Background: FliY is a flagellar rotor protein of the CheC phosphatase family.
Results: The FliY structure resembles that of the rotor protein FliM but contains two active centers for CheY dephosphorylation.
Conclusion: FliY incorporates properties of the FliM/FliN rotor proteins and the CheC/CheX phosphatases to serve multiple functions in the flagellar switch.
Significance: FliY distinguishes flagellar architecture and function in different types of bacteria.
Keywords: Bacterial Protein Phosphatases, Bacterial Signal Transduction, Cell Motility, Chemotaxis, Protein-protein Interactions, X-ray Crystallography, Flagella Rotor
Abstract
Rotating flagella propel bacteria toward favorable environments. Sense of rotation is determined by the intracellular response regulator CheY, which when phosphorylated (CheY-P) interacts directly with the flagellar motor. In many different types of bacteria, the CheC/CheX/FliY (CXY) family of phosphatases terminates the CheY-P signal. Unlike CheC and CheX, FliY is localized in the flagellar switch complex, which also contains the stator-coupling protein FliG and the target of CheY-P, FliM. The 2.5 Å resolution crystal structure of the FliY catalytic domain from Thermotoga maritima bears strong resemblance to the middle domain of FliM. Regions of FliM that mediate contacts within the rotor compose the phosphatase active sites in FliY. Despite the similarity between FliY and FliM, FliY does not bind FliG and thus is unlikely to be a substitute for FliM in the center of the switch complex. Solution studies indicate that FliY dimerizes through its C-terminal domains, which resemble the Escherichia coli switch complex component FliN. FliY differs topologically from the E. coli chemotaxis phosphatase CheZ but appears to utilize similar structural motifs for CheY dephosphorylation in close analogy to CheX. Recognition properties and phosphatase activities of site-directed mutants identify two pseudosymmetric active sites in FliY (Glu35/Asn38 and Glu132/Asn135), with the second site (Glu132/Asn135) being more active. A putative N-terminal CheY binding domain conserved with FliM is not required for binding CheY-P or phosphatase activity.
Introduction
Bacterial chemotaxis, the movement of cells in response to the surrounding environment, is achieved through a coordinated network of over twenty proteins that link receptors in the cytoplasmic membrane with the flagellar motor (1). Central to the network, the intracellular messenger protein CheY undergoes receptor-regulated phosphorylation on a conserved aspartate residue by the histidine kinase CheA and then binds to the flagellar rotor to change its direction of rotation (1, 2). To maintain an optimal concentration of phosphorylated CheY (CheY-P)2 for signal transmission and adaptation, phosphatases are required (3–6). For example, in Escherichia coli, the phosphatase CheZ decreases the CheY-P lifetime from ∼20 s to ∼200 milliseconds (1). CheZ is generally found in proteobacteria; other types of bacteria such as those of the genus Thermotoga and Bacillus do not have CheZ but instead encode phosphatases of the CheC/CheX/FliY (CXY) family (see Fig. 1A) (7–10).
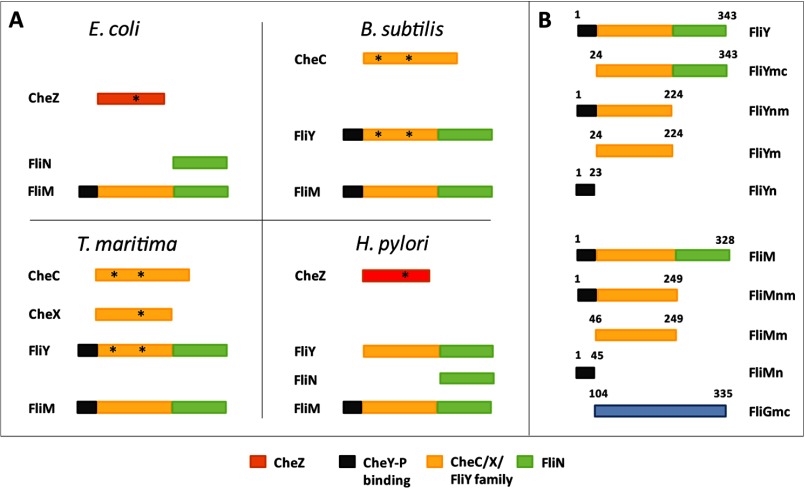
FIGURE 1.
A, domain organization of CheC family and related proteins in E. coli, H. pylori, T. maritima, and B. subtilis. Structurally unrelated CheY phosphatases E. coli CheZ (orange) and H. pylori (red) are found in proteobacteria. The asterisk indicates active centers. B, schematic representation of different constructs of FliY, FliM, and FliG from T. maritima used in this study. Black shading denotes CheY-P binding domain, yellow shading denotes CheC homology domain, and green shading represents the rotor protein FliN homology domain.
Structure function studies have been carried out on CheC and CheX (6–10), but not FliY, the last member of the family to have its crystallographic structure determined. The CXY family contains a consensus sequence D/S-X3-E-X2-N-X22-P that defines the phosphatase active site, with CheC and FliY having two such repeats and CheX only one (7–9, 11). CheX dimerization generates two active sites per dimer (11), but the recent crystal structure of the complex between CheX and BeF3−-activated CheY3 from Borrelia burgdorferi shows CheY3 bound to a single subunit of CheX, which suggests that binding CheY-P may dissociate the CheX dimer (12). In CheC and CheX, the invariant Glu residue in the consensus sequence (in boldface type above) is essential for binding CheY-P, whereas the invariant Asn residue (also in boldface type) is critical for the phosphatase activity (see Fig. 2A) (11, 12). These residues structurally mimic the conserved Asp and Gln residues essential for phosphatase activity in CheZ (13). In the structure of the CheX·CheY3·BeF3−·Mg2+ complex, the helix bearing the conserved Glu/Asn residues on CheX adopts a perpendicular orientation with respect to CheY3 α1 (12). The Glu/Asn residues themselves participate in an extensive hydrogen bond network with the aspartyl-phosphate and surrounding residues of CheY. The Asn residue hydrogen bonds with an ordered water molecule, which is positioned for in-line attack of the phosphate mimic BeF3−. Alone, CheC is a weak phosphatase (11, 14), but its affinity for CheY-P increases in complex with CheD, a chemoreceptor deamidase (11, 14–16). Formation of the CheD·CheC complex is proposed to allow levels of CheY-P to influence receptor modification state by sequestering CheD (15, 17).
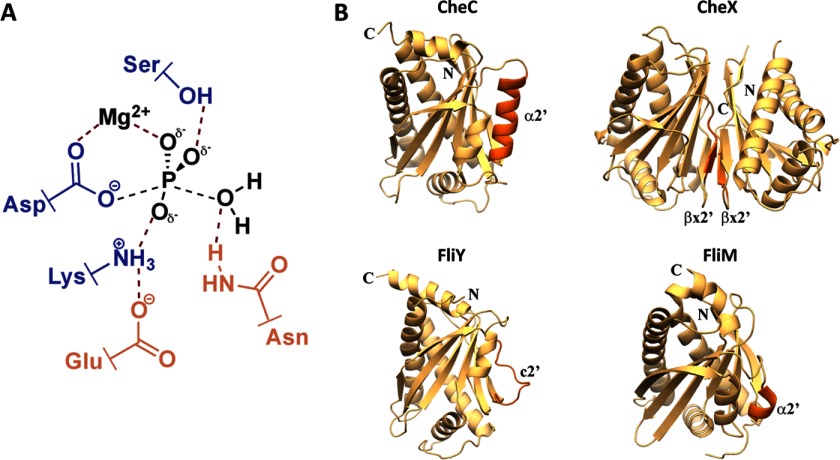
FIGURE 2.
