Background: Macrophage M2b polarization conferred by self-DNA immunization initiates and propagates lupus nephritis.
Results: Knockdown of DNA-dependent activator of interferon-regulatory factors (DAI) ameliorates SLE syndrome via blunting macrophage M2b polarization.
Conclusion: DAI functions as a DNA sensor in self-DNA-induced macrophage M2b polarization and lupus nephritis.
Significance: We disclose the mechanism by which self-DNA induces macrophage M2b polarization and lupus nephritis.
Keywords: Autoimmune Diseases, Calcium, Inflammation, Innate immunity, Macrophages, DAI (DLM-1/ZBP1), DNA Sensor, Lupus Nephritis, Macrophage Polarization, Systemic Lupus Erythematosus (SLE)
Abstract
DNA-dependent activator of interferon-regulatory factors (DAI) functions as a cytoplasmic DNA sensor that activates the innate immune system. We previously found that activated lymphocyte-derived self-apoptotic DNA (ALD-DNA) immunization led to pathological macrophage activation and M2b polarization, which could initiate and propagate murine lupus nephritis. However, the specific DNA sensor(s) as well as underlying molecular mechanisms involved in ALD-DNA-induced macrophage M2b polarization in systemic lupus erythematosus (SLE) disease remains unknown. In this study, we reported that DAI expression was significantly increased in SLE patients as well as in lupus mice. Gain- and loss-of-function studies revealed that DAI was involved in ALD-DNA-induced macrophage activation and M2b polarization. Moreover, ALD-DNA notably induced dimerization/oligomerization of DAI and consequently activation of nuclear factor κB (NF-κB) and interferon regulatory factor 3 (IRF3) signaling pathways via calcium signaling, resulting in macrophage activation and M2b polarization. More importantly, blockade of DAI in vivo or selective knockdown of DAI in macrophages could ameliorate SLE syndrome via blunting macrophage M2b polarization and inhibiting inflammatory response in lupus mice. Our results suggest that DAI could function as a DNA sensor and a regulator in ALD-DNA-induced macrophage M2b polarization and lupus nephritis, providing the possible molecular mechanisms involved in ALD-DNA-induced macrophage M2b polarization in SLE disease and making DAI as a potential therapeutic target for the treatment of SLE.
Introduction
Nucleic acids, exposed in a cell by infection or by incomplete clearance during cell apoptosis, can evoke immune responses in many autoimmune diseases (1–5). Systemic lupus erythematosus (SLE)2 is one of the prototypical inflammatory diseases to be linked to the aberrant DNA recognition and the associated autoimmune responses (6–8). In this disease, self-DNA, which is released from improperly cleared apoptotic cells could trigger the innate immune activation and mediate the onset and propagation of an aggressive adaptive immune response, leading to the additional production of autoantibodies and apoptosis, indicating that apoptotic DNA and its recognition by the innate immune system triggers autoreactive Ig production and autoimmune responses in SLE disease (9–12).
The removal of apoptotic cell debris including self-DNA is a dedicated function of phagocytic cells, particularly macrophages (13). In pathologies in which apoptotic cell clearance is disrupted, as in SLE, redundant nuclear antigens direct macrophage activation and aggravate chronic inflammation (14). As in human lupus, kidney disease, especially lupus nephritis, is a major cause of morbidity, which is generally thought to be triggered by deposition of autoantibodies and the subsequent leukocyte infiltration and inflammation (15, 16). Emerging data reveal that renal infiltrating macrophages (Mϕ) are prominent within the inflamed kidneys, which contributes to tissue damage in lupus nephritis by mediating many processes associated with inflammation, proteinuria, complement activation, and excessive tissue remodeling (17–20). Recently, accumulating data demonstrate that renal macrophage infiltration is associated with poor disease outcome in SLE disease (21, 22).
In our previous study (23), by immunizing syngeneic female BALB/c with activated lymphocyte-derived apoptotic DNA (ALD-DNA), mice developed an emblematical SLE syndrome, indicating that ALD-DNA might serve as an important self-immunogen to trigger the autoimmune responses, which eventually lead to the pathogenesis of SLE (24). Further study demonstrated that ALD-DNA immunization led to macrophage infiltration and aberrant activation and M2b polarization in murine renal tissues, which mediated the onset and aggravation of SLE disease (25, 26), suggesting that ALD-DNA-induced aberrant activation and M2b polarization of macrophage plays a crucial pathogenic role in ALD-DNA-mediated lupus nephritis and autoimmune response.
Innate immune responses to self-antigen in SLE are often triggered by self-DNA which, under certain circumstances, can gain assess to the cytoplasm and trigger inflammation (8). Self-DNA recognition by the innate immune system has a central role in the vicious cycle in SLE disease (8, 27). However, in addition to membrane-type Toll-like receptors (TLRs), the concrete cytoplasmic DNA sensor(s) involved in the self-DNA-induced autoimmune response in SLE remains to be investigated (28–39). More recently, DNA-dependent activator of IFN-regulatory factors (DAI) (also referred to as DLM-1/ZBP1; we refer to it as DAI hereafter for convenience) was reported to be the first molecule that might function as a cytoplasmic DNA sensor related to protective and pathologic immune responses (40–42). Whether DAI is involved in ALD-DNA-induced pathological activation of macrophages needs to be investigated.
Calcium (Ca2+) functions as a major second messenger that controls a wide range of important cellular processes in macrophages through the interactions that Ca2+ makes (cross-talk) with other signaling pathways (43). Emerging studies reveal that many signal-transducing pathways, such as Toll-like receptor, Fc receptor (FcR), or complement receptor binding, lead to calcium fluxes within monocytes and macrophages through the calcium channel or from mobilization from intracellular stores (44, 45). Calcium signaling, therefore, appears to be highly integrated into the modulation of macrophage activity. In the current study we sought to determine whether calcium signaling is involved in DAI-mediated signaling in macrophage activation.
We report here that DAI expression is predominantly increased in SLE patients as well as in ALD-DNA-induced lupus mice. ALD-DNA could induce the dimerization/oligomerization of DAI and consequently activate DAI signaling pathways via regulating calcium signaling, thus resulting in macrophage aberrant activation and lupus nephritis, implying the possible mechanisms for the recognition and regulation of ALD-DNA-induced pathological macrophage activation in the context of SLE disease.
EXPERIMENTAL PROCEDURES
Mice
Six- to 8-week-old female BALB/c mice were obtained from the Experimental Animal Center of Chinese Academy of Sciences (Shanghai, People's Republic of China) and maintained in pathogen-free housing. Animals were handled according to the Guide for the Care and Use of Medical Laboratory Animals (Ministry of Health, P.R. China) and with the ethical approval of the Shanghai Medical Laboratory Animal Care and Use Committee as well as the Ethical Committee of Fudan University.
Human Samples
A total of 30 SLE patients (28 female, 2 male; age 38 ± 15 years, disease duration 76.32 ± 66.10 months (mean ± S.D.)), 25 acute bacterial pneumonia patients (23 female, 2 male; age 30 ± 12 years), 20 tuberculosis patients (18 female, 2 male; age 34 ± 13 years), 26 asthma patients (23 female, 3 male; age 36 ± 10 years), 25 type-I diabetes patients (22 female, 3 male; age 40 ± 14 years), and 30 healthy controls (26 female, 4 male; age 35 ± 14 years (mean ± S.D.)), collected from Renji Hospital and Zhongshan Hospital, were included in the study. All of the controls were matched with the SLE patients for age, sex, and race. All SLE patients fulfilled the American College of Rheumatology classification criteria for SLE. SLE activity was assessed with the SLE Disease Activity Index (SLEDAI). All participants are from Chinese Han population. The SLE patients with concurrent infections were excluded from the study. Peripheral blood samples were collected from each subject in tubes containing acid citrate dextrose formula A. Use of human tissues with informed consent was approved by the Institutional Review Board of Fudan University.
Plasmids and Reagents
Small interfering RNAs (siRNAs) against the DAI gene DAI siRNA (siDAI), the corresponding control siRNA (siControl), FLAG-tagged DAI, HA-tagged DAI, the vector controls, MSCVpac-FLAG-DAI, the retroviral vector controls, pcDNA3-DAI (pDAI), and pcDNA3 vector were kindly provided by Prof. Tadatsugu Taniguchi (University of Tokyo, Tokyo, Japan). DAI shRNA (shDAI) and the corresponding control shRNA (shControl) were obtained from Santa Cruz Biotechnology (Santa Cruz). Nuclear factor κB (NF-κB) luciferase reporter plasmids were obtained from Stratagene (Stratagene). Interferon regulatory factor 3 (IRF3) reporter plasmids were kind gifts from Dr. Takashi Fujita (Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan). The plasmid pRL-SV40 containing the Renilla luciferase gene was purchased from Promega (Promega). The pharmacological reagents including the cell-permeable cytosolic calcium chelator O,O′-Bis(2-aminophenyl)ethyleneglycol-N,N,N′,N′-tetraacetic acid, tetraacetoxymethyl ester (BAPTA-AM) (blocking cytosolic calcium), EGTA (blocking entrance of extracellular calcium), cyclosporin A (CsA, disrupting mitochondrial calcium), 7-chloro-3,5-dihydro-5-phenyl-1H-4,1-benzothiazepine-2-on (CGP37157, an inhibitor of the mitochondrial sodium-calcium pump), and calcium mobilizing agents valinomycin and thapsigargin were obtained from Tocris Bioscience (Bristol).
Cell Culture and Transfection
RAW264.7 cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen) in a 5% CO2 incubator at 37 °C. For generation of bone marrow-derived macrophages (BMDMs), bone marrow cells were harvested from uninfected, normal BALB/c mice and filtered through nylon mesh. Bone marrow cells were cultured in L929 cell-conditioned medium at a density of 3 × 105 cells/ml of medium and maintained in a 5% CO2 incubator at 37 °C as described previously (46, 47). Six days after the initial bone marrow cells culture, the medium was changed and the purity of F4/80+ cells was more than 90%, as determined by flow cytometry (FACSCalibur; BD Biosciences, San Jose, CA). PA317 cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS on a 100-mm dish at a concentration of 2 × 105 cells/ml for 24 h and transfected with the MSCVpac-FLAG-DAI plasmid by electroporation as described previously (48, 49). The culture supernatants of transfected PA317 cells were harvested and stored at −70 °C. RAW264.7 cells were infected with the retrovirus from the culture supernatants for 48 h, and then selected with 3.0 μg/ml of puromycin (Amresco) for stable DAI expression clones.
