Background: Ketoconazole binds to and antagonizes pregnane X receptor (PXR) activation.
Results: Yeast high throughput screens of PXR mutants define a unique region for ketoconazole binding.
Conclusion: Ketoconazole genetically interacts with specific PXR surface residues.
Significance: A yeast-based genetic method to discover novel nuclear receptor interactions with ligands that associate with surface binding sites is suggested.
Keywords: Drug Development, Drug Metabolism, Mutagenesis, Nuclear Receptors, Protein Drug Interactions, Proteomics, Antagonist, Ketoconazole, Pregnane X Receptor, Yeast Two-hybrid
Abstract
The pregnane X receptor (PXR) is a master regulator of xenobiotic metabolism, and its activity is critical toward understanding the pathophysiology of several diseases, including inflammation, cancer, and steatosis. Previous studies have demonstrated that ketoconazole binds to ligand-activated PXR and antagonizes receptor control of gene expression. Structure-function as well as computational docking analysis suggested a putative binding region containing critical charge clamp residues Gln-272, and Phe-264 on the AF-2 surface of PXR. To define the antagonist binding surface(s) of PXR, we developed a novel assay to identify key amino acid residues on PXR based on a yeast two-hybrid screen that examined mutant forms of PXR. This screen identified multiple “gain-of-function” mutants that were “resistant” to the PXR antagonist effects of ketoconazole. We then compared our screen results identifying key PXR residues to those predicted by computational methods. Of 15 potential or putative binding residues based on docking, we identified three residues in the yeast screen that were then systematically verified to functionally interact with ketoconazole using mammalian assays. Among the residues confirmed by our study was Ser-208, which is on the opposite side of the protein from the AF-2 region critical for receptor regulation. The identification of new locations for antagonist binding on the surface or buried in PXR indicates novel aspects to the mechanism of receptor antagonism. These results significantly expand our understanding of antagonist binding sites on the surface of PXR and suggest new avenues to regulate this receptor for clinical applications.
Introduction
The pregnane X receptor (PXR),3 is a master regulator of xenobiotic metabolism. Because its original cloning and characterization (1, 2), PXR has been implicated in a host of pathophysiologic consequences in vivo (e.g. cancer drug resistance and potentiation of malignancy, clinically important adverse drug interactions, development of hypertriglyceridemia and nonalcoholic hepatic steatosis, inflammation and accentuation of drug toxicities) (for review, see Refs. 3 and 4). Thus, although there has been significant progress in the identification of agonist ligands for PXR and their structure activity relationships (5), there have been limited descriptions of drug-like PXR antagonists (6–15). Such small molecule antagonists need to be devoid of cellular toxicity, with limited off-target effects if they are to have potential for clinical application (3, 16).
Although natural ligands possessing PXR antagonist properties exist (e.g. sulforaphane, ketoconazole, ET743) (8, 10, 15), it remains unclear how and where the antagonists bind to and exert actions on PXR. The critical question is whether there are one or more “antagonist-binding pockets” capable of binding ligands outside the ligand binding pocket (11, 12). Takeshita et al. (17) first described the antagonist effect of ketoconazole on ligand-activated PXR. Although ketoconazole is a weak activator of PXR, in the presence of a strong agonist ligand (e.g. rifampicin) it also acts as a moderate antagonist (8). Our laboratory specifically demonstrated that in the human PXR scintillation proximity assay, the IC50 for ketoconazole was 74.4 μm (Kb ∼ 55.3 μm) (8). These values indicated that at biologically effective concentrations ranging from 6 to 25 μm, it was unlikely that ketoconazole could effectively compete with ligands (e.g. rifampicin) for binding to the ligand binding pocket of PXR. Hence, these results suggested that ketoconazole might act outside this pocket or in another domain or site on PXR. One unique site for interaction was the surface formed upon PXR activation (i.e. the AF2 interaction surface) that could directly or indirectly influence surface interactions with co-activators (e.g. SRC-1 (steroid receptor coactivator 1)). Preliminary docking studies suggested that the AF2 interaction surface could potentially bind to ketoconazole and other non-azole PXR antagonists, and a ligand based pharmacophore suggested these molecules may map to similar features (11, 12). These combined studies defined two pockets and potential AF2 surface residues that could bind ketoconazole.
Based on these observations and the fact that mutations outside the PXR ligand binding domain (LBD) can result in PXR unresponsive to ketoconazole antagonism (18), we developed a novel high throughput yeast based two-hybrid assay to study ketoconazole binding residues on PXR (19, 20). We specifically focused on this genetic approach as isolation and purification of full-length and/or mutant PXR has been extremely difficult and this limitation would hamper a structural approach to solving this question. Single mutations on PXR, especially on the AF2 surface defined by hydrophobic groove formed by α helices 3, 4, 5, and 12 directly above the ligand binding pocket on the surface of the receptor, invariably result in inactive mutants. Based on crystal structure considerations of stabilization of αAF (H12), we embarked on creating mutational libraries that would rescue the effect of single mutations in this region. Indeed, we have shown that rescue or gain-of-function second mutations can be made for the study of the ketoconazole binding surface on PXR (18). On this principle, we adopted and developed a high throughput yeast screen of PXR mutants interacting with its coactivator, SRC-1 (Supplemental Fig. S1). In this screen, which was adapted for a compound known to be cytotoxic to yeast, we were able to demonstrate key mutations on PXR that were enriched in clones unable to bind to ketoconazole. We conclude that the original residues are direct interaction residues with ketoconazole and are important for the inhibitory actions of the drug on PXR. Furthermore, we confirmed these findings in mammalian systems. Thus, we highlight a novel method toward detecting residues important for ligand action on nuclear receptor surfaces.
EXPERIMENTAL PROCEDURES
Cell Lines, Materials, and Reagents
Cell culture media and PCR reagents were purchased from Invitrogen unless indicated otherwise. The Saccharomyces cerevisiae strain was CTY10–5d (MATa ade2 trp1-901 leu2-3,112 his3-200 gal4−gal80− URA3::lexA-lacZ), which contains an integrated GAL1-lacZ gene with a lexA operator (21). Escherichia coli XL-1 Blue (Agilent Technologies, Santa Clara, CA) was used to amplify the mutant cDNA library. The cell line CV-1 was from the American Type Culture Collection (ATCC, Manassas, VA) and cultured according to ATCC recommendations. Charcoal adsorbed fetal bovine serum, DMSO, rifampicin, ketoconazole, and 5-bromo-4-chloro-3-indoyl β-d-galactosidase (X-gal) were from Sigma; [3H]ketoconazole 10 Ci/ml (ART0794) was obtained from American Radiolabeled Chemicals, Inc. (St Louis, MO). Antibodies for immunoblots were obtained as follows: PXR (H-160), LexA (D-19), SRC-1 (M-20), and Gal4AD (C-10) were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). E. coli BL21 was used to express glutathione S-transferase fusion protein (GST protein). pSG5-PXR, pGL3-cyp3A4-luc, and other plasmids used for PXR transactivation and mammalian two-hybrid assays have been described elsewhere (8, 9, 35).
Construction of Ketoconazole-resistant Yeast
ERG11 (sterol 14α-demethylase) is an established target for ketoconazole. Indeed, loss of ERG11 (by homologous recombination or development of mutations) results in nonviable yeast. These effects may be rescued by concomitant presence of suppressor mutation in ERG3 (sterol Δ5,6-desaturase) (22). To obtain viable yeast cells resistant to ketoconazole that did not carry transporter alterations as a cause of azole resistance (23–25), we developed novel strains of CTY10–5d yeast by first deleting ERG3 (erg3Δ) and then introducing an additional deletion in ERG11 (erg3Δ/erg11Δ) genes by homologous recombination (supplemental Fig. S2 and Experimental Procedures) (26).
Drug Sensitivity (Spot) Assay
Sensitivity to ketoconazole was tested by spotting serial dilutions of yeast culture onto plates containing different concentrations of ketoconazole (27, 28). The transformants were pre-grown in yeast extract/agar/peptone/dextrose (YAPD) broth to late-exponential phase and then re-inoculated into fresh medium to a cell concentration of 5 × 106 cells/ml. The optical density was measured at 600 nm (A600), and the number of cells/ml of culture was determined after the yeast was incubated for 6–7 h at 30 °C. Serial dilutions in sterile water containing 107, 106, 105, and 104 cells/ml were spotted (2 μl of each dilution per plate) onto YAPD solid plates containing either solvent or ketoconazole. The plates were incubated at 30 °C for 48 h before minimum inhibitory concentration (MIC) determination.
MIC Estimations
The MIC of ketoconazole was defined as the minimum inhibitory concentration of ketoconazole at which no growth was observed when 2 μl of the second dilution (i.e. 106 cells/ml) of the culture was spotted onto plates containing ketoconazole (28).
Ketoconazole Accumulation by S. cerevisiae
The net rate of ketoconazole accumulation by early exponential-phase S. cerevisiae cells was measured as described previously for fluconazole (29). Briefly, in this filter-based assay, yeast cells were grown to a density of 107 cells/ml, centrifuged, and resuspended in PBS. [3H]Ketoconazole and unlabeled ketoconazole were added to cells to give a final ketoconazole concentration and specific radioactivity of 100 nm and 7.4 GBq/ml, respectively. The cells were incubated at 30 °C with shaking at 170 rpm. At various times, triplicate samples of 3 ml each were removed and filtered in a Millipore vacuum manifold with Whatman GF/C filters (Sigma) that had been presoaked in 100 mm unlabeled (cold) ketoconazole. The filters were washed 4 times with 4 ml of PBS containing 100 mm unlabeled ketoconazole and were transferred to scintillation vials. The filters were dried at 37 °C for 60 min before scintillation fluid (10 ml) was added. The vials were capped and left at room temperature overnight before measuring radioactivity in a Packard liquid scintillation analyzer, Tri-CARB 2900TR, Packard (Meriden, CT). Notably, in this assay ketoconazole accumulation was shown to reach equilibrium by 60 min (data not shown), and cells were, therefore, analyzed before this time period. The details of the assay have been described previously (27). Controls included heat-killed yeast cells and blank filters to determine and subtract out nonspecific drug binding to cells and filters that did not exceed 10% of input radioactivity. Accumulated [3H]ketoconazole picomoles were calculated by dividing dpm by specific activity (1 μCi = 2.2 × 106 dpm).
