the hydrophobic residues at the α2-α3 helices are shaded in green. Similarly, the acidic residues at the α1 and α4 helices and the basic residues at the α2 and α3 helices of the ASC PYD are shaded red and blue, respectively. Conserved residues among the PYHIN-only proteins are marked with orange boxes. The AIM2-specific residues within the PYHIN proteins are marked with black boxes.
Protein Structure Docking
Docking of the AIM2 PYD (Protein Data Bank code 3VD8) with the ASC PYD (Protein Data Bank code 1UCP) or the AIM2 HIN domain (Protein Data Bank code 3RN2) was performed with the web server ClusPro (48), taking into account both shape complementarity and electrostatic charge interactions. An interactive docking program, Hex (v6.3) (49), was also used for comparison. No preidentification of residues within or outside the contact surface was specified. The choice of either domain as the stationary receptor resulted in a similar mode of interactions for the AIM2 PYD (see Figs. 5 and 6): the top docking models consistently placed the α2 helix of the AIM2 PYD at the PYD-PYD and PYD-HIN interfaces. The top ranked docking model for the PYD-HIN complex was subjected to energy minimization using the relax mode of the Rosetta (v3.4) program (50).
FIGURE 5.
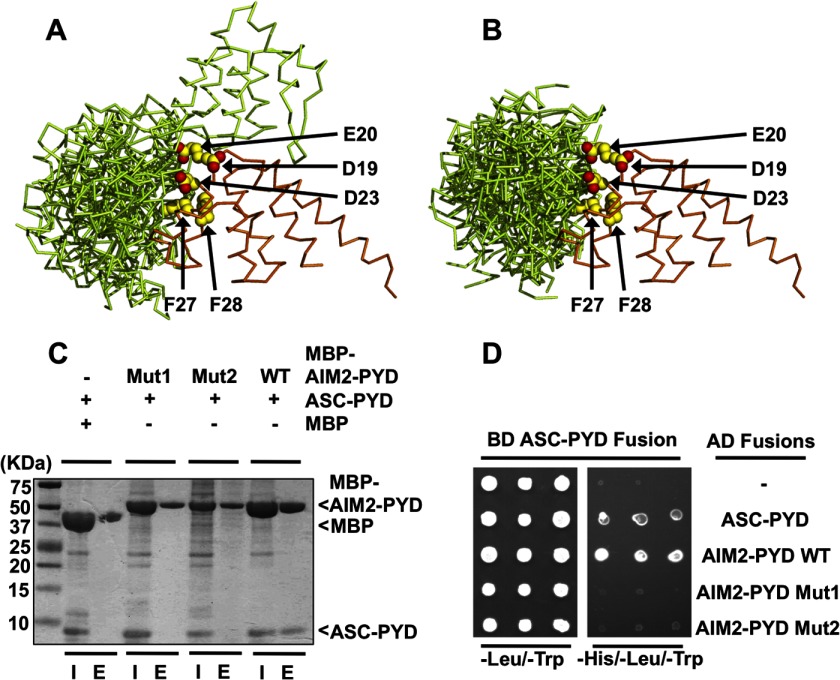
Analysis of the AIM2 PYD and ASC PYD association. A, superposition of the top 10 docking models of the AIM2 PYD-ASC PYD complex by the ClusPro server with the AIM2 PYD as the stationary receptor. The AIM2 PYD and ASC PYD are colored orange and green, respectively. The AIM2 residues Asp-19, Glu-20, Asp-23, Phe-27, and Phe-28 are shown as yellow spheres. B, superposition of the top 10 docking models of the AIM2 PYD-ASC PYD complex with the ASC PYD as the stationary receptor. The 10 models were superimposed on their AIM2 PYDs. C, MBP pulldown assay for the WT and mutant AIM2 PYD association with the ASC PYD. “I” denotes input sample, and “E” denotes elution sample. D, yeast two-hybrid analysis of the AIM2 PYD and ASC PYD interaction. Yeast cells co-expressing GAL4 DNA-binding domain (BD)-ASC PYD fusion and GAL4 activation domain (AD) fusions were grown on agar plates lacking leucine and tryptophan (−Leu/−Trp) for transformant growth and lacking histidine, leucine, and tryptophan (−His/−Leu/−Trp) for detecting PYD-PYD interaction. Three individual clones for each combination were plated. − denotes empty vector control.
FIGURE 6.
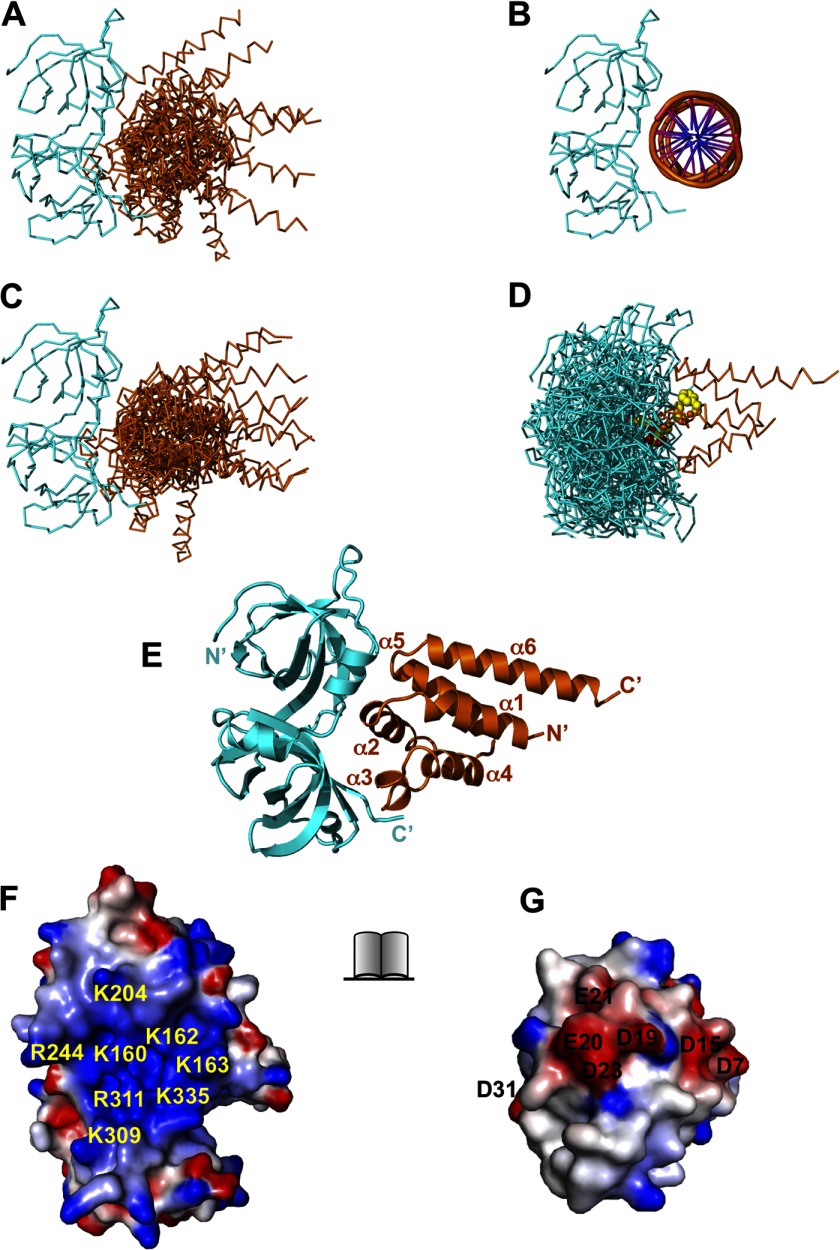
Docking of the AIM2 PYD and HIN domains. A, superposition of the top 10 docking models of the AIM2 PYD-HIN domain complex with the HIN domain as the stationary receptor by the ClusPro server. The AIM2 PYD and HIN domain are colored orange and cyan, respectively. B, the structure of the AIM2 HIN-DNA complex (Protein Data Bank code 3RN2) is shown for comparison. C, superposition of the top 10 docking models of the AIM2 PYD-HIN complex with the PYD as the stationary receptor by the ClusPro server. The 10 models were superimposed on their HIN domains. D, the original 10 docking models for C are superimposed on their PYDs with the AIM2 residues Asp-19, Glu-20, Asp-23, Phe-27, and Phe-28 shown as yellow spheres. E, the top scoring model from A was energy-minimized with Rosetta and is shown in ribbons. The PYD and HIN domain are colored orange and cyan, respectively. The PYD helices and termini are labeled. The open book view of the PYD-HIN interface is shown as electrostatic surface in F (HIN) and G (PYD). The basic residues in HIN are labeled in yellow, and acidic residues in PYD are labeled in black.