DNA Preparation
ALD-DNA and unactivated lymphocyte-derived DNA (UnALD-DNA) were prepared with murine splenocytes that were generated from surgically resected spleens of 6- to 8-week-old female BALB/c mice and cultured with or without concanavalin A (Sigma) in vitro as previously described (25). In brief, for generation of ALD-DNA, splenocytes were seeded at 2 × 106 cells/ml in 75-cm2 cell culture flask and cultured in the presence of concanavalin A (5 μg/ml) for 6 days to induce apoptosis. The apoptotic cells were stained with FITC-labeled Annexin V (BD Biosciences) and propidium iodide (Sigma), and sorted using a FACSAria (BD Biosciences). Genomic DNAs from syngeneic apoptotic splenocytes were treated with S1 nuclease (Takara Bio, Shiga, Japan) and proteinase K (Sigma), and then purified using the DNeasy Blood & Tissue Kits (Qiagen, Valencia, CA) according to the manufacturer's instructions. UnALD-DNA was prepared with unactivated (resting) splenocytes and extracted using the same methods. To exclude contaminations with LPS, sterile endotoxin-free plasticware and reagents were used for DNA preparation. DNA samples were also monitored for low level of endotoxin by the Limulus amoebocyte lysate assay (BioWhittaker) according to the manufacturer's instructions. The concentration of DNA was determined by detection of the absorbance (A) at 260 nm. The apoptotic DNA ladder of ALD-DNA was confirmed by agarose gel electrophoresis. In the in vitro experiments of macrophage activation analysis, ALD-DNA or UnALD-DNA were transfected into the macrophages with PEITM (Polyplus Transfection) according to the manufacturer's instructions unless otherwise noted.
Generation of SLE Murine Model
To generate the SLE murine model, 6- to 8-week-old syngeneic female BALB/c mice were divided into several groups of 8–10 mice and subcutaneously injected on the back with 0.2 ml of an emulsion containing ALD-DNA (50 μg/mouse) in PBS plus equal volumes of CFA (Sigma) at week 0, and followed by two booster immunizations of ALD-DNA (50 μg/mouse) emulsified with IFA (Sigma) at weeks 2 and 4 for a total of 3 times as previously described (50). Mice receiving an equal volume of PBS plus CFA or IFA, or UnALD-DNA (50 μg/mouse) plus CFA or IFA were used as controls. Mice were bled from the retro-orbital sinus prior to immunization and at 2-week intervals until 3 months after the initial immunization. 8 or 12 weeks later, mice were sacrificed and surgically resected spleens and kidneys were collected for further analysis.
Gene Silencing in Vitro and in Vivo
To block DAI expression in macrophages in vitro, RAW264.7 cells and BMDMs were transfected with siDAI using a Mouse Macrophage Nucleofector Kit (Amaxa) according to the manufacturer's instructions. The cells were then used for subsequent assays after incubation for 48 h in the presence of puromycin (4.0 μg/ml; Sigma). siDAI was used to suppress endogenous DAI expression; nonsense sequence was used as siControl. BMDMs were transfected with shDAI or shControl and cells stably expressing shDAI were isolated by puromycin selection according to the manufacturer's instructions. In vivo transfection of peritoneal cells with siRNA using TransIT-TKO reagent (Takara Mirus) was performed to block the DAI expression in peritoneal macrophages in vivo as described (51). The next day after siRNA treatment, the peritoneal macrophages were collected and purified for further cellular function analysis. The control siRNA was confirmed not to have any affect on DAI expression. Real-time PCR and Western blot analysis were performed to determine the knockdown effect of DAI. No cytotoxic effect of siRNA was observed on macrophages or on mice. To block the DAI expression in lupus mice, 6–8-week-old female BALB/c mice were randomized to inject with siDAI or siControl using in vivo jetPEITM according to the manufacturer's instructions (Polyplus Transfection) every other 3 days for 6 weeks (52). 24 h after the initial siDAI or siControl treatment, the mice were immunized with ALD-DNA (50 μg/mouse), UnALD-DNA (50 μg/mouse), or PBS for 3 times in 4 weeks as previously described (25). 8 or 12 weeks after the initial immunization, mice were sacrificed and surgically resected spleens and kidneys were collected for further cellular function and tissue histology analysis.
Real-time PCR Analysis
Total RNA was extracted from peripheral blood mononuclear cell (PBMC), cultured cells, peritoneal macrophages, renal macrophages, or lymphocytes of tissues with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. The cDNA was synthesized with PrimeScript RT reagent kit (Takara Bio). The expression of the genes encoding DAI, TNF-α, IL-6, IL-10, and monocyte chemoattractant protein-1 (MCP-1) was quantified by real-time PCR using a Lightcycler 480 and SYBR Green system (Roche Diagnostic Systems) following the manufacturer's protocol (53).
ELISA and Nitrite Analysis
To assess protein levels of mouse TNF-α, IL-1β, IL-6, IL-10, IL-12, MCP-1 (eBioscience), and IFN-β (R&D Systems), ELISAs were performed with relative ELISA Kits according to the manufacturer's instructions. Nitrite derived from NO was determined with the Griess Reagent System (Promega) in macrophage-conditioned medium according to the manufacturer's instructions.
Western Blotting and Co-immunoprecipitation
Protein extraction from cultured cells or tissues, Western blot analysis, and co-immunoprecipitation were carried out as previously described (54, 55). Antibodies used here were obtained from Santa Cruz and included those against β-actin, GAPDH, DAI, FLAG, HA, and IgG-HRP.
Flow Cytometry Analysis and Cell Sorting
Renal macrophages, dendritic cells, T cells, and B cells were sorted from nephritic single-cell suspensions using a FACSAria (BD Biosciences) with FITC-labeled anti-F4/80 (eBioscience), phycoerythrin-labeled anti-CD11b (BD Biosciences), APC-labeled anti-CD11c (BD Biosciences), FITC-labeled anti-CD4, and phycoerythrin-labeled anti-CD19, respectively. The purity of cells was more than 90%, as determined by flow cytometry (FACSCalibur; BD Biosciences).
Luciferase Reporter Assay
Luciferase activity were performed as described previously (55). In brief, HEK293 cells were cotransfected with the mixture of 0.1 μg of NF-κB luciferase reporter plasmid (pNF-κB-Luc; Stratagene, La Jolla, CA) or 0.1 μg of IRF3 luciferase reporter plasmid (pIRF3-Luc), 0.1 μg of pRL-SV40 containing Renilla luciferase gene (pRL-SV40-Renilla-Luc; Promega, Madison, WI), with or without the indicated amounts of pcDNA3-DAI (pDAI) or pcDNA3 vector using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Total amounts of plasmid DNA were equalized with empty control vector. After 36 h, cells were left untreated or treated with the indicated amounts of UnALD-DNA or ALD-DNA for another 12 h. Luciferase activities were determined using the Luciferase Reporter Assay System according to the manufacturer's instructions (Promega, Madison, WI) (55). Luciferase activities were calculated as the percentage of the empty pcDNA3 vector and the values were normalized to the activity of Renilla luciferase.
Intracellular Calcium Measurement
Macrophages were incubated with 2 mm Fluo-4-acetoxymethyl ester (AM) (Molecular Probes, Eugene, OR) in Ca2+-free Hanks' balanced salt solution in the dark at room temperature for 30 min as previously described (56, 57). Cells were then washed twice and resuspended in Hanks' balanced salt solution buffer. Intracellular calcium was monitored at excitation of 496 nm and emission of 526 nm using SpectraMax Gemini XS dual-scanning microplate spectrofluorometer (Molecular Devices).
Anti-dsDNA Antibody and Proteinuria Examination
Anti-dsDNA antibodies in serum of mice were determined by ELISA as described previously (23). In brief, ELISA plates (Costar, Cambridge, MA) were pretreated with protamine sulfate (Sigma) and then coated with calf thymus dsDNA (Sigma). After incubation with mouse serum, the levels of anti-dsDNA Abs were detected with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Southern Biotechnology Associates, Birmingham, AL). Tetramethylbenzidine substrate was used to develop colors and absorbance at 450 nm was measured on a microplate reader (Bio-Tek ELX800, Bio-Tek Instruments, Winooski, VT). Proteinuria of the mice was measured with the BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's instructions.
Adoptive Transfer of Macrophages
Mice were immunized with ALD-DNA or PBS plus Freund's adjuvant as described in the “Generation of SLE murine model.” Macrophages stably expressing shDAI (shDAI macrophages) were retrieved and then injected intravenously into recipient mice (2.5 × 106 cells per mouse) at 0, 2, and 4 weeks after the initial immunization for a total of three times.
Pathological Analysis
Murine renal tissues were surgical resected and fixed in 4% paraformaldehyde (Sigma), processed, and embedded in paraffin. H&E staining of renal tissue sections were performed according to the manufacturer's instructions and assessed by a pathologist blinded to treatment group. The kidney score of glomerulonephritis was determined using the ISN/RPS2003 classification. Fluorescent staining of cryosections was used for IC deposition analysis in the glomeruli. Sections were fixed in acetone for 10 min and incubated with FITC-conjugated goat anti-mouse IgG (H+L chain specific) Abs (Sigma) for 30 min. Pictures were acquired with Nikon SCLIPSS TE2000-S microscope (Nikon, Melville, NY) equipped with ACT-1 software (Nikon). Original magnification was ×200.
Statistical Analysis
Experimental data were presented as mean ± S.E. of at least three independent replicates using GraphPad Prism 5 (GraphPad Software, La Jolla, CA) and assessing comparisons between different groups by the Student's t test, one-way analysis of variance. The statistical significance level was set as *, p < 0.05, **, p < 0.01, and ***, p < 0.001.