Drug Binding and Cold Competition Assay
The ketoconazole binding experiments were performed as described previously (23). Recombinant GST fusion proteins were expressed in E. coli and purified using glutathione-Sepharose beads. Beads with GST proteins (10 μg) were incubated with 0.2 mm radiolabeled ketoconazole ([3H]ketoconazole, specific radioactivity 10 Ci/mmol) in drug binding buffer (10 mm K2HPO4, 10 mm KH2PO4, pH 7.0, 2 mm EDTA, 50 mm NaCl, 1 mm DTT, 0.5 mm CHAPS, 10% glycerol, and protease inhibitors) at 4 °C for 3 h. For cold competition, an unlabeled 1000-fold (1000×) excess of ketoconazole was added. The beads were then washed briefly in 500 μl of ice-cold drug binding buffer three times. The washed beads were resuspended in 100 μl of drug binding buffer and added to 5 ml of scintillation fluid (Fisher). After 10 min, scintillation counting was performed using a liquid scintillation analyzer, Tri-CARB 2900TR. All experiments were performed with at least three replicates.
Rh123 (Rhodamine 123) Efflux Assay
To determine broadly whether efflux pump activity was altered in genetically modified yeast, Rh123 retention was assayed. Briefly, ERG3/ERG11, erg3Δ, erg3Δ/erg11Δ yeast cells from YAPD cultures in the exponential growth phase (OD600 ∼ 0.5) were collected after centrifugation (3000 × g, 5 min at 20 °C) and washed 3 times with water. The cells were resuspended at a concentration of 0.5 × 106 to 1.0 × 107 cells/ml in PBS and incubated with 10 μm Rh123 at 37 °C for 30/60 min and centrifuged at 12,000 × g in a microcentrifuge. The resulting pellet was washed twice, resuspended in 200 μl of PBS, and transferred to a 96-well plate. The fluorescence of the reaction mixture was recorded with a spectrofluorimeter (excitation and emission wavelengths of 485 and 538 nm, respectively). To determine whether cells assayed for Rh123 retention assay were metabolically active, we measured their metabolic activities using the Live/Dead kit based on FUN-1 (2-chloro-4-[2,3-dihydro-3-methyl-{benzo-1,3-thiazol-2-yl}-methylidene]-1-phenylquinolinium iodide; Molecular Probes Inc., Eugene, OR) by following the manufacturer's instructions. FUN-1 is a membrane-permeant nucleic acid binding asymmetric halogenated cyanine dye that gives rise to cylindrical intravacuolar structures in metabolically active yeast cells. ERG3/ERG11, erg3Δ, erg3Δ/erg11Δ cells (107 cells/ml) were incubated with FUN-1 for 45 min at 37 °C, and fluorescence was estimated with a spectrofluorimeter (excitation and emission wavelengths, 485 and 585 nm, respectively). Rh123 retention by the cells was expressed as fluorescence accumulated per unit of metabolic activity (30, 31).
Yeast Growth Rate
Briefly, yeast cell (ERG3/ERG11, erg3Δ, erg3Δ/erg11Δ, blank) concentrations were measured by OD660 nm every hour over a period of 24 h (microplate reader, Biotech Synergy, San Diego, CA). The data were analyzed according to published methods with slight modifications (32, 33). Briefly, we used Prism 4.0a for Macintosh (2003) to analyze natural log transformed OD660-nm ratios (D660 at time t (OD) divided by initial D660 (ODi) values, denoted as ln(OD/ODi)) as a function of time (h). Subsequently, we performed a nonlinear curve fit using the pre-specified “sigmoidal dose-response (variable slope)” equation with initial x and y constraints equaling zero. The maximal growth rate (μm) is defined as the maximal slope of the Ln curve, and the cell doubling time at μm is defined by the equation ln 2/μm. λ, lag-time, was defined as the value corresponding to the intersection of the maximal slope of the ln curve with the x axis (SigmaPlot 9.0). The same experiment was repeated using the exact protocol specified in the prior paper in which yeast concentrations were measured by OD660 nm every 10 min over a period of 48 h (32). The OD plots show similar trends (supplemental Fig. S3).
Construction of PXR and SRC-1 Fusions in Yeast Vectors
Human PXR LBD (107–434 amino acids) was obtained by PCR amplification of pSG5-hPXR plasmid (8) using the primer pairs: forward, 5′-ACC GGATCCCGATGAAGAAGGAGATGATCATGTCC-3′, and reverse, 5′-AGAGTCGACTCAGCTACCTGTGATGCC-3′. The PCR product was directionally ligated to pSH2–1 at BamHI/SalI sites within the vector (pSH-PXR). Human SRC-1 full-length (1–1401 amino acids) was obtained by PCR amplification of pCMX-SRC1 plasmid (provided by Dr. Michael G. Rosenfeld, UCLA, Los Angeles, CA) using the primer pairs: forward, 5′-TATAGC GGCCGCATGAGTGGCCTCGGGGACAGTTCATCC-3′, and reverse, 5′-GCGGTCGACTTATTCAGTCAGTAGCTG-3′. The PCR product was directionally ligated to pGADNOT at NotΙ/SalΙ sites of vector (pGADNOT-SRC-1). A reverse two-hybrid pair was also constructed: pSH-SRC-1 and pGADNOT-PXR. Primers for PCR amplification of full-length SRC-1 included: forward, 5′-ATATGTCGACAAATGAGTGGCCTCGGGGACAGTTCATCC-3′, and reverse, 5′-GCGGTCGACTTATTCAGTCAGTAGCTG-3′. The PCR product was ligated to pSH2–1 within a SalΙ site present in the vector. Primers for PCR amplification of PXR LBD included: forward, 5′-AAGCGGCCGCATGAAGAAGGAGATGATCATGTCCGACGAG-3′, and reverse, 5′-AGAGTCGACTCAGCTACCTGTGATGCC-3′. The PCR product was directionally ligated to pGADNOT at NotΙ/SalΙ sites of vector. The authenticity of all derived plasmids were verified by sequencing.
Construction of PXR Mutant Library for Reverse Two-hybrid Screening
The Genemorph II random mutagenesis kit from Agilent Technologies was used to construct PXR mutants for the yeast two-hybrid assay. This kit is based on a novel error-prone PCR enzyme mixture (Mutazyme I polymerase + Taq polymerase) that ensures a “less” biased mutagenesis profile that balances mutations rates of A and Ts versus G and Cs. We used a titrated amount of template plasmid DNA (750–1000 ng) and 30 PCR reaction cycles to ∼0–4.5 mutations per kb of DNA. Primers for error-prone PCR included forward, 5′- ACCGGATCCCGATGAAGAAGGAGATGATCATGTCC-3′, and reverse, 5′-AGAGTCGACTCAGCTACCTGTGATGCC-3′. The PCR products were shotgun-cloned into BamHΙ and SalΙ sites within the pSH2–1 vector. The transformants were pooled as a library, and plasmids were purified using the MaxiPrep kit (Qiagen).
Plasmids Construction and Mutagenesis
pSG5-PXR, VP16-PXR, and GST-PXR LBD expression vector pGEX-6P1-PXR used in this paper were described previously (8). Site-directed mutagenesis of the hPXR constructs was performed using the QuikChange Site-Directed Mutagenesis kit (Agilent Technologies).
Yeast Two-hybrid Assays
Yeast Strain erg3Δ/erg11Δ was co-transformed with 1 μg of either pSH-PXR or pooled pSH-PXR mutant library and 1 μg of pGADNOT-SRC-1 or 1 μg each of pGADNOT-PXR and pSH-SRC-1, respectively. Transformed yeast was plated on Leu-/His-YPD selective agar plates with or without ketoconazole (25 μm). X-gal colony filter lift assays and β-galactosidase assays were performed (see below), and select colonies were picked for plasmid isolation using Zymoprep Yeast Plasmid Miniprep Kit II (Zymo Research, Irvine, CA). Plasmid DNAs were then amplified by PCR using primer pair forward, 5′-ACCGGATCCCGATGAAGAAGGAGATGATCATGTCC-3′, and reverse, 5′-AGAGTCGACTCAGCTACCTGTGATGCC-3′. Plasmid DNA and PCR products were sequenced to identify the mutations.
X-Gal Filter Assay and Liquid β-Gal Assay
X-gal assays were performed as previously described (9, 21). Briefly, appropriately sized circular nitrocellulose membranes (Schleicher & Schuell) were overlaid on yeast colonies within plates and allowed to soak completely. After complete soaking, the nitrocellulose was lifted off plates carefully to avoid smearing the colonies and placed in −80 °C for 15 min. The nitrocellulose was subsequently placed at room temperature for 10 min. The nitrocellulose was placed cell-side up on the two layers of Whatman No. 3MM filter paper that was soaked with ∼3 ml of X-gal solution (20 mg/ml) in a Petri dish. The liquid β-gal assay was performed from three independent transformants by using O-nitrophenyl-β-d-galactopyranoside; Sigma) as described by Vojtek, A. et al. (34).