The CXY phosphatase family. A, proposed transition state for CheY-P dephosphorylation (adapted from Ref. 12) highlighting the role of the essential Glu and Asn side chains (orange) and contributing residues from CheY (blue). B, structural comparison within the CXY family and FliMm. Secondary structural elements of each protein are shown as ribbons. The α2′/βx′/c2′ regions differentiate the members from each other (dark orange).
Many non-proteobacteria encode the third member of this phosphatase family, FliY (7, 9, 14, 18). For example, FliY is found in pathogenic Gram-positive bacteria from the genera Bacillus or Helicobacter and pathogenic spirochetes from the genera Leptospira and Treponema. Lower pathogenicity of Leptospira has been associated with inactivation of fliY gene (19). FliY is thought to localize in the switch complex that composes the flagellar C-ring (7, 9, 14, 18). In E. coli, the switch complex contains many copies of FliG, FliM, and FliN (see Fig. 1 for domain designations and relationships of rotor proteins and phosphatases). FliG connects the Cytoplasmic ring (C-ring) to the Membrane and Supramembranous ring (MS-ring) and interacts with the stator to allow rotation (20). Structures of FliG from various organisms (20–24) reveal three globular domains linked by flexible linkers. FliM is sandwiched between FliG and FliN in the C-ring and is directly involved in binding CheY-P (25–27). The middle and C-terminal domains of FliG interact with FliM (24, 28). FliM contains an amino-terminal CheY-binding motif (FliMn) that recruits CheY-P to the motor (25–27). NMR studies have shown that in addition to CheY-P binding to the N-terminal motif, it also interacts with the middle domain of FliM, albeit weakly (29). The crystal structure of the middle domain of FliM (FliMm) reveals a topology similar to CheC (30). The conserved GGXG motif is essential for binding FliG as evident in the FliG·FliM complex structure (24, 31). The C-terminal domain of FliM interacts with the third rotor protein, FliN (25, 27, 32). FliN represents the donut-shaped structure present at the bottom of the C-ring membrane distal in EM reconstructions (32, 33). Recent cross-linking studies in E. coli show that CheY interacts with FliN and in doing so transmits conformational signals to FliG (34). Crystal structures of the C-terminal two thirds of Thermotoga maritima FliN reveal a tightly intertwined dimer formed mostly of β-sheets (32). Nonetheless, E. coli FliN is a tetramer in solution, which is assumed to be the assembly state found in the C-ring (32).
The fliY gene was first identified and characterized in Bacillus subtilis as a multidomain protein, with an amino-terminal CheY-P binding domain (FliYn), a middle domain (FliYm) similar to FliM, and a C-terminal domain similar to FliN (FliYc, Fig. 1) (18, 30). The deletion mutant FliYΔ6–15 of B. subtilis FliY cannot bind CheY-P (35). The large middle domain (FliYm) has structural homology to the CheC phosphatase family. It has been suggested that CheC/CheX phosphatases and the FliM/FliY flagellar rotor proteins all evolved from a common ancestor (36). FliY conserves the dephosphorylation sites of CheC and CheX but FliM does not. B. subtilis FliY has greater phosphatase activity than CheC alone (14). Some bacteria such as B. subtilis and T. maritima do not contain a separate FliN, presumably deriving this function solely from FliY; whereas in other bacteria, such as Helicobacter pylori FliY and FliN are expressed as separate proteins (37). B. subtilis FliY can complement the motility defect in a Salmonella FliN mutant (18), which suggests redundant functions for FliY and FliN, as well as localization of FliY in the C-ring. The importance of having the primary CheY-P phosphatase localized to the switch is currently not well understood.
Herein we report the crystallographic structure of T. maritima FliYm, characterize its phosphatase activity, and investigate the interaction properties of full-length FliY. The structure reveals how a variable α/β/coil 2′ region diverges among the CXY family members to impart specific functions. We verify that both putative active sites of FliY bind phosphorylated CheY have CheY phosphatase activity but that the second site is more active than the first. FliYn does not increase the binding affinity of CheY-P, and FliYm does not appear to associate with FliG. These findings have implications for the function and architecture of the flagellar rotor in non-enteric bacteria.
EXPERIMENTAL PROCEDURES
Protein Preparation
The genes encoding T. maritima FliY (residues 1–343), FliYmc (residues 24–343), FliYnm (residues 1–224), FliYm (residues 24–224), CheA, CheY were PCR cloned from T. maritima genomic DNA (obtained from the American Type Culture Collection) into the vector pET28a (Novagen) and expressed with a His6 tag in an E. coli strain BL21-DE3. Cells were induced with 100 μm isopropyl 1-thio-β-d-galactopyranoside at OD of 0.6 and grown at 37 °C for 6–10 h. Point mutations on FliYnm were introduced by QuikChange or overlap PCR and verified by sequencing.
Proteins were purified with nickel-nitrilotriacetic acid affinity chromatography and the His6 tags were subsequently cleaved with thrombin. Samples were then run on size exclusion chromatography (Superdex 75 or Superdex 200; Pharmacia Biotech) and concentrated in GF buffer (50 mm Tris, pH 7.5, 150 mm NaCl).
Selenomethionine-substituted FliYm was grown in minimal media supplemented with 16 amino acids. 100 mg of l-selenomethionine was added to 2 liters of media. Cells were induced with 100 μm isopropyl 1-thio-β-d-galactopyranoside at OD of 0.6 and grown overnight at room temperature. The protein purification carried out as described above, except that 10 mm DTT was added to all the buffers.
Crystallization and Data Collection
Crystals were obtained from vapor diffusion of 2-μl drops containing 1 μl of reservoir solution (0.2 m ammonium sulfate, 30% w/v PEG 8000 (Hampton Screen)) and 1 μl of the FliYm protein in GF buffer. A single anomalous diffraction data set was collected at the peak wavelength of selenium (0.97670 Å) at the Cornell High Energy Synchrotron Source, station A1. Crystals were soaked in the cryoprotectant (15% glycerol) briefly before flash cooling in a N2 cold stream.
Structural Determination and Refinement
Diffraction data were scaled with HKL2000 (38), and the structure was determined with SOLVE (39). The initial model produced by automatic chain-tracing in SOLVE provided the foundation to build the complete model manually with XFIT (40), which was then refined with CNS (41). Water molecules were added with water picking algorithms in CNS and adjusted manually amid cycles of refinement.
