Background: Biotin ligase tagging with ZO-1 was applied to identify a more complete tight junction proteome.
Results: Identical but also different proteins and functional networks were identified near the N and C ends of ZO-1.
Conclusion: The ends of ZO-1 are embedded in different functional subcompartments of the tight junction.
Significance: Biotin tagging with ZO-1 expands the tight junction proteome and defines subcompartments of the junction.
Keywords: Adherens Junction, Mass Spectrometry (MS), Membrane, Proteomics, Tight Junctions, ZO-1
Abstract
The proteins and functional protein networks of the tight junction remain incompletely defined. Among the currently known proteins are barrier-forming proteins like occludin and the claudin family; scaffolding proteins like ZO-1; and some cytoskeletal, signaling, and cell polarity proteins. To define a more complete list of proteins and infer their functional implications, we identified the proteins that are within molecular dimensions of ZO-1 by fusing biotin ligase to either its N or C terminus, expressing these fusion proteins in Madin-Darby canine kidney epithelial cells, and purifying and identifying the resulting biotinylated proteins by mass spectrometry. Of a predicted proteome of ∼9000, we identified more than 400 proteins tagged by biotin ligase fused to ZO-1, with both identical and distinct proteins near the N- and C-terminal ends. Those proximal to the N terminus were enriched in transmembrane tight junction proteins, and those proximal to the C terminus were enriched in cytoskeletal proteins. We also identified many unexpected but easily rationalized proteins and verified partial colocalization of three of these proteins with ZO-1 as examples. In addition, functional networks of interacting proteins were tagged, such as the basolateral but not apical polarity network. These results provide a rich inventory of proteins and potential novel insights into functions and protein networks that should catalyze further understanding of tight junction biology. Unexpectedly, the technique demonstrates high spatial resolution, which could be generally applied to defining other subcellular protein compartmentalization.
Introduction
Tight junctions of epithelial and endothelial cells form charge- and size-selective barriers that regulate the paracellular movement of ions and solutes (1, 2) and function in cell polarity (3) and cytoskeletal regulation (4). To date, about 40 proteins have been localized to the tight junction (5), but identification approaches have not been systematic, and the list is likely to be incomplete. Trying to identify a complete and informative set of these proteins is complicated by the difficulty of isolating the tight junction. Biochemical fractionation of junction-enriched membrane fractions allowed identification of a number of critical tight junction proteins, including, for example, ZO-1 (6), occludin (7), and claudins (8, 9) among others. Co-immunoprecipitation with known proteins has identified a few additional components (e.g. ZO-2 (10) and ZO-3 (11)). However, most protein assignments to the junction have been made by co-immunolocalization with ZO-1 or occludin, two of the hallmark tight junction proteins. Because these approaches depend somewhat on serendipity, it seems unlikely that the full set of relevant tight junction proteins has been identified. In addition, the functional tight junction probably includes many proteins not strictly limited to the junction, including, for example, actin, myosin, kinases, phosphatases, and signaling and trafficking proteins. Some of these may be transiently but critically associated with junctions; for others, only a small fraction of the total cell amount may be physically at the tight junction.
With the goal of identifying a more complete set of tight junction-associated proteins and to begin to define junction-associated protein networks, we took advantage of a recently published technique (12) to identify proximal proteins in living cells. In this method, a biotin ligase engineered to have lowered substrate specificity is fused to a protein of interest and expressed in cells. When exposed to additional biotin, the ligase portion of the fusion protein releases highly reactive BioAMP, which reacts readily with primary amines (protein N termini and ϵ-amino groups of lysine residues) on proximal neighboring proteins. Biotin-tagged proteins can be captured on Streptavidin beads and purified for proteomic or other analyses. Roux et al. (12) demonstrated that fusion of biotin ligase to the nuclear membrane protein lamin A tagged both known and novel nuclear membrane constituents. To apply this method to the tight junction, we fused biotin ligase with ZO-1, which is a functionally important, well characterized scaffolding protein (13, 14) and a ubiquitous component of tight junctions. ZO-1 is a 220-kDa multidomain protein member of the MAGUK (membrane-associated guanylate kinase) homolog family. Its N-terminal half contains three PDZ domains, an SH32 domain, and a region with homology to guanylate kinase (15). The first PDZ domain is the binding site for the strand-forming claudin family of proteins (16); PDZ2 is the site for heterodimerization with the ZO-1 homolog, ZO-2 (17); and PDZ3 is the binding site for the adhesive Ig superfamily tight junction protein, JAM (junctional adhesion molecule) (18). The guanylate kinase domain is the binding site for occludin (19, 20). The C-terminal end of ZO-1 contains an actin filament binding site (21) and interacts with the signaling and cytoskeletal adaptor protein, cingulin (22). Because these and other unique protein-protein interactions have already been defined for the N- and C-terminal halves of ZO-1, we separately fused biotin ligase to each end of ZO-1 to ask whether the radius of activity of the fused ligase (12) was sufficiently limited to allow selective tagging of proteins proximal to each end. Overall, we expected that the ZO-1 biotin ligase fusions might allow us to identify new proteins and functional networks at the junction and possibly near each end of ZO-1.
