Background: Mutation of Trp-40 in the Cel7A cellobiohydrolase from Trichoderma reesei (TrCel7A) causes a loss of crystalline cellulose-degrading ability.
Results: Mutant W40A showed reduced specific activity for crystalline cellulose and diffused the cellulose chain from the entrance of the active site tunnel.
Conclusion: Trp-40 is essential for chain end loading to initiate processive hydrolysis of TrCel7A.
Significance: The mechanisms of crystalline polysaccharide degradation are clarified.
Keywords: Biodegradation, Biofuel, Carbohydrate-binding Protein, Cellulase, Glycoside Hydrolases, Molecular Dynamics, Protein Engineering, Trichoderma reesei, Cellobiohydrolase, Crystalline Cellulose Degradation
Abstract
The glycoside hydrolase family 7 cellobiohydrolase Cel7A from Trichoderma reesei is one of the best studied cellulases with the ability to degrade highly crystalline cellulose. The catalytic domain and the cellulose-binding domain (CBD) are both necessary for full activity on crystalline substrates. Our previous high-speed atomic force microscopy studies showed that mutation of Trp-40 at the entrance of the catalytic tunnel drastically decreases the ability to degrade crystalline cellulose. Here, we examined the activities of the WT enzyme and mutant W40A (with and without the CBD) for various substrates. Evaluation and comparison of the specific activities of the enzymes (WT, W40A, and the corresponding catalytic subunits (WTcat and W40Acat)) adsorbed on crystalline cellulose indicated that Trp-40 is involved in recruiting individual substrate chains into the active site tunnel to initiate processive hydrolysis. This was supported by molecular dynamics simulation study, i.e. the reducing end glucose unit was effectively loaded into the active site of WTcat, but not into that of W40Acat, when the simulation was started from subsite −7. However, when similar simulations were carried out starting from subsite −5, both enzymes held the substrate for 50 ns, indicating that the major difference between WTcat and W40Acat is the length of the free chain end of the substrate required to allow initiation of processive movements; this also reflects the difference between crystalline and amorphous celluloses. The CBD is important for enhancing the enzyme population on crystalline substrate, but it also decreases the specific activity of the adsorbed enzyme, possibly by attaching the enzyme to non-optimal places on the cellulose surface and/or hindering processive hydrolysis.
Introduction
Cellulose is a linear polymer of β1,4-linked d-glucose and is the main component of the plant cell wall. The major cellulase produced by the cellulolytic ascomycete Trichoderma reesei is the glycoside hydrolase (GH)4 family 7 cellobiohydrolase (TrCel7A). TrCel7A has a two-domain structure composed of a catalytic domain and a CBM1 (carbohydrate-binding module 1) family cellulose-binding domain (CBD) connected by a glycosylated linker peptide (1). The three-dimensional x-ray structure of the TrCel7A catalytic domain with cello-oligosaccharides has been determined, revealing a long active site tunnel formed by loops that extend from the central β-sandwich fold and that are stabilized by 10 disulfide bridges (2). This tunnel contains nine glucosyl-binding sites (−7 to +2), and the catalytic site (−1/+1) is located at the far end, leading to production of cellobiose from the reducing end of the cellulose chain. The tunnel is aligned by hydrogen bond-forming residues, and in addition, four tryptophan residues provide hydrophobic stacking interactions for the glucosyl units at the tunnel entrance (Trp-40), at the center of the tunnel (Trp-38), and around the catalytic sites (Trp-367 and Trp-376), as shown in Fig. 1. It is believed that the tight holding of the substrate and the asymmetry of the active site tunnel favor effective and continuous hydrolysis of crystalline cellulose in a processive mode (2, 3).
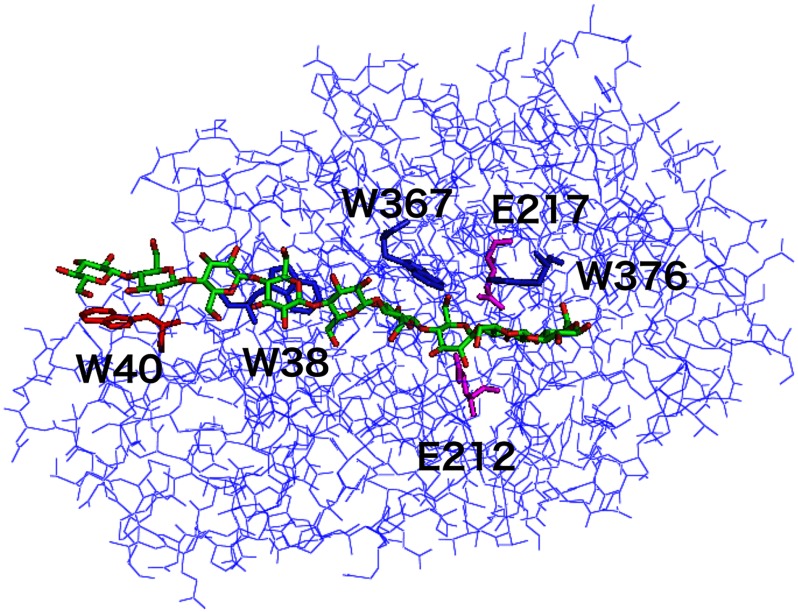
FIGURE 1.
Three-dimensional structure of the catalytic domain of TrCel7A cellobiohydrolase. The α-carbon chain structure of the TrCel7A catalytic domain is shown, together with a modeled continuous cellononaose (Glc-9) chain in the active site tunnel (Protein Data Bank code 8CEL). Trp-40 at subsite −7 is shown in red, and Glu-212, which acts as the catalytic nucleophile between subsites −1 and +1 and was used as a reference position in the molecular dynamics simulations, is shown in magenta.
