Abstract
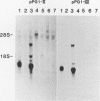
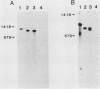
We have examined genomic sequences and mRNA species hybridizing to a cDNA clone of a yolk sac carcinoma chondroitin sulfate proteoglycan designated PG19. Genomic blot hybridizations with cDNAs covering the majority of the PG19 mRNA sequence revealed 15 to 17 gene fragments. Similar analysis with probes representing either the propeptide or the combined core protein COOH-terminal domain and 3' untranslated sequences revealed single genomic fragments indicating that a single gene codes for the PG19 proteoglycan. Genomic blot analysis with cDNA sequences coding for the serine-glycine repeat of the core protein identified the same gene fragments observed with the entire PG19 cDNA, indicating that this coding region is homologous with sequences present in multiple genes. The same probes were also used to examine mRNA expression. In addition to the PG19 mRNA, several PG19-related mRNAs could be seen. These PG19-related mRNAs had homology with the serine-glycine coding sequence of the PG19 cDNA. These mRNAs may be coding for proteoglycans. The mRNA coding for PG19 appeared to be uniquely expressed in parietal yolk sac and mast cell lineages. The PG19 mRNA existed in different forms in parietal yolk sac and mast cell lines due to cell-type-specific differences in the length of the 5' untranslated sequences. These results indicate that expression of the PG19 proteoglycan gene is regulated both in terms of cell-type-specific transcription and selection of a transcriptional start site.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alfonso-Pizarro A., Carlson D. P., Ross J. Subcellular localization of RNAs in transfected cells: role of sequences at the 5' terminus. Nucleic Acids Res. 1984 Nov 26;12(22):8363–8380. doi: 10.1093/nar/12.22.8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan M., Lanyon W. G., Paul J. Multiple origins of transcription in the 4.5 Kb upstream of the epsilon-globin gene. Cell. 1983 Nov;35(1):187–197. doi: 10.1016/0092-8674(83)90221-0. [DOI] [PubMed] [Google Scholar]
- Axelsson I., Heinegård D. Characterization of chondroitin sulfate-rich proteoglycans from bovine corneal stroma. Exp Eye Res. 1980 Jul;31(1):57–66. doi: 10.1016/0014-4835(80)90090-1. [DOI] [PubMed] [Google Scholar]
- Benyajati C., Spoerel N., Haymerle H., Ashburner M. The messenger RNA for alcohol dehydrogenase in Drosophila melanogaster differs in its 5' end in different developmental stages. Cell. 1983 May;33(1):125–133. doi: 10.1016/0092-8674(83)90341-0. [DOI] [PubMed] [Google Scholar]
- Bos J. L., Verlaan-de Vries M., Jansen A. M., Veeneman G. H., van Boom J. H., van der Eb A. J. Three different mutations in codon 61 of the human N-ras gene detected by synthetic oligonucleotide hybridization. Nucleic Acids Res. 1984 Dec 11;12(23):9155–9163. doi: 10.1093/nar/12.23.9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon M. A., Oldberg A., Pierschbacher M., Ruoslahti E. Molecular cloning and sequence analysis of a chondroitin sulfate proteoglycan cDNA. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1321–1325. doi: 10.1073/pnas.82.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon M. A., Shiga M., Ruoslahti E. Identification from cDNA of the precursor form of a chondroitin sulfate proteoglycan core protein. J Biol Chem. 1986 Sep 25;261(27):12534–12537. [PubMed] [Google Scholar]
- Brennan M. J., Oldberg A., Hayman E. G., Ruoslahti E. Effect of a proteoglycan produced by rat tumor cells on their adhesion to fibronectin-collagen substrata. Cancer Res. 1983 Sep;43(9):4302–4307. [PubMed] [Google Scholar]
- Brennan M. J., Oldberg A., Pierschbacher M. D., Ruoslahti E. Chondroitin/dermatan sulfate proteoglycan in human fetal membranes. Demonstration of an antigenically similar proteoglycan in fibroblasts. J Biol Chem. 1984 Nov 25;259(22):13742–13750. [PubMed] [Google Scholar]
- Couchman J. R., Woods A., Hök M., Christner J. E. Characterization of a dermatan sulfate proteoglycan synthesized by murine parietal yolk sac (PYS-2) cells. J Biol Chem. 1985 Nov 5;260(25):13755–13762. [PubMed] [Google Scholar]
