Background: Hha facilitates H-NS-mediated silencing of foreign genes in bacteria.
Results: Two Hha monomers bind opposing faces of the H-NS N-terminal dimerization domain.
Conclusion: Hha binds the dimerization domain of H-NS and may contact DNA via positively charged surface residues.
Significance: The structure of Hha and H-NS in complex provides a mechanistic model of how Hha may affect gene regulation.
Keywords: Bacterial Genetics, DNA-binding Protein, Gene Regulation, Gene Transfer, Microbial Pathogenesis, Microbiology
Abstract
The bacterial nucleoid-associated proteins Hha and H-NS jointly repress horizontally acquired genes in Salmonella, including essential virulence loci encoded within Salmonella pathogenicity islands. Hha is known to interact with the N-terminal dimerization domain of H-NS; however, the manner in which this interaction enhances transcriptional silencing is not understood. To further understand this process, we solved the x-ray crystal structure of Hha in complex with the N-terminal dimerization domain of H-NS (H-NS(1–46)) to 3.2 Å resolution. Two monomers of Hha bind to symmetrical sites on either side of the H-NS(1–46) dimer. Disruption of the Hha/H-NS interaction by the H-NS site-specific mutation I11A results in increased expression of the Hha/H-NS co-regulated gene hilA without affecting the expression levels of proV, a target gene repressed by H-NS in an Hha-independent fashion. Examination of the structure revealed a cluster of conserved basic amino acids that protrude from the surface of Hha on the opposite side of the Hha/H-NS(1–46) interface. Hha mutants with a diminished positively charged surface maintain the ability to interact with H-NS but can no longer regulate hilA. Increased expression of the hilA locus did not correspond to significant depletion of H-NS at the promoter region in chromatin immunoprecipitation assays. However, in vitro, we find Hha improves H-NS binding to target DNA fragments. Taken together, our results show for the first time how Hha and H-NS interact to direct transcriptional repression and reveal that a positively charged surface of Hha enhances the silencing activity of H-NS nucleoprotein filaments.
Introduction
Horizontal gene transfer is widely recognized as the dominant source of genetic diversity among free living microbes and is a crucial aspect of bacterial adaptation and survival. Horizontally acquired sequences, such as pathogenicity islands, typically display lower GC content than the host genome average, but the reason for this bias is unknown (1–3). To control the expression of newly acquired sequences, several bacterial species encode regulatory proteins that bind to genomic regions with lower GC content than the host genome and repress transcription. These regulatory proteins have been termed xenogeneic silencing proteins, and to date, three families have been identified: the H-NS family of the γ-proteobacteria, the MvaT proteins from the genus Pseudomonas, and the Lsr2 family from Actinomycetales (4–8). Most well studied are the H-NS proteins from Escherichia coli and Salmonella enterica sv. Typhimurium (S. Typhimurium) where genome-wide ChIP-chip and microarray studies have identified the set of genes under H-NS repression (4, 6, 9). In S. Typhimurium, H-NS regulates over 400 genes, including Salmonella pathogenicity islands 1–5 (SPI1 to -5)4 (4, 5). To recognize foreign genes, both H-NS and Lsr2 employ a prokaryotic AT-hook DNA binding motif that preferentially inserts into the narrow minor groove of AT-rich sequences (10). In this manner, these proteins bind newly acquired sequences and prevent the potentially harmful effects of their uncontrolled expression (11).
The mechanism by which H-NS down-regulates gene expression has not been determined. Critical to this process, however, is the ability of H-NS to self-associate and form extended protein filaments along target sequences (12–15). H-NS self-association is achieved through two distinct dimerization domains located at the N terminus and central regions of the protein, whereas the C-terminal domain of H-NS is responsible for the DNA binding activity (16–18). Nucleoprotein filament formation is thought to occur in a cooperative manner whereby H-NS DNA binding initiates form a single high affinity site and is extended by polymerization of local H-NS molecules along the DNA (19).
At many loci, H-NS alone is insufficient for full transcriptional silencing, and members of the Hha/YdgT family of co-regulatory proteins are also required (20, 21). Hha and YdgT are small (8-kDa) protein paralogues that enhance repression at a subset of the H-NS regulon. Unlike H-NS, Hha-like proteins are found exclusively among enteric bacteria, and thus several species, such as Vibrio cholera and Bordetella pertussis, that encode H-NS homologues lack an associated Hha protein. In Salmonella, hha mutants display mild growth defects and increased expression of genes encoded within SPI1, responsible for bacterial invasion of epithelial cells, as well as genes encoded within SPI2 required for intracellular survival (22–24). As a result, hha mutants exhibit an initial hyperinvasive phenotype in cell culture and are attenuated for virulence in competitive murine infection models (22, 24). The loss of ydgT has minimal effects on Salmonella strains encoding a functional Hha protein; however, hha/ydgT double mutants display greater losses in fitness and virulence than the additive effects of the single mutants (24, 25). Hha and YdgT appear to be highly redundant with regard to their function, and given that hha mutations result in more dramatic phenotypes than deleting ydgT, Hha is thought to play a more dominant regulatory role, and YdgT is considered as a “backup” molecule (26), although it is unclear what the purpose of such a backup would be.
Microarray analysis found that over 1000 genes are misregulated in S. Typhimurium strains lacking both hha and ydgT (Δhha/ΔydgT) (21). A comparison of the Δhha/ΔydgT microarray data with the set of genes misregulated in a Δhns strain of Salmonella indicates a significant, but incomplete, overlap. The gene expression profiles of the Δhha/ΔydgT and Δhns strains are particularly well correlated with respect to the up-regulation of genes encoded within all five pathogenicity islands (SPI1 to -5). In contrast, ancestral genes regulated by H-NS, such as proV, which encodes a glycine betaine transporter, and rcsA, which activates colanic acid synthesis, are unaffected by the loss of hha/ydgT (21).
The overlap between genes controlled by Hha/YdgT and those controlled by H-NS is likely to be the result of direct interactions between these two families of proteins. During its initial characterization, Hha was found to co-purify from E. coli BL21 (DE3) with the native H-NS protein (27). The ability to interact with H-NS is conserved among other Hha family members, including YdgT and the Hha ortholog from Yersinia enterolitica, YmoA (26, 28). Mapping of the Hha/H-NS interaction interface by two-dimensional heteronuclear single quantum correlation experiments revealed that Hha binds to the N-terminal dimerization domain of H-NS (29, 30). This finding has led to speculation that Hha-like proteins indirectly exert regulatory control by altering the oligomerization properties of H-NS. The notion that Hha and YdgT act through H-NS to modulate gene expression is supported by experimental evidence that Hha/H-NS complex formation is required for silencing of the hlyCABD operon in E. coli, which encodes the α-hemolysin toxin (28, 31). Complicating this model are reports that Hha can independently bind to specific regulatory sequences within the hilA promoter from S. Typhimurium and the esp operon of enterohemorrhagic E. coli (22, 23, 32, 33). Despite extensive characterization of Hha-like molecules for over 2 decades, a clear understanding of how these proteins influence gene expression and why certain H-NS-repressed loci require Hha-co-regulation whereas others do not is still lacking.
To address the mechanism of Hha-mediated gene silencing, we have determined the crystal structure of Hha from S. Typhimurium in complex with the N-terminal dimerization domain of H-NS. Two molecules of Hha are shown to bind opposite surfaces of the truncated H-NS dimer, with minimal structural rearrangements occurring upon complex formation. Mutations in H-NS that disrupt the Hha/H-NS interaction result in the up-regulation of some H-NS-repressed genes, with the extent of up-regulation being specific to the particular mutation. Conserved positively charged residues on the surface of Hha are positioned in the same orientation as the predicted orientation of the DNA binding domain of H-NS. Hha variants harboring mutations within this positively charged surface maintain the ability to interact with H-NS but can no longer silence Hha-regulated genes. In vivo, Hha mutants defective in gene silencing have no effect on H-NS DNA binding, whereas in vitro we find that both wild-type Hha and the silencing-defective mutants enhance H-NS shifting of AT-rich PCR fragments. Taken together, our results support a model whereby Hha molecules coat opposite surfaces of H-NS protein filaments and mediate gene silencing through a positively charged cluster of surface-exposed residues.
EXPERIMENTAL PROCEDURES
Plasmid Construction
A list of the plasmids employed in this study is available in Table 1. The hns coding sequence was amplified from Salmonella LT2 genomic DNA and restriction-digested with NdeI and XhoI enzymes. The resulting fragment was ligated into the corresponding sites the pET21b vector, and the plasmid generated, pSSA2, was used for overexpression of the H-NS protein with a C-terminal His6 tag. The plasmids used for expression and purification of H-NSI11A (pSSA40) and H-NSR12H (pSSA41) were generated by site-directed mutagenesis of pSSA2. The hha and ydgT loci from Salmonella LT2 were PCR-amplified and cloned into the NdeI and BamHI restriction sites of vector pET15b, creating the plasmids pSSA1 and pSSA43. These plasmids were used for the overexpression and purification of Hha and YdgT with N-terminal His6 tags and included a thrombin cleavage site for removal of the His6 tags from the respective proteins. The plasmids used for expression and purification of HhaR14A, HhaR16A, HhaR17A, HhaR26A, HhaK30A, HhaR14A/R17A, and HhaD48N were made by site-directed mutagenesis of plasmid pSSA1.
TABLE 1.