RESULTS
DAI Expression Is Significantly Increased in SLE Patients as well as in Lupus Mice
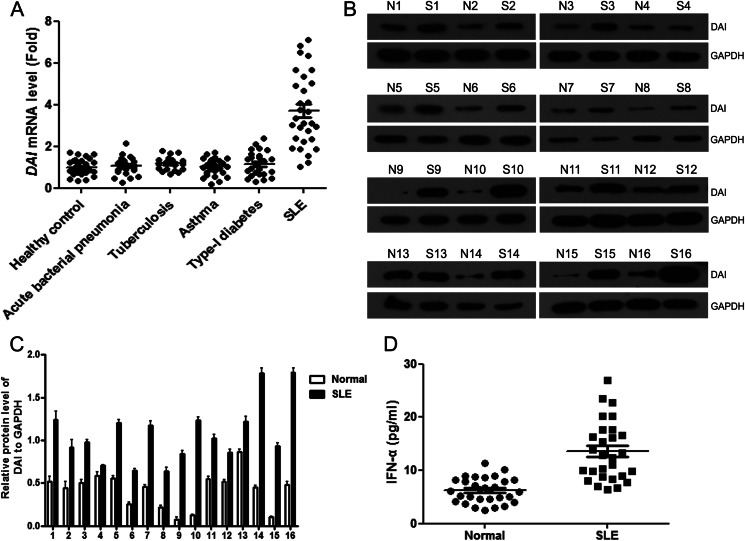
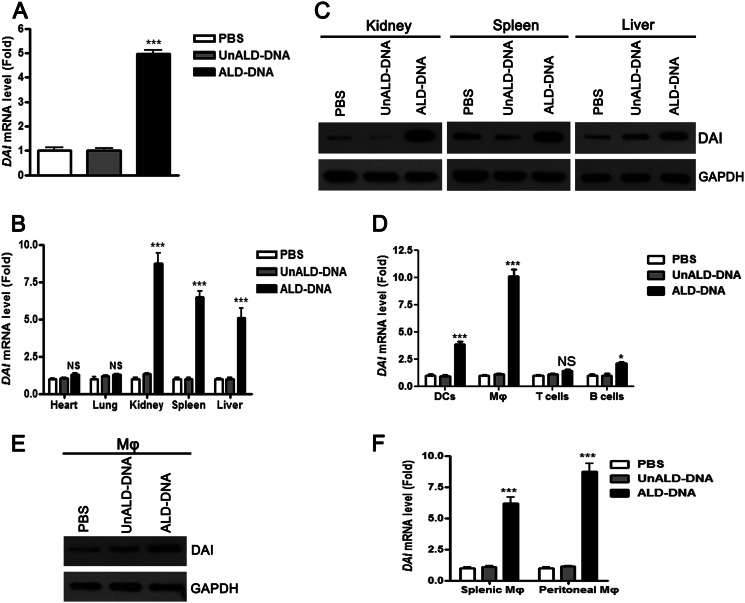
To investigate whether DAI was involved in SLE disease, real-time PCR analysis was performed to detect the levels of DAI in PBMCs of SLE patients and the matched controls. It was found that DAI expression was significantly increased in SLE patients as compared with healthy normal controls and patients with acute inflammation from infection (acute bacterial pneumonia), chronic inflammation from infection (tuberculosis), allergic reaction (asthma), or other chronic autoimmune disease (type I diabetes) (Fig. 1A). These increased mRNA levels of DAI in PBMCs of SLE patients were confirmed at the protein level by Western blot (Fig. 1, B and C). DAI is an IFN-inducible gene and increases type I IFN responses (40). Therefore, levels of IFN-α in serum of SLE patients were determined. It was found that levels of IFN-α were significantly increased in patient sera as compared with healthy normal controls (Fig. 1D). To further elucidate the expression of DAI in lupus mice, real-time PCR analysis was performed to detect the levels of DAI in PBMCs of ALD-DNA-immunized lupus mice. It was found that DAI expression was also notably increased in the PBMCs of SLE mice as compared with controls (Fig. 2A). To characterize the expression of DAI in lupus mice, lymphocytes from various tissues of lupus mice including hearts, lungs, kidneys, spleens, and livers were obtained to evaluate the levels of DAI. The expression of DAI only increased in lymphocytes obtained from kidneys, spleens, and livers but not in those from hearts and lungs, which were confirmed in protein levels (Fig. 2, B and C). As lupus nephritis was a major cause of morbidity in SLE patients, kidney tissues were selected to analyze the expression of DAI in dendritic cells, macrophages, T cells, and B cells. It was found that DAI expression was remarkably up-regulated in renal macrophages and dendritic cells, and highest in macrophages (Fig. 2D). Western blot analysis further confirmed the increased expression of DAI in renal macrophages (Fig. 2E). Moreover, real-time PCR analysis revealed that DAI expression was also significantly increased in splenic and peritoneal macrophages (Fig. 2F). These results suggest that DAI expression is notably increased in SLE patients and in lupus mice.
FIGURE 1.
DAI expression is notably increased in PBMCs of SLE patients. A, real-time PCR analysis of DAI mRNA levels in PBMCs from healthy normal controls (n = 30), patients with acute bacterial pneumonia (n = 25), tuberculosis (n = 20), asthma (n = 26), type-I diabetes (n = 25), and SLE (n = 30). B, Western blot analysis of DAI protein levels in PBMCs from SLE patients (S) and relative healthy normal controls (N). Data are representative of results obtained in three independent experiments, n = 16. C, graphical representations of band intensities in B. Expression of DAI was normalized to GAPDH expression. D, levels of IFN-α in serum of SLE patients and healthy normal controls were determined by ELISA. Data are mean ± S.E. of three independent experiments.
FIGURE 2.
DAI expression is remarkably up-regulated in ALD-DNA-immunized lupus mice. Six- to eight-week-old female BALB/c mice were immunized subcutaneously with PBS, UnALD-DNA (50 μg/mouse), or ALD-DNA (50 μg/mouse) for a total of 3 times in 4 weeks. 8 weeks later, mice were sacrificed and surgically resected hearts, lungs, kidneys, spleens, and kidneys were collected for further analysis. A, DAI mRNA levels in PBMCs from ALD-DNA-immunized lupus mice and controls were analyzed by real-time PCR. B, DAI mRNA levels in lymphocytes of hearts, lungs, kidneys, spleens, and livers from ALD-DNA-immunized lupus mice and controls were analyzed by real-time PCR. C, DAI protein levels in lymphocytes of kidneys, spleens, and livers from ALD-DNA-immunized lupus mice and controls were analyzed by Western blot. D, DAI mRNA levels in dendritic cells (DC), Mϕ (macrophages), T cells, and B cells of kidneys from ALD-DNA-immunized lupus mice and controls were analyzed by real-time PCR. E, DAI protein levels in renal macrophages from ALD-DNA-immunized lupus mice and controls were determined by Western blot. F, DAI mRNA levels in splenic Mϕ and peritoneal Mϕ from ALD-DNA-immunized lupus mice and controls were analyzed by real-time PCR.
DAI Expression Is Significantly Up-regulated in Macrophages upon ALD-DNA Stimulation
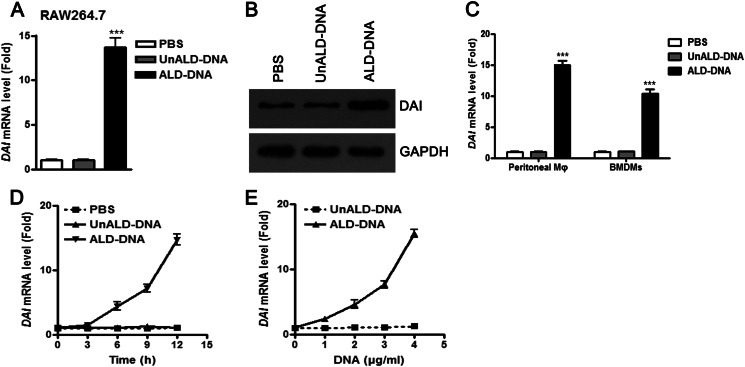
To determine whether ALD-DNA could up-regulate DAI expression, real-time PCR analysis was performed to detect the levels of DAI in macrophages stimulated with ALD-DNA. Expression of DAI was significantly enhanced in ALD-DNA-induced macrophages, which was further confirmed in protein levels by Western blot analysis (Fig. 3, A and B). Besides, ALD-DNA could also increase DAI expression in peritoneal macrophages and BMDMs (Fig. 3C). Moreover, real-time PCR analysis showed that ALD-DNA was able to increase DAI expression in macrophages in time- and dose-dependent manners (Fig. 3, D and E). These results demonstrate that ALD-DNA could increase DAI expression in macrophages.
FIGURE 3.
DAI expression is significantly up-regulated in macrophages upon ALD-DNA stimulation in vitro. A and B, RAW264.7 cells were stimulated with PBS, UnALD-DNA (4 μg/ml), or ALD-DNA (4 μg/ml). A, 12 h later, DAI mRNA levels in the macrophages were detected by real-time PCR. B, 24 h later, DAI protein levels in the macrophages were determined by Western blot. Data are representative of results obtained in three independent experiments. C, peritoneal macrophages and BMDMs were stimulated with PBS, UnALD-DNA (4 μg/ml), and ALD-DNA (4 μg/ml) for 12 h. DAI mRNA levels in the macrophages were detected by real-time PCR. D and E, DAI mRNA levels in RAW264.7 cells stimulated with ALD-DNA (4 μg/ml) for the indicated time (D) or with increasing amounts of ALD-DNA for 12 h (E) were determined by real-time PCR. Data are mean ± S.E. of three independent experiments. ***, p < 0.001.
Knockdown of DAI Blunts ALD-DNA-induced Macrophage Activation
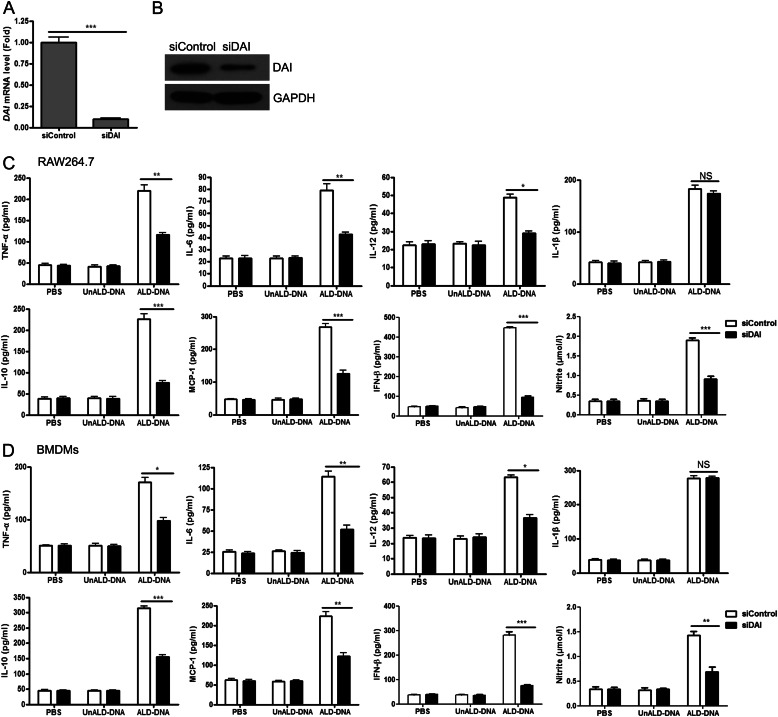
Our previous study revealed that ALD-DNA could induce macrophage activation and M2b polarization in vitro and in vivo (25, 26). To ascertain the role of DAI in ALD-DNA-induced macrophage activation, we examined the expression of activation markers in siDAI-treated macrophages. Real-time PCR and Western blot analysis first confirmed that siDAI transfection could notably inhibit DAI expression in macrophages in the presence of ALD-DNA stimulation (Fig. 4, A and B). ELISA further showed that increased levels of TNF-α, IL-6, IL-12, IL-10, MCP-1, IFN-β, and nitrite (the inducible NO synthase, iNOS) but not IL-1β in ALD-DNA-induced RAW264.7 cells was reversed through knockdown of endogenous DAI by siDAI (Fig. 4C). Similar effects on inflammatory cytokine expression were also obtained in primary mouse macrophages (Fig. 4D). These results suggest that DAI might contribute to ALD-DNA-induced macrophage activation and M2b polarization.
