Background: Collagen IX is an integral cartilage extracellular matrix component important in skeletal development and joint function.
Results: Proteomic analysis and validation studies revealed novel alterations in collagen IX null cartilage.
Conclusion: Matrilin-4, collagen XII, thrombospondin-4, fibronectin, βig-h3, and epiphycan are components of the in vivo collagen IX interactome.
Significance: We applied a proteomics approach to advance our understanding of collagen IX ablation in cartilage.
Keywords: Cartilage, Chondrocytes, Collagen, Extracellular Matrix, Mass Spectrometry (MS), Proteomics, Chondrodysplasia
Abstract
The cartilage extracellular matrix is essential for endochondral bone development and joint function. In addition to the major aggrecan/collagen II framework, the interacting complex of collagen IX, matrilin-3, and cartilage oligomeric matrix protein (COMP) is essential for cartilage matrix stability, as mutations in Col9a1, Col9a2, Col9a3, Comp, and Matn3 genes cause multiple epiphyseal dysplasia, in which patients develop early onset osteoarthritis. In mice, collagen IX ablation results in severely disturbed growth plate organization, hypocellular regions, and abnormal chondrocyte shape. This abnormal differentiation is likely to involve altered cell-matrix interactions but the mechanism is not known. To investigate the molecular basis of the collagen IX null phenotype we analyzed global differences in protein abundance between wild-type and knock-out femoral head cartilage by capillary HPLC tandem mass spectrometry. We identified 297 proteins in 3-day cartilage and 397 proteins in 21-day cartilage. Components that were differentially abundant between wild-type and collagen IX-deficient cartilage included 15 extracellular matrix proteins. Collagen IX ablation was associated with dramatically reduced COMP and matrilin-3, consistent with known interactions. Matrilin-1, matrilin-4, epiphycan, and thrombospondin-4 levels were reduced in collagen IX null cartilage, providing the first in vivo evidence for these proteins belonging to the collagen IX interactome. Thrombospondin-4 expression was reduced at the mRNA level, whereas matrilin-4 was verified as a novel collagen IX-binding protein. Furthermore, changes in TGFβ-induced protein βig-h3 and fibronectin abundance were found in the collagen IX knock-out but not associated with COMP ablation, indicating specific involvement in the abnormal collagen IX null cartilage. In addition, the more widespread expression of collagen XII in the collagen IX-deficient cartilage suggests an attempted compensatory response to the absence of collagen IX. Our differential proteomic analysis of cartilage is a novel approach to identify candidate matrix protein interactions in vivo, underpinning further analysis of mutant cartilage lacking other matrix components or harboring disease-causing mutations.
Introduction
In cartilage, the macromolecular organization of the extracellular matrix (ECM)4 is essential for tissue mechanostability during development and throughout adult life. The two major supramolecular assemblies of the cartilage ECM are composed of fibrillar collagen and an extrafibrillar network that is predominantly aggrecan and hyaluronan. The interaction between these macromolecular networks is stabilized by mutual binding to adaptor protein complexes within an ill-defined perifibrillar compartment. Disruption of these macromolecular networks or the components that bridge them compromises the mechanostability of cartilage and underlies a broad spectrum of inherited skeletal dysplasias (1, 2).
Collagen IX is proposed to stabilize the fibrillar and proteoglycan networks via lateral association with collagen II/XI and outward projection of the N-terminal COL3-NC4 domain from the fibril surface. This conformation directs the organization of the fibrillar network, stabilizes the individual fibrils, and permits interaction of the N-terminal domains with other perifibrillar matrix components such as matrilin-3 and cartilage oligomeric matrix protein (COMP) (3). In humans, autosomal dominant mutations in genes encoding components of this collagen IX·COMP·matrilin-3 adaptor protein complex lead to skeletal defects. In-frame deletions in any of the three collagen IX genes cause multiple epiphyseal dysplasia, which is associated with irregular epiphyses of the long bones, moderate disproportionate dwarfism, and osteoarthritis-like cartilage degeneration (4). Mutated collagen IX chains that are retained in intracellular inclusions compromise endoplasmic reticulum function (5), whereas secreted chains may exert a dominant-negative effect on the assembly and function of the cartilage ECM, as shown for mutant COMP secreted into the matrix (6).
Further insight into the role of these perifibrillar components has been gained through gene targeting studies in mice (7). The absence of collagen IX disturbs the organization of the pre- and perinatal growth plate. The columnar arrangement of chondrocytes parallel to the long bone axis is altered and large hypocellular areas with irregular proteoglycan deposition are found in the central regions of the growth plate. These alterations appear to affect chondrocyte survival, leading to growth retardation and short stature (8, 9). At later stages in juvenile and adult mice the phenotype becomes attenuated but at 6 months an osteoarthritis-like phenotype develops, characterized by proteoglycan depletion and loss of intact collagen II (10). Hence, alteration of collagen IX homeostasis affects the ECM architecture, the differentiation and survival of chondrocytes, and the matrix-chondrocyte cross-talk. Similar pathological events are observed in mice expressing dominant mutations in COMP and matrilin-3, underlining the stabilizing role of these three components as a functional protein complex (11, 12).
The detailed characterization of these essential perifibrillar components is clearly an important step toward elucidating the molecular mechanisms underlying human chondrodysplasias, as well as identification of novel disease gene targets. Recently, fibronectin was identified as a new ligand for the collagen IX NC4 domain using a yeast two-hybrid screen and the two proteins were shown to co-localize in articular cartilage (13). Using affinity chromatography combined with mass spectrometry, Brown et al. (14) identified potential interactions between bovine fetal cartilage proteins and the recombinant amino-terminal domain of collagen XI. To date, however, a proteomic analysis of the perifibrillar “interactome,” which in vivo comprises direct or indirect physical, functional and genetic interactions, has not been performed. We recently developed the methodology for differential profiling of proteins extracted from wild-type mouse cartilage, using postnatal 3-day (P3) and 21-day (P21) femoral heads (15). Despite the limited quantity of tissue available, this in vivo model of cartilage development enables changes in protein expression to be assessed within the complex physiological environment of native cartilage.
In this study we used solubility-based protein fractionation in combination with label-free quantitative mass spectrometry to assess the response to collagen IX ablation at the proteomic level. Among the significant differences between wild-type and knock-out cartilage were known interaction partners of collagen IX, demonstrating the applicability of this approach to other mouse mutants lacking cartilage constituents. In addition, we found several novel members of the collagen IX interactome and validated our findings using immunoblotting and immunohistochemistry. These results not only extend our understanding of the perifibrillar adaptor protein complexes but provide broader insight into the biomolecular response to collagen IX ablation in mice.
EXPERIMENTAL PROCEDURES
Cartilage Dissection and Protein Extraction
Femoral head cartilage was obtained from P3 and P21 wild-type C57/Bl6, COMP-deficient (16) and collagen IX-deficient mice (10) after sacrifice in accordance with Institutional Animal Ethics guidelines. We used three biological replicates per genotype per time point, where each replicate sample comprised femoral heads pooled from either four P3 mice or three P21 mice. Cartilage was dissected by dislocation of the hip joint, fracture at the femoral neck, and removal of the ligamentum teres at the insertion site. The tissue was rinsed in PBS, frozen on dry ice, and stored at −80 °C until required. The P3 and P21 femoral head cartilage was pulverized using a liquid nitrogen-cooled tissue grinder, transferred to Eppendorf tubes containing 100 μl of Tris acetate buffer, pH 8.0, 10 mm EDTA, and 0.1 unit of chondroitinase ABC and deglycosylated for 6 h at 37 °C. Sequential protein extracts were prepared using a nondenaturing buffer (1 m NaCl in 100 mm Tris acetate, pH 8.0) followed by a chaotropic guanidine buffer (4 m GdnHCl, 65 mm DTT, 10 mm EDTA in 50 mm sodium acetate, pH 5.8) and centrifugal ultrafiltration (100 kDa cut-off) as described (15). Protein extracts were precipitated with 9 volumes of ethanol, washed twice in 70% (v/v) ethanol, and resuspended in 150 μl of solubilization buffer containing 7 m urea, 2 m thiourea, 4% CHAPS, and 30 mm Tris, pH 8.0. Protein concentrations were estimated using the Bradford assay (Pierce).
SDS-PAGE, Two-dimensional Electrophoresis, and Immunoblotting
Sample aliquots equivalent to 2% of the protein yield were resolved through 4–12% acrylamide BisTris NuPAGE gels (Invitrogen). Protein bands were detected by silver staining (15) to identify any genotype-specific differences. Proteins were separated by two-dimensional electrophoresis using pH 3–10 isoelectric focusing strips (Bio-Rad) and 3–12% NuPAGE Zoom gels (Invitrogen) as described previously (17). Proteins were visualized by silver staining and overlaid pseudo-colored images of representative gels were generated using the automated spot matching and warping software Delta2D (Decodon). The complete set of two-dimensional electrophoresis gel images is available in supplemental Fig. S1. To verify differences in protein abundance by immunoblotting, 5-μg sample aliquots were resolved by NuPAGE and proteins were transferred to low-fluorescence PVDF (GE Healthcare). Rabbit polyclonal antibodies to fibronectin (Millipore), TGF-β-induced protein βig-H3 (polyclonal antibody kindly donated by Dr Mark Gibson (18)), and collagen α1(VI) (polyclonal 1027) were used at a dilution of 1:1000 and mouse antibodies to GAPDH (Abcam monoclonal number 6C5) and acetylated α-tubulin (Sigma mouse Ig2B monoclonal number T6793) were used at 1:2500. Blots were probed with Cy5-conjugated anti-rabbit or Cy3-conjugated anti-mouse antibodies at 1:2500, detected by laser scanning (Typhoon 9410, GE Healthcare) and bands were quantified relative to gel loading control proteins using ImageQuant (GE Healthcare).