Plasmids used in this study
| Plasmid | Vector | Description or use | Reference or source |
|---|---|---|---|
| pSSA1 | pET15b | Expression of His6-tagged Hha | This study |
| pSSA2 | pET21b | Expression of His6-tagged H-NS | Ref. 62 |
| pSSA33 | pCDF1b | Expression of H-NS (1–46) | This study |
| pSSA34 | pCDF1b | Expression of H-NS (1–64) | This study |
| pSSA35 | pCDF1b | Expression of H-NS (1–80) | This study |
| pSSA37 (pHNSI11A) | pHSG576 | Complementation with FLAG-tagged H-NSI11A | This study |
| pSSA38 (pHNSR12H) | pHSG576 | Complementation with FLAG-tagged H-NSR12H | This study |
| pSSA40 | pET21b | Expression of H-NSI11A | This study |
| pSSA41 | pET21b | Expression of H-NSR12H | This study |
| pSSA42 (pHhaWT) | pHSG576 | Complementation with HhaWT | This study |
| pSSA43 | pET15b | Expression of His6-tagged YdgT | This study |
| pSSA44 | pET15b | Expression of His6-tagged HhaR14A | This study |
| pSSA45 | pET15b | Expression of His6-tagged HhaR16A | This study |
| pSSA46 | pET15b | Expression of His6-tagged HhaR17A | This study |
| pSSA47 | pET15b | Expression of His6-tagged HhaR26A | This study |
| pSSA48 | pET15b | Expression of His6-tagged HhaK30A | This study |
| pSSA49 (pHhaR14A) | pHSG576 | Complementation with HhaR14A | This study |
| pSSA50 | pHSG576 | Complementation with HhaR16A | This study |
| pSSA51 (pHhaR17A) | pHSG576 | Complementation with HhaR17A | This study |
| pSSA52 (pHhaR26A) | pHSG576 | Complementation with HhaR26A | This study |
| pSSA53 | pHSG576 | Complementation with HhaK30A | This study |
| pSSA54 | pET15b | Expression of His6-tagged HhaR14AR17A | This study |
| pSSA55 | pET15b | Expression of His6-tagged HhaD48N | This study |
| pSSA56 (pHhaR14/R17A) | pHSG576 | Complementation with HhaR14A/R17A | This study |
| pSSA57 (pHhaD48N) | pHSG576 | Complementation with HhaD48N | This study |
| pSSA58 | pCDF1b | Expression of H-NSFLAG | This study |
| pSSA59 | pCDF1b | Expression of H-NSI11AFLAG | This study |
| pSSA60 | pCDF1b | Expression of H-NSR12HFLAG | This study |
| pSSA61 | pCDF1b | Expression of H-NSR12AFLAG | This study |
| pWN423 | pHSG576 | Empty vector with FLAG tag | Ref. 1 |
| pWN425 (pHNSWT) | pHSG576 | Complementation with FLAG-tagged H-NS | Ref. 1 |
The H-NS truncation mutants H-NS(1–46), H-NS(1–64), and H-NS(1–80) were cloned into the vector pCDF1b (Novagen), which contains a compatible replication origin and antibiotic resistance cassette for co-expression studies with the pET15b vector. The pCDF1b vector was digested with the restriction enzymes XhoI and NdeI, which eliminated the sequence encoding the His6 tag from the vector backbone, thereby allowing for the overexpression of untagged H-NS(1–46), H-NS(1–64), and H-NS(1–80) proteins from plasmids pSSA33, pSSA34, and pSSA35, respectively. It should be noted that in order to facilitate cloning into the NcoI site of pCDF1b, the second residue of H-NS, serine, was mutated to a glycine, and this substitution is present in the H-NS(1–46) protein co-crystallized with Hha. For the nickel resin pull-down assays, H-NSWT, H-NSI11A and H-NSR12H were PCR-amplified from the plasmids pSSA2, pSSA40, and pSSA41, respectively, with primers encoding a FLAG tag at the C-terminal end. The PCR fragments were digested with NcoI and XhoI restriction enzymes and ligated into the vector pCDF1b, generating the plasmids pSSA58 (H-NSWT), pSSA59 (H-NSI11A), and pSSA60 (H-NSR12H).
To construct an hha complementation plasmid, the hha gene from Salmonella 14028s and ∼300 bp of the upstream promoter sequence were cloned into the PstI site of the low copy vector pHSG576 (34). Transformants were screened for the presence of the hha genes, and the native promoter sequence was inserted in the opposite orientation to the lacI promoter. The resulting plasmid, pSSA42 (pHhaWT), restored various phenotypes of the Δhha strain. The plasmids pSSA49 (pHhaR14A), pSSA50 (pHhaR16A), pSSA51 (pHhaR17A), pSSA52 (pHhaR26A), pSSA53 (pHhaK30A), pSSA56 (pHhaR14R17A), and pSSA57 (pHhaD48N) were generated by site-directed mutagenesis of pSSA42. The sequences of all the plasmids generated in this study were confirmed by Sanger Sequencing at the TCAG Sequencing Facility (Center for Applied Genomics, Hospital for Sick Children).
Site-directed Mutagenesis
Plasmid templates were PCR-amplified with New England Biolabs Phusion high fidelity DNA polymerase according to the manufacturer's instructions (50 ng of plasmid template was added to a 50-μl PCR). 6 μl of New England Biolabs buffer 4, 1 μl of DpnI enzyme, and 3 μl of H2O were added directly to the PCRs to eliminate the parental plasmid. Following a 2-h incubation at 37 °C, 2 μl of the reaction mixtures were transformed into New England Biolabs Turbo Competent E. coli cells.
Protein Expression and Purification
Hha, H-NS, and YdgT expression constructs were transformed into the E. coli BL21 (DE3) strain, and the transformants were selected on Luria-Bertani (LB) agar plates supplemented with 100 μg/ml ampicillin. The resulting strains were grown in liquid culture until an optical density at 600 nm of 0.6 was reached. Isopropyl 1-thio-β-d-galactopyranoside was added to a final concentration of 1 mm, and the induced expression cultures were grown for an additional 16 h at 15 °C with shaking. Cells from 1-liter cultures were harvested by centrifugation at 2500 × g for 30 min and resuspended in 25 ml of cell lysis buffer (10 mm Tris, pH 8.0, 500 mm NaCl, 10 mm imidazole, and 5 mm β-mercaptoethanol). The cell suspensions were lysed by sonication, and the insoluble cellular debris was removed by centrifugation at 13,000 × g for 45 min. Qiagen Ni2+ resin was equilibrated in cell lysis buffer and incubated with the cell lysates for 15 min at 4 °C on a rocking platform. The cell lysate/Ni2+ resin mixture was applied to a gravity flow column and washed with 50 ml of wash buffer (10 mm Tris, pH 8.0, 500 mm NaCl, 30 mm imidazole, and 5 mm β-mercaptoethanol). The proteins of interest were eluted from the Ni2+ resin with 15 ml of elution buffer (10 mm Tris, pH 8.0, 500 mm NaCl, 250 mm imidazole, and 5 mm β-mercaptoethanol).
Following Ni2+ chromatography, all samples were further purified by gel filtration chromatography over a Superdex 200 16/60 column from GE Healthcare, pre-equilibrated with storage buffer (25 mm HEPES, pH 7.5, 150 mm NaCl, 5 mm DTT, and 5% (v/v) glycerol). Protein-containing fractions were pooled and analyzed by SDS-polyacrylamide gel electrophoresis. Thrombin was added to purified Hha protein samples for removal of the N-terminal His6 tag.
Co-expression assays were performed as described above with the following exceptions. Plasmids encoding the H-NS variants and Hha or YdgT were co-transformed into a Δhns strain of BL21 (DE3). The transformants were selected on LB agar plates supplemented with 100 μg/ml ampicillin (for selections of pET vectors) and 50 μg/ml streptomycin (for selection of pCDF1b constructs). Following Ni2+ purification, eluates were analyzed by SDS-PAGE.
Crystallization and Structure Determination
Selenomethionyl-incorporated H-NS(1–46)/Hha complex was concentrated to 10 mg/ml by spin ultrafiltration (10 kDa molecular weight cut-off; Millipore) and screened against commercially available sparse matrix crystal screens (MCSG1-4, Microlytic). Crystal trials were set up in 48-well VDX plates using the hanging drop vapor diffusion technique. Protein and crystallization solutions were mixed in a 1:1 ratio with a final drop size of 3 μl suspended over 250 μl of crystallization solution and stored at 20 °C.
Crystals of selenomethionyl-incorporated Hha/H-NS(1–46) complex appeared in condition 89 of the MCSG1 screen (0.2 m potassium chloride, 20% (w/v) PEG 3350) after 5 days. Crystals grown in this condition were singular and grew to dimensions of ∼500 × 100 × 100 μm. Crystals were cryoprotected in the crystallization solution supplemented with 20% (v/v) ethylene glycol prior to flash freezing in liquid nitrogen. X-ray diffraction data were collected at beamline X29A, National Synchrotron Light Source, Brookhaven National Laboratory. Low resolution (180 images of 2° Δϕ oscillation) and high resolution (360 images of 1.0° Δϕ oscillation) data sets were collected on an ADSC Q315 CCD detector with a 350-mm crystal-to-detector distance and an exposure time of 0.4 s/image. The data were merged, integrated, and scaled using the HKL2000 software program (35). A total of six (of 10) selenium sites were located using HKL2MAP (36), and density-modified phases were calculated using SOLVE/RESOLVE (37). The resulting density-modified Se-SAD (selenium-single-wavelength anomalous diffraction) map was interpretable and allowed for manual model building using both the previously solved crystal structure of S. Typhimurium H-NS(1–83) (PDB code 3NR7) and the NMR ensemble of E. coli Hha (PDB code 1JW2) as guides. Subsequent model adjustments were made manually in COOT (38) between iterative rounds of refinement, which was carried out using PHENIX.REFINE (39). Due the high anisotropy of the diffraction exhibited by crystals of the Hha/H-NS(1–46) complex, the structure factors were subjected to anisotropic scaling using the UCLA diffraction anisotropy server (40). Using an F/σ(F) cut-off of 3.0, anisotropic analysis indicated that the diffraction along a* extended to 2.9 Å, whereas diffraction along b* and c* extended to 3.2 and 3.1Å, respectively. Ellipsoidal truncation of the data followed by anisotropic scaling resulted in higher quality electron density maps and a lower overall Rfree during refinement. The final model was refined to an Rwork/Rfree of 28.3 and 33.1%.
Molecular images of the H-NS(1–46)/Hha were generated with the UCSF Chimera software package (41).
Electrophoretic Mobility Shift Assays
A 300-bp fragment of the ssrA or hilA promoter regions were PCR-amplified and gel-purified. Various concentrations of purified H-NSWT, H-NSI11A, and H-NSR12H were combined with 40 ng of DNA and binding buffer (15 mm HEPES, pH 8.0, 40 mm KCl, 1 mm EDTA, 0.5 mm DTT, 5% (v/v) glycerol). After a 30-min incubation at room temperature, 4 μl of 6× Fermentas loading dye was added to each 20-μl reaction. The DNA-protein complexes were separated by gel electrophoresis, carried out for 2.5 h at 70 V on a 6% native polyacrylamide gel at 4 °C (buffered with Tris acetate EDTA). The gels were stained with SYBR Green for 20 min at room temperature and imaged.
Mobility shift assays performed with purified Hha, H-NS, and the hilA promoter fragment were carried out with a binding buffer containing 250 mm KCl instead of 40 mm. The increased salt concentration reduces the affinity of H-NS for the DNA fragment and facilitates Hha-dependent enhancement of DNA binding.
Gel Filtration Chromatography
Purified H-NSWT, H-NSI11A, and H-NSR12H were dialyzed overnight at 4 °C in to the column running buffer (25 mm Tris, pH 7.5, 150 mm NaCl, 2 mm DTT, 2.5 mm EDTA, and 5% (v/v) glycerol). Protein samples were applied to a Tricorn Superdex 200 10/300 GL column pre-equilibrated with the same running buffer at concentrations of 25, 75, and 100 μm. Prior to the sample runs, the column was calibrated with molecular mass protein standards in the range of 18.2–440 kDa.