Phosphatase Assays
CheA (12–30 μm) and CheY (33–300 μm) were premixed with 5 μl of TKM buffer (50 mm Tris, pH 7.5, 200 mm KCl, 10 mm MgCl2) with various volumes of GF buffer. The samples were incubated with 2 μl of an ATP solution (15 μl of an 11 μm cold ATP solution, 3–8 μl of [γ-32P]ATP (3000 Ci/mmol, PerkinElmer Life Sciences)) made to a total volume of 75 μl with filtered nanopure water. For the control containing no FliY, the sample was quenched at 15 min with 25 μl of 3×SDS buffer containing 50 mm EDTA, pH 8. Once the samples were incubated for 15 min, 2.5 μl of FliY (5 or 10 μm, native or mutants) was added to a total volume of 25 μl per sample and quenched at various time points with the same SDS buffer as the control. The proteins were separated with 4–20% Tris-glycine SDS-PAGE at 120 V for 2 h. The proteins were affixed with water, then stained with Coomassie blue for 10 min, and destained with water for 3 h. The gels were dried in a GelAir Drying System for 3 h and placed in a cassette, and the film was exposed for a minimum of 24 h before visualization using a STORM PhosphoImager.
Multiangle Light Scattering
Size-exclusion chromatography coupled with multiangle light scattering was used to study the molar mass of the various protein fragments. Proteins (2 mg/ml) were run at room temperature on the column (BioSep-SEC-S 3000 column (Phenomenex)) pre-equilibrated with GF buffer. Analysis and molecular weight determination was carried out with Wyatt technologies ASTRA. Bovine serum albumin (Sigma) was used as a control for data quality.
Pulldown Assays
Assays were carried out in binding buffer (25 mm HEPES, pH 7.5, 500 mm NaCl, and 50 mm imidazole). Proteins were incubated in 30 μl of nickel-nitrilotriacetic acid with the binding buffer for 30 min at room temperature. The beads were washed with binding buffer thrice and once with binding buffer containing 1% Triton X-100 to minimize nonspecific binding. 2×SDS loading dye was added to the resin, boiled for 5 min at 90 °C, and centrifuged at 13,000 rpm for 5 min. The supernatant was used for SDS-PAGE analysis. To demonstrate the binding of various constructs of FliY (100 μm) to CheY (75 μm) and CheY-P (75 μm), pulldown assays were performed with His-tagged proteins as described previously (42, 43) with minor modifications. The samples that required phosphorylation of CheY, 20 mm acetyl phosphate (Sigma-Aldrich) in the presence of 20 mm MgCl2·6H2O was added for incubation and wash steps to the binding buffer to ensure complete phosphorylation of CheY.
RESULTS
Gene Structure of T. maritima FliY
In the annotated genome of T. maritima two adjacent flagellar genes demarked by an authentic frameshift have been designated as FliY1 and FliY2 (gene ID 897481 and 897793, respectively), FliY1 corresponds to FliYnm, whereas FliY2 corresponds to E. coli FliN. FliY2 was cloned and characterized, and its structure was determined (32). As annotated, all reading frames indicate a stop codon between FliY1 and FliY2. However, PCR cloning from genomic DNA and subsequent sequencing revealed an additional guanine nucleotide (no. 479) between the two reading frames that abrogates the stop codon and allowed for expression of a fused FliY1-FliY2. The composite full-length FliY (343 residues) was expressed in E. coli and found to be fully soluble and well behaved (supplemental Fig. 1).
FliYm Is an α/β Globular Protein with a Fold Similar to CheC and CheX
The structure of T. maritima FliYm (residues 24–223) was determined at 2.5 Å resolution by single anomalous diffraction of selenomethionine substituted protein (Table 1). The two FliY molecules in the asymmetric unit have a nearly identical structure with a main chain Cα root mean square deviation of 0.6 Å. The structure of FliYm shares a pseudo symmetric topology with the phosphatases CheC and CheX and the rotor protein FliMm (Fig. 2B). In all cases, the first half of the protein relates to the second half both in sequence and structure by a pseudo-2-fold rotation axis roughly perpendicular to the central β-sheet (supplemental Fig. 2). The FliYm α/β-globular fold comprises five α-helices and six β-strands. The six β-strands (β1-β2′-β3′-β3-β2-β1′; primes denote symmetry-related features) form a continuous antiparallel β-sheet and are very similar in structure to those of the other family members. Two long helices (α1 and α1′) pack against and run diagonal to the central β-sheet, and two medium length helices (α3 and α3′) cap the hydrophobic core generated by the β-sheet and α1/α1′. The fifth small helix (α2) packs on the opposite face of the β-sheet as α1/α1′ (Fig. 2B). Notably, the symmetry-related feature to α2 (hereafter referred to as c2′), is not a helix, but rather an extended loop that has β-like geometry, yet the main-chain does not hydrogen bond with β1′, the neighboring β-strand. In CheC, this region is helical (α2′) and pseudosymmetric to α2, whereas in CheX, this region also displays pseudosymmetry but rather forms a β-strand (βx2′/βx2) that mediates CheX dimerization by allowing for a continuous seven-stranded β-sheet across the dimer interface. Thus, FliY appears to be a hybrid between CheC and CheX, where α2 is CheC-like, but the symmetry-related region, c2′, is not helical and instead more closely resembles βx2′ in CheX. As predicted from sequence alignments of FliYm with CheX and CheC (11), FliYm lacks a Gly residue following β1′ that would allow for the extended loop to align with the β-sheet and become βx2′. Thus, FliYm remains monomeric and shows greater overall resemblance to CheC than to CheX (supplemental Fig. 3).
TABLE 1.
Data collection, phasing, and refinement statistics
| Data collection | |
|---|---|
| Space group | P212121 |
| Unit cell (Å) | a = 49.75, b = 85.89, c = 119.94 |
| Resolution range (Å) | 50–2.5 (2.54–2.50)a |
| Rmergeb | 0.090 (0.157)a |
| I/σI | 19.4 (13.6)a |
| Completeness (%) | 99.9 (99.3)a |
| Redundancy | 7.4 (6.8)a |
| Phasing FOMc | 0.37 (0.39)a |
| Refinement | |
| No. of reflections | 33,692 |
| Rwork/Rfree | 0.213/0.261 |
| No. of atoms | |
| Residues | 200 (24–223) for chain A, 198 (24–220) for chain B |
| Water | 175 |
| B-factors (Å2) | |
| Wilson | 27.1 |
| Main chain | 25.2 |
| Side chain | 28.8 |
| Water | 30.2 |
| Geometry (r.m.s.d.)c | |
| Bond lengths (Å) | 0.007 |
| Bond angles | 1.3° |
a Highest resolution range for compiling statistics.
b Rmerge = ΣΣi|Ii − 〈I〉|/ΣΣiIi.
c r.m.s.d., root mean square deviation; FOM, figure of merit.
Association State of FliY
Multiangle light scattering coupled to size-exclusion chromatography indicated that full-length FliY behaves as a single species in solution (Table 2), with a molar mass of ∼80 kDa, which is consistent with a dimer (subunit molecular mass, 37.8 kDa). However, FliYnm and FliYm were both monomeric with molar masses of 22.2 and 19.8 kDa, respectively (Table 2 and supplemental Fig. 4). Thus, the C terminus of FliY mediates dimerization, which is consistent with the dimeric state of FliYc in solution and in crystal structures, where it folds as an intertwined, domain-swapped dimer (32).
TABLE 2.