Protein Identification by In-gel Digestion and Tandem Mass Spectrometry
Proteins were excised using a sterile scalpel blade, destained, and subjected to in-gel digestion with 250 ng of sequencing grade trypsin (Sigma) using standard methods (19). Peptides were resolved and analyzed using an XCT Plus ion trap mass spectrometer with on-line HPLC-Chip cube interface (Agilent, Palo Alto, CA) and MS/MS spectra were searched against the Mus musculus subset of the SwissProt database (version 2012_03) using Mascot version 2.2.06 (Matrix Science). S-Carboxamidomethylation of cysteine was specified as a fixed modification and methionine oxidation as a variable modification. Charge states of +1, +2, and +3 were accepted. Parent ion and fragment ion mass tolerances of 1.2 and 0.6 Da, respectively, were used. Enzyme cleavage was set to trypsin, allowing for a maximum of three missed cleavages. Where database searches returned multiple protein identifications the highest-scoring match was accepted.
HPLC-LTQ-Orbitrap Tandem Mass Spectrometry
Protein samples were analyzed by capillary HPLC-MS/MS using an LTQ-Orbitrap XL (ThermoFisher Scientific). Equal volumes of each fraction (90 μl/extract) were sequentially reduced, alkylated, and trypsin digested as previously described (20). Approximately 50 μg of protein/biological sample (E0 + E1 + E2) was processed. Normalization of total spectral counts for each biological sample was used to compensate for minor differences in protein concentration, variation in trypsin digestion efficiency between digests, and peptide loading onto the HPLC. Aliquots of tryptic peptides equivalent to 25% of the in-solution digests were loaded onto a 300 μm × 5-mm C18 trap column (Dionex Acclaim® PepMapTM μ-Precolumn) controlled by UltiMate 3000 HPLC system (Dionex) and separated on a Vydac Everest C18 300 Å, 150 μm × 150-mm analytical column (Alltech) using an acetonitrile gradient described previously (20). The LTQ-Orbitrap was fitted with a dynamic nano-electrospray ion source (Proxeon) containing a 30-μm inner diameter uncoated silica emitter (New Objective). The LTQ-Orbitrap XL was controlled using Xcalibur 2.0 software (ThermoFisher Scientific) and operated in data-dependent acquisition mode whereby the survey scan was acquired in the Orbitrap with a resolving power set to 60,000 (at 400 m/z). MS/MS spectra were concurrently acquired in the LTQ mass analyzer on the eight most intense ions from the FT survey scan. Charge state filtering, where unassigned precursor ions were not selected for fragmentation, and dynamic exclusion (repeat count 1, repeat duration 30 s, exclusion list size 500) were used. Fragmentation conditions in the LTQ were: 35% normalized collision energy, activation q of 0.25, 30-ms activation time, and minimum ion selection intensity of 500 counts.
Database Searching
The acquired MS/MS data were analyzed using Mascot version 2.2.06 (Matrix Science). Proteome Discoverer version 1.3 (ThermoFisher Scientific) was used to extract tandem mass spectra from Xcalibur raw files and submit searches to an in-house Mascot server according to the parameters: S-carboxamidomethylation of cysteine residues specified as a fixed modification and deamidation of asparagine and glutamine, hydroxylation of proline, and oxidation of methionine specified as variable modifications. Parent ion tolerance of 10 ppm and fragment ion mass tolerances of 0.8 Da were used and enzyme cleavage was set to trypsin, allowing for a maximum of two missed cleavages. The database searched consisted of 46,137 sequences, comprising the UniProt complete proteome set for M. musculus (45,889 sequences downloaded on May 23rd, 2011) and sequences for common contaminants (downloaded from the Max Plank Institute). The automatic Mascot decoy database search was performed for all datasets.
Criteria for Protein Identification
The Mascot search results were loaded into Scaffold version 3.2 to assign probabilities to peptide and protein matches (21). Peptide-spectrum matches were accepted if the peptide was assigned a probability greater than 0.95 as specified by the Peptide Prophet algorithm (22). Based on the fit of the data to the predicted distributions of correct and incorrect matches, only peptide-spectrum matches for charge states +1, +2, and +3 were accepted. Protein identifications were accepted if the protein contained two or more unique peptide sequences and the protein was assigned a probability >0.99 by the Protein Prophet algorithm (23). This threshold will constrain the protein false discovery rate (FDR) to <1%. The minimal list of proteins that satisfy the principal of parsimony is reported.
Statistical Analysis of LTQ-Orbitrap Mass Spectrometry Data
Statistical analysis was performed essentially as described (15). Briefly, MS/MS data for the fractionated samples were recombined in silico using the Scaffold MudPIT function. Global normalization was applied to the unweighted spectral counts and fold-changes were estimated using the mean of the normalized counts for the collagen IX knock-out relative to the wild-type, with a pseudo count of one added to allow for missing values. The β-binomial test (24) was used to assess differences between the knock-out and wild-type samples, and a q-value threshold (25) of <0.1 was applied to identify proteins with a significant difference in abundance between wild-type and collagen IX knock-out cartilage.
Generation of a Collagen IX Protein Interaction Network
The official gene symbols corresponding to the list of proteins with significant differences in abundance were analyzed using Ingenuity Pathway Analysis (IPA). The IPA Core Analysis with default parameters was used to generate a molecular network from the IPA Knowledge Base. Using the IPA “Keep” tool, only relationship types of “membership” and “protein-protein interaction” were retained. Proteins not identified in our analysis were removed.
ELISA Style Ligand Binding Assay
Collagen IX was extracted from cultures of embryonic chicken chondrocytes and purified as previously described (26). Collagen IX was diluted in TBS and 50 μl (10 mg/ml) were coated overnight at room temperature onto 96-well plates (NuncMaxisorb). After washing with TBS supplemented with 0.05% Tween 20, plates were blocked for 1 h at room temperature with TBS containing 5% low-fat milk powder. His myc-tagged matrilin-3 and -4 were affinity purified from the cell culture supernatant of transfected 293 EBNA cells (27) and thrombospondin-4 was purchased from R&D Systems (Wiesbaden-Nordenstadt, Germany). Proteins were diluted in blocking solution to the concentrations indicated and incubated in coated wells for 2 h at room temperature. To ascertain the level of nonspecific binding, increasing concentrations of ligand were incubated in uncoated but blocked wells. Measurements were always performed in duplicate. After extensive washing with TBS supplemented with 0.05% Tween 20, bound ligands were detected with specific rabbit antibodies directed against matrilin-3 and -4 and guinea pig antibodies against thrombospondin-4 (28), respectively. Bound primary antibodies were detected by corresponding horseradish peroxidase-conjugated secondary antibodies (DakoCytomation) and tetramethylbenzidine as substrate. After stopping the reaction with 10% sulfuric acid, absorption was measured at 450 nm.
RNA Isolation and Quantification
RNA was isolated by acid guanidinium thiocyanate/phenol/chloroform extraction as previously described (29) and the quality of total RNA samples was confirmed by capillary electrophoresis according to the manufacturer's specifications (Agilent). RNA samples were reverse transcribed into cDNA using the Superscript III 1°-strand Synthesis kit (Invitrogen). Pre-existing primer-probe pairs of the Roche universal probe library were employed for Matn4 and Thbs4. Quantitative real-time PCR (qPCR) were performed on the DNAengine-Opticon2 system (Bio-Rad). The efficiency of the qPCR was determined and relative expression levels were calculated by the Δ-ΔCT method (30) using actin for normalization.
Immunofluorescence
For immunohistochemical analysis, P3 wild-type and collagen IX null femoral heads were isolated, fixed overnight in 4% paraformaldehyde in phosphate-buffered saline, and decalcified in 0.5% EDTA for 3 weeks. Paraffin sections (5 μm) were cut, pre-treated with hyaluronidase and Triton X-100, and then stained with antibodies as described (31). Immunohistochemical analysis was performed with the following antibodies: rabbit polyclonal antibodies against matrilin-1, matrilin-3, or matrilin-4 (1:1000, (27)), collagen XII (32), and fibronectin (Chemicon Ab number 2033). Corresponding goat anti-rabbit secondary antibodies coupled to Alexa Fluor 488 (2 μg/ml, Invitrogen/Molecular Probes) were applied. Bright field as well as fluorescence images were taken (Nikon Eclipse TE2000-U Microscope) and analyzed using the NIS-Elements software (Nikon).
RESULTS
Analysis of Femoral Head Cartilage Protein Fractions
Collagen IX ablation causes age-dependent pathological changes in chondrocyte organization in growth plate cartilage and the redistribution/loss of ECM components (8, 9, 33). To further characterize these defects we investigated the global effect of collagen IX ablation on protein expression in femoral head cartilage at two postnatal developmental time points. Proteins were extracted from wild-type and collagen IX deficient 3-day (P3) and 21-day (P21) cartilage using 1 m NaCl followed by 4 m GdnHCl, a method developed to fractionate the proteins and facilitate proteomic analysis (15, 17). We first used SDS-PAGE to resolve the extracts and silver staining to detect differences in protein abundance between wild-type and collagen IX-deficient cartilage (Fig. 1). The NaCl-soluble fraction (E0) showed no obvious differences in any of the protein bands at either developmental stage, whereas several genotype-specific alterations were evident in the low (E1) and high (E2) molecular mass GdnHCl-soluble fractions. Analysis of these proteins by ion trap tandem mass spectrometry (MS/MS) identified two known collagen IX-binding proteins (supplemental Table S1, A). COMP was reduced in collagen IX null P21 E1 extracts and matrilin-3 was reduced in collagen IX null P21 E2 extracts. These results confirm previous reports showing the absence of collagen IX results in depletion of these two interacting proteins in vitro (34, 35) and in vivo (8). A third protein band, which was clearly and consistently reduced in the collagen null P3 E1 extracts, was identified as matrilin-1. This is consistent with the interaction between matrilin-1 A-domains and collagen IX shown in vitro (36) and provides the first evidence for the association of collagen IX and matrilin-1 under native conditions in vivo.
