Background: The role of Smad-independent TGF-β signaling in craniofacial development is poorly elucidated.
Results: In craniofacial mesenchymal cells, Tak1 regulates both R-Smad C-terminal and linker region phosphorylation in TGF-β signaling.
Conclusion: Tak1 plays an irreplaceable role in craniofacial ecto-mesenchyme during embryogenesis.
Significance: Understanding the mechanisms of TGF-β signaling contributes to knowledge of pathogenetic mechanisms underlying common craniofacial birth defects.
Keywords: Bone Morphogenetic Protein (BMP), Craniofacial Development, Embryo, SMAD Transcription Factor, Transforming Growth Factor beta (TGFbeta), Craniofacial Birth Defects, Linker Region Phosphorylation
Abstract
Although the importance of TGF-β superfamily signaling in craniofacial growth and patterning is well established, the precise details of its signaling mechanisms are still poorly understood. This is in part because of the concentration of studies on the role of the Smad-dependent (so-called “canonical”) signaling pathways relative to the Smad-independent ones in many biological processes. Here, we have addressed the role of TGF-β-activated kinase 1 (Tak1, Map3k7), one of the key mediators of Smad-independent (noncanonical) TGF-β superfamily signaling in craniofacial development, by deleting Tak1 specifically in the neural crest lineage. Tak1-deficient mutants display a round skull, hypoplastic maxilla and mandible, and cleft palate resulting from a failure of palatal shelves to appropriately elevate and fuse. Our studies show that in neural crest-derived craniofacial ecto-mesenchymal cells, Tak1 is not only required for TGF-β- and bone morphogenetic protein-induced p38 Mapk activation but also plays a role in agonist-induced C-terminal and linker region phosphorylation of the receptor-mediated R-Smads. Specifically, we demonstrate that the agonist-induced linker region phosphorylation of Smad2 at Thr-220, which has been shown to be critical for full transcriptional activity of Smad2, is dependent on Tak1 activity and that in palatal mesenchymal cells TGFβRI and Tak1 kinases mediate both overlapping and distinct TGF-β2-induced transcriptional responses. To summarize, our results suggest that in neural crest-derived ecto-mesenchymal cells, Tak1 provides a critical point of intersection in a complex dialogue between the canonical and noncanonical arms of TGF-β superfamily signaling required for normal craniofacial development.
Introduction
Members of the TGF-β superfamily are involved in many normal and pathologic events including development, inflammation, fibrosis, and cancer (1). TGF-βs and bone morphogenetic proteins (BMPs)3 form two important subgroups within the superfamily (2, 3). They bind to largely subfamily-specific membrane-spanning complexes of Type I and Type 2 receptors resulting in activation of components of intracellular Smad-dependent and Smad-independent pathways. In the Smad-dependent pathway, receptor-regulated Smads (R-Smads) become phosphorylated at their C termini via type I receptor kinase activity, complex with Smad4, and accumulate in the nucleus where they act as transcriptional co-regulators (2, 3). Initially it was thought that TGF-βs signal exclusively through TGF-β type I and II receptors (TGFβRI and TGFβRII) to activate TGF-β R-Smads 2 and 3 and similarly BMPs signal through specific Type I and II receptors to activate BMP R-Smads 1, 5, and 8 (4). However, a recent report showed that at least in certain specific cell lines, TGF-βs can also signal via mixed receptor complexes, resulting in activation of BMP R-Smads (5).
In addition to C-terminal phosphorylation at a conserved SSXS sequence, more recent research has shown that R-Smads can also be phosphorylated in the linker region between their Mad homology domains, which is important for both full activation and cessation of activation (6, 7). Specific residues become phosphorylated as the result of distinct pathways: antagonists, such as EGF, which enable the activated R-Smads to remain in the cytoplasm and be degraded (8); and agonist-induced linker phosphorylation that is required for maximal transcriptional activity of R-Smad-Smad4 complexes, as well as for rapid turnover of corresponding R-Smads (7, 9). In this “action turnover switch” model, the Thr/Ser residues adjacent to proline-rich sequences in R-Smads (Thr-220 in Smad2) are phosphorylated by nuclear Cdks creating a docking site for transcriptional co-regulators (Pin1 in the case of TGF-β R-Smads and Yap in the case of Bmp R-Smads). Moreover, it has been suggested that this phosphorylation may enable subsequent phosphorylation of other linker region residues, such as Ser-250 in Smad2, providing a docking site for ubiquitin ligases, such as Nedd4L, targeting R-Smads for proteasome-mediated degradation (9, 10).
In addition to Smad-dependent (canonical) TGF-β/BMP signaling, ligands in the TGF-β superfamily can also activate Smad-independent (noncanonical) pathways, resulting in activation of the Mapk signaling (4). A critical step in initiation of the noncanonical pathway is activation of TGF-β activated kinase-1 (Tak1, Map3k7) by Traf6-mediated polyubiquitination (11, 12). Other studies have shown that Tak1 is required for fine-tuning of BMP effects during bone development, as well as for TGF-β-induced NF-κB and JNK activation, and also mediates cytokine-induced Ikk activation (13–15).
Although the canonical pathway tends to be treated as the more important, recent studies suggest that the Smad-independent TGF-β signaling plays a more significant role in vivo than previously thought (16, 17). In a mouse model of Marfan's syndrome in which Tgf-β signaling is elevated, the aortic aneurysm phenotypes are made worse by reducing the canonical pathway component Smad4, and the phenotypes are made less severe by attenuation of noncanonical pathways (16). These results show not only that the noncanonical pathway is of importance in disease mechanisms but also that the two pathways somehow interact with one another. Mutations in TGF-β type I and type II receptors (TGFBR1 and TGFBR2) associated with Marfan's syndrome and Loeys-Dietz syndrome lead to an increase in TGF-β signaling activity in the absence of normal canonical signaling (18). The importance of maintaining the correct balance between the pathways for normal developmental events is implied by the presence of craniofacial defects including cleft palate in patients suffering from Loeys-Dietz syndrome (19) and in a mouse model when Tgfbr2 is genetically removed from the neural crest cell population (which forms the principal source of mesenchymal cells in the secondary palate precursor structures, the palatal shelves), resulting in cleft palate in the presence of excessive TGF-β-induced noncanonical signaling (17).
In light of the important role played by the Smad-independent pathway implied by these studies, as well as the well established importance on Tgf-β and Bmp signaling in normal craniofacial development in mouse models (17), we examined the role of Tak1 in craniofacial neural crest development by deleting Tak1 function in premigratory neural crest cells using the Wnt1-Cre driver line. The mutant mice display hypoplastic facial structures and cleft palate, which is caused by a delayed palatal shelf elevation. We found that Tak1 is required for appropriate activation of both p38 Mapk (noncanonical pathway) and TGF-β/Bmp R-Smads (canonical pathway) in the neural crest-derived craniofacial ecto-mesenchyme. We also show that Tak1 deficiency results in attenuated TGF-β R-Smad linker region phosphorylation and that Tak1 kinase mediates both distinct and overlapping agonist-induced transcriptional responses. Collectively, these results imply that in neural crest-derived mesenchymal cells, Tak1 mediates both canonical and noncanonical arms of the TGF-β superfamily signaling.
EXPERIMENTAL PROCEDURES
Mice
Tak1-flox mice were generated by flanking a critical exon 2 with asymmetric loxP sites (Lox66 and Lox71) in opposing orientations (see Fig. 1; details will be described elsewhere). Wnt1-Cre (from the Jackson Laboratories) and Tgfbr1FX (kindly provided by S. Karlsson) mice have been described earlier (20, 21). Tak1FX+/Wnt1-Cre+ male mice were crossed with Tak1FXFX female mice to obtain timed pregnancies. The presence of a vaginal plug was designated as embryonic day 0 (E0). DNA for genotyping was prepared from yolk sac or from tail tissue using DirectPCR lysis reagents (Viagen Biotech). Mouse lines were maintained in mixed genetic backgrounds. All experiments involving the use of animals were approved by the Institutional Animal Use and Care Committee at the University of Michigan at Ann Arbor.
FIGURE 1.