Reverse Transcription Quantitative PCR
Cultures for RNA analysis were grown to an A600 nm of 0.5–0.6 at 37 °C, and 0.5 ml of culture was combined with 1 ml of RNAprotect reagent (Qiagen). The samples were incubated at room temperature for 30 min, and the cells were harvested by centrifugation at 4600 × g for 10 min. Total RNA was purified using the Aurum Total RNA minikit (Bio-Rad) followed by reverse transcription using an iScript cDNA synthesis kit (Bio-Rad). The resulting cDNA was analyzed by real-time quantitative PCR with gene-specific primers and the SsoFast Evagreen Supermix (Bio-Rad) according to the manufacturer's instructions.
Chromatin Immunoprecipitation Assays
Samples from the same cultures used in the RNA analysis (45 ml) were fixed with 1% (v/v) formaldehyde for 15 min at room temperature. The cross-linking reactions were quenched by the addition of glycine to a final concentration of 130 mm. Cells were pelleted by centrifugation (6170 × g for 10 min) and washed twice with ice-cold phosphate-buffered saline solution (PBS). Total cellular genomic DNA was sheared by sonication using a biorupter sonicating water bath (Diagnode) until DNA fragments of ∼500 bp or smaller were achieved. The cell lysates were immunoprecipitated overnight with anti-FLAG antibody (Sigma catalogue no. F1804) and protein G-agarose beads (Calbiochem). The samples were incubated at 65 °C for 5 h to break the DNA-protein cross-links. The DNA fragments that co-precipitated with H-NSFLAG were quantified by real-time quantitative PCR as described above.
Western Blot
Cell pellets were resuspended in cell lysis buffer containing 9.32 m urea, 2.67 m thiourea, 40 mm Tris, and 86.78 mm CHAPS (pH 8.5). Cells were lysed by sonication, and the total protein concentrations were quantified using a Bradford assay (Bio-Rad). 10 μg of total protein was combined with 2× SDS-PAGE loading dye and run on a 12% polyacrylamide SDS BisTris gel. Transfer to a nitrocellulose membrane was performed with the Bio-Rad semidry electrophoretic transfer cell at 15 V for 1 h. The membrane was blocked at 4 °C overnight in TBST (1× Tris-buffered saline, 0.05% Tween 20) with 5% skim milk powder. The membrane was probed with anti-FLAG M2 antibody (Sigma) diluted 1:1000 in TBST with 5% (w/v) skim milk for 1 h at room temperature followed by goat anti-rabbit secondary antibody conjugated with horseradish peroxidase (diluted 1:10,000 in TBST with 5% milk) for 1 h at room temperature. DnaK was probed as a loading control using a mouse primary antibody (1:1000 in TBST with 5% milk) followed by a goat anti-mouse secondary antibody conjugated with horseradish peroxidase (1:10,000 in TBST with 5% milk).
RESULTS
Purification and Crystallization of the Hha/H-NS(1–46) Complex
To generate large quantities of Hha and H-NS for crystallographic studies, we employed a co-expression system previously described to increase the solubility properties of Hha (42). Hha was overexpressed with an N-terminal His6 tag (HhaHis6) from a pET15b vector in tandem with an untagged version of H-NS from pCDF1b. During purification over nickel resin, untagged H-NS co-eluted with HhaHis6. To avoid the heterogeneous nature of the oligomerization properties of H-NS, multiple C-terminal H-NS truncation constructs were cloned into the vector pCDF1b, each of which contained the N-terminal dimerization domain of H-NS responsible for Hha binding. HhaHis6 co-purified with the H-NS constructs containing residues 1–46 (H-NS(1–46)) and residues 1–64 (H-NS(1–64)); however, no association was detected between HhaHis6 and the construct encompassing H-NS residues 1–80 (data not shown). The complex consisting of HhaHis6 and H-NS(1–46) was chosen as the primary candidate for crystallization because it was highly soluble and eluted from a Superdex S75 gel filtration column as a single population, separated from free Hha, with an apparent stoichiometry of 1:1 (data not shown). Following gel filtration chromatography, the His6 tag was cleaved from the N terminus of Hha by the addition of thrombin, and the resulting Hha/H-NS(1–46) sample crystallized in multiple conditions from a screening suite formulated by the Midwest Center for Structural Genomics. Selenomethionine-incorporated Hha/H-NS(1–46) was prepared in a similar manner, and the 3.2 Å structure was determined using the single wavelength anomalous dispersion technique (Table 2).
TABLE 2.
X-ray data collection and refinement statistics
| H-NS1–46/Hha | |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.979 |
| Space group | P21212 |
| Cell dimensions | |
| a, b, c (Å) | 77.9, 82.9, 47.1 |
| α, β, γ (degrees) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.0–2.9 (3.0–2.9)a |
| Total no. of reflections | 117,015 |
| Total no. of unique reflections | 7129 |
| Rmerge (%)b | 9.8 (69.7)a |
| I/σI | 23.9 (4.3)a |
| Completeness (%) | 99.9 (100.0)a |
| Redundancy | 16.4 (14.0)a |
| Refinement | |
| Resolution | |
| a*, b*, c* (Å)c | 47.1–2.9, 47.1–3.2, 47.1–3.1 |
| Rwork/ Rfree (%)d | 28.3/33.1 |
| No. of atoms, protein | 1683 |
| Average B-factor, protein (Å2) | 69.2 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (degrees) | 1.420 |
| Ramachandran plot (%)e | |
| Total favored | 97.2 |
| Total allowed | 99.5 |
| Coordinate error (Å)f | 0.59 |
a Values in parentheses correspond to the highest resolution shell.
b Rmerge = ΣΣ|I(k) − 〈I〉|/ΣI(k), where I(k) and 〈I〉 represent the diffraction intensity values of the individual measurements and the corresponding mean values. The summation is over all unique measurements.
c Resolution limits recommended by the UCLA diffraction anisotropy server (40).
d Rwork = Σ‖Fobs| − k|Fcalc‖/|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively. Rfree is the sum extended over a subset of reflections (5%) excluded from all stages of the refinement.
e As calculated using MOLPROBITY (59).
f Maximum likelihood-based coordinate error, as determined by PHENIX (39).
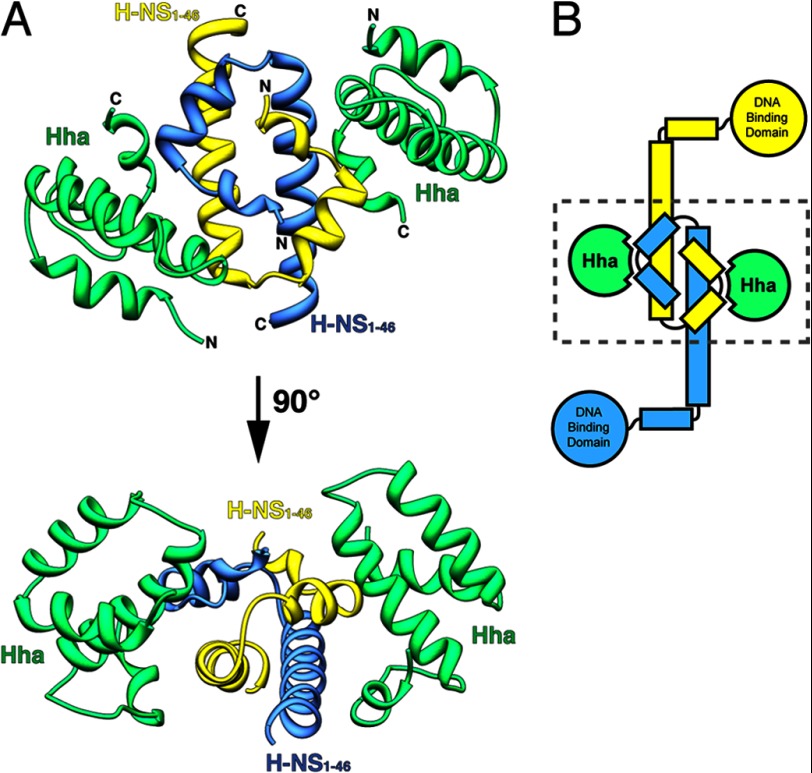
The structure of Hha in complex with H-NS(1–46) reveals that two Hha monomers bind to opposite faces of the H-NS(1–46) dimer (Fig. 1). Individual structures of both Hha and H-NS N-terminal domains were previously solved by NMR and x-ray crystallography. The solution structure of E. coli Hha determined by the Structural Genomics Consortium reveals that the protein consists of four α-helices (43). Helices 1–3 associate into a bundle through hydrophobic interactions, whereas the fourth short helix turns back on the distal end of helix 3. Numerous studies have reported the structure of the H-NS N-terminal dimerization domain, including structures from E. coli, S. Typhimurium, and V. cholera (16, 44–46). According to these structures, the first 46 residues of H-NS consist of two short α-helices and a third elongated helix that forms a coiled-coil motif upon dimerization. Superposition of the Hha/H-NS(1–46) complex with the previously solved isolated structures indicates that no significant structural perturbations of Hha/H-NS(1–46) occur when the complex is formed. A comparison of Hha from the Hha/H-NS(1–46) complex with the monomeric solution structure of Hha (PDB code 1JW2) shows significant similarity between the two structures, with a root mean square (r.m.s.) deviation of 1.9 Å over 67 equivalent Cα atoms. Likewise, the H-NS(1–46) dimer portion the Hha/H-NS(1–46) complex overlaps with the oligomeric crystal structure of H-NS(1–83) from S. Typhimurium (PDB code 3NR7) over 41 equivalent Cα positions with an r.m.s. deviation of 0.7 Å.
FIGURE 1.
Structure of Hha in complex with the N-terminal dimerization domain of H-NS (H-NS(1–46)). A, Hha and H-NS(1–46) crystallized in a 1:1 ratio with molecules of Hha binding either side of the H-NS(1–46) dimer. Hha is shown in green, and the H-NS(1–46) monomers are colored yellow and blue. The N and C termini are indicated in black. B, schematic representation of an H-NS dimer bound to two molecules of Hha. The dashed box indicates the region crystallized.
The relative orientation of the H-NS monomers within the homodimer has been a somewhat controversial topic (47). A solution structure of the H-NS(1–57) dimer from S. Typhimurium suggested that the monomers oligomerize in a parallel topology, with the two C-terminal “tails” pointing in the same direction away from the N-terminal dimerized “head” (45). Conversely, three independent studies have reported an antiparallel conformation for the H-NS dimer, where the C-terminal tail ends of H-NS helices 3 project outwards in opposite directions from the dimerized head region (16, 44, 46). A recent molecular dynamics study of both the antiparallel and parallel H-NS structures revealed that the antiparallel domain is more stable than the parallel conformation under most conditions (47). In our structure of the Hha/H-NS(1–46) complex, the two monomers of H-NS(1–46) are present in an antiparallel conformation. Notably, each Hha molecule contacts both monomeric subunits within the H-NS(1–46) dimer in a fashion that can only be supported when the H-NS dimer is arranged in the antiparallel conformation.