FIGURE 4.
Knockdown of DAI blocks ALD-DNA-induced macrophage activation in vitro. A and B, RAW264.7 cells were transfected with a plasmid vector encoding siControl or siDAI, stimulated with ALD-DNA (4 μg/ml), and then subjected to real-time PCR (A) and Western blot analysis (B) to evaluate the expression of DAI. Data in B are representative of results obtained in three independent experiments. C and D, RAW264.7 cells (C) and BMDMs (D) were transfected with siControl or siDAI, and treated with PBS, UnALD-DNA (4 μg/ml), or ALD-DNA (4 μg/ml) for 24 h. Levels of TNF-α, IL-6, IL-12, IL-1β, IL-10, MCP-1, IFN-β, and nitrite (the inducible NO synthase, iNOS) in the culture supernatants of macrophages were measured by ELISA. Data are mean ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, not significant.
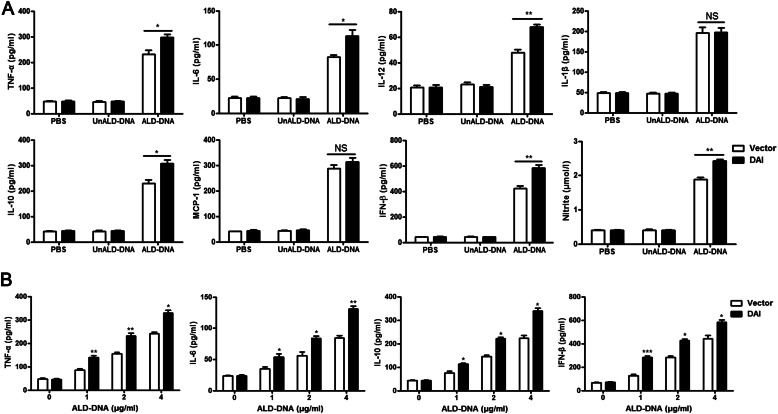
Overexpression of DAI Promotes ALD-DNA-induced Macrophage Activation
To further investigate the effect of DAI on ALD-DNA-induced macrophage activation and M2b polarization, stable DAI-expressing RAW264.7 (RAW-DAI) and control (RAW-Vector) cells were produced and stimulated with ALD-DNA. ELISA analysis for the production of inflammatory markers induced by ALD-DNA showed remarkably increased production of TNF-α, IL-6, IL-12, IL-10, IFN-β, and nitrite (the inducible NO synthase, iNOS) but no significant change of IL-1β and MCP-1 production in RAW-DAI cells compared with those in RAW-Vector cells (Fig. 5, A and B). Taken together, these results suggest that increasing DAI expression could enhance ALD-DNA-induced macrophage activation and M2b polarization.
FIGURE 5.
Overexpression of DAI promotes ALD-DNA-induced macrophage activation. RAW-Vector and RAW-DAI cells were stimulated with ALD-DNA, UnALD-DNA, or PBS. A, 24 h after stimulation, levels of TNF-α, IL-6, IL-12, IL-1β, IL-10, MCP-1, IFN-β, and nitrite (the inducible NO synthase, iNOS) in the culture supernatants were measured by ELISA. B, RAW-Vector and RAW-DAI cells were stimulated with increasing amounts of ALD-DNA for 24 h, and cytokine expression levels of TNF-α, IL-6, IL-10, and IFN-β in the culture supernatants were measured by ELISA. Data are mean ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, not significant.
ALD-DNA Induces Dimerization/Oligomerization of DAI and Consequently Activates DAI Signaling Pathway
In many cell types, DNA may serve as a scaffold to mediate the formation of a tandem array of DAI molecules, which then recruit and activate downstream signaling molecules, such as NF-κB and IRF3 (40, 58). To test this hypothesis, HA-tagged and FLAG-tagged DAI were cotransfected into HEK293 cells and stimulated with ALD-DNA. Cell extracts were then prepared and subjected to immunoprecipitation. As shown in Fig. 6A, co-immunoprecipitation was observed between the two distinctly labeled DAI molecules, reaching a maximum at 2 h after ALD-DNA stimulation. Moreover, ALD-DNA induced dimerization/oligomerization of DAI in a dose-dependent manner (Fig. 6B). To further reveal the molecular mechanisms involved in DAI-mediated macrophage activation induced by ALD-DNA, we determined the effect of DAI on the activity of NF-κB and IRF3 in ALD-DNA-treated cells. It was found that overexpression of DAI in ALD-DNA-treated cells could activate NF-κB and IRF3 in a dose-dependent manner (Fig. 6, C-F). However, knockdown of DAI could significantly inhibit the activity of NF-κB and IRF3 in ALD-DNA-induced macrophages (Fig. 6, G and H). These results indicate that ALD-DNA could induce dimerization/oligomerization of DAI and consequently activate DAI signaling pathways.
FIGURE 6.
ALD-DNA induces dimerization/oligomerization of DAI and consequently activates DAI signaling pathways. A and B, dimer/oligomer formation of DAI by ALD-DNA stimulation. HA-tagged DAI (HA-DAI) and FLAG-tagged DAI (FLAG-DAI) were transiently coexpressed in HEK293 cells. The cells were stimulated with ALD-DNA (4 μg/ml) for the indicated periods (A) or stimulated with increasing amounts of ALD-DNA for 2 h (B) and analyzed by immunoprecipitation (IP) with anti-HA antibody, followed by immunoblotting with anti-FLAG (upper) and anti-HA (lower) antibodies. C, HEK293 cells were infected with 0.1 μg of pNF-κB-Luc, plus increasing amounts of pDAI, then left stimulated with UnALD-DNA (1 μg/ml) or ALD-DNA (1 μg/ml). Luciferase activities were measured and normalized to Renilla luciferase activities. D, HEK293 cells were infected with 0.1 μg of pNF-κB-Luc, plus 0.1 μg of pDAI, then left stimulated with increasing amounts of UnALD-DNA or ALD-DNA. Luciferase activities were measured and normalized to Renilla luciferase activities. E, HEK293 cells were infected with 0.1 μg of pIRF3-Luc, plus increasing amounts of pDAI, then left stimulated with UnALD-DNA (1 μg/ml) or ALD-DNA (1 μg/ml). Luciferase activities were measured and normalized to Renilla luciferase activities. F, HEK293 cells were infected with 0.1 μg of pIRF3-Luc, plus 0.1 μg of pDAI, then left stimulated with increasing amounts of UnALD-DNA or ALD-DNA. Luciferase activities were measured and normalized to Renilla luciferase activities. G and H, macrophages treated with siDAI or siControl were infected with 0.1 μg of pNF-κB-Luc (G) or pIRF3-Luc (H), plus 0.1 μg of pDAI, then left stimulated with UnALD-DNA (1 μg/ml) or ALD-DNA (1 μg/ml). Luciferase activities were measured and normalized to Renilla luciferase activities. Data are representative of results obtained in three independent experiments.
Calcium Signaling Orchestrates DAI-mediated Macrophage Activation Induced by ALD-DNA
Emerging studies reveal that calcium signaling is involved in macrophage survival and activation (44, 59, 60). To determine whether calcium signaling was required for ALD-DNA-induced macrophage activation, RAW264.7 cells were treated with the cell-permeable cytosolic calcium chelator BAPTA-AM (blocking cytosolic calcium), EGTA (blocking entrance of extracellular calcium), CsA (disrupting mitochondrial calcium), or CGP37157 (an inhibitor of the mitochondrial sodium-calcium pump). It was found that the activity of NF-κB and IRF3 induced by ALD-DNA was blocked by BAPTA-AM, CsA, and CGP37157 treatments but not EGTA treatment, suggesting that ALD-DNA-induced activation of NF-κB and IRF3 involves cytosolic calcium activation and ALD-DNA likely acts on mitochondria calcium control (Fig. 7, A and B). Next studies were done to determine whether DAI acts on intracellular calcium to trigger ALD-DNA-induced NF-κB and IRF3 activation. It was found that BAPTA-AM, CsA, and CGP37157 but not EGTA inhibited DAI-mediated activation of NF-κB and IRF3 and cytokine secretion induced by ALD-DNA (Fig. 7, C-E). However, BAPTA-AM, CsA, and CGP37157 had no significant effect on NF-κB and IRF3 activity and cytokine secretion in siDAI-treated macrophages (Fig. 7, C-E). Furthermore, the activity of NF-κB and IRF3 and cytokine secretion in DAI target siRNA-expression macrophages could be recovered by reagents that increase the cytosolic calcium with calcium mobilizing agents valinomycin and thapsigargin (Fig. 7, F–H). Moreover, it was found that the levels of cytosolic calcium were regulated by DAI expression in response to ALD-DNA stimulation (Fig. 7I). These data indicate that DAI alters cytosolic calcium regulation in ALD-DNA-induced macrophages, leading to cytokine production in macrophages. As calcium/calmodulin-dependent protein kinase II (CaMKII) is the major downstream effector of calcium, we further determine the expression of CaMKII. Although the expression of the α isoform of CaMKII (CaMKII-α) remained almost unchanged (data not shown), a marked increase of CaMKII-α phosphorylation (Thr-286) in macrophages could be observed after stimulation with ALD-DNA (Fig. 7J). However, this increased effect could be abrogated by siDAI treatment (Fig. 7J). This raises the possibility that the calcium/CaMKII pathway may be potentially involved in DAI signaling in ALD-DNA-induced macrophages. A previous study (61) reported that DAI and TRIF recruited RIP1 in a pathway that activated NF-κB. We further determined the effect of calcium signaling on RIP1 expression. It was found that BAPTA-AM, CsA, and CGP37157 treatment could significantly inhibit RIP1 expression (Fig. 7K and data not shown). All these data indicate that ALD-DNA could induce DAI-mediated macrophage activation via the calcium/CaMKII pathway.