FIGURE 1.
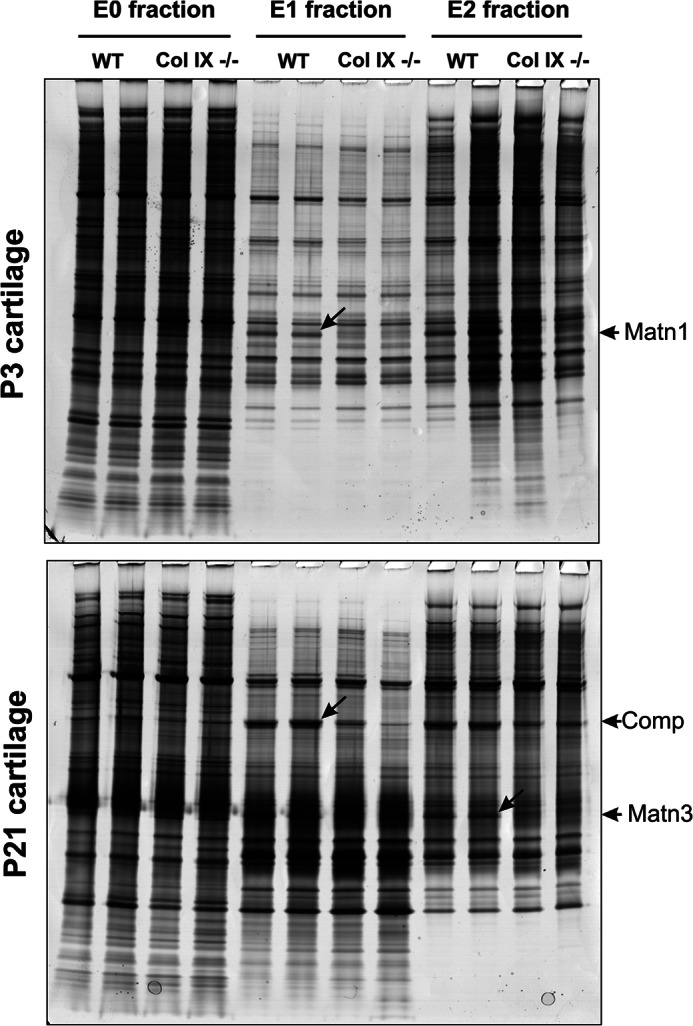
Analysis of sequential extracts of P3 and P21 mouse femoral head cartilage by SDS-PAGE and identification of differentially abundant proteins. Cartilage proteins were partitioned into three fractions based on differential solubility and molecular mass. The 1 m NaCl extract (E0 fraction) was not partitioned further, whereas the 4 m GdnHCl extract was separated into nominal high and low mass E1 and E2 fractions. Aliquots of replicate sequential extracts equivalent to 2% of the protein yield were resolved by 4–12% NuPAGE. Proteins that were clearly different between wild-type (WT) and collagen IX null (Col IX−/−) cartilage extracts, marked by diagonal arrows, were excised, digested with trypsin, and identified by tandem mass spectrometry on an HPLC-interfaced Agilent XCT Plus 3-dimensional ion trap. The differentially abundant proteins were identified as matrilin-1 (Matn1), cartilage oligomeric matrix protein (Comp), and matrilin-3 (Matn3) as indicated by arrows at the gel right-hand margin.
LTQ-Orbitrap Tandem Mass Spectrometry and Statistical Analysis
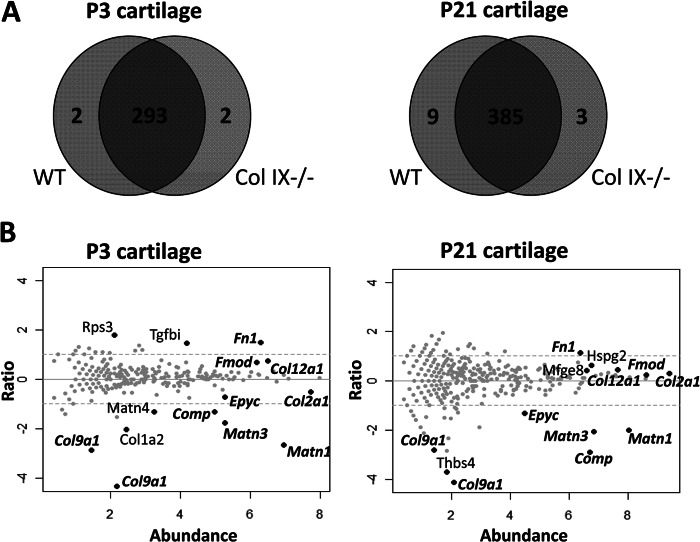
To achieve more comprehensive analysis of the effect of collagen IX ablation we applied a proteomic approach previously used to quantify protein expression changes in wild-type cartilage development (15). Three biological replicates used to prepare independent sequential extracts were prepared from P3 and P21 femoral head cartilage of wild-type and collagen IX null mice. The 36 protein samples were trypsin digested in solution and peptides were resolved by capillary HPLC and analyzed using a high-resolution LTQ-Orbitrap hybrid tandem mass spectrometer. Database searching identified 297 proteins in the P3 dataset (24,101 peptide-spectrum matches with a FDR of 0.78%). Similarly, 397 proteins were identified in P21 cartilage (41,764 peptide-spectrum matches with a FDR of 0.92%). The complete set of peptides identified in this study is listed in supplemental Table S2 and the complete list of proteins in supplemental Table S3. The relationship between proteins identified in wild-type and collagen IX-deficient cartilage is shown in Fig. 2A. To identify statistically significant differences between wild-type and collagen IX null cartilage, each protein was assigned a p value using the β-binomial test and q values were estimated to correct for multiple comparisons. At a FDR threshold of q < 0.1, the number of differentially abundant proteins between wild-type and collagen IX-deficient cartilage was 14 at P3 and 13 at P21, with a single false positive expected for each comparison (supplemental Table S3).
FIGURE 2.
LTQ-Orbitrap analysis of proteins extracted from wild-type and collagen IX-deficient cartilage. A, representation of the proteins identified in P3 and P21 cartilage. The Venn diagrams illustrate the overlap in proteins identified in wild-type (WT) and collagen IX null (Col IX−/−) cartilage extracts from P3 and P21 femoral heads. The numbers of proteins identified with two or more peptide sequences are indicated. B, relative abundance of the proteins identified in wild-type (WT) and collagen IX-deficient (NULL) cartilage. Spectral count data for each protein is plotted using the x axis to represent abundance (½ × log2 [NULL × WT]) and the y axis for fold-change (log2 [NULL/WT]). Proteins increased or decreased in abundance in collagen IX-deficient cartilage are represented by data points above or below the y axis, respectively. Black data points represent the proteins that are significantly altered between the genotypes and are annotated using official gene symbols. Proteins that are differentially abundant in both P3 and P21 cartilage are labeled in bold italics.
Protein Abundance Differences between Wild-type and Collagen IX Null Cartilage
Normalized counts for the proteins identified in wild-type and collagen IX null cartilage were plotted as ratio versus abundance using ½ × log2 [NULL × WT] to express protein abundance and log2 [NULL/WT] to express the protein ratio. Proteins with a significant difference in abundance (q < 0.1) were annotated on the plots (Fig. 2B). With the exception of one ribosomal subunit (Rps3) all the significant proteins were secreted components of the cartilage pericellular or extracellular matrix, indicating that the major effect of collagen IX ablation was altered organization or stability of the ECM. As would be anticipated, peptides mapping to collagen α1(IX) were only detected in wild-type cartilage, the gene symbol Col9a1 referring to UniProt accession number Q05722 and the unreviewed entry Q9D0D2. A further seven matrix components were differentially abundant between wild-type and collagen IX null cartilage at both developmental stages, cross-validating the two data sets. COMP (2.5- and 7.4-fold), matrilin-3 (3.4- and 4.2-fold), matrilin-1 (4.0- and 6.4-fold), and epiphycan (1.7- and 2.5-fold) were significantly reduced in collagen IX-deficient cartilage, the fold-changes specifying differences between wild-type and collagen IX null cartilage at P3 and P21, respectively. In contrast, fibronectin (2.2- and 2.8-fold), collagen XII (1.4- and 1.7-fold), and fibromodulin (1.2- and 1.6-fold) were detected at increased levels in the collagen IX knock-out compared with wild-type cartilage.
Several additional matrix components were identified as differentially abundant in P3 or P21 cartilage only. In addition to matrilin-1 and matrilin-3, levels of matrilin-4 were also significantly reduced in collagen IX-deficient cartilage at P3 only. Although the cross-linked fractions of the fibrillar collagens were not analyzed in this study, the extracted fraction of collagen α2(I) chains were also reduced in P3 collagen IX null cartilage. The final protein that was significantly altered in P3 cartilage was the TGFβ-induced protein βig-h3 (Tgfbi), which increased 2.7-fold in the collagen IX knock-out.