Multiangle light scattering data
Mw, weight average molar mass; Mn, number average molar mass.
| Predicted subunit molar mass (kDa) | Observed molar mass (kDa) | Polydispersity Mw/Mn | |
|---|---|---|---|
| FliY fulllength | 37.8 | 80.3 (1%) | 1.012 |
| FliYnm | 24.4 | 22.2 (4%) | 1.005 |
| FliYm | 21.9 | 19.8 (3%) | 1.002 |
FliY Is a CheY Phosphatase
T. maritima FliY actively dephosphorylates CheY as monitored under conditions of steady-state phosphotransfer from CheA with [γ-32P]ATP. CheA is included in the assay because of the instability of CheY-P, which requires its in situ production (11, 14, 42, 43). FliY does not affect CheA autophosphorylation (Fig. 3A, lanes 7 and 8). The rate of FliY phosphatase activity greatly exceeds the rate of CheA phosphorylation of CheY when 33 μm CheY is treated with 12 μm CheA and 5 μm FliY or 5 μm FliYnm (Fig. 3A). At CheY levels as great as 300 μm, sub-stoichiometric amounts of FliY (5 μm) dephosphorylate nearly all available CheY-P, and thus, the reaction requires catalytic turnover by FliY. FliYmc, FliYnm, and FliYm behave very similarly to FliY under similar conditions, and thus, the N- and C-terminal domains of FliY have little effect on phosphatase activity (Fig. 3B). Furthermore, FliY dimerization, which is disrupted in the absence of FliYc, is not important for dephosphorylation of CheY-P.
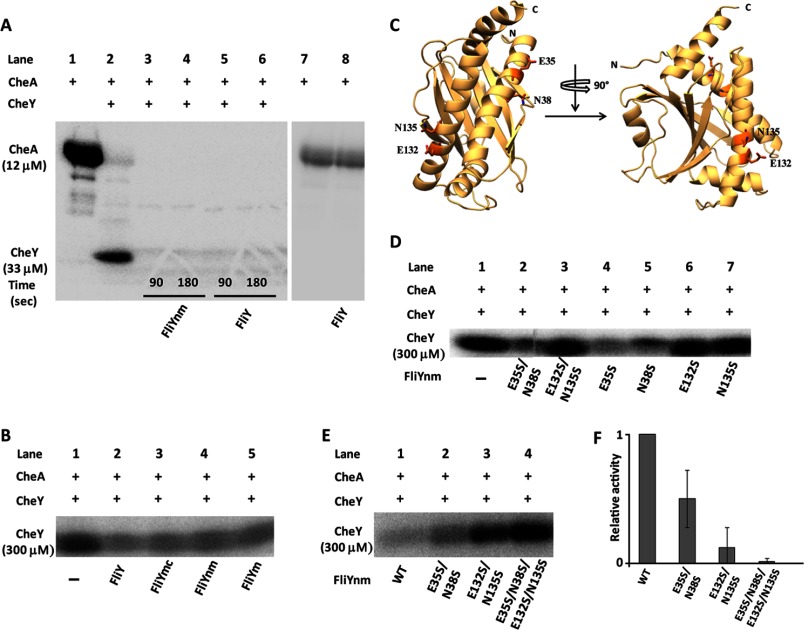
FIGURE 3.
FliY phosphatase activity. A, autophosphorylated T. maritima CheA (12 μm; lane 1) transfers 32P to 33 μm CheY (lanes 2–6). 5 μm FliYnm (lanes 3 and 4) or 5 μm FliY (lanes 5 and 6) dephosphorylates CheY-P. 90 s and 180 s time points were measured after FliY addition. FliY does not affect CheA autophosphorylation (lanes 7 and 8). B, effect of N-terminal CheY binding domain (FliYn) on CheY-32P dephosphorylation: All reactions contained 30 μm CheA and 300 μm CheY. Bands correspond to CheY-32P after transfer from CheA, measured at 45 s after addition of 5 μm FliY (lane 2), 5 μm FliYmc (lane 3), 5 μm FliYnm (lane 4), and 5 μm FliYm (lane 5). C, ribbon diagram of the FliYm topology with active site Glu and Asn residues shown as sticks. D, effects of FliY double and single mutants on phosphatase activity measured 45 s after addition of 5 μm FliY variants. E, effect of FliYnm mutants on CheY-32P dephosphorylation. Reactions contained 30 μm CheA and 300 μm CheY. Bands correspond to CheY-32P after transfer from CheA measured 5 min after addition of 10 μm FliYnm (WT and variants). F, relative phosphatase activities of FliY mutants measured as the ratio of CheY-P dephosphorylated per unit time relative to WT activity. The experiments were performed in quadruplicate; error bars indicate S.D. For ease of visualization, the gel images have different contrast ratios.
Based on homology to CheX and CheC (11), FliY has two potential active centers that are composed from conserved residues containing acidic and amide side chains. α1 and α1′ harbor the putative catalytic residues Glu35/Asn38 and Glu132/Asn135, respectively (Fig. 3C). To evaluate the importance of each active site for CheY-P hydrolysis, phosphatase activity was measured in FliYnm variants where Glu35, Asn38, Glu132, and Asn135 were mutated to Ser individually, in pairs, and in totality. The time points and relative concentrations of each protein were optimized such that a comparison could be made between levels of CheY-P under steady state phosphorylation by CheA. The N35S/E38S/N132S/E135S mutant had no phosphatase activity (Fig. 3E). When each active site was examined separately, the E35S/N38S mutant had reduced phosphatase activity compared with WT FliYnm; however, the phosphatase activity of E132S/N135S was severely impaired. The single mutants E132S and N135S had similar, albeit lesser effects (Fig. 3D). Thus, both putative active centers contribute to FliY phosphatase activity; however, the second Glu132/Asn135 site is dominant.
To study the contributions of the FliY domains on phosphatase activity, we performed the dephosphorylation assay with various FliY variants (FliY, FliYmc, FliYnm, and FliYm). The full-length and FliY domain fragments were all able to dephosphorylate CheY-P with similar activity provided that they contained the FliY middle domain (Fig. 3B). Thus, FliY dimerization, which is disrupted in the absence of FliYc, does not largely influence phosphatase activity and, surprisingly, neither does the putative N-terminal CheY binding domain (FliYn). We then tested whether purified FliY required the N-terminal domain to interact with CheY in pulldown experiments. Strong interaction between FliY and affinity-tagged CheY is only observed when CheY is phosphorylated by the phosphate donor acetyl phosphate. This interaction did not depend on the presence of FliYn (Fig. 4A). In contrast, the switch protein FliMm alone showed no interaction with CheY-P or CheY; however, FliMnm, which contains the N-terminal peptide, bound both CheY-P and CheY and bound the former most strongly.