MBP Pulldown
Purified MBP or MBP-tagged wild type or mutant AIM2 PYD samples (200 μg) were immobilized with 50 μl of amylose beads (New England Biolabs, Ipswich, MA) in buffer B (20 mm HEPES-Na, pH 7.4, 100 mm NaCl). After three washing steps using buffer B, 200 μg of AIM2 HIN protein was incubated with the beads followed by three washing steps with buffer B. The bound proteins were eluted with 50 μl of buffer B plus 10 mm maltose and analyzed by SDS-PAGE.
Yeast Two-hybrid Assay
The GAL4-based Matchmaker yeast two-hybrid system from Clontech was used to examine the interaction between the AIM2 PYD and the ASC PYD. The PYD coding sequences were cloned into the pGBKT7 plasmid encoding DNA-binding domain and the pGADT7 plasmid encoding activation domain. Combinations of DNA-binding domain and activation domain plasmid pairs were transformed into yeast strain AH109. The cells were plated on agar plates of minimum synthetic dropout medium without leucine and tryptophan (−Leu/−Trp) to select for transformants. Single colonies were then picked and replicated onto −His/−Leu/−Trp plates. Growth of the colonies at 30 °C was recorded 72 h later.
Isothermal Titration Calorimetry (ITC)
The association between the AIM2 PYD and HIN domain was measured using an ITC200 calorimeter (Microcal, Piscataway, NJ) at 288 K. All protein samples were dialyzed extensively in buffer B. The wild type or mutant MBP-AIM2 PYD at 0.3 mm was titrated into ∼200 μl of purified AIM2 HIN domain at 30 μm. The binding isotherm data were analyzed with the program Origin using a single site binding model after subtracting the buffer dilution background.
Fluorescence Polarization Assay
A 5′,6-fluorescein-labeled 20-mer DNA oligo with a sequence of 5′-ccatcaaagagagaaagagc-3′ (Integrated DNA Technologies, Coralville, IA) was dissolved in buffer B and annealed with its reverse complement DNA oligo. A 4 nm concentration of the probe was mixed with 100 nm purified AIM2 HIN domain, and increasing concentrations of the wild type or mutant MBP-AIM2 PYD were added to the above DNA-HIN complex. The mixtures were then aliquoted in triplets into black 96-well plates, and the fluorescence polarization was measured with a Paradigm spectrometer (Molecular Devices, Sunnyvale, CA). Data were analyzed using the program Prism (GraphPad, San Diego, CA).
RESULTS
Sequence Comparison of the Human PYDs
There are a total of 22 PYD-containing proteins in the human genome (13), including the PYHIN proteins, the NLR family members NLRP1–14, the pyrin protein encoded by the Mediterranean fever gene, the adapter protein ASC, and PYD-only proteins (POPs) POP1/ASC2 and POP2. A sequence alignment of these PYDs guided by the known structures reveals that several conserved residues are shared between the PYHIN PYDs and other PYDs (Fig. 1). These residues are mostly hydrophobic with the exception of Lys-26 at the α2 helix (see below). Divergent sequences outside of these conserved residues place the PYHIN proteins at a separate clade in a phylogenetic tree (supplemental Fig. 1). For example, the PYHIN PYDs contain shorter α2–3 loops compared with the other PYDs, and their amino- and carboxyl-terminal regions harbor conserved residues among the PYHIN proteins only (Fig. 1, orange boxes). Furthermore, the AIM2 PYD sequence further diverges from those of the other PYHIN proteins with the AIM2-specific residues located throughout the PYD sequence (Fig. 1, black boxes). Notably, the AIM2-specific residues at the α2 helix contribute to the distinct pattern of surface charge and hydrophobicity patch at the PYD as described below. To understand the contribution of these AIM2-specific residues to its distinct structure and in turn its unique ability to associate with the adapter ASC, we initiated structural studies of the AIM2 PYD.
The AIM2 PYD Adopts a Six-helix Bundle Structure
Our initial efforts to crystallize the AIM2 PYD were hampered by its tendency to form severe aggregates upon overexpression, perhaps partially reflecting its function as a protein-protein interaction module. We overcame this challenge using an MBP fusion strategy reported previously to be successful for other recalcitrant crystallization targets (35, 36, 51). The MBP-PYD fusion protein was expressed, purified to high homogeneity, and crystallized. Crystallographic data collection, model building, and refinement statistics are presented in Table 1. One MBP-PYD fusion protein is present in the crystallographic asymmetric unit (supplemental Fig. 2A). There are four hydrogen bonds and 10 van der Waals contacts between the α1 and α6 helices of the AIM2 PYD and the MBP-linker region, which may stabilize the relative positions of the MBP and PYD, thus facilitating crystallization (supplemental Fig. 2B).
TABLE 1.