The processivity of cellobiohydrolases has been estimated by measurement of the ratio of released cellobiose to the sum of released glucose and cellotriose (4–6), by product analysis of a mixture of cellulose and p-nitrophenyl lactoside (7), or by the use of labeled cellulose substrates (8, 9). Recently, we demonstrated the sliding movement of single TrCel7A molecules on a highly crystalline cellulose surface by real-time monitoring with a high-speed atomic force microscope (10–12). We showed that an inactive mutant did not slide on a cellulose surface and also that the CBD was unnecessary for the sliding movement. These studies showed that the sliding movement reflected processive hydrolysis by the TrCel7A catalytic domain. Moreover, the atomic force microscopy results demonstrated that the TrCel7A W40A mutant could not slide on highly crystalline cellulose, even though it was active toward non-crystalline substrates. We interpreted these results as showing that not only catalysis but also chain end recognition is important for effective sliding on and degradation of crystalline cellulose.
In this study, we prepared four samples of TrCel7A, i.e. WT, W40A, and the corresponding catalytic subunits (WTcat and W40Acat) without the CBD. We then characterized their hydrolytic activity and binding properties for defined crystalline and amorphous cellulose substrates to examine the contributions of Trp-40 and the CBD to cellulose degradation. We also carried out molecular dynamics simulations of WTcat and W40Acat to elucidate the mechanism through which Trp-40 promotes processive hydrolysis of cellulose chains.
EXPERIMENTAL PROCEDURES
Construction of Mutant-encoding Plasmid and Transformation of T. reesei
Escherichia coli strain DH5α (Promega) was used as the cloning host for DNA construction. The pEM-F5 plasmid containing the cel7A gene (cDNA) under the control of its own promoter was used as an expression vector (13), and the T. reesei ALKO 3413 strain (Roal Ltd.) lacking the genes coding for endogenous Cel7A and Cel7B was used as the host for production of the mutant enzyme. For selection of T. reesei transformants, the hygromycin selection plasmid pRLMex30 was used (14). The mutant expression plasmid was constructed by overlap extension PCR, and the DNA sequence of the whole mutated area was confirmed by DNA sequencing. Transformation and choice of the most productive transformant were performed basically as described previously (15).
Isolation of TrCel7A and Mutant Enzymes
The production and purification of intact TrCel7A and the catalytic module of TrCel7A were performed as described previously (16). The TrCel7A W40A mutant was produced in a Chemap LF 20 fermenter (working volume of 15 liters). The cultivation medium used contained 60 g/liter whey powder, 30 g/liter spent grain, 5.0 g/liter KH2PO4, and 5 g/liter (NH4)2SO4. Inocula (spore suspensions in 50% glycerol at −80 °C) were grown on the same medium with only 20 g/liter carbon source and 10 g/liter spent grain, buffered with 15 g/liter KH2PO4, in two stages of 4× 50 ml (3 days, 200 rpm, 28 °C) and 5× 200 ml (10% (v/v) transfer, 2 days, 200 rpm, 28 °C). The fermentation lasted for 192 h, and the other cultivation conditions were as follows: temperature, 28 °C; pH 4–5 (lower limit controlled by the addition of NH4OH and upper limit controlled by the addition of H3PO4); dissolved oxygen, >30% (agitation at 400–800 rpm); and aeration, 5 liters/min. Foaming was controlled by the automatic addition of Struktol J 633 polyoleate antifoaming agent (Schill+Seilacher GmbH). After fermentation, the biomass was separated by centrifugation at 4000 × g for 30 min at 4 °C. The culture filtrate contained at least 1 g/liter W40A protein as judged by Western blot analysis (see below).
Purification of the W40A mutants was started by adjusting the clarified Trichoderma culture filtrate (300 ml) in a Bio-Gel P-6 gel filtration column to 50 mm sodium acetate buffer (pH 5.6). The sample (containing both intact W40A and its catalytic domain (W40Acat)) was then applied to a DEAE-Sepharose FF column equilibrated with 50 mm sodium acetate (pH 5.6). The enzyme was eluted with a linear gradient of 0–0.5 m NaCl in the equilibration buffer. The fractions containing the highest activity for 4-methylumbelliferyl β-d-lactoside (MULac) were pooled and adjusted to pH 6.0. Ammonium sulfate was added to 0.65 m, and the sample was loaded onto a phenyl-Sepharose FF column equilibrated with 50 mm potassium phosphate (pH 6.0) containing 0.65 m ammonium sulfate. The unadsorbed proteins, including the W40A catalytic module (W40Acat), were eluted with the equilibration buffer, and the adsorbed W40A protein was eluted with a decreasing linear gradient to 50 mm potassium phosphate buffer (pH 6.0). Analysis of the fractions was performed using MULac as a substrate. SDS-PAGE and Western blotting were also employed as described previously (15). The purest fractions containing intact two-domain W40A and W40Acat were separately combined into two pools. The buffer of both pools was exchanged to 50 mm sodium acetate buffer (pH 5.0) in a Bio-Gel P-6 column equilibrated with the same buffer. The fractions containing the highest protein concentration were pooled, concentrated, and used for the biochemical characterization described below. The results from SDS-PAGE of the purified enzymes are shown in supplemental Fig. S1. The contaminating activities for 4-methylumbelliferyl β-d-glucoside and hydroxyethylcellulose were also checked as described previously (17), and the protein preparations were estimated to contain <0.1% β-glucosidase or endoglucanase contamination.
The protein concentration of TrCel7A preparations was measured in terms of UV absorption at 280 nm. For the WT enzyme, ϵ = 83,000 m−1 cm−1 was used, and for WTcat, ϵ = 80,000 m−1 cm−1 (based on quantitative amino acid analysis) was used.5 For W40A and W40Acat, theoretical ϵ values of 77,500 and 74,500 m−1 cm−1 were used, respectively; these were calculated from the measured values for the wild-type proteins by subtracting ϵ = 5,500 m−1 cm−1 for a tryptophan residue (18).