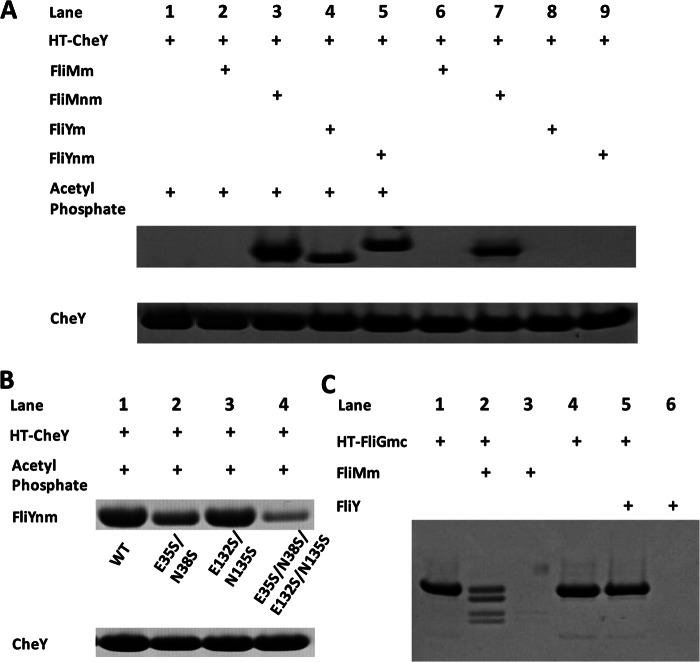
FIGURE 4.
Interaction between CheY and FliY domains. A, pulldown assay of FliYnm (100 μm) and FliYm (100 μm) with His-tagged CheY (75 μm) in presence of acetyl phosphate where indicated. Shown is His-tagged CheY in presence of acetyl phosphate (lane 1). Negative and positive controls are shown: interaction of FliMm (lane 2) and FliMnm (lane 3), respectively, with CheY in presence of acetyl phosphate. FliYm (lane 4) and FliYnm (lane 5) interacted strongly with CheY only in presence of acetyl phosphate. No interaction was observed between CheY and FliMm (lane 6), FliYm (lane 8), or FliYnm (lane 9) in the absence of acetyl phosphate. Reduced interaction was seen with FliMnm in the absence of acetyl phosphate (lane 7). B, pulldown assay of FliYnm (100 μm, lane 1) and variants (100 μm, lane 2, E35S/N38S; lane 3, E132S/N135S; lane 4, E35S/N38S/E132S/N135S) with His-tagged CheY (75 μm) in the presence of acetyl phosphate demonstrate that both active sites bind CheY-P. C, pulldown assay of FliY (80 μm) with His-tagged FliGmc (40 μm, lane 5). Positive control is as follows: His-tagged FliGmc pulldown of FliMm (80 μm, lane 2). Controls of FliMm without tag (lane 3) and FliY without tag (lane 6) show no interaction with the affinity beads. Upper pair of bands in lane 2 represents FliGmc ± His6 tag, whereas a lower pair of bands represents FliMm and an N-terminal cleavage product. The presence of untagged FliGmc in the pulldown indicates a larger than dimeric complex formed by FliGmc and FliMm.
Mutation studies showed that each FliY active site binds CheY-P (Fig. 4B). Alteration of residues in both active sites (E35S/N38S/E132S/N135S) greatly reduced binding but did not abrogate it entirely (Fig. 4B). This indicates that the Ser replacements can still mediate some interaction with CheY-P or that other peripheral residues are also involved in binding. Binding to CheY-P was increased when either the first or second active site was restored in the absence of the other; however, restoration of the first active site alone showed the greatest increase in CheY-P binding (E132S/N135S; Fig. 4B). Thus, the more active dephosphorylation center (Glu132/Asn135) exhibits lower apparent CheY-P affinity than the less active center (E35/N38). Given the very low affinity of unphosphorylated CheY for FliY, the weaker apparent binding at the second site may reflect its ability to turnover substrate more rapidly.
Interaction of FliY with FliG
The C-terminal domain of FliY is homologous to FliN (and FliMc), and thus, FliY is thought to be located in the flagellar rotor (14, 18). Indeed, B. subtilis FliY was able to complement a S. typhimurium fliN amber mutant and restore motility (44). Furthermore, some FliY sequences conserve with FliM residues known to form an important loop for engaging FliG (MGGXGE; supplemental Fig. 2). In pulldown assays, FliMm shows a strong interaction with affinity-tagged T. maritima FliGmc (Fig. 4C). But, FliY shows no such interaction with FliGmc (Fig. 4C). We also investigated the interaction between FliGmc and FliYnm by site-specific spin labeling and pulsed dipolar electron spin resonance spectroscopy (data not shown). Nitroxide spin labels on FliMnm and FliGmc provided interaction distances that agreed with the FliG-FliM complex crystal structure (24). However, spin labels at a similar position on FliY did not yield any observable interaction with spin-labeled FliG. These experiments suggest that FliYm cannot replace FliMm in its association with FliG and therefore has a unique position in the flagellar rotor.
DISCUSSION
Comparison with CheC/CheX and FliMm
FliYm maintains the topology of the CheC/CheX family, although the overall resemblance to CheC is greater than to CheX (supplemental Fig. 3). The most notable difference is that FliYm does not have defined secondary structure between residues 165–178 (c2′), which corresponds to α2′ in CheC and βx2′ in CheX (supplemental Fig. 2) (11). The α2′ helix in CheC mediates binding to the activator CheD (15) and βx2′ mediates dimerization in CheX (11). In FliM, the small α2′ helix mediates contact to the α1/α1′ face of a neighboring FliM subunit within the rotor (30). The α1/α1′ helices containing the active site residues of FliYm are similar in size as to those of CheC and superposition of the conserved motifs on α1/α1′ from CheC and FliYm show similar spatial orientation of the residue side chains (supplemental Fig. 3). Structural variation in the c2′ region of FliY suggests that FliY has a distinct architectural role from FliM and does not undergo associations similar to those of CheC and CheX.
Phosphatase Activity of FliY
FliY has been assigned to be the primary CheY phosphatase in B. subtilis, despite the presence of CheC (14, 16). Unlike B. subtilis, Thermotoga encodes all three members of the CheC phosphatase family, and T. maritima CheC and CheX have been previously demonstrated to dephosphorylate CheY-P (11). FliY also dephosphorylates CheY-P, which establishes three distinct chemotaxis phosphatases in T. maritima, despite the presence of only one CheY homolog per genome. Sequence conservation with CheX, CheC (and CheZ, see below) suggested the possibility of two active sites on FliY. The double mutant of the second putative active site (E132S/N135S) has low observable phosphatase activity (Fig. 3, D and E). The E35S/N38S mutant also has reduced phosphatase activity but not to the same extent as E132S/N135S. The combined effect of mutating both active sites is roughly cumulative (Fig. 3F). The low phosphatase activity of the first site, although apparent in Fig. 3, D and E, may be masked somewhat by the coupled nature of the assay. CheY-P hydrolyzes relatively quickly (t½ = 30–150 s (46)), and thus, conditions were used where the mutant activities would distinguish steady-state levels of CheY-P in the presence of CheA. In the case of the E35S/N38S double mutant, CheY phosphorylation by CheA competes with FliY-catalyzed dephosphorylation, but the overall dephosphorylation rate has been reduced compared with WT due to loss of the first active site. Why FliY contains two active centers of differing activity is unclear, although two different sites may facilitate the optimum deactivation rate of CheY in the confines of the flagellar switch complex, where many copies of CheY-P simultaneously contribute to switching the rotation sense (47, 48).