X-ray diffraction data collection and structural refinement statistics
r.m.s.d., root mean square deviation.
| Unit cell | |
| a, b, c (Å) | 61.6, 91.9, 100.3 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 50-2.07 (2.10-2.07)a |
| No. of reflections (total/unique) | 245,865/35,164 |
| Redundancy | 7.0 (6.0)a |
| Completeness (%) | 99.0 (97.9)a |
| I/σ(I) | 12.4 (2.2)a |
| Rmerge (%)b | 11.9 (64.6)a |
| Rpim (%)c | 4.9 (28.9)a |
| Refinement | |
| Resolution (Å) | 50-2.07 |
| No. of protein atoms | 3,688 |
| No. of solvent/heteroatoms | 390 |
| r.m.s.d. bond lengths (Å) | 0.003 |
| r.m.s.d. bond angles (°) | 0.694 |
| Rwork (%)d | 18.21 |
| Rfree (%)e | 22.12 |
| Ramachandran plot favored/disallowedf | 97.5/0.0 |
| Protein Data Bank code | 3VD8 |
a Numbers in parentheses correspond to the last resolution shell.
b Rmerge = ΣhΣi|Ii(h) − I(h)|/ΣhΣiIi(h) where Ii(h) and I(h) are the ith and mean measurement of the intensity of reflection h.
c Rpim = Σh[(1/n − 1)½]Σi|Ii(h) −I(h)|/ΣhΣiIi(h) where Ii(h) and I(h) are the ith and mean measurement of the intensity of reflection h, and n is the redundancy of reflection h.
d Rwork = Σh‖Fobs (h)| − |Fcalc(h)‖/Σh|Fobs(h)| where Fobs(h) and Fcalc(h) are the observed and calculated structure factors, respectively. No I/σ cutoff was applied.
e Rfree is the R value obtained for a test set of reflections consisting of a randomly selected 5% subset of the data set excluded from refinement.
f Values from MolProbity server.
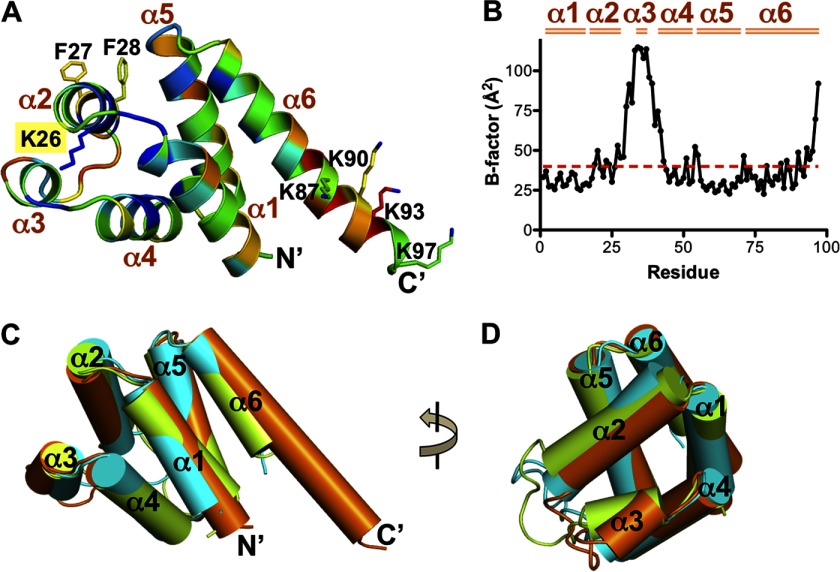
The AIM2 PYD is well resolved in the electron density maps (supplemental Fig. 2C). It has a globular structure of a six-helix bundle similar to other members of the death domain superfamily (Figs. 1 and 2A). Typical of the known PYD structures, the AIM2 PYD has a short α3 helix (33). This helix and the connecting loops possess higher temperature factors compared with the bulk of the domain, suggesting higher static or dynamic mobility (Fig. 2B). A search of structural homologues using the Dali server (52) revealed that the AIM2 PYD is most similar to that from ASC and NLRP3 with Z-scores of 12.4 and 12.3, respectively (Table 2). Superposition of these three PYD structures illustrates that the six helices from each can be superimposed well with variations in the orientation and length of the helices (Fig. 2, C and D). In particular, the α6 helix of the AIM2 PYD is the longest among the known PYD structures with the most variable sequences (Figs. 1 and 2, A and C). The functional significance of this long α6 helix is currently unclear.
FIGURE 2.
Overall structure of the AIM PYD. A, ribbon diagram of the AIM2 PYD structure colored based on sequence conservation scores calculated by the ConSurf server (47). The color is from blue (conserved) to red (non-conserved). Residues Lys-26, Phe-27, and Phe-28 at the α2 helix and Lys-87, Lys-90, Lys-93, and Lys-97 at the α6 helix are shown as ball-and-stick models. The six α helices and both termini are labeled. B, a plot of the temperature factors for the AIM2 PYD residues. The average temperature factor of the domain is indicated as a red dotted line, and the six α helices are marked above the plot. C, superposition of the PYD structures from AIM2 (orange), ASC (green), and NLRP3 (cyan) with each helix represented as a cylinder. D, a 90° rotation of the view in C.
TABLE 2.
Comparison of the AIM2 PYD structure with other PYD structures by the Dali server
Protein Data Bank codes are in parentheses. r.m.s.d., root mean square deviation; MNDA, myeloid cell nuclear differentiation antigen.
| Z-score | r.m.s.d. | Aligned residues | Identity | |
|---|---|---|---|---|
| Å | % | |||
| hASCa (1UCP) | 12.4 | 1.6 | 83 | 19 |
| hNLRP3a (3QF2) | 12.3 | 1.8 | 82 | 27 |
| hPOP1a (2HM2) | 12.2 | 1.5 | 80 | 23 |
| hMNDAa (2BDG) | 12.1 | 3.8 | 92 | 28 |
| hNLRP4a (4EWI) | 11.1 | 1.6 | 78 | 27 |
| hNLRP7a (2KM6) | 10.8 | 2.4 | 85 | 20 |
| mNLRP10b (2DO9) | 10.7 | 3.6 | 89 | 24 |
| hNLRP12a (2L6A) | 10.3 | 1.6 | 78 | 23 |
| mP205b (2YU0) | 9.5 | 4.1 | 84 | 24 |
| hNLRP1a (1PN5) | 8.1 | 2.4 | 76 | 20 |
a Human protein.
b Mouse protein.