Degradation of MULac
Enzyme activities for MULac were determined by monitoring the amount of released methylumbelliferone at various substrate concentrations (0–3.0 mm) in 50 mm sodium acetate buffer (pH 5.0) at 27 °C. The enzyme concentrations used were 0.3 μm for WT, WTcat, W40A, and W40Acat. The enzyme reactions were stopped by the addition of 0.5 m Na2CO3, and liberation of methylumbelliferone was measured as described by Voutilainen et al. (19). The data obtained were fitted to the Michaelis-Menten equation. To calculate the kinetic parameters for substrate inhibition, the following equation was used for curve fitting: v = kcatE0S/(Km + S + S2/Km2), where E0 is the enzyme concentration, S is the substrate concentration, Km is the Michaelis constant, Km2 is the substrate inhibition constant, and kcat is the catalytic constant. All kinetic parameters were calculated using DeltaGraph 5.5.5 (Red Rock Software) or KaleidaGraph 3.6.4 (Synergy Software).
Cellulose Preparation
Highly crystalline cellulose (HCC) was prepared from green alga Cladophora spp. as described previously (20). Phosphoric acid-swollen cellulose (PASC) was prepared from Avicel (Funakoshi Corp.) as follows. Avicel was mixed with 85% (w/w) phosphoric acid, and a completely clear solution was obtained by smashing with a glass stick. After overnight incubation at 4 °C, cellulose was regenerated in water, and a suspension was prepared in a high-speed blender. The cellulose suspension was washed with water and stored at 4 °C. The crystallinity of the cellulose substrates was estimated by x-ray powder diffraction (21) and showed that HCC is ∼99% crystalline, whereas PASC is non-crystalline.
Cellulose Hydrolysis
The hydrolytic activities for HCC and PASC were determined by shaking each enzyme (final concentration of 1.4 μm) and 0.1% substrate at 27 °C in 50 mm sodium acetate (pH 5.0). Hydrolysis was evaluated at nine different time points up to 24 h, and the formation of soluble reducing sugars was determined with the p-hydroxybenzoic acid hydrazide method using cellobiose as a standard (22). The amount of released cello-oligosaccharides was also determined by high-performance anion-exchange chromatography at three or four time points basically as described previously (23). The standards were glucose (Fluka, Buchs, Switzerland); cellobiose, cellotetraose, and cellopentaose (Serva, Heidelberg, Germany); and cellotriose and cellohexaose (Associates Cape Cod, Inc.). In all cases, cellobiose was the major soluble product formed. Plots of the cellobiose produced were fitted to an exponential and linear equation, q(t) = a·(1 − exp(−b·t)) + c·t, and a differential equation, v(t) = a·b·exp(−b·t) + c, where a, b, and c are constants, t is the incubation time, q(t) is the product concentration, and v(t) is the production velocity.
Adsorption Analysis
Adsorption on HCC and PASC (0.1%, w/v) was measured under conditions similar to those used for hydrolysis with 1.4 μm enzyme at pH 5.0 and 27 °C. After 1 and 2 h of incubation, the mixtures were filtered through a Millex-GV 0.22-μm membrane (Millipore) to terminate the reaction. Then, 700 μl of 50 mm sodium acetate (pH 5.0) was added to 300 μl of filtrate, and the protein concentration was measured using a Varian Cary Eclipse spectrofluorometer. A separate standard curve was prepared for each protein. The binding experiments were done in triplicate. The amount of adsorbed enzyme was calculated from the initial protein concentration, and the remaining cellulose contents at 1- and 2-h time points were calculated from the amount of cellobiose produced.
Molecular Dynamics Simulation
The initial coordinates of the TrCel7A catalytic domain were taken from Protein Data Bank structure 8CEL (2), and the structure of the W40A mutant was created by in silico mutation. The protonation states at pH 7.0 were determined using the PDB2PQR server (24), where the 10 disulfide bonds described in the Protein Data Bank were assigned to the relevant cysteine residues. To compare the behavior of the cellulose chain at the tunnel entrances of WTcat and W40Acat, the reducing end glucose (head) of cellononaose was initially positioned at subsite −7 (other parts were created automatically), which is the nonreducing end (tail) position in the original Protein Data Bank structure. The systems were then fully solvated with explicit solvent (including crystal water), and 19 Na+ counterions were added to obtain electrostatic neutrality. All simulations were performed using the AMBER 11 package (25). The AMBER ff03 force field was used for proteins. We employed the general AMBER force field with the AM1-BCC partial charges for the N-terminal pyroglutamic acid, the GLYCAM 06 force field for cellononaose, and the TIP3P model for water molecules. The systems were energetically minimized for 300 steepest descent steps and equilibrated for 1 ns with gradually reducing restraints. Finally, two 50-ns production runs were performed with different initial velocities for each system. The temperature and pressure were controlled using the Berendsen rescaling method (26), and the long-range electrostatic forces were calculated using the particle mesh Ewald method (27). The trajectory analysis was conducted using the AMBER module ptraj, and the snapshot structures were visualized with VMD (28). In addition, the molecular dynamics simulations starting form subsite −5 were performed using same methods as described above for both WTcat and W40Acat.
RESULTS
Hydrolysis of Soluble Substrate
The kinetic constants (kcat, Km, and Km2) of the four enzyme variants (WT, WTcat, W40A, and W40Acat) with a soluble substrate (MULac) were measured, and the results are shown in Table 1. There is no significant difference detected in the values. The W40A mutant seems to have a slightly increased substrate inhibition constant (Km2), which hints that the MULac substrate has also nonproductive binding modes involving subsite −7.
TABLE 1.
Kinetic constants measured on MULac at pH 5.0 and 27 °C
| Enzyme | kcat | Km | kcat/Km | Km2a |
|---|---|---|---|---|
| min−1 | mm | min−1 mm−1 | mm | |
| WT | 29.5 ± 1.7 | 0.358 ± 0.03 | 82 ± 1.5 | 4.70 ± 1.6 |
| WTcat | 24.9 ± 1.6 | 0.335 ± 0.03 | 74 ± 1.6 | 6.22 ± 2.7 |
| W40A | 19.8 ± 1.5 | 0.318 ± 0.03 | 62 ± 1.6 | 7.07 ± 4.1 |
| W40Acat | 21.7 ± 1.3 | 0.293 ± 0.02 | 74 ± 1.7 | 7.77 ± 4.2 |
a Km2 is the substrate inhibition constant.