Conversely, thrombospondin-4 was identified as differentially abundant between wild-type and collagen IX null cartilage at P21 only. Noticeably, no peptides corresponding to thrombospondin-4 were detected in the collagen IX knock-out. Two further components were significantly increased in the collagen IX knock-out P21 cartilage only. Lactadherin (Mfge8), a cell-surface glycoprotein that is expressed but with no reported function in cartilage (37) was increased 1.3-fold and perlecan (Hspg2), a known constituent of the chondrocyte pericellular environment, was increased 1.5-fold.
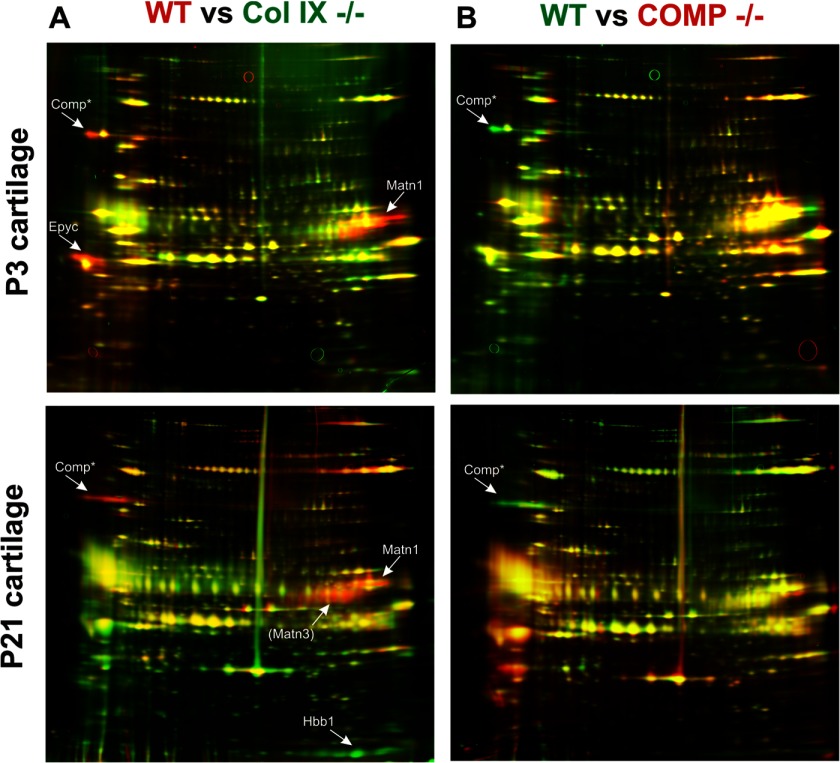
Comparison of Wild-type, Collagen IX Null, and COMP Null Cartilage by Two-dimensional Electrophoresis and Immunoblotting
The results of our proteomic analysis indicate that the major effect of collagen IX ablation is altered abundance of pericellular and extracellular components and provide new evidence for collagen IX-dependent interactions in cartilage. Although COMP is a known collagen IX-binding protein, the effect of COMP ablation is more subtle than the collagen IX knock-out, with only a mild disruption in chondrocyte columnar organization and no effect on the distribution of matrilin-3 (33). One question that arises from our data is whether any of the perturbations in the collagen IX knock-out ECM were caused by reduced COMP levels. We were able to address this by comparing wild-type, collagen IX null, and COMP null cartilage extracts. As a comprehensive analysis of the COMP knock-out was beyond the scope of this report, we focused on the cartilage fraction enriched in ECM components such as collagen VI, biglycan, decorin, fibromodulin, and the matrilins (17), using two-dimensional electrophoresis and identification of protein spots by mass spectrometry (Fig. 3, supplemental Fig. S1 and Table 1B). The reduced intensities of protein spots corresponding to COMP, matrilin-1, matrilin-3, and epiphycan were consistent with the differences observed by LTQ-Orbitrap analysis of wild-type and collagen IX null cartilage. However, little effect on matrilin-1, matrilin-3, or epiphycan levels in the COMP null cartilage was observed. The minor effect of COMP ablation on the GdnHCl-soluble ECM fraction provides further evidence for collagen IX, rather than COMP, as the functionally more important component in the perifibrillar complex.
FIGURE 3.
Differential two-dimensional electrophoresis analysis of guanidine-extracted proteins from wild-type, collagen IX-deficient, and COMP-deficient cartilage. The ECM-enriched (E1) fraction of wild-type (WT), collagen IX-deficient (Col IX−/−), and COMP-deficient (COMP−/−) cartilage was resolved by two-dimensional electrophoresis, silver stained, and the pseudo-colored images aligned and overlaid using Delta 2D software (Decodon). Proteins that showed clear and consistent genotype-specific differences were identified by tandem mass spectrometry. These proteins, epiphycan (Epyc), matrilin-1 (Matn1), matrilin-3 (Matn3), and hemoglobin (Hbb1), were labeled with arrows. Matn3 is annotated in parentheses to indicate this protein was identified on the basis of a single peptide. The annotation of the COMP protein spot is based on matched migration with COMP previously identified by two-dimensional electrophoresis and mass spectrometry (17). A, proteins more abundant in wild-type cartilage appear as red protein spots and proteins more abundant in collagen IX-deficient cartilage appear as green spots. B, proteins more abundant in wild-type cartilage appear as green protein spots and proteins more abundant in COMP-deficient cartilage appear as red spots.
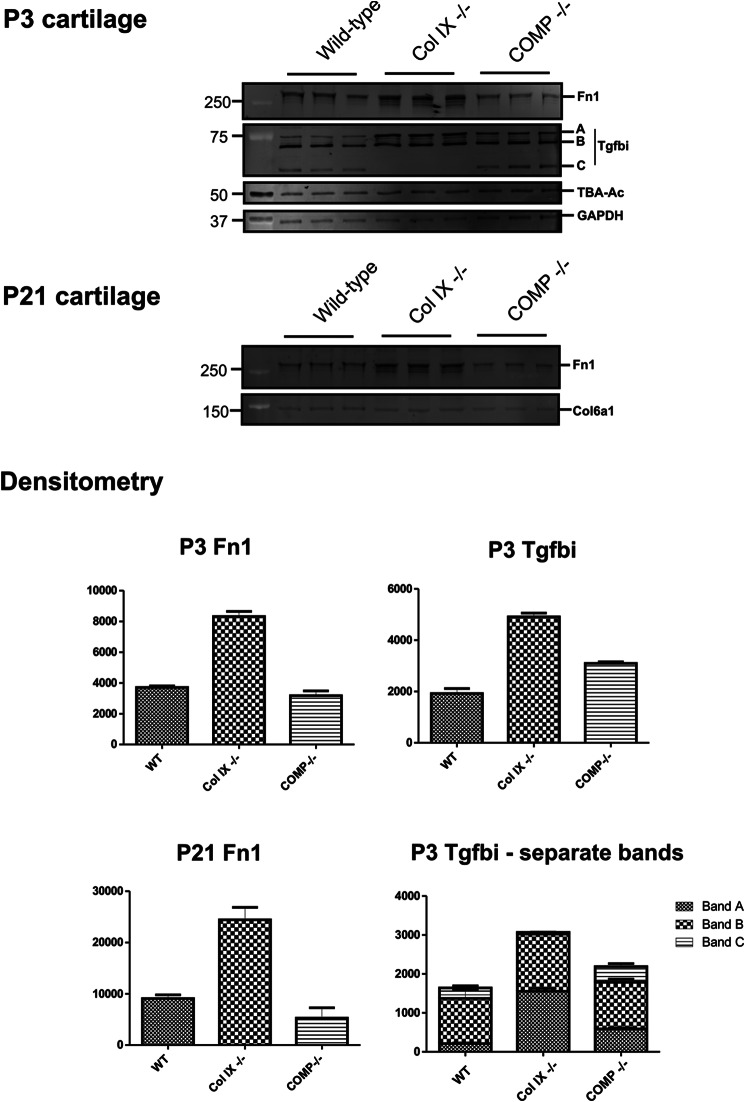
Two proteins increased in collagen IX-deficient cartilage (fibronectin and Tgfbi) are products of TGFβ-mediated signaling in chondrocytes in vitro and in vivo (38–40). To expand further on the results of our proteomic analysis, extracts prepared from wild-type, collagen IX null, and COMP null mice were analyzed by immunoblotting using antibodies to fibronectin and Tgfbi (Fig. 4). We found increased levels of fibronectin in P3 and P21 femoral head cartilage of collagen IX null mice, consistent with the LTQ-Orbitrap analysis. In addition, this alteration was genotype-specific, as COMP ablation had no measurable effect on fibronectin levels. Immunoblot analysis of Tgfbi revealed both quantitative and qualitative differences between the genotypes. The secreted form of Tgfbi migrates as a protein band of ∼75 kDa, corresponding to the predicted molecular mass of the intact protein lacking only its signal peptide (41). In wild-type and COMP null replicate cartilage extracts, Tgfbi migrated as the secreted protein plus two smaller isoforms. Processed Tgfbi isoforms were previously observed in co-cultures of ovarian cancer and peritoneal cells (41), however, our analysis provides the first evidence of Tgfbi processing in cartilage. Interestingly, the smaller of the two processed isoforms (band C) was not detected in the collagen IX-deficient cartilage, whereas the intact isoform (band A) was detected at correspondingly higher levels, suggesting that both the expression and processing of Tgfbi are affected in the collagen IX knock-out. Compared with wild-type P3 cartilage, total levels of Tgfbi were increased only in collagen IX-deficient cartilage and did not differ substantially in the COMP knock-out. However, quantitation of the individual bands by densitometry revealed a small but consistent increase in the level of intact Tgfbi in the COMP knock-out compared with the wild-type.