Mammalian Transactivation and Two-hybrid Assays
Transactivation assays were performed in CV-1 cells. Cells were transfected with various reporters, the expression vectors, and SV40-Renilla plasmid was cotransfected as transfection control. For mammalian two-hybrid assays, the expression plasmids Gal4DB-SRC-1-RID (receptor interaction domain) or Gal4DB-SMRT-1-RID and VP16-hPXR were cotransfected with Tk-MH100 × 4-Luc reporter plasmid and SV40-Renilla control vector in CV-1 cells. Twenty-four hours after transfection, cells were treated with drugs and harvested 48 h. The luciferase activity was detected using the dual-luciferase reporter assay system and 20/20 Luminometer from Promega, Madison, WI (8, 35). Statistical analysis was performed with Prism (Version 4.0) using the nonparametric Student's t test. Nonlinear regression (curve fit) analysis was performed using the Sigmoidal dose-response (variable slope) equation.
Protein Pulldown Assays
The GST-SRC-1-RID fusion protein was expressed in E. coli BL21 cells and purified using glutathione-Sepharose (GE Healthcare) as described previously (8, 35). Verification of intact protein synthesis was obtained on 12% SDS-PAGE gels. Full-length human PXR in pSG5 vector was translated in vitro in the presence of [35S]methionine using the TNT-coupled reticulocyte lysate system (Promega) according to the manufacturer's instructions. Purified GST fusion protein (5 μg) was incubated with 5 μl of in vitro translated 35S-labeled protein with moderate shaking at 4 °C overnight in NETN (20 mmol/liter Tris, pH 8.0, 100 mmol/liter NaCl, 1.0 mmol/liter EDTA, 0.5% Nonidet P-40) and in the presence of 0.2% DMSO, 10 μm rifampicin, or 10 μm rifampicin plus 25 μm ketoconazole. GST was used as a negative control. The bound protein was washed 3 times with NETN, and the beads were collected by centrifugation at 3000 rpm for 5 min. The bound protein was eluted into SDS sample buffer and subjected to 12% SDS-PAGE, and the gel was exposed to phosphorimaging.
Immunoblot
Yeast cells were resuspended in distilled water containing 0.2 m NaOH and incubated for 5 min at room temperature, The cells were pelleted and resuspended in SDS sample buffer. After boiling and brief centrifugation, ∼6 μl of supernatant was typically loaded per lane of 10% SDS mini gel. At the completion of electrophoresis, proteins were transferred to nitrocellulose membranes (Protran BA79, Whatman). Western blotting was performed using anti-PXR antibody (H-160), anti-LexA antibody (d-19), anti-SRC-1 antibody (M-20), and anti-Gal4-AD antibody (C-10) from Santa Cruz Biotechnology.
Docking
The 1NRL structure was downloaded from the Protein Data Bank, and protein was prepared using the “prepare protein” protocol in Discovery Studio 3.5 (Accelrys, San Diego, CA) for rigid docking as described previously (11, 12). Ketoconazole was prepared using the “prepare ligands” protocol. A binding site sphere of 14 Å diameter was created using Ser-208 as the center, and then ketoconazole was docked into the 1NRL structure using LibDock (36) with Fast conformation generation, energy threshold 20 kcal/mol, steepest descent minimization, CHARMm forcefield. Ligand conformations were then manually assessed where the higher the LigDockScore, the higher the predicted affinity for the protein. Two-dimensional interaction maps were generated to show proposed ligand-protein interactions.
RESULTS
The erg3Δ/erg11Δ Yeast Strain Is Resistant to Ketoconazole
The growth characteristics of ERG3/ERG11, erg3Δ, and erg3Δ/erg11Δ indicate that all these cell types have nearly similar growth characteristics (cell doubling time ∼2.7, 2.8, and 2.4 h, lag time (λ) ∼1.4, 2.1, and 2.6 h, respectively) (supplemental Fig. S3). erg3Δ yeast cells are more sensitive to the cytotoxic effects of ketoconazole than the ERG3/ERG11, yet erg3Δ/erg11Δ remains resistant to ketoconazole at concentrations exceeding 41 μm (Fig. 1A). ERG3/ERG11 is sensitive to ketoconazole (MIC ∼9 μm); however, erg3Δ yeast are more sensitive to ketoconazole (MIC ∼6 μm), whereas erg3Δ/erg11Δ yeast (MIC ∼>41 μm, data not shown) are relatively more resistant to ketoconazole when compared with ERG3/ERG11 yeast (-fold resistant ∼>5). Because the MDR (multidrug resistance) efflux pump, Pdr5p, and other transporters are known to mediate azole resistance in yeast (24, 25, 28), we used Rh123 as a known substrate of Pdr5p (38, 39) to determine energy-dependent transport differences between ERG3/ERG11, erg3Δ, and erg3Δ/erg11Δ yeast strains. Our results indicated that at OD660 ∼ 0.5, the retention of Rh123 by metabolically active yeast was significantly higher at 30 min for the erg3Δ/erg11Δ compared with either ERG3/ERG11 or erg3Δ strain; however, this difference was not observed at 60 min (Fig. 1B). Next, we tested whether there were differences in the net accumulation (uptake) of [3H]ketoconazole (with cells analyzed at 30 and 60 min) in our yeast strains. At 30 and 60 min, the amount of [3H]ketoconazole retained in erg3Δ/erg11Δ was higher than ERG3/ERG11 yeast (Fig. 1C). These studies indicate that the mutant yeast strain is amenable for yeast two-hybrid analysis using ketoconazole.
FIGURE 1.
erg3Δ/erg11Δ yeast strain is resistant to ketoconazole. A, serial dilution (cells/ml) spotting of ERG3/ERG11, erg3Δ, or erg3Δ/erg11Δ yeast on YAPD solid plates containing indicated concentration (μm) of ketoconazole is shown. B, Rh123 retention was measured at 30 min and 60 min and expressed as fluorescence accumulated per unit of metabolic (live cell) activity. C, ketoconazole accumulation in yeast was measured using [3H]ketoconazole-specific activity detected at 30 and 60 min. Specific activity was converted to pmol and normalized to mg dry filter weight as previously published (28). Drug vehicle (control), 0.2% DMSO. Histograms represent the mean ± S.D. of three independent experiments each performed in triplicate. Rh, rhodamine.
Ketoconazole Disrupts Wild-type But Not Mutant PXR Association with Coactivator, SRC-1
To determine whether the erg3Δ/erg11Δ was suitable for use in yeast two-hybrid screens, we performed an assay in this strain to see if we could detect a colorimetric readout of the association of PXR and steroid receptor coactivator-1 (SRC-1) as previously published using the ERG3/ERG11 strain (9). Because yeast has significant sterol production, it has previously been shown that lacZ expression in yeast can be induced without the need for additional exogenous ligand (9, 40). We found that lacZ expression (blue colonies) was also induced in the erg3Δ/erg11Δ yeast strain transformed with PXR and SRC-1; however, there was no induction of LacZ expression (white colonies) in yeast transformed with empty vectors, PXR, or SRC-1 individually (Fig. 2A). From our previous work with ketoconazole and PXR antagonism in mammalian cells, in the presence of a strong PXR agonist ligand we observed PXR antagonism at ketoconazole concentrations of 10 μm and above (8). Therefore, for subsequent studies in yeast, we arbitrarily chose a concentration 2.5× the minimal concentration required to antagonize ligand-activated PXR. To determine whether ketoconazole (25 μm) disrupted PXR and SRC-1 interaction in erg3Δ/erg11Δ yeast, replica plates containing ketoconazole were soaked with nitrocellulose, and X-gal filter lift and β-galactosidase liquid assays were performed. We show that ketoconazole disrupts PXR and SRC-1 interactions in yeast as all the colonies from the replica filter were now white (which was also shown by the significantly reduced β-galactosidase activity by liquid enzymatic assays) (Fig. 2B). We previously showed in mammalian assays that the PXR mutant (T248E/K277Q) can be activated by a strong ligand (e.g. rifampicin) but is immune to the antagonistic effects of ketoconazole (18). Similarly, when we engineered the PXR double mutant (T248E/K277Q) in the yeast plasmid and then performed yeast transformations with SRC-1, we were able to show that the colonies exposed to ketoconazole still retain LacZ expression (Fig. 2C). The transformed yeast strains have been shown to express PXR, LexA, and SRC-1 protein in the presence or absence of ketoconazole (Fig. 2D).
FIGURE 2.
Ketoconazole disrupts wild-type but not mutant PXR association with coactivator, SRC-1. A, erg3Δ/erg11Δ strain was transformed with the indicated plasmids, and plated colonies were subjected to X-gal lift assay (left panel) and β-gal liquid assay (right panel). Lane 1, pSH empty vector + pGADNOT empty vector; lane 2, pSH-PXR + pGADNOT empty vector; lane 3, pSH empty vector + pGADNOT-SRC-1; lane 4, pSH-PXR + pGADNOT-SRC-1; lane 5, pSH-INI-1 + pGADNOT-c-Myc (positive control). B, shown is a erg3Δ/erg11Δ colony replica in plates containing vehicle (0.2% DMSO; lane 1) or ketoconazole (25 μm; lane 2). An X-gal lift assay (left panel) and β-gal liquid assay (right panel) were then performed. C, erg3Δ/erg11Δ colony replica in plates containing vehicle (lane 1 and 3) or ketoconazole (25 μm; lanes 2 and 4) is shown. T248E/K277Q indicates specific PXR mutant. D, shown are immunoblots of specific proteins as indicated from yeast colonies (lanes 1 and 2, pSH-PXR + pGADNOT-SRC-1; lanes 3 and 4,pSH-PXR T248E/K277Q mutant + pGADNOT-SRC-1) randomly picked from plates containing vehicle (lanes 1 and 3) or ketoconazole (lanes 2 and 4). E, erg3Δ/erg11Δ strain was transformed with the indicated plasmids, and plated colonies were subjected to an X-gal lift assay (left panel) and β-gal liquid assay (right panel). Lane 1, pSH empty vector + pGADNOT empty vector; lane 2, pSH-SRC-1 + pGADNOT empty vector; lane 3, pSH empty vector + pGADNOT-PXR; lane 4, pSH-SRC-1 + pGADNOT-PXR; lane 5, pSH-INI-1 + pGADNOT-c-Myc (positive control). F, procedures were as in B and C. erg3Δ/erg11Δ colony replica in plates containing vehicle (lane 1) or ketoconazole (25 μm; lane 2) are shown. X-gal lift assay (left panel) and β-gal liquid assay (right panel) were then performed.