- Culp L. A., Murray B. A., Rollins B. J. Fibronectin and proteoglycans as determinants of cell-substratum adhesion. J Supramol Struct. 1979;11(3):401–427. doi: 10.1002/jss.400110314. [DOI] [PubMed] [Google Scholar]
- Didsbury J. R., Theil E. C., Kaufman R. E., Dickey L. F. Multiple red cell ferritin mRNAs, which code for an abundant protein in the embryonic cell type, analyzed by cDNA sequence and by primer extension of the 5'-untranslated regions. J Biol Chem. 1986 Jan 15;261(2):949–955. [PubMed] [Google Scholar]
- Doege K., Fernandez P., Hassell J. R., Sasaki M., Yamada Y. Partial cDNA sequence encoding a globular domain at the C terminus of the rat cartilage proteoglycan. J Biol Chem. 1986 Jun 25;261(18):8108–8111. [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Fransson L. A., Carlstedt I., Cöster L., Malmström A. Binding of transferrin to the core protein of fibroblast proteoheparan sulfate. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5657–5661. doi: 10.1073/pnas.81.18.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg D. A. Isolation and partial characterization of the Drosophila alcohol dehydrogenase gene. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5794–5798. doi: 10.1073/pnas.77.10.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindlay G. J., Lanyon W. G., Allan M., Paul J. Alternative sites of transcription initiation upstream of the canonical cap site in human gamma-globin and beta-globin genes. Nucleic Acids Res. 1984 Feb 24;12(4):1811–1820. doi: 10.1093/nar/12.4.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultmark D., Klemenz R., Gehring W. J. Translational and transcriptional control elements in the untranslated leader of the heat-shock gene hsp22. Cell. 1986 Feb 14;44(3):429–438. doi: 10.1016/0092-8674(86)90464-2. [DOI] [PubMed] [Google Scholar]
- Jackson F. R., Bargiello T. A., Yun S. H., Young M. W. Product of per locus of Drosophila shares homology with proteoglycans. Nature. 1986 Mar 13;320(6058):185–188. doi: 10.1038/320185a0. [DOI] [PubMed] [Google Scholar]
- Kanwar Y. S., Farquhar M. G. Presence of heparan sulfate in the glomerular basement membrane. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1303–1307. doi: 10.1073/pnas.76.3.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koda J. E., Bernfield M. Heparan sulfate proteoglycans from mouse mammary epithelial cells. Basal extracellular proteoglycan binds specifically to native type I collagen fibrils. J Biol Chem. 1984 Oct 10;259(19):11763–11770. [PubMed] [Google Scholar]
- Krusius T., Ruoslahti E. Primary structure of an extracellular matrix proteoglycan core protein deduced from cloned cDNA. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7683–7687. doi: 10.1073/pnas.83.20.7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrogan M., Simonsen C. C., Smouse D. T., Farnham P. J., Schimke R. T. Heterogeneity at the 5' termini of mouse dihydrofolate reductase mRNAs. Evidence for multiple promoter regions. J Biol Chem. 1985 Feb 25;260(4):2307–2314. [PubMed] [Google Scholar]
- McKnight S. L., Gavis E. R., Kingsbury R., Axel R. Analysis of transcriptional regulatory signals of the HSV thymidine kinase gene: identification of an upstream control region. Cell. 1981 Aug;25(2):385–398. doi: 10.1016/0092-8674(81)90057-x. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema T. R., Jr, Hascall V. C., Eisenstein R. Characterization of bovine aorta proteoglycan extracted with guanidine hydrochloride in the presence of protease inhibitors. J Biol Chem. 1979 Feb 25;254(4):1312–1318. [PubMed] [Google Scholar]
- Oldberg A., Hayman E. G., Ruoslahti E. Isolation of a chondroitin sulfate proteoglycan from a rat yolk sac tumor and immunochemical demonstration of its cell surface localization. J Biol Chem. 1981 Nov 10;256(21):10847–10852. [PubMed] [Google Scholar]
- Oldberg A., Linney E., Ruoslahti E. Molecular cloning and nucleotide sequence of a cDNA clone coding for the cell attachment domain in human fibronectin. J Biol Chem. 1983 Sep 10;258(17):10193–10196. [PubMed] [Google Scholar]