Hydrolysis of Amorphous Cellulose
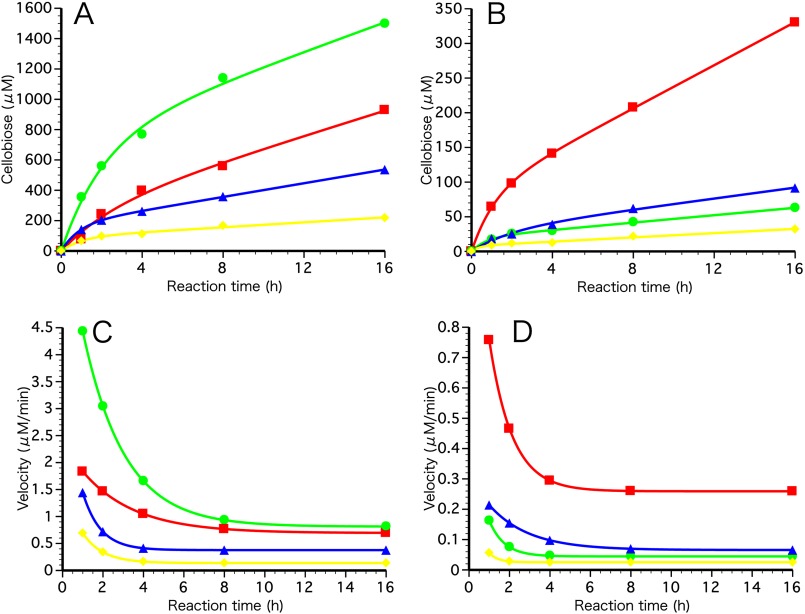
To study the hydrolysis of insoluble cellulose in detail, we first used amorphous cellulose (PASC) as a substrate and determined the amount of bound protein as well as the velocity of cellobiose production, which was calculated from the progress curves. As shown in Fig. 2A, all variants showed hydrolytic activity on PASC, and furthermore, the W40A mutant enzyme was >2-fold more efficient than the WT enzyme in terms of cellobiose production (Table 2). The specific activities of the bound enzymes were also determined by dividing the cellobiose production rates by the amount of adsorbed enzyme at 1- and 2-h time points (Table 2). Because WT and W40A bound equally well to PASC, the specific activity of adsorbed W40A was 2.7 (1 h) and 2.2 (2 h) times greater than that of WT, indicating that Trp-40 slows down the hydrolysis of amorphous cellulose. Although the single-domain W40Acat version had quite low activity for PASC, its specific activity was similarly 2.7 (1 h) and 2.6 (2 h) times higher than that of WTcat, indicating that the negative effect of Trp-40 is independent of the presence or absence of the CBD. However, as shown in Table 2, W40Acat was significantly less well adsorbed on PASC compared with WT, W40A, and WTcat.
FIGURE 2.
Progress curves and product (cellobiose) velocity curves of cellulose hydrolysis by WT, W40A, WTcat, and W40Acat. PASC (A) and HCC (Cladophora spp.; B) were incubated at 27 °C in 50 mm sodium acetate (pH 5.0) with 1.4 μm each enzyme at a 0.1% (w/v) substrate concentration. Duplicate assays were performed at each time point. Soluble products were determined by the p-hydroxybenzoic acid hydrazide method using cellobiose as a standard as described under “Experimental Procedures.” Red squares, WT; green circles, W40A; blue triangles, WTcat; yellow diamonds, W40Acat. Cellobiose production velocities were calculated from progress curves on PASC (C) and HCC (D).
TABLE 2.
Hydrolysis velocity and adsorption of the TrCel7A enzyme variants on HCC and PASC measured at 27 °C in 50 mm sodium acetate (pH 5.0)
The enzyme concentration was 1.4 μm, and the substrate concentration was 0.1% (w/v).
| Cellulose substrate | Hydrolysis velocitya | Bound enzymeb | Specific activity of adsorbed enzymec |
|---|---|---|---|
| μm min−1 | nmol/mg of cellulose | min−1 | |
| PASC | |||
| WT | |||
| 1-h time point | 1.8 ± 1.1 | 1.4 | 1.3 |
| 2-h time point | 1.5 ± 0.6 | 1.5 | 1.1 |
| W40A | |||
| 1-h time point | 4.4 ± 1.1 | 1.3 | 3.5 |
| 2-h time point | 3.0 ± 0.5 | 1.6 | 2.4 |
| WTcat | |||
| 1-h time point | 1.4 ± 0.0 | 1.1 | 1.3 |
| 2-h time point | 0.72 ± 0.01 | 1.2 | 0.6 |
| W40Acat | |||
| 1-h time point | 0.69 ± 0.10 | 0.20 | 3.5 |
| 2-h time point | 0.34 ± 0.02 | 0.24 | 1.7 |
| HCC | |||
| WT | |||
| 1-h time point | 0.76 ± 0.02 | 0.62 | 1.2 |
| 2-h time point | 0.47 ± 0.01 | 0.56 | 0.9 |
| W40A | |||
| 1-h time point | 0.16 ± 0.00 | 0.30 | 0.5 |
| 2-h time point | 0.08 ± 0.01 | 0.41 | 0.2 |
| WTcat | |||
| 1-h time point | 0.21 ± 0.06 | ∼0.05d | 4.2 |
| 2-h time point | 0.15 ± 0.02 | ∼0.07d | 2.1 |
| W40Acat | |||
| 1-h time point | 0.06 ± 0.01 | 0.10 | 0.6 |
| 2-h time point | 0.03 ± 0.01 | 0.15 | 0.3 |
a Hydrolysis velocity, shown as cellobiose produced per min, was calculated at the 1- and 2-h time points from the progress curves shown in Fig. 2.
b The bound enzyme concentration was determined from the supernatant after 1 and 2 h of incubation with HCC or PASC as described under “Experimental Procedures” and then dividing this concentration by the remaining cellulose amount.
c The specific activity of the adsorbed enzyme was calculated by dividing the hydrolysis rate at the 1- and 2-h time points by the bound enzyme concentration. The uncertainty in the specific activity values was estimated to be 20% based on three repeated experiments.
d The error was estimated to be 50% because the measured concentration was <0.1 μm. Therefore, the amounts of adsorbed WTcat on HCC are given as rough estimates.