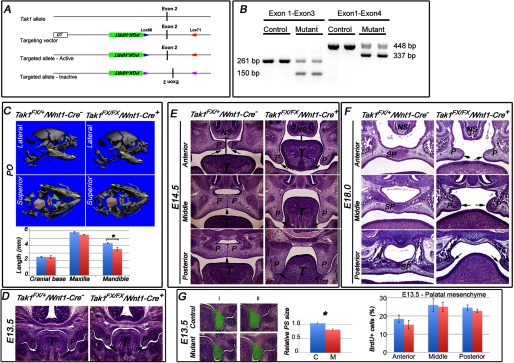
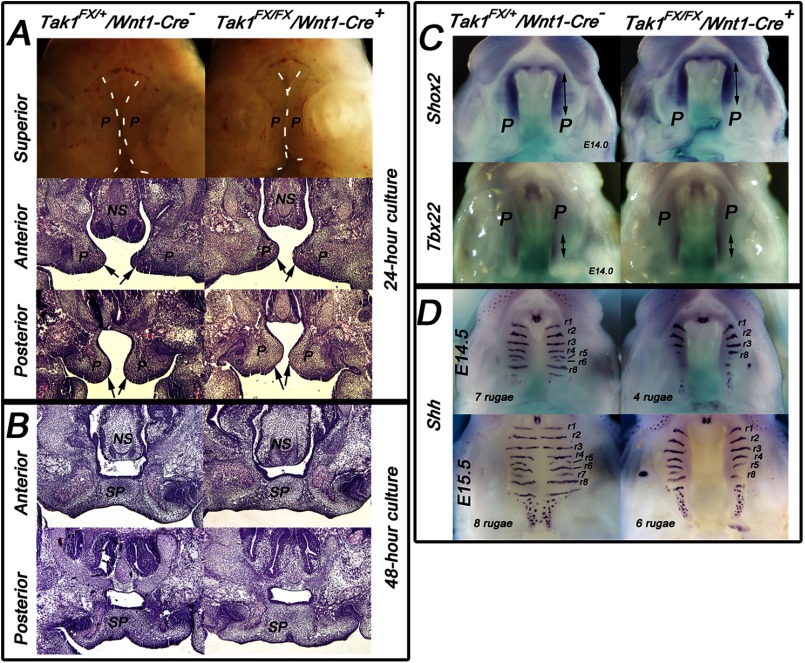
Deletion of Tak1 in neural crest cells leads to mandibular hypoplasia and cleft palate. A, schematic presentation of the gene-targeting strategy. “Flipping” of exon 2, which encodes the kinase domain of Tak1, causes alternative splicing between exons 1 and 3 (B) inactivating the function of Tak1. B, RT-PCR analysis of RNA samples harvested from prefusion (at E14) palatal shelves of control (Tak1FX/+/Wnt1-Cre−) and mutant (Tak1FX/FX/Wnt1-Cre+) embryos. Left panel, exon 1-specific sense and exon 3-specific antisense primers result in amplification of 261-bp wild-type and 150-bp mutated PCR products, respectively. Exon 1-specific sense and exon 4-specific antisense primers result in amplification of 448-bp wild-type and 337-bp mutated PCR products, respectively. C, lateral and superior μCT images of control (Tak1FX/+/Wnt1-Cre−) and mutant (Tak1FX/FX/Wnt1-Cre+) heads at P0. The yellow arrows in the lateral images depict the length of the mandible. The yellow arrows in the superior views depict the length of the maxilla. The red arrows in the superior images depict the length of the cranial base, and the green arrows illustrate palatine bones in a control (fused palate) and mutant (cleft palate). The bar graph shows lengths of the cranial base, maxilla, and mandible of controls (blue columns) and mutants (red columns) (n = 3). D–F, histological comparison of palatal phenotypes of controls and Tak1/Wnt1-Cre mutants: frontal sections at anterior, middle, and posterior levels at E14.5 (E), E18.0 (F), and the level of the first molar at E13.5 (D). The black arrows point to tips of palatal shelves, the black arrowheads point to the midline seam (in a control), and the black vertical lines depict the distance between the tongue and nasal septum. G, left-hand panel shows sections from two independent control and mutant samples at the level of the first mandibular molar. The green highlighting illustrates the area measured to compare palatal shelf size between controls and mutants. Middle panel, bar graph shows relative quantification of palatal shelf (PS) size in controls (C, blue columns) and mutants (M, red columns) (n = 3). Right panel, bar graph summarizes results from BrdU incorporation assays on controls (blue columns) and Tak1/Wnt1-Cre mutants (red columns) palatal shelves at E13.5 (n = 4). Error bars, S.E. *, p < 0.05. NS, nasal septum; P, palatal shelf; SP, secondary palate.
Genotyping
Tak1FXFX mice were genotyped by PCR using the following primer sequences (annealing at 60 °C): Tak1FX sense: 5′-gataccttacactggggacca-3′ and Tak1FX antisense 5′-ggcattcagttgtggagcatt-3′. Wnt1-Cre and Tgfbr1FX mice were genotyped as previously described (20, 21).
Conventional RT-PCR
To assess the recombination efficiency of the floxed Tak1 locus, the total RNAs were isolated from prefusion palatal shelves harvested at E14.0 or maxillary and mandibular first pharyngeal arch at E11 (RNeasy mini kit; Qiagen), and cDNAs synthesized (Omniscript reverse transcriptase, Qiagen) according to manufacturer's protocols. The following primers were used: Tak1 exon 1-specific sense primer, 5′-GGGGATCATGTCGACAGCCTC-3′; Tak1 exon 3-specific antisense primer, 5′-GTTCACACGTGACAACTGCCG-3′; and Tak1 exon 4-specific antisense primer, 5′-GCATGCTGTGCAGGTAAGCCA-3′. β-Actin was used as a quality and loading control: sense primer, 5′-GTGGGCCGGTCTAGGCACCAA-3′; and antisense primer, 5′CGGTTGCCTTAGGGTTCA-GG-3′.
Real Time Quantitative PCR
Total RNAs were isolated, and cDNAs were synthesized as outlined above. Real time quantitative PCR experiments were carried out using Universal Probe Library-based assays (Roche Applied Science) with gene-specific primer sequences generated by the manufacturer's online algorithm and TaqMan Universal PCR master mix (Applied Biosystems). 30-μl assays were quantified using the ABI7300 PCR and detection system (Applied Biosystems) and analyzed using 7500 System v1.2.2 software.
Histology, in Situ Hybridization, Cell Death, and Proliferation Assays
For histological analyses, the tissues were processed, sectioned, and stained by hematoxylin and eosin according to standard protocols. For whole mount in situ hybridization, the tissues were fixed in 4% buffered formaldehyde for 12–16 h and dehydrated though a graded methanol series (20, 50, 70, 95, and 100%) containing PBST. Antisense RNA probes were synthesized with NTP digoxigenin RNA labeling mix (Roche Applied Science) following the manufacturer's instructions. Probe templates for Shox2, Tbx22, and Shh were obtained from Y.-P. Chen (22), R. Jiang (23), and S. Bellusci (24), respectively. A probe template for Tgfb3 was prepared as described (25). Apoptotic cells were detected using a TUNEL assay (Dead End from Promega) following the manufacturer's instructions. For cell proliferation analyses, cell proliferation labeling reagent (RPN201; Amersham Biosciences) was used. BrdU-positive cells were detected using anti-BrdU antibody (RPN202; Amersham Biosciences). Fluorescent images were viewed on an Olympus BX51 microscope and documented using an Olympus DP71 camera.
MicroCT Analyses
Specimens were embedded in 1% agarose, placed in a 19-mm-diameter tube, and scanned over the entire length of the skull using a microCT system (μCT100; Scanco Medical, Bassersdorf, Switzerland). The scan settings were: voxel size 10 μm, medium resolution, 55 kVp, 109 μA, 0.5 mm AL filter, and integration time 500 ms. Images were created using the manufacturer's evaluation software and a fixed global threshold to segment bone from non-bone. Skull shape was analyzed by measuring the distance from the supraoccipital bone to the anterior end of the frontal bone (length) and from the top of the parietal bone to the cranial base (height) and calculating the height/length ratio. Mandibular length was measured as shown in Fig. 1.
Western Blotting
Tissues or cells were lysed in 2× Laemmli sample buffer (26) and quantified by Quant-It protein assay system (Invitrogen); samples (5 μg of protein per lane) were run on NuPage 4–12% Bis-Tris gradient gels (Invitrogen) and transferred by “iBlot dry blotting” (Invitrogen) onto nitrocellulose filters. Immunoblotting and detection were done according to standard protocols. Documentation and quantification was accomplished by using the UVP BioSpectrum AC imaging system. The antibodies used were: pTak1 (antibody 9339; Cell Signaling), Smad2 (antibody 5339; Cell Signaling), pSmad2-C (antibody 3101; Cell Signaling), Smad1 (antibody 6944; Cell Signaling), pSmad1/5/8-C (antibody 9511; Cell Signaling), p-p38 Mapk (antibody 4511; Cell Signaling), p-Jnk (antibody 4668; Cell Signaling), p42/44 Erk (antibody 4376; Cell Signaling), β-actin (antibody A1978; Sigma-Aldrich), pSmad2-L(S250) (antibody 35741; Immuno-Biological Laboratories), pSmad2-L(T220) and pSmad3-L(T179) (antibody 28087; Immuno-Biological Laboratories), histone H3 (antibody 4499; Cell Signaling), and PSMA2 (antibody 2455; Cell Signaling).
Primary Craniofacial Mesenchymal Cell Cultures
Mandibular and maxillary processes of the first mandibular arch and prefusion palatal shelves were dissected from E11.0 and E14.0 embryos, respectively. The cultures were established as described by Thomas et al. (27). The neural crest origin and purity of cultures was confirmed by establishing cultures of embryos that carried both the Wnt1-Cre transgene and were positive for the R26-lacZ reporter (Fig. 3B). β-Galactosidase staining was carried out as described (28). After 7–10 days, the cells were stimulated with growth factors (10 ng/ml TGF-β2 or 100 ng/ml BMP2) for 0, 10, or 40 min and harvested for Western blot analyses. Chemical inhibitors flavopiridol (Cdk inhibitor; 1 or 10 μm; Sigma-Aldrich), UO126 (Erk1/2 (and Jnk) inhibitor; 10 μm; Sigma-Aldrich), SB431542 (TGFβRI inhibitor; 10 μm; Sigma-Aldrich), 5Z-7-oxozeaenol (Tak1 inhibitor; 2 μm; eMolecules), and SB202190 (p38 Mapk inhibitor; 10 μm; Sigma-Aldrich) were added to cells 60 min prior to TGF-β2 or BMP2 addition. The cells were not starved of serum prior to these treatments. Some cultures were transduced with replication-deficient recombinant adenoviral preparations using a multiplicity of infection of 1000. Ad-Gfp and Ad-Cre (titer, 4 × 1012 viral particles/ml) were obtained from the University of Michigan Biomedical Research Core Facility. For immunofluorescence, cultured cells were fixed in 4% buffered formaldehyde (5 min) and stained using anti-Smad2 antibody (antibody 5339; Cell Signaling). Binding was detected using Alexa Fluor 594 goat anti-rabbit secondary antibody (Invitrogen), and the slides were mounted in Vectashield with DAPI (Vector Labs Inc.). Fluorescent images were viewed and documented as outlined above. In some experiments, nuclear and cytoplasmic fractions were separated using the NE-PER nuclear and cytoplasmic extraction kit (kit 78833; Thermo Scientific) according to the manufacturer's instructions.
FIGURE 3.