Because SRC-1 is a coactivator (and was cloned into the pGADNot vector), we wanted to test whether SRC-1 could activate lacZ expression when cloned into the pSH vector system and whether this would change the activation profile and/or affect the leakiness of the yeast two-hybrid assay. Using our redesigned plasmids we performed two-hybrid assays in erg3Δ/erg11Δ yeast. As before, we show that lacZ expression (blue colonies) was also induced in erg3Δ/erg11Δ yeast strain transformed with PXR and SRC-1; however, there was no induction of LacZ expression (white colonies) in yeast transformed with empty vectors, PXR, or SRC-1 individually (Fig. 2E). In the presence of ketoconazole, the PXR and SRC-1 interactions in yeast are disrupted as all the colonies from the replica filter are now white. Similarly, when we engineer the PXR double mutant (T248E/K277Q) in this yeast plasmid and then perform erg3Δ/erg11Δ yeast transformations with SRC-1, we are able to show that the colonies exposed to ketoconazole still retain lacZ expression (Fig. 2F). These results suggest that for the purposes of yeast screening of PXR and SRC-1 interactions, there is no advantage for use of a specific cloning vector system for bait or prey. Thus, further yeast two-hybrid studies were all performed using the plasmids pSH-PXR and pGADNot-SRC-1.
High-throughput PXR Mutation Screen Reveals Specific Ketoconazole Binding Residues on PXR
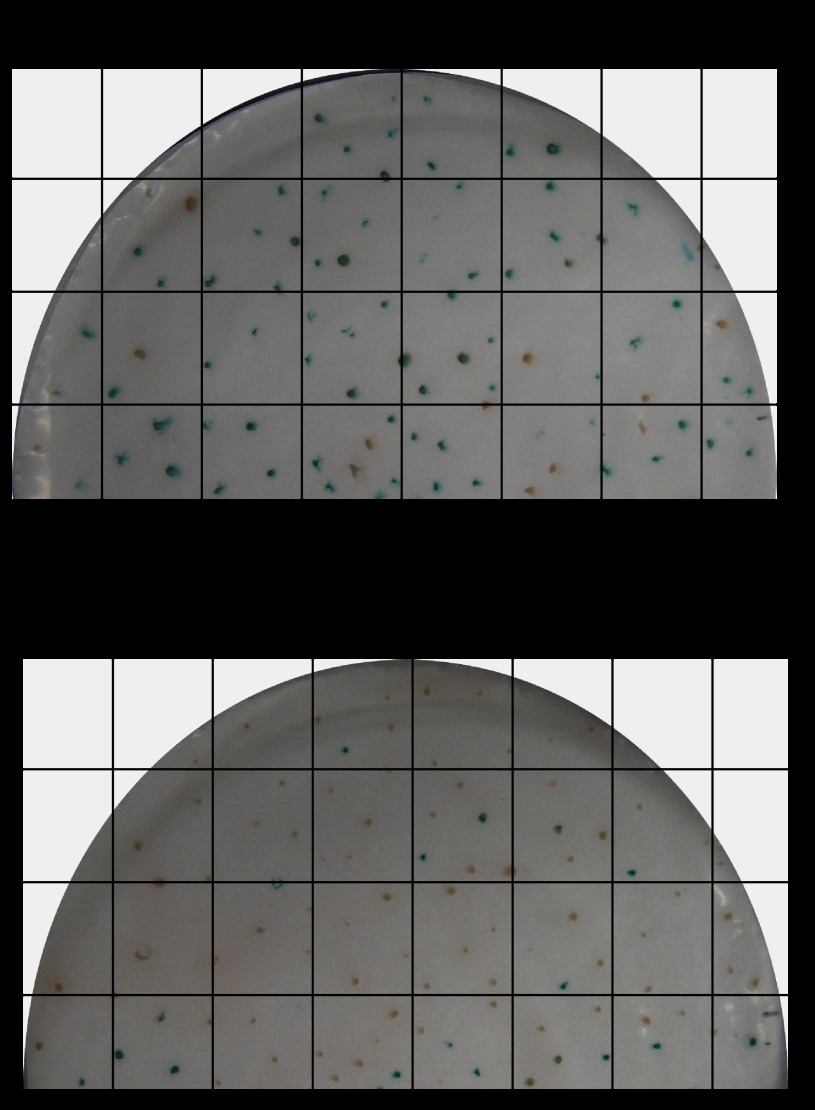
The yeast screen was performed using a pooled PXR mutant library generated by random mutagenesis. As represented in Fig. 3, the yeast transformants on the no-drug (−ketoconazole)-containing plates were on average represented as blue colonies (n = 81 blue/94 total screened; ∼86%) and white colonies (n = 13 white/94 total screened; ∼14%). On any given screen the transformants on ketoconazole containing (+ketoconazole) plates were represented as blue colonies (n = 16 blue/93 total screened; ∼17%) and white colonies (n = 77 white/93 total screened; ∼83%). It was estimated that ∼81% of blue colonies on ketoconazole minus plates transitioned to white colonies on ketoconazole plus plates. Thus, approximately ∼17% of blue colonies in the ketoconazole minus plate were still represented as blue colonies on ketoconazole plus plates (Fig. 3). These results indicate that the mutator frequency was ideal for generation of sufficient numbers of PXR mutants that could be activated under basal conditions. Furthermore, the presence of ∼17% ketoconazole minus blue colonies on ketoconazole plus plates suggests that it would be feasible to assess for ketoconazole binding residues on PXR.
FIGURE 3.
High-throughput PXR mutation yeast screen. pSH-PXR mutant cDNA library and pGADNOT-SRC-1 co-transformed into erg3Δ/erg11Δ strain were replica plated in vehicle (0.2% DMSO) (A)- or ketoconazole (25 μm) (B)-containing plates. An X-gal assay was then performed. The grid shows the method used for colony counts and sampling of the corresponding blue/white colonies in the two plates.
Next, we performed 21 yeast transformations and screens, and with each round of yeast transformations and plating we picked (n = 10–25 colonies: 10–25 blue, 10–25 white) from the ketoconazole plus plates for plasmid isolation, PCR amplification, and sequencing. In all, we picked (n = 284 colonies: 108 blue, 176 white) from the ketoconazole plus plates for plasmid isolation, amplification, and sequencing. Note that for all plasmid amplification steps, we had a control plasmid of known sequence (pSG5-PXR) that was also amplified to detect whether our amplification step caused additional mutations. None were detected (data not shown). The following mutations were observed as they occurred two or more times on the corresponding blue colony screen: S208W (∼36%), Q272H (∼16%), F264T (∼30%), and F264W (∼18%) (Table 1). There were several mutations that were detected once in the blue colonies; however, on closer inspection when the yeast assays were repeated with measurements using the liquid β-gal assay, these all were indeed white colonies (data not shown). Additionally, several weakly positive blue colonies in the ketoconazole plus plates revealed mutations that were then studied in mammalian systems (transcription and mammalian two-hybrid assays) and we found to still be inhibited by ketoconazole (Table 2). The following mutations were observed as they occurred two or more times on the white colony screen (blue in ketoconazole minus and white in ketoconazole plus plates): E270W (8.7%), E282Q (2.4%), K259E (2.4%), E270G (1.6%), and L424D (2.4%) (Table 2). Indeed, these mutants could still be inhibited by ketoconazole when studied in mammalian systems (transcription and mammalian two-hybrid assays). Finally, on a screen of 50 white colonies from both the ketoconazole minus and plus plates, the following mutations were observed as they occurred two or more times: R287K (10%), P268H (6%), F264T/L424D (6%). Indeed, these mutants were also found to be inactive to PXR ligand, rifampicin, using mammalian systems (transcription and mammalian two-hybrid assays) (data not shown). Interestingly, the double mutant F264T/L424D is a revertant mutation, as mutant F264T was immune to antagonist of the effects of ketoconazole, yet L424D influenced the antagonist binding properties of the F264T mutant, thus making the double mutant re-sensitized to ketoconazole antagonism. This implies that Leu-424 may also be remotely involved in the interaction with ketoconazole (see Fig. 5 and Table 2).
TABLE 1.
Characterization summary of PXR (blue colony) mutants in vitro and in vivo
Keto, ketoconazole; +, no interaction with ketoconazole; −, inhibitory interaction with ketoconazole; Binding, [3H]ketoconazole binds PXR; No binding, no evidence for [3H]ketoconazole binding PXR. β-Gal liquid assays indicated using the mean units.

* P > 0.1.
TABLE 2.
Characterization summary of PXR (white colony) mutants in vitro and in vivo
Keto, ketoconazole; +, no interaction with ketoconazole; −, inhibitory interaction with ketoconazole; Binding, [3H]ketoconazole binds PXR; No binding, no evidence for [3H]ketoconazole binding PXR. β-Gal liquid assay indicated using the mean units.