FIGURE 7.
Calcium signaling orchestrates DAI-mediated macrophage activation induced by ALD-DNA. A and B, HEK293 cells were infected with 0.1 μg of pNF-κB-Luc (A) or pIRF3-Luc (B), plus 0.1 μg of pDAI for 36 h. The cells were pretreated with EGTA (1 mm), BAPTA-AM (50 μm), CsA (3 μg/ml), or CGP37157 (10 μm) for 2 h, then left stimulated with UnALD-DNA (1 μg/ml) or ALD-DNA (1 μg/ml) for another 12 h. Luciferase activities were measured and normalized to Renilla luciferase activities. C and D, the siControl or siDAI-expressing macrophages were infected with 0.1 μg of pNF-κB-Luc (C) or pIRF3-Luc (D) for 36 h. The cells were pretreated with EGTA (1 mm), BAPTA-AM (50 μm), CsA (3 μg/ml), or CGP37157 (10 μm) for 2 h, then stimulated with ALD-DNA (1 μg/ml) for another 12 h. Luciferase activities were measured and normalized to Renilla luciferase activities. E, the siControl or siDAI-expressing macrophages were pretreated with EGTA (1 mm), BAPTA-AM (50 μm), CsA (3 μg/ml), or CGP37157 (10 μm) for 2 h, then stimulated with ALD-DNA (1 μg/ml) for another 24 h. ELISA analysis was performed to detect the levels of TNF-α, IL-6, and IFN-β. F and G, the siControl or siDAI-expressing macrophages were infected with 0.1 μg of pNF-κB-Luc (F) or pIRF3-Luc (G) for 36 h. The cells were pretreated with valinomycin (1 nm) or thapsigargin (20 nm) for 2 h, then stimulated with ALD-DNA (1 μg/ml) for another 12 h. Luciferase activities were measured and normalized to Renilla luciferase activities. H, the siControl or siDAI-expressing macrophages were pretreated with valinomycin (1 nm) or thapsigargin (20 nm) for 2 h, then stimulated with ALD-DNA (1 μg/ml) for another 24 h. ELISA analysis was performed to detect the levels of TNF-α, IL-6, and IFN-β. Data are mean ± S.E. of three independent experiments. **, p < 0.01; ***, p < 0.001; NS, not significant. I, the siControl or siDAI-expressing macrophages were stimulated with ALD-DNA (1 μg/ml) for 30 min. The levels of intracellular calcium were measured. J, the siControl or siDAI-expressing macrophages were stimulated with ALD-DNA (1 μg/ml) for 30 min. Western blot analysis was performed to determine the levels of CaMKII-α phosphorylation (T286). K, the RAW264.7 cells were pretreated with BAPTA-AM (50 μm) for 2 h, then left stimulated with ALD-DNA (1 μg/ml) for another 24 h. Western blot analysis was performed to determine the levels of RIP1. Data are representative of results obtained in three independent experiments.
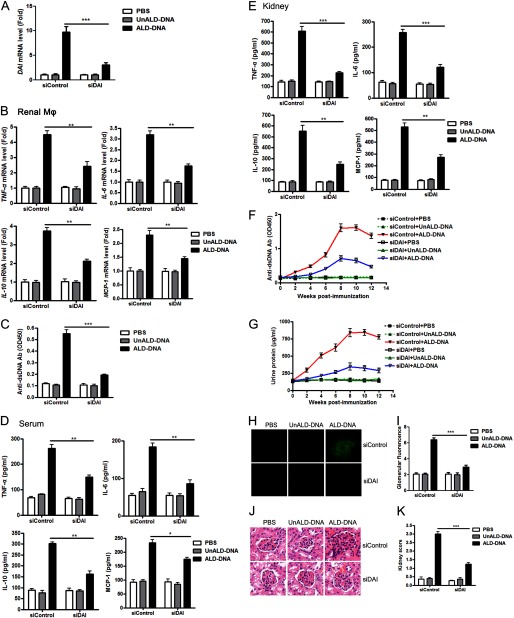
Decreased DAI Expression in Vivo Impairs Inflammatory Response of Macrophages against ALD-DNA
To further confirm the role of DAI in ALD-DNA-induced macrophage activation, the peritoneal macrophages obtained from siDAI-treated mice were stimulated with ALD-DNA. Real-time PCR analysis first confirmed that siDAI treatment could notably inhibit DAI expression in peritoneal macrophages in the presence of ALD-DNA stimulation (Fig. 8A). Consistent with the above mentioned results, ELISA analysis showed that increased expression of proinflammatory cytokines TNF-α, IL-6, and MCP-1 in ALD-DNA-induced macrophages was reversed by knockdown of endogenous DAI via DAI-specific siRNA (Fig. 8B). IL-10, an anti-inflammatory cytokine, has been reported to be increased in SLE patients and its serum level correlates with disease activity (62–66). So IL-10 levels were also determined by ELISA. It was found that increased IL-10 levels were reversed by siDAI treatment (Fig. 8B). Moreover, to clarify the effect of decreased DAI on the macrophage antigen-presenting ability, we performed ELISA analysis for anti-dsDNA Ab production by B cells cocultured with macrophages from siDAI-treated mice combined with ALD-DNA induction. Anti-dsDNA Ab production by B cells was severely decreased by coculturing with siDAI-treated peritoneal macrophages compared with siControl-treated cells (Fig. 8C). Taken together, these data indicate that DAI might play an important role in ALD-DNA-mediated macrophage activation in vivo.
FIGURE 8.
Knockdown of DAI by siRNA in vivo hampers the ALD-DNA-induced macrophage activation. In vivo transfection of peritoneal cells with siDAI was performed to block the DAI expression in peritoneal macrophages in mice. A and B, the peritoneal macrophages purified from siDAI or siControl-treated mice were stimulated with ALD-DNA (4 μg/ml) in vitro. A, 12 h later, the mRNA level of DAI was analyzed by real-time PCR. B, 24 h later, levels of TNF-α, IL-6, IL-10, and MCP-1 in the culture supernatants was analyzed by ELISA. Data are mean ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001. C, peritoneal macrophages purified from siDAI- or siControl-treated mice were cocultured with CD4+ T cells and CD19+ B cells isolated from the SLE mice, and stimulated with ALD-DNA for 6 days. Anti-dsDNA IgG levels in the culture supernatants were evaluated by ELISA. Data are mean ± S.E. of three independent experiments, n = 4. ***, p < 0.001.
Blockade of DAI in Vivo Ameliorates SLE Syndrome Accompanied with Dampened Macrophage Activation and Decreased Inflammatory Response in Lupus Mice
To test the hypothesis that inhibition of DAI could ameliorate the SLE syndrome in the lupus murine model by suppressing ALD-DNA-induced macrophage activation, we performed siDAI treatment in lupus mice. Decreased DAI mRNA level and lower mRNA levels of the activation markers were found in renal macrophages from siDAI-treated SLE mice versus those from siControl-treated SLE mice (Fig. 9, A and B). To clarify the effect of decreased DAI on macrophage antigen-presenting ability, we performed ELISA analysis for anti-dsDNA Ab production by B cells cocultured with macrophages from siDAI-treated lupus mice combined with ALD-DNA induction. Anti-dsDNA Ab production by B cells was severely reduced by coculturing with macrophages from siDAI-treated SLE mice as compared with those from siControl-treated SLE mice (Fig. 9C). ELISA analysis showed decreased levels of inflammatory cytokines in serum and kidney tissues of siDAI-treated lupus mice (Fig. 9, D and E). Furthermore, remarkably reduced anti-dsDNA Abs (Fig. 9F), decreased urine protein levels (Fig. 9G), reduced immune complex (IC) deposition (Fig. 9, H and I), alleviated renal pathology (Fig. 9J), and decreased kidney score (Fig. 9K) were found in siDAI-treated lupus mice (Fig. 9, F–K). These data demonstrate that siDAI treatment could alleviate nephritis in the lupus murine model accompanied with blunting macrophage activation and inhibiting inflammatory response.
FIGURE 9.
Knockdown of DAI in vivo alleviates SLE syndrome accompanied with blunted renal macrophage activation and decreased inflammatory response in lupus mice. ALD-DNA-immunized lupus mice were treated with siDAI or siControl. PBS- and UnALD-DNA-immunized mice were used as controls. A, real-time PCR analysis of the DAI mRNA level in renal macrophages purified from siDAI-treated lupus mice or siControl-treated lupus mice at week 12 after initial immunization. B, at week 12 after initial immunization, mRNA levels of TNF-α, IL-6, IL-10, and MCP-1 in the renal macrophages purified from the mice were evaluated by real-time PCR. C, at week 12 after the initial immunization, the purified renal macrophages from the siDAI-treated or siControl-treated lupus mice were cocultured with CD4+ T cells and CD19+ B cells from the SLE mice, then stimulated with ALD-DNA for 6 days. Levels of anti-dsDNA IgG in the culture supernatants were analyzed by ELISA. D, at week 12, levels of TNF-α, IL-6, IL-10, and MCP-1 in serum of the mice were determined by ELISA. E, at week 12, kidney tissue was collected and homogenized, the expressions of TNF-α, IL-6, IL-10, and MCP-1 were determined by ELISA. F, serum anti-dsDNA Ab level every 2 weeks were measured by ELISA. G, urine protein levels of mice were assessed by the BCA Protein Assay kit. Data in A-G are mean ± S.E. of three independent experiments, n = 8. H, 12 weeks after the initial immunization, glomerular immune deposition was detected by direct immunofluorescence for IgG in frozen kidney section of mice. Representative images (magnification ×200) of 10 mice are shown for each group. I, mean glomerular fluorescence intensity (arbitrary units) was determined for IgG in siDAI-treated lupus mice and siControl-treated lupus mice at week 12 after the initial immunization, n = 10. **, p < 0.01. J, 12 weeks after initial immunization, nephritic pathology was evaluated by H&E staining of renal tissues. Images (magnification ×200) are representative of at least 10 mice in each group. K, the kidney score was assessed using paraffin sections stained with H&E in J. ***, p < 0.001.