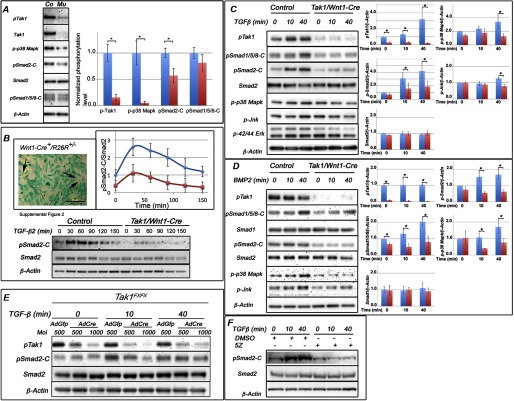
Activation of R-Smads and Mapks is less in Tak1/Wnt1-Cre embryos. A, representative immunoblot from prefusion palatal shelves harvested from control and Tak1/Wnt1-Cre mutant embryos at E14.0 and analyzed for phosphorylation of Tak1, Smad1/5/8 (BMP Smads), Smad2 (TGF-β Smad), and p38 Mapk. Co, control sample; Mu, Tak1/Wnt1-Cre mutant sample. The bar graph shows relative quantification of pTak1, p38 Mapk, pSmad2-C, and pSmad1/5/8-C in controls (blue columns) and mutants (red columns) (normalized to β-actin; n = 4). B, primary palatal mesenchymal cell cultures were established (see “Experimental Procedures”) from prefusion palatal shelves (at E14.0) of mouse embryos carrying both the Wnt1-Cre transgene and the R26Rlacz reporter (top left). Cells stained blue are derived from the neural crest. The black arrows point to a few negatively staining cells. Scale bar, 200 μm. A representative immunoblot of primary palatal mesenchymal cells stimulated with TGF-β2 and analyzed for C-terminal Smad2 phosphorylation is shown. Top right, the diagram illustrates the differences in amount of normalized Smad2-C phosphorylation between control (blue data points and blue line) and Tak1/Wnt1-Cre mutant (red data points and red line) cells. The values were normalized to Smad2. C, representative immunoblot from primary palatal mesenchymal cells stimulated with TGF-β2 (10 ng/ml), and protein lysates were analyzed for Tak1, C-terminal Smad1/5/8, C-terminal Smad2, total Smad2, p38 Mapk, Jnk, and Erk1/2 phosphorylation. The bar graphs show relative quantification of pTak1, p38 Mapk, pSmad2-C, pJnk, and Smad2 in controls (blue columns) and mutants (red columns) (normalized to β-actin; n = 3). D, representative immunoblot of primary palatal mesenchymal cells stimulated with BMP2 (100 ng/ml) and analyzed for Tak1 phosphorylation, C-terminal Smad1/5/8 phosphorylation, total Smad1, C-terminal Smad2 phosphorylation, total Smad2, p38 Mapk phosphorylation, and Jnk phosphorylation. The bar graphs show relative quantification of pTak1, p38 Mapk, pSmad1/5/8-C, and total Smad1 in controls (blue columns) and mutants (red columns) (normalized to β-actin; n = 3). Error bars, S.E. *, p < 0.05. E, representative immunoblot of primary Tak1FX/FX palatal mesenchymal cells transduced with AdGfp (control) or AdCre (mutant). The transduced cells were stimulated with TGF-β and analyzed for Tak1 phosphorylation, C-terminal Smad2 phosphorylation, total Smad2, and β-actin. Moi, multiplicity of infection. F, representative immunoblot of O9-1 neural crest stem cells stimulated with TGF-β2 in the presence or absence of the Tak1 kinase inhibitor 5Z-7-oxozeaenol (5Z) and analyzed for C-terminal Smad2 phosphorylation, total Smad2, and β-actin.
Neural Crest Stem Cell Cultures
The neural crest stem cell line O9-1 was obtained from M. Ishii. Undifferentiated cells were cultured in the presence of leukemia inhibitory factor and basic FGF in SNL cell-conditioned medium as described (29).
Roller Bottle Organ Cultures
Heads from embryos were collected at E13.5, and mandibles, tongues, and brains were removed in PBS. The resulting mid-face samples were cultured for 24–48 h at 37 °C in roller bottles (60 rotations/min) in serum-free BGJb medium without penicillin and streptomycin. The bottles were gassed at the beginning of the culture and every 12 h by gently bubbling the medium for 2 min with O2/CO2 (95%/5%). The palatal cultures were fixed, sectioned, and stained as described above.
Microarray Analyses
Palatal mesenchymal cells were isolated and aliquoted to 4 groups as shown in Table 1. Confluent cultures were incubated in the presence of inhibitors (TGFβRI inhibitor SB431542, 10 μm; Tak1 inhibitor 5Z-7-oxozeaenol, 2 μm) for 1 h and then stimulated with TGF-β2 (10 ng/ml) for 2 h. Total RNAs were then isolated using an RNeasy kit (Qiagen). Concentration and purity were determined by spectrophotometry, and RNA integrity was confirmed by running aliquots on 1% agarose gel. 500 ng of total RNA of each sample was amplified and biotin-labeled according to the GeneChip 3′ IVT expression kit user manual (Affymetrix). Microarray analysis was performed with the GeneChip mouse genome 430 2.0 array (Affymetrix). The biotin-labeled RNAs were fragmented, and 10 μg of each was hybridized to a GeneChip at 45 °C for 16 h. Labeled bacterial RNAs of known concentrations were spiked in hybridization to generate an internal standard and to allow normalization between the chips. The chips were washed and stained with streptavidin R-phycoerythrin (Molecular Probes). After scanning the chips by GeneChip Scanner 3000 7G System, the data were analyzed by Affymetrix GeneChip-related software packages GCOS, Data Mining Tool, and Affymetrix web database “NetAffx.” Microarray data were submitted to the GEO repository under accession number GSE45491.
TABLE 1.
| Sample | Treatment | Stimulation |
|---|---|---|
| 1 | None | None |
| 2 | None | TGF-β2 (10 ng/ml; 120 min) |
| 3 | SB431542 (10 μm; 60 min) | TGF-β2 (10 ng/ml; 120 min) |
| 4 | 5Z-7-Oxozeaenol (2 μm; 60 min) | TGF-β2 (10 ng/ml; 120 min) |
RESULTS
Neural Crest-specific Tak1 Mutants Display Mandibular Hypoplasia and Cleft Palate
A recent study demonstrated that haploinsufficiency of Tak1 in neural crest cells rescued the cleft palate phenotype of Tgfbr2 mutant mice (17). However, the role of Tak1 itself in craniofacial development is currently poorly known. To address this question, we crossed mice homozygous for the floxed Tak1 allele (Tak1FXFX; for details, see Fig. 1A) with transgenic Wnt1-Cre mice that also were heterozygous for the floxed Tak1 allele (Tak1FX+/Wnt1-Cre+). The expected Mendelian proportion of Tak1FXFX/Wnt1-Cre+ mice (hereafter referred to as Tak1/Wnt1-Cre mutants) survived to birth (n = 16 of 66) but then died within 24 h.
We used micro-CT analysis to compare the structure of control and Tak1/Wnt1-Cre mutant littermate heads at P0 in detail. The craniofacial skeleton appeared generally stunted in mutants (Fig. 1C). Many of craniofacial bones are largely derived from neural crest cells, including cranial base, mandible, maxilla, and frontal skull (29). Although the average lengths of control and Tak1/Wnt1-Cre mutant cranial bases did not differ, those of the mandible and maxilla were ∼20 and 5% shorter, respectively, in the mutants (Fig. 1C). The skulls in mutants were also shorter than in controls (length versus height ratio 1.74 ± 0.04 (average ± S.E.) and 1.49 ± 0.02 in controls and mutants, respectively; n = 3). The mutants also displayed cleft secondary palate with high penetrance (13 of 16, 80%).
To investigate the mechanism underlying the palatal defect, we collected mutant and control embryos at defined time points and analyzed palatal phenotype in detail by histology (Fig. 1, D–F). At E13.5, the palatal shelves in both control and mutants were still growing vertically toward the floor of the mouth, parallel to the sides of, and separated by, the tongue, which was almost square in cross-section (Fig. 1D). By E14.5 (Fig. 1E), in controls the tongue had changed shape in cross-section, becoming wider than it was tall. It was no longer interposed between the palatal shelves, which had elevated to point toward one another parallel to the top of the tongue, and commenced fusion with one another along the midline of the oral cavity in the mid- and posterior regions, although a “seam” of adhered epithelia from each shelf still remained. In contrast, in Tak1/Wnt1-Cre mutants, the tongue remained interposed between the palatal shelves, lying close to the nasal septum, and was still relatively square in cross-section. The shelves had failed to elevate very much. By E18 (Fig. 1F), in controls the secondary palates had fused completely along their entire length: the seam formed where their epithelia had met in the midline had disappeared, so their mesenchymal cores were confluent, and differentiation into cartilage commenced. In most Tak1/Wnt1-Cre mutants examined, secondary palate formation had failed, leaving a cleft from anterior to posterior. Although the tongue was no longer completely interposed between the palatal shelves, which had elevated, the shelves had failed to adhere to or fuse with one another, although some cartilage formation had occurred. Although now above the tongue in anterior and mid-positions, the underside of the shelves and the top of the tongue were very close compared with the same structures in controls.
Because components of the head were smaller in mutants than controls at birth, we examined the relative size of palatal shelves in littermates at E13.5 and found that shelves in mutants were ∼20% smaller in cross-sectional area than those in controls (Fig. 1G, left panel). BrdU incorporation assays on prefusion palatal shelf sections from littermates showed a lower average value in mutant versus control cells at the anterior level (not significantly different), but no difference at the mid and posterior levels (Fig. 1G, right panel). To determine whether the smaller mutant shelf area at E13.5 was the result of an earlier lower cell proliferation rate, we performed the same assay at E11 on mandibular primordia (first pharyngeal arch mesenchyme), but no significant difference between control and mutant tissues was detected (data not shown). No differences in cell death were detected at E11 or E13.5 (data not shown).
Because Tak1/Wnt1-Cre mutants displayed a distinct reduction in craniofacial size at birth (Fig. 1), likely resulting in a hypoplastic oral cavity, and the position and shape of the tongue appeared abnormal in mutants during palatogenesis, we wondered whether the failure in palatal shelf elevation could be caused by the failure of the tongue to “descend” appropriately. To address this, at E13.5 (when palatal shelves are still growing vertically) we cultured mouse embryonic heads from which we had removed the mandible and tongue. This was performed in roller bottles under chemically defined conditions. In both control and mutant samples, sufficient elevation of palatal shelves occurred after 24 h that, after a further 24-h culture, fusion occurred, with equivalent mesenchymal confluence regardless of genotype (Fig. 2, A and B). This suggests that in Tak1/Wnt1-Cre mutants the tongue obstructed palatal shelf elevation in vivo, contributing to the pathogenesis of cleft palate in Tak1/Wnt1-Cre mutants in vivo (44).