FIGURE 5.
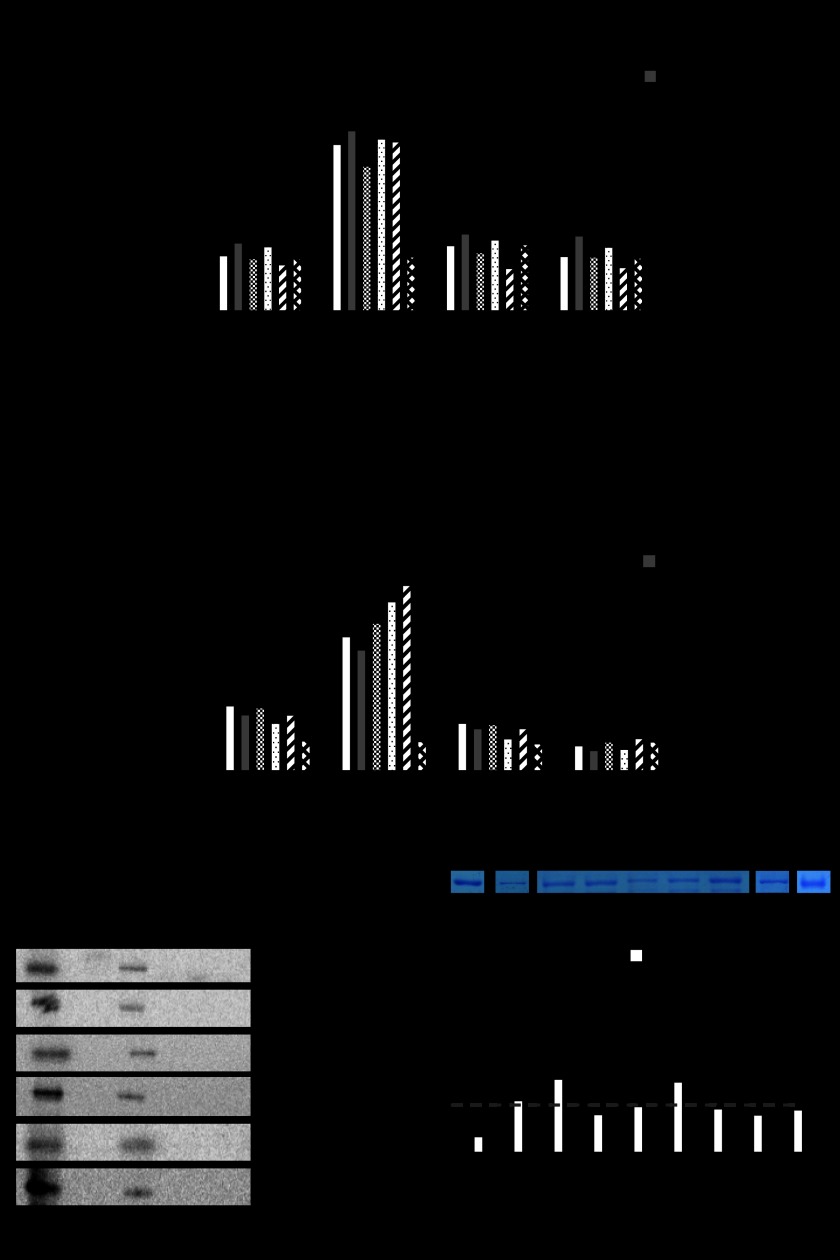
Mammalian characterization of PXR mutations identified on yeast screen (white colonies). A, shown is a mammalian hPXR (or hPXR mutant) transactivation assay (as illustrated) in CV-1 cells in the presence or absence of rifampicin, ketoconazole or both. B, shown is a mammalian two-hybrid assay (as illustrated) in CV-1 cells performed in the presence or absence of drugs as in A. A and B, histograms represent the mean ± S.D. of two independent assays each performed in triplicate. C, shown is a GST pulldown assay using 35S-labeled human PXR (or mutants as indicated) and GST or GST-SRC-1 (RID) in the presence of rifampicin (10 μm) or rifampicin (10 μm) and ketoconazole (25 μm) as indicated. The Input lane represents 10% of the protein in the binding assay. D, shown is ketoconazole binding with cold competition. Glutathione-Sepharose beads with GST fusion PXR (or mutants) protein fragments as indicated were used for direct binding assay. Coomassie Blue-stained pure GST fusion proteins are shown in the upper panel. Note: proteins are not in the same gel. Direct binding assay was performed using [3H]ketoconazole (black bars) and the cold competition assay (white bars) was performed using [3H]ketoconazole and unlabeled excess ketoconazole (1000× cold). Histograms represent the mean ± S.D. of at least two experiments, each performed in triplicate. Black arrow, background binding CPM; RLU, relative light units; hPXR, human PXR; SRC-1(RID), SRC-1 receptor interacting domain, UASG, upstream activating sequences for Gal4; VP16, VP16 activation domain; DB, DNA binding; Tk, thymidine kinase; MRP2, MRP2 promoter; aa, amino acids.
The mutations obtained from our yeast two-hybrid analysis were then reanalyzed in mammalian transcription, protein-protein interaction (GST pulldown) and protein binding assays. We were able to demonstrate that the recurring mutants found in the corresponding blue colonies within the ketoconazole plus plates, S208W, Q272H, F264T, and F264W, were all able to transactivate with a human PXR ligand, rifampicin, in both the mammalian transcription (Table 1; Fig. 4A) as well as the two-hybrid (Fig. 4B) assay. The same trends were observed in protein pulldown assays (Fig. 4C) and [3H]ketoconazole protein binding studies (Fig. 4D). The mutant Q272H was of particular interest, given that an analog of ketoconazole (FLB-1) lacking the imidazole group was able to antagonize PXR activation and, therefore, suggested specific interactions between the imidazole moiety and histidine (11, 41, 42). To clarify whether ketoconazole indeed interacts with residues Ser-208, Gln-272, and Phe-264 on PXR, we chose to perform [3H]ketoconazole protein binding studies. Indeed, as predicted by our protein pulldown studies, whereas ketoconazole binds to wild-type PXR protein, it is unable to bind to the PXR mutants (Fig. 4D). Residual [3H]ketoconazole binding CPM values, after cold competition, are likely due to nonspecific but high avidity binding to amino acid residues 107–204. We were also able to demonstrate that the following mutations observed on the white colony screen, E270W, E282Q, K259E, E270G, and L424D, were all able to be transactivated with human PXR ligand, rifampicin, but antagonized with ketoconazole in both the mammalian transcription (Fig. 5A) as well as the two-hybrid (Fig. 5B) assay. The same trends were observed in protein pulldown assays (Fig. 5C) and [3H]ketoconazole protein binding studies (Fig. 5D).
FIGURE 4.
Mammalian characterization of PXR mutations identified on yeast screen (blue colonies). A, a mammalian hPXR (or hPXR mutant) transactivation assay (as illustrated) in CV-1 cells in the presence or absence of rifampicin, ketoconazole, or both is shown. B, a mammalian two-hybrid assay (as illustrated) in CV-1 cells performed in the presence or absence of drugs was as in A. A and B, histograms represent the mean ± S.D. of two independent assays each performed in triplicate. C, shown is a GST pulldown assay using 35S-labeled human PXR (or mutants as indicated) and GST or GST-SRC-1 (RID) in the presence of rifampicin (10 μm) or rifampicin (10 μm) and ketoconazole (25 μm) as indicated. The Input lane represents 10% of the protein in the binding assay. D, ketoconazole binding with cold competition is shown. Glutathione-Sepharose beads with GST fusion PXR (or mutants) protein fragments as indicated were used for direct binding assay. Coomassie Blue-stained pure GST fusion proteins are shown in the upper panel. Note: proteins are not in the same gel. Direct binding assay was performed using [3H]ketoconazole (black bars), and cold competition assay (white bars) assay was performed using [3H]ketoconazole and unlabeled excess ketoconazole (1000× cold). Histograms represent the mean ± S.D. of at least two experiments each performed in triplicate. Black arrow, background binding CPM; RLU, relative light units; hPXR, human PXR; SRC-1(RID), SRC-1 receptor interacting domain, UASG, upstream activating sequences for Gal4; VP16, VP16 activation domain; DB, DNA binding; Tk, thymidine kinase; MRP2, MRP2 promoter; aa, amino acids.