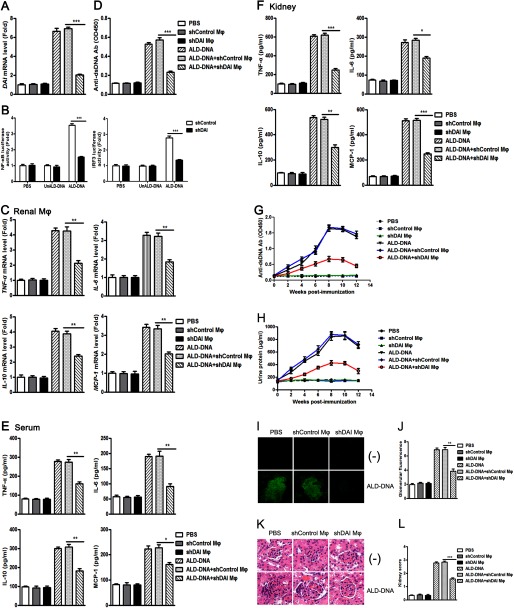
Knockdown of DAI in Macrophages Ameliorates SLE Syndrome via Suppressing Macrophage Activation and Inhibiting Inflammatory Response in Lupus Mice
To further test the hypothesis that inhibition of DAI could ameliorate the SLE syndrome in lupus mice by suppressing ALD-DNA-induced macrophage activation, we injected ex vivo programmed shDAI-macrophages (treated by shDAI) or shControl-macrophages (treated by shControl) into lupus mice. Decreased DAI mRNA level (Fig. 10A), reduced NF-κB and IRF3 activity (Fig. 10B), and lower mRNA levels of the activation markers (Fig. 10C) were found in renal macrophages from lupus mice adoptively transferred with shDAI-macrophages versus those from lupus mice adoptively transferred with shControl-macrophages. To clarify the effect of decreased DAI on the macrophage antigen-presenting ability, we performed ELISA analysis for anti-dsDNA Ab production by B cells cocultured with macrophages from lupus mice adoptively transferred with shDAI-macrophages combined with ALD-DNA induction. Anti-dsDNA Ab production by B cells was severely reduced by being cocultured with macrophages from lupus mice adoptively transferred with shDAI-macrophages (Fig. 10D). ELISA analysis showed decreased levels of inflammatory cytokines in serum and kidney tissues of lupus mice adoptively transferred with shDAI-macrophages (Fig. 10, E and F). Furthermore, remarkably reduced anti-dsDNA Abs (Fig. 10G), decreased urine protein levels (Fig. 10H), reduced immune complex deposition (Fig. 10, I and J), alleviated renal pathology (Fig. 10K), and decreased kidney score (Fig. 10L) were found in lupus mice adoptively transferred with shDAI-macrophages (Fig. 10, G–L). These data demonstrate that selective knockdown of DAI in macrophages could alleviate nephritis in lupus mice through blunting macrophage activation and inhibiting the inflammatory response.
FIGURE 10.
Adoptive transfer of macrophages stably expressing shDAI alleviates lupus nephritis. Macrophages stably expressing shDAI (shDAI Mϕ) were retrieved and 2.5 × 106 injected (i.v.) into mice at weeks 0, 2, and 4 after the initial ALD-DNA immunization. Mice injected with macrophages expressing shControl (shControl Mϕ) were used as control. A, real-time PCR analysis of the DAI mRNA level in renal macrophages purified from shDAI macrophage-treated lupus mice or shControl macrophage-treated lupus mice at week 12 after the initial immunization. B, the renal macrophages purified from lupus mice adoptively transferred with shDAI-macrophages were infected with 0.1 μg of pNF-κB-Luc or pIRF3-Luc for 36 h. The cells were stimulated with ALD-DNA (1 μg/ml), UnALD-DNA (1 μg/ml), or PBS for another 12 h. Luciferase activities were measured and normalized to Renilla luciferase activities. C, at week 12 after the initial immunization, mRNA levels of TNF-α, IL-6, IL-10, and MCP-1 in the renal macrophages purified from shDAI macrophage-treated lupus mice or shControl macrophage-treated lupus mice were evaluated by real-time PCR. D, at week 12 after the initial immunization, the purified renal macrophages from the shDAI macrophage-treated lupus mice or shControl macrophage-treated lupus mice were cocultured with CD4+ T cells and CD19+ B cells from the SLE mice, then stimulated with ALD-DNA for 6 days. Levels of anti-dsDNA IgG in the culture supernatants were analyzed by ELISA. E, at week 12, levels of TNF-α, IL-6, IL-10, and MCP-1 in serum of the mice were determined by ELISA. F, at week 12, kidney tissues were collected and homogenized, the expression of TNF-α, IL-6, IL-10, and MCP-1 were determined by ELISA. G, serum anti-dsDNA Ab level every 2 weeks were measured by ELISA. H, urine protein levels of mice were assessed by the BCA Protein Assay kit. Data in A-H are mean ± S.E. of three independent experiments, n = 8. I, 12 weeks after the initial immunization, glomerular immune deposition was detected by direct immunofluorescence for IgG in frozen kidney sections of mice. Representative images (magnification ×200) of 10 mice are shown for each group. J, mean glomerular fluorescence intensity (arbitrary units) was determined for IgG in shDAI macrophage-treated lupus mice or shControl macrophage-treated lupus mice at week 12 after the initial immunization, n = 10. **, p < 0.01. K, 12 weeks after the initial immunization, nephritic pathology was evaluated by H&E staining of renal tissues. Images (magnification ×200) are representative of at least 10 mice in each group. L, the kidney score was assessed using paraffin sections stained with H&E in J. ***, p < 0.001.
DISCUSSION
The data presented herein demonstrate for the first time, to our knowledge, that DAI was identified as an essential DNA sensor and regulator of ALD-DNA-induced macrophage aberrant activation and attendant functions in SLE disease. We found that DAI expression was predominantly increased in SLE patients as well as in ALD-DNA-immunized lupus mice. ALD-DNA led to dimerization/oligomerization of DAI and consequently activated DAI signaling pathways via calcium signaling, thus resulting in pathological macrophage activation and SLE disease. These results suggest that DAI might contribute to the ALD-DNA-induced pathogenic immune response mediated by macrophages, which could be useful for deciphering the mechanisms of ALD-DNA-induced macrophage activation and M2b polarization in the context of SLE disease and other self-DNA-mediated autoimmune disease.
It is well established that DAI, the first molecular to be reported that might function as a cytoplasmic DNA receptor, is involved in the regulation of innate immune response when exposed to DNA (40, 67). Self-DNA has long been regarded as the key nucleic antigen to trigger the inflammatory autoimmune response in SLE disease (68, 69). In our previous study, we demonstrated that immunizing syngenetic female BALB/c with self-ALD-DNA could induce macrophage activation and M2b polarization, which initiated the subsequent adaptive immune response, thus resulting in the onset and development of SLE disease, indicating that ALD-DNA could serve as a critical self-antigen to trigger autoimmune response in SLE disease (25). We hypothesize that DAI might be a putative DNA sensor and regulator that contributed to the ALD-DNA-induced immune response. We provided several lines of evidence that supported this notion. First, DAI expression was notably increased in ALD-DNA-induced lupus mice. Second, ALD-DNA induced the up-regulation of DAI expression in macrophages in vitro and in vivo. By knockdown or overexpression of DAI, we found that DAI was involved in ALD-DNA-induced macrophage activation and M2b polarization. Third, ALD-DNA stimulation led to dimerization/oligomerization of DAI and activation of DAI signaling pathways via calcium signaling. More importantly, knockdown of DAI attenuated the ALD-DNA-induced macrophage activation and alleviated SLE disease. These results demonstrated that the predominantly activated DAI signaling induced by ALD-DNA triggered and regulated the macrophage aberrant activation and M2b polarization in SLE disease. However, the concrete mechanisms involved in the binding of DAI to ALD-DNA remain to be revealed.
Although DAI was the first identified DNA sensor responsible for activation of the innate immune response (8), it was clear from the present results that there might be an additional cytosolic DNA sensor existing for ALD-DNA-induced macrophage activation, as evidenced by partially reduced inflammatory cytokine levels and no significant change of IL-1β production in siDAI-treated macrophages or macrophages obtained from siDAI-treated mice (data not shown). Our results demonstrated further the complex nature of the DNA-induced activation of innate immunity. Further studies on the identification of another ALD-DNA sensor(s) are required to be investigated. Moreover, a previous study revealed that DAI differentially contributed to the initiation of innate immune response, depending on the cell type and the microenvironment, as evidenced by the different roles of DAI in L929 cells, mouse embryonic fibroblasts, or human cells etc. (41, 70, 71). Here we found that DAI was involved in the ALD-DNA-induced immune response in RAW264.7 cells, BMDMs, and peritoneal macrophages. Whether DAI contributed to self-apoptotic DNA-mediated autoimmune response in human macrophages and in SLE patients remain to be revealed. Finally, how DAI and other DNA sensor(s) are involved in the ALD-DNA-induced pathologic immune response in SLE disease awaits further investigation. In this context, analysis of DAI knock-out mice, which were immunized with ALD-DNA, may be useful in helping to clarify the role of DAI in the ALD-DNA-induced pathogenic autoimmune response. But the present study demonstrated that other existing DNA sensor(s) contributed to ALD-DNA-mediated macrophage activation, analysis of the DAI knock-out mice could not elucidate the concrete role of DAI until the other involved DNA sensor(s) and its function in ALD-DNA-induced immune response were identified.
Calcium is a ubiquitous intracellular signal responsible for controlling numerous cellular processes, and found to be important for the functions of macrophages (59, 60). Recent studies revealed that calcium was involved in regulating diverse cellular responses via cross-talking with other signaling pathways including Toll-like receptor, Fc receptor, and complement receptor induced signaling (44, 45). In this study, we demonstrated that BAPTA-AM, CsA, and CGP37157 treatments, but not EGTA treatment, could inhibit NF-κB and IRF3 activity in ALD-DNA-stimulated macrophages, whereas the NF-κB and IRF3 activity in DAI-specific siRNA-expressing macrophages can be recovered by reagents that increase cytosolic calcium, indicating that the binding of DAI to ALD-DNA can trigger the elevation of intracellular calcium in macrophages, and in turn calcium promotes inflammatory cytokine production via activating NF-κB and IRF3. Therefore, positive cross-talk with the calcium signaling pathway is required for full activation of DAI-mediated responses in macrophages. However, the molecular mechanisms underlying the cross-talk between DAI and the calcium pathway in macrophage activation needs further investigation.