FIGURE 2.
Palatal shelves of Tak1/Wnt1-Cre mutants elevate and fuse in vitro, but show delayed rugal formation in vivo. A and B, control (left panels) and mutant (right panels) embryos were harvested at E13.5, and dissected midfacial segments (without mandible, tongue, and brain) were cultured under chemically defined conditions in roller bottles for 24 h (A) or 48 h (B). The arrows point to the tips of elevated palatal shelves in both control and mutant samples cultured for 24 h (n = 3). C and D, comparison of Shox2 and Tbx22 (C, E14.0) and Shh (D, E14.5 and E15.5) expression in control and Tak1/Wnt1-Cre mutant palatal regions. The double-headed arrows in C illustrate the length of the positive signal. Rugal stripes (positive Shh expression) were identified and numbered (r1–r8) according to Ref. 43. E–H, n = 3. NS, nasal septum; P, palatal shelves; SP, secondary palate.
Previous studies have shown that TGF-β superfamily signaling is involved in the anterior-posterior patterning of the secondary palate (22, 30–32). Therefore, we compared expression patterns of established anterior (Shox2) and posterior (Tbx22) mesenchymal markers between Tak1/Wnt1-Cre mutant and control littermates using whole mount in situ hybridization. These experiments did not demonstrate detectable differences in Shox2 and Tbx22 expression patterns or intensities between controls and mutants (Fig. 2C), suggesting that in Tak1/Wnt1-Cre mutants, the anterior-posterior patterning is not grossly affected.
Mesenchymal Bmp signaling has been shown to be important for maintenance of Shh expression in the rugae of the palatal epithelium (30), which in turn plays a critical role in regulation of growth and patterning along the palatal oro-nasal axis (33, 34). Our experiments demonstrate that, unlike palatal mesenchyme-specific Bmpr1a mutant shelves (35), there was no reduction in Shh expression in the primary palate of Tak1/Wnt1-Cre mutants. However, whereas the amount of Shh expression in individual ruga was not decreased, at the time of palatal fusion (E14.5–E15.5), a marked delay in rugal formation suggests that the rate of expansion of the secondary palate was reduced (Fig. 2D).
In summary, Tak1 deficiency in the craniofacial neural crest resulted in facial (particularly mandibular) and palatal shelf hypoplasia and cleft palate. Palatal shelf elevation was delayed, and the tongue was abnormal in position and shape in mutants immediately prior to the normal stage of palatal fusion. No difference between control and Tak1/Wnt1-Cre mutants in anterior-posterior patterning of prefusion palatal shelves was detected by marker gene analysis, but a delay in ruga pattern formation suggests abnormal morphogenesis or maturation there. In rolling culture, removal of the tongue and mandible resulted in mutant palatal shelf elevation, and fusion occurred in both control and mutant heads.
Both Canonical and Noncanonical TGF-β Superfamily Signaling Pathways Are Affected in Tak1/Wnt1-Cre Mutants
Neural crest cells make a contribution to all the craniofacial structures identified as showing abnormalities in Tak1/Wnt1-Cre mutants, including palatal shelves, tongue, and mandible. Of these, the palatal shelf mesenchyme is the most uniformly composed of neural crest cells. To identify signaling processes influenced by Tak1 deficiency in vivo, we harvested prefusion palatal shelves from control and Tak1/Wnt1-Cre embryos and analyzed their protein for Tak1, phosphorylation of Tak1, and its downstream signaling target p38 Mapk, the TGF-β signaling target Smad2, and BMP signaling targets Smad1/5/8 (Fig. 3A). The level of Tak1 and Tak1 C-terminal phosphorylation at Ser-412 was significantly less in Tak1/Wnt1-Cre mutant samples, demonstrating an efficient Cre-induced recombination in the Tak1 locus. The level of p38 Mapk phosphorylation was also markedly and consistently less in these mutant palatal shelves when compared with corresponding control samples. Similarly, C-terminal Smad2 phosphorylation was consistently lower in mutant shelves, whereas there were no differences in Smad2 protein levels. C-terminal Smad1/5/8 phosphorylation was lower on average in mutant samples but not significantly so. These results show that Tak1 deficiency leads to decreased signaling activity in both noncanonical and canonical TGF-β signaling pathways.
Tak1 Mediates C-terminal Smad Phosphorylation in Craniofacial Mesenchymal Cells
To gain a deeper insight into processes regulated by Tak1-mediated signaling in postmigratory neural crest cells, we established control and Tak1/Wnt1-Cre mutant craniofacial mesenchymal primary cultures from both prefusion palatal shelves (at E14) and first pharyngeal arches, which give rise to maxilla and mandible (at E11), and analyzed their ability to respond to TGF-β stimulation (Fig. 3, B and C). As expected, control mesenchymal cells isolated from the secondary palate showed an increase in Tak1 Ser-412 phosphorylation in response to TGF-β stimulation and beyond a clearly detectable base-line Tak1 phosphorylation level (Fig. 3C). In Tak1/Wnt1-Cre cells, there was no detectable Tak1 phosphorylation, with or without TGF-β stimulation. A modest increase in phosphorylation of Smad-independent pathway targets p38 Mapk, Jnk, and Erk1/2 was induced by TGF-β2 in control cells, but in mutant cells no such change was observed (Fig. 3C). TGF-β2 stimulation induced C-terminal phosphorylation of both TGF-β Smads (Smad2) and BMP Smads (Smad1/5/8) in control palatal mesenchymal cells. In mutant cells, Smad2 C-terminal also became phosphorylated in response to TGF-β2 stimulation, but less so than in control cells (Fig. 3, B and C), and BMP Smads did not show detectable C-terminal phosphorylation. In all samples, Smad2 protein levels remained stable for 90 min after TGF-β stimulation (Fig. 3B). Similar results were obtained using cells isolated from first pharyngeal arches (which also contribute to mandible and tongue) at E11, using a neural crest stem cell line in conjunction with the Tak1 kinase inhibitor (5Z-7-oxozeaenol) (Fig. 3F and data not shown).
In summary, the relative pattern of phosphorylation of downstream targets shown by control and mutant palatal shelf primary culture cells in response to TGF-β stimulation is similar to that found between control and mutant E14 palatal shelves recovered in vivo. Because Tak1 has also been shown to be involved in BMP signaling, we stimulated both the control and Tak1/Wnt1-Cre mutant ecto-mesenchymal cells with BMP2 and monitored the phosphorylation of signaling molecules as above (Fig. 3D). Unlike TGF-β2 and as expected, BMP2 was not able to induce Tak1 phosphorylation at Ser-412 in control cells, nor did mutant cells show detectable phospho-Tak1 levels. Control cells were responsive to BMP2 stimulation in their progressive increase in C-terminal BMP-Smad (Smad 1/5/8) phosphorylation, but Smad2 phosphorylation was not altered. In mutant cells, base-line C-terminal Smad1/5/8 phosphorylation levels were lower, and their responsiveness to BMP2 stimulation was reduced, when compared with control cells. Collectively, the results of these experiments suggest that in neural crest-derived mesenchymal cells, Tak1 contributes more to TGF-β than BMP-induced C-terminal phosphorylation of the corresponding R-Smads. BMP2 induced progressive p38 Mapk and Jnk phosphorylation in controls cells, but no increase was detected in mutant cells.
To provide additional evidence for the role of Tak1 in C-terminal phosphorylation of Smad2, we transduced palatal mesenchymal cells carrying the homozygous Tak1-flox allele with recombinant adenoviruses expressing the Cre recombinase and analyzed the transduced and control cells (transduced with AdGfp) for Tak1 and Smad2 phosphorylation (Fig. 3E). These results confirm that the reduction in C-terminal phosphorylation of Smad2 is an immediate result of Tak1 deficiency rather than just a long term adaptation to severely reduced Tak1 levels in our Tak1/Wnt1-Cre mutants.
To conclude, we show that Tak1 is required for appropriate activation of both canonical and noncanonical TGF-β and BMP signaling pathways in neural crest-derived mesenchymal cells. Moreover, in the presence of Tak1, TGF-β2 was able to induce activation of both TGF-β and BMP R-Smads, i.e. Smad2 and Smad1/5/8, respectively.
Agonist-induced Linker Region Phosphorylation Is Affected in Palatal Mesenchymal Cells Deficient in Tak1 Mutants
Several studies have shown that in R-Smads the intervening sequence that links the DNA-binding domain and transcriptional domain, the so-called linker region, becomes phosphorylated by a set of divergent stimuli (including FGFs, EGFs, and stress signals) via Mapks and also by members of the TGF-β superfamily (36–38). This agonist-induced linker region phosphorylation (ALP) at Thr-220 in Smad2 (Thr-179 in Smad3) located immediately upstream of a PY motif is of particular interest, because it was recently suggested that, in addition to priming Smads for turnover, phosphorylation of this threonine residue is required for a full transcriptional activity of Smad complexes (7, 9). Therefore, we stimulated both control and Tak1-deficient primary mesenchymal cells (from both the first pharyngeal arch and the palate) with TGF-β2 and analyzed phosphorylation of these key residues (Smad2-L(T220) and Smad3-L(T179)) and of Smad2-L(S250) using phospho-residue-specific antibodies. Our results demonstrate that in control cells TGF-β2 induced efficient phosphorylation of the Smad2 linker region at Ser-250 and Thr-220 and at Thr-179 in Smad3, whereas cells deficient in Tak1 failed to show comparable induction of ALP (Fig. 4A). As before, practically identical results were obtained using cells isolated from first pharyngeal arches at E11.0 (data not shown). Moreover, O9-1 neural crest stem cells showed similar increases in TGF-β2-induced linker region phosphorylation at Smad2-L(T220) and Smad3-L(T179) and showed that this induction was largely inhibited by 5Z-7-oxozeaenol (Fig. 4A, lower panel).