Both FliY active sites show a high degree of symmetry in sequence and structure and they both bind the substrate CheY-P, but not the product CheY. Conservative mutations of suspected catalytic residues in either site lower phosphatase activity, but mutations in both sites are needed to abolish activity. Thus, it is very likely that FliY has two independent catalytic centers. The greater activity of the Glu132/Asn135 site compared with the Glu35/Asn38 site most likely stems from the peripheral regions surrounding the consensus motifs. Either single mutant E132S or N135S showed nearly the same loss in activity as the double mutant E132S/N135S (Fig. 3D). However, the N38S mutation has a larger affect on dephosphorylation than E35S. The requirement of Asn at positions 38 and 135 is most likely due to their role in aligning a catalytic water molecule, as seen in the structure of CheX:CheY3 (12). Substitution of the conserved Asn residues from both active sites in T. maritima and B. subtilis CheC (with activating CheD present) removed all activity, whereas the double Glu mutant could dephosphorylate CheY-P to some extent (11, 16).
Implications for Interaction with CheY
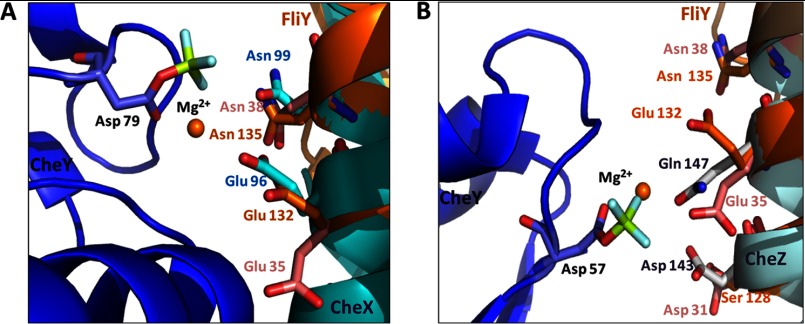
The FliY active sites are very similar to the active site of CheX and CheZ as revealed by the crystallographic structures of those proteins in complex with a phospho-mimic of CheY (CheY·BeF3−·Mg2+) (12). Superimposition of the α1 and α1′ helices of T. maritima FliYm with those of CheX in the B. burgdorferi CheX·CheY3·BeF3−·Mg2+ cocrystal structure (Protein Data Bank code 3HZH) generated a clash free model with the Glu35 (or Glu132) and Asn38 (or Asn135) aligned with Glu96 and Asn99 of CheX, respectively (Fig. 5A). Although the side chain of FliY Glu132 is directed away from the predicted location of the CheY phosphate in the structure, the presence of CheY-P likely produces the proper position (12). CheZ also projects acid and amide-containing side chains from an α-helix to bind and hydrolyze the CheY phosphoryl group (13). Superposition of α1 and α1′ helices of T. maritima FliY on to the active site helix of E. coli CheZ·CheY·BeF3−·Mg2+ (Protein Data Bank code 1KMI) led to a clash-free model that superimposed FliY Asp31 (or Ser128) to Asp143 of CheZ, but it did not align the essential T. maritima FliY Asn38 (or Asn135) near the active site (Fig. 5B). Shifting down the FliY helices to superimpose Asn38 (or Asn135) with Gln147 of CheZ generated some clashes and mismatched the side chain lengths. Thus, the chemical mechanism of hydrolysis is likely similar in CXY and CheZ phosphatases (12), although the reactive residues are supplied somewhat differently (11).
FIGURE 5.
Structural comparison of FliY to CheX and CheZ. Comparison of the active center residues of FliY (site 1 in pink, site 2 in orange) and B. burgdorferi CheX (cyan) in complex with CheY·BeF3−·Mg2+ (bule) (A) and E. coli CheZ (light blue) in complex with CheY·BeF3−·Mg2+ (B).
The N-terminal CheY Binding Domain Is Not Essential for Phosphatase Activity
FliY has an N-terminal CheY binding peptide that is homologous to the N terminus of FliM. Crystal structures of activated CheY in complex with the FliMn peptide have shown that upon CheY phosphorylation, Tyr106 changes conformation from an exposed to a partially buried position to facilitate the packing of the helical FliMn binding motif against the α4-β5-α5 region of CheY (49–51). The residues involved in binding CheY are well conserved in FliM and FliY and a similar motif is found in the C terminus of CheZ (supplemental Fig. 5) (52, 53). Isothermal titration calorimetric measurements demonstrated a much higher affinity interaction for T. maritima CheY binding to FliMnm compared with FliMm (11). The affinity between CheY and FliMnm is increased by orders of magnitude in the presence of the phosphormimic BeF3−. We have also demonstrated here that CheY-P binds to FliMnm more strongly than does CheY and that neither CheY-P nor CheY binds to FliMm under these conditions (Fig. 4A). Given that the N-terminal peptides of FliM and FliY have conserved binding regions (supplemental Fig. 5), it is surprising that the in vitro phosphatase activity of FliY does not depend on FliYn (Fig. 3B). Indeed, FliYnm or FliYm show the same degree of binding to CheY-P, which is contrary to what has been observed for the B. subtilis proteins (14). Thus, FliYn is not necessary to recruit CheY-P to the FliY active site, although it may interact weakly with CheY in the phosphorylated form (the binding of which would be masked by the strong direct binding to FliYm in Fig. 4A). Notably, if CheY were bound to FliYn in the same mode as found in the CheY-FliM peptide cocrystal structures, the short linker between FliYn and FliYm would prevent the CheY aspartyl phosphate residue from accessing the FliY active center. The short linker of about nine residues between the conserved CheY binding motif of FliYn and the central domain (α1) in T. maritima FliY compared with the >20 residues linkers of B. subtilis FliY, E. coli FliM, and T. maritima FliM (supplemental Fig. 5) likely restrict CheY-P from accessing the dephosphorylation centers of T. maritima FliY. Thus, binding of T. maritima FliYn to CheY may be more important for switching the rotor direction (in assistance to FliMn) than dephosphorylation of CheY-P.