FIGURE 4.
Validation of proteomics data by fluorescent Western blotting and densitometry. GdnHCl-soluble extracts (E2 fraction) of P3 and P21 wild-type, Col IX−/−, and COMP−/− mouse femoral head cartilage were resolved by SDS-PAGE and probed with antibodies to the TGFβ-induced protein Tgfbi, fibronectin (Fn1). In P21 samples collagen VI (Col6a1) was used as a loading control, as similar levels were observed in the two-dimensional electrophoresis comparison of wild-type, collagen IX-deficient, and COMP-deficient P21 cartilage. In P3 cartilage, GAPDH, and acetylated α-tubulin (TBA-Ac) were used as housekeeping proteins. Background-subtracted volumes for each protein were calculated by densitometry, normalized to the values obtained for the housekeeping proteins to correct for minor loading differences between biological replicates, and plotted as mean values (n = 3) on the y axis, where error bars are mean ± S.E.
Evidence for the Direct Interaction between Matrilin-4, but Not Thrombospondin-4, and Collagen IX
Two proteins that were detected at decreased levels in collagen IX-deficient cartilage relative to wild-type cartilage were thrombospondin-4 and matrilin-4. The altered abundance of these proteins may be caused by the loss of a key binding partner as shown previously for matrilin-3 (8, 35) or reduced gene expression in collagen IX null cartilage. To distinguish between these possibilities, we first assessed the direct interaction of collagen IX with matrilin-4 and thrombospondin-4 by solid phase binding assays (Fig. 5A). We observed a dose-dependent interaction between collagen IX and matrilin-4 with a binding affinity of 0.5 nm. In parallel, we used matrilin-3 as a positive control for our assay (data not shown) and confirmed the previously described interaction with an apparent Kd in the low nanomolar range (35). In contrast, no association between thrombospondin-4 and collagen IX was observed under the conditions used. This suggests that the loss of matrilin-4 from cartilage lacking collagen IX is due to disruption of a specific interaction between collagen IX and matrilin-4, whereas decreased levels of thrombospondin-4 are more likely the result of reduced gene expression. To confirm this we used quantitative real-time PCR to compare the relative mRNA levels of matrilin-4 and thrombospondin-4 in wild-type and collagen IX null cartilage. Although expression of matn4 was not significantly altered, we observed a modest but significant reduction in thbs4 expression in collagen IX-deficient cartilage compared with the wild-type (Fig. 5B).
FIGURE 5.
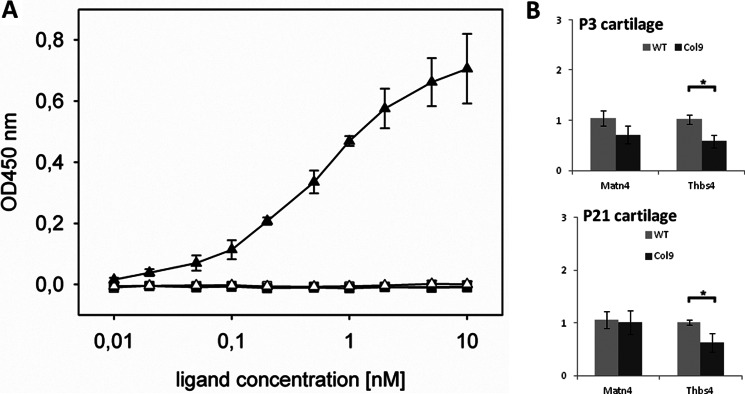
Investigation of collagen IX protein binding properties and the relative expression of thrombospondin-4 and matrilin-4 mRNA in wild-type and collagen IX-deficient cartilage. A, binding of soluble matrilin-4 (closed triangles) and thrombospondin-4 (closed squares), to immobilized collagen IX (500 ng/well) was determined using ELISA-style solid phase assays. Uncoated but blocked wells were used as negative controls (open triangles). The two ligands were used at the concentrations indicated and matrilin-4 or thrombospondin-4 specific antibodies were used to detect the level of bound ligand. Duplicate measurements were performed at each concentration and the mean values of seven independent assays for matrilin-4 and three independent assays for thrombospondin-4 are plotted. The resulting saturation curve for matrilin-4 was used to calculate an approximate Kd value of 0.5 nm. B, relative expression level of matrilin-4 (matn4) and thrombospondin-4 (thbs4) in femoral head cartilage of P3 and P21 wild-type and collagen IX-deficient cartilage was determined by qPCR. Results are shown relative to wild-type mRNA expression levels using β actin for normalization. Measurements were performed in triplicate on each biological replicate (n = 6) and the error bars represent the mean ± S.E. Significant differences between the expression levels detected in wild-type and collagen IX were determined using the unpaired two-tailed Student's t test (* = p < 0.05).
Altered Abundance and Distribution of Cartilage Matrix Components in Collagen IX-deficient Cartilage
Previous studies have shown altered distribution and abundance of known collagen IX-interacting proteins in the tibia of collagen IX-deficient mice, with overall loss of matrilin-3 and the re-distribution of COMP to an abnormal hypocellular region at the center of the epiphysis (8). To evaluate region-specific differences in protein abundance in wild-type and collagen IX-deficient femoral head cartilage, significantly altered matrix components that we identified using proteomics were further investigated by immunofluorescence (Fig. 6). In wild-type P3 cartilage immunofluorescence signals for matrilin-1, -3, and -4 were distributed evenly throughout the tissue (Fig. 6, A–C). In addition to the severely reduced staining for all three matrilins in the collagen IX knock-out we noted some interesting regional differences. Staining for matrilin-4 was reduced to background levels throughout the tissue, whereas diffuse matrix staining for matrilin-3 was observed in and surrounding the hypocellular region. Unlike matrilin-3 and -4, a specific signal for matrilin-1 was detected around the few chondrocytes remaining in the hypocellular region. Overall these results suggest that collagen IX is essential for normal distribution of the three matrilins expressed in epiphyseal cartilage.
FIGURE 6.
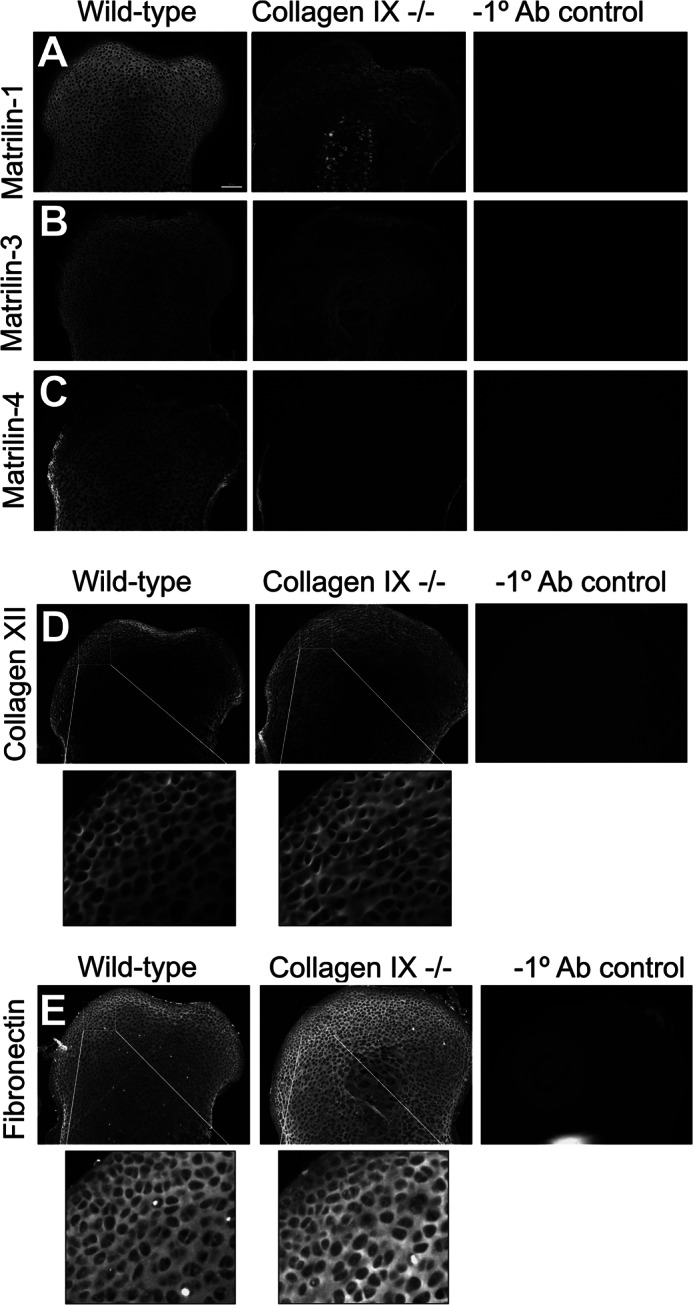
Region-specific changes in matrix protein distribution analyzed by immunofluorescence. The protein distribution of matrilin-1 (A), matrilin-3 (B), matrilin-4 (C), collagen XII (D), and fibronectin (E) in the femoral head cartilage of wild-type and collagen IX-deficient P3 mice was determined by immunostaining using in-plane matched paraffin sections. Images from each antibody series, including wild-type negative controls lacking primary antibody were captured using identical microscope and software settings. The boxed regions in D and E were magnified digitally to highlight region-specific differences in staining intensity. Images are representative of biological replicates of wild-type and collagen IX-deficient mice (n = 5). Scale bars, 100 μm.
