Background: TMEM16A and 16B work as Cl− channel, whereas 16F works as phospholipid scramblase. The function of other TMEM16 members is unknown.
Results: Using TMEM16F−/− cells, TMEM16C, 16D, 16F, 16G, and 16J were shown to be lipid scramblases.
Conclusion: Some TMEM16 members are divided into two Cl− channels and five lipid scramblases.
Significance: Learning the biochemical function of TMEM16 family members is essential to understand their physiological role.
Keywords: Calcium, Lipid Transport, Phosphatidylcholine, Phosphatidylserine, Plasma Membrane, Cl− Channel, TMEM16, Phospholipid Scramblase, Tissue-specific Expression
Abstract
Asymmetrical distribution of phospholipids between the inner and outer plasma membrane leaflets is disrupted in various biological processes. We recently identified TMEM16F, an eight-transmembrane protein, as a Ca2+-dependent phospholipid scramblase that exposes phosphatidylserine (PS) to the cell surface. In this study, we established a mouse lymphocyte cell line with a floxed allele in the TMEM16F gene. When TMEM16F was deleted, these cells failed to expose PS in response to Ca2+ ionophore, but PS exposure was elicited by Fas ligand treatment. We expressed other TMEM16 proteins in the TMEM16F−/− cells and found that not only TMEM16F, but also 16C, 16D, 16G, and 16J work as lipid scramblases with different preference to lipid substrates. On the other hand, a patch clamp analysis in 293T cells indicated that TMEM16A and 16B, but not other family members, acted as Ca2+-dependent Cl− channels. These results indicated that among 10 TMEM16 family members, 7 members could be divided into two subfamilies, Ca2+-dependent Cl− channels (16A and 16B) and Ca2+-dependent lipid scramblases (16C, 16D, 16F, 16G, and 16J).
Introduction
Phospholipids and glycosphingolipids are distributed asymmetrically in plasma membrane leaflets, with phosphatidylserine (PS)5 and phosphatidylethanolamine (PE) in the inner leaflet, and phosphatidylcholine (PC), galactosylceramide (GalCer), and glucosylceramide mainly in the outer leaflet (1, 2). The lipid asymmetry is disrupted in various processes, including apoptotic cell death (3); activated platelets (4); red blood cell aging (5); pyrenocyte formation in definitive erythropoiesis (6); fusion of macrophages, myocytes, or cytotrophoblasts (7–9); and sperm capacitation (10).
Distribution of lipids in plasma membranes is regulated by three types of lipid transporters: flippases, floppases, and scramblases. Flippases, also called ATP-dependent aminophospholipid translocases, transport aminophospholipids from the extracellular leaflet to the cytoplasmic side (1, 11). The type IV P-type ATPases (P4-ATPase), a subfamily of the P-type ATPase multispan transmembrane proteins, are strong candidates for flippases (12). Floppases are transporters that move a wide range of lipids from the cytosolic to the extracellular leaflet in an ATP-dependent manner. The ATP-binding cassette (ABC) ATPase, particularly ABCA1, has been proposed as a floppase (13), but ABCA1-deficient cells exhibit no defects in transbilayer phospholipid movement (14), arguing against this role.
Once established, the phospholipid distribution between the outer and inner leaflets is not easily disrupted; ATP-dependent translocase inactivation alone does not appear sufficient to cause the rapid PS exposure seen in apoptotic cell death and platelet activation. Thus, a phospholipid scramblase that bidirectionally and nonspecifically transports phospholipids in response to Ca2+ has been proposed (15). Using a liposome reconstitution system with synthetic phospholipids, Basse et al. (16) purified a 37-kDa protein from human erythrocytes and named it phospholipid scramblase (PLSCR). Its cDNA was then isolated (17). However, because the Ca2+-induced PS exposure is normal in PLSCR1−/− cells (18), PLSCR function as a phospholipid scramblase has been challenged (15, 19).
By repeatedly selecting cell populations that efficiently exposed PS in response to Ca2+ ionophore, we recently established a subline of mouse pro-B cell line (Ba/F3) that constitutively exposes PS (20). The Ba/F3 subline harbors a mutated form of TMEM16F protein, a protein carrying eight transmembrane regions with cytoplasmic N and C termini. Ba/F3 cells carrying the mutated form of TMEM16F constitutively exposed PS and PE and internalized PC and sphingomyelin. We thus proposed TMEM16F as a phospholipid scramblase (20). Confirming that TMEM16F is a Ca2+-dependent phospholipid scramblase, recessive TMEM16F mutations were identified in human patients with Scott syndrome (20, 21), which is known to result from a phospholipid-scrambling defect; these patients suffer from impaired blood clotting. However, it is not clear whether TMEM16F is involved in other processes, such as apoptotic cell death or cell fusion. Two of the 10 TMEM16 family members, TMEM16A and 16B, are Ca2+-dependent Cl− channels (22–24); this raises a question of whether TMEM16F is likewise a Cl− channel and whether any other TMEM16 family members are phospholipid scramblases.
In this report, we established an immortalized fetal thymocyte (IFET) cell line from fetal thymus of mice carrying a floxed TMEM16F allele. IFETs express TMEM16F, 16H, and 16K and expose PS in response to a Ca2+ ionophore. Deleting TMEM16F in the IFETs completely abolished their ability to expose PS in response to Ca2+ ionophore. On the other hand, Fas ligand (FasL) treatment efficiently induced PS exposure in the TMEM16F-deficient cells. In the presence of TMEM16C, 16D, 16F, 16G, and 16J, TMEM16F−/− IFETs responded to Ca2+ ionophore by scrambling phospholipids and galactosylceramide, whereas other family members did not. However, two family members, TMEM16A and 16B, but not others showed Ca2+-dependent Cl− channel activity. These results, together with their tissue-specific expression, suggest that the TMEM16 family members have distinct functions in lipid scrambling and Cl− channel functions in various biological processes.
EXPERIMENTAL PROCEDURES
Materials and Cell Lines
Leucine zipper-tagged human FasL was produced in COS-7 cells as described (25). One unit of FasL is defined as the activity that kills 1.0 × 105 mouse WR19L cells expressing Fas (W3 cells) in 4 h. A caspase inhibitor, Q-VD-Oph, was purchased from R&D Systems (Minneapolis, MN). IFETs were maintained in RPMI 1640 medium containing 10% FCS (Nichirei Bioscience, Tokyo, Japan) and 50 μm β-mercaptoethanol. HEK293T and Plat-E cells (26) were cultured in DMEM containing 10% FCS.
cDNA Cloning
Mouse TMEM16F cDNA (NCBI: NM_175344) was described (20). Mouse cDNAs for TMEM16A (GenBank BC062959.1), 16B (GenBank BC033409.1), and 16G (GenBank BC116706.1) were from DNAFORM (Yokohama, Japan). Mouse cDNAs for TMEM16C (NCBI NM_ 001128103.1), 16D (Ensemble ENSMUST 00000070175), and 16K (NCBI NM_133979.2) were cloned from brain tissue by RT-PCR, and cDNAs for TMEM16E (NCBI NM_177694.5), 16H (NCBI NM_001164679.1), and 16J (NCBI NM_178381.3) were isolated from the skeletal muscle, thymus, and stomach, respectively. All cDNAs were verified by sequencing. The following primers were used to isolate TMEM16 cDNAs (the extra sequence for the restriction enzyme is underlined): TMEM16A, 5′-ATATGGATCCACCATGAGGGTCCCCGAGAAGTA and 5′-ATATGAATTCCAGCGCGTCCCCATGGTACT; TMEM16B, 5′-ATATGAATTCCGCATGCACTTTCACGACAACCA and 5′- ATATGAATTCTACATTGGTGTGCTGGGACC; TMEM16C, 5′-ATATGGATCCAAAATGGTCCACCACTCAGGCTC and 5′-ATATCAATTGAGGCCATTCATGGTGAATAG; TMEM16D, 5′-ATATAGATCTAAAATGGAGGCCAGCTCTTCTGG and 5′- ATATCAATTGTGGCCACTCATTGTGATGTG; TMEM16E, 5′-ATATGGATCCGAGATGGTGGAGCAGGAAGGCTT and 5′-ATATCAATTGGACTGTAGTTTTAGCCTTCA; TMEM16G, 5′-ATATAGATCTGACATGCTGCGGGGGCAAGCGCG and 5′-ATATGAATTCGCCTCCGGTAACCCCTACTG; TMEM16H, 5′-ATATAGATCTGCCATGGCCGAGGCGGCTTCGGG and 5′-ATATGAATTCAGGCCTGTGACCTGCGTCCT; TMEM16J, 5′-ATATGAATTCAGCATGCAGGATGATGAGAGTTC and 5′-ATATCAATTGTACATCCGTGCTCCTGGAAC; TMEM16K, 5′-ATATGGATCCAAGATGAGAGTGACTTTATCAAC and 5′-ATATCAATTGGGTAGCTTCCTTCCCATCTT. Because the native mouse cDNAs for TMEM16C, 16D, and 16E produced a low level of proteins in mammalian cells, sequences with enhanced mRNA stability and translational efficiency were custom ordered from GENEART (Regensburg, Germany) (supplemental Figs. S1–S3).