The Highly Conserved Lys-26 Residue Stabilizes the α3 Helix
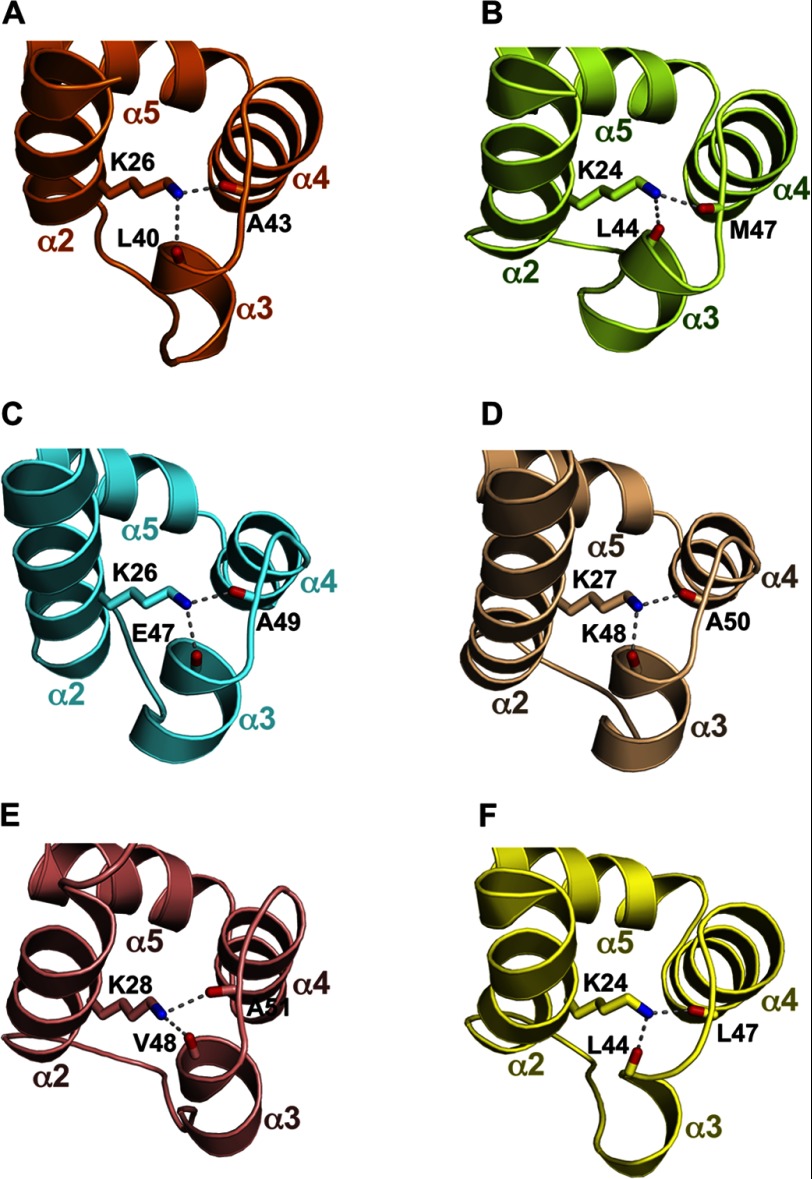
Among the death domain superfamily members, PYDs have uniquely short α3 helices connected to flexible loops (33). We therefore wondered how the conformation of the α3 helices is stabilized. We noticed that the α2 helices of PYDs contain highly conserved lysine residues (Figs. 1 and 2A). The side chain of this residue in the AIM2 PYD points toward the α3 helix (Fig. 2A), resulting in exposure of only 8% of its surface to the solvent. The poor surface exposure suggests that Lys-26 may not be involved in domain-domain interactions. What then is the function of this conserved lysine residue? Analysis of the PYD structures from AIM2 (Protein Data Bank code 3VD8), ASC (Protein Data Bank code 1UCP), NLRP3 (Protein Data Bank code 3QF2), NLRP4 (Protein Data Bank code 4EWI), NLRP7 (Protein Data Bank code 2KM6), and POP1 (Protein Data Bank code 2HM2) reveals that this conserved α2 helix lysine residue buttresses the α3 helix through hydrogen bonds with two main chain carbonyl oxygens (i.e. those of Leu-40 and Ala-43 from AIM2), thereby stabilizing the conformation of α3 and its connecting loops (Fig. 3, A–F). In addition, the only non-conserved residues at this position (Gln in NLRP1 and Arg in NLRP6) are all capable of forming similar hydrogen bonds. Because the α2-α3 helices of the DD, DED, and CARD have all been implicated in homotypic interactions (20, 21, 53, 54), it is possible that the conserved Lys-26 residue may indirectly facilitate PYD-PYD interactions through stabilizing the α3 helix.
FIGURE 3.
The conserved lysine residue at the α2 helix buttresses the α3 helix. The region near the conserved lysine residue and the α3 helix are shown for the AIM2 PYD in A (orange), ASC PYD in B (green), NLRP3 PYD in C (cyan), NLRP4 PYD in D (wheat), NLRP7 PYD in E (salmon), and POP1 PYD in F (yellow). Hydrogen bonds are indicated with gray dotted lines.
Surface of the AIM2 PYD Exhibits Distinct Pattern of Electrostatic Charges and Hydrophobicity
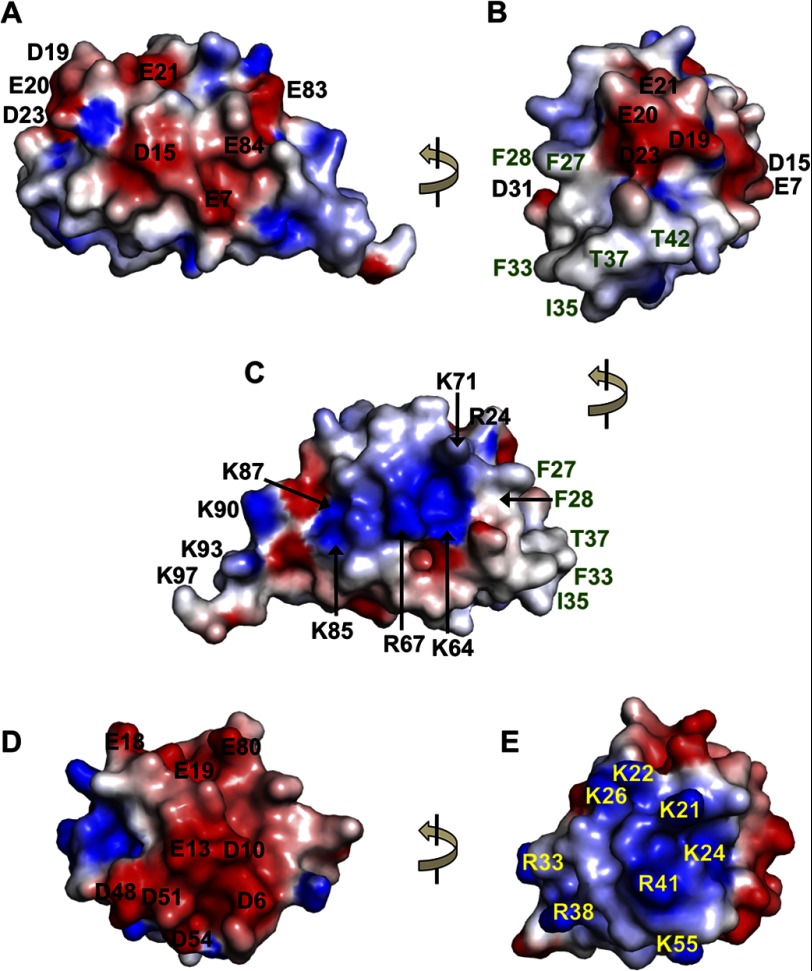
Besides the Lys-26 residue that is largely buried, most of the charged AIM2 PYD residues are exposed and contribute to a bipolar distribution of electrostatic charges (Fig. 4, A–C) similar to other known PYD structures. However, the pattern of charge distribution is distinct for the AIM2 PYD with dominant acidic residues at the α1-α2 helices including residues Glu-7, Asp-15, Asp-19, Glu-20, Glu-21, and Asp-23 (Figs. 1 and 4, A and B) and primarily basic residues Lys-64, Arg-67, Lys-71, Lys-79, Arg-80, Lys-85, Lys-87, Lys-90, Lys-93, and Lys-97 at the α5-α6 helices (Figs. 2A and 4C). In agreement with this distinct pattern of charge distribution, residues Lys-64 and Lys-85 are conserved among the PYHIN proteins only (Fig. 1), and residues Glu-20 and Glu-21 are AIM2-specific residues (Fig. 1 and supplemental Fig. 3, A and B). In comparison, the ASC PYD displays a different charge distribution with acidic α1 and α4 helices and basic α2-α3 helices (Fig. 4, D and E).