Mutations of H-NS at Residues Ile-11 and Arg-12 Abolish the Hha/H-NS Interaction without Affecting the Capacity of H-NS to Bind DNA and Self-associate in Vitro
Two models of Hha-mediated gene regulation have been proposed. In one model, Hha exerts regulatory control solely through its interaction with H-NS (28, 31). In the second model, Hha can bind DNA and influence transcription independently of H-NS (22, 23, 32, 33). To establish whether or not Hha-mediated gene silencing requires an interaction with H-NS, we first identified and characterized H-NS mutants incapable of binding Hha. These “Hha-blind” H-NS mutants were subsequently tested for their ability to silence Hha/H-NS-repressed genes (see below). If Hha requires H-NS to modulate gene expression, we predicted that disrupting the H-NS/Hha interaction would have a similar effect on gene expression as deleting hha.
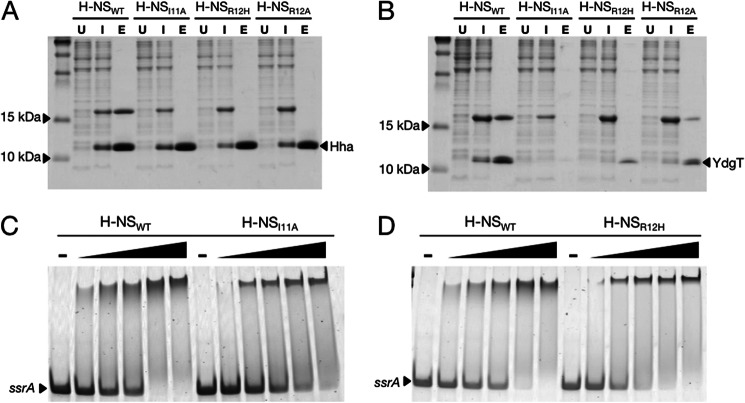
A number of H-NS residues involved in binding Hha were previously identified by Garcia et al. (30) by performing heteronuclear single quantum correlation NMR experiments with 15N-labeled H-NS(1–46). Garcia et al. (30) reported that residues within a conserved motif in helix 2 of the H-NS N-terminal dimerization domain were perturbed upon titration of Hha and that mutating H-NS residue Arg-12 to His (H-NSR12H) disrupted the Hha/H-NS interaction. The H-NSR12H mutant had originally been identified in a genetic screen as a point mutant that resulted in the loss of H-NS silencing in vivo (18). To expand upon these findings, we performed site-directed mutagenesis within the Hha recognition motif of H-NS helix 2 to isolate additional mutations that would abolish the Hha/H-NS interaction but would not otherwise compromise the function of H-NS. Hha/H-NS complex formation was measured by co-expressing His6-tagged Hha with FLAG-tagged H-NS variants in a BL21 (DE3) Δhns strain. Following purification by nickel chromatography, the ability of the FLAG-tagged H-NS variants to co-purify with HhaHis6 was detected by SDS-PAGE of the nickel column eluate. In addition to the H-NSR12H mutant, an H-NS mutant harboring an Ile-11 to Ala substitution (H-NSI11A) failed to co-purify with HhaHis6 (Fig. 2A). We also assessed the interaction between the Hha paralogue YdgT and the H-NS mutants using the co-expression system. YdgTHis6 appears to be susceptible to degradation when it cannot interact with H-NS, as indicated by the absence of a Coomassie-stained band corresponding to the molecular weight of YdgT after co-purification with either H-NSI11A or H-NSR12H (Fig. 2B). Unlike Hha, YdgT maintains some interaction with the H-NS point mutant H-NSR12A, suggesting that the YdgT/H-NS interaction is differentially affected by mutations that abolish Hha/H-NS complex formation. Indeed, the solution structure of the YdgT protein from E. coli (PDB code 2JQT) indicates that YdgT lacks the short C-terminal α-helix, equivalent to helix 4 in Hha, that interacts with the H-NS residue Arg-12 according to our Hha/H-NS(1–46) complex structure (48).
FIGURE 2.
H-NS mutations at residues Ile-11 and Arg-12 disrupt the Hha/H-NS and YdgT/H-NS interactions without affecting DNA binding activity in vitro. A, Coomassie-stained SDS-PAGE of His6-tagged Hha co-expressed with FLAG-tagged H-NSWT, H-NSI11A, H-NSR12H, and H-NSR12A. Samples from co-expression cultures were taken prior to induction with isopropyl 1-thio-β-d-galactopyranoside (lanes marked U (for “uninduced”)), 16 h after the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside (lanes marked I (for “induced”)), and after purification by nickel chromatography (lanes marked E (for “eluate”)). His6-tagged Hha copurifies with H-NSWT but not the point mutants H-NSI11A, H-NSR12H, and H-NSR12A. B, nickel resin purification of His6-tagged YdgT after co-expression with the same H-NS constructs in A. C and D, purified H-NSWT, H-NSI11A, and H-NSR12H were added to a 300-bp PCR fragment from the ssrA promoter region at concentrations of 150, 200, 300, 400, and 500 nm. The dash above four of the lanes indicates that no protein was added to the samples. Protein-DNA binding reactions were separated on a 6% polyacrylamide gel by native gel electrophoresis and stained using SYBR Green nucleic acid stain. The H-NS point mutants H-NSI11A and H-NSR12H shift the ssrA promoter fragment at similar concentrations as H-NSWT.
Prior to characterizing the H-NSI11A and H-NSR12H mutants in vivo, we first verified that these mutations do not adversely affect the DNA binding and oligomerization properties of H-NS in vitro. H-NS DNA binding activity was assessed using an electrophoretic mobility shift assay (EMSA). Purified H-NSWT, H-NSI11A, and H-NSR12H were combined with a 300-bp PCR fragment from the promoter region of ssrA, a gene located in SPI2 and a known target of H-NS repression. The binding reactions were separated on a native polyacrylamide gel, and the DNA-protein complexes were visualized with SYBR Green nucleic acid stain. The addition of 300–500 nm H-NSWT, H-NSI11A, or H-NSR12H to the reactions resulted in complete shifting of the ssrA promoter fragment to higher molecular weight complexes (Fig. 2, C and D). H-NS is well documented to bind DNA in a cooperative manner, which accounts for the sudden shifting of ssrA over a narrow concentration range (12, 19). The EMSAs indicate that H-NSI11A and H-NSR12H bind DNA at concentrations similar to that of wild-type H-NS; however, some qualitative differences in their band shifting patterns were reproducibly observed. The lower mobility protein-DNA complexes generated by H-NSWT appear as broad diffuse bands, whereas the protein-DNA complexes generated by H-NSI11A and H-NSR12H appear as discrete, well defined bands.
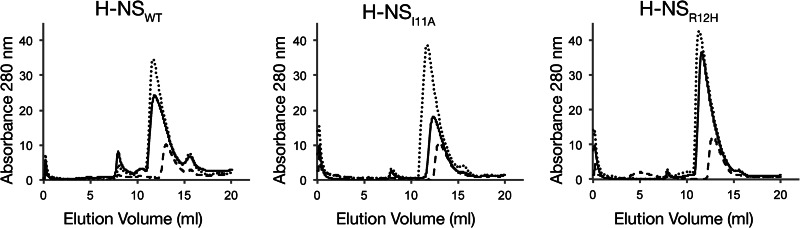
In solution, H-NS exhibits concentration-dependent oligomerization in the absence of DNA that can be detected by gel filtration chromatography (49). The oligomerization properties of H-NSWT, H-NSI11A, and H-NSR12H were compared by applying the purified proteins to a calibrated Tricorn Superdex 200 column at the concentrations of 25, 75, and 100 μm. Because H-NS self-associates into elongated helices, the molecular mass values determined by gel filtration are likely to overestimate the true size of the average H-NS oligomer in the sample. The 25, 75, and 100 μm H-NSWT samples eluted from the S200 gel filtration column with an average calculated molecular mass of 158, 275, and 301 kDa, respectively (Table 3 and Fig. 3). Similarly, the average calculated molecular masses following gel filtration for H-NSI11A at the same initial concentrations were 158, 213, and 295 kDa. Compared with H-NSWT and H-NSI11A, H-NSR12H consistently eluted from the S200 column with slightly higher apparent molecular masses of 177, 309, and 354 kDa. Although H-NSI11A and H-NSR12H both display concentration-dependent oligomerization properties, the gel filtration results suggest that H-NSR12H may self-associate into more extended protein filaments than H-NSWT and H-NSI11A.
TABLE 3.
Oligomerization properties of H-NS variants measured by gel filtration chromatography
| Protein | Concentration | Calculated molecular weight |
|---|---|---|
| μm | kDa | |
| H-NSWT | 25 | 158 |
| 75 | 275 | |
| 100 | 301 | |
| H-NSI11A | 25 | 158 |
| 75 | 213 | |
| 100 | 295 | |
| H-NSR12H | 25 | 177 |
| 75 | 309 | |
| 100 | 354 |
FIGURE 3.
H-NSI11A and H-NSR12H self-associate in a concentration-dependent manner. Nickel-purified H-NSWT, H-NSI11A, and H-NSR12H were applied to a Tricorn Superdex 200 10/300 GL column at concentrations of 25 μm (dashed lines), 75 μm (solid lines), and 100 μm (dotted lines). The molecular weight of the average protein complex in each sample was calculated according to prior calibration with molecular weight protein standards between 18.2 and 440 kDa (Table 3). As the sample concentration was increased, H-NSWT, H-NSI11A, and H-NSR12H eluted from the column as higher molecular weight oligomeric complexes.
Disruption of the Hha/H-NS Interaction in Vivo Results in Misregulation of Select H-NS-repressed Genes
To clarify whether or not Hha can influence gene expression outside of its interaction with H-NS, we investigated the regulatory consequences of disrupting the Hha/H-NS complex in vivo. The H-NS mutants hnsI11A and hnsR12H as well as hnsWT were cloned into a low copy vector encoding a C-terminal FLAG epitope tag and the native hns promoter. The plasmids generated, pHNSWT, pHNSI11A, and pHNSR12H, were subsequently transformed into an hns mutant strain of S. Typhimurium. The Δhns background used in this study harbors an additional mutation within the rpoS gene consisting of a 5-residue in-frame deletion. This mutation was acquired after laboratory passage of S. Typhimurium and reduces rpoS activity and thus significantly improves bacterial tolerance of the hns deletion (11, 50). S. Typhimurium Δhns strains harboring pHNSWT, pHNSI11A, pHNSR12H, and the empty plasmid control, pWN423, were grown to mid-logarithmic phase (A600 of 0.6), and samples were removed for gene expression analysis. Total RNA was purified and reverse transcribed with degenerate primers, and the transcript levels of three H-NS-regulated genes were assessed by qPCR. Two of the genes analyzed, hilA and ssrA, are positive regulators of SPI1 and SPI2, respectively, and were previously shown to require both Hha and H-NS for silencing (21). proV is reportedly silenced by H-NS in an Hha-independent fashion and thus serves as a negative control.