Establishment of TMEM16F−/− IFET Cell Line
TMEM16F conditionally targeted mice were generated by UNITECH (Chiba, Japan) as a custom order. In brief, a neo-loxP cassette carrying the PGK promoter-driven neo gene and flanked by FRT sequences was inserted into intron 3 of the TMEM16F gene (Fig. 1A). A 1.0-kb DNA fragment containing exon 2 was replaced with a fragment carrying the corresponding sequence and a loxP sequence. The diphtheria toxin A fragment (DT-A) driven by the thymidine kinase (tk) promoter, was inserted at the 5′ end of the vector. Mouse Bruce-4 ES cells were transfected with the targeting vector by electroporation, and G418-resistant clones were screened for homologous recombination by PCR. Positive clones were injected into blastocysts to generate TMEM16F+/NeoFRT mice. The TMEM16F+/NeoFRT mice were crossed with CAG-FLPe transgenic mice to remove the Neo cassette (27). Offspring were backcrossed to wild-type C57BL/6 mice to remove the CAG-FLPe transgene, generating TMEM16F+/flox mice. Mice were housed in a specific pathogen-free facility at Kyoto University, and all animal experiments were carried out in accordance with protocols approved by Kyoto University.
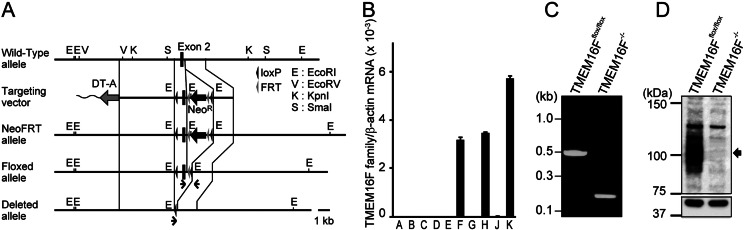
FIGURE 1.
Establishment of TMEM16F−/− IFET cell line. A, schematic representation of wild-type and mutant TMEM16F alleles together with the targeting vector. Recognition sites for EcoRI (E), EcoRV (V), KpnI (K), and SmaI (S) in the flanking region of exon 2 (filled boxes) are indicated. In the target vector, a 1.0-kb DNA fragment carrying exon 2 and its flanking region were replaced by a 2.7-kb fragment carrying two loxP sequences (filled arrowheads) and PGK-neo (NeoR) flanked by FRT sequences (gray arrowheads). Diphtheria toxin A fragment (DT-A) driven by the tk promoter was inserted at the 5′ site of the vector. In NeoFRT allele, the TMEM16F chromosomal gene was replaced by the targeting vector. In Floxed allele, the FRT-flanked NeoR gene was removed by FLPe recombinase. In deleted allele, the loxP-flanked exon 2 of TMEM16F gene was deleted by Cre recombinase. Primers used in C are indicated by arrows. Scale bar, 1.0 kb. B, real-time PCR analysis for mRNA of TMEM16F family members in IFETs. An IFET cell line was established from TMEM16Fflox/flox fetal thymocytes. TMEM16A–16H, 16J, and 16K mRNA in TMEM16Fflox/flox IFETs was quantified by real-time PCR and expressed relative to β-actin mRNA. The experiment was carried out for three times, and the average value was plotted ± S.D. (error bars). C, deletion of TMEM16F exon 2 in the IFET cell line. TMEM16Fflox/flox IFETs were infected by Cre-bearing adenovirus to establish TMEM16F−/− IFET cells. Chromosomal DNA from TMEM16Fflox/flox and TMEM16F−/− IFETs was analyzed by PCR with the primers indicated in A. D, Western blots for TMEM16F in TMEM16Fflox/flox and TMEM16F−/− IFETs. Cell lysates (10 μg of proteins) were separated by 7.5% SDS-PAGE and blotted with rabbit anti-TMEM16F serum (upper panel) or anti-α-tubulin antibody (lower panel). Molecular mass standards (Precision Plus Standard; Bio-Rad) are shown in kDa at left.
IFET cell lines were established as described (28). In brief, TMEM16F+/flox mice were intercrossed, and fetal thymocytes were obtained at embryonic day 14.5. Thymocytes were cultured in DMEM containing 10% FCS, 1× nonessential amino acids, 10 mm Hepes-NaOH buffer (pH 7.4), and 50 μm β-mercaptoethanol. Retroviruses carrying genes for H-rasV12 and c-myc were produced in Plat-E cells with pCX4 vector (29), concentrated by centrifugation, and attached to RetroNectin-coated plates (Takara Bio, Kyoto, Japan). Thymocytes were attached to the retrovirus-coated plate by centrifugation at 400 × g for 5 min and cultured in medium containing 5 ng/ml mouse IL-7 (PeproTech, Rocky Hill, NJ) (30). The resultant IFETs were infected with 1 × 105 pfu/ml Adeno-Cre (Takara Bio) and cloned by limited dilution. Clones carrying the TMEM16F−/− allele were selected by PCR with following primers: wild-type specific sense primer, CTCCAGAGTTTGTAAGTAACACAT; mutant specific sense primer, CAGTCATCGATGAATTCATAACTT; and common antisense primer, AAGACTGATTTCCAAGGTTATCGAA.
Transformation of TMEM16F−/− IFETs
Mouse TMEM16 cDNAs were inserted into pMXs puro c-FLAG (20) to express proteins tagged with FLAG at the C terminus. Retrovirus was produced in Plat-E cells and used to infect TMEM16F−/− IFETs. Stable transformants were selected in medium containing 2 μg/ml puromycin. Mouse Fas cDNA (GenBank NM_007987) was introduced into IFETs by retrovirus-mediated transformation, and its expression was confirmed by flow cytometry with an anti-Fas mAb (Jo2) (MBL, Nagoya, Japan).
Real-time PCR
Total RNA was reverse-transcribed using Superscript III reverse-transcriptase (Invitrogen) or a High Capacity RNA-to-cDNATM kit (Applied Biosystems). Aliquots of the products were amplified in a reaction mixture containing LightCycler® 480 SYBR Green I Master (Roche Diagnostics). Primers used for real-time PCR were as follows: TMEM16A, 5′-ACCCCGACGCCGAATGCAAG and 5′-GCTGGTCCTGCCTGACGCTG; 16B, 5′-GAGGCGCACACCTGGGTCAC and 5′-ATGGGGCGTGGATCCGGACA; 16C, 5′-GCCAGCAATTGCCAACCCCG and 5′-GCAGTCCGACTCCTCCAGCTCT; 16D, 5′-ACAGGCATGCTCTTCCCCGC and 5′-GCGATCACTGCTCGGCGTCT; 16E, 5′-AGCAGCTCCAGCTTCGGCCT and 5′-TTCACGCTCTGCAGGGTGGC; 16F, 5′-CCCACCTTTGGATCACTGGA and 5′-TCGTATGCTTGTCTTTTCCT; 16G, 5′-ACATGTGCCCGCTGTGCTCC and 5′-GGGCCGAGGCCTCTCCTCAA; 16H, 5′-TGGAGGAGCCACGTCCCCAG and 5′-GCGGGGCAGACCCTTCACAC; 16J, 5′-GCTGTGGTGGTGACTGGGGC and 5′-CCAGGCGCGTGGATTTCCCA; 16K, 5′-TGGGGGCAGAAGCAGTCGGT and 5′-GGCCTGTGGGTAGCCAGGGAT; β-actin, 5′-TGTGATGGTGGGAATGGGTCAG and 5′-TTTGATGTCACGCACGATTTCC. The mRNA was quantified at the point where LightCycler System detected the upstroke of the exponential phase of PCR accumulation with the respective linearized plasmid DNA as reference.