With the caveat that this method is dependent on both the presence of and access to lysines on target proteins, our results suggest that the use of biotin ligase tagging from ZO-1 provides a more complete set of functionally relevant proteins and thus insights into potentially pertinent networks than previous methods. Proteins identified by this method include many recognized tight junction and adherens junction proteins. We also find many signaling, adhesion, cytoskeletal, polarity, and trafficking proteins that probably play roles in different aspects of junction regulation. Consistent with the idea that this method provides a high degree of spatial resolution, a number of proteins are biotinylated exclusively or predominantly by biotin ligase fused to either the N or C terminus of ZO-1. Finally, along with the expected tight junction proteins, we identify a number of proteins that might not have been predicted based on current understanding of the tight junction; study of these proteins may provide new insights into the role of ZO-1 and functions of the tight junction.
EXPERIMENTAL PROCEDURES
Constructs
Myc-biotin ligase plasmid (pcDNA3.1 mycBioID) was a gift of Kyle Roux (Addgene plasmid 35700); the Myc-biotin ligase insert (BL) was excised and subcloned into pTRE2hyg (BD Biosciences); full-length human ZO-1 was subcloned 3′ to the biotin ligase coding region (BL-ZO-1) using the In-Fusion PCR-based cloning kit (BD Biosciences). ZO-1-BL was made by cloning full-length ZO-1 5′ to the biotin ligase coding sequence and moving the Myc tag to the C-terminal end of the fusion protein. Human EGFP-RN-tre (US6NL) was a kind gift from Dr. Letizia Lanzetti (University of Turin, Italy). Mouse EFR3A from Open Biosystems (clone ID 3495608) was subcloned into EGFP-N1 using InFusion-based PCR cloning (Clontech). All constructs were verified by DNA sequencing.
Cell Culture, Immunoblots, and Immunofluorescence
Tet-off MDCK II cells (BD Biosciences) were cultured under standard conditions in DMEM (4.5 g/liter glucose), 10% fetal bovine serum, and penicillin/streptomycin. Transfections with BL-, BL-ZO-1-, ZO-1-BL-, EGFP-RN-tre-, and GFP-EFR3A-encoding plasmids were performed by nucleofection (Lonza, Allendale, NJ). GFP-expressing cells were analyzed by immunofluorescence 48 h after transfection, and stable antibiotic-resistant ZO-1 fusions and biotin ligase-expressing cells were selected using hygromycin (250 μg/ml). Stable clones were screened for transgene expression by immunoblot using a Myc antibody (9B11, Cell Signaling Technology (Danvers, MA)) with anti-γ-tubulin antibody (11317, Abcam (Cambridge, MA)) used as a loading control. All immunoblots were performed as described previously (23). The rat monoclonal anti-ZO-1 antibody (23) does not recognize human ZO-1. ZO-2 (catalog no. 38-9100), claudin-2 (catalog no. 32-5600), and occludin (catalog no. 33-1500) antibodies were from Invitrogen; CK1 (KC1A) antibody (catalog no. 2655) was from Cell Signaling Technology; and cingulin antibody was a kind gift from Dr. Sandra Citi (University of Geneva, Geneva, Switzerland). The antibody against TOCA-1 (ABS70, FBPL1) was from Millipore (Temecula, CA). Secondary antibodies for immunoblot were from Rockland (Gilbertsville, PA), and secondary antibodies for immunofluorescence were from Jackson Immunoresearch (West Grove, PA) except for Streptavidin 568 (Invitrogen).
Localization of transgene products was assessed by immunofluorescence microscopy. Stably expressing cells were cultured in the absence of doxycycline for 7–10 days on Transwell permeable supports (0.4-μm polyester membrane, 12-mm inserts, Corning); transiently transfected cells were plated on glass coverslips and tested 48 h post-transfection. Immunofluorescence was performed as described previously (24); unless otherwise noted in the figure legends, cells were fixed with ethanol. Images were taking using a Zeiss LSM UV confocal microscope, ×40 oil lens, and images were generated using Zeiss Zen software. Contrast adjustment and montages were made using Adobe Photoshop.
Culture of Cells in Low Calcium
Cells were washed three times with Ca2+- and Mg2+-free PBS and incubated overnight in SMEM supplemented with 50 μm biotin (Sigma-Aldrich), 2% dialyzed fetal bovine serum, and penicillin/streptomycin. After 15–16 h, cells were processed for immunofluorescence, immunoblot, or proteomic analysis.