In the absence of H-NS, the steady-state levels of proV, hilA, and ssrA transcripts were increased 40-fold or more compared with the strain complemented with pHNSWT (Fig. 4A). Introduction of the H-NSR12H mutant also resulted in increased transcript levels of all three genes, suggesting that the R12H mutation interferes with H-NS function beyond its inability to interact with Hha. In contrast, the H-NSI11A mutant maintained wild type levels of proV transcript but derepressed hilA and ssrA by 145- and 9.5-fold compared with H-NSWT levels. The H-NSI11A construct more closely mimicked the gene expression pattern of the S. Typhimurium Δhha strain, although the extent to which hilA and ssrA were misregulated varied. hilA expression in the Δhha strain increased 181-fold compared with H-NSWT levels and increased 147-fold in the pHNSI11A strain. On the other hand, ssrA expression was 10-fold higher in the strain harboring H-NSI11A compared with 2.5-fold in the Δhha strain (Fig. 4A). These results are consistent with the notion that an association between Hha and H-NS is required to achieve full transcriptional silencing of hilA under the conditions tested.
FIGURE 4.
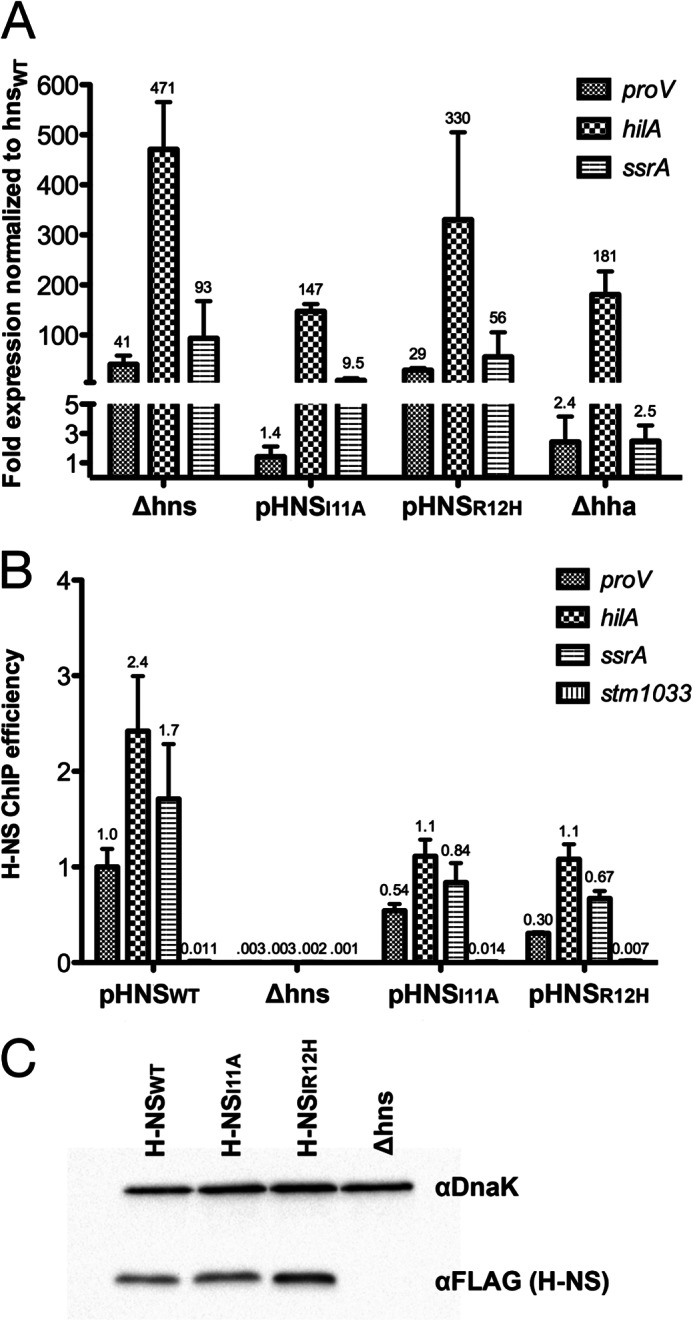
H-NS mutants I11A and R12H differentially misregulate gene targets in S. Typhimurium. A, reverse transcription qPCR was performed on samples from S. Typhimurium Δhns strains harboring pHNSWT, pHNSI11A, pHNSR12H, an empty plasmid control (Δhns), and a Δhha strain. Total RNA was purified and reverse transcribed, and the resulting cDNA was quantified by qPCR with primers against proV, hilA, and ssrA. Transcript levels were graphed as -fold expression relative to the Δhns strain expressing pHNSWT, and the error bars represent the S.D. from three biological replicates. B, H-NS enrichment was measured by ChIP qPCR at the proV, hilA, ssrA, and stm1033 promoter regions with samples from the same strains used in A. stm1033 is a region previously shown to be unbound by H-NS. H-NS enrichment is expressed as the immunoprecipitation efficiencies (percentage recovery after immunoprecipitation compared with initial input). Error bars, S.D. on three biological replicates. C, Western blot analysis of H-NS expression levels from the complemented Δhns strains used in A and B. H-NS was probed with α-FLAG monoclonal antibody, and α-DnaK served as a loading control.
To determine if abrogating the Hha/H-NS interaction affects H-NS DNA binding, ChIP qPCR was performed on the S. Typhimurium Δhns strains harboring pHNSWT, pHNSI11A, and pHNSR12H. Cultures were grown to mid-logarithmic phase and fixed with formaldehyde, the DNA was sheared by sonication, and H-NS/DNA complexes were immunoprecipitated by the addition of an anti-FLAG antibody. qPCR was used to measure the immunoprecipitation efficiency (i.e. percentage recovery after immunoprecipitation compared with initial input) of the proV, hilA, and ssrA genes. Overall, the mutants H-NSI11A and H-NSR12H displayed a partial loss of DNA binding activity compared with H-NSWT (Fig. 4B). The H-NSI11A immunoprecipitation efficiencies for proV, hilA, and ssrA were reduced by 1.8-, 2.2-, and 2-fold relative to H-NSWT, respectively. Similarly, compared with H-NSWT, the H-NSR12H immunoprecipitation efficiencies were reduced by ∼3.3-fold at proV, 2.2-fold at hilA, and 2.5-fold at ssrA. stm1033, a gene that does not bind H-NS in vivo, displayed a level of enrichment with H-NSWT, H-NSI11A, and H-NSR12H that was over 25-fold lower than the bona fide binding targets (4). We note that occupancy at the proV, hilA, and ssrA promoter regions by H-NS and its mutant variants does not directly correlate with the gene expression data. Despite a similar loss in the DNA binding ability of the H-NSI11A and H-NSR12H mutants, proV expression levels were 29-fold higher in the strain harboring pHNSR12H compared with the pHNSI11A- and pHNSWT-expressing strains. These data indicate that occupancy of DNA by H-NS per se is not sufficient to promote silencing and that subtle variations in nucleoid structure and local occupancy, which cannot be measured by the ChIP method we employed, are likely to play an equally important role in gene regulation.
To exclude the possibility of variant expression levels from the plasmids pHNSWT, pHNSI11A, and pHNSR12H, intracellular H-NS levels were quantified by Western blot analysis. Samples taken from the same cultures used in the ChIP assay (harboring pHNSWT, pHNSI11A, and pHNSR12H) were normalized for total protein concentration, separated by SDS-PAGE, and transferred to a nitrocellulose membrane. H-NS was probed with α-FLAG antibody and a α-DnaK antibody served as a loading control (Fig. 4C). Whereas the expression levels of H-NSWT and H-NSI11A were comparable, H-NSR12H expression was somewhat higher. H-NS is known to repress its own promoter, and the H-NSR12H mutation may impair the ability of H-NS to autoregulate its expression (51). We conclude that the inability of H-NSI11A and H-NSR12H to silence hilA and ssrA is not due to lower levels of expression.
Conserved Positively Charged Residues on the Surface of Hha Are Critical for Gene Silencing
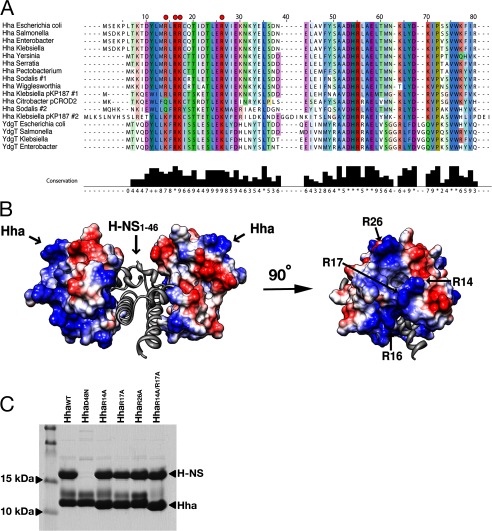
Sequence alignment of Hha and YdgT proteins from various Enterobactericiae indicates that several charged residues are highly conserved (Fig. 5A). An electrostatic surface representation of the Hha solution structure shows that positively charged and negatively charged residues cluster on opposing surfaces of the molecule (Fig. 5B). Overall, Hha is a modestly basic protein but contains 11 acidic residues, 10 of which are highly conserved among the Hha family of proteins. In a previous study carried out by de Alba et al. (52), the authors systematically mutated each of the 10 conserved aspartate and glutamate residues of Hha to asparagine and glutamine, respectively, and evaluated the effects of the mutations on the Hha/H-NS interaction. Only two of the 10 Hha residues mutated, Glu-25 and Asp-48, had significant diminishing effects on the Hha/H-NS association, and one of the mutants generated, E53Q, formed insoluble aggregates. Consistent with these findings, the Hha/H-NS(1–46) structure reveals that the negatively charged surface of Hha, containing residues Glu-25 and Asp-48, takes part in the interaction interface with the H-NS(1–46) dimer. Although the negatively charged surface of Hha is partially buried upon complex formation with H-NS(1–46), a cluster of positively charged residues (Arg-14, Arg-16, Arg-17, Arg-26, and Lys-30) projects out from the surface of Hha, pointing away from the Hha/H-NS(1–46) interaction interface (Fig. 5B).
FIGURE 5.