Western Blotting
Cells were lysed in radioimmuneprecipitation assay buffer (50 mm Hepes-NaOH buffer (pH 8.0) containing 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, 150 mm NaCl, and protease inhibitor mixture (cOmplete, mini, Roche Diagnostics)). After removing debris, cell lysates were mixed with 5× SDS sample buffer (200 mm Tris-HCl (pH 6.8), 10% SDS, 25% glycerol, 5% β-mercaptoethanol, and 0.05% bromphenol blue), incubated at room temperature for 30 min, and separated by 10% SDS-PAGE (Bio Craft, Tokyo, Japan). After proteins were transferred to a PVDF membrane (Millipore), membranes were probed with HRP-conjugated mouse anti-FLAG M2 (Sigma-Aldrich), and peroxidase activity was detected using a Western Lightning®-ECL system (PerkinElmer Life Sciences).
To prepare rabbit antibody against mouse TMEM16F, the N-terminal region of mouse TMEM16F (amino acids 1–289) was fused to glutathione S-transferase (GST) in a pGEX-5X-1 vector (GE Healthcare). The recombinant protein was produced in Escherichia coli, purified with glutathione-Sepharose, and used to immunize rabbits at Takara Bio as a custom order. Western blotting with the rabbit anti-TMEM16F and HRP-labeled goat anti-rabbit Ig (Dako, Copenhagen, Denmark) was carried out as described above using Immunoreaction Enhancer Solution (Can Get Signal®; Toyobo Life Science, Tokyo, Japan).
Analysis of PS Exposure
The Ca2+-induced PS exposure was examined as described (20). In brief, 5 × 105 cells were stimulated at 20 °C with 3.0 μm A23187 in 500 μl of 10 mm Hepes-NaOH buffer (pH 7.4) containing 140 mm NaCl, 2.5 mm CaCl2, and 5 μg/ml propidium iodide, and 1000-fold-diluted Cy5-labeled annexin V (Bio Vision, Milpitas, CA) and applied to the injection chamber of a FACSAria (BD Biosciences) set at 20 °C.
Internalization of NBD-PC and NBD-GalCer
Cells (106) were stimulated at 15 °C with 250 nm A23187 in 1 ml of Hanks' balanced salt solution (Invitrogen) containing 1 mm CaCl2, with a fluorescent probe, 100 nm NBD-PC (Avanti Polar Lipids, Alabaster, AL), or 250 nm NBD-GalCer) (Avanti Polar Lipids). Aliquots (150 μl) were mixed with 150 μl of Hanks' balanced salt solution containing 5 mg/ml fatty-acid free BSA (Sigma-Aldrich) and 500 nm Sytox Blue (Molecular Probes) and analyzed by FACSAria.
Induction of Apoptosis
Apoptosis was induced with FasL as described (25). In brief, IFETs expressing mouse Fas were treated with 60 units/ml FasL at 37 °C for 2 h, and PS exposure was determined by flow cytometry with Cy5-annexin V. To detect activated caspase 3, cells were fixed at 37 °C for 10 min in PBS containing 1% paraformaldehyde, permeabilized with 90% methanol at −20 °C, and stained with rabbit mAb against active caspase 3 (Cell Signaling). Cells were then incubated with Alexa Fluor 488-labeled goat anti-rabbit IgG (Invitrogen) and analyzed by FACSAria.
Electrophysiology
TMEM16 sequences, FLAG-tagged at C terminus, were inserted into pEF-BOS-EX (31). HEK293T cells (2.5 × 105) were co-transfected with 1.0 μg of TMEM16 expression vector and 0.1 μg of pMAX-EGFP (Lonza Group, Basel, Switzerland) using FuGENE6 (Promega). At 24 h after transfection, cells were reseeded on glass coverslips coated with fibronectin (Sigma-Aldrich). Within 24 h after reseeding, whole cell recordings of cells expressing EGFP were performed using a patch clamp amplifier (Axopatch 200B; Molecular Devices) as described (23, 32). The extracellular solution contained 140 mm NaCl, 5 mm KCl, 2 mm CaCl2, 1 mm MgCl2, 30 mm glucose, and 10 mm Hepes-NaOH (pH 7.4). The intracellular solution contained 140 mm NaCl, 1.12 mm EGTA, 1 mm CaCl2, 30 mm glucose, and 10 mm Hepes-NaOH (pH 7.4). The free Ca2+ concentration (500 nm) was calculated with WEBMAXC software.
RESULTS
Establishment of TMEM16F−/− Fetal Thymocyte Cell Lines
Ca2+-dependent PS exposure is reduced by knocking down TMEM16F mRNA and accelerated by TMEM16F overexpression, suggesting that TMEM16F is a phospholipid scramblase (20). To demonstrate TMEM16F involvement in Ca2+-dependent phospholipid scrambling and to determine whether TMEM16F plays a role in exposing PS to the cell surface during apoptotic cell death, we established from fetal thymus tissue a TMEM16F-deficient mouse cell line that expresses a small number of TMEM16 family members, including TMEM16F (see below).
A targeting vector in which exon 2 of TMEM16F gene was flanked by loxP sequences was used to replace the TMEM16F allele in a mouse embryonic stem cell (ES) line from a C57BL/6 background (Fig. 1A). Mice carrying the floxed allele were generated from the ES clone and intercrossed. Embryos were genotyped at embryonic day 14.5, and fetal TMEM16Fflox/flox thymus cells were infected with retroviruses carrying H-rasV12 and c-myc to establish IFET cell lines. Flow cytometry analysis showed that IFETs expressed Thy-1 weakly and CD44 strongly but did not express CD4 or CD8; this indicated that they were derived from a T cell lineage at an early developmental stage. A real-time RT-PCR analysis showed that IFETs expressed TMEM16F, 16H, and 16K (Fig. 1B). Next, IFETs were infected with adenovirus carrying the CRE recombinase gene, and cells missing exon 2 of the TMEM16F gene were cloned (Fig. 1C). Removing exon 2 causes a frameshift and truncates TMEM16F protein at the N-terminal region. Accordingly, Western blotting with an anti-TMEM16F antibody showed broad bands around 120 kDa in TMEM16Fflox/flox but not TMEM16F−/− IFETs (Fig. 1D). An apparent molecular mass of TMEM16F detected by SDS-PAGE is slightly larger than the expected molecular mass for TMEM16F (106 kDa), which may be explained by glycosylation because mouse TMEM16F carry 6 putative N-glycosylation sites (Asn-Xaa-Ser/Thr).
Requirement of TMEM16F for Ca2+-induced, but Not Apoptotic PS Exposure
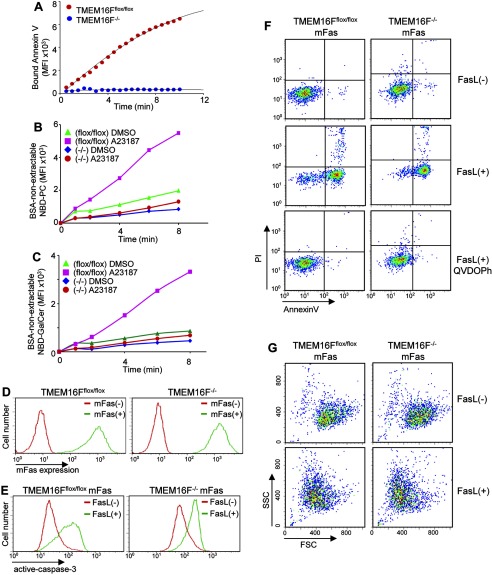
TMEM16Fflox/flox IFETs treated at 20 °C with a Ca2+ ionophore A23187 quickly exposed PS (Fig. 2A); however, this exposure was completely absent in TMEM16−/− IFETs. Similarly, the treatment of TMEM16Fflox/flox but not TMEM16F−/− IFETs with A23187 caused rapid PE exposure, detected by binding of RO09-0198 peptide (20) (data not shown). We then examined the role of TMEM16F in lipid internalization and found that TMEM16Fflox/flox but not TMEM16−/− IFETs internalized NBD-PC and NBD-GalCer upon Ca2+ ionophore treatment (Fig. 2, B and C). These results indicated that TMEM16F is responsible for Ca2+-dependent lipid scrambling in IFETs.
FIGURE 2.