Purification of Biotinylated Proteins for Mass Spectrometry and Immunoblots
MDCK II cells stably expressing transgenes were cultured for 7–10 days in 150-mm dishes; nine dishes were used for each proteomic analysis. Affinity capture of biotinylated proteins was performed slightly modified from Roux et al. (12). Biotin (50 μm) was added to dishes 15–16 h before cells were collected. After three washes with PBS, cells were scraped in PBS, pelleted, and lysed in a total of 5 ml of radioimmune precipitation assay buffer (1% Triton X-100, 0.5% deoxycholate, 0.2% SDS, 50 mm Tris, pH 7.5, 150 mm NaCl with protease inhibitors). Samples were sonicated, incubated on ice for 10 min, resonicated, and centrifuged for 20 min at 12,000 × g to remove insoluble material. Supernatants were transferred to fresh tubes containing 500 μl of (slurry) prewashed Dynabeads MyOne Streptavidin C1 and incubated for 4 h at 4 °C in an end-over-end mixer. Beads were washed for 8–10 min twice with 2% SDS; once with 0.1% deoxycholate, 1% Triton X-100, 500 mm NaCl, 1 mm EDTA, 50 mm Hepes, pH 7.3; and once with 250 mm LiCl, 1 mm EDTA, 0.5% deoxycholate, 0.5% Nonidet P-40, 10 mm Tris, pH 8.0, followed by two washes in 50 mm Tris, pH 7.5, 50 mm NaCl. Bound proteins were eluted by a 10-min incubation at 98 °C in biotin-saturated 4× SDS sample buffer (8% SDS, 250 mm Tris, pH 6.8, 0.57 m mercaptoethanol, 40% glycerol). Eluted proteins were subjected to SDS-PAGE, and gels were stained briefly with SimplyBlue Safe Stain (Invitrogen). Lanes were excised and divided into 12–16 bands, destained, reduced, alkylated, and digested overnight with trypsin (Promega, V511A Sequencing grade). Eluted peptides were purified on ZipTips (C18, Millipore) and transferred into sample vials (Agilent, Santa Clara, CA) for mass spectrometry.
Mass Spectrometry
Liquid chromatography tandem mass spectrometry was performed using an Eksigent nanoLC-Ultra 1D Plus system (Dublin, CA) coupled to an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) using collision-induced dissociation fragmentation. Peptides were first loaded onto a Zorbax 300SB-C18 trap column (Agilent, Palo Alto, CA) at a flow rate of 6 μl/min for 6 min and then separated on a reversed-phase PicoFrit analytical column (New Objective, Woburn, MA) using a 40-min linear gradient of 5–40% acetonitrile in 0.1% formic acid at a flow rate of 250 nl/min. LTQ-OrbitrapVelos settings were as follows: spray voltage, 1.5 kV; full MS mass range, m/z 300–2000. The LTQ-OrbitrapVelos was operated in a data-dependent mode (i.e. one MS1 high resolution (60,000) scan for precursor ions followed by six data-dependent MS2 scans for precursor ions above a threshold ion count of 500 with collision energy of 35%).
MASCOT Database Search
The raw file generated from the LTQ OrbitrapVelos was analyzed using Proteome Discoverer version 1.3 software (Thermo Fisher Scientific, LLC) using our six-processor Mascot cluster at the National Institutes of Health (version 2.3) search engine. The following search criteria were set to the following: databases, Swiss Institute of Bioinformatics taxonomy (SwissProt) (Homo sapiens (human) and Canis familiaris (dog)) and National Center for Biotechnology Information (NCBI) RefSeq database (Canis lupis); enzyme, trypsin; miscleavages, 2; variable modifications, oxidation (M), deamidation (NQ), acetyl (protein N terminus); fixed modification, carbamidomethyl (C); MS peptide tolerance 20 ppm; MS/MS tolerance as 0.8 Da. Post-database search, the peptides were filtered for a false discovery rate of 1% and rank 1 peptides (unique to one protein). Proteins identified will have at least two unique peptide hits based on the above criteria, but also we accepted a unique peptide hit only if the mascot peptide score was above a score of 40 and more than one peptide spectrum match (PSM) for that given peptide.
All samples were analyzed in triplicate. Although peptides were screened against the SwissProt human and dog databases and the NCBI RefSeq dog database, we only used the results from the human database for further analysis. The SwissProt dog database is extremely small, and although the NCBI RefSeq dog database gave us more proteins, the most abundant proteins were the same as those identified in the human database. More important, the SwissProt human database is better curated and more completely annotated, facilitating functional classification. As well, a larger number of proteins in the RefSeq dog database are uncharacterized, including, for example, JCAD (25). For purposes of comparison, the RefSeq protein lists are included as supplemental File S7. After lists of all proteins were compiled, keratins and other epidermal proteins, histones, and endogenously biotinylated carboxylases were discarded. Additionally, for the purposes of functional analysis, we removed ribosomal proteins, because they were also prevalent in the samples from cells expressing the biotin ligase alone. Although it is possible that their biotinylation may reveal a hitherto unappreciated function for ZO-1, given the extended period of biotinylation, it seemed more likely that they represent co-translational biotinylation, and thus they were not considered functionally relevant. The ribosomal proteins are included as a separate list in supplemental File S6. Criteria for inclusion in the final protein lists were appearance in at least two or three sample runs. UniProt descriptors and literature searches were used to classify proteins into functional categories. To compare protein amounts from different mass spectrometry analyses, PSM values for individual proteins were normalized by division with the total PSMs for each run. This normalized PSM value for each run was used to generate the results presented in Fig. 4. Differences between samples were determined by analysis of variance followed by Dunnett's test (GraphPad Prism version 5).