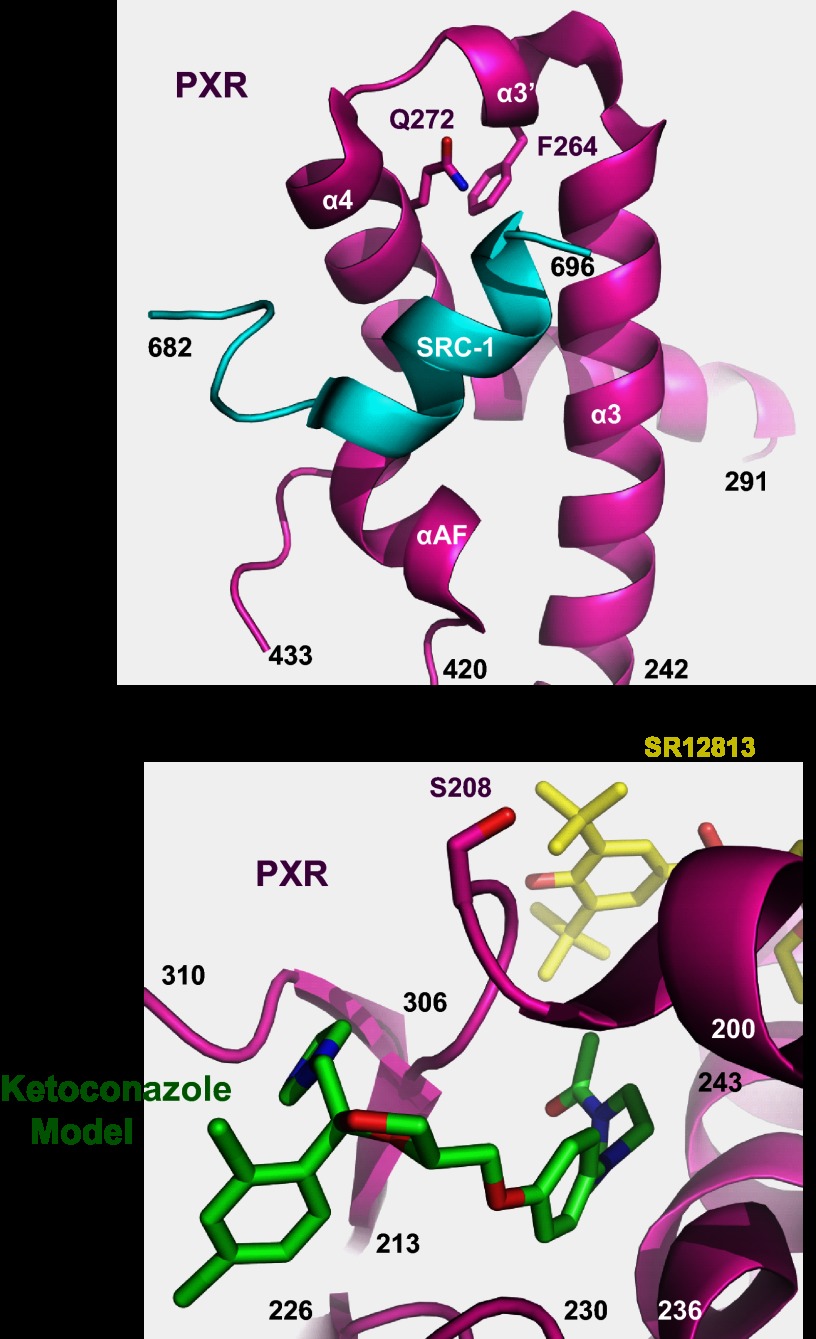
Visualization of Residues Identified in This Study and Ketoconazole Docked Near Ser-208
Fig. 6 shows residues in the 1NRL structure that were identified in this study by a yeast two-hybrid assay. Gln-272 and Phe-264 are close to the coactivator SRC-1 binding groove, and ketoconazole interacts with Gln-272 and Phe-264, which is thought to result in the disruption of PXR-SRC-1 interaction (Fig. 6A). Ser-208 is clearly shown to be distant from the AF-2 domain on the opposite side of the PXR structure (Fig. 6B). We, therefore, docked ketoconazole at this location to determine if this was a likely additional antagonist binding site. Fifteen (16) docked poses of ketoconazole were obtained; the best scoring (LibDock score 129.4) occupied a channel leading into the LBD (supplemental Fig. S5A) with the azole ring solvent exposed but directed toward Ser-208 (supplemental Fig. S5B). Eight poses had ketoconazole similarly positioned in this channel. The second best docked pose (LibDock score 110.16) presented ketoconazole on the surface of PXR occupying a cleft (supplemental Fig. S6A) also in close proximity to Ser-208 (supplemental Fig. S6B). In both binding poses Ser-208 is involved in van der Waals interactions with ketoconazole.
FIGURE 6.
Structural localization of ketoconazole interacting PXR residues identified by yeast two-hybrid assay. A, shown is the location of Phe-264 (F264) and Gln-272 (Q272) in the human PXR LBD crystal structure (magenta) relative to the experimentally resolved binding site of the LXXLL motif of human SRC-1 (cyan). B, shown is the location of Ser-208 in the human PXR LBD crystal structure (magenta) relative to the experimentally resolved binding site for the ligand SR-12813 (yellow). Additionally, the position of a computationally modeled ketoconazole molecule (green) observed near Ser-208 is also shown.
DISCUSSION
Nuclear receptor alternate-site modulators, specifically those defining novel pharmacophores, is the next leap forward in the discovery of newer high affinity ligands with receptor-specific action (43–49). Our method describes a novel technique to determine genetic interactions of protein residues with ligands/antagonists. This approach is likely to have its greatest utility for deciphering protein residue-ligand interactions for proteins/enzymes that are not amenable to classical structural biology techniques (e.g. crystal structure elucidation). When combined with docking models to predicted allosteric sites, this method promises to deliver additional evidence supporting those particular interactions. Indeed, our method provides the means to identify additional sites hitherto undiscovered by conventional approaches.
Drug resistance in yeast can be obtained by mutations that prevent the uptake of drug (permeability mutations), result in rapid export (export mutations), or affect the drug target gene or a gene in the downstream/upstream of the target gene to overcome the deleterious effect (pathway mutations). For the purpose of isolating mutants of PXR resistant to ketoconazole, we need yeast strains that allow the uptake of the drug to facilitate studies on protein-protein interactions and yet are viable in the presence of the drug. Therefore, permeability and export mutations are not desirable, but pathway mutations of yeast are. It has been previously reported that yeast strains that harbor mutations in ERG3 and ERG11 genes are viable and exhibit significant resistance to ketoconazole and its analogs (MIC, 10–100-fold greater than parental strain) (50, 51). Currently available yeast two-hybrid strains do not harbor these mutations. Therefore, we introduced mutations of ERG3 and ERG11 into yeast strain CTY5–10d. The mutant yeast performed robustly in the yeast two-hybrid assays and, when the results were coupled to docking studies, provided a meaningful interpretation of residues that would likely interact with ketoconazole. To our surprise, multiple additional modes of binding were determined presenting novel opportunities for antagonist discovery.
Using the yeast-two hybrid method we have identified important residues for ketoconazole interactions on PXR. Four of these residues had been predicted earlier, and docking had suggested ketoconazole interfered with co-activator binding (11). The current study identified Ser-208 as a new ketoconazole interacting amino acid that is distant from the AF-2 (Fig. 6B) and thus presents a second potential antagonist location deserving of further study and site-directed mutagenesis. Using docking, we propose two potential regions close to Ser-208 on the surface of the protein (supplemental Figs. S5 and S6). One site is a channel leading to the LBD in which ketoconazole can reach into the LBD and potentially interfere with agonist binding (supplemental Fig. S5). A second location is a surface cleft (supplemental Fig. S6), which is solvent-exposed and may interfere with protein-protein interactions, specifically homodimerization, as it is close to the homodimer interface (52). Preventing homodimerization is known to decrease receptor activity (53). Neither of these two locations near Ser-208 has been previously identified as important for PXR antagonism. Non-azole analogs of ketoconazole may also dock in these same locations (12). However, other smaller antagonists fit preferably in the AF-2 region. Identification of these new sites close to Ser-208, an amino acid identified as important for ketoconazole binding, suggests the further potential to design site-specific antagonists. Further analysis of whether interfering with co-activator binding, homodimerization, or agonist binding in the LBD is the preferred form of antagonism may need to proceed in parallel with the assessment of which may be more amenable to drug-like design. Also it should be noted that we used the 1NRL structure because it was originally co-crystallized with SRC-1 and was the subject of our previous docking efforts (11, 12). In these previous studies the agonist was removed, whereas in the current study it was present to prevent ketoconazole from binding in the LBD. It is likely that different crystal structures (with different agonists bound) may reveal other potential sites of antagonist binding as well impacting which site may be preferred.
One obstacle in this assay is the presence of false positive blue colonies, which may not always indicate ketoconazole-resistant PXR mutations. The lacZ expression could be regained in the presence of ketoconazole due to mutation in the PXR that renders the protein constitutively active or make it independent of SRC-1 interaction. Such mutants of PXR can be easily distinguished from the true ketoconazole-resistant mutants by testing the ability of the LexA-DB-PXR to self-activate lacZ expression, i.e. these mutants will yield blue colonies in the absence of GAL4AC-SRC-1. Such mutants could then be separated from the panel of true ketoconazole-resistant mutants. In our screen of 108 blue colonies on keto-plus plates, the recurring mutants S208W, Q272H, F264T, and F264W were reintroduced into yeast in the absence of GAL4AC-SRC-1. There was no evidence for self-activation of lacZ expression (data not shown). As a corollary, the mutants were also introduced into yeast using the GAL4AC plasmid in the absence of LexA-DB-SRC-1. These experiments also showed no evidence for self-activation of lacZ (data not shown). It is still conceivable that mutants that appear at a frequency below that of our threshold (i.e. occurrence once in the entire assay) may still provide important alternate residues that interact with ketoconazole. However, because our focus was on defining high probability binding sites as those residues defining the site that would be overrepresented in such a screen, we did not fully investigate mutants appearing is a very low frequency. It is conceivable that there could be some false positives due to constitutive activity of the mutation itself. Furthermore, feasible methods for objective screening based on colorimetric density evaluations might be required to optimize this assay for high throughput use in drug discovery (54).
Another approach to verification of true-positive ketoconazole PXR mutants would be to verify whether interactions of PXR with its co-repressor (i.e. SMRT) is lost due to the mutation (8, 35). Indeed, mammalian two-hybrid assays using PXR and SMRT plasmids showed significantly reduced activation of mutant PXR-SMRT interactions (supplemental Fig. S4).