An indispensable role of TMEM16F for Ca2+-induced but not apoptotic PS exposure. A, Ca2+ ionophore-induced PS exposure. TMEM16Fflox/flox and TMEM16F−/− IFETs were treated at 20 °C with 3.0 μm A23187 in the presence of Cy5-labeled annexin V. Annexin V binding to the cells was monitored by flow cytometry for 10 min and expressed in mean fluorescence intensity (MFI). B and C, Ca2+ ionophore-induced lipid internalization. TMEM16Fflox/flox and TMEM16F−/− IFETs were treated at 15 °C with 250 nm A23187 in the presence of 100 nm NBD-PC (B) or 250 nm NBD-GalCer (C). Using aliquots of the reaction mixture, the BSA-nonextractable level of NBD-PC or NBD-GalCer in the Sytox Blue-negative population was determined at the indicated time by FACSAria and expressed in mean fluorescence intensity. D, transformation of IFETs with mouse Fas. TMEM16Fflox/flox and TMEM16F−/− IFETs were infected with a retrovirus carrying mouse Fas and were stained with a PE-labeled hamster mAb against mouse Fas (green). The staining profile of parental cells is also shown (red). E–G, FasL-induced apoptosis. Fas-expressing TMEM16Fflox/flox and TMEM16F−/− IFETs were treated at 37 °C for 2 h with 60 units/ml FasL in the absence or presence of 50 μm Q-VD-OPh. In E, the cells were permeabilized with 90% methanol and stained with rabbit anti-active caspase 3 followed by incubation with Alexa Fluor 488-labeled goat anti-rabbit IgG. In F, cells were stained with Cy5-labeled annexin V and propidium iodide (PI) and analyzed by FACSAria. In G, cells were analyzed by FACSAria before and after FasL treatment; the forward-scattered light (FSC) and side-scattered light profiles (SSC) are shown.
In agreement with a previous report showing that Fas is not expressed in T cells at early developmental stages (33), IFETs do not express Fas (Fig. 2D). When IFETs were transformed with mouse Fas, FasL efficiently activated caspase 3 (Fig. 2E), and the cells quickly responded by exposing PS (Fig. 2F). A TMEM16F-null mutation did not affect either FasL-induced PS exposure or caspase activation (Fig. 2, E and F). In cells undergoing apoptosis, cell size decreases and cellular granularity increases (34). Treating the TMEM16Fflox/flox and TMEM16F−/− IFETs with FasL decreased the cell size (forward-scattered light) and increased the cellular granularity (side-scattered light) to the same extent (Fig. 2G). Therefore, we concluded that caspase-dependent apoptotic PS exposure and cell shrinkage take place independently of TMEM16F.
Ability of TMEM16 Family Members to Expose PS
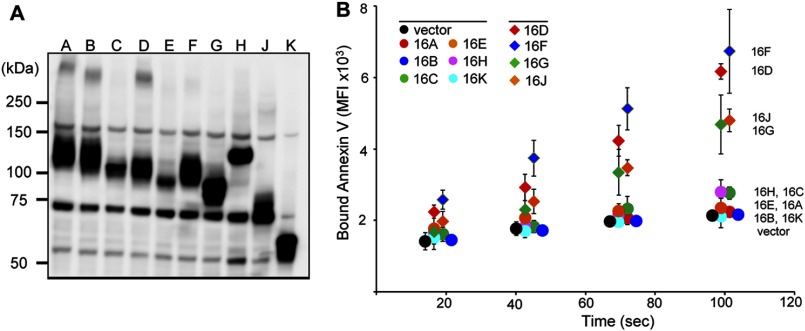
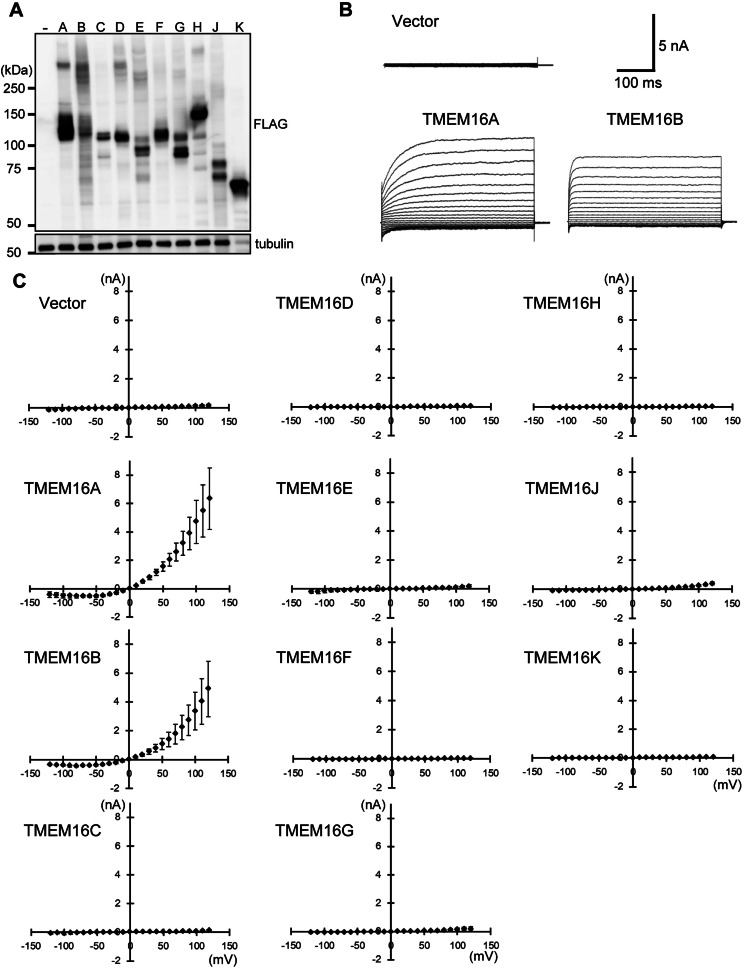
The 10 TMEM16 family members have similar topologies and 20–60% amino acid sequence identity (35, 36). To examine the ability of TMEM16 family members to scramble phospholipids, we transformed TMEM16F−/− IFETs, in which the Ca2+-dependent lipid scramblase activity is completely lost, with mouse retroviral vectors carrying FLAG-tagged TMEM16 family members. Because the expression plasmids for TMEM16C, 16D, and 16E with their endogenous sequences produced very low protein levels in IFETs, their sequences were modified to optimize the mRNA stability and translation efficiency. Western blotting with an anti-FLAG mAb detected a specific band for each TMEM16 family member (Fig. 3A). Except for TMEM16K, their apparent molecular mass, detected by SDS-PAGE, is larger than the calculated molecular mass, which may be explained by glycosylation because these members carry one to six N-glycosylation sites. On the other hand, the apparent molecular mass (65 kDa) of TMEM16K, which does not have a putative N-glycosylation site, was significantly smaller than its estimated molecular mass (76 kDa). Some membrane proteins are known to behave anomalously in SDS-PAGE (37), and TMEM16K may belong to this category. The Western blots also showed that most of the TMEM16 family members were expressed at similar levels, except that the TMEM16E level was 3–5 times lower, and TMEM16K level 5–10 times higher than those of other family members (Fig. 3A). As expected, Ca2+ ionophore treatment efficiently induced TMEM16F−/− IFET transformants expressing TMEM16F to expose PS (Fig. 3B). The TMEM16D as well as TMEM16G and 16J transformants also exposed PS upon Ca2+ treatment, although the ability of TMEM16G or 16J to enhance the PS exposure was weaker than that of TMEM16F and 16D. On the other hand, no or little PS-exposing activity was detected with TMEM16A, 16B, 16C, 16E, 16H, and 16K. Similarly, TMEM16F−/− IFETs lost the ability to internalize NBD-PS, and this activity was rescued strongly by transforming the cells with TMEM16D, 16F, and 16J, and weakly by 16G, whereas IFET transformants expressing TMEM16C and 16E did not internalize NBD-PS (data not shown).
FIGURE 3.
Ca2+-dependent PS exposure by TMEM16 family members. The ten TMEM16 family members were FLAG-tagged at the C terminus and introduced into TMEM16F−/− IFETs to establish stable transformants. A, Western blotting. TMEM16 protein expression in each transformant was analyzed by Western blotting with an anti-FLAG mAb. Note that the amount of TMEM16K lysate protein analyzed was one-eighth that of the others. B, Ca2+-induced PS exposure by TMEM16 family members. TMEM16F−/− IFETs transformed with the indicated TMEM16 family member were stimulated with 3.0 μm A23187. Annexin V binding was monitored with a FACSAria at 20 °C for 2 min and expressed in MFI. The experiments were carried out three times, and the average values were plotted ± S.D. (error bars).