FIGURE 4.
Electrostatic surface of the PYDs. A, electrostatic charge surface of the AIM2 PYD is displayed on a scale of −5 kT/e (red) to 5 kT/e (blue). The acidic residues are labeled in black. The view is the same as in Fig. 2A with the α1 helix facing the viewer. B, a 90° rotation of the view in A with the α2-α3 helices facing the viewer. The acidic residues are labeled in black, and the non-charged residues are labeled in green. C, electrostatic charge surface of the AIM2 PYD with its α5 helix facing the viewer. The view is rotated ∼180° from that in A. D, the electrostatic charge surface of the ASC PYD is shown with the same view as in A with the α1 helix facing the viewer. The acidic residues are labeled in black. E, a 90° rotation of the view in D and the same as in B with the α2-α3 helices facing the viewer. The basic residues are labeled in yellow.
Adjacent to the acidic residues at the AIM2 PYD α2 helix are several hydrophobic and non-charged residues that are exposed to the solvent. These include residues Phe-27, Phe-28, Phe-33, Ile-35, Thr-37, and Thr-42 (Figs. 1 and 4, B and C). Similar to the acidic residues Glu-20 and Glu-21 of the α2 helix, the hydrophobic residues Phe-27 and Phe-28 are conserved among the AIM2 proteins from different species but not among the other PYHIN proteins (Fig. 1 and supplemental Fig. 3, A and B), suggesting that they may contribute to AIM2-specific functions. Intriguingly, similar hydrophobic residues at the α2 helix of the FADD DED were shown previously to be essential for its binding of the caspase-8 DED (53, 55), and a recent report on the NLRP3 PYD structure suggested that hydrophobic and charged residues may mediate the NLRP3 inflammasome assembly (28). Could these AIM2-specific hydrophobic and charged residues also play a role in mediating PYD-PYD interactions?
Polar and Hydrophobic Interactions May Mediate the AIM2 PYD-ASC PYD Association
To investigate the potential roles of the α2 helix residues in mediating AIM2-ASC association, we performed docking studies of the AIM2 PYD and ASC PYD using the ClusPro protein-protein docking server V2.0 (48). The results show that the α2 helix of the AIM2 PYD is indeed located at the PYD-PYD interface with the α2 helix acidic residues Asp-19, Glu-20, and Asp-23 and hydrophobic residues Phe-27 and Phe-28 within contact distance of the ASC PYD (Fig. 5, A and B). This suggests that the α2 helix of the AIM2 PYD may be involved in mixed polar and hydrophobic interactions with the ASC PYD.
To examine the role of the AIM2 PYD α2 helix in mediating its interaction with the ASC PYD, we performed an MBP pulldown assay. Although the wild type MBP-AIM2 PYD associated with the ASC PYD as expected, mutation of either the hydrophobic residues (F27A and F28A; designated “Mut1”) or acidic residues (D19A, E20A, E21A, and D23A; “Mut2”) abolished this association (Fig. 5C). This result was further confirmed with a yeast two-hybrid assay (Fig. 5D), suggesting that both hydrophobic and acidic residues at the AIM2 PYD α2 helix are essential for its interaction with the ASC PYD.
Electrostatic Interactions Mediate the PYD-HIN Association
The AIM2 receptor is composed of a PYD and a HIN domain, the latter responsible for the recognition of dsDNA in a sequence-independent manner (34). Our previous work using an AIM2 PYD model built from the ASC PYD structure suggested that in the absence of the dsDNA ligand the AIM2 receptor resides in an autoinhibited state with its PYD and HIN domain forming an intramolecular complex. To further characterize this intramolecular domain interaction, we sought to investigate the PYD-HIN interaction through docking of their crystal structures using the ClusPro docking server and compare the results with those from the interactive docking program Hex (49) that we used previously.