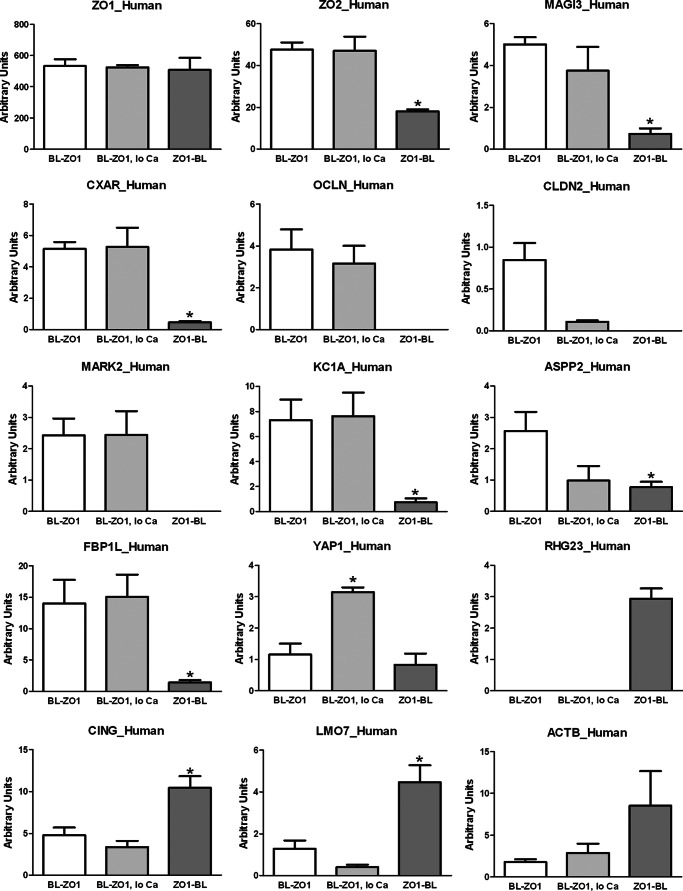
FIGURE 4.
Comparison of the relative amounts of specific proteins recovered from cells expressing biotin ligase fused to the N-terminal end of ZO-1 incubated in normal (BL-ZO1) or low calcium (BL-ZO1, lo Ca) or to the C-terminal end of ZO-1 (ZO1-BL). ZO-1 was recovered equally from all samples (top left), whereas differing amounts of other proteins were recovered from MDCK cells treated with 50 μm biotin for 15 h and expressing the different constructs. Normalized PSMs were calculated as described under “Experimental Procedures”; n = 3 for each bar, mean ± S.E (error bars). *, p < 0.05 by analysis of variance followed by Dunnett's test.
RESULTS
The Biotin Ligase ZO-1 Fusion Proteins Localize to Tight Junctions
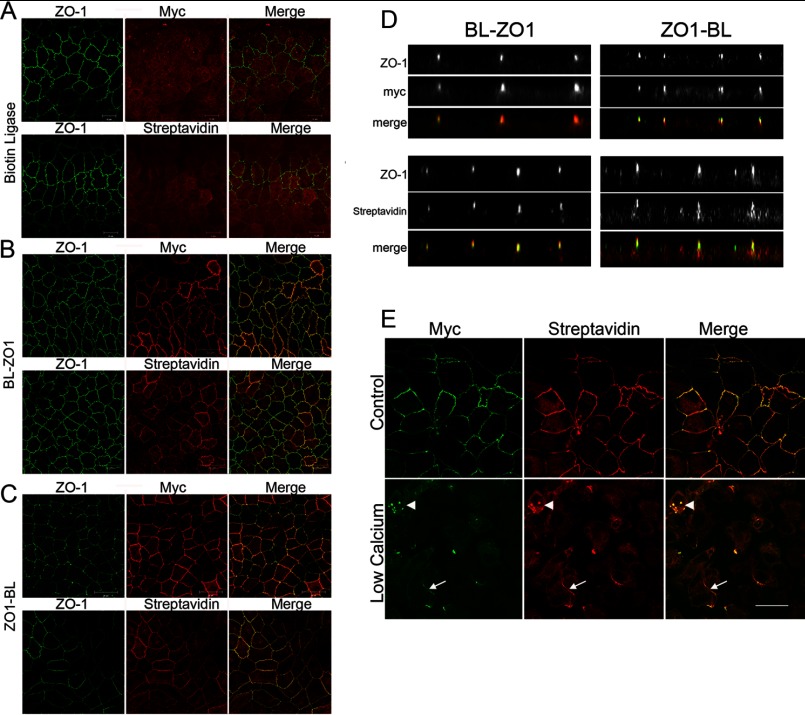
As a first step in the analysis of proteins proximal to ZO-1, we assessed how well our fusion proteins colocalized with endogenous ZO-1 compared with an unfused Myc-tagged biotin ligase protein alone. Immunofluorescence microscopy demonstrated that Myc-tagged biotin ligase was diffusely distributed in MDCK II cells (Fig. 1A, top panels). In contrast, the Myc-tagged biotin ligase fused to either the N-terminal (BL-ZO-1; Fig. 1B, top panels) or C-terminal end (ZO-1-BL; Fig. 1C, top panels) showed that both constructs were largely colocalized with endogenous ZO-1 at tight junctions. Without the addition of biotin to the medium, expression of biotin ligase fusion proteins results in only modest levels of cellular biotinylation; the level of biotinylation is greatly increased when 50 μm biotin is added to the cell culture medium for 8–24 h (not shown) (12). When the transgene-expressing cells were treated with 50 μm biotin for 15 h and the distribution of biotinylated proteins was detected with fluorescent streptavidin, biotinylated proteins were diffusely distributed in cells expressing biotin ligase alone (Fig. 1A, bottom panels), whereas most of the biotinylated proteins in cells expressing biotin ligase fused to either the N-terminal (Fig. 1B, bottom panels) or C-terminal (Fig. 1C, bottom panels) end of ZO-1 were associated with the tight junction. The colocalization of the biotin ligase ZO-1 fusion proteins and biotinylated proteins can be seen more clearly in z-sections (Fig. 1D). Both N-terminal biotin ligase-ZO-1 (Fig. 1D, top left panels) and the streptavidin-labeled biotinylated proteins were concentrated with ZO-1 (Fig. 1D, bottom left panels). Similarly, the C-terminal fusion protein, Myc epitope-tagged ZO-1-biotin ligase, was colocalized with endogenous ZO-1 (Fig. 1D, top right panels), as was most of the biotinylated protein (Fig. 1D, bottom right panels). Although the strongest streptavidin signal was associated with ZO-1, there was always some streptavidin signal found more diffusely within the cells. However, the concentration of streptavidin labeling at the tight junction suggests that the majority of labeled proteins, subsequently identified by mass spectrometry, are ZO-1 tight junction “neighbors.”