Ability of TMEM16 Family Members to Scramble Lipids
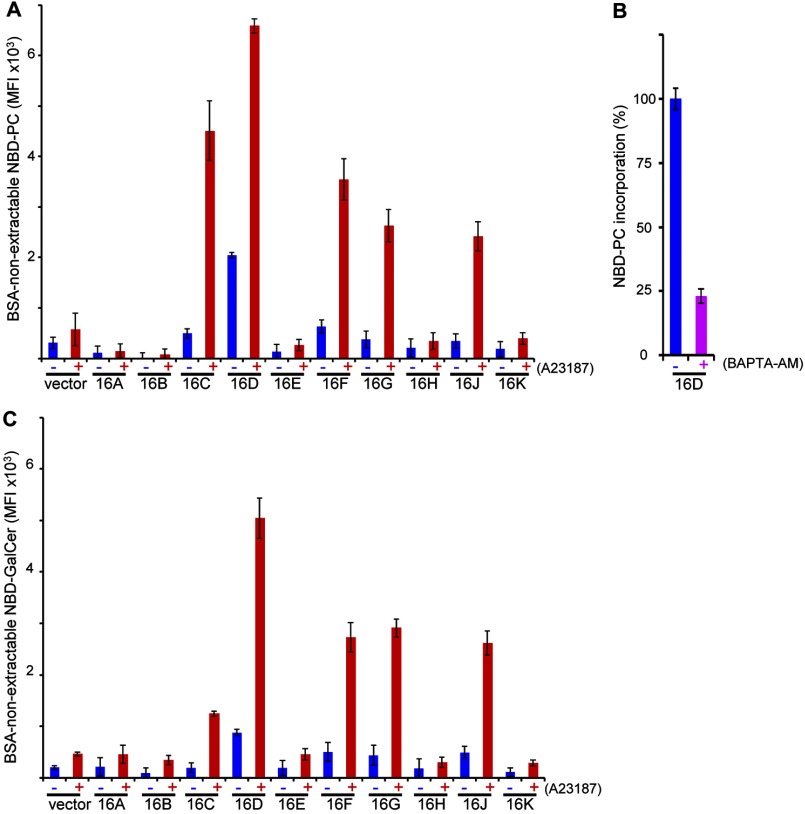
TMEM16F scrambled not only PS and PE, but also other lipids (Fig. 2). To examine the lipid scramblase activity of other TMEM16 family members, TMEM16F−/− IFETs expressing TMEM16 family members were incubated with a fluorescent probe, NBD-PC or NBD-GalCer. As shown in Fig. 4A, the TMEM16F−/− IFETs expressing TMEM16D constitutively, or without A23187 treatment, internalized NBD-PC, and this internalization was strongly enhanced by the A23187 treatment. The A23187-induced NBD-PC uptake with the TMEM16D transformants was stronger than that observed with the 16F transformants. Pretreatment of TMEM16D transformants with BAPTA-AM, a cell-permeable Ca2+ chelator, reduced the NBD-PC uptake observed without Ca2+ ionophore (Fig. 4B), suggesting that the endogenous cellular level of Ca2+ is sufficient to activate the scrambling activity of TMEM16D. As with PS exposure, the A23187 treatment did not induce NBD-PC uptake in IFETs expressing TMEM16A, 16B, 16E, 16H, or 16K (Fig. 4A). However, cells expressing TMEM16C, 16G, or 16J did internalize NBD-PC when treated with Ca2+ ionophore.
FIGURE 4.
Ca2+-dependent internalization of NBD-PC and NBD-GalCer by TMEM16 family members. A and C, ability of TMEM16 family members to internalize NBD-PC and NBD-GalCer. TMEM16F−/− IFETs transformed with the indicated TMEM16 family member were treated at 15 °C with (+) or without (−) 250 nm A23187 in the presence of 100 nm NBD-PC for 4 min (A) or 250 nm NBD-GalCer for 5 min (C), and the internalized, or BSA-non extractable NBD-PC or NBD-GelCer, was quantified by FACSAria and expressed in MFI. B, requirement of Ca2+ for the constitutive internalization of NBD-PC by TMEM16D. The TMEM16D transformants of TMEM16F−/− IFETs were treated with 40 μm BAPTA-AM for 30 min in Ca2+-free RPMI 1640 medium and incubated at 15 °C for 8 min in Hanks' balanced salt solution containing 1 mm CaCl2 and 100 nm NBD-PC. The internalized NBD-PC was determined as above and expressed as percentage of the internalized NBD-PC obtained without BAPTA-AM treatment. All experiments in A–C were carried out three times, and the average values were plotted ± S.D. (error bars).
A similar result was obtained using NBD-GalCer as a substrate. When treated with A23187, TMEM16F−/− transformants expressing TMEM16F incorporated NBD-GalCer, but those expressing TMEM16A, 16B, 16E, 16H, or 16K did not (Fig. 4C). Cells expressing TMEM16D constitutively incorporated NBD-GalCer, and this uptake was enhanced by A23187 treatment. The cells expressing TMEM16C, 16G, or 16J also internalized NBD-GalCer, although the ability of TMEM16C to internalize NBD-GalCer was weaker compared with others. These results suggested that TMEM16C, 16D, 16F, 16G, and 16J scramble various phospholipids and glycosphingolipids with some different substrate preference.
Chloride Channel Activity of TMEM16 Family Members
TMEM16A and 16B are Ca2+-dependent Cl− channels (22–24). To determine whether there are any other TMEM16 family Cl− channels and whether the scramblase activity of TMEM16 family members depends on Cl− channel activity, human 293T cells were co-transfected with the TMEM16 expression plasmid and a vector expressing GFP (Fig. 5A). The Ca2+-dependent chloride channel activity in GFP-positive cells was then determined by whole cell patch clamp analysis (23). We chose 293T cell line as host cells because it has little Ca2+-dependent Cl− channel activity (Fig. 5B) and was used successfully to show that TMEM16A and 16B act as Cl− channels (22–24).
FIGURE 5.
Ca2+-dependent Cl− channel activity of TMEM16 family members. A, expression of TMEM16 family members in HEK293T cells. HEK293T cells were transfected with a pEF-BOS-EX vector carrying cDNA for the FLAG-tagged TMEM16 family member. Two days later, the expression level of each TMEM16 member was analyzed by Western blotting with anti-FLAG and anti-α-tubulin mAbs. Note that the amount of TMEM16K lysate protein analyzed was one-eighth that of the others. B, Ca2+ ionophore-induced TMEM16A and 16B Cl− channel activity. HEK293T cells were co-transfected with a pEF-BOS-EX vector carrying TMEM16A or 16B cDNA, and pMAX-EGFP. Two days later, the Cl− channel activity of EGFP-positive cells was examined by electrophysiology. The pipette (intracellular) solution contained 500 nm free Ca2+. Representative whole cell membrane currents elicited at −120 to +120 mV in 10 millivolt steps are shown for vector-, TMEM16A-, and 16B-transfected cells. The holding membrane potential was maintained at 0 mV. C, outward rectification of the Cl− current by TMEM16 family members. HEK293T cells were co-transfected with pMAX-EGFP and pEF-BOS-EX vector for the indicated TMEM16 family member, and electrophysiology was carried out as described above. Membrane currents were measured at the indicated voltage pulses (mV). Experiments were independently done three to five times, and the average values were plotted against the applied membrane potential ± S.D. (error bars).
In the patch clamp analysis, increasing the intracellular free Ca2+ in the pipette solution to 500 nm yielded large outward rectifying currents in cells expressing TMEM16A or 16B (Fig. 5, B and C). In contrast, other TMEM16 family members induced little if any Ca2+-dependent current in 293T cells, and the effect of increasing the pipette solution Ca2+ concentration from 500 nm to 5 μm was negligible (data not shown). Therefore, we concluded that within the TMEM16 family, only TMEM16A and 16B act as Ca2+-dependent Cl− channels and that the phospholipid scrambling activity of TMEM16C, 16D, 16F, 16G, and 16J is independent of Cl− channel activity.