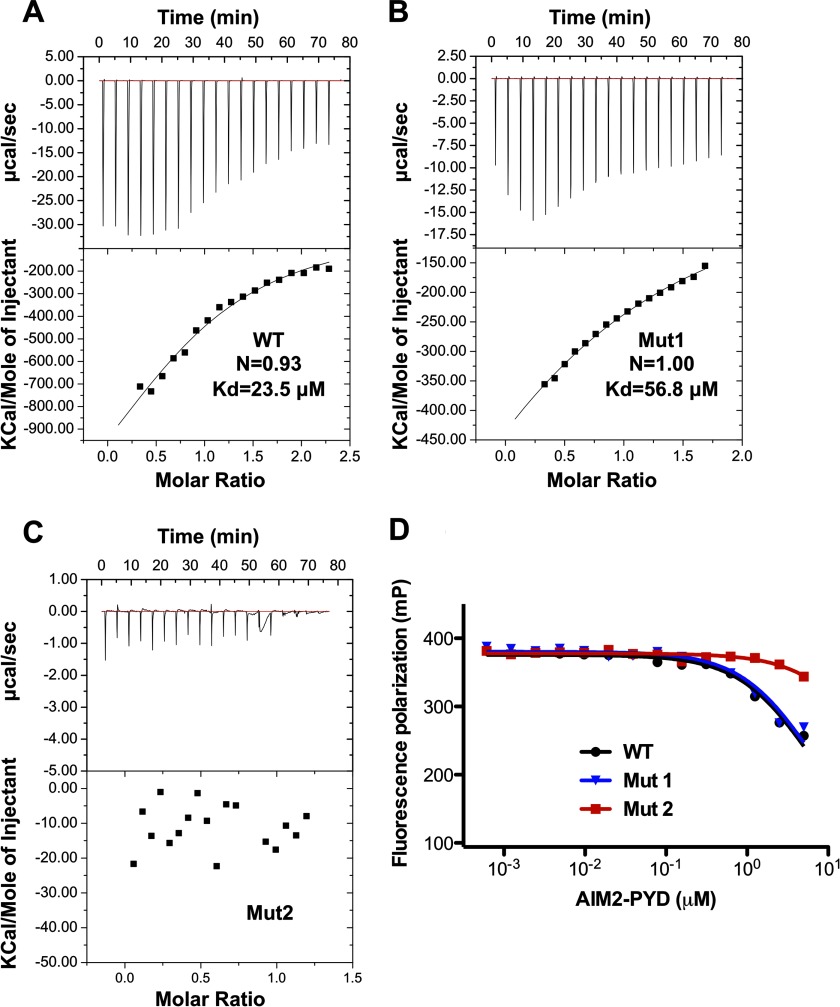
The top 10 solutions from the ClusPro server placed the AIM2 PYD at the concave surface of the HIN domain overlapping with its DNA-binding surface (Fig. 6, A and B). Most of the docking models placed the negatively charged α2 helix of PYD at the center of its interface with the HIN domain, whereas the positively charged α6 helix was located away from the interface in agreement with our previous docking and mutagenesis studies (34). Importantly, such docking models were essentially recapitulated regardless of whether the HIN or PYD was chosen as the stationary receptor molecule (Fig. 6, C and D) and further confirmed using the interactive docking program Hex (supplemental Fig. 4). The top ranked docking model from ClusPro was energy-minimized using the program Rosetta (v3.4) (50). The resulting structure shows that the PYD-HIN interface is dominated by electrostatic interactions between basic residues from the HIN domain DNA-binding surface and acidic residues from the α1-α2 helices of PYD (Fig. 6, E–G). To confirm this interaction, we performed ITC studies to analyze the interaction of the AIM2 PYD and HIN domain in solution. The wild type AIM2 PYD binds the AIM2 HIN domain with a dissociation constant (Kd) of 23.5 μm (Fig. 7A). Mutation of the hydrophobic residues (Mut1) only marginally affected the binding with a Kd of 56.8 μm (Fig. 7B). In contrast, mutation of the acidic residues at the α2 helix (Mut2) abolished the interaction (Fig. 7C). In agreement, Mut1 retained its ability to compete for HIN domain binding by DNA, whereas Mut2 largely lost this inhibitory function (Fig. 7B). These data suggest that the acidic residues are essential for the PYD-HIN interaction, whereas the adjacent hydrophobic residues may not be in direct contact with the DNA-binding surface of the HIN domain.
FIGURE 7.
Binding of the AIM2 PYD and HIN domains. A, the association of the AIM2 HIN and wild type (A), Mut1 (F27A/F28A; B), and Mut2 (D19A/E20A/E21A/D23A; C) were analyzed by ITC. D, inhibition of the AIM2 HIN-DNA interaction by the wild type and mutant AIM2 PYDs as analyzed by fluorescence polarization. mP, millipolarization units.
DISCUSSION
The AIM2 receptor is a critical innate immune sensor in the cytosol that responds to invasion by certain DNA viruses and bacteria and plays a role in the autoimmune disorder psoriasis. A fundamental aspect of the AIM2 function is the ability of its PYD to interact with that of the adapter ASC and form an inflammasome. In this study, we determined the AIM2 PYD crystal structure, which reveals a six-helix bundle typical of the death domain superfamily. The AIM2 PYD features distinct charge distributions with acidic α1-α2 helices and basic α5-α6 helices. The α2 helix also contains surface-exposed hydrophobic residues Phe-27 and Phe-28 that are unique to AIM2 PYD. A highly conserved Lys-26 residue at the α2 helix is largely buried and forms hydrogen bonds with main chain carbonyl oxygens within and adjacent to the short α3 helix. This may stabilize the conformation of the α3 helix to facilitate its participation in PYD-PYD interactions. Such hydrogen bonding-mediated stabilization of the α3 helices appears to be unique for the PYDs as most of the reported DD, DED, and CARD structures use hydrophobic contacts between the α2 and α3 helices (20, 21, 23, 54).
What are the implications of the above structural features for the AIM2 PYD-ASC PYD interactions? Three types of associations have been identified for the death domain fold, all of which involve electrostatic interactions (56). The type I interaction has been reported for DD (20, 21), DED (53), and CARD (54), whereas type II and type III interactions have only been reported for DD (20, 21, 57). As no PYD-PYD complex structure has been reported, it is currently unknown which of the three types or a novel type of interaction may mediate the PYD-PYD association. One mode of interaction predicted by our docking studies illustrates that the AIM2 PYD α2 helix binds the ASC PYD through acidic and hydrophobic residues unique to the AIM2 PYD, and this was confirmed by our MBP pulldown and yeast two-hybrid studies. This is also in agreement with reports that hydrophobic and/or charged residues may mediate FADD DED and caspase-8 DED association as well as NLRP3 PYD and ASC PYD association (28, 53, 55).
Analysis of the complementary charge distribution between the AIM2 PYD and ASC PYD suggests other possible modes of interaction: the acidic α1 helix of the former (Fig. 4A) may interact with the basic α2-α3 helices of the latter (Fig. 4E), and the positively charged α5 helix of the AIM2 PYD (Fig. 4C) may associate with the negatively charged α4 helix of the ASC PYD (Fig. 4D). These predicted types of interactions are not mutually exclusive: oligomeric death domain signaling platforms such as the PIDDosome (20) and MyDDosome (21) are assembled through several binding sites at multiple interfaces. Much of the details on the oligomeric PYD complexes will require future experimental characterization of the inflammasome structures.
Structural studies of the AIM2 inflammasome have so far been focused on its HIN domain (34, 58). Based on the electrostatic nature of the AIM2 HIN domain association with dsDNA, we hypothesized that an intramolecular interaction between the HIN domain and PYD retains the receptor in an autoinhibited state in the absence of ligand binding (34). The direct interaction of the PYD and HIN domain was confirmed by an in vitro pulldown assay (34). In the current work, our docking studies using the crystal structures of the PYD and HIN domain clearly demonstrated strong preference of the two domains to interact through their respective charged surfaces (Fig. 6). We further validated this autoinhibition model using ITC studies and fluorescence polarization inhibition assays. Because the PYD and HIN domain are covalently linked, their high effective local concentrations may promote the intramolecular association that prevents undesirable activation of AIM2. Similar autoinhibition mechanisms have been observed for other multidomain receptors such as retinoic acid-inducible gene 1 (RIG-I) (59) and Apaf-1 (60). It appears that autoinhibition is a common regulatory mechanism for multidomain immune receptors to safeguard against spurious activation of excessive inflammatory responses. Our docking models of the AIM2 PYD-ASC PYD and AIM2 PYD-AIM2 HIN complexes reveal that overlapping negatively charged surface at the AIM2 PYD may be involved in its association with both partner domains. Such mutually exclusive interactions may serve to ensure that AIM2 interacts with the downstream adapter ASC only upon activation by the dsDNA ligand.