FIGURE 1.
ZO-1 biotin ligase fusion proteins but not biotin ligase alone colocalize with endogenous ZO-1. A, neither biotin ligase alone (myc, top middle) nor biotinylated proteins in the same cells (Streptavidin, bottom middle) colocalize (Merge, right panels) with endogenous ZO-1 (left panels) in MDCK cells. In contrast, both biotin ligase fused to the N terminus (B, BL-ZO1, myc) and C terminus (C, ZO1-BL) of ZO-1 colocalized (Merge, B and C, top right) with endogenous ZO-1 (B and C, top left panels; the monoclonal antibody to ZO-1 recognizes the canine but not the human protein). In addition, the majority of biotinylated proteins (Streptavidin, B and C, bottom middle panels) were also colocalized (B and C, bottom right) with endogenous ZO-1 (B and C, bottom left). D (left panels), confocal z-sections reveal that both BL-ZO-1 (myc) and biotinylated proteins (Streptavidin) are concentrated (Merge) with endogenous ZO-1 (left). D (right panels), similarly, ZO-1-BL (myc) and biotinylated proteins are also concentrated (Merge) with ZO-1 (right), although there is also some non-junctional immunofluorescence associated with both the transgene and the biotinylated proteins. E, MDCK cells expressing biotin ligase-ZO-1 were cultured in normal (top panels) or low calcium (bottom panels) medium supplemented with 50 μm biotin for 15 h. Immunofluorescent localization of transgene (Myc) and biotinylated proteins (Streptavidin) revealed that incubation in low calcium resulted in the loss of most of the tight junction-associated protein, although remnants remain (arrows); some Myc and streptavidin signal appears vesicular (arrowheads), whereas most is diffuse. Bar, 20 μm.
To test whether the profile of proximal proteins changed following structural disassembly of the junction, we cultured MDCK cells in low calcium medium; this is known to result in ZO-1 endocytosis and loss from tight junctions (26). As expected, when MDCK cells expressing N-terminal biotin ligase-ZO-1 fusion protein were cultured overnight in low calcium in the presence of biotin, the transgene was mostly lost from tight junctions (Fig. 1E, compare top and bottom left panels), although remnants of junctional plaques remained (white arrows). Most of the biotin ligase fusion protein was distributed diffusely throughout the cell, with concentration evident in occasional vesicular structures (white arrowheads). The bulk of the biotinylated proteins as detected with fluorescent streptavidin (Fig. 1E, middle panels) also relocalized from the plasma membrane to both a vesicular and diffuse intracellular distribution.
Coomassie-stained Protein Gels of Samples from Cells Expressing BL-ZO-1 and ZO-1-BL Reveal Differences in the Biotinylation Patterns
To examine the proteins biotinylated by the biotin ligase alone and the ligase fused to the N- or C-terminal end of ZO-1, expressing cells were incubated with biotin for 15 h; in addition, one set of cells was cultured with biotin in low calcium medium overnight. Cells were lysed, and biotinylated proteins were purified on streptavidin beads; purified proteins were separated by SDS-PAGE and stained with Coomassie Brilliant Blue (Fig. 2). All samples contained complex mixtures of proteins that were dependent on the presence of the specific transgene and on the addition of biotin (not shown). The heaviest signal in each lane (arrowheads) marked the position of the transgene, indicating that the fusion protein is most effective at self-biotinylation. Inspection of the pattern of Coomassie-stained bands reveals both similarities and differences among the different conditions, with the most different pattern being in the biotin ligase alone and the most similar N-terminal biotin ligase-ZO-1 (BL-ZO-1) cultured in normal and low calcium (BL-ZO-1, low Ca2+). The pattern of staining with Coomassie was quite similar among the three protein preparations for each construct and in the low calcium samples, suggesting that both the biotinylation and the extraction methods were very reproducible (not shown).