Expression of TMEM16 Family Members in Mouse Tissues
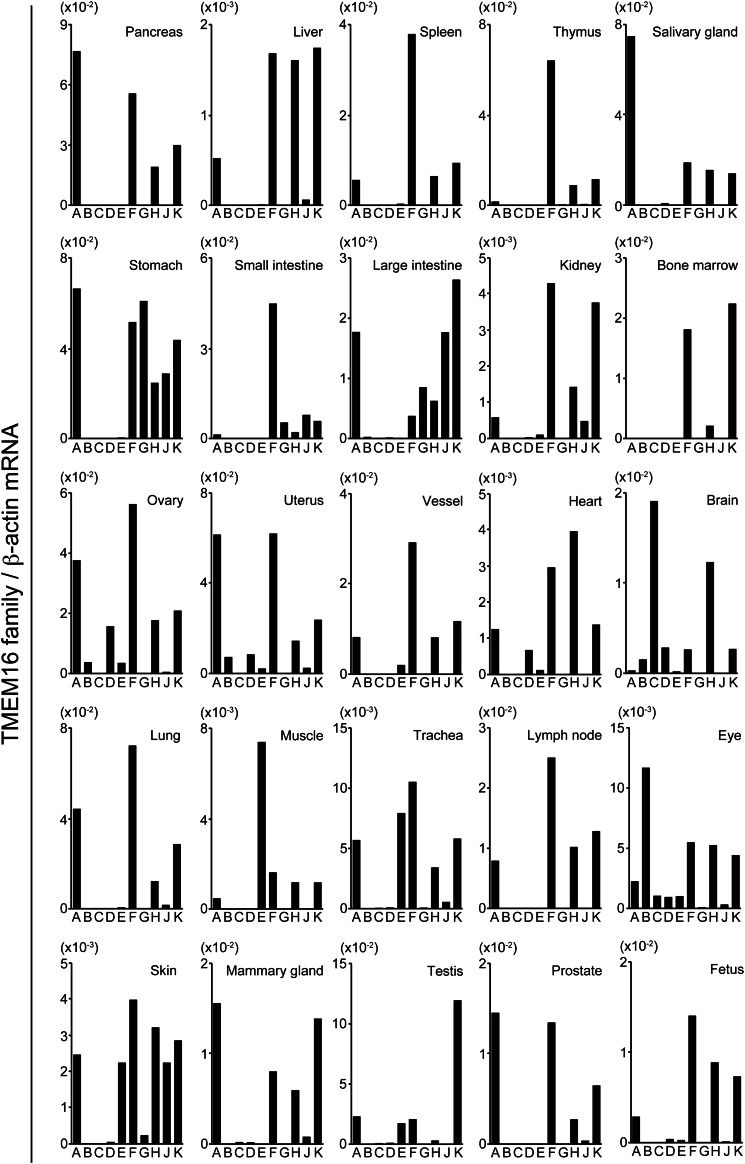
Real-time PCR analysis of TMEM16 mRNA in various mouse tissues showed that each tissue expressed only a limited number of TMEM16 family members (Fig. 6). Of the two Cl− channels of the TMEM16 family, TMEM16A and 16B, we found that TMEM16B was strongly expressed in brain and eye tissues, but weakly expressed or absent in tissues where TMEM16A was strongly expressed, such as the pancreas, liver, salivary glands, stomach, lung, skin, and mammary glands. Of the five lipid scramblases of TMEM16 family, 16C, 16D, 16F, 16G, and 16J, TMEM16F was ubiquitously expressed in various tissues. Whereas other scramblases were present only in a few tissues, TMEM16C and 16J were strongly expressed in the brain and skin, respectively, whereas 16D was found at a low level in a few tissues such as the brain, ovary, heart, and eyes, and 16G and 16J were found in the stomach and intestines. Of the TMEM16 proteins that did not show scramblase or Cl− channel activity, 16H and 16K were expressed ubiquitously in various tissues, whereas 16E was expressed only in the muscle and skin.
FIGURE 6.
Real-time PCR analysis for TMEM16 family member mRNA in mouse tissues. RNA was prepared from the indicated mouse tissues, and mRNA levels quantified by real-time PCR were expressed relative to β-actin mRNA for each TMEM16 family member.
DISCUSSION
Various biological processes require that PS, normally located in the inner plasma membrane leaflets, be exposed to the outer leaflet in response to Ca2+ (11, 15). Previously, our search for a Ca2+-activated phospholipid scramblase identified TMEM16F, a protein with eight transmembrane regions (20). Here we showed that TMEM16F-deficient cells failed to expose PS in response to Ca2+. However, FasL-induced cell shrinkage, PS exposure, and caspase activation were all comparable between the wild-type and TMEM16F−/− IFET cells; this agrees with our previous results that a constitutively active form of TMEM16F has no effect on FasL-induced apoptotic cell death (38). This is also consistent with the report by Williamson et al. (39) that an EBV-transformed cell line established from a Scott patient exposed PS in response to apoptotic signals but not in response to Ca2+, and it supports the proposal that there are two distinct pathways for PS exposure in platelets (40). On the other hand, Martins et al. (41) recently suggested that TMEM16F is involved in apoptotic cell death, based on cell shrinkage in a study using the siRNA and the TMEM16F inhibitor CaCCinh-AO1. Whether these apparently different results are due to different methods and cell lines remains to be clarified.
IFETs expressed TMEM16F, 16H, and 16K; of these only 16F acted as a Ca2+-dependent phospholipid scramblase. This result was consistent with our finding that TMEM16F-deficient IFETs failed to expose PS when treated with Ca2+ ionophore. However, our results are inconsistent with a previously reported finding that Ca2+ is required to expose PS during apoptotic cell death (42). Preliminary analysis with several cell lines suggests that the Ca2+ requirement for apoptotic PS exposure depends on cell lines; Ca2+ is required for apoptotic PS exposure in mouse T cell lymphoma WR19L, but not in Ba/F3 and IFETs.6 How this is regulated may become clear only when the scramblase responsible for apoptotic PS exposure is identified.
There are ten TMEM16 family members, all of which have eight transmembrane regions with cytosolic N and C termini (35, 36). When TMEM16A and 16B were found to be Cl− channels, it was thought that other family members would have the same function. This family was therefore designated as the “anoctamin” or “ano” (anion and octa) family (24, 35, 43). However, except for one report of Cl− channel activity in TMEM16F, 16G, and 16K (44), Cl− channel activity has only been detected for TMEM16A and 16B (Ref. 45 and this work). Duran et al. (45) suggested that other family members might not act as Cl− channels because of their intracellular localization. Here, we showed that five TMEM16 family members, 16C, 16D, 16F, 16G, and 16J, can scramble phospholipids and/or glycosphigolipids, with different preference to lipids. TMEM16G and 16F are present in the plasma membrane (20),7 and it is likely that other TMEM16 family scramblases are also located in the plasma membrane. On the other hand, TMEM16 family members (16E, 16H, and 16K) that have neither scramblase activity nor Cl− channel activity may not be present in the plasma membrane. In fact, immunohistochemical analysis with hamster mAb against TMEM16E and 16K showed that they are localized in endoplasmic reticulum or Golgi apparatus.8 We found here that TMEM16C scrambles PC and GalCer, but not PS, suggesting a substrate specificity for TMEM16 family members. Identification of the active site and its mutational analysis will be necessary to understand how this specificity is generated.
The ABC transporters and P-type ATPases encompass transporters that regulate lipid efflux and influx, respectively, in an ATP-dependent manner (46, 47). These families include channels for cationic and anionic ions. We showed here that TMEM16 family members, which are activated by Ca2+, also can be divided into lipid scramblases (16C, 16D, 16F, 16G, and 16J) and Cl− channels (16A and 16B). The amino acid sequence in the first cytoplasmic region is well conserved among TMEM16 family members and suggests its common role such as Ca2+ signaling (38, 48). The third extracellular loop, which forms a “pore region” and plays an important role in the Cl− channel activity (24), is also well conserved among the family members. Whether this region is involved in the lipid scrambling remains to be determined. In any case, it will be an interesting challenge to determine how some TMEM16 members work as a Cl− channel, whereas others work as lipid scramblases.
The amino acid sequences of TMEM16E that showed neither scramblase nor Cl− channel activity is 49.2% identical with that of TMEM16F; this is higher than the 40.7% identity between lipid scramblase TMEM16D and 16F. This, along with TMEM16E localization to intracellular vesicles (49), suggests that it may be a lipid scramblase that works at intracellular vesicles. TMEM16E mutations cause human diseases gnathodiaphyseal dysplasia, Miyoshi muscular dystrophy 3, and muscular dystrophy, limb-girdle, type 2L (50, 51) in bone and muscle, and understanding whether TMEM16E functions as a Cl− channel or lipid scramblase will be important to understanding the pathology of these diseases. TMEM16H and 16K, which also have neither the scramblase nor Cl− channel activity, are primordial members of the TMEM16 family, and their orthologs can be found in Caenorrhabditis elegans and Drosophila. To understand how the loss-of-function mutations of the TMEM16K gene cause cerebellar ataxia in humans (52), it will be necessary to elucidate the TMEM16K biochemical function.