Acknowledgments
We thank Dr. Lars C. Pedersen at the NIEHS, National Institutes of Health for the MBP fusion plasmids, Dr. David S. Waugh at the NCI, National Institutes of Health for the tobacco etch virus protease expression construct, and Dr. Gerhard Wagner at Harvard Medical School for the GB1 encoding plasmid. We thank Dr. Tinghe Wu for help with the ASC PYD expression construct and Dr. D. Eric Anderson at the Mass Spectrometry facility of the NIDDK, National Institutes of Health for technical support. We are grateful to the beam line scientists at the GM/CA-CAT, Advanced Photon Source for support, which is funded in whole or in part with federal funds from the NCI (Grant Y1-CO-1020) and NIGMS (Grant Y1-GM-1104), National Institutes of Health.
This work was supported, in whole or in part, by a National Institutes of Health grant from the Division of Intramural Research, NIAID (to T. S. X.).

This article contains supplemental Figs. 1–4.
The atomic coordinates and structure factors (code 3VD8) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- AIM2
- absent in melanoma 2
- PYD
- pyrin domain
- ASC
- apoptosis-associated speck-like protein containing a CARD
- CARD
- caspase recruitment domain
- HIN
- hematopoietic interferon-inducible nuclear
- PYHIN
- pyrin and hematopoietic interferon-inducible nuclear
- NLR
- NOD-like receptor
- NOD
- nucleotide-binding oligomerization domain
- DD
- death domain
- DED
- death effector domain
- MBP
- maltose-binding protein
- ITC
- isothermal titration calorimetry
- POP
- PYD-only protein
- FADD
- Fas-associated via death domain.
REFERENCES
- 1. Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D. R., Latz E., Fitzgerald K. A. (2009) AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fernandes-Alnemri T., Yu J.-W., Datta P., Wu J., Alnemri E. S. (2009) AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458, 509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roberts T. L., Idris A., Dunn J. A., Kelly G. M., Burnton C. M., Hodgson S., Hardy L. L., Garceau V., Sweet M. J., Ross I. L., Hume D. A., Stacey K. J. (2009) HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science 323, 1057–1060 [DOI] [PubMed] [Google Scholar]
- 4. Bürckstümmer T., Baumann C., Blüml S., Dixit E., Dürnberger G., Jahn H., Planyavsky M., Bilban M., Colinge J., Bennett K. L., Superti-Furga G. (2009) An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 10, 266–272 [DOI] [PubMed] [Google Scholar]
- 5. Rathinam V. A., Jiang Z., Waggoner S. N., Sharma S., Cole L. E., Waggoner L., Vanaja S. K., Monks B. G., Ganesan S., Latz E., Hornung V., Vogel S. N., Szomolanyi-Tsuda E., Fitzgerald K. A. (2010) The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11, 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fernandes-Alnemri T., Yu J.-W., Juliana C., Solorzano L., Kang S., Wu J., Datta P., McCormick M., Huang L., McDermott E., Eisenlohr L., Landel C. P., Alnemri E. S. (2010) The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol. 11, 385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sauer J.-D., Witte C. E., Zemansky J., Hanson B., Lauer P., Portnoy D. A. (2010) Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe 7, 412–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim S., Bauernfeind F., Ablasser A., Hartmann G., Fitzgerald K. A., Latz E., Hornung V. (2010) Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur. J. Immunol. 40, 1545–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ge J., Gong Y.-N., Xu Y., Shao F. (2012) Preventing bacterial DNA release and absent in melanoma 2 inflammasome activation by a Legionella effector functioning in membrane trafficking. Proc. Natl. Acad. Sci. U.S.A. 109, 6193–6198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dombrowski Y., Peric M., Koglin S., Kammerbauer C., Göss C., Anz D., Simanski M., Gläser R., Harder J., Hornung V., Gallo R. L., Ruzicka T., Besch R., Schauber J. (2011) Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci. Transl. Med. 3, 82ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Unterholzner L., Keating S. E., Baran M., Horan K. A., Jensen S. B., Sharma S., Sirois C. M., Jin T., Latz E., Xiao T. S., Fitzgerald K. A., Paludan S. R., Bowie A. G. (2010) IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11, 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ludlow L. E., Johnstone R. W., Clarke C. J. (2005) The HIN-200 family: more than interferon-inducible genes? Exp. Cell Res. 308, 1–17 [DOI] [PubMed] [Google Scholar]
- 13. Kwon D., Yoon J. H., Shin S. Y., Jang T. H., Kim H. G., So I., Jeon J. H., Park H. H. (2012) A comprehensive manually curated protein-protein interaction database for the death domain superfamily. Nucleic Acids Res. 40, D331–D336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cridland J. A., Curley E. Z., Wykes M. N., Schroder K., Sweet M. J., Roberts T. L., Ragan M. A., Kassahn K. S., Stacey K. J. (2012) The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC Evol. Biol. 12, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bertin J., DiStefano P. S. (2000) The PYRIN domain: a novel motif found in apoptosis and inflammation proteins. Cell Death Differ. 7, 1273–1274 [DOI] [PubMed] [Google Scholar]
- 16. Martinon F., Hofmann K., Tschopp J. (2001) The pyrin domain: a possible member of the death domain-fold family implicated in apoptosis and inflammation. Curr. Biol. 11, R118–20 [DOI] [PubMed] [Google Scholar]
- 17. Staub E., Dahl E., Rosenthal A. (2001) The DAPIN family: a novel domain links apoptotic and interferon response proteins. Trends Biochem. Sci. 26, 83–85 [DOI] [PubMed] [Google Scholar]
- 18. Pawłowski K., Pio F., Chu Z., Reed J. C., Godzik A. (2001) PAAD—a new protein domain associated with apoptosis, cancer and autoimmune diseases. Trends Biochem. Sci. 26, 85–87 [DOI] [PubMed] [Google Scholar]
- 19. Fairbrother W. J., Gordon N. C., Humke E. W., O'Rourke K. M., Starovasnik M. A., Yin J. P., Dixit V. M. (2001) The PYRIN domain: a member of the death domain-fold superfamily. Protein Sci. 10, 1911–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Park H. H., Logette E., Raunser S., Cuenin S., Walz T., Tschopp J., Wu H. (2007) Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell 128, 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin S.-C., Lo Y.-C., Wu H. (2010) Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature 465, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Acehan D., Jiang X., Morgan D. G., Heuser J. E., Wang X., Akey C. W. (2002) Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol. Cell 9, 423–432 [DOI] [PubMed] [Google Scholar]