FIGURE 2.
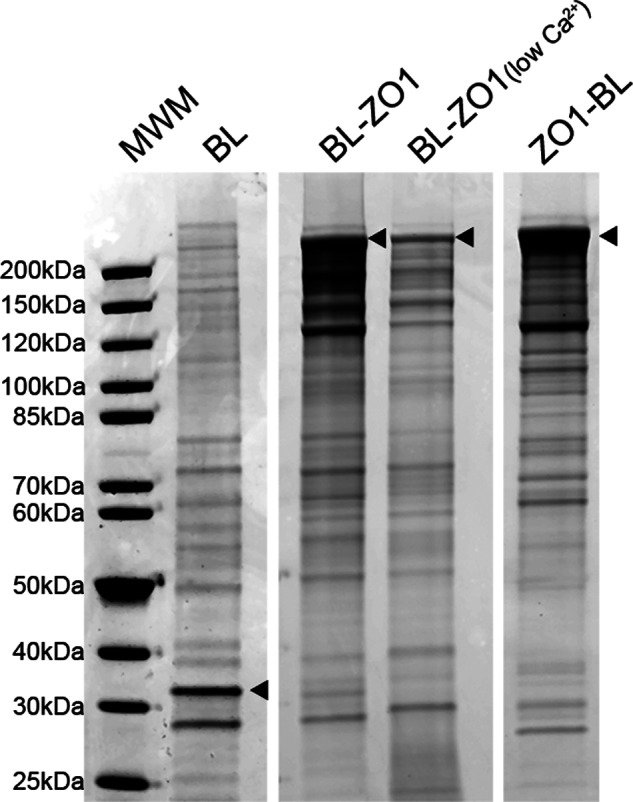
Coomassie-stained SDS-PAGE reveals that streptavidin-purified biotinylated proteins from MDCK cells expressing different transgenes show differing protein patterns. Shown are proteins purified from biotin-treated cells expressing biotin ligase alone (BL), biotin ligase fused to the N terminus of ZO-1 (BL-ZO1), cells expressing the same construct but incubated in low calcium medium overnight (BL-ZO1, low Ca2+), and cells expressing biotin ligase fused to the C terminus of ZO-1 (ZO1-BL). The positions of the transgenes are marked with arrowheads; the protein pattern for BL-ZO-1 in normal, and low calcium is quite similar. Triplicate samples gave very similar protein patterns. MWM, molecular weight markers.
Proteomic Analysis of Biotinylated Proteins Reveals Differences in Protein Functional Categories among BL-, BL-ZO-1-, and ZO-1-BL-expressing Cells
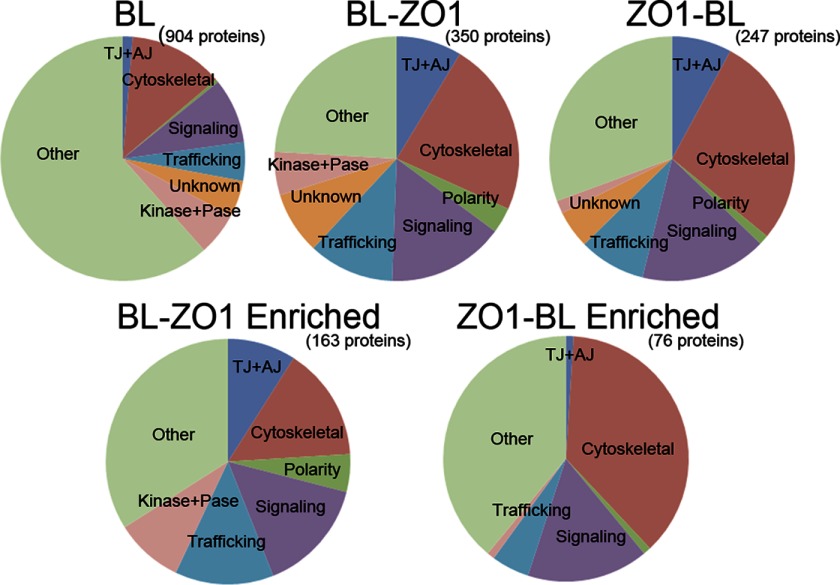
The entire lanes from triplicate Coomassie-stained gels for each condition were cut into equal sized bands and used for proteomic analysis by mass spectrometry. Triplicate analyses of cells expressing BL alone identified peptides assigned to 906 unique proteins (supplemental File S1), whereas triplicate analyses of cells expressing BL-ZO-1, BL-ZO-1 cultured in low calcium, and ZO-1-BL identified 350, 224, and 247 proteins, respectively, in at least two of three separate tagging experiments (supplemental Files S2–S4). For reference, the transcriptome of rat renal proximal tubule includes ∼9000 transcripts (27), suggesting that BL tagging is not random but is quite selective for a small reproducible set (2–3%) of the total proteome.