In conclusion, our study demonstrates that ALD-DNA induces the dimerization/oligomerization of DAI, and then activation of DAI signaling pathways via calcium signaling, thus resulting in macrophage aberrant activation and SLE disease. These findings imply the possible mechanisms involved in the recognition and regulation of ALD-DNA-induced pathological innate immune response mediated by macrophages in the context of SLE disease.
Acknowledgments
We thank Dr. Tadatsugu Taniguchi (University of Tokyo, Japan) and Dr. Takashi Fujita (Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan) for the gifts of plasmids.
This work was supported by grants of National Natural Science Foundation of China Grants 30890141, 31100629, 31270863, and 81273300, Major State Basic Research Development Program of China Grant 2013CB530501, Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT, IRT1075), A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), Shanghai STC Grant 10JC1401400, and Postdoctoral Science Foundation of China Grant 2012T50397.
- SLE
- systemic lupus erythematosus
- DAI
- DNA-dependent activator of IFN-regulatory factors
- IFN
- interferon
- ALD-DNA
- activated lymphocyte-derived DNA
- NF-κB
- nuclear factor κB
- IRF3
- interferon regulatory factor 3
- Mϕ
- macrophages
- shRNA
- short hairpin RNA
- shDAI
- DAI short hairpin RNA
- shControl
- control short hairpin RNA
- BMDM
- bone marrow-derived macrophage
- UnALD-DNA
- unactivated lymphocyte-derived DNA
- siDAI
- DAI-specific siRNA
- siControl
- control siRNA vector
- MCP-1
- monocyte chemoattractant protein-1
- BAPTA-AM
- O,O′-bis(2-aminophenyl)ethylene glycol-N,N,N′,N′-tetraacetic acid, tetraacetoxymethyl ester
- CsA
- cyclosporin A
- PBMC
- peripheral blood mononuclear cell
- CaMKII
- calcium/calmodulin-dependent protein kinase II.
REFERENCES
- 1. Okabe Y., Kawane K., Akira S., Taniguchi T., Nagata S. (2005) Toll-like receptor-independent gene induction program activated by mammalian DNA escaped from apoptotic DNA degradation. J. Exp. Med. 202, 1333–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Casciola-Rosen L. A., Anhalt G., Rosen A. (1994) Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J. Exp. Med. 179, 1317–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mevorach D., Zhou J. L., Song X., Elkon K. B. (1998) Systemic exposure to irradiated apoptotic cells induces autoantibody production. J. Exp. Med. 188, 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Savill J., Dransfield I., Gregory C., Haslett C. (2002) A blast from the past. Clearance of apoptotic cells regulates immune responses. Nat. Rev. Immunol. 2, 965–975 [DOI] [PubMed] [Google Scholar]
- 5. Wen Z. K., Xu W., Xu L., Cao Q. H., Wang Y., Chu Y. W., Xiong S. D. (2007) DNA hypomethylation is crucial for apoptotic DNA to induce systemic lupus erythematosus-like autoimmune disease in SLE-non-susceptible mice. Rheumatology 46, 1796–1803 [DOI] [PubMed] [Google Scholar]
- 6. Finke D., Randers K., Hoerster R., Hennig H., Zawatzky R., Marion T., Brockmann C., Klempt-Giessing K., Jacobsen K., Kirchner H., Goerg S. (2007) Elevated levels of endogenous apoptotic DNA and IFN-α in complement C4-deficient mice. Implications for induction of systemic lupus erythematosus. Eur. J. Immunol. 37, 1702–1709 [DOI] [PubMed] [Google Scholar]
- 7. Rahman A., Isenberg D. A. (2008) Systemic lupus erythematosus. N. Engl. J. Med. 358, 929–939 [DOI] [PubMed] [Google Scholar]
- 8. Hornung V., Latz E. (2010) Intracellular DNA recognition. Nat. Rev. Immunol. 10, 123–130 [DOI] [PubMed] [Google Scholar]
- 9. Terada K., Hirose S., Okuhara E. (1992) Production of antibodies specific for double stranded antigen DNA cloned from immune complexes in plasma of a SLE patient. Biochem. Biophys. Res. Commun. 183, 797–802 [DOI] [PubMed] [Google Scholar]
- 10. Vinuesa C. G., Goodnow C. C. (2002) Immunology. DNA drives autoimmunity. Nature 416, 595–598 [DOI] [PubMed] [Google Scholar]
- 11. Cohen P. L., Caricchio R., Abraham V., Camenisch T. D., Jennette J. C., Roubey R. A., Earp H. S., Matsushima G., Reap E. A. (2002) Delayed apoptotic cell clearance and lupus-like autoimmunity in mice lacking the c-mer membrane tyrosine kinase. J. Exp. Med. 196, 135–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ravichandran K. S., Lorenz U. (2007) Engulfment of apoptotic cells. Signals for a good meal. Nat. Rev. Immunol. 7, 964–974 [DOI] [PubMed] [Google Scholar]
- 13. Erwig L. P., Henson P. M. (2008) Clearance of apoptotic cells by phagocytes. Cell Death Differ. 15, 243–250 [DOI] [PubMed] [Google Scholar]
- 14. Chung E. Y., Kim S. J., Ma X. J. (2006) Regulation of cytokine production during phagocytosis of apoptotic cells. Cell Res. 16, 154–161 [DOI] [PubMed] [Google Scholar]
- 15. Triantafyllopoulou A., Franzke C. W., Seshan S. V., Perino G., Kalliolias G. D., Ramanujam M., van Rooijen N., Davidson A., Ivashkiv L. B. (2010) Proliferative lesions and metalloproteinase activity in murine lupus nephritis mediated by type I interferons and macrophages. Proc. Natl. Acad. Sci. U.S.A. 107, 3012–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roszer T., Menéndez-Gutiérrez M. P., Lefterova M. I., Alameda D., Nuñez V., Lazar M. A., Fischer T., Ricote M. (2011) Autoimmune kidney disease and impaired engulfment of apoptotic cells in mice with macrophage peroxisome proliferator-activated receptor γ or retinoid X receptor α deficiency. J. Immunol. 186, 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iwata Y., Boström E. A., Menke J., Rabacal W. A., Morel L., Wada T., Kelley V. R. (2012) Aberrant macrophages mediate defective kidney repair that triggers nephritis in lupus-susceptible mice. J. Immunol. 188, 4568–4580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bethunaickan R., Berthier C. C., Ramanujam M., Sahu R., Zhang W., Sun Y., Bottinger E. P., Ivashkiv L., Kretzler M., Davidson A. (2011) A unique hybrid renal mononuclear phagocyte activation phenotype in murine systemic lupus erythematosus nephritis. J. Immunol. 186, 4994–5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wada T., Yokoyama H., Su S. B., Mukaida N., Iwano M., Dohi K., Takahashi Y., Sasaki T., Furuichi K., Segawa C., Hisada Y., Ohta S., Takasawa K., Kobayashi K., Matsushima K. (1996) Monitoring urinary levels of monocyte chemotactic and activating factor reflects disease activity of lupus nephritis. Kidney Int. 49, 761–767 [DOI] [PubMed] [Google Scholar]
- 20. Schiffer L., Bethunaickan R., Ramanujam M., Huang W., Schiffer M., Tao H., Madaio M. P., Bottinger E. P., Davidson A. (2008) Activated renal macrophages are markers of disease onset and disease remission in lupus nephritis. J. Immunol. 180, 1938–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hill G. S., Delahousse M., Nochy D., Rémy P., Mignon F., Méry J. P., Bariéty J. (2001) Predictive power of the second renal biopsy in lupus nephritis. Significance of macrophages. Kidney Int. 59, 304–316 [DOI] [PubMed] [Google Scholar]
- 22. Sean Eardley K., Cockwell P. (2005) Macrophages and progressive tubulointerstitial disease. Kidney Int. 68, 437–455 [DOI] [PubMed] [Google Scholar]
- 23. Qiao B., Wu J., Chu Y. W., Wang Y., Wang D. P., Wu H. S., Xiong S. D. (2005) Induction of systemic lupus erythematosus-like syndrome in syngeneic mice by immunization with activated lymphocyte-derived DNA. Rheumatology 44, 1108–1114 [DOI] [PubMed] [Google Scholar]
- 24. Walport M. J. (2000) Lupus, DNase and defective disposal of cellular debris. Nat. Genet. 25, 135–136 [DOI] [PubMed] [Google Scholar]
- 25. Zhang W., Xu W., Xiong S. (2010) Blockade of Notch1 signaling alleviates murine lupus via blunting macrophage activation and M2b polarization. J. Immunol. 184, 6465–6478 [DOI] [PubMed] [Google Scholar]
- 26. Zhang W., Xu W., Xiong S. (2011) Macrophage differentiation and polarization via phosphatidylinositol 3-kinase/Akt-ERK signaling pathway conferred by serum amyloid P component. J. Immunol. 187, 1764–1777 [DOI] [PubMed] [Google Scholar]
- 27. Barbalat R., Ewald S. E., Mouchess M. L., Barton G. M. (2011) Nucleic Acid recognition by the innate immune system. Annu. Rev. Immunol. 29, 185–214 [DOI] [PubMed] [Google Scholar]
- 28. Marshak-Rothstein A. (2006) Toll-like receptors in systemic autoimmune disease. Nat. Rev. Immunol. 6, 823–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ishii K. J., Akira S. (2006) Innate immune recognition of, and regulation by, DNA. Trends Immunol. 27, 525–532 [DOI] [PubMed] [Google Scholar]
- 30. Keating S. E., Baran M., Bowie A. G. (2011) Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol. 32, 574–581 [DOI] [PubMed] [Google Scholar]
- 31. Chiu Y. H., Macmillan J. B., Chen Z. J. (2009) RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138, 576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stetson D. B., Ko J. S., Heidmann T., Medzhitov R. (2008) Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ablasser A., Bauernfeind F., Hartmann G., Latz E., Fitzgerald K. A., Hornung V. (2009) RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 10, 1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goubau D., Rehwinkel J., Reis e Sousa C. (2010) PYHIN proteins. Center stage in DNA sensing. Nat. Immunol. 11, 984–986 [DOI] [PubMed] [Google Scholar]