Fibronectin has recently been identified as a collagen IX-interacting protein based on in vitro binding and co-localization in articular cartilage (13). Unlike the matrilins, which were depleted in the collagen IX knock-out, fibronectin was markedly increased relative to wild-type cartilage (Fig. 6E). Immunofluorescence staining revealed a more intense signal for fibronectin in all regions of the P3 collagen IX-deficient cartilage, apart from the surface layer. One further protein that was more abundant in the collagen IX knock-out was collagen XII, a fibril-associated collagen with interrupted triple helices (FACIT collagen) that interacts with both collagen I and collagen II. Similar to the developing rat epiphyses (42), collagen XII was localized to the presumptive articular surface in the wild-type mouse femoral head, whereas collagen XII was observed throughout the epiphyses of the collagen IX knock-out, with the exception of the hypocellular region (Fig. 6D). The increased expression and matrix-associated staining for collagen XII in the collagen IX null cartilage provides the first evidence of a potential compensatory mechanism by another FACIT collagen.
IPA Analysis of Differentially Abundant Proteins
We used IPA to identify relationships between the set of proteins identified as differentially abundant between collagen IX knock-out and wild-type cartilage. A single network was identified using IPA Core Analysis of the 15 ECM proteins (Fig. 7). Direct connections to collagen IX included the well established interacting protein matrilin-3 (8, 35) and more recently described interactions such as fibronectin (13). Although most of the nodes had one or more protein connections, two of the 15 differentially abundant proteins, epiphycan and lactadherin, were “orphan” proteins with no network connectivity.
FIGURE 7.
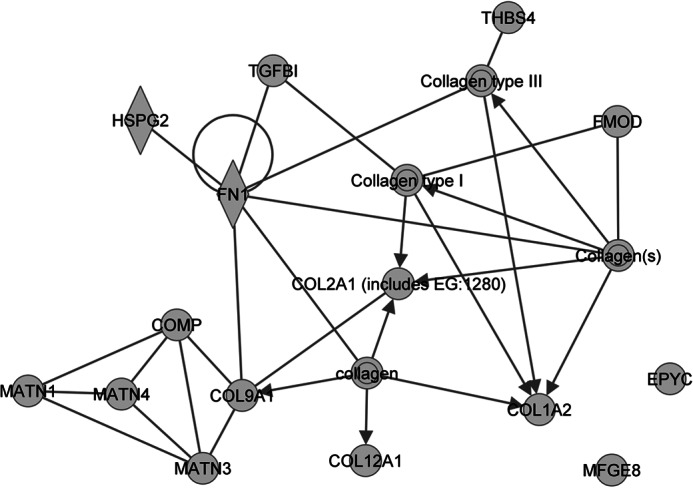
Interaction network generated using the 15 ECM proteins identified as differentially abundant between collagen IX knock-out and wild-type cartilage. Ingenuity pathway analysis was used to identify relationships between the 15 differentially abundant ECM proteins. In the network, proteins are represented using nodes and relationships between two proteins are represented using edges, where an edge represents a protein-protein interaction, and edges with arrowheads represent membership to a group or complex. All edges are supported by at least one literature reference. The gene products belonging to the collagen protein family are represented by an IPA network in supplemental Fig. S2.
DISCUSSION
Collagen IX was the first of many cartilage structural proteins to be investigated using targeted gene manipulation in mice (10, 43). These original studies revealed the role of collagen IX in maintaining cartilage integrity and were predictive of the degenerative changes in patients with multiple epiphyseal dysplasia (4). Here we used proteomics to gain a more comprehensive view of the collagen IX null phenotype and to further understand collagen IX function in vivo. A striking feature of our comparison between wild-type and collagen IX null cartilage is the overlap between the differentially expressed proteins identified at the two stages of cartilage development. In both P3 and P21 femoral head cartilage, the top five significant proteins ranked by q value were matrilin-1, matrilin-3, collagen α1(IX), COMP, and fibronectin. This data confirms the key interactions between collagen IX, matrilin-3, and COMP, and provides new evidence for the association of collagen IX with matrilin-1 and fibronectin in vivo. Other significant alterations in collagen IX null cartilage were detected either in P3 or P21 extracts. Most of these proteins are either enriched at P21 relative to P3 (thrombospondin-4 and lactadherin) or enriched at P3 relative to P21 (Tgfbi and matrilin-4) in normal femoral head cartilage (15). This highlights the benefit of using developmental stages with distinct tissue architecture and matrix composition. Other differentially expressed proteins, including collagen XII and epiphycan, had also not been previously associated with collagen IX. The significance of these results is discussed below.
Development of the mouse femoral head involves maturation of the articular cartilage and calcification of the underlying epiphyseal cartilage (15). In normal epiphyses, collagen XII expression is restricted to the articular surface in a reciprocal pattern to that of matrilin-1 (42). However, we detected elevated levels of collagen XII in the collagen IX null cartilage and further analysis by immunohistochemistry revealed collagen XII staining throughout the underlying region normally lacking collagen XII. A similar role to collagen IX has been proposed for collagen XII in the organization and stability of collagen fibrils, via lateral association of the COL1 domains (44). In addition to collagen II, there is evidence for a partial overlap in other ligands for these two FACIT collagens, for example, fibromodulin (45, 46) and COMP (47). However, the distinct sequence and domain structure of the collagen IX and collagen XII perifibrillar domains (44, 48) are likely to confer different properties, in particular the interaction with integrins and perifibrillar matrix components. Although up-regulation of collagen XII signals a potential compensation response to loss of collagen IX, the major phenotypic changes in collagen IX null cartilage suggests distinct properties and nonredundant function of these two fibril-associated collagens. It would be interesting to further examine the functional relationship between collagens IX and XII in double-deficient mice.
The collagen IX protein interaction network in Fig. 7 places the cohort of differentially abundant proteins in the context of previously observed interactions. From this network it is clear that the overall consequence of collagen IX ablation is the altered abundance of cartilage pericellular and ECM proteins. These perturbations may be caused by the loss of a direct or indirect binding partner, a feedback mechanism that affects mRNA expression or altered proteolytic turnover of matrix components. A comprehensive analysis of the regulatory mechanisms involved in all the protein abundance changes observed is beyond the scope of this report. However, our results indicate differential regulation of thrombospondin-4 at the gene expression level, whereas matrilin-4 was identified as a likely binding partner for collagen IX based on solid phase interaction in vitro.
Whereas most nodes of the network are known cartilage constituents with multiple connections, such as fibronectin, two have no interacting partners. One of these orphan proteins is epiphycan, a small leucine-rich repeat proteoglycan that has an expression pattern largely restricted to developing growth plate and articular cartilage (49). Small leucine-rich repeat proteoglycans interact with collagen chains during fibril assembly and the mild effect of epiphycan ablation on skeletal development is consistent with a role in interfibrillar matrix assembly or stability. To date, no direct binding partners of epiphycan have been identified, although the more severe osteoarthritis-like cartilage degeneration in Bgn/Epyc double-deficient mice indicates a genetic interaction of the two small leucine-rich repeat proteoglycans (50). The reduced abundance of epiphycan in P3 and P21 collagen IX null cartilage is the first evidence for a direct/indirect physical or genetic interaction with collagen IX.
The differential expression of lactadherin (Mfge8), the second orphan protein, in the collagen IX null cartilage was another unanticipated result. Our two-dimensional electrophoresis analysis of P21 femoral head cartilage previously identified a series of lactadherin isoforms that were preferentially extracted in the GdnHCl-soluble fraction (17), consistent with tight integration with other matrix components. Lactadherin is associated with the cell periphery via αv integrin and is reported to promote neovascularization of endothelial cells (51). However, little is known about the function of this glycoprotein in cartilage. Pericellular staining for lactadherin in developing articular cartilage and prehypertrophic chondrocytes in the growth plate led to the hypothesis that the role of lactadherin in cartilage is location dependent (37). Analysis of lactadherin distribution in wild-type and collagen IX null femoral head cartilage will identify whether the observed increase in lactadherin abundance is specific to the growth plate or articular cartilage.
The TGFβ-induced protein Tgfbi (βig-h3 or RGD-CAP) is a widely expressed secreted protein that has been implicated in the differentiation of chondrocytes and precartilaginous precursors (40, 52). Although the function of Tgfbi is not fully understood, the inverse relationship between Tgfbi expression and chondrocyte maturation suggests an inhibitory effect on hypertrophy and matrix mineralization (40, 53). Tgfbi is attached at periodic intervals with collagen VI microfibrils extracted under native conditions (18) and Tgfbi·biglycan complexes enhance the formation of collagen VI networks in vitro (54), supporting a role for Tgfbi in matrix organization. In addition, Tgfbi interacts with fibronectin and is thought to facilitate cell-matrix interactions via an integrin-binding RGD motif (55). Validation of our proteomics analysis by Western blotting confirmed that fibronectin and Tgfbi, both of which are TGFβ-regulated proteins, were expressed at elevated levels in collagen IX null cartilage but were unaffected in COMP null cartilage. The loss of ECM integrity and changes in chondrocyte organization in the collagen IX knock-out may therefore disturb the normal pattern of TGFβ signaling in developing cartilage. Furthermore, we observed altered post-translational processing of Tgfbi, a proteolytic event that is likely to abolish integrin binding due to loss of the RGD-containing C terminus (41). Thus both the levels and functionality of Tgfbi appear to be profoundly altered in cartilage lacking collagen IX.