FIGURE 4.
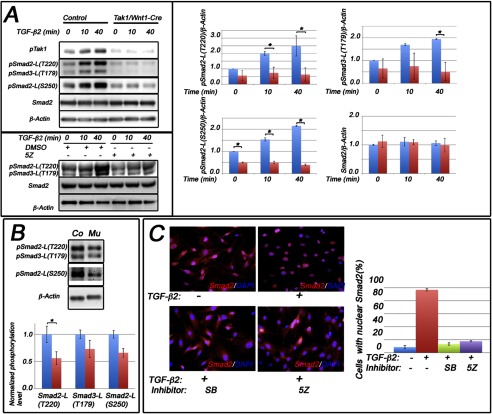
Tak1 is required for appropriate Smad2 nuclear localization and agonist-induced Smad2/Smad3 linker-region phosphorylation in palatal mesenchymal cells. A, upper panel, primary palatal mesenchymal cells were stimulated with TGF-β2. Shown are representative Western blots of protein lysates analyzed for Smad2 linker region phosphorylation at Thr-220 (pSmad2-L(T220)) and Ser-250 (pSmad2-L(S250)) and for Smad3 linker region phosphorylation at Thr-179 (pSmad3-L(T179)). The bar graphs show relative quantification of pSmad2-L(T220), pSmad3-L(T179), and pSmad2-L(S250) in controls (blue columns) and mutants (red columns) (normalized to β-actin; n = 3). A, lower panel, representative immunoblot of O9-1 neural crest stem cells stimulated with TGF-β2 in the presence or absence of the Tak1 kinase inhibitor 5Z-7-oxozeaenol (5Z) or vehicle (dimethyl sulfoxide, DMSO) and analyzed for Smad2 linker region phosphorylation at Thr-220 (pSmad2-L(T220)), Thr-179 (pSmad3-L(T179)), total Smad2, and β-actin. B, representative immunoblot of prefusion palatal shelves harvested from control (Co) and Tak1/Wnt1-Cre mutant (Mu) embryos at E14.0 and analyzed for linker region phosphorylation at Thr-220 (pSmad2-L(T220)) and Ser-250 (pSmad2-L(S250)) and for Smad3 linker region phosphorylation at Thr-179 (pSmad3-L(T179)). The bar graph shows relative quantification of agonist-induced linker region phosphorylation in controls (blue columns) and mutants (red columns) (normalized to β-actin; n = 3). C, representative images showing subcellular localization of Smad2 in unstimulated cells (upper left), cells stimulated with TGF-β2 (upper right), cells stimulated with TGF-β2 in the presence of the TGFβRI kinase inhibitor (SB431542 (SB), lower left), and cells stimulated with TGF-β2 in the presence of Tak1 inhibitor (5Z-7-oxozeaenol (5Z), lower right). Red, immunostaining for Smad2; blue, nuclear counterstaining with DAPI. The bar graph shows quantification of nuclear localization of Smad2 (n = 3; error bars, S.E.).
To investigate whether these in vitro results were reflective of palatal shelf mesenchyme in vivo, we prepared protein lysates directly from prefusion palatal shelves of control and Tak1/Wnt1-Cre mutant embryos and analyzed them for the Smad2/3 linker region phosphorylation. Endogenous levels of Smad2 linker phosphorylation at Thr-220 and Ser-250 and Smad3 phosphorylation at Thr-179 were all significantly lower in mutant palatal samples than in those of controls (Fig. 4B).
It has previously been shown that in several established cell lines, e.g., HEK293T and HaCaT, Thr-220 in the linker region of Smad2 is directly phosphorylated by nuclear cyclin-dependent kinases Cdk8 and Cdk9 (7, 9). Therefore, we examined the effect of Tak1 inactivation on subcellular localization of total Smad2 both on palatal mesenchymal cells (harvested at E14) (Fig. 4C) and on first pharyngeal arch mesenchymal cells (harvested at E11; data not shown). In both cell types, TGF-β2-induced Smad2 nuclear accumulation was effectively inhibited by the Tak1 kinase inhibitor (5Z-7-oxozeaenol) and by the TGFβRI kinase inhibitor (SB431542) (Fig. 4C and data not shown). Tak1 kinase inhibitor also inhibited both TGF-β2-induced Smad2-C-terminal and linker region phosphorylation at Smad2-L(T220), Smad3-L(T179) and Smad2-L(S250) (Fig. 5A). Certain Tak1-mediated ALP was also dependent on the presence of the TGFβRI protein and the kinase activity of TGFβRI, because both the Cre-mediated Tgfbr1 deletion and TGFβRI inhibition by the SB431542 could attenuate TGF-β2-induced phosphorylation at Smad2-L(T220)/Smad3-L(T179) (Fig. 5, A and B). As expected, agonist-induced phosphorylation of Smad2 at the C terminus was also inhibited by the physical loss of Tgfbr1 and by SB431542 (Fig. 5, A and B). In palatal mesenchymal cells, flavopiridol (Cdk inhibitor) inhibited ALP at Thr-220 and Ser-250 (Smad2) (Fig. 5A), whereas UO126 (Erk1/2 and Jnk inhibitor) could effectively prevent TGF-β2-induced Smad2-L(S250) phosphorylation but could only weakly inhibit ALP at Thr-220 (Fig. 5A). The effect of SB202190 (p38 Mapk inhibitor) on phosphorylation at Smad2-L(T220) was very similar to that of UO126 (data not shown). Our cell fractionation experiments confirmed that most of the Smad2 molecules with TGF-β2-induced phosphorylation at Thr-220 could be detected in the nucleus and that this phosphorylation was effectively inhibited by flavopiridol (Fig. 5C). In summary, our results show that in craniofacial mesenchymal cells Tak1 deficiency led to reduced agonist-induced C-terminal phosphorylation and decreased nuclear accumulation of R-Smads and that agonist-induced linker phosphorylation of Smad2 both at Thr-220 (which is at least partially mediated by Cdks) and at Ser-250 (mediated by Erk1/2 and/or Jnk) was dependent on the functional Tak1 protein (Fig. 5, D and E).
FIGURE 5.
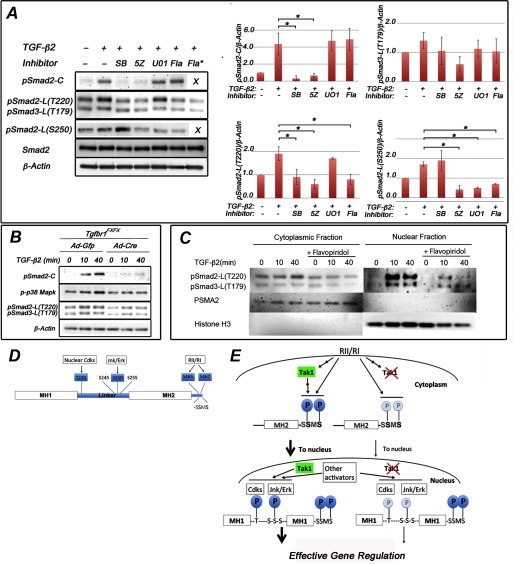
Smad2 linker-region at Thr-220 is phosphorylated by nuclear kinases in palatal mesenchymal cells. A, representative immunoblot of primary palatal mesenchymal cells stimulated with TGF-β2 in the presence of SB431542 (SB), 5Z-7-oxozeaenol (5Z), UO126 (UO1), and flavopiridol (Fla; 1 and 10 μm (asterisk)). Protein lysates were analyzed for Smad2 C-terminal phosphorylation, Smad2 linker region phosphorylation (at Thr-220 and Ser-250), and Smad3 linker region phosphorylation (at Thr-179). The bar graphs show relative quantification of pSmad2-C, pSmad-L(T220), pSmad3-L(T179), and pSmad2-L(S250) (normalized to β-actin; n = 3). Error bars, S.E. *, p < 0.05. B, representative immunoblot of primary palatal mesenchymal cells from Tgfbr1FX/FX embryos were transduced with Ad-Gfp or Ad-Cre, the cells were stimulated with TGF-β2, and protein lysates were analyzed for C-terminal Smad2 phosphorylation, p38 Mapk phosphorylation, and linker region phosphorylation at Smad2-L(T220) and at Smad3-L(T179). β-actin was loading control. C, palatal mesenchymal cells were stimulated by TGF-β2 in the presence and absence of flavopiridol. Cytosolic and nuclear lysates were analyzed for Smad2 linker region phosphorylation at Thr-220 (pSmad2-L(T220)) and for Smad3 linker region phosphorylation at Thr-179 (pSmad3-L(T179)). Antibodies against PSMA2 and histone H3 were used to assess the purity of cytosolic and nuclear fractions (middle and bottom panels). D, schematic representation of Smad2 depicting Thr/Ser residues phosphorylated by nuclear Cdks and Jnk/Erk in the linker region and by TGFβRI (RI) at the C terminus examined in this study. E, schematic summary illustrating the role(s) of Tak1 in phosphorylation of Smad2 (left side of diagram) in which Smad2 C-terminal phosphorylation levels were greatly enhanced in response to TGF-β stimulation when Tak1 was present, as well as Tgfbr1. Cdk-dependent phosphorylation of Thr-220 occurs in the nucleus. Phosphorylation of Ser-250 is Mapk-dependent but also inhibited by Cdk inhibitor, which may be due to some dependence on the prior phosphorylation of Thr-220 in the model of Aragón et al. (9). Deletion of Tak1 results in reduced C-terminal phosphorylation, leading to a reduced nuclear accumulation of Smad2, and reduced linker region phosphorylation (right side of diagram), which would be expected to reduce effectiveness in gene regulation according to the model of Aragón et al. (9).