In addition to isolation of ketoconazole-resistant mutants of PXR, this assay could be used to isolate intramolecular revertants of ketoconazole-resistant mutants. Intramolecular revertants are second site suppressors (a second mutation independent of the first keto-resistant mutation) of ketoconazole-resistant mutants that render the mutant PXR sensitive to ketoconazole (e.g. F264T/L424D). Such mutations are valuable in determining the exact residues and binding pockets of ketoconazole and in determining the mechanism.
Our approach provides a powerful new method for isolation of genetic interaction allosteric ligand-protein residues for proteins not amenable to conventional structural biology and/or proteomic approaches. The recent development of allosteric site modulators of nuclear receptors has paved the way for new, more potent and less toxic drugs that might enter the clinic in the future. For example, conventional anti-androgens targeting AR invariably fail in the clinic due to the development of resistance that are largely attributed to AR signaling (54, 55). AR has the unique property of binding large peptide motifs, thus, implicating utility for size-exclusion principles in drug discovery to selectively target AR or other NRs (56). Indeed, compounds (e.g. amphipathic benzene coactivator binding inhibitors) have been discovered with IC50 values as low as 1.9 μm that selectively target AR over ER (57). These compounds are active in LNCaP cells harboring the AR T877A mutation (rendering agonist activity rather than antagonist activity for hydroxyflutamide), which is present in ∼30% of patients with metastatic prostate cancer (58). More recently, novel synthetic compounds with peroxisome proliferator-activated receptor-γ agonist activity have been screened with a unique mode of action; that is, complete lack of classical transcriptional induction but with a site-selective block of Cdk5-mediated phosphorylation that renders peroxisome proliferator-activated receptor-γ active in adipocytes (44). One inhibitor (i.e. SR1664) site and tissue selective actions on peroxisome proliferator-activated receptor-γ results in potent anti-diabetic activity without causing fluid retention and weight that are typical side effects of conventional LBD targeting peroxisome proliferator-activated receptor-γ drugs (44). Similar efforts could be developed for PXR for which modulators of activity through post-translational modifications (e.g. phosphorylation, acetylation) have been shown to occur with discrete enzymes (37, 59, 60).
Acknowledgments
S. Ekins kindly acknowledges Accelrys for providing Discovery Studio. We acknowledge Drs. Sandhya Kortagere, Drexel University, Philadelphia, PA, Gregory Prelich, Albert Einstein College of Medicine, Bronx, NY, and Jay E. Wrobel, Fox Chase Chemical Diversity Center, Doylestown, PA for helpful discussions and insight.
This work was supported, in whole or in part, by National Institutes of Health Grants CA127231. This work was also supported by a Damon Runyon Foundation Clinical Investigator Award (CI 1502) (to S. M.).

This article contains supplemental Experimental Procedures and Figs. S1–S6.
- PXR
- pregnane X receptor
- SRC-1
- steroid receptor coactivator 1
- LBD
- ligand binding domain
- MIC
- minimum inhibitory concentration
- Rh123
- rhodamine 123
- AR
- androgen receptor
- NRs
- nuclear receptors
- ER
- estrogen receptor.
REFERENCES
- 1. Kliewer S. A., Moore J. T., Wade L., Staudinger J. L., Watson M. A., Jones S. A., McKee D. D., Oliver B. B., Willson T. M., Zetterström R. H., Perlmann T., Lehmann J. M. (1998) An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92, 73–82 [DOI] [PubMed] [Google Scholar]
- 2. Blumberg B., Sabbagh W., Jr., Juguilon H., Bolado J, Jr., van Meter C. M., Ong E. S., Evans R. M. (1998) SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 12, 3195–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Biswas A., Mani S., Redinbo M. R., Krasowski M. D., Li H., Ekins S. (2009) Elucidating the “Jekyll and Hyde” nature of PXR. The Case for Discovering Antagonists. Pharm. Res. 26, 1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pondugula S. R., Mani S. (2013) Pregnane xenobiotic receptor in cancer pathogenesis and therapeutic response. Cancer Lett. 328, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ekins S., Kortagere S., Iyer M., Reschly E. J., Lill M. A., Redinbo M. R., Krasowski M. D. (2009) Challenges predicting ligand-receptor interactions of promiscuous proteins. The nuclear receptor PXR. PLoS Comput. Biol. 5, e1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xue Y., Chao E., Zuercher W. J., Willson T. M., Collins J. L., Redinbo M. R. (2007) Crystal structure of the PXR-T1317 complex provides a scaffold to examine the potential for receptor antagonism. Bioorg. Med. Chem. 15, 2156–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carnahan V. E., Redinbo M. R. (2005) Structure and function of the human nuclear xenobiotic receptor PXR. Curr. Drug Metab. 6, 357–367 [DOI] [PubMed] [Google Scholar]
- 8. Huang H., Wang H., Sinz M., Zoeckler M., Staudinger J., Redinbo M. R., Teotico D. G., Locker J., Kalpana G. V., Mani S. (2007) Inhibition of drug metabolism by blocking the activation of nuclear receptors by ketoconazole. Oncogene 26, 258–268 [DOI] [PubMed] [Google Scholar]
- 9. Wang H., Li H., Moore L. B., Johnson M. D., Maglich J. M., Goodwin B., Ittoop O. R., Wisely B., Creech K., Parks D. J., Collins J. L., Willson T. M., Kalpana G. V., Venkatesh M., Xie W., Cho S. Y., Roboz J., Redinbo M., Moore J. T., Mani S. (2008) The phytoestrogen coumestrol is a naturally occurring antagonist of the human pregnane X receptor. Mol. Endocrinol. 22, 838–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Synold T. W., Dussault I., Forman B. M. (2001) The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat. Med. 7, 584–590 [DOI] [PubMed] [Google Scholar]
- 11. Ekins S., Chang C., Mani S., Krasowski M. D., Reschly E. J., Iyer M., Kholodovych V., Ai N., Welsh W. J., Sinz M., Swaan P. W., Patel R., Bachmann K. (2007) Human pregnane X receptor antagonists and agonists define molecular requirements for different binding sites. Mol. Pharmacol. 72, 592–603 [DOI] [PubMed] [Google Scholar]
- 12. Ekins S., Kholodovych V., Ai N., Sinz M., Gal J., Gera L., Welsh W. J., Bachmann K., Mani S. (2008) Computational discovery of novel low micromolar human pregnane x receptor antagonists. Mol. Pharmacol. 74, 662–672 [DOI] [PubMed] [Google Scholar]
- 13. Lei F. (2009) Studying the specificity of hPXR antagonists using a panel of cell-based assays. M.Sc. thesis, The University of Tennessee Health Science Center, TN [Google Scholar]
- 14. Healan-Greenberg C., Waring J. F., Kempf D. J., Blomme E. A., Tirona R. G., Kim R. B. (2008) A human immunodeficiency virus protease inhibitor is a novel functional inhibitor of human pregnane X receptor. Drug Metab. Dispos. 36, 500–507 [DOI] [PubMed] [Google Scholar]
- 15. Zhou C., Poulton E. J., Grün F., Bammler T. K., Blumberg B., Thummel K. E., Eaton D. L. (2007) The dietary isothiocyanate sulforaphane is an antagonist of the human steroid and xenobiotic nuclear receptor. Mol. Pharmacol. 71, 220–229 [DOI] [PubMed] [Google Scholar]
- 16. Mani S., Dou W., Redinbo M. R. (2013) PXR antagonists and implications for drug metabolism. Drug Metab. Rev. 45, 60–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takeshita A., Taguchi M., Koibuchi N., Ozawa Y. (2002) Putative role of the orphan nuclear receptor SXR (steroid and xenobiotic receptor) in the mechanism of CYP3A4 inhibition by xenobiotics. J. Biol. Chem. 277, 32453–32458 [DOI] [PubMed] [Google Scholar]
- 18. Wang H., Huang H., Li H., Teotico D. G., Sinz M., Baker S. D., Staudinger J., Kalpana G., Redinbo M. R., Mani S. (2007) Activated pregnenolone X-receptor is a target for ketoconazole and Its analogs. Clin. Cancer Res. 13, 2488–2495 [DOI] [PubMed] [Google Scholar]
- 19. Das S., Kalpana G. V. (2009) Reverse two-hybrid screening to analyze protein-protein interaction of HIV-1 viral and cellular proteins. Methods Mol. Biol. 485, 271–293 [DOI] [PubMed] [Google Scholar]
- 20. Kim J. Y., Park O. G., Lee J. W., Lee Y. C. (2007) One- plus two-hybrid system, a novel yeast genetic selection for specific missense mutations disrupting protein/protein interactions. Mol. Cell Proteomics 6, 1727–1740 [DOI] [PubMed] [Google Scholar]
- 21. Kalpana G. V., Goff S. P. (1993) Genetic analysis of homomeric interactions of human immunodeficiency virus type 1 integrase using the yeast two-hybrid system. Proc. Natl. Acad. Sci. U.S.A. 90, 10593–10597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bard M., Lees N. D., Turi T., Craft D., Cofrin L., Barbuch R., Koegel C., Loper J. C. (1993) Sterol synthesis and viability of erg11 (cytochrome P450 lanosterol demethylase) mutations in Saccharomyces cerevisiae and Candida albicans. Lipids 28, 963–967 [DOI] [PubMed] [Google Scholar]
- 23. Thakur J. K., Arthanari H., Yang F., Pan S. J., Fan X., Breger J., Frueh D. P, Gulshan K., Li D. K., Mylonakis E., Struhl K., Moye-Rowley W. S., Cormack B. P, Wagner G., Näär A. M. (2008) A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452, 604–609 [DOI] [PubMed] [Google Scholar]