Hydrolysis of HCC
In contrast to PASC, cellobiose production by WTcat, W40A, and W40Acat was significantly decreased compared with WT when HCC was used as a substrate (Fig. 2B). The results indicate important roles for both Trp-40 and the CBD in the hydrolysis of crystalline cellulose. As shown in Table 2, the velocities of cellobiose production by W40A and WTcat on HCC were reduced to ∼20 and 30% of that by WT, respectively. The specific activity of bound W40A was 45% (1 h) or 21% (2 h) of that of WT, whereas WTcat showed a specific activity that was 4.4 (1 h) or 2.6 (2 h) times higher than that of WT. The adsorption of W40A was 48% (1 h) or 73% (2 h) of that of WT. These results indicate that Trp-40 may play a role in initiating the degradation of HCC as well as in the processive hydrolysis of HCC, whereas the CBD hinders hydrolysis.
Molecular Dynamics Simulation
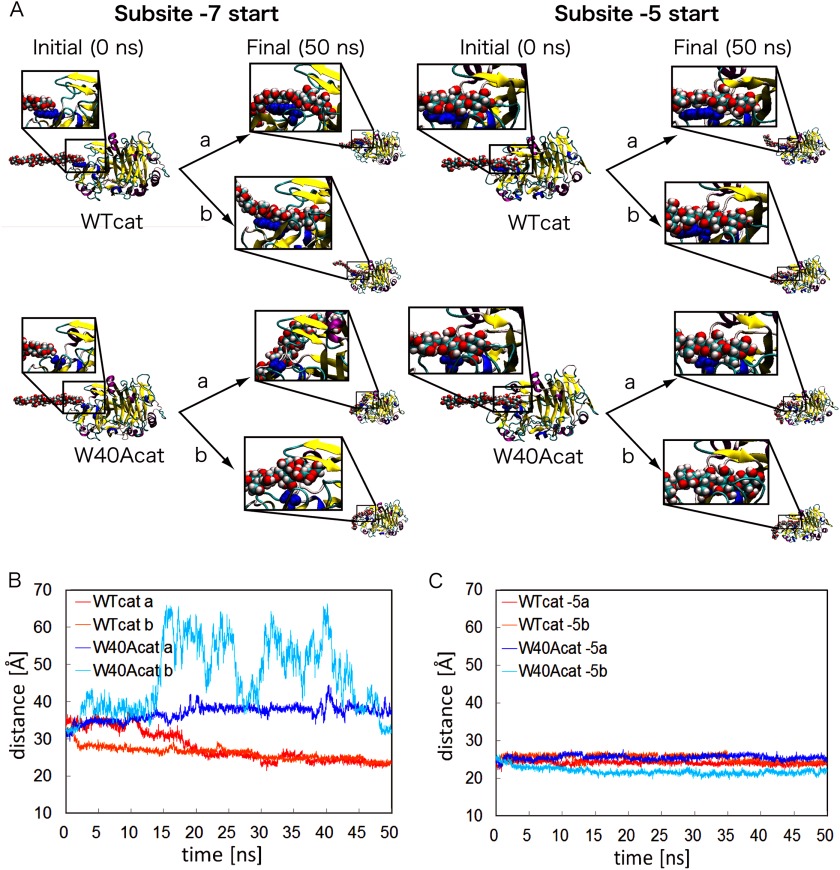
To examine the function of Trp-40 at the atomic level, molecular dynamics simulations were carried out for WTcat and W40Acat with cellononaose, with the reducing end glucose initially positioned at Trp-40 or Ala-40. Two 50 ns simulation runs were performed for each system (Fig. 3A), and the distances between the reducing end glucose and the catalytic carboxylic acid Glu-212 are plotted in Fig. 3B. In the case of WTcat, cellononaose entered the catalytic tunnel and finally penetrated ∼11 Å within the tunnel, approximately two glucose units, toward the catalytic residue Glu-212 during the first simulation (supplemental Video S1). The cellononaose finally entered from subsite −7 to subsite −5. The second trial gave similar results, although the approach toward Glu-212 was much faster than in the first trial (supplemental Video S2). In contrast, in the case of W40Acat, the cellononaose did not move into the active site tunnel but diffused away from the alanine residue at subsite −7. The substrate chain bound on the back side of the catalytic domain in the first trial and on one of the loops forming the active site tunnel in the second trial (supplemental Videos S3 and S4). On the other hand, when the reducing end of the chain was initially positioned at subsite −5, both W40Acat and WTcat kept the cellononaose around the initial position, as shown in Fig. 3A. In the simulations of WTcat, glucose rings were stuck to subsites −5, −6, and −7 (supplemental Videos S5 and S6), which is similar to the simulations starting from subsite −7. However, the simulation of W40Acat starting from subsite −5 showed that the interactions at subsites −5 and −6 were strong enough to keep the cellononaose chain in the tunnel, which is very different form the simulations starting from subsite −7 (supplemental Videos S7 and S8). Interestingly, in the second trial of W40Acat, the cellononaose processed into the subsite tunnel much deeper than with WTcat (Fig. 3C). Consequently, these results indicate that Trp-40 at subsite −7 may assist entry of the cellulose into the subsite tunnel with a much shorter length of the free chain end, which initiates the processive reaction of TrCel7A.
FIGURE 3.
Initial and final snapshots of WTcat and W40Acat with cellononaose and the distances between cellononaose and the catalytic site in the molecular dynamics simulations. The reducing end glucose of cellononaose was initially placed at subsite −7 or −5 of WTcat and W40Acat. Simulations were performed twice (trials a and b) for each enzyme. Initial and final structures are depicted in the close-up views (A). The distances between the catalytic residue Glu-212 and the reducing end glucose of cellononaose during the simulations for subsites −7 and −5 are plotted in B and C, respectively.