- 23. Qi S., Pang Y., Hu Q., Liu Q., Li H., Zhou Y., He T., Liang Q., Liu Y., Yuan X., Luo G., Li H., Wang J., Yan N., Shi Y. (2010) Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell 141, 446–457 [DOI] [PubMed] [Google Scholar]
- 24. Martinon F., Burns K., Tschopp J. (2002) The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 10, 417–426 [DOI] [PubMed] [Google Scholar]
- 25. Liepinsh E., Barbals R., Dahl E., Sharipo A., Staub E., Otting G. (2003) The death-domain fold of the ASC PYRIN domain, presenting a basis for PYRIN/PYRIN recognition. J. Mol. Biol. 332, 1155–1163 [DOI] [PubMed] [Google Scholar]
- 26. de Alba E. (2009) Structure and interdomain dynamics of apoptosis-associated speck-like protein containing a CARD (ASC). J. Biol. Chem. 284, 32932–32941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hiller S., Kohl A., Fiorito F., Herrmann T., Wider G., Tschopp J., Grütter M. G., Wüthrich K. (2003) NMR structure of the apoptosis- and inflammation-related NALP1 pyrin domain. Structure 11, 1199–1205 [DOI] [PubMed] [Google Scholar]
- 28. Bae J. Y., Park H. H. (2011) Crystal structure of NALP3 protein pyrin domain (PYD) and its implications in inflammasome assembly. J. Biol. Chem. 286, 39528–39536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eibl C., Grigoriu S., Hessenberger M., Wenger J., Puehringer S., Pinheiro A. S., Wagner R. N., Proell M., Reed J. C., Page R., Diederichs K., Peti W. (2012) Structural and functional analysis of the NLRP4 pyrin domain. Biochemistry 51, 7330–7341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pinheiro A. S., Proell M., Eibl C., Page R., Schwarzenbacher R., Peti W. (2010) Three-dimensional structure of the NLRP7 pyrin domain: insight into pyrin-pyrin-mediated effector domain signaling in innate immunity. J. Biol. Chem. 285, 27402–27410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pinheiro A. S., Eibl C., Ekman-Vural Z., Schwarzenbacher R., Peti W. (2011) The NLRP12 pyrin domain: structure, dynamics, and functional insights. J. Mol. Biol. 413, 790–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Natarajan A., Ghose R., Hill J. M. (2006) Structure and dynamics of ASC2, a pyrin domain-only protein that regulates inflammatory signaling. J. Biol. Chem. 281, 31863–31875 [DOI] [PubMed] [Google Scholar]
- 33. Kersse K., Verspurten J., Vanden Berghe T., Vandenabeele P. (2011) The death-fold superfamily of homotypic interaction motifs. Trends Biochem. Sci. 36, 541–552 [DOI] [PubMed] [Google Scholar]
- 34. Jin T., Perry A., Jiang J., Smith P., Curry J. A., Unterholzner L., Jiang Z., Horvath G., Rathinam V. A., Johnstone R. W., Hornung V., Latz E., Bowie A. G., Fitzgerald K. A., Xiao T. S. (2012) Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity 36, 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ullah H., Scappini E. L., Moon A. F., Williams L. V., Armstrong D. L., Pedersen L. C. (2008) Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana. Protein Science 17, 1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moon A. F., Mueller G. A., Zhong X., Pedersen L. C. (2010) A synergistic approach to protein crystallization: combination of a fixed-arm carrier with surface entropy reduction. Protein Sci. 19, 901–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 38. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 40. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 41. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang H., Guranovic V., Dutta S., Feng Z., Berman H. M., Westbrook J. D. (2004) Automated and accurate deposition of structures solved by x-ray diffraction to the Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 60, 1833–1839 [DOI] [PubMed] [Google Scholar]
- 44. Honig B., Nicholls A. (1995) Classical electrostatics in biology and chemistry. Science 268, 1144–1149 [DOI] [PubMed] [Google Scholar]
- 45. Lee B., Richards F. M. (1971) The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 55, 379–400 [DOI] [PubMed] [Google Scholar]
- 46. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. (2010) ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kozakov D., Hall D. R., Beglov D., Brenke R., Comeau S. R., Shen Y., Li K., Zheng J., Vakili P., Paschalidis I. Ch., Vajda S. (2010) Achieving reliability and high accuracy in automated protein docking: Cluspro, PIPER, SDU, and stability analysis in CAPRI rounds 13–19. Proteins 78, 3124–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ritchie D. W., Venkatraman V. (2010) Ultra-fast FFT protein docking on graphics processors. Bioinformatics 26, 2398–2405 [DOI] [PubMed] [Google Scholar]
- 50. Kuhlman B., Dantas G., Ireton G. C., Varani G., Stoddard B. L., Baker D. (2003) Design of a novel globular protein fold with atomic-level accuracy. Science 302, 1364–1368 [DOI] [PubMed] [Google Scholar]
- 51. Potter J. A., Randall R. E., Taylor G. L. (2008) Crystal structure of human IPS-1/MAVS/VISA/Cardif caspase activation recruitment domain. BMC Struct. Biol. 8, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Holm L., Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang J. K., Wang L., Zheng L., Wan F., Ahmed M., Lenardo M. J., Wu H. (2005) Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition. Mol. Cell 20, 939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Qin H., Srinivasula S. M., Wu G., Fernandes-Alnemri T., Alnemri E. S., Shi Y. (1999) Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature 399, 549–557 [DOI] [PubMed] [Google Scholar]
- 55. Eberstadt M., Huang B., Chen Z., Meadows R. P., Ng S. C., Zheng L., Lenardo M. J., Fesik S. W. (1998) NMR structure and mutagenesis of the FADD (Mort1) death-effector domain. Nature 392, 941–945 [DOI] [PubMed] [Google Scholar]
- 56. Weber C. H., Vincenz C. (2001) A docking model of key components of the DISC complex: death domain superfamily interactions redefined. FEBS Lett. 492, 171–176 [DOI] [PubMed] [Google Scholar]
- 57. Xiao T., Towb P., Wasserman S. A., Sprang S. R. (1999) Three-dimensional structure of a complex between the death domains of Pelle and Tube. Cell 99, 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sung M. W., Watts T., Li P. (2012) Crystallographic characterization of mouse AIM2 HIN-200 domain bound to a 15 bp and an 18 bp double-stranded DNA. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68, 1081–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kowalinski E., Lunardi T., McCarthy A. A., Louber J., Brunel J., Grigorov B., Gerlier D., Cusack S. (2011) Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 147, 423–435 [DOI] [PubMed] [Google Scholar]
- 60. Riedl S. J., Li W., Chao Y., Schwarzenbacher R., Shi Y. (2005) Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature 434, 926–933 [DOI] [PubMed] [Google Scholar]