Tak1 as a TGF-β Signal Transducer in Neural Crest-derived Facial Mesenchymal Cells
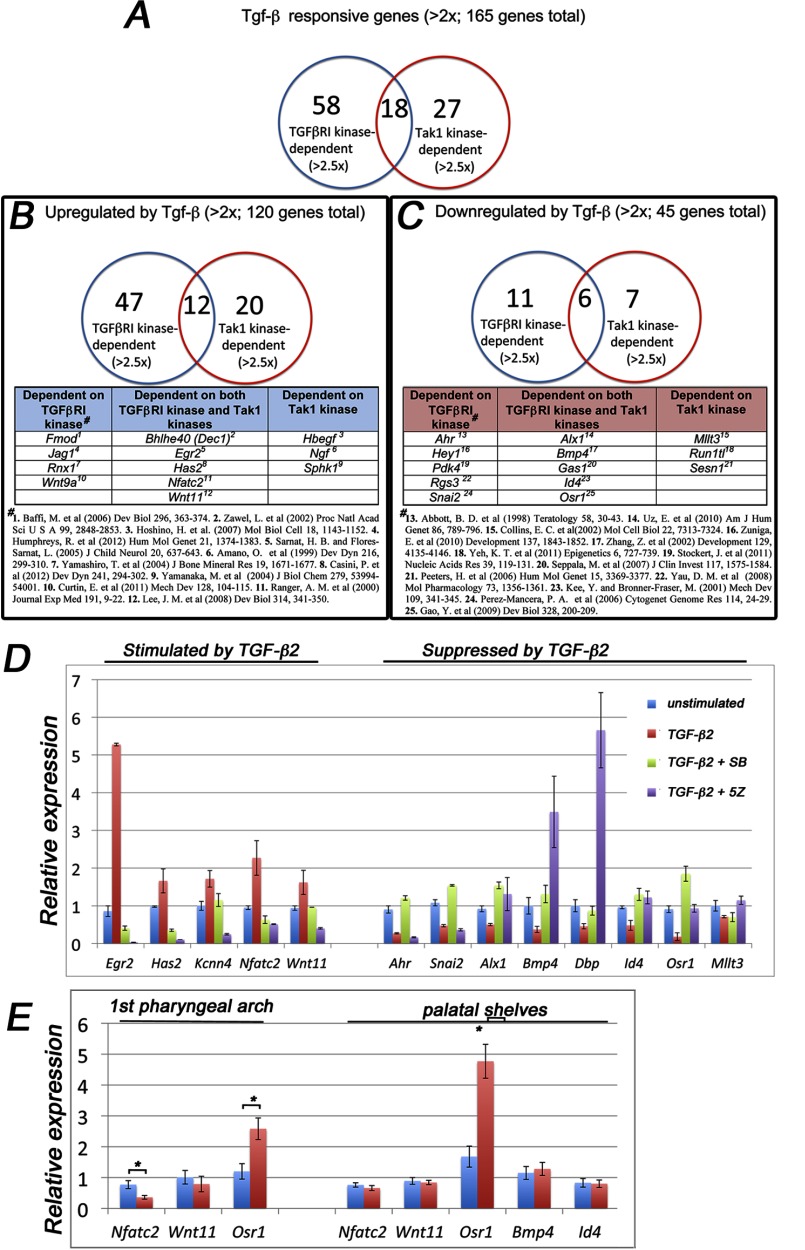
Previous studies have demonstrated that TGF-β-induced Tak1 activation is dependent on the presence of TGFβRI protein and is mediated by an adaptor protein Traf6 (11, 12). However, whether or not this activation is dependent on the TGFβRI kinase activity is still controversial (11, 12). As shown above, the presence of either TGFβRI or its kinase activity is sufficient for agonist-induced Tak1-mediated linker region phosphorylation of Smad2 in ecto-mesenchymal cells (Fig. 5, A and B). To better define the relationship between Tak1 kinase-mediated and TGFβRI kinase-mediated transcriptional responses, we compared expression profiles of TGF-β2-responsive genes in primary palatal mesenchymal cells in the presence of inhibitors for either TGFβRI kinase or Tak1 kinase using a genome-wide transcriptomic analysis (Fig. 6). Of TGF-β2-responsive genes (>2-fold change; p < 0.05), 58 were dependent on TGFβRI kinase, 27 were dependent on Tak1 kinase and 18 were dependent on both (>2.5-fold change; p < 0.05) (Fig. 6A). Among the up-regulated genes (120 genes; >2-fold change; p < 0.05), 47 were TGFβRI kinase-dependent, 20 were Tak1 kinase-dependent, and 12 were dependent on both (>2.5-fold change; p < 0.05) (Fig. 6B). Among the down-regulated genes (45 genes; >2-fold change; p < 0.05), 11 were TGFβRI kinase-dependent, 7 were Tak1 kinase-dependent, and 6 were dependent on both (>2.5-fold change; p < 0.05) (Fig. 6C). Differential expression of several Tak1 kinase/TGFβRI kinase-dependent genes was confirmed by real time RT-PCR (Fig. 5D). When more stringent criteria were used and only the genes that displayed >3-fold expression change when stimulated with TGF-β2 were analyzed, 10 genes were dependent on both TGFβRI and Tak1 kinase activities, whereas 8 genes and 1 gene were dependent on only TGFβRI or only Tak1 kinase activities, respectively (data not shown).
FIGURE 6.
Tak1 and TGFβRI mediate both distinct and overlapping gene responses in palatal mesenchymal cells. Total RNAs from unstimulated palatal mesenchymal cells, from cells stimulated with TGF-β2, from cells treated with the TGFβRI kinase inhibitor (SB431542 (SB), 10 μm, 60 min prior stimulation) and stimulated with TGF-β2 (2 h), and cells treated with Tak1 inhibitor (5Z-7-oxozeaenol (5Z), 2 μm, 60 min prior stimulation) and stimulated with TGF-β2 (2 h) were subjected to genome-wide transcriptomic analysis on the MOE 430A 2.0 microarray. A, Venn diagram showing a summary of the number TGFβRI kinase- and Tak1 kinase-dependent TGF-β2 responsive genes (>2-fold change; p < 0.05). B, summary of number of TGFβRI kinase- and Tak1 kinase-dependent genes up-regulated by TGF-β2 in palatal mesenchymal cells (five examples of genes are shown in each group). C, summary of number of TGFβRI kinase- and Tak1 kinase-dependent genes that are down-regulated by TGF-β2 (3–5 examples of genes in each group). # in B and C denotes references highlighting relevance of a listed gene either in TGF-β signaling or in craniofacial/neural crest biology. D, real time RT-PCR quantification of differentially expressed genes. Blue columns, no inhibitor, no TGF-β2; red columns, no inhibitor, stimulated with TGF-β2; green columns, 10 μm SB431542 60 min prior stimulation with TGF-β2; purple column, 2 μm 5Z-7-oxozeaenol 60 min prior to stimulation with TGF-β2. E, bar graph showing relative real time RT-PCR quantification of selected TGF-β-responsive Tak1-dependent genes (identified by the microarray screen) in the craniofacial mesenchyme. Blue columns, control; red columns, Tak1/Wnt1-Cre mutant (n = 3). Error bars, S.E. *, p < 0.05. 1st pharyngeal arch, first pharyngeal arch harvested at E11.0; palatal shelves, prefusion palatal shelves harvested at E14.0.
To examine whether any of the kinase-dependent gene expression findings reflected those between control and Tak1-deficient cells in vivo, we harvested NC-derived craniofacial mesenchymal tissues (both mandibular arch at E11 and palatal shelves at E14) from control and Tak1/Wnt1-Cre mutant embryos and compared expression levels of TGF-β-responsive genes using real time RT-PCR (Fig. 6E). Among the analyzed genes, Nfatc2, which was induced by TGF-β2 in cultured palatal mesenchymal cells dependent on both kinases, was clearly down-regulated in Tak1/Wnt1-Cre mutant first pharyngeal arch tissues at E11 and slightly decreased, although not significantly, in mutant palatal shelves at E14. Similarly, Osr1, which was repressed by TGF-β2 in cultured palatal mesenchymal cells dependent on both kinases, was expressed at higher levels both in Tak1/Wnt1-Cre mutant first pharyngeal arch (2.2-fold higher) and prefusion palatal shelf tissues (2.8-fold higher) than in corresponding control tissues. Any differences in average expression of other genes tested, e.g., Wnt11 and Bmp4, were relatively modest but some still showed tendencies consistent with the findings made in mesenchymal cell culture assays in vitro.
To conclude, TGFβRI and Tak1 kinases mediated both distinct and overlapping transcriptional responses in palatal mesenchymal cells, when stimulated by TGF-β2. In this cell type, agonist-induced Tak1 activation could occur independently of TGFβRI kinase activity.
DISCUSSION
Although Tak1, a member of the Mapk kinase kinase subfamily, was originally shown to mediate TGF-β signaling pathways (39), its physiological role in TGF-β superfamily signaling in vivo, particularly during embryogenesis, is still poorly elucidated. Here we show that Tak1 plays a critical nonredundant role in craniofacial development, and that, in embryonic neural crest-derived craniofacial cells, Tak1 is required not only for Smad-independent TGF-β superfamily signaling but also for maximal ligand-induced C-terminal and linker phosphorylation of R-Smads.
Role of Tak1 in Craniofacial Development
Previous studies have shown that TGF-β and Bmp signaling plays an essential role in the neural crest-derived palatal mesenchyme (35, 40, 41). Of particular note is a recent report demonstrating that cleft palate in Tgfbr2/Wnt1-Cre mutants results from TGF-β2-induced activation of an alternative pathway via TGFβRI and TGFβRIII that induces activation of Tak1 and hence p38 Mapk (17). This aberrant activation results in attenuated expression of Fgf9 and Pitx2, leading to reduced mesenchymal cell proliferation and cleft palate (42). These results raised the questions of what role neural crest-expressed Tak1 normally plays in craniofacial development and in the activation of signaling pathway components downstream of TGF-β2 signaling.