DISCUSSION
The protein-carbohydrate interactions of carbohydrate-active enzymes are characterized by a network of hydrogen bonds and hydrophobic stacking interactions involving aromatic amino acid residues (29, 30). Stacking interactions seem to be especially important in the case of enzymes and individual protein domains that act at the surface of insoluble substrates, e.g. carbohydrate-binding modules (31), chitinases (32), and cellulases (33). TrCel7A has a two-domain structure composed of a catalytic domain and a CBD, both of which have aromatic amino acid residues that serve to align the substrate-binding area and form hydrophobic stacking interactions with the glucosyl units of cellulose (2, 31). In processive cellulose hydrolysis, the TrCel7A catalytic domain slides along the cellulose chain and releases the product (cellobiose) from subsites +1 and +2 and then proceeds to the next cellobiose unit from the reducing end. To investigate this process, we focused on the roles of TrCel7A Trp-40 and CBD in cellulose degradation. Trp-40 is located in an overhang at the entrance to the active site tunnel, and in the productive binding mode, it makes contact with the sugar ring (2).
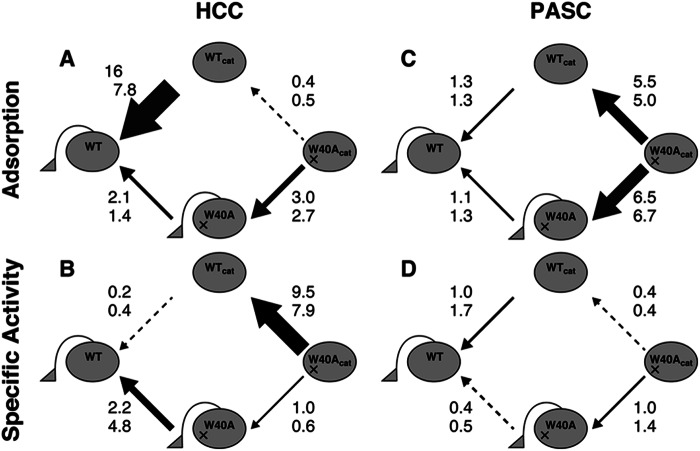
With soluble MULac as a substrate, no significant difference in activity was detected, as expected (Table 1). Elimination of the CBD did not significantly affect the PASC hydrolysis by the WT enzyme, whereas mutation of Trp-40 to Ala caused a clear increase in cellobiose production from amorphous substrates (Table 2). Because PASC is an insoluble substrate, the adsorption behavior was also compared and is summarized in Scheme 1. The results indicate that the CBD had a very small effect, if any, on the adsorption of the WT enzyme on PASC, which was also an expected result. However, W40Acat bound less strongly to PASC compared with WTcat and the CBD-containing mutant (W40A). This suggests that adsorption of the TrCel7A catalytic domain on amorphous cellulose is dependent on Trp-40. The CBD can, however, compensate for the binding defect caused by the W40A mutation. Concerning the specific activity of the adsorbed enzymes on PASC, Trp-40 had a negative effect, as shown in Scheme 1. We assume that this is due to the stacking of a substrate at subsite −7, which interferes with the cellulose chain slide within the tunnel and with the dissociation of the catalytic domain from a cellulose chain.
SCHEME 1.
Effect of CBD and Trp-40 on adsorption and specific activity of the adsorbed enzyme (velocity/adsorption) during HCC and PASC hydrolysis. The numbers next to the arrows indicate the ratios calculated from the 1-h (upper) and 2-h (lower) data. The amount of adsorbed enzyme for HCC (A) and PASC (C) and the specific activity for HCC (B) and PASC (D) were calculated from Table 1. The values of the front edge of the arrows were divided by the values of the tail edge of the arrows.
In contrast, the hydrolysis of HCC showed completely different behavior compared with MULac and PASC. Enzyme adsorption was clearly promoted by the CBD, whereas Trp-40 had less of an effect on the overall binding. However, the specific activity of the adsorbed enzyme on HCC was greatly enhanced by the presence of Trp-40 but hindered by the CBD. This is an important finding because there is likely to be a very sensitive balance between Trp-40 and the CBD affecting the processes of processivity and adsorption. Considering the molecular dynamics simulation results, Trp-40 should have a particularly important role when a limited number of reducing end glucose units are available to the active site. The essential difference between amorphous and crystalline celluloses is an availability of the free chain ends. Therefore, HCC has only a small number of glucose units free at the chain end, whereas PASC has longer chain ends available. The free reducing end of the chain might not reach a deep position of the subsite without Trp-40, which is critical for HCC degradation. In contrast, the chain ends of PASC easily reach subsite −5, and Trp-40 is unnecessary for chain loading. These results are also supported by a previous molecular dynamics study that calculated the binding energy of each subsite (3, 34). Concerning the role of the CBD, the population of the adsorbed enzyme is clearly larger in the presence of the CBD, as can be seen when comparing the adsorption of WTcat and WT. In contrast, the CBD lowers the specific activity of the adsorbed enzyme. There may be two reasons for this. 1) The CBD may restrict the intramolecular processive movement of the enzyme on cellulose surface. 2) The CBD may lead to the nonproductive binding of enzyme on places where the catalytic module does not find a free chain end. In our high-speed atomic force microscopy study, we observed the apparent movement of WTcat on HCC and suggested that the major contribution of the CBD is simply to increase the concentration of adsorbed enzyme on the crystalline substrate (10). The present experiments with WT and WTcat further suggest that CBD-mediated adsorption involves nonproductive binding and/or disturbance of the hydrolytic activity, which is seen as reduced specific activity of the adsorbed enzyme.
In our previous high-speed atomic force microscopy work, we observed a “wriggling” movement of the W40A mutant enzyme on the HCC surface and speculated that this is due to an inability to recruit the substrate chain into the active site tunnel (10). Our molecular dynamics simulation results presented here also support this hypothesis, and the biochemical data suggest a function of Trp-40 in initiating the processive sliding movement on HCC. The importance of this Trp residue in crystalline cellulose degradation is further supported by the fact that this residue is conserved among the sequences of GH7 family cellobiohydrolases (CAZy Database) as shown in supplemental Fig. S2 (35). Furthermore, there is a Trp residue at the subsite entrance that is conserved in GH6 family cellobiohydrolases. It has been shown previously that T. reesei GH6 family Cel6A cellobiohydrolase variants with this corresponding Trp residue mutated to alanine or aspartic acid have clearly decreased hydrolysis activity on bacterial microcrystalline cellulose but not PASC (36). This suggests that a tryptophan residue located at the entrance of the subsite is essential for substrate loading in all cellobiohydrolases that degrade crystalline celluloses.