Seeking biologic insight into the tagged proteins, we initially analyzed protein lists using standard functional annotation categories based on gene ontology terms with common Web-based tools (e.g. DAVID Bioinformatics Resources version 6.7 (28), IPA-Ingenuity® Systems). However, because there is some misannotation in these databases and we had a relatively small number of proteins and were asking in some cases about known interactions at cell junctions, we found that it was more informative to construct our own functional categories based on both UniProt (29) and searching for functional characterizations in the primary literature. Using these modified functional categories (Fig. 3, top left chart), the control background of proteins tagged in BL-expressing cells was predominantly involved in cell cycle regulation, DNA and RNA processing, transcription, and energy metabolism (“other” category). A significant fraction was also identified in cytoskeletal, signaling, and trafficking pathways as well as kinases or phosphatases. A small fraction (1.3%) was identified as tight or adherens junction proteins; 5% were of unknown function.
FIGURE 3.
Functional analysis of proteins recovered from MDCK cells expressing biotin ligase alone and ZO-1 biotin ligase fusion proteins. Top, streptavidin-purified proteins identified by mass spectrometry from cells expressing biotin ligase alone (left chart) or biotin ligase fused to the N terminus (Biotin Ligase-ZO1, middle) or C terminus of ZO-1 (ZO1-Biotin Ligase, right). Functional classification revealed similar distribution for the two ZO-1 constructs, whereas the largest fraction of proteins tagged from cells expressing biotin ligase alone were classified as “other,” including proteins involved in DNA and RNA synthesis, metabolism, etc. TJ+AJ, tight junction and adherens junction proteins. Bottom, in contrast, functional analysis of proteins recovered exclusively or >3-fold enriched (as determined by the averaged normalized PSMs) from ZO-1-BL-expressing (left) and BL-ZO-1-expressing (right) cells reveals differences in several categories, including tight junction and adherens junction, signaling, polarity, and other membrane proteins and kinases and phosphatases. For the purposes of this analysis, DVL1 and DVL3 were included with polarity proteins because of their interactions with VANG1, VANG2, and MARK2.
In contrast (Fig. 3, top middle), proteins tagged in BL-ZO-1-expressing cells contained a higher fraction of tight and adherens junction proteins; those identified as cytoskeletal, polarity, or signaling protein; and a similar fraction of kinases, phosphatases, and proteins with unknown function. Protein categories tagged in cells expressing ZO-1-BL (Fig. 3, top right) had a functional distribution similar to those tagged by BL-ZO-1 but with small differences in the fractions of some categories. Cells grown in low calcium were nearly identical to cells expressing the same vector in normal calcium (not shown).
Both ZO-1 biotin ligase fusion proteins together tagged over 400 ZO-1 “neighbors” (supplemental Files S2–S4). However, it was obvious by lining proteins up by total spectral counts that some proteins were more heavily represented than others in the total group of tagged proteins. These heavily tagged proteins could be grouped in several categories. Some proteins were heavily tagged both in cells expressing biotin ligase alone and in ZO-1 fusion protein-expressing cells (e.g. filamin A, myosin 2A, and cortactin). Although both myosin 2A (30) and cortactin (31) have been reported to be important at cell junctions, their high recovery from cells expressing the biotin ligase alone does not allow us to infer any specific role that is enriched at the junction. We chose instead to concentrate on proteins that were exclusively present or more abundant (at least 3-fold enriched; see “Experimental Procedures”) in samples from cells expressing the ZO-1 fusions compared with cells expressing BL alone. We recognize, however, that this approach could result in our discarding some proteins that could be relevant to the tight junction but are found elsewhere in the cell.
Proteins exclusive to or more abundant in ZO-1 fusion protein-expressing cells included many expected tight junction- and adherens junction-associated proteins (Table 1); this list comprises ZO-1, -2, and -3, transmembrane proteins, and other scaffolding and cytoskeletal proteins. More interesting was a second group that included some proteins that we would not have predicted to be heavily tagged by ZO-1 (a selected list is included as Table 2); these include some polarity and/or Wnt-signaling proteins, cytoskeletal proteins, and signaling proteins. Several of these have been associated with tight or adherens junctions in one or two publications, but they do not appear in review articles on tight junction proteins (5).
TABLE 1.