- Oldberg A., Ruoslahti E. Interactions between chondroitin sulfate proteoglycan, fibronectin, and collagen. J Biol Chem. 1982 May 10;257(9):4859–4863. [PubMed] [Google Scholar]
- Parthasarathy N., Spiro R. G. Characterization of the glycosaminoglycan component of the renal glomerular basement membrane and its relationship to the peptide portion. J Biol Chem. 1981 Jan 10;256(1):507–513. [PubMed] [Google Scholar]
- Radhakrishnamurthy B., Smart F., Dalferes E. R., Jr, Berenson G. S. Isolation and characterization of proteoglycans from bovine lung. J Biol Chem. 1980 Aug 25;255(16):7575–7582. [PubMed] [Google Scholar]
- Reddy P., Jacquier A. C., Abovich N., Petersen G., Rosbash M. The period clock locus of D. melanogaster codes for a proteoglycan. Cell. 1986 Jul 4;46(1):53–61. doi: 10.1016/0092-8674(86)90859-7. [DOI] [PubMed] [Google Scholar]
- Robinson H. C., Horner A. A., Hök M., Ogren S., Lindahl U. A proteoglycan form of heparin and its degradation to single-chain molecules. J Biol Chem. 1978 Oct 10;253(19):6687–6693. [PubMed] [Google Scholar]
- Robinson J., Viti M., Hök M. Structure and properties of an under-sulfated heparan sulfate proteoglycan synthesized by a rat hepatoma cell line. J Cell Biol. 1984 Mar;98(3):946–953. doi: 10.1083/jcb.98.3.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai S., Tanaka T., Kosher R. A., Tanzer M. L. Cloning and sequence analysis of a partial cDNA for chicken cartilage proteoglycan core protein. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5081–5085. doi: 10.1073/pnas.83.14.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakashita S., Engvall E., Ruoslahti E. Basement membrane glycoprotein laminin binds to heparin. FEBS Lett. 1980 Jul 28;116(2):243–246. doi: 10.1016/0014-5793(80)80654-5. [DOI] [PubMed] [Google Scholar]
- Sant A. J., Cullen S. E., Giacoletto K. S., Schwartz B. D. Invariant chain is the core protein of the Ia-associated chondroitin sulfate proteoglycan. J Exp Med. 1985 Dec 1;162(6):1916–1934. doi: 10.1084/jem.162.6.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H. S., Bargiello T. A., Clark B. T., Jackson F. R., Young M. W. An unusual coding sequence from a Drosophila clock gene is conserved in vertebrates. Nature. 1985 Oct 3;317(6036):445–448. doi: 10.1038/317445a0. [DOI] [PubMed] [Google Scholar]
- Singer R. H., Penman S. Messenger RNA in HeLa cells: kinetics of formation and decay. J Mol Biol. 1973 Aug 5;78(2):321–334. doi: 10.1016/0022-2836(73)90119-8. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Suzuki S., Pierschbacher M. D., Hayman E. G., Nguyen K., Ohgren Y., Ruoslahti E. Domain structure of vitronectin. Alignment of active sites. J Biol Chem. 1984 Dec 25;259(24):15307–15314. [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou A. P., Lai C., Danielson P., Noonan D. J., Sutcliffe J. G. Structural characterization of a heterogeneous family of rat brain mRNAs. Mol Cell Biol. 1986 Mar;6(3):768–778. doi: 10.1128/mcb.6.3.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich A., Shine J., Chirgwin J., Pictet R., Tischer E., Rutter W. J., Goodman H. M. Rat insulin genes: construction of plasmids containing the coding sequences. Science. 1977 Jun 17;196(4296):1313–1319. doi: 10.1126/science.325648. [DOI] [PubMed] [Google Scholar]
- Van Beveren C., van Straaten F., Curran T., Müller R., Verma I. M. Analysis of FBJ-MuSV provirus and c-fos (mouse) gene reveals that viral and cellular fos gene products have different carboxy termini. Cell. 1983 Apr;32(4):1241–1255. doi: 10.1016/0092-8674(83)90306-9. [DOI] [PubMed] [Google Scholar]
- Woodley D. T., Rao C. N., Hassell J. R., Liotta L. A., Martin G. R., Kleinman H. K. Interactions of basement membrane components. Biochim Biophys Acta. 1983 Dec 27;761(3):278–283. doi: 10.1016/0304-4165(83)90077-6. [DOI] [PubMed] [Google Scholar]
- Young R. A., Hagenbüchle O., Schibler U. A single mouse alpha-amylase gene specifies two different tissue-specific mRNAs. Cell. 1981 Feb;23(2):451–458. doi: 10.1016/0092-8674(81)90140-9. [DOI] [PubMed] [Google Scholar]