In this study we identified a cohort of differentially abundant proteins in mouse cartilage lacking collagen IX, a key component of the perifibrillar protein adaptor complex, using label-free proteomics. In addition to the proteins that were identified as statistically significant, further changes in the collagen IX knock-out are consistent with previous results (9, 56). Specifically, collagen α2(IX) chains were 2.9- and 4.3-fold reduced in P3 and P21 collagen IX null cartilage, respectively, whereas collagen α1(X) chains were 2.5-fold increased. Other matrix components and cell adhesion proteins that were detected at elevated levels in the collagen IX null cartilage included cilp2, chondroadherin, osteopontin, and SPARC. Validation studies on these candidates may reveal additional changes in matrix composition and lead to further insight into collagen IX knock-out cartilage phenotype. Importantly, we have found both expected and novel changes that expand our knowledge of the collagen IX interactome and demonstrate proof-of-principal for further proteomics studies in mouse cartilage knock-out and disease models.
Acknowledgments
We thank Mats Paulsson for critical reading of this manuscript and Paul O'Donnell (Bio21 institute, University of Melbourne) for assistance in analyzing excised proteins by mass spectrometry. Access to proteomic infrastructure funding in the QIMR Protein Discovery Centre was made possible through the Australian Government National Collaborative Infrastructure Scheme (NCRIS) provided via Bioplatforms Australia and the Queensland State Government.
This work was supported by grants from the National Health and Medical Research Council of Australia, the Murdoch Childrens Research Institute, the Victorian Government's Operational Infrastructure Support Program, and Deutsche Forschungsgemeinschaft Grants BR2304/5-1, BR2304/7-1, and ZA561/2-1.

This article contains Tables S1–S3 and Figs. S1–S2.
- ECM
- extracellular matrix
- COMP
- cartilage oligomeric matrix protein
- FDR
- false discovery rate
- IPA
- ingenuity pathway analysis
- P3
- 3 days postnatal
- P21
- 21 days postnatal
- MS/MS
- tandem mass spectrometry
- qPCR
- quantitative real-time PCR
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- GdnHCl
- guanidine HCl.
REFERENCES
- 1. Newman B., Wallis G. A. (2003) Skeletal dysplasias caused by a disruption of skeletal patterning and endochondral ossification. Clin. Genet. 63, 241–251 [DOI] [PubMed] [Google Scholar]
- 2. Warman M. L., Cormier-Daire V., Hall C., Krakow D., Lachman R., LeMerrer M., Mortier G., Mundlos S., Nishimura G., Rimoin D. L., Robertson S., Savarirayan R., Sillence D., Spranger J., Unger S., Zabel B., Superti-Furga A. (2011) Nosology and classification of genetic skeletal disorders. 2010 revision. Am. J. Med. Genet. A. 155A, 943–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eyre D. R., Weis M. A., Wu J. J. (2006) Articular cartilage collagen. An irreplaceable framework? Eur. Cell. Mater. 12, 57–63 [DOI] [PubMed] [Google Scholar]
- 4. Briggs M. D., Chapman K. L. (2002) Pseudoachondroplasia and multiple epiphyseal dysplasia. Mutation review, molecular interactions, and genotype to phenotype correlations. Hum. Mutat. 19, 465–478 [DOI] [PubMed] [Google Scholar]
- 5. Bönnemann C. G., Cox G. F., Shapiro F., Wu J. J., Feener C. A., Thompson T. G., Anthony D. C., Eyre D. R., Darras B. T., Kunkel L. M. (2000) A mutation in the α3 chain of type IX collagen causes autosomal dominant multiple epiphyseal dysplasia with mild myopathy. Proc. Natl. Acad. Sci. U.S.A. 97, 1212–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schmitz M., Becker A., Schmitz A., Weirich C., Paulsson M., Zaucke F., Dinser R. (2006) Disruption of extracellular matrix structure may cause pseudoachondroplasia phenotypes in the absence of impaired cartilage oligomeric matrix protein secretion. J. Biol. Chem. 281, 32587–32595 [DOI] [PubMed] [Google Scholar]
- 7. Aszódi A., Bateman J. F., Gustafsson E., Boot-Handford R., Fässler R. (2000) Mammalian skeletogenesis and extracellular matrix. What can we learn from knockout mice? Cell Struct. Funct. 25, 73–84 [DOI] [PubMed] [Google Scholar]
- 8. Blumbach K., Niehoff A., Paulsson M., Zaucke F. (2008) Ablation of collagen IX and COMP disrupts epiphyseal cartilage architecture. Matrix Biol. 27, 306–318 [DOI] [PubMed] [Google Scholar]
- 9. Dreier R., Opolka A., Grifka J., Bruckner P., Grässel S. (2008) Collagen IX-deficiency seriously compromises growth cartilage development in mice. Matrix Biol. 27, 319–329 [DOI] [PubMed] [Google Scholar]
- 10. Fässler R., Schnegelsberg P. N., Dausman J., Shinya T., Muragaki Y., McCarthy M. T., Olsen B. R., Jaenisch R. (1994) Mice lacking α1 (IX) collagen develop noninflammatory degenerative joint disease. Proc. Natl. Acad. Sci. U.S.A. 91, 5070–5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leighton M. P., Nundlall S., Starborg T., Meadows R. S., Suleman F., Knowles L., Wagener R., Thornton D. J., Kadler K. E., Boot-Handford R. P., Briggs M. D. (2007) Decreased chondrocyte proliferation and dysregulated apoptosis in the cartilage growth plate are key features of a murine model of epiphyseal dysplasia caused by a matn3 mutation. Hum. Mol. Genet. 16, 1728–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Piróg-Garcia K. A., Meadows R. S., Knowles L., Heinegård D., Thornton D. J., Kadler K. E., Boot-Handford R. P., Briggs M. D. (2007) Reduced cell proliferation and increased apoptosis are significant pathological mechanisms in a murine model of mild pseudoachondroplasia resulting from a mutation in the C-terminal domain of COMP. Hum. Mol. Genet. 16, 2072–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parsons P., Gilbert S. J., Vaughan-Thomas A., Sorrell D. A., Notman R., Bishop M., Hayes A. J., Mason D. J., Duance V. C. (2011) Type IX collagen interacts with fibronectin providing an important molecular bridge in articular cartilage. J. Biol. Chem. 286, 34986–34997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brown R. J., Mallory C., McDougal O. M., Oxford J. T. (2011) Proteomic analysis of Col11a1-associated protein complexes. Proteomics 11, 4660–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wilson R., Norris E. L., Brachvogel B., Angelucci C., Zivkovic S., Gordon L., Bernardo B. C., Stermann J., Sekiguchi K., Gorman J. J., Bateman J. F. (2012) Changes in the chondrocyte and extracellular matrix proteome during postnatal mouse cartilage development. Mol. Cell. Proteomics 11.1, mcp.M111.014159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Svensson L., Aszódi A., Heinegård D., Hunziker E. B., Reinholt F. P., Fässler R., Oldberg A. (2002) Cartilage oligomeric matrix protein-deficient mice have normal skeletal development. Mol. Cell. Biol. 22, 4366–4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wilson R., Bateman J. F. (2008) A robust method for proteomic characterization of mouse cartilage using solubility-based sequential fractionation and two-dimensional gel electrophoresis. Matrix Biol. 27, 709–712 [DOI] [PubMed] [Google Scholar]
- 18. Hanssen E., Reinboth B., Gibson M. A. (2003) Covalent and non-covalent interactions of βig-h3 with collagen VI. βig-h3 is covalently attached to the amino-terminal region of collagen VI in tissue microfibrils. J. Biol. Chem. 278, 24334–24341 [DOI] [PubMed] [Google Scholar]
- 19. Wilson R., Belluoccio D., Bateman J. F. (2008) Proteomic analysis of cartilage proteins. Methods 45, 22–31 [DOI] [PubMed] [Google Scholar]
- 20. Wilson R., Diseberg A. F., Gordon L., Zivkovic S., Tatarczuch L., Mackie E. J., Gorman J. J., Bateman J. F. (2010) Comprehensive profiling of cartilage extracellular matrix formation and maturation using sequential extraction and label-free quantitative proteomics. Mol. Cell. Proteomics 9, 1296–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Searle B. C. (2010) Scaffold. A bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 10, 1265–1269 [DOI] [PubMed] [Google Scholar]
- 22. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 23. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 24. Pham T. V., Piersma S. R., Warmoes M., Jimenez C. R. (2010) On the β-binomial model for analysis of spectral count data in label-free tandem mass spectrometry-based proteomics. Bioinformatics 26, 363–369 [DOI] [PubMed] [Google Scholar]