Implications for Rotor Assembly
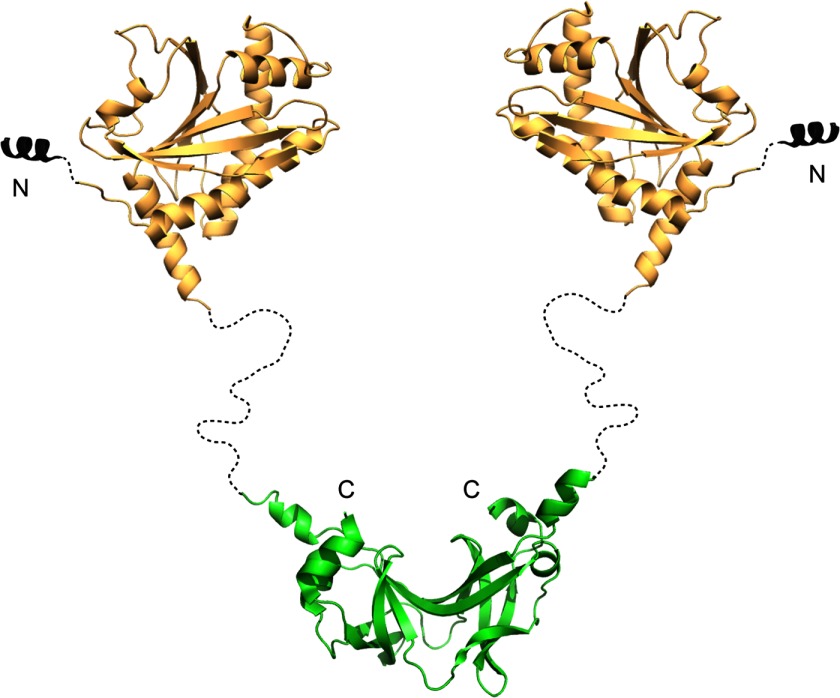
The structure of the flagellar rotor in bacteria genera such as Thermotoga or Bacillus may be different from that in Escherichia and Salmonella due to the presence of FliY. Previous studies have shown that T. maritima FliN forms intertwined dimers (32), which is consistent with FliY forming a dimer mediated by FliYc (i.e. FliN). Computational methods (with Psipred; (54, 55)) predict that the ∼30-residue region connecting FliYm and FliYc does not have defined secondary structure and would allow substantial flexibility between the C-terminal and middle domains (Fig. 6). EM images and biochemical studies localize FliN to the bottom of the C-ring in Salmonella (32). Given the size of FliY (more than twice that of E. coli FliN), we predict FliYc to be also present as a rigid structure at the base of the C-ring with the FliYnm region extending out of the base. The inability of FliYm to bind FliGmc suggests that it does not substitute for FliMm in the center of the rotor. If the additional FliYm domain were to be fixed in position, it may partly account for the larger diameter of the rotor in FliY-containing bacterial such as Treponema primitia (56) and the extra electron density clearly visible at the bottom of the C-ring in Leptospira interrogans (45). The structure of T. maritima FliYm provides an atomic resolution model for a key component of the flagellar rotor of many bacteria and will aid the further exploration of rotor architecture in these species.
FIGURE 6.
Model of FliY. Model derives from crystal structures of the middle domain (Protein Data Bank code 4HYN) and the C-terminal ∼100 residues (Protein Data Bank code 1YAB) Secondary structure prediction suggests that the long linker region is unstructured. The color pattern as defined in Fig. 1.
Acknowledgments
We thank the Cornell Synchrotron Light Source for access to data collection facilities and Hendrick Szurmant for pointing out the possibility of a full-length FliY/N protein in T. maritima.
This work was supported by National Institutes of Health, NIGMS Grant GM064664.

This article contains supplemental Figs. 1–5 and additional references.
The atomic coordinates and structure factors (code 4HYN) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- CheY-P
- phosphorylated CheY
- FliMn
- amino-terminal CheY-binding motif
- FliMm
- middle domain of FliM.
REFERENCES
- 1. Wadhams G. H., Armitage J. P. (2004) Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5, 1024–1037 [DOI] [PubMed] [Google Scholar]
- 2. Bilwes A. M., Park S. Y., Quezada C. M., Simon M. I., Crane B. R. (2003) Structure and Function of CheA, the Histidine Kinase Central to Bacterial Chemotaxis in Histidine Kinases in Signal Transduction (Inouye M., Dutta R., eds), pp. 48–74, Academic Press, San Diego [Google Scholar]
- 3. Clausznitzer D., Oleksiuk O., Løvdok L., Sourjik V., Endres R. G. (2010) Chemotactic response and adaptation dynamics in Escherichia coli. PLoS Comput. Biol. 6, e1000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sourjik V., Berg H. C. (2002) Receptor sensitivity in bacterial chemotaxis. Proc. Natl. Acad. Sci. U.S.A. 99, 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kuo S. C., Koshland D. E., Jr. (1987) Roles of CheY and CheZ gene products in controlling flagellar rotation in bacterial chemotaxis of Escherichia coli. J. Bacteriol. 169, 1307–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Silversmith R. E., Guanga G. P., Betts L., Chu C., Zhao R., Bourret R. B. (2003) CheZ-mediated dephosphorylation of the Escherichia coli chemotaxis response regulator CheY: role for CheY glutamate 89. J Bacteriol 185, 1495–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Muff T. J., Ordal G. W. (2008) The diverse CheC-type phosphatases: chemotaxis and beyond. Mol. Microbiol. 70, 1054–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Szurmant H., Ordal G. W. (2004) Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 68, 301–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Silversmith R. E. (2010) Auxiliary phosphatases in two-component signal transduction. Curr. Opin. Microbiol. 13, 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wuichet K., Zhulin I. B. (2010) Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 3, ra50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Park S. Y., Chao X., Gonzalez-Bonet G., Beel B. D., Bilwes A. M., Crane B. R. (2004) Structure and function of an unusual family of protein phosphatases: the bacterial chemotaxis proteins CheC and CheX. Mol. Cell 16, 563–574 [DOI] [PubMed] [Google Scholar]
- 12. Pazy Y., Motaleb M. A., Guarnieri M. T., Charon N. W., Zhao R., Silversmith R. E. (2010) Identical phosphatase mechanisms achieved through distinct modes of binding phosphoprotein substrate. Proc. Natl. Acad. Sci. U.S.A. 107, 1924–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao R., Collins E. J., Bourret R. B., Silversmith R. E. (2002) Structure and catalytic mechanism of the E. coli chernotaxis phosphatase CheZ. Nat. Struct. Biol. 9, 570–575 [DOI] [PubMed] [Google Scholar]
- 14. Szurmant H., Muff T. J., Ordal G. W. (2004) Bacillus subtilis CheC and FliY are members of a novel class of CheY-P-hydrolyzing proteins in the chemotactic signal transduction cascade. J. Biol. Chem. 279, 21787–21792 [DOI] [PubMed] [Google Scholar]
- 15. Chao X., Muff T. J., Park S. Y., Zhang S., Pollard A. M., Ordal G. W., Bilwes A. M., Crane B. R. (2006) A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation. Cell 124, 561–571 [DOI] [PubMed] [Google Scholar]
- 16. Muff T. J., Ordal G. W. (2007) The CheC phosphatase regulates chemotactic adaptation through CheD. J. Biol. Chem. 282, 34120–34128 [DOI] [PubMed] [Google Scholar]
- 17. Glekas G. D., Plutz M. J., Walukiewicz H. E., Allen G. M., Rao C. V., Ordal G. W. (2012) Elucidation of the multiple roles of CheD in Bacillus subtilis chemotaxis. Mol. Microbiol. 86, 743–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bischoff D. S., Ordal G. W. (1992) Identification and characterization of FliY, a novel component of the Bacillus subtilis flagellar switch complex. Mol. Microbiol. 6, 2715–2723 [DOI] [PubMed] [Google Scholar]
- 19. Liao S., Sun A., Ojcius D. M., Wu S., Zhao J., Yan J. (2009) Inactivation of the fliY gene encoding a flagellar motor switch protein attenuates mobility and virulence of Leptospira interrogans strain Lai. BMC Microbiol. 9, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brown P. N., Hill C. P., Blair D. F. (2002) Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 21, 3225–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lloyd S. A., Whitby F. G., Blair D. F., Hill C. P. (1999) Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature 400, 472–475 [DOI] [PubMed] [Google Scholar]