In conclusion, this results of this study indicate that the CBD of TrCel7A serves to increase the enzyme concentration on the HCC surface, but at the expense of reducing the specific activity of the adsorbed enzyme. The main function of the conserved Trp-40 residue at the edge of the subsite tunnel appears to be recruitment of cellulose chain reducing ends into the active site tunnel and initiation of processive hydrolysis of the cellulose chain on HCC. This is important because long free cellulose chain ends are not frequently available on HCC, but again, there is a tradeoff due to the stacking interaction of Trp-40, which impedes progression of the cellulose chain within the active site tunnel. Consequently, removal of Trp-40 leads to more efficient degradation of less crystalline substrates.
Acknowledgments
We thank Michael Bailey, Harry Boer, Tiina Hartikainen, Tarmo Pellikka, Tiina Liljankoski, and Riitta Suihkonen (VTT Technical Research Centre of Finland) for help in production, purification, and initial characterization of the TrCel7A W40A mutant enzymes.
This work was supported by Grant-in-aid for Innovative Areas 24114008 from the Japanese Ministry of Education, Culture, Sports, and Technology (MEXT) (to K. I.); Grant-in-aid for Scientific Research (B) 24380089 and Grants-in-aid for Scientific Research for Young Scientists (A) 19688016, (A) 21688023, and (B) 17780245 (to K. I.) and Grant-in-aid for JSPS Fellows 257574 (to A. N.) and 08J07454 (to T. T.) from the Japan Society for the Promotion of Science (JSPS); and a grant from the Advanced Low Carbon Technology Research and Development Program (ALCA) of the Japan Science and Technology Agency (JST) (to K. I.)

This article contains supplemental Figs. S1 and S2 and Videos S1–S8.
A. Koivula, unpublished data.
- GH
- glycoside hydrolase
- TrCel7A
- T. reesei Cel7A
- CBD
- cellulose-binding domain
- MULac
- 4-methylumbelliferyl β-d-lactoside
- HCC
- highly crystalline cellulose
- PASC
- phosphoric acid-swollen cellulose.
REFERENCES
- 1. Abuja P. M., Schmuck M., Pilz I., Tomme P., Claeyssens M., Esterbauer H. (1988) Structural and functional domains of cellobiohydrolase I from Trichoderma reesei. Eur. Biophys. J. 15, 339–342 [Google Scholar]
- 2. Divne C., Ståhlberg J., Teeri T. T., Jones T. A. (1998) High-resolution crystal structures reveal how a cellulose chain is bound in the 50 Å long tunnel of cellobiohydrolase I from Trichoderma reesei. J. Mol. Biol. 275, 309–325 [DOI] [PubMed] [Google Scholar]
- 3. Mulakala C., Reilly P. J. (2005) Hypocrea jecorina (Trichoderma reesei) Cel7A as a molecular machine: a docking study. Proteins 60, 598–605 [DOI] [PubMed] [Google Scholar]
- 4. Klyosov A. A. (1990) Trends in biochemistry and enzymology of cellulose degradation. Biochemistry 29, 10577–10585 [DOI] [PubMed] [Google Scholar]
- 5. Medve J., Karlsson J., Lee D., Tjerneld F. (1998) Hydrolysis of microcrystalline cellulose by cellobiohydrolase I and endoglucanase II from Trichoderma reesei: adsorption, sugar production pattern, and synergism of the enzymes. Biotechnol. Bioeng. 59, 621–634 [PubMed] [Google Scholar]
- 6. von Ossowski I., Ståhlberg J., Koivula A., Piens K., Becker D., Boer H., Harle R., Harris M., Divne C., Mahdi S., Zhao Y., Driguez H., Claeyssens M., Sinnott M. L., Teeri T. T. (2003) Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A. A comparison with Phanerochaete chrysosporium Cel7D. J. Mol. Biol. 333, 817–829 [DOI] [PubMed] [Google Scholar]
- 7. Jalak J., Väljamäe P. (2010) Mechanism of initial rapid rate retardation in cellobiohydrolase-catalyzed cellulose hydrolysis. Biotechnol. Bioeng. 106, 871–883 [DOI] [PubMed] [Google Scholar]
- 8. Kipper K., Väljamäe P., Johansson G. (2005) Processive action of cellobiohydrolase Cel7A from Trichoderma reesei is revealed as ‘burst’ kinetics on fluorescent polymeric model substrates. Biochem. J. 385, 527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kurasin M., Väljamäe P. (2011) Processivity of cellobiohydrolases is limited by the substrate. J. Biol. Chem. 286, 169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Igarashi K., Koivula A., Wada M., Kimura S., Penttilä M., Samejima M. (2009) High speed atomic force microscopy visualizes processive movement of Trichoderma reesei cellobiohydrolase I on crystalline cellulose. J. Biol. Chem. 284, 36186–36190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Igarashi K., Uchihashi T., Koivula A., Wada M., Kimura S., Okamoto T., Penttilä M., Ando T., Samejima M. (2011) Traffic jams reduce hydrolytic efficiency of cellulase on cellulose surface. Science 333, 1279–1282 [DOI] [PubMed] [Google Scholar]
- 12. Igarashi K., Uchihashi T., Koivula A., Wada M., Kimura S., Penttilä M., Ando T., Samejima M. (2012) Visualization of cellobiohydrolase I from Trichoderma reesei moving on crystalline cellulose using high-speed atomic force microscopy. Methods Enzymol. 510, 169–182 [DOI] [PubMed] [Google Scholar]
- 13. Margolles-Clark E., Hayes C. K., Harman G. E., Penttilä M. (1996) Improved production of Trichoderma harzianum endochitinase by expression in Trichoderma reesei. Appl. Environ. Microbiol. 62, 2145–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mach R. L., Schindler M., Kubicek C. P. (1994) Transformation of Trichoderma reesei based on hygromycin B resistance using homologous expression signals. Curr. Genet. 25, 567–570 [DOI] [PubMed] [Google Scholar]