- 25. Käll L., Storey J. D., Noble W. S. (2009) QVALITY. Non-parametric estimation of q-values and posterior error probabilities. Bioinformatics 25, 964–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blaschke U. K., Eikenberry E. F., Hulmes D. J., Galla H. J., Bruckner P. (2000) Collagen XI nucleates self-assembly and limits lateral growth of cartilage fibrils. J. Biol. Chem. 275, 10370–10378 [DOI] [PubMed] [Google Scholar]
- 27. Klatt A. R., Nitsche D. P., Kobbe B., Macht M., Paulsson M., Wagener R. (2001) Molecular structure, processing, and tissue distribution of matrilin-4. J. Biol. Chem. 276, 17267–17275 [DOI] [PubMed] [Google Scholar]
- 28. Kim D. S., Li K. W., Boroujerdi A., Peter Yu Y., Zhou C. Y., Deng P., Park J., Zhang X., Lee J., Corpe M., Sharp K., Steward O., Eroglu C., Barres B., Zaucke F., Xu Z. C., Luo Z. D. (2012) Thrombospondin-4 contributes to spinal sensitization and neuropathic pain states. J. Neurosci. 32, 8977–8987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chomczynski P., Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159 [DOI] [PubMed] [Google Scholar]
- 30. Pfaffl M. W. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Groma G., Grskovic I., Schael S., Ehlen H. W., Wagener R., Fosang A., Aszodi A., Paulsson M., Brachvogel B., Zaucke F. (2011) Matrilin-4 is processed by ADAMTS-5 in late Golgi vesicles present in growth plate chondrocytes of defined differentiation state. Matrix Biol. 30, 275–280 [DOI] [PubMed] [Google Scholar]
- 32. Izu Y., Sun M., Zwolanek D., Veit G., Williams V., Cha B., Jepsen K. J., Koch M., Birk D. E. (2011) Type XII collagen regulates osteoblast polarity and communication during bone formation. J. Cell Biol. 193, 1115–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Posey K. L., Hankenson K., Veerisetty A. C., Bornstein P., Lawler J., Hecht J. T. (2008) Skeletal abnormalities in mice lacking extracellular matrix proteins, thrombospondin-1, thrombospondin-3, thrombospondin-5, and type IX collagen. Am. J. Pathol. 172, 1664–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Holden P., Meadows R. S., Chapman K. L., Grant M. E., Kadler K. E., Briggs M. D. (2001) Cartilage oligomeric matrix protein interacts with type IX collagen, and disruptions to these interactions identify a pathogenetic mechanism in a bone dysplasia family. J. Biol. Chem. 276, 6046–6055 [DOI] [PubMed] [Google Scholar]
- 35. Budde B., Blumbach K., Ylöstalo J., Zaucke F., Ehlen H. W., Wagener R., Ala-Kokko L., Paulsson M., Bruckner P., Grässel S. (2005) Altered integration of matrilin-3 into cartilage extracellular matrix in the absence of collagen IX. Mol. Cell. Biol. 25, 10465–10478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fresquet M., Jowitt T. A., Stephen L. A., Ylöstalo J., Briggs M. D. (2010) Structural and functional investigations of Matrilin-1 A-domains reveal insights into their role in cartilage ECM assembly. J. Biol. Chem. 285, 34048–34061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yoshimi M., Miyaishi O., Nakamura S., Shirasawa S., Kamochi H., Miyatani S., Ikawa Y., Shinomura T. (2005) Identification of genes preferentially expressed in articular cartilage by suppression subtractive hybridization. J. Med. Dent. Sci. 52, 203–211 [PubMed] [Google Scholar]
- 38. Kutsuna T., Inoue H., Takeda H., Takahashi T., Yamamoto H., Miura H., Higashiyama S. (2011) Fibronectin regulates proteoglycan production balance in transforming growth factor-β1-induced chondrogenesis. Int. J. Mol. Med. 28, 829–834 [DOI] [PubMed] [Google Scholar]
- 39. Takeda H., Inoue H., Kutsuna T., Matsushita N., Takahashi T., Watanabe S., Higashiyama S., Yamamoto H. (2010) Activation of epidermal growth factor receptor gene is involved in transforming growth factor-β-mediated fibronectin expression in a chondrocyte progenitor cell line. Int. J. Mol. Med. 25, 593–600 [DOI] [PubMed] [Google Scholar]
- 40. Ohno S., Doi T., Tsutsumi S., Okada Y., Yoneno K., Kato Y., Tanne K. (2002) RGD-CAP (βig-h3) is expressed in precartilage condensation and in prehypertrophic chondrocytes during cartilage development. Biochim. Biophys. Acta 1572, 114–122 [DOI] [PubMed] [Google Scholar]
- 41. Ween M. P., Lokman N. A., Hoffmann P., Rodgers R. J., Ricciardelli C., Oehler M. K. (2011) Transforming growth factor-β-induced protein secreted by peritoneal cells increases the metastatic potential of ovarian cancer cells. Int. J. Cancer 128, 1570–1584 [DOI] [PubMed] [Google Scholar]
- 42. Gregory K. E., Keene D. R., Tufa S. F., Lunstrum G. P., Morris N. P. (2001) Developmental distribution of collagen type XII in cartilage. Association with articular cartilage and the growth plate. J. Bone Miner. Res. 16, 2005–2016 [DOI] [PubMed] [Google Scholar]
- 43. Nakata K., Ono K., Miyazaki J., Olsen B. R., Muragaki Y., Adachi E., Yamamura K., Kimura T. (1993) Osteoarthritis associated with mild chondrodysplasia in transgenic mice expressing α1(IX) collagen chains with a central deletion. Proc. Natl. Acad. Sci. U.S.A. 90, 2870–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gordon M. K., Gerecke D. R., Dublet B., van der Rest M., Olsen B. R. (1989) Type XII collagen. A large multidomain molecule with partial homology to type IX collagen. J. Biol. Chem. 264, 19772–19778 [PubMed] [Google Scholar]
- 45. Tillgren V., Onnerfjord P., Haglund L., Heinegård D. (2009) The tyrosine sulfate-rich domains of the LRR proteins fibromodulin and osteoadherin bind motifs of basic clusters in a variety of heparin-binding proteins, including bioactive factors. J. Biol. Chem. 284, 28543–28553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Font B., Eichenberger D., Rosenberg L. M., van der Rest M. (1996) Characterization of the interactions of type XII collagen with two small proteoglycans from fetal bovine tendon, decorin and fibromodulin. Matrix Biol 15, 341–348 [DOI] [PubMed] [Google Scholar]
- 47. Agarwal P., Zwolanek D., Keene D. R., Schulz J. N., Blumbach K., Heinegård D., Zaucke F., Paulsson M., Krieg T., Koch M., Eckes B. (2012) Collagen XII and XIV, new partners of cartilage oligomeric matrix protein in the skin extracellular matrix suprastructure. J. Biol. Chem. 287, 22549–22559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gordon M. K., Gerecke D. R., Dublet B., van der Rest M., Sugrue S. P., Olsen B. R. (1990) The structure of type XII collagen. Ann. N.Y. Acad. Sci. 580, 8–16 [DOI] [PubMed] [Google Scholar]
- 49. Johnson H. J., Rosenberg L., Choi H. U., Garza S., Höök M., Neame P. J. (1997) Characterization of epiphycan, a small proteoglycan with a leucine-rich repeat core protein. J. Biol. Chem. 272, 18709–18717 [DOI] [PubMed] [Google Scholar]
- 50. Nuka S., Zhou W., Henry S. P., Gendron C. M., Schultz J. B., Shinomura T., Johnson J., Wang Y., Keene D. R., Ramírez-Solis R., Behringer R. R., Young M. F., Höök M. (2010) Phenotypic characterization of epiphycan-deficient and epiphycan/biglycan double-deficient mice. Osteoarthritis Cartilage 18, 88–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Silvestre J. S., Théry C., Hamard G., Boddaert J., Aguilar B., Delcayre A., Houbron C., Tamarat R., Blanc-Brude O., Heeneman S., Clergue M., Duriez M., Merval R., Lévy B., Tedgui A., Amigorena S., Mallat Z. (2005) Lactadherin promotes VEGF-dependent neovascularization. Nat. Med. 11, 499–506 [DOI] [PubMed] [Google Scholar]
- 52. Hashimoto K., Noshiro M., Ohno S., Kawamoto T., Satakeda H., Akagawa Y., Nakashima K., Okimura A., Ishida H., Okamoto T., Pan H., Shen M., Yan W., Kato Y. (1997) Characterization of a cartilage-derived 66-kDa protein (RGD-CAP/βig-h3) that binds to collagen. Biochim. Biophys. Acta 1355, 303–314 [DOI] [PubMed] [Google Scholar]
- 53. Ohno S., Tanaka N., Ueki M., Honda K., Tanimoto K., Yoneno K., Ohno-Nakahara M., Fujimoto K., Kato Y., Tanne K. (2005) Mechanical regulation of terminal chondrocyte differentiation via RGD-CAP/βig-h3 induced by TGF-β. Connect. Tissue Res. 46, 227–234 [DOI] [PubMed] [Google Scholar]
- 54. Reinboth B., Thomas J., Hanssen E., Gibson M. A. (2006) βig-h3 interacts directly with biglycan and decorin, promotes collagen VI aggregation, and participates in ternary complexing with these macromolecules. J. Biol. Chem. 281, 7816–7824 [DOI] [PubMed] [Google Scholar]
- 55. Billings P. C., Whitbeck J. C., Adams C. S., Abrams W. R., Cohen A. J., Engelsberg B. N., Howard P. S., Rosenbloom J. (2002) The transforming growth factor-β-inducible matrix protein βig-h3 interacts with fibronectin. J. Biol. Chem. 277, 28003–28009 [DOI] [PubMed] [Google Scholar]
- 56. Hagg R., Hedbom E., Möllers U., Aszódi A., Fässler R., Bruckner P. (1997) Absence of the α1(IX) chain leads to a functional knock-out of the entire collagen IX protein in mice. J. Biol. Chem. 272, 20650–20654 [DOI] [PubMed] [Google Scholar]