Here we report that Tak1 is required for normal facial and mandibular growth and palatogenesis in vivo and that it mediates both Smad-dependent and Smad-independent TGF-β and Bmp signaling in neural crest cells in vitro. The overall craniofacial phenotypes of Tak1/Wnt1-Cre mutants were relatively mild when compared with those of the corresponding Tgfbr1, Tgfbr2, or Bmpr1a mutants. Unlike in Tgfb receptor mutants, palatal mesenchymal cell proliferation in general was only slightly affected in Tak1 mutants, because we did not detect any significant differences at the time points examined. Although overactivation of Tak1 in the Tgfbr2/Wnt1-Cre mutants led to a reduction in cell proliferation (16), removal of Tak1 in our Tak1/Wnt1-Cre mutants resulted in moderate hypoplasia. Because we could not detect increased apoptosis between embryonal days 11–14, generalized hyperplasia in NC-derived tissues as the result of the absence of Tak1 does not seem to occur.
Normal formation of the secondary palate requires distinct developmental events; not just formation and growth of two palatal shelves but their elevation, physical contact with one another, fusion, loss of epithelial seam, and differentiation into cartilage and bone. Of these steps, shelf elevation and patterning (ruga formation) showed developmental delay in Tak1Wnt1Cre mutants. It was recently shown that a Turing-type reaction-diffusion mechanism establishes the normal pattern of rugae (43). Although the delay in ruga formation may indicate an intrinsic defect in palatogenesis, it also is possible that a failure of the tongue to descend disturbs the shelf tissue proportions, altering the distribution of signals for ruga formation and slowing their increase in number. An essential role for Tak1 in palatal shelves themselves in the processes of palatal shelf elevation onwards is hard to define because these processes occurred indistinguishably in control and Tak1/Wnt1-Cre mutant heads in rolling culture in vitro when tongue and mandible were absent, but their morphology looked different from normal palatal shelves in vivo (Fig. 1B). This result, along with histological observations, suggests that the failure of the tongue to descend sufficiently in mutants most likely contributes to the cleft palate phenotype in vivo by physically impeding shelf elevation. Consistent with these findings, Song et al. have recently shown that Tak1 plays an important role in tongue development by controlling Fgf10 expression (44).
At the protein level, Tak1/Wnt1-Cre mutant tissues showed reduced levels of activation of both Smad-independent and -dependent pathways rather than an up-regulation of Smad-dependent pathway signaling. As our studies on primary culture cells also showed, Tak1 functions as a modulator of signal strength rather than an “on/off switch” for the Smad-independent pathway in the NC-derived craniofacial mesenchyme. Our results do not exclude that altered TGF-β/Bmp-independent Tak1-mediated signaling in these cells may also contribute to the relatively mild phenotype.
Role of Tak1 in TGF-β-induced Signaling Events
Our studies of the dependence of TGF-β2-induced signaling on Tak1 in embryonic neural crest cells have revealed important details not only of phosphorylation of components of signaling pathways but of their cell type-dependent variation. We show that in NC-derived craniofacial mesenchymal cells, Tak1 mediates both TGF-β and BMP-induced activation of their corresponding R-Smads. Shim et al. (45) showed that in chondrocytes Tak1 can phosphorylate the C-terminal residues of an R-Smad (Smad1) only as part of the Bmp signaling pathway. Moreover, we show that in neural crest cells, TGF-β stimulation is able to stimulate BMP R-Smad activation directly and that this process is, in part, mediated by Tak1. In fact, a recent study on established immortalized cell lines (of epithelial, endothelial, and mesenchymal origins) suggested that this unconventional TGF-β-induced Bmp R-Smad activation is required for a subset of critical TGF-β-induced cellular functions (5). Although our studies show that this unconventional R-Smad activation takes place in primary NC-derived craniofacial mesenchymal cells in vitro, it remains to be shown whether it plays a role in craniofacial ectomesenchyme during embryogenesis in vivo.
Linker regions in both TGF-β and BMP R-Smads are Ser/Thr-rich and are known to be phosphorylated by several different kinases, e.g., GSK, Mapks, and Cdks, and it has been suggested that these post-translational modifications have both activatory and inhibitory regulatory functions in TGF-β signaling (38). Here we show that palatal and pharyngeal arch mesenchymal cells deficient in Tak1 show reduced agonist-induced linker region phosphorylation both at Thr-220 and Ser-250 in Smad2 in vitro and that particularly Thr-220 phosphorylation in the Smad2 linker region is reduced in neural crest cell-specific Tak1 mutants in vivo. Moreover, our results suggest that in NC-derived mesenchymal cells, Thr-220 and Ser-250 are phosphorylated by Cdks and Mapks, respectively. These findings differ from those made in fibroblast and melanoma cell lines, which showed that Mapks rather than Cdks are the primary kinases responsible for the linker region phosphorylation (46, 47). Therefore, there seems to be distinct cell type-specific differences between the potency of particular kinases phosphorylate the specific residues.
It was recently suggested that ALP at Smad2-L(T220) plays a role in the so-called “action-turnover switch” function of R-Smads and may be required both for maximal TGF-β-induced transcriptional activity and efficient termination of the signal in immortalized cell lines (9, 10). Our present results show that, in primary craniofacial mesenchymal cells, Tak1, by functioning as an amplifier of C-terminal R-Smad phosphorylation, could have indirectly regulated ALP at Smad Thr220 because deficient Tak1 activity would result in reduced ligand-induced R-Smad activation and hence decreased R-Smad nuclear accumulation and less linker-region phosphorylation at Thr-220 by nuclear Cdks (Fig. 5E). However, our results do not exclude the possibility that Tak1 could also contribute to linker region phosphorylation by other mechanisms, such as regulating activity of other downstream kinases (Fig. 5E). Endogenous levels of Smad2-C phosphorylation and Smad Thr-220 phosphorylation were also reduced in Tak1/Wnt1Cre mutant palatal shelves, implying that Tak1 may modulate TGF-β superfamily signaling in vivo in the same way.
Interdependence of TGFβRI and Tak1 Signaling?
Two recent studies addressed the mechanism of TGF-β-induced Tak1 activation in HEK 293 cells (11, 12). Although these studies agree that a ligand-induced interaction between TGFβRI and Traf6 results in Traf6 autoubiquitination and subsequent Tak1 activation, they did not agree on a role for TGFβRI kinase activity in TGF-β-induced Tak1 activation. Here we show that, in craniofacial mesenchymal cells, the presence of TGFβRI or its kinase activity is sufficient for Tak1-mediated signaling events downstream of TGF-β2. To explore whether all the Tak1-dependent TGF-β transcriptional responses are dependent on the TGFβRI kinase activity, we performed a genome-wide transcriptomic analysis on prefusion palatal mesenchymal cells stimulated with TGF-β2 in the presence or absence of either TGFβRI or Tak1 kinase inhibitors. This assay demonstrated that many of the TGF-β-induced transcriptional responses (both stimulatory and repressive) are simultaneously dependent on both TGFβRI kinase and Tak1 kinase activities and that surprisingly few genes, e.g., Mllt3, respond to TGF-β2 stimulation if TGFβRI kinase activity is inhibited.
In conclusion, our combined evidence implies that Tak1 has a novel multimodal role in the craniofacial neural crest-derived mesenchyme in regulating activation of both TGF-β- and Bmp-induced Smad-dependent and Smad-independent signaling processes. Tak1 deficiency in NC-derived mesenchymal cells leads to attenuation of both canonical and noncanonical TGF-β and Bmp signaling, which contribute to relatively subtle but consistent growth distortions in craniofacial structures. This discoordination in craniofacial growth likely results in delayed elevation of palatal shelves, which will never form a contact in the midline, resulting in cleft secondary palate.
Acknowledgments
We thank Saverio Bellusci, Rulang Jiang, and YiPing Chen for in situ hybridization probes, Stefan Karlsson for Tgfbr1FX mice, YiPing Chen for sharing unpublished data, Taocong Jin for help with microarray data analyses, Michelle Lynch for Micro-CT analyses, and Sean Edwards and Joseph Helman for support during the study.
This work was supported, in whole or in part, by National Institutes of Health Grants R01DE020843 and ES071003-11 (to Y. M.), RO1 DE013085 and HL074862 (to V. K.), and S10RR026475-01 (to the University of Michigan μCT Core Facility).
- BMP
- bone morphogenetic protein
- TGFβR
- TGF-β type I and II receptor
- En
- embryonic day n
- ALP
- agonist-induced linker region phosphorylation
- NC
- neural crest.