- 24. White T. C., Marr K. A., Bowden R. A. (1998) Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11, 382–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ghannoum M. A., Rice L. B. (1999) Antifungal agents. Mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 12, 501–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C. (1993) A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21, 3329–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mukhopadhyay K., Kohli A., Prasad R. (2002) Drug susceptibilities of yeast cells are affected by membrane lipid composition. Antimicrob. Agents Chemother. 46, 3695–3705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaur R., Bachhawat A. K. (1999) The yeast multidrug resistance pump, Pdr5p, confers reduced drug resistance in erg mutants of Saccharomyces cerevisiae. Microbiology 145, 809–818 [DOI] [PubMed] [Google Scholar]
- 29. Clark F. S., Parkinson T., Hitchcock C. A, Gow N. A. (1996) Correlation between rhodamine 123 accumulation and azole sensitivity in Candida species. Possible role for drug efflux in drug resistance. Antimicrob. Agents Chemother. 40, 419–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mukherjee P. K., Chandra J., Kuhn D. M., Ghannoum M. A. (2003) Mechanism of fluconazole resistance in Candida albicans biofilms. Phase-specific role of efflux pumps and membrane sterols. Infect. Immun. 71, 4333–4340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Millard P. J., Roth B. L., Thi H. P., Yue S. T., Haugland R. P. (1997) Development of the FUN-1 family of fluorescent probes for vacuole labeling and viability testing of yeasts. Appl. Environ. Microbiol. 63, 2897–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Toussaint M., Conconi A. (2006) High-throughput and sensitive assay to measure yeast cell growth. a bench protocol for testing genotoxic agents. Nat. Protoc. 1, 1922–1928 [DOI] [PubMed] [Google Scholar]
- 33. Toussaint M., Levasseur G., Gervais-Bird J., Wellinger R. J., Elela S. A, Conconi A. (2006) A high-throughput method to measure the sensitivity of yeast cells to genotoxic agents in liquid cultures. Mutat. Res. 606, 92–105 [DOI] [PubMed] [Google Scholar]
- 34. Vojtek A. B., Hollenberg S. M., Cooper J. A. (1993) Mammalian Ras interacts directly with the serine/threonine kinase. Cell 74, 205–214 [DOI] [PubMed] [Google Scholar]
- 35. Mani S., Huang H., Sundarababu S., Liu W., Kalpana G., Smith A. B., Horwitz S. B. (2005) Activation of the steroid and xenobiotic receptor (human pregnane X receptor) by nontaxane microtubule-stabilizing agents. Clin. Cancer Res. 11, 6359–6369 [DOI] [PubMed] [Google Scholar]
- 36. Rao S. N., Head M. S., Kulkarni A., LaLonde J. M. (2007) Validation studies of the site-directed docking program LibDock. J. Chem. Inf. Model. 47, 2159–2171 [DOI] [PubMed] [Google Scholar]
- 37. Dong H., Lin W., Wu J., Chen T. (2010) Flavonoids activate pregnane x receptor-mediated CYP3A4 gene expression by inhibiting cyclin-dependent kinases in HepG2 liver carcinoma cells. BMC. Biochem. 11, 23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Egner R., Bauer B. E., Kuchler K. (2000) The transmembrane domain 10 of the yeast Pdr5p ABC antifungal efflux pump determines both substrate specificity and inhibitor susceptibility. Mol. Microbiol. 35, 1255–1263 [DOI] [PubMed] [Google Scholar]
- 39. Egner R., Rosenthal F. E., Kralli A., Sanglard D., Kuchler K. (1998) Genetic separation of FK506 susceptibility and drug transport in the yeast Pdr5 ATP-binding cassette multidrug resistance transporter. Mol. Biol. Cell 9, 523–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Masuyama H., Suwaki N., Tateishi Y., Nakatsukasa H., Segawa T., Hiramatsu Y. (2005) The pregnane X receptor regulates gene expression in a ligand- and promoter-selective fashion. Mol. Endocrinol. 19, 1170–1180 [DOI] [PubMed] [Google Scholar]
- 41. Das B. C., Madhukumar A. V., Anguiano J., Kim S., Sinz M., Zvyaga T. A., Power E. C., Ganellin C. R., Mani S. (2008) Synthesis of novel ketoconazole derivatives as antagonists of the human Pregnane X Receptor (PXR; NR1I2l also termed SXR, PAR). Bioorg. Med. Chem. Lett. 18, 3974–3977 [DOI] [PubMed] [Google Scholar]
- 42. Venkatesh M., Wang H., Cayer J., Leroux M., Salvail D., Das B., Wrobel J. E., Mani S. (2011) In vivo and in vitro characterization of a first-in-class novel azole analog that targets pregnane X receptor activation. Mol. Pharmacol. 80, 124–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moore T. W., Mayne C. G., Katzenellenbogen J. A. (2010) Minireview. Not picking pockets. Nuclear receptor alternate-site modulators (NRAMs). Mol. Endocrinol. 24, 683–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Choi J. H., Banks A. S., Kamenecka T. M., Busby S. A., Chalmers M. J., Kumar N., Kuruvilla D. S., Shin Y., He Y., Bruning J. B., Marciano D. P., Cameron M. D., Laznik D., Jurczak M. J., Schürer S. C., Vidović D., Shulman G. I., Spiegelman B. M., Griffin P. R. (2011) Antidiabetic actions of a non-agonist PPARγ ligand blocking Cdk5-mediated phosphorylation. Nature 477, 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Eglen R., Reisine T. (2011) Drug discovery and the human kinome. Recent trends. Pharmacol. Ther. 130, 144–156 [DOI] [PubMed] [Google Scholar]
- 46. Koole C., Wootten D., Simms J., Valant C., Sridhar R., Woodman O. L., Miller L. J., Summers R. J., Christopoulos A., Sexton P. M. (2010) Allosteric ligands of the glucagon-like peptide 1 receptor (GLP-1R) differentially modulate endogenous and exogenous peptide responses in a pathway-selective manner. Implications for drug screening. Mol. Pharmacol. 78, 456–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Buzón V., Carbó L. R., Estruch S. B., Fletterick R. J., Estébanez-Perpiñá E. (2012) A conserved surface on the ligand binding domain of nuclear receptors for allosteric control. Mol. Cell. Endocrinol. 348, 394–402 [DOI] [PubMed] [Google Scholar]
- 48. Joseph J. D., Wittmann B. M., Dwyer M. A., Cui H., Dye D. A., McDonnell D. P., Norris J. D. (2009) Inhibition of prostate cancer cell growth by second-site androgen receptor antagonists. Proc. Natl. Acad. Sci. U.S.A. 106, 12178–12183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bokoch M. P., Zou Y., Rasmussen S. G., Liu C. W., Nygaard R., Rosenbaum D. M., Fung J. J., Choi H. J., Thian F. S., Kobilka T. S., Puglisi J. D., Weis W. I., Pardo L., Prosser R.S., Mueller L., Kobilka B. K. (2010) Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature 463, 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Geber A., Hitchcock C. A., Swartz J. E., Pullen F. S., Marsden K. E., Kwon-Chung K. J., Bennett J. E. (1995) Deletion of the Candida glabrata ERG3 and ERG11 genes. Effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob. Agents Chemother. 39, 2708–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mellado E., Garcia-Effron G., Buitrago M. J., Alcazar-Fuoli L., Cuenca-Estrella M., Rodriguez-Tudela J. L. (2005) Targeted gene disruption of the 14-α sterol demethylase (cyp51A) in Aspergillus fumigatus and its role in azole drug susceptibility. Antimicrob. Agents Chemother. 49, 2536–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Teotico D. G., Frazier M. L., Ding F., Dokholyan N. V., Temple B. R., Redinbo M. R. (2008) Active nuclear receptors exhibit highly correlated AF-2 domain motions. Comput. Biol. 4, e1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Noble S. M., Carnahan V. E., Moore L. B., Luntz T., Wang H., Ittoop O. R., Stimmel J. B., Davis-Searles P. R., Watkins R. E., Wisely G. B., LeCluyse E., Tripathy A., McDonnell D. P., Redinbo M. R. (2006) Human PXR forms a tryptophan zipper-mediated homodimer. Biochemistry 45, 8579–8589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Serebriiskii I. G., Mitina O., Pugacheva E. N., Benevolenskaya E., Kotova E., Toby G. G., Khazak V., Kaelin W. G., Chernoff J., Golemis E. A. (2002) Detection of peptides, proteins, and drugs that selectively interact with protein targets. Genome Res. 12, 1785–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sharifi N. (2010) New agents and strategies for the hormonal treatment of castration-resistant prostate cancer. Expert. Opin. Investig. Drugs. 19, 837–846 [DOI] [PubMed] [Google Scholar]
- 56. Chang C. Y., McDonnell D. P. (2005) Androgen receptor-cofactor interactions as targets for new drug discovery. Trends Pharmacol. Sci. 26, 225–228 [DOI] [PubMed] [Google Scholar]
- 57. Gunther J. R., Parent A. A., Katzenellenbogen J. A. (2009) Alternative inhibition of androgen receptor signaling. Peptidomimetic pyrimidines as direct androgen receptor/coactivator disruptors. ACS Chem. Biol. 4, 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen Y., Sawyers C. L., Scher H. I. (2008) Targeting the androgen receptor pathway in prostate cancer. Curr Opin. Pharmacol. 8, 440–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Biswas A., Pasquel D., Tyagi R. K., Mani S. (2011) Acetylation of pregnane X receptor protein determines selective function independent of ligand activation. Biochem. Biophys. Res. Commun. 406, 371–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Staudinger J. L., Xu C., Biswas A., Mani S. (2011) Post-translational modification of pregnane x receptor. Pharmacol. Res. 64, 4–10 [DOI] [PMC free article] [PubMed] [Google Scholar]