The PS exposure to the cell surface plays an important role not only in apoptotic cell death and platelet activation, but also in cell fusion (8, 53), neutrophil turnover (54), and the interaction of lymphocytes with antigen-presenting cells (55, 56). In most of these processes, the PS exposure depends on Ca2+, suggesting that a TMEM16 family member is involved. However, of the five TMEM family members (16C, 16D, 16F, 16G, and 16J) that can scramble lipids, only 16F is expressed in muscle, lymphocytes, and bone marrow. In fact, Ehlen et al. recently showed that TMEM16F−/− osteoblasts lost the ability to scramble PS, leading to the decreased deposition of mineral in the bone tissues (57). It will be interesting to examine whether the other tissues such as muscles and immune system develop normally in TMEM16F−/− mice or in patients with Scott syndrome.
Acknowledgments
We thank Dr. H. Ohmori (Department of Physiology, Graduate School of Medicine, Kyoto University) for valuable advice on electrophysiology, Dr. T. Akagi (KAN Research Institute, Kobe, Japan) for pCX4 vector, E. Imanishi for technical assistance, and M. Fujii for secretarial assistance.
This work was supported in part by grants-in-aid from the Ministry of Education, Science, Sports, and Culture in Japan and Research Grants from the Naito Foundation and Nakajima Foundation (to J. S.).

This article contains supplemental Figs. S1–S3.
J. Suzuki and S. Nagata, unpublished results.
K. Ishihara and S. Nagata, unpublished results.
S. Gyobu, K. Ishihara, J. Suzuki, and S. Nagata, unpublished results.
- PS
- phosphatidylserine
- ABC
- ATP-binding cassette
- FasL
- Fas ligand
- GalCer
- galactosylceramide
- IFET
- immortalized fetal thymocyte
- NBD-GalCer
- N-[6-[(7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl]-d-galactosyl-β1–1′-sphingosine
- NBD-PC
- 1-oleoyl-2-(6-((7-nitro-2–1,3-benzoxadiazol-4-yl)amino)hexanoyl)-sn-glycero-3-phosphocholine
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- Q-VD-OPh
- quinolyl-valyl-O-methylaspartyl-[-2,6-difluorophenoxy]-methyl ketone.
REFERENCES
- 1. Balasubramanian K., Schroit A. (2003) Aminophospholipid asymmetry: a matter of life and death. Annu. Rev. Physiol. 65, 701–734 [DOI] [PubMed] [Google Scholar]
- 2. van Meer G., Voelker D. R., Feigenson G. W. (2008) Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nagata S., Hanayama R., Kawane K. (2010) Autoimmunity and the clearance of dead cells. Cell 140, 619–630 [DOI] [PubMed] [Google Scholar]
- 4. Zwaal R. F., Comfurius P., Bevers E. M. (1998) Lipid-protein interactions in blood coagulation. Biochim. Biophys. Acta 1376, 433–453 [DOI] [PubMed] [Google Scholar]
- 5. Boas F. E., Forman L., Beutler E. (1998) Phosphatidylserine exposure and red cell viability in red cell aging and in hemolytic anemia. Proc. Natl. Acad. Sci. U.S.A. 95, 3077–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yoshida H., Kawane K., Koike M., Mori Y., Uchiyama Y., Nagata S. (2005) Phosphatidylserine-dependent engulfment by macrophages of nuclei from erythroid precursor cells. Nature 437, 754–758 [DOI] [PubMed] [Google Scholar]
- 7. Sessions A., Horwitz A. (1983) Differentiation-related differences in the plasma membrane phospholipid asymmetry of myogenic and fibrogenic cells. Biochim. Biophys. Acta 728, 103–111 [DOI] [PubMed] [Google Scholar]
- 8. Helming L., Gordon S. (2009) Molecular mediators of macrophage fusion. Trends Cell Biol. 19, 514–522 [DOI] [PubMed] [Google Scholar]
- 9. Adler R. R., Ng A. K., Rote N. S. (1995) Monoclonal antiphosphatidylserine antibody inhibits intercellular fusion of the choriocarcinoma line, JAR. Biol. Reprod. 53, 905–910 [DOI] [PubMed] [Google Scholar]
- 10. Gadella B. M., Harrison R. A. (2002) Capacitation induces cyclic adenosine 3′,5′-monophosphate-dependent, but apoptosis-unrelated, exposure of aminophospholipids at the apical head plasma membrane of boar sperm cells. Biol. Reprod. 67, 340–350 [DOI] [PubMed] [Google Scholar]
- 11. Leventis P. A., Grinstein S. (2010) The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 39, 407–427 [DOI] [PubMed] [Google Scholar]
- 12. Folmer D. E., Elferink R. P., Paulusma C. C. (2009) P4 ATPases: lipid flippases and their role in disease. Biochim. Biophys. Acta 1791, 628–635 [DOI] [PubMed] [Google Scholar]
- 13. Oram J. F., Vaughan A. M. (2000) ABCA1-mediated transport of cellular cholesterol and phospholipids to HDL apolipoproteins. Curr. Opin. Lipidol. 11, 253–260 [DOI] [PubMed] [Google Scholar]
- 14. Williamson P., Halleck M. S., Malowitz J., Ng S., Fan X., Krahling S., Remaley A. T., Schlegel R. A. (2007) Transbilayer phospholipid movements in ABCA1-deficient cells. PLoS ONE 2, e729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bevers E. M., Williamson P. L. (2010) Phospholipid scramblase: an update. FEBS Lett. 584, 2724–2730 [DOI] [PubMed] [Google Scholar]
- 16. Bassé F., Stout J. G., Sims P. J., Wiedmer T. (1996) Isolation of an erythrocyte membrane protein that mediates Ca2+-dependent transbilayer movement of phospholipid. J. Biol. Chem. 271, 17205–17210 [DOI] [PubMed] [Google Scholar]
- 17. Zhou Q., Zhao J., Stout J. G., Luhm R. A., Wiedmer T., Sims P. J. (1997) Molecular cloning of human plasma membrane phospholipid scramblase: a protein mediating transbilayer movement of plasma membrane phospholipids. J. Biol. Chem. 272, 18240–18244 [DOI] [PubMed] [Google Scholar]
- 18. Zhou Q., Zhao J., Wiedmer T., Sims P. J. (2002) Normal hemostasis but defective hematopoietic response to growth factors in mice deficient in phospholipid scramblase 1. Blood 99, 4030–4038 [DOI] [PubMed] [Google Scholar]
- 19. Sahu S. K., Gummadi S. N., Manoj N., Aradhyam G. K. (2007) Phospholipid scramblases: an overview. Arch. Biochem. Biophys. 462, 103–114 [DOI] [PubMed] [Google Scholar]
- 20. Suzuki J., Umeda M., Sims P. J., Nagata S. (2010) Calcium-dependent phospholipid scrambling by TMEM16F. Nature 468, 834–838 [DOI] [PubMed] [Google Scholar]
- 21. Castoldi E., Collins P. W., Williamson P. L., Bevers E. M. (2011) Compound heterozygosity for 2 novel TMEM16F mutations in a patient with Scott syndrome. Blood 117, 4399–4400 [DOI] [PubMed] [Google Scholar]
- 22. Caputo A., Caci E., Ferrera L., Pedemonte N., Barsanti C., Sondo E., Pfeffer U., Ravazzolo R., Zegarra-Moran O., Galietta L. (2008) TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science 322, 590–594 [DOI] [PubMed] [Google Scholar]
- 23. Schroeder B. C., Cheng T., Jan Y. N., Jan L. Y. (2008) Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell 134, 1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang Y. D., Cho H., Koo J. Y., Tak M. H., Cho Y., Shim W. S., Park S. P., Lee J., Lee B., Kim B. M., Raouf R., Shin Y. K., Oh U. (2008) TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature 455, 1210–1215 [DOI] [PubMed] [Google Scholar]
- 25. Shiraishi T., Suzuyama K., Okamoto H., Mineta T., Tabuchi K., Nakayama K., Shimizu Y., Tohma J., Ogihara T., Naba H., Mochizuki H., Nagata S. (2004) Increased cytotoxicity of soluble Fas ligand by fusing isoleucine zipper motif. Biochem. Biophys. Res. Commun. 322, 197–202 [DOI] [PubMed] [Google Scholar]
- 26. Morita S., Kojima T., Kitamura T. (2000) Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 27. Kanki H., Suzuki H., Itohara S. (2006) High-efficiency CAG-FLPe deleter mice in C57BL/6J background. Exp. Anim. 55, 137–141 [DOI] [PubMed] [Google Scholar]