Highly conserved, positively charged residues protrude from the surface of Hha when in complex with H-NS(1–46). A, an alignment of diverse Hha-like and YdgT-like molecules from selected enteric bacterial species or their plasmids. Residues are colored according to conservation. The conserved, positively charged residues targeted for mutagenesis are indicated with red dots. Alignments were performed using the default settings on the COBALT server at NCBI (60), and results were displayed using Jalview (61). B, surface electrostatic representation of Hha in complex with H-NS(1–46). The 3.2 Å resolution of the Hha/H-NS(1–46) complex did not enable modeling of several surface-exposed side chains; therefore, the Hha solution structure (PDB code 1JW2) was aligned to Hha from the Hha/H-NS(1–46) complex (r.m.s. deviation = 1.9 Å over 67 Cα atoms). Positively charged residues are colored blue, and negatively charged residues are colored red. Surface-exposed basic residues of Hha that point away from H-NS(1–46) are labeled. C, Coomassie-stained SDS-PAGE after co-expression and Ni2+ purification of FLAG-tagged H-NSWT and His6-tagged HhaWT, HhaD48N, HhaR14A, HhaR17A, HhaR26A, and HhaR1A/R17A. All of the Hha mutants maintained an interaction with H-NSWT with the exception of the D48N point mutation, previously reported to disrupt the interaction.
The model of H-NS nucleoprotein filament formation presented by Arold et al. (16) predicts that the arginine-rich surface of Hha is oriented in the same direction as the H-NS DNA binding domain. Given our Hha/H-NS complex structure, we therefore hypothesized that Hha may be providing H-NS with an additional DNA binding surface and enhancing the formation of H-NS/DNA filaments at suboptimal target sequences. To test this hypothesis, we mutated several of the conserved residues on the surface of Hha to alanine, generating the constructs HhaR14A, HhaR16A, HhaR17A, HhaR26A, and HhaK30A and the double mutant HhaR14A/R17A. The Hha mutants were overexpressed with N-terminal His6 tags and purified over nickel resin. Most of the mutants displayed solubility properties similar to those of HhaWT, with the exception of HhaR16A and HhaK30A, which had a tendency to form insoluble aggregates. The remaining four mutants, HhaR14A, HhaR17A, HhaR26A, and HhaR14A/R17A, were confirmed to retain HhaWT secondary structure characteristics by circular dichroism (CD) spectroscopy (data not shown). To verify that the surface arginine mutations do not interfere with the ability of Hha to interact with H-NS, Hha/H-NS complex formation was assessed using the previously described co-expression/nickel resin pull-down assay. The point mutant HhaD48N, which was shown to disrupt the Hha/H-NS interaction by de Alba et al. (52), served as a negative control. His6-tagged HhaR14A, HhaR17A, HhaR26A, HhaR14A/R17A, and HhaD48N were co-expressed with FLAG-tagged H-NS in an hns mutant strain of BL21 (DE3). With the exception of HhaD48N, all of the Hha mutants tested co-purified with H-NS (Fig. 5C).
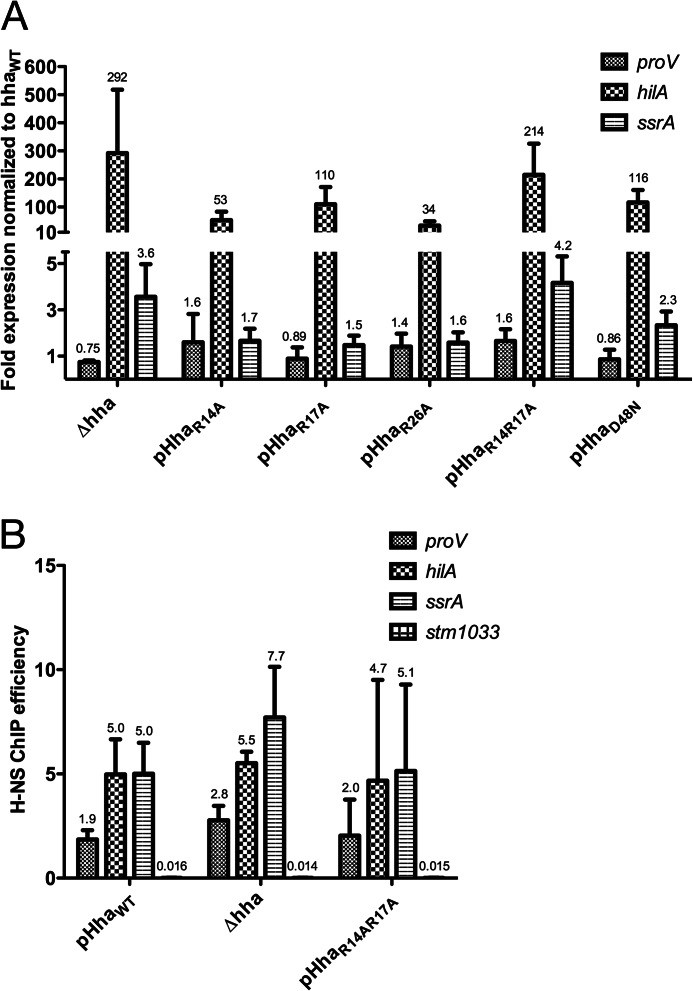
S. Typhimurium strains lacking Hha are characterized by a decreased growth rate and increased expression of several H-NS-repressed loci. The ability of the Hha surface-exposed arginine mutants to complement the Δhha defects was assessed by reverse transcription qPCR.
Transcript levels of proV, hilA, and ssrA from S. Typhimurium Δhha strains harboring plasmids expressing HhaWT, HhaR14A, HhaR17A, HhaR26A, HhaR14A/R17A, and HhaD48N under control of the native hha promoter were determined. All of the Hha mutants tested were deficient in silencing hilA but maintained HhaWT levels of proV, whereas ssrA was affected to varying extents (Fig. 6A). The HhaR14AR17A double mutant was most deficient in transcriptional silencing. hilA and ssrA expression were 214- and 4.2-fold up-regulated in the strain harboring pHhaR14A/R17A, relative to HhaWT levels, which are roughly comparable with expression levels in the Δhha strain harboring the empty plasmid control. It should be noted that the -fold expression values presented in Fig. 6A have been normalized to a Δhha strain complemented with HhaWT, whereas the values presented in Fig. 4A have been normalized to a Δhns strain complemented with H-NSWT, and this accounts for the different expression levels reported for Δhha in the two figures. These findings are in agreement with the proposed model whereby the positively charged surface of Hha stabilizes Hha/H-NS/DNA filaments and is therefore critical for transcriptional repression.
FIGURE 6.
Positively charged residues of the surface of Hha are critical for function. A, transcript analysis was performed on a Δhha strain of S. Typhimurium harboring an empty plasmid control (Δhha) or pHhaWT, pHhaR14A, pHhaR17A, pHhaR26A, or pHhaR14AR17A. Expression levels of proV, hilA, and ssrA were normalized to the Δhha strain harboring pHhaWT. All of the Hha variants tested resulted in significant up-regulation of hilA, whereas the expression profile of the HhaR14A/R17A double mutant most closely paralleled the Δhha strain. B, ChIP qPCR was performed with samples from the pHhaWT, Δhha, and pHhaR14A/R17A strains. H-NS enrichment at the proV, hilA, ssrA, and stm1033 promoter regions is expressed as ChIP efficiencies. Error bars, S.D. on three biological replicates. Significant differences between the ChIP efficiencies of H-NS from the pHhaWT, Δhha, and pHhaR14A/R17A strains were not noted.
If Hha provides H-NS with an additional DNA binding surface, we would expect to observe a decrease in H-NS DNA binding when the positively charged surface of Hha is mutated or in strains lacking Hha altogether. To further explore this model, we performed ChIP qPCR on samples taken from the same cultures used in the reverse transcription qPCR assay. H-NS enrichment at the proV, hilA, and ssrA promoter regions was measured and expressed as immunoprecipitation frequencies (Fig. 6B). Surprisingly, no significant changes in H-NS enrichment were observed between the strains expressing HhaWT, HhaR14A/R17A, and the empty plasmid control Δhha. The implications of these findings are discussed below.
Hha Enhances H-NS DNA Binding in Vitro
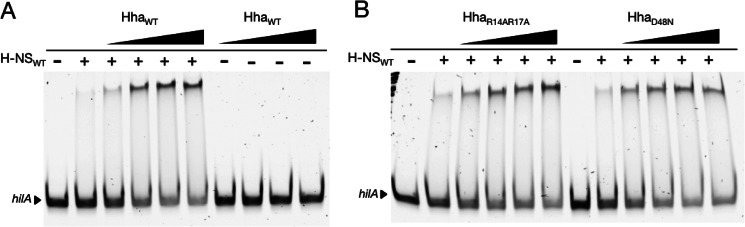
We tested the effect of Hha on the ability of H-NS to bind DNA in vitro by performing EMSAs. A linear 300-bp PCR product from the hilA promoter region was incubated with various concentrations of purified Hha and H-NS. As the concentration of Hha in the binding reactions was increased while the concentration of H-NS was held constant, the hilA fragment shifted to a lower mobility protein-DNA complex (Fig. 7A). The addition of Hha in the absence of H-NS did not affect the mobility of the hilA fragment. Therefore, we observe that Hha enhanced H-NS DNA binding in vitro but could not form stable protein-DNA complexes in the absence of H-NS. This enhancement was most noticeable when the concentration of H-NS was kept low (300 nm) and the salt component of the binding reactions was increased to 250 mm in order to lessen the DNA binding capability of H-NS (i.e. we notice no additional effect of Hha under conditions where H-NS has already fully shifted the promoter fragment). Notably, EMSAs performed with the purified Hha arginine mutants revealed that H-NS DNA binding to hilA was also enhanced by the addition of HhaR14A/R17A, although this mutant was completely defective for gene silencing in vivo (Fig. 7B).
FIGURE 7.
Hha augments H-NS DNA binding in vitro. A, electrophoretic mobility shift assays performed with a 300-bp PCR product from the hilA promoter region. No protein was added to the samples marked with minus signs. The plus signs signify 300 nm H-NS in the binding reaction. The addition of HhaWT to the samples indicated at concentrations of 300, 600, 900, and 1200 nm resulted in increased shifting of the hilA fragment. When HhaWT is combined with the hilA fragment in the absence of H-NS, no shifting is observed. B, EMSAs performed with HhaR14R17A and HhaD48N also result in improved shifting of the hilA fragment. The same protein concentrations from A were applied.