- 15. Ståhlberg J., Divne C., Koivula A., Piens K., Claeyssens M., Teeri T. T., Jones T. A. (1996) Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei. J. Mol. Biol. 264, 337–349 [DOI] [PubMed] [Google Scholar]
- 16. Suuräkki A., Tenkanen M., Siika-aho M., Niku-paavola M.-L., Viikari L., Buchert J. (2000) Trichoderma reesei cellulases and their core domains in the hydrolysis and modification of chemical pulp. Cellulose 7, 189–209 [Google Scholar]
- 17. Tomme P., Van Tilbeurgh H., Pettersson G., Van Damme J., Vandekerckhove J., Knowles J., Teeri T., Claeyssens M. (1988) Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur. J. Biochem. 170, 575–581 [DOI] [PubMed] [Google Scholar]
- 18. Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Voutilainen S. P., Boer H., Linder M. B., Puranen T., Rouvinen J., Vehmaanperä J., Koivula A. (2007) Heterologous expression of Melanocarpus albomyces cellobiohydrolase Cel7B, and random mutagenesis to improve its thermostability. Enzyme Microb. Technol. 41, 234–243 [Google Scholar]
- 20. Igarashi K., Wada M., Hori R., Samejima M. (2006) Surface density of cellobiohydrolase on crystalline celluloses. A critical parameter to evaluate enzymatic kinetics at a solid-liquid interface. FEBS J. 273, 2869–2878 [DOI] [PubMed] [Google Scholar]
- 21. Wada M., Okano T., Sugiyama J. (2001) Allomorphs of native crystalline cellulose I evaluated by two equatorial d-spacings. J. Wood Sci. 47, 124–128 [Google Scholar]
- 22. Voutilainen S. P., Puranen T., Siika-Aho M., Lappalainen A., Alapuranen M., Kallio J., Hooman S., Viikari L., Vehmaanperä J., Koivula A. (2008) Cloning, expression, and characterization of novel thermostable family 7 cellobiohydrolases. Biotechnol. Bioeng. 101, 515–528 [DOI] [PubMed] [Google Scholar]
- 23. Tenkanen M., Makkonen M., Perttula M., Viikari L., Teleman A. (1997) Action of Trichoderma reesei mannanase on galactoglucomannan in pine kraft pulp. J. Biotechnol. 57, 191–204 [DOI] [PubMed] [Google Scholar]
- 24. Dolinsky T. J., Nielsen J. E., McCammon J. A., Baker N. A. (2004) PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Case D. A., Darden T. A., Cheatham T. E., 3rd, Simmerling C. L., Wang J., Duke R. E., Luo R., Walker R. C., Zhang W., Merz K. M., Roberts B., Wang B., Hayik S., Roitberg A., Seabra G., Kolossváry I., Wong K. F., Paesani F., Vanicek J., Liu J., Wu X., Brozell S. R., Steinbrecher T., Gohlke H., Cai Q., Ye X., Wang J., Hsieh M.-J., Cui G., Roe D. R., Mathews D. H., Seetin M. G., Sagui C., Babin V., Luchko T., Gusarov S., Kovalenko A., Kollman P. A. (2010) AMBER 11, University of California, San Francisco [Google Scholar]
- 26. Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., DiNola A., Haak J. R. (1984) Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 [Google Scholar]
- 27. Darden T., York D., Pedersen L. (1993) Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 28. Humphrey W., Dalke A., Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- 29. Quiocho F. A. (1986) Carbohydrate-binding proteins: tertiary structures and protein-sugar interactions. Annu. Rev. Biochem. 55, 287–315 [DOI] [PubMed] [Google Scholar]
- 30. Vyas N. K. (1991) Atomic features of protein-carbohydrate interactions. Curr. Opin. Struct. Biol. 1, 732–740 [Google Scholar]
- 31. Linder M., Mattinen M. L., Kontteli M., Lindeberg G., Ståhlberg J., Drakenberg T., Reinikainen T., Pettersson G., Annila A. (1995) Identification of functionally important amino acids in the cellulose-binding domain of Trichoderma reesei cellobiohydrolase I. Protein Sci. 4, 1056–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zakariassen H., Aam B. B., Horn S. J., Vårum K. M., Sørlie M., Eijsink V. G. H. (2009) Aromatic residues in the catalytic center of chitinase A from Serratia marcescens affect processivity, enzyme activity, and biomass converting efficiency. J. Biol. Chem. 284, 10610–10617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ishiguro M., Hori T., Ishida T., Yoshida M., Takabatake K., Kaneko S., Igarashi K., Samejima M. (2010) Molecular cloning of cDNAs encoding two glycoside hydrolase family 7 cellobiohydrolases from the basidiomycete Flammulina velutipes. Plant Biotechnol. 27, 273–281 [Google Scholar]
- 34. GhattyVenkataKrishna P., Alekozai E. M., Beckham G. T., Schulz R., Crowley M. F., Uberbacher E. C., Cheng X. (2013) Initial recognition of a cellodextrin chain in the cellulose-binding tunnel may affect cellobiohydrolase directional specificity. Biophys. J. 104, 904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Davies G. J., Ducros V., Lewis R. J., Borchert T. V., Schülein M. (1997) Oligosaccharide specificity of a family 7 endoglucanase: insertion of potential sugar-binding subsites. J. Biotechnol. 57, 91–100 [DOI] [PubMed] [Google Scholar]
- 36. Koivula A., Kinnari T., Harjunpää V., Ruohonen L., Teleman A., Drakenberg T., Rouvinen J., Jones T. A., Teeri T. T. (1998) Tryptophan 272: an essential determinant of crystalline cellulose degradation by Trichoderma reesei cellobiohydrolase Cel6A. FEBS Lett. 429, 341–346 [DOI] [PubMed] [Google Scholar]