- 22. Lee L. K., Ginsburg M. A., Crovace C., Donohoe M., Stock D. (2010) Structure of the torque ring of the flagellar motor and the molecular basis for rotational switching. Nature 466, 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Minamino T., Imada K., Kinoshita M., Nakamura S., Morimoto Y. V., Namba K. (2011) Structural insight into the rotational switching mechanism of the bacterial flagellar motor. PLOS Biol. 9, e1000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paul K., Gonzalez-Bonet G., Bilwes A. M., Crane B. R., Blair D. (2011) Architecture of the flagellar rotor. EMBO J. 30, 2962–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mathews M. A., Tang H. L., Blair D. F. (1998) Domain analysis of the FliM protein of Escherichia coli. J. Bacteriol. 180, 5580–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sockett H., Yamaguchi S., Kihara M., Irikura V. M., Macnab R. M. (1992) Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J. Bacteriol. 174, 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Toker A. S., Macnab R. M. (1997) Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J. Mol. Biol. 273, 623–634 [DOI] [PubMed] [Google Scholar]
- 28. Brown P. N., Terrazas M., Paul K., Blair D. F. (2007) Mutational analysis of the flagellar protein FliG: Sites of interaction with FliM and implications for organization of the switch complex. J. Bacteriol. 189, 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dyer C. M., Vartanian A. S., Zhou H., Dahlquist F. W. (2009) A molecular mechanism of bacterial flagellar motor switching. J. Mol. Biol. 388, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Park S. Y., Lowder B., Bilwes A. M., Blair D. F., Crane B. R. (2006) Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor. Proc. Natl. Acad. Sci. U.S.A. 103, 11886–11891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vartanian A. S., Paz A., Fortgang E. A., Abramson J., Dahlquist F. W. (2012) Structure of flagellar motor proteins in complex allows for insights into motor structure and switching. J. Biol. Chem. 287, 35779–35783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brown P. N., Mathews M. A., Joss L. A., Hill C. P., Blair D. F. (2005) Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J. Bacteriol. 187, 2890–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thomas D. R., Francis N. R., Xu C., DeRosier D. J. (2006) The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar typhimurium. J. Bacteriol. 188, 7039–7048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sarkar M. K., Paul K., Blair D. (2010) Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 107, 9370–9375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Szurmant H., Bunn M. W., Cannistraro V. J., Ordal G. W. (2003) Bacillus subtilis hydrolyzes CheY-P at the location of its action, the flagellar switch. J. Biol. Chem. 278, 48611–48616 [DOI] [PubMed] [Google Scholar]
- 36. Kirby J. R., Kristich C. J., Saulmon M. M., Zimmer M. A., Garrity L. F., Zhulin I. B., Ordal G. W. (2001) CheC is related to the family of flagellar switch proteins and acts independently from CheD to control chemotaxis in Bacillus subtilis. Mol. Microbiol. 42, 573–585 [DOI] [PubMed] [Google Scholar]
- 37. Lowenthal A. C., Hill M., Sycuro L. K., Mehmood K., Salama N. R., Ottemann K. M. (2009) Functional analysis of the Helicobacter pylori flagellar switch proteins. J. Bacteriol. 191, 7147–7156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Otwinowski A., Minor W. (1997) Processing of X-ray diffraction data in oscillation mode. Methods Enzymol. 276, 307–325 [DOI] [PubMed] [Google Scholar]
- 39. Terwilliger T. C., Berendzen J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McRee D. E. (1999) XtalView Xfit - A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 41. Brunger A. T. (2007) Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 42. Muff T. J., Foster R. M., Liu P. J., Ordal G. W. (2007) CheX in the three-phosphatase system of bacterial chemotaxis. J. Bacteriol. 189, 7007–7013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Muff T. J., Ordal G. W. (2007) Assays for CheC, FliY, and CheX as representatives of response regulator phosphatases. Methods Enzymol. 423, 336–348 [DOI] [PubMed] [Google Scholar]
- 44. Bischoff D. S., Ordal G. W. (1992) Bacillus subtilis chemotaxis: a deviation from the Escherichia coli paradigm. Mol. Microbiol. 6, 23–28 [DOI] [PubMed] [Google Scholar]
- 45. Raddi G., Morado D. R., Yan J., Haake D. A., Yang X. F., Liu J. (2012) Three-dimensional structures of pathogenic and saprophytic leptospira species revealed by cryo-electron tomography. J. Bacteriol. 194, 1299–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Swanson R. V., Sanna M. G., Simon M. I. (1996) Thermostable chemotaxis proteins from the hyperthermophilic bacterium Thermotoga maritima. J. Bacteriol. 178, 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Khan S., Pierce D., Vale R. D. (2000) Interactions of the chemotaxis signal protein CheY with bacterial flagellar motors visualized by evanescent wave microscopy. Curr. Biol. 10, 927–930 [DOI] [PubMed] [Google Scholar]
- 48. Sowa Y., Berry R. M. (2008) Bacterial flagellar motor. Q. Rev. Biophys. 41, 103–132 [DOI] [PubMed] [Google Scholar]
- 49. Lee S. Y., Cho H. S., Pelton J. G., Yan D., Henderson R. K., King D. S., Huang L., Kustu S., Berry E. A., Wemmer D. E. (2001) Crystal structure of an activated response regulator bound to its target. Nat. Struct. Biol. 8, 52–56 [DOI] [PubMed] [Google Scholar]
- 50. Lee S. Y., Cho H. S., Pelton J. G., Yan D., Berry E. A., Wemmer D. E. (2001) Crystal structure of activated CheY - Comparison with other activated receiver domains. J. Biol. Chem. 276, 16425–16431 [DOI] [PubMed] [Google Scholar]
- 51. Dyer C. M., Quillin M. L., Campos A., Lu J., McEvoy M. M., Hausrath A. C., Westbrook E. M., Matsumura P., Matthews B. W., Dahlquist F. W. (2004) Structure of the constitutively active double mutant cheY(D13K) Y-106W alone and in complex with a flim peptide. J. Mol. Biol. 342, 1325–1335 [DOI] [PubMed] [Google Scholar]
- 52. Silversmith R. E. (2005) High mobility of carboxyl-terminal region of bacterial chemotaxis phosphatase CheZ is diminished upon binding divalent cation or CheY-P substrate. Biochemistry 44, 7768–7776 [DOI] [PubMed] [Google Scholar]
- 53. Guhaniyogi J., Robinson V. L., Stock A. M. (2006) Crystal structures of beryllium fluoride-free and beryllium fluoride-bound CheY in complex with the conserved C-terminal peptide of CheZ reveal dual binding modes specific to CheY conformation. J. Mol. Biol. 359, 624–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Buchan D. W., Ward S. M., Lobley A. E., Nugent T. C., Bryson K., Jones D. T. (2010) Protein annotation and modelling servers at University College London. Nucleic Acids Res. 38, W563–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 56. Murphy G. E., Leadbetter J. R., Jensen G. J. (2006) In situ structure of the complete Treponema primitia flagellar motor. Nature 442, 1062–1064 [DOI] [PubMed] [Google Scholar]