DISCUSSION
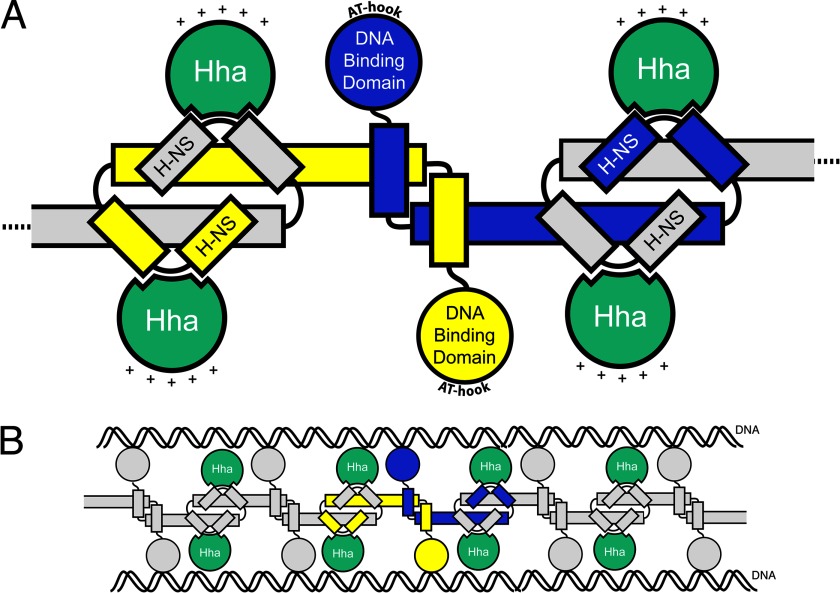
Recent structural studies have provided important insight into how H-NS-like proteins silence the expression of foreign sequences. We now know that H-NS binds AT-rich xenogeneic genes based on their inherent topographical distinctions from the host chromosome through a prokaryotic AT-hook motif located in a loop region of the H-NS C-terminal DNA binding domain (10). The manner in which H-NS self-associates was clarified by the crystal structure showing H-NS polymerized into extended superhelical filaments (4). These higher ordered H-NS filaments are thought to assemble along target sequences and repress transcription by mechanisms that remain unclear. One major question is how the Hha/YdgT co-repressors modulate H-NS activity through direct interactions with H-NS nucleoprotein complexes. In this study, we present the 3.2 Å crystal structure of Hha in complex with the N-terminal domain of H-NS. We identified a mutation in the N-terminal dimerization domain of H-NS (I11A) that abolishes the Hha/H-NS interaction while retaining other properties of H-NS required for normal repression of the proV locus. Replacing H-NSWT with H-NSI11A in S. Typhimurium results in a similar gene expression pattern as observed in the Δhha strain, suggesting that Hha mediates its effects on transcription primarily through its interaction with H-NS. This structure provides a plausible model of how Hha could assemble into an H-NS nucleoprotein filament (Fig. 8).
FIGURE 8.
Model of Hha interaction with H-NS. A, diagram of four Hha molecules (green) bound to two H-NS dimers. Monomers of the H-NS dimer are represented in yellow and blue. The AT-hook motif in the H-NS DNA binding domain interacts with the minor groove of target DNA sequences. The relative orientation of the positively charged surface of Hha is shown with plus signs. B, model for how Hha could be arranged in the H-NS nucleoprotein complex in “bridging” mode.
An electrostatic representation of the Hha solution structure shows an asymmetrical charge distribution that relates to specific functions. The predominantly basic surface of Hha points away from H-NS(1–46), whereas a combination of basic and acidic Hha residues interact with H-NS. de Alba et al. (52) identified residues Glu-25 and Asp-48 from Hha as critical for the Hha/H-NS interaction, and our structure confirms that these residues are located within the Hha/H-NS(1–46) interaction interface. A genetic screen of YdgT from E. coli, the Hha paralogue alternatively named Cnu, also implicated electrostatic interactions as the dominant attractive forces between Hha and H-NS (53). Mutagenesis of conserved basic residues on the surface of Hha opposite to the Hha/H-NS(1–46) interaction interface identified variants (HhaR14A, HhaR17A, HhaR26A, and HhaR14A/R17A) that retain the ability to interact with H-NS but are deficient in silencing hilA. Elimination of both Arg-14 and Arg-17 from Hha resulted in a higher -fold derepression of hilA than the additive effects of either single change. Our results demonstrate that positively charged residues of Hha that protrude from the surface of the Hha/H-NS(1–46) complex are essential for regulatory control and suggest that Hha could potentially provide an additional interaction surface for the nucleoprotein complex.
We consistently observe that mutations in Hha or point mutations in the dimerization domain of H-NS result in substantial up-regulation of hilA transcription but have little effect on H-NS occupancy at the same region in vivo when measured by ChIP. The lack of profound H-NS depletion at target promoters in the absence of Hha suggests that Hha is not required for global H-NS DNA binding in vivo. Our data instead point to a model where significant changes in gene expression can be directed by subtle changes in local H-NS occupancy or nucleoid structure. This model is consistent with data regarding Salmonella pagC gene expression, where it was observed that SlyA, a transcriptional activator, could co-occupy the pagC promoter region with H-NS during transcription (54). We propose that Hha facilitates the formation of repressive protein-DNA complexes with subtle features that cannot be detected by ChIP. It is possible that productive silencing complexes involve multiple interactions between Hha, H-NS, and DNA and that disrupting a single interaction is sufficient to abolish silencing without affecting binding to DNA per se. Features that cannot be discerned by ChIP include whether H-NS forms either bridges or filaments that have been detected by atomic force microscopy but currently have not been assigned a functional relevance in vivo (55–57). An alternative and not exclusive model is that Hha could alter the oligomerization properties of H-NS. By binding both H-NS monomers, Hha may provide additional stability to the N-terminal H-NS dimer and increase H-NS self-association. Such a model would explain why the in vitro DNA binding assays performed with purified components found that Hha can increase the ability of H-NS to bind DNA under a narrow range of conditions. Finally, it is possible that the fact that Hha facilitates H-NS binding in EMSAs does not point to the function of Hha in silencing and that Hha contributes to silencing in other ways, perhaps by interacting with RNA polymerase.
The structure of the Hha/H-NS(1–46) complex presented in this work confirms the anti-parallel arrangement of H-NS and shows that each H-NS dimer provides two Hha binding surfaces. The 1:1 ratio of Hha to H-NS is in line with our observations following gel filtration chromatography and Coomassie SDS-PAGE (data not shown). Our observations contradict a previous fluorescence anisotropy study that derived a model involving a 1:2 ratio of Hha to H-NS(1–64) (29). Although this study was performed with the construct H-NS(1–64), a separate report by the same group whereby Hha was titrated into 15N-labeled H-NS(1–46) also concluded that one molecule of Hha associates with each H-NS dimer (30). We also find there are no large scale structural conformation changes in either Hha or H-NS(1–46) upon complex formation, which contrasts with strong structural perturbations observed in the 1H-15N heteronuclear single quantum correlation spectra of Hha upon titration of H-NS(1–64) (29). The Hha residues whose signals were most perturbed included surface-exposed residues as well as hydrophobic residues deeply buried in the interfaces between helices 1 and 3 and helices 2 and 3. In contrast, an overlay of the Hha structure from our Hha/H-NS(1–46) complex with the unbound solution structure revealed significant overlap (r.m.s. deviation of 1.9 Å) and the absence of any major structural rearrangements.
More sensitive assays are required to discern the manner by which Hha modifies the silencing activity of H-NS, such as single molecule manipulations of DNA previously used to measure H-NS-induced DNA bridging (58). Such experiments would help address if the nucleoprotein structure at genes that require Hha for silencing differs from the structures observed at genes, such as proV, that are Hha-independent. Other outstanding questions that merit investigation include determining the regulatory signals that control Hha/YdgT expression and investigating the silencing properties of H-NS molecules from species, such as Vibrio and Bordetella, that do not encode the Hha/YdgT co-silencers.
The Navarre laboratory is supported by an Operating Grant and New Investigator Award from the Canada Institutes for Health Research (CIHR) (MOP-86683 and MSH-87729) and a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (RGPIN 386286-10). This work was also supported by CIHR Grant 13337 (to P. L. H.). The National Synchrotron Light Source Beam line X29 is supported by the United States Department of Energy Office of Biological and Environmental Research and the National Institutes of Health National Center for Research Resources.
The atomic coordinates and structure factors (code 4ICG) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SPI
- Salmonella pathogenicity island
- PDB
- Protein Data Bank
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- r.m.s.
- root mean square.