- 35. Unterholzner L., Keating S. E., Baran M., Horan K. A., Jensen S. B., Sharma S., Sirois C. M., Jin T., Latz E., Xiao T. S., Fitzgerald K. A., Paludan S. R., Bowie A. G. (2010) IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11, 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang P., An H., Liu X., Wen M., Zheng Y., Rui Y., Cao X. (2010) The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a β-catenin-dependent pathway. Nat. Immunol. 11, 487–494 [DOI] [PubMed] [Google Scholar]
- 37. Zhang Z., Yuan B., Bao M., Lu N., Kim T., Liu Y. J. (2011) The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 12, 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Papatriantafyllou M. (2011) Innate immunity. AT-rich DNA trapped in the cytoplasm. Nat. Rev. Immunol. 11, 569. [DOI] [PubMed] [Google Scholar]
- 39. Fernandes-Alnemri T., Yu J. W., Datta P., Wu J., Alnemri E. S. (2009) AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458, 509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Takaoka A., Wang Z., Choi M. K., Yanai H., Negishi H., Ban T., Lu Y., Miyagishi M., Kodama T., Honda K., Ohba Y., Taniguchi T. (2007) DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505 [DOI] [PubMed] [Google Scholar]
- 41. Wang Z., Choi M. K., Ban T., Yanai H., Negishi H., Lu Y., Tamura T., Takaoka A., Nishikura K., Taniguchi T. (2008) Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc. Natl. Acad. Sci. U.S.A. 105, 5477–5482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lladser A., Mougiakakos D., Tufvesson H., Ligtenberg M. A., Quest A. F., Kiessling R., Ljungberg K. (2011) DAI (DLM-1/ZBP1) as a genetic adjuvant for DNA vaccines that promotes effective antitumor CTL immunity. Mol. Ther. 19, 594–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Berridge M. J., Lipp P., Bootman M. D. (2000) The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 1, 11–21 [DOI] [PubMed] [Google Scholar]
- 44. Liu X., Yao M., Li N., Wang C., Zheng Y., Cao X. (2008) CaMKII promotes TLR-triggered proinflammatory cytokine and type I interferon production by directly binding and activating TAK1 and IRF3 in macrophages. Blood 112, 4961–4970 [DOI] [PubMed] [Google Scholar]
- 45. Sutterwala F. S., Noel G. J., Clynes R., Mosser D. M. (1997) Selective suppression of interleukin-12 induction after macrophage receptor ligation. J. Exp. Med. 185, 1977–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li K., Xu W., Guo Q., Jiang Z., Wang P., Yue Y., Xiong S. (2009) Differential macrophage polarization in male and female BALB/c mice infected with coxsackievirus B3 defines susceptibility to viral myocarditis. Circ. Res. 105, 353–364 [DOI] [PubMed] [Google Scholar]
- 47. Lumeng C. N., Bodzin J. L., Saltiel A. R. (2007) Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest. 117, 175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sato M., Suemori H., Hata N., Asagiri M., Ogasawara K., Nakao K., Nakaya T., Katsuki M., Noguchi S., Tanaka N., Taniguchi T. (2000) Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunity 13, 539–548 [DOI] [PubMed] [Google Scholar]
- 49. Takaoka A., Yanai H., Kondo S., Duncan G., Negishi H., Mizutani T., Kano S., Honda K., Ohba Y., Mak T. W., Taniguchi T. (2005) Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature 434, 243–249 [DOI] [PubMed] [Google Scholar]
- 50. Zhang W., Wu J., Qiao B., Xu W., Xiong S. (2011) Amelioration of lupus nephritis by serum amyloid P component gene therapy with distinct mechanisms varied from different stage of the disease. PLoS One 6, e22659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Amarzguioui M., Lundberg P., Cantin E., Hagstrom J., Behlke M. A., Rossi J. J. (2006) Rational design and in vitro and in vivo delivery of Dicer substrate siRNA. Nat. Protoc. 1, 508–517 [DOI] [PubMed] [Google Scholar]
- 52. Chen M., Zhang W., Xu W., Zhang F., Xiong S. (2011) Blockade of TLR9 signaling in B cells impaired anti-dsDNA antibody production in mice induced by activated syngenic lymphocyte-derived DNA immunization. Mol. Immunol. 48, 1532–1539 [DOI] [PubMed] [Google Scholar]
- 53. Xu J., Yun X., Jiang J., Wei Y., Wu Y., Zhang W., Liu Y., Wang W., Wen Y., Gu J. (2010) Hepatitis B virus X protein blunts senescence-like growth arrest of human hepatocellular carcinoma by reducing Notch1 cleavage. Hepatology 52, 142–154 [DOI] [PubMed] [Google Scholar]
- 54. Liu H., Xu J., Zhou L., Yun X., Chen L., Wang S., Sun L., Wen Y., Gu J. (2011) Hepatitis B virus large surface antigen promotes liver carcinogenesis by activating the Src/PI3K/Akt pathway. Cancer Res. 71, 7547–7557 [DOI] [PubMed] [Google Scholar]
- 55. Xu J., Liu H., Chen L., Wang S., Zhou L., Yun X., Sun L., Wen Y., Gu J. (2012) Hepatitis B virus X protein confers resistance of hepatoma cells to anoikis by up-regulating and activating p21-activated kinase 1. Gastroenterology 143, 199–212 [DOI] [PubMed] [Google Scholar]
- 56. Grynkiewicz G., Poenie M., Tsien R. Y. (1985) A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 260, 3440–3450 [PubMed] [Google Scholar]
- 57. Jiang Z., Yin X., Jiang Q. (2011) Natural forms of vitamin E and 13′-carboxychromanol, a long-chain vitamin E metabolite, inhibit leukotriene generation from stimulated neutrophils by blocking calcium influx and suppressing 5-lipoxygenase activity, respectively. J. Immunol. 186, 1173–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ishii K. J., Coban C., Kato H., Takahashi K., Torii Y., Takeshita F., Ludwig H., Sutter G., Suzuki K., Hemmi H., Sato S., Yamamoto M., Uematsu S., Kawai T., Takeuchi O., Akira S. (2006) A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat. Immunol. 7, 40–48 [DOI] [PubMed] [Google Scholar]
- 59. Tano J. Y., Vazquez G. (2011) Requirement for non-regulated, constitutive calcium influx in macrophage survival signaling. Biochem. Biophys. Res. Commun. 407, 432–437 [DOI] [PubMed] [Google Scholar]
- 60. Wang L., Tassiulas I., Park-Min K. H., Reid A. C., Gil-Henn H., Schlessinger J., Baron R., Zhang J. J., Ivashkiv L. B. (2008) “Tuning” of type I interferon-induced Jak-STAT1 signaling by calcium-dependent kinases in macrophages. Nat. Immunol. 9, 186–193 [DOI] [PubMed] [Google Scholar]
- 61. Kaiser W. J., Upton J. W., Mocarski E. S. (2008) Receptor-interacting protein homotypic interaction motif-dependent control of NF-κB activation via the DNA-dependent activator of IFN regulatory factors. J. Immunol. 181, 6427–6434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Llorente L., Zou W., Levy Y., Richaud-Patin Y., Wijdenes J., Alcocer-Varela J., Morel-Fourrier B., Brouet J. C., Alarcon-Segovia D., Galanaud P., Emilie D. (1995) Role of interleukin 10 in the B lymphocyte hyperactivity and autoantibody production of human systemic lupus erythematosus. J. Exp. Med. 181, 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Csiszár A., Nagy G., Gergely P., Pozsonyi T., Pócsik E. (2000) Increased interferon-γ (IFN-γ), IL-10 and decreased IL-4 mRNA expression in peripheral blood mononuclear cells (PBMC) from patients with systemic lupus erythematosus (SLE). Clin. Exp. Immunol. 122, 464–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hagiwara E., Gourley M. F., Lee S., Klinman D. K. (1996) Disease severity in patients with systemic lupus erythematosus correlates with an increased ratio of interleukin-10:interferon-γ-secreting cells in the peripheral blood. Arthritis Rheum. 39, 379–385 [DOI] [PubMed] [Google Scholar]
- 65. Houssiau F. A., Lefebvre C., Vanden Berghe M., Lambert M., Devogelaer J. P., Renauld J. C. (1995) Serum interleukin 10 titers in systemic lupus erythematosus reflect disease activity. Lupus 4, 393–395 [DOI] [PubMed] [Google Scholar]
- 66. Park Y. B., Lee S. K., Kim D. S., Lee J., Lee C. H., Song C. H. (1998) Elevated interleukin-10 levels correlated with disease activity in systemic lupus erythematosus. Clin. Exp. Rheumatol. 16, 283–288 [PubMed] [Google Scholar]
- 67. Yanai H., Savitsky D., Tamura T., Taniguchi T. (2009) Regulation of the cytosolic DNA-sensing system in innate immunity. A current view. Curr. Opin. Immunol. 21, 17–22 [DOI] [PubMed] [Google Scholar]
- 68. Rieber M., Urbina C., Rieber M. S. (1989) DNA on membrane receptors. A target for monoclonal anti-DNA antibody induced by a nucleoprotein shed in systemic lupus erythematosus. Biochem. Biophys. Res. Commun. 159, 1441–1447 [DOI] [PubMed] [Google Scholar]
- 69. Nagata S., Hanayama R., Kawane K. (2010) Autoimmunity and the clearance of dead cells. Cell 140, 619–630 [DOI] [PubMed] [Google Scholar]
- 70. Lippmann J., Rothenburg S., Deigendesch N., Eitel J., Meixenberger K., van Laak V., Slevogt H., N′Guessan P D., Hippenstiel S., Chakraborty T., Flieger A., Suttorp N., Opitz B. (2008) IFNβ responses induced by intracellular bacteria or cytosolic DNA in different human cells do not require ZBP1 (DLM-1/DAI). Cell Microbiol. 10, 2579–2588 [DOI] [PubMed] [Google Scholar]
- 71. Ishii K. J., Kawagoe T., Koyama S., Matsui K., Kumar H., Kawai T., Uematsu S., Takeuchi O., Takeshita F., Coban C., Akira S. (2008) TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature 451, 725–729 [DOI] [PubMed] [Google Scholar]