REFERENCES
- 1. Blobe G. C., Schiemann W. P., Lodish H. F. (2000) Role of transforming growth factor β in human disease. N. Engl. J. Med. 342, 1350–1358 [DOI] [PubMed] [Google Scholar]
- 2. Massagué J. (1998) TGF-β signal transduction. Annu. Rev. Biochem. 67, 753–791 [DOI] [PubMed] [Google Scholar]
- 3. Derynck R., Zhang Y., Feng X. H. (1998) Smads. Transcriptional activators of TGF-β responses. Cell 95, 737–740 [DOI] [PubMed] [Google Scholar]
- 4. Derynck R., Zhang Y. E. (2003) Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 425, 577–584 [DOI] [PubMed] [Google Scholar]
- 5. Daly A. C., Randall R. A., Hill C. S. (2008) Transforming growth factor β-induced Smad1/5 phosphorylation in epithelial cells is mediated by novel receptor complexes and is essential for anchorage-independent growth. Mol. Cell. Biol. 28, 6889–6902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Matsuzaki K., Kitano C., Murata M., Sekimoto G., Yoshida K., Uemura Y., Seki T., Taketani S., Fujisawa J., Okazaki K. (2009) Smad2 and Smad3 phosphorylated at both linker and COOH-terminal regions transmit malignant TGF-β signal in later stages of human colorectal cancer. Cancer Res. 69, 5321–5330 [DOI] [PubMed] [Google Scholar]
- 7. Alarcón C., Zaromytidou A. I., Xi Q., Gao S., Yu J., Fujisawa S., Barlas A., Miller A. N., Manova-Todorova K., Macias M. J., Sapkota G., Pan D., Massagué J. (2009) Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell 139, 757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kretzschmar M., Liu F., Hata A., Doody J., Massagué J. (1997) The TGF-β family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes Dev. 11, 984–995 [DOI] [PubMed] [Google Scholar]
- 9. Aragón E., Goerner N., Zaromytidou A. I., Xi Q., Escobedo A., Massagué J., Macias M. J. (2011) A Smad action turnover switch operated by WW domain readers of a phosphoserine code. Genes Dev. 25, 1275–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao S., Alarcón C., Sapkota G., Rahman S., Chen P. Y., Goerner N., Macias M. J., Erdjument-Bromage H., Tempst P., Massagué J. (2009) Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-β signaling. Mol. Cell 36, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sorrentino A., Thakur N., Grimsby S., Marcusson A., von Bulow V., Schuster N., Zhang S., Heldin C. H., Landström M. (2008) The type I TGF-β receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat. Cell Biol. 10, 1199–1207 [DOI] [PubMed] [Google Scholar]
- 12. Yamashita M., Fatyol K., Jin C., Wang X., Liu Z., Zhang Y. E. (2008) TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-β. Mol. Cell 31, 918–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang C., Deng L., Hong M., Akkaraju G. R., Inoue J., Chen Z. J. (2001) TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412, 346–351 [DOI] [PubMed] [Google Scholar]
- 14. Gingery A., Bradley E. W., Pederson L., Ruan M., Horwood N. J., Oursler M. J. (2008) TGF-β coordinately activates TAK1/MEK/AKT/NFκB and SMAD pathways to promote osteoclast survival. Exp. Cell Res. 314, 2725–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hoffmann A., Preobrazhenska O., Wodarczyk C., Medler Y., Winkel A., Shahab S., Huylebroeck D., Gross G., Verschueren K. (2005) Transforming growth factor-β-activated kinase-1 (TAK1), a MAP3K, interacts with Smad proteins and interferes with osteogenesis in murine mesenchymal progenitors. J. Biol. Chem. 280, 27271–27283 [DOI] [PubMed] [Google Scholar]
- 16. Holm T. M., Habashi J. P., Doyle J. J., Bedja D., Chen Y., van Erp C., Lindsay M. E., Kim D., Schoenhoff F., Cohn R. D., Loeys B. L., Thomas C. J., Patnaik S., Marugan J. J., Judge D. P., Dietz H. C. (2011) Noncanonical TGFβ signaling contributes to aortic aneurysm progression in Marfan syndrome mice. Science 332, 358–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iwata J., Hacia J. G., Suzuki A., Sanchez-Lara P. A., Urata M., Chai Y. (2012) Modulation of noncanonical TGF-β signaling prevents cleft palate in Tgfbr2 mutant mice. J. Clin. Invest. 122, 873–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cardoso S., Robertson S. P., Daniel P. B. (2012) TGFBR1 mutations associated with Loeys-Dietz syndrome are inactivating. J. Recept. Signal. Transduct. Res. 32, 150–155 [DOI] [PubMed] [Google Scholar]
- 19. Loeys B. L., Chen J., Neptune E. R., Judge D. P., Podowski M., Holm T., Meyers J., Leitch C. C., Katsanis N., Sharifi N., Xu F. L., Myers L. A., Spevak P. J., Cameron D. E., De Backer J., Hellemans J., Chen Y., Davis E. C., Webb C. L., Kress W., Coucke P., Rifkin D. B., De Paepe A. M., Dietz H. C. (2005) A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat. Genet. 37, 275–281 [DOI] [PubMed] [Google Scholar]
- 20. Larsson J., Goumans M. J., Sjöstrand L. J., van Rooijen M. A., Ward D., Levéen P., Xu X., ten Dijke P., Mummery C. L., Karlsson S. (2001) Abnormal angiogenesis but intact hematopoietic potential in TGF-β type I receptor-deficient mice. EMBO J. 20, 1663–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Danielian P. S., Muccino D., Rowitch D. H., Michael S. K., McMahon A. P. (1998) Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 8, 1323–1326 [DOI] [PubMed] [Google Scholar]
- 22. Yu L., Gu S., Alappat S., Song Y., Yan M., Zhang X., Zhang G., Jiang Y., Zhang Z., Zhang Y., Chen Y. (2005) Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development 132, 4397–4406 [DOI] [PubMed] [Google Scholar]
- 23. Liu W., Lan Y., Pauws E., Meester-Smoor M. A., Stanier P., Zwarthoff E. C., Jiang R. (2008) The Mn1 transcription factor acts upstream of Tbx22 and preferentially regulates posterior palate growth in mice. Development 135, 3959–3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bellusci S., Furuta Y., Rush M. G., Henderson R., Winnier G., Hogan B. L. (1997) Involvement of Sonic hedgehog (Shh) in mouse embryonic lung growth and morphogenesis. Development 124, 53–63 [DOI] [PubMed] [Google Scholar]
- 25. Dudas M., Nagy A., Laping N. J., Moustakas A., Kaartinen V. (2004) Tgf-β3-induced palatal fusion is mediated by Alk-5/Smad pathway. Dev. Biol. 266, 96–108 [DOI] [PubMed] [Google Scholar]
- 26. Harlow E., Lane D. (1988) Antibodies: A Laboratory Manual, pp. 481–482, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York [Google Scholar]
- 27. Thomas P. S., Kim J., Nunez S., Glogauer M., Kaartinen V. (2010) Neural crest cell-specific deletion of Rac1 results in defective cell-matrix interactions and severe craniofacial and cardiovascular malformations. Dev. Biol. 340, 613–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nagy A., Gertsenstein M., Vintersten K., Behringer R. (2003) Manipulating the Mouse Embryo: A Laboratory Manual, pp. 687–691, 3rd Ed., Academic Press, New York [Google Scholar]
- 29. Ishii M., Arias A. C., Liu L., Chen Y. B., Bronner M. E., Maxson R. E. (2012) A stable cranial neural crest cell line from mouse. Stem Cells Dev. 21, 3069–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Z., Song Y., Zhao X., Zhang X., Fermin C., Chen Y. (2002) Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development 129, 4135–4146 [DOI] [PubMed] [Google Scholar]
- 31. Hilliard S. A., Yu L., Gu S., Zhang Z., Chen Y. P. (2005) Regional regulation of palatal growth and patterning along the anterior-posterior axis in mice. J. Anat. 207, 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu W., Sun X., Braut A., Mishina Y., Behringer R. R., Mina M., Martin J. F. (2005) Distinct functions for Bmp signaling in lip and palate fusion in mice. Development 132, 1453–1461 [DOI] [PubMed] [Google Scholar]
- 33. Han J., Mayo J., Xu X., Li J., Bringas P., Jr., Maas R. L., Rubenstein J. L., Chai Y. (2009) Indirect modulation of Shh signaling by Dlx5 affects the oral-nasal patterning of palate and rescues cleft palate in Msx1-null mice. Development 136, 4225–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rice R., Spencer-Dene B., Connor E. C., Gritli-Linde A., McMahon A. P., Dickson C., Thesleff I., Rice D. P. (2004) Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J. Clin. Invest. 113, 1692–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baek J. A., Lan Y., Liu H., Maltby K. M., Mishina Y., Jiang R. (2011) Bmpr1a signaling plays critical roles in palatal shelf growth and palatal bone formation. Dev. Biol. 350, 520–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Massagué J., Seoane J., Wotton D. (2005) Smad transcription factors. Genes Dev. 19, 2783–2810 [DOI] [PubMed] [Google Scholar]
- 37. Wang G., Matsuura I., He D., Liu F. (2009) Transforming growth factor-β-inducible phosphorylation of Smad3. J. Biol. Chem. 284, 9663–9673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wrighton K. H., Feng X. H. (2008) To (TGF)β or not to (TGF)β. Fine-tuning of Smad signaling via post-translational modifications. Cell. Signal. 20, 1579–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yamaguchi K., Shirakabe K., Shibuya H., Irie K., Oishi I., Ueno N., Taniguchi T., Nishida E., Matsumoto K. (1995) Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science 270, 2008–2011 [DOI] [PubMed] [Google Scholar]
- 40. Ito Y., Yeo J. Y., Chytil A., Han J., Bringas P., Jr., Nakajima A., Shuler C. F., Moses H. L., Chai Y. (2003) Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development 130, 5269–5280 [DOI] [PubMed] [Google Scholar]
- 41. Dudas M., Kim J., Li W. Y., Nagy A., Larsson J., Karlsson S., Chai Y., Kaartinen V. (2006) Epithelial and ectomesenchymal role of the type I TGF-β receptor ALK5 during facial morphogenesis and palatal fusion. Dev. Biol. 296, 298–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Iwata J., Tung L., Urata M., Hacia J. G., Pelikan R., Suzuki A., Ramenzoni L., Chaudhry O., Parada C., Sanchez-Lara P. A., Chai Y. (2012) Fibroblast growth factor 9 (FGF9)-pituitary homeobox 2 (PITX2) pathway mediates transforming growth factor β (TGFβ) signaling to regulate cell proliferation in palatal mesenchyme during mouse palatogenesis. J. Biol. Chem. 287, 2353–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Economou A. D., Ohazama A., Porntaveetus T., Sharpe P. T., Kondo S., Basson M. A., Gritli-Linde A., Cobourne M. T., Green J. B. (2012) Periodic stripe formation by a Turing mechanism operating at growth zones in the mammalian palate. Nat. Genet. 44, 348–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Song Z., Liu C., Iwata J., Gu S., Suzuki A., Sun C., He W., Shu R., Li L., Chai Y., Chen Y. (2013) Mice with Tak1 deficiency in neural crest lineage exhibit cleft palate associated with abnormal tongue development. J. Biol. Chem. 288, 10440–10450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shim J. H., Greenblatt M. B., Xie M., Schneider M. D., Zou W., Zhai B., Gygi S., Glimcher L. H. (2009) TAK1 is an essential regulator of BMP signalling in cartilage. EMBO J. 28, 2028–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hough C., Radu M., Doré J. J. (2012) Tgf-β induced Erk phosphorylation of Smad linker region regulates Smad signaling. PLoS One 7, e42513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cohen-Solal K. A., Merrigan K. T., Chan J. L., Goydos J. S., Chen W., Foran D. J., Liu F., Lasfar A., Reiss M. (2011) Constitutive Smad linker phosphorylation in melanoma. A mechanism of resistance to transforming growth factor-β-mediated growth inhibition. Pigment Cell Melanoma Res. 24, 512–524 [DOI] [PMC free article] [PubMed] [Google Scholar]