REFERENCES
- 1. Lawrence J. G., Ochman H. (1997) Amelioration of bacterial genomes. Rates of change and exchange. J. Mol. Evol. 44, 383–397 [DOI] [PubMed] [Google Scholar]
- 2. Groisman E. A., Ochman H. (1996) Pathogenicity islands. Bacterial evolution in quantum leaps. Cell 87, 791–794 [DOI] [PubMed] [Google Scholar]
- 3. Daubin V., Lerat E., Perrière G. (2003) The source of laterally transferred genes in bacterial genomes. Genome Biol. 4, R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Navarre W. W., Porwollik S., Wang Y., McClelland M., Rosen H., Libby S. J., Fang F. C. (2006) Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313, 236–238 [DOI] [PubMed] [Google Scholar]
- 5. Lucchini S., Rowley G., Goldberg M. D., Hurd D., Harrison M., Hinton J. C. (2006) H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2, e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grainger D. C., Hurd D., Goldberg M. D., Busby S. J. (2006) Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 34, 4642–4652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Castang S., McManus H. R., Turner K. H., Dove S. L. (2008) H-NS family members function coordinately in an opportunistic pathogen. Proc. Natl. Acad. Sci. U.S.A. 105, 18947–18952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gordon B. R., Imperial R., Wang L., Navarre W. W., Liu J. (2008) Lsr2 of Mycobacterium represents a novel class of H-NS-like proteins. J. Bacteriol. 190, 7052–7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oshima T., Ishikawa S., Kurokawa K., Aiba H., Ogasawara N. (2006) Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 13, 141–153 [DOI] [PubMed] [Google Scholar]
- 10. Gordon B. R., Li Y., Cote A., Weirauch M. T., Ding P., Hughes T. R., Navarre W. W., Xia B., Liu J. (2011) Structural basis for recognition of AT-rich DNA by unrelated xenogeneic silencing proteins. Proc. Natl. Acad. Sci. U.S.A. 108, 10690–10695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Navarre W. W., McClelland M., Libby S. J., Fang F. C. (2007) Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 21, 1456–1471 [DOI] [PubMed] [Google Scholar]
- 12. Badaut C., Williams R., Arluison V., Bouffartigues E., Robert B., Buc H., Rimsky S. (2002) The degree of oligomerization of the H-NS nucleoid structuring protein is related to specific binding to DNA. J. Biol. Chem. 277, 41657–41666 [DOI] [PubMed] [Google Scholar]
- 13. Stella S., Spurio R., Falconi M., Pon C. L., Gualerzi C. O. (2005) Nature and mechanism of the in vivo oligomerization of nucleoid protein H-NS. EMBO J. 24, 2896–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dame R. T., Wyman C., Goosen N. (2000) H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 28, 3504–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Spurio R., Falconi M., Brandi A., Pon C. L., Gualerzi C. O. (1997) The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 16, 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arold S. T., Leonard P. G., Parkinson G. N., Ladbury J. E. (2010) H-NS forms a superhelical protein scaffold for DNA condensation. Proc. Natl. Acad. Sci. U.S.A. 107, 15728–15732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shindo H., Ohnuki A., Ginba H., Katoh E., Ueguchi C., Mizuno T., Yamazaki T. (1999) Identification of the DNA binding surface of H-NS protein from Escherichia coli by heteronuclear NMR spectroscopy. FEBS Lett. 455, 63–69 [DOI] [PubMed] [Google Scholar]
- 18. Ueguchi C., Suzuki T., Yoshida T., Tanaka K., Mizuno T. (1996) Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J. Mol. Biol. 263, 149–162 [DOI] [PubMed] [Google Scholar]
- 19. Bouffartigues E., Buckle M., Badaut C., Travers A., Rimsky S. (2007) H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat. Struct. Mol. Biol. 14, 441–448 [DOI] [PubMed] [Google Scholar]
- 20. Baños R. C., Vivero A., Aznar S., García J., Pons M., Madrid C., Juárez A. (2009) Differential regulation of horizontally acquired and core genome genes by the bacterial modulator H-NS. PLoS Genet 5, e1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vivero A., Baños R. C., Mariscotti J. F., Oliveros J. C., García-del Portillo F., Juárez A., Madrid C. (2008) Modulation of horizontally acquired genes by the Hha-YdgT proteins in Salmonella enterica serovar Typhimurium. J. Bacteriol. 190, 1152–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fahlen T. F., Wilson R. L., Boddicker J. D., Jones B. D. (2001) Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 183, 6620–6629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Olekhnovich I. N., Kadner R. J. (2007) Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J. Bacteriol. 189, 6882–6890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Silphaduang U., Mascarenhas M., Karmali M., Coombes B. K. (2007) Repression of intracellular virulence factors in Salmonella by the Hha and YdgT nucleoid-associated proteins. J. Bacteriol. 189, 3669–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Coombes B. K., Wickham M. E., Lowden M. J., Brown N. F., Finlay B. B. (2005) Negative regulation of Salmonella pathogenicity island 2 is required for contextual control of virulence during typhoid. Proc. Natl. Acad. Sci. U.S.A. 102, 17460–17465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paytubi S., Madrid C., Forns N., Nieto J. M., Balsalobre C., Uhlin B. E., Juárez A. (2004) YdgT, the Hha paralogue in Escherichia coli, forms heteromeric complexes with H-NS and StpA. Mol. Microbiol. 54, 251–263 [DOI] [PubMed] [Google Scholar]
- 27. Nieto J. M., Madrid C., Prenafeta A., Miquelay E., Balsalobre C., Carrascal M., Juárez A. (2000) Expression of the hemolysin operon in Escherichia coli is modulated by a nucleoid-protein complex that includes the proteins Hha and H-NS. Mol. Gen. Genet 263, 349–358 [DOI] [PubMed] [Google Scholar]
- 28. Nieto J. M., Madrid C., Miquelay E., Parra J. L., Rodríguez S., Juárez A. (2002) Evidence for direct protein-protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of proteins. J. Bacteriol. 184, 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. García J., Cordeiro T. N., Nieto J. M., Pons I., Juárez A., Pons M. (2005) Interaction between the bacterial nucleoid associated proteins Hha and H-NS involves a conformational change of Hha. Biochem. J. 388, 755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. García J., Madrid C., Juárez A., Pons M. (2006) New roles for key residues in helices H1 and H2 of the Escherichia coli H-NS N-terminal domain. H-NS dimer stabilization and Hha binding. J. Mol. Biol. 359, 679–689 [DOI] [PubMed] [Google Scholar]
- 31. García J., Madrid C., Cendra M., Juárez A., Pons M. (2009) N9L and L9N mutations toggle Hha binding and hemolysin regulation by Escherichia coli and Vibrio cholerae H-NS. FEBS Lett. 583, 2911–2916 [DOI] [PubMed] [Google Scholar]
- 32. Olekhnovich I. N., Kadner R. J. (2006) Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enterica. J. Mol. Biol. 357, 373–386 [DOI] [PubMed] [Google Scholar]
- 33. Sharma V. K., Zuerner R. L. (2004) Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 186, 7290–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Takeshita S., Sato M., Toba M., Masahashi W., Hashimoto-Gotoh T. (1987) High-copy-number and low-copy-number plasmid vectors for lacZ α-complementation and chloramphenicol- or kanamycin-resistance selection. Gene 61, 63–74 [DOI] [PubMed] [Google Scholar]
- 35. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 36. Pape T., Schneider T. R. (2004) HKL2MAP. A graphical user interface for phasing with SHELX programs. J. Appl. Crystallogr. 37, 843–844 [Google Scholar]
- 37. Terwilliger T. C., Berendzen J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 39. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Strong M., Sawaya M. R., Wang S., Phillips M., Cascio D., Eisenberg D. (2006) Toward the structural genomics of complexes. Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 42. Pons J. I., Rodríguez S., Madrid C., Juárez A., Nieto J. M. (2004) In vivo increase of solubility of overexpressed Hha protein by tandem expression with interacting protein H-NS. Protein Expr. Purif. 35, 293–297 [DOI] [PubMed] [Google Scholar]
- 43. Yee A., Chang X., Pineda-Lucena A., Wu B., Semesi A., Le B., Ramelot T., Lee G. M., Bhattacharyya S., Gutierrez P., Denisov A., Lee C. H., Cort J. R., Kozlov G., Liao J., Finak G., Chen L., Wishart D., Lee W., McIntosh L. P., Gehring K., Kennedy M. A., Edwards A. M., Arrowsmith C. H. (2002) An NMR approach to structural proteomics. Proc. Natl. Acad. Sci. U.S.A. 99, 1825–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bloch V., Yang Y., Margeat E., Chavanieu A., Augé M. T., Robert B., Arold S., Rimsky S., Kochoyan M. (2003) The H-NS dimerization domain defines a new fold contributing to DNA recognition. Nat. Struct. Biol. 10, 212–218 [DOI] [PubMed] [Google Scholar]
- 45. Esposito D., Petrovic A., Harris R., Ono S., Eccleston J. F., Mbabaali A., Haq I., Higgins C. F., Hinton J. C., Driscoll P. C., Ladbury J. E. (2002) H-NS oligomerization domain structure reveals the mechanism for high order self-association of the intact protein. J. Mol. Biol. 324, 841–850 [DOI] [PubMed] [Google Scholar]
- 46. Cerdan R., Bloch V., Yang Y., Bertin P., Dumas C., Rimsky S., Kochoyan M., Arold S. T. (2003) Crystal structure of the N-terminal dimerisation domain of VicH, the H-NS-like protein of Vibrio cholerae. J. Mol. Biol. 334, 179–185 [DOI] [PubMed] [Google Scholar]
- 47. Takagi S., Masuoka K., Uchida N., Ishiwata K., Araoka H., Tsuji M., Yamamoto H., Kato D., Matsuhashi Y., Kusumi E., Ota Y., Seo S., Matsumura T., Matsuno N., Wake A., Miyakoshi S., Makino S., Ohashi K., Yoneyama A., Taniguchi S. (2009) High incidence of haemophagocytic syndrome following umbilical cord blood transplantation for adults. Br. J. Haematol. 147, 543–553 [DOI] [PubMed] [Google Scholar]
- 48. Bae S. H., Liu D., Lim H. M., Lee Y., Choi B. S. (2008) Structure of the nucleoid-associated protein Cnu reveals common binding sites for H-NS in Cnu and Hha. Biochemistry 47, 1993–2001 [DOI] [PubMed] [Google Scholar]
- 49. Smyth C. P., Lundbäck T., Renzoni D., Siligardi G., Beavil R., Layton M., Sidebotham J. M., Hinton J. C., Driscoll P. C., Higgins C. F., Ladbury J. E. (2000) Oligomerization of the chromatin-structuring protein H-NS. Mol. Microbiol. 36, 962–972 [DOI] [PubMed] [Google Scholar]
- 50. Zhou Y., Gottesman S. (2006) Modes of regulation of RpoS by H-NS. J. Bacteriol. 188, 7022–7025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ueguchi C., Kakeda M., Mizuno T. (1993) Autoregulatory expression of the Escherichia coli hns gene encoding a nucleoid protein. H-NS functions as a repressor of its own transcription. Mol. Gen. Genet 236, 171–178 [DOI] [PubMed] [Google Scholar]
- 52. de Alba C. F., Solórzano C., Paytubi S., Madrid C., Juarez A., García J., Pons M. (2011) Essential residues in the H-NS binding site of Hha, a co-regulator of horizontally acquired genes in Enterobacteria. FEBS Lett. 585, 1765–1770 [DOI] [PubMed] [Google Scholar]
- 53. Yun S. H., Ji S. C., Jeon H. J., Wang X., Lee Y., Choi B. S., Lim H. M. (2012) A mutational study of Cnu reveals attractive forces between Cnu and H-NS. Mol. Cells 33, 211–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Perez J. C., Latifi T., Groisman E. A. (2008) Overcoming H-NS-mediated transcriptional silencing of horizontally acquired genes by the PhoP and SlyA proteins in Salmonella enterica. J. Biol. Chem. 283, 10773–10783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dame R. T., Wyman C., Goosen N. (2001) Structural basis for preferential binding of H-NS to curved DNA. Biochimie 83, 231–234 [DOI] [PubMed] [Google Scholar]
- 56. Dame R. T., Wyman C., Wurm R., Wagner R., Goosen N. (2002) Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J. Biol. Chem. 277, 2146–2150 [DOI] [PubMed] [Google Scholar]
- 57. Dame R. T., Luijsterburg M. S., Krin E., Bertin P. N., Wagner R., Wuite G. J. (2005) DNA bridging. A property shared among H-NS-like proteins. J. Bacteriol. 187, 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dame R. T. (2008) Single-molecule micromanipulation studies of DNA and architectural proteins. Biochem. Soc. Trans. 36, 732–737 [DOI] [PubMed] [Google Scholar]
- 59. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity. All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Papadopoulos J. S., Agarwala R. (2007) COBALT. Constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079 [DOI] [PubMed] [Google Scholar]
- 61. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009) Jalview Version 2. A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ali S. S., Beckett E., Bae S. J., Navarre W. W. (2011) The 5.5 protein of phage T7 inhibits H-NS through interactions with the central oligomerization domain. J. Bacteriol. 193, 4881–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]