- 28. Imao T., Nagata S. (2013) Apaf-1- and caspase-8-independent apoptosis. Cell Death Differ. 20, 343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Akagi T., Sasai K., Hanafusa H. (2003) Refractory nature of normal human diploid fibroblasts with respect to oncogene-mediated transformation. Proc. Natl. Acad. Sci. U.S.A. 100, 13567–13572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Watson J. D., Morrissey P. J., Namen A. E., Conlon P. J., Widmer M. B. (1989) Effect of IL-7 on the growth of fetal thymocytes in culture. J. Immunol. 143, 1215–1222 [PubMed] [Google Scholar]
- 31. Murai K., Murakami H., Nagata S. (1998) Myeloid-specific transcriptional activation by murine myeloid zinc finger protein-2. Proc. Natl. Acad. Sci. U.S.A. 95, 3461–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuba H., Yamada R., Ohmori H. (2003) Evaluation of the limiting acuity of coincidence detection in nucleus laminaris of the chicken. J. Physiol. 552, 611–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ogasawara J., Suda T., Nagata S. (1995) Selective apoptosis of CD4 CD8 thymocytes by the anti-Fas antibody. J. Exp. Med. 181, 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dive C., Gregory C. D., Phipps D. J., Evans D. L., Milner A. E., Wyllie A. H. (1992) Analysis and discrimination of necrosis and apoptosis (programmed cell death) by multiparameter flow cytometry. Biochim. Biophys. Acta 1133, 275–285 [DOI] [PubMed] [Google Scholar]
- 35. Galietta L. (2009) The TMEM16 protein family: a new class of chloride channels? Biophys. J. 97, 3047–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Duran C., Hartzell H. C. (2011) Physiological roles and diseases of tmem16/anoctamin proteins: are they all chloride channels? Acta Pharmacol. Sin. 32, 685–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rath A., Glibowicka M., Nadeau V. G., Chen G., Deber C. M. (2009) Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Segawa K., Suzuki J., Nagata S. (2011) Constitutive exposure of phosphatidylserine on viable cells. Proc. Natl. Acad. Sci. U.S.A. 108, 19246–19251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Williamson P., Christie A., Kohlin T., Schlegel R. A., Comfurius P., Harmsma M., Zwaal R. F., Bevers E. M. (2001) Phospholipid scramblase activation pathways in lymphocytes. Biochemistry 40, 8065–8072 [DOI] [PubMed] [Google Scholar]
- 40. Schoenwaelder S. M., Yuan Y., Josefsson E. C., White M. J., Yao Y., Mason K. D., O'Reilly L. A., Henley K. J., Ono A., Hsiao S., Willcox A., Roberts A. W., Huang D. C., Salem H. H., Kile B. T., Jackson S. P. (2009) Two distinct pathways regulate platelet phosphatidylserine exposure and procoagulant function. Blood 114, 663–666 [DOI] [PubMed] [Google Scholar]
- 41. Martins J. R., Faria D., Kongsuphol P., Reisch B., Schreiber R., Kunzelmann K. (2011) Anoctamin 6 is an essential component of the outwardly rectifying chloride channel. Proc. Natl. Acad. Sci. U.S.A. 108, 18168–18172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hampton M. B., Vanags D. M., Pörn-Ares M. I., Orrenius S. (1996) Involvement of extracellular calcium in phosphatidylserine exposure during apoptosis. FEBS Lett. 399, 277–282 [DOI] [PubMed] [Google Scholar]
- 43. Hartzell H. C., Yu K., Xiao Q., Chien L. T., Qu Z. (2009) Anoctamin/TMEM16 family members are Ca2+-activated Cl− channels. J. Physiol. 587, 2127–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schreiber R., Uliyakina I., Kongsuphol P., Warth R., Mirza M., Martins J. R., Kunzelmann K. (2010) Expression and function of epithelial anoctamins. J. Biol. Chem. 285, 7838–7845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Duran C., Qu Z., Osunkoya A. O., Cui Y., Hartzell H. C. (2012) ANOs 3–7 in the anoctamin/Tmem16 Cl− channel family are intracellular proteins. Am. J. Physiol. Cell Physiol. 302, C482–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Palmgren M. G., Nissen P. (2011) P-type ATPases. Annu. Rev. Biophys. 40, 243–266 [DOI] [PubMed] [Google Scholar]
- 47. Chen T. Y., Hwang T. C. (2008) CLC-0 and CFTR: chloride channels evolved from transporters. Physiol. Rev. 88, 351–387 [DOI] [PubMed] [Google Scholar]
- 48. Ferrera L., Caputo A., Ubby I., Bussani E., Zegarra-Moran O., Ravazzolo R., Pagani F., Galietta L. (2009) Regulation of TMEM16A chloride channel properties by alternative splicing. J. Biol. Chem. 284, 33360–33368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mizuta K., Tsutsumi S., Inoue H., Sakamoto Y., Miyatake K., Miyawaki K., Noji S., Kamata N., Itakura M. (2007) Molecular characterization of GDD1/TMEM16E, the gene product responsible for autosomal dominant gnathodiaphyseal dysplasia. Biochem. Biophys. Res. Commun. 357, 126–132 [DOI] [PubMed] [Google Scholar]
- 50. Bolduc V., Marlow G., Boycott K. M., Saleki K., Inoue H., Kroon J., Itakura M., Robitaille Y., Parent L., Baas F., Mizuta K., Kamata N., Richard I., Linssen W. H., Mahjneh I., de Visser M., Bashir R., Brais B. (2010) Recessive mutations in the putative calcium-activated chloride channel anoctamin 5 cause proximal LGMD2L and distal MMD3 muscular dystrophies. Am. J. Hum. Genet. 86, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tsutsumi S., Kamata N., Vokes T. J., Maruoka Y., Nakakuki K., Enomoto S., Omura K., Amagasa T., Nagayama M., Saito-Ohara F., Inazawa J., Moritani M., Yamaoka T., Inoue H., Itakura M. (2004) The novel gene encoding a putative transmembrane protein is mutated in gnathodiaphyseal dysplasia (GDD). Am. J. Hum. Genet. 74, 1255–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vermeer S., Hoischen A., Meijer R. P., Gilissen C., Neveling K., Wieskamp N., de Brouwer A., Koenig M., Anheim M., Assoum M., Drouot N., Todorovic S., Milic-Rasic V., Lochmüller H., Stevanin G., Goizet C., David A., Durr A., Brice A., Kremer B., van de Warrenburg B. P., Schijvenaars M. M., Heister A., Kwint M., Arts P., van der Wijst J., Veltman J., Kamsteeg E. J., Scheffer H., Knoers N. (2010) Targeted next-generation sequencing of a 12.5-Mb homozygous region reveals ANO10 mutations in patients with autosomal-recessive cerebellar ataxia. Am. J. Hum. Genet. 87, 813–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. van den Eijnde S. M., van den Hoff M. J., Reutelingsperger C. P., van Heerde W. L., Henfling M. E., Vermeij-Keers C., Schutte B., Borgers M., Ramaekers F. C. (2001) Transient expression of phosphatidylserine at cell-cell contact areas is required for myotube formation. J. Cell Sci. 114, 3631–3642 [DOI] [PubMed] [Google Scholar]
- 54. Stowell S. R., Karmakar S., Arthur C. M., Ju T., Rodrigues L. C., Riul T. B., Dias-Baruffi M., Miner J., McEver R. P., Cummings R. D. (2009) Galectin-1 induces reversible phosphatidylserine exposure at the plasma membrane. Mol. Biol. Cell 20, 1408–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Del Buono B. J., White S. M., Williamson P. L., Schlegel R. A. (1989) Plasma membrane lipid organization and the adherence of differentiating lymphocytes to macrophages. J. Cell. Physiol. 138, 61–69 [DOI] [PubMed] [Google Scholar]
- 56. Fischer K., Voelkl S., Berger J., Andreesen R., Pomorski T., Mackensen A. (2006) Antigen recognition induces phosphatidylserine exposure on the cell surface of human CD8+ T cells. Blood 108, 4094–4101 [DOI] [PubMed] [Google Scholar]
- 57. Ehlen H. W., Chinenkova M., Moser M., Munter H. M., Krause Y., Gross S., Brachvogel B., Wuelling M., Kornak U., Vortkamp A. (2013) Inactivation of anoctamin-6/Tmem16f, a regulator of phosphatidylserine scrambling in osteoblasts, leads to decreased mineral deposition in skeletal tissues. J. Bone Miner. Res. 28, 246–259 [DOI] [PubMed] [Google Scholar]