Tight junction and adherens junction proteins tagged by biotin ligase fused to ZO-1
| UniProt ID | Name | Localization/Function (UniProt) | Reference |
|---|---|---|---|
| ACTN | Actin | Involved in various types of cell motility, concentrated at adherens junctions | 68 |
| AFAD | Afadin, Af6, MLLT4 | Interacts with actin, nectins, essential for nectin/e-cadherin association, ZO-1 (possibly indirect) | 69 |
| CGNL1 | Paracingulin, JACOP | Involved in Rho signaling, implicated in junction assembly | 70 |
| CING | Cingulin | Involved in Rho signaling, response to mucosal injury, CLDN2 expression | 71 |
| CLDN2 | Claudin-2 | Integral membrane-sealing protein of tight junction, interacts with PDZ1 of ZO proteins | 72 |
| ClDN3 | Claudin-3 | Integral membrane sealing protein of tight junction, interacts with PDZ1 of ZO proteins | 9 |
| CTNA1 | α-Catenin | Adherens junction protein | 73 |
| CTND1 | δ-Catenin, p120 catenin | Adherens junction protein | 74 |
| CXAR | Coxsackie and adenovirus receptor, CAR | Involved in cell-cell adhesion | 32 |
| JAM1 | Junctional adhesion molecule A, JAM-A, F11R | Involved in cell-cell adhesion, interacts with PAR3 | 75 |
| MAGI1 | MAGUK, WW, and PDZ domain-containing protein 1 | Scaffolding protein, interacts with actin, synapdopodin, CXAR, SH3K1 in neurons | 76 |
| MAGI3 | MAGUK, WW, and PDZ domain-containing protein 3 | Scaffolding protein, involved in Wnt signaling, LPA receptor signaling | 77 |
| OCLN | Occludin | Integral membrane protein of tight junction, interacts with ZO-1 via GUK domain | 7 |
| PAR3L | Partitioning-defective 3 homolog B | Tight junction/polarity protein | 58 |
| PLAK | Junction plakoglobin, γ-catenin | Adherens junction protein | 73 |
| PVRL2 | Polio virus-related receptor 2, nectin-2 | Involved in cell-cell adhesion | 78 |
| SHRM2 | Shroom2 | Involved in morphology changes, binds actin | 79 |
| SYNPO | Synaptopodin | Actin-associated protein, interacts with MAGI1 | 80 |
| ZO1 | Tight junction protein ZO-1 | Scaffolding protein of tight junctions, also localized to adherens junctions in non-epithelial cells | 15 |
| ZO2 | Tight junction protein ZO-2 | Scaffolding protein of tight junctions, also localized to nucleus, forms heterodimers with ZO-1 | 10 |
| ZO3 | Tight junction protein ZO-3 | Peripheral membrane protein of tight junctions, interacts with ZO-1 | 11 |
TABLE 2.
Selected non-tight junction proteins tagged by biotin ligase fused to ZO-1
| UniProt ID | Name | Localization/Function (UniProt) | Reference |
|---|---|---|---|
| PP1A | Ser/Thr protein phosphatase 1 α catalytic subunit | Protein phosphatase, interacts with and regulates phosphorylation of PAR3, subset colocalizes with ZO-1, influences TJ form | 59 |
| ANXA2 | Annexin A2 | Involved in tight junction assembly | 81 |
| ARF6 | ADP-ribosylation factor 6 | A GEF for ARF6 affecting kinetics of TJ assembly | 82 |
| ASPP2 | Apoptosis-stimulating of 53 protein 2, IRSp53 | Regulator of cell growth, polarity protein | 36, 37 |
| BAIP2a | Brain-specific angiogenesis inhibitor 1-associated protein 2 | Adapter protein that links membrane-bound small G-proteins to cytoplasmic effector protein, phosphorylated by MARK2, involved in cell/matrix/polarity signaling by MARK2 | 53 |
| BIN3 | Bridging integrator 3 | Cytoskeletal protein, BAR domain-containing | 83 |
| COF1a | Cofilin-1 | Regulates actin dynamics, PAR3 mediates inhibition of LIM kinases regulates cofilin phosphorylation and TJ assembly | 84 |
| DC1L2 | Dynein component | Dynein interacts with β-catenin, may tether microtubule at adherens junction | 85 |
| DVL1,2 | Dishevelled-1, -2 | Wnt signaling, planar cell polarity, affects cell contact maturation | 60 |
| EPS8a | EGF-R pathway substrate 8 | Regulates cell and barrier function in testis | 86 |
| FBP1L | Formin-binding protein-like 1, TOCA-1 | Cytoskeletal protein, F-bar, involved in endocytosis and CDC42-induced actin polymerization, localizes to cell junctions in C. elegans | 38 |
| FGD4 | FYVE, RhoGEF, and pleckstrin homology domain-containing protein 4 | Cytoskeleton, activates CDC42 | 87 |
| JCAD | Junction protein associated with coronary artery disease | Associated with adherens junctions in endothelial cells | 25 |
| KC1A | Casein kinase 1 α | Serine/threonine protein kinase, phosphorylates OCLN, co-localizes with e-cadherin | 34 |
| LAP2 | LAP2, Erbin, ERBB2-interacting protein | Interacts with δ-catenin and ARVCF | 88 |
| LMO7 | Lim domain-only protein 7 | Involved in nectin/e-cadherin interaction | 44 |
| LPP | Lipoma preferred partner | Polarity protein, interacts with Scrib | 57 |
| MARK2 | Serine/threonine-protein kinase MARK2, Par1b | Serine/threonine protein kinase, involved in microtubule dynamics and cell polarity, phosphorylates DVL, BAIP2 | 89 |
| NHS | Nance-Horan syndrome protein | One isoforms colocalizes with ZO-1, may play role with SCRIB /VANG1 in polarity | 90, 91 |
| PAR3L/D3 | Partitioning-defective homolog 3B/3 | Localizes to tight junctions, PAR3L does not bind aPKC | 58 |
| RAB5A,B,C | Ras-related protein 5A, -B, -C | GTPase, early endosomes, plasma membrane fusion | 92, 93 |
| RASF8 | Ras-associated domain-containing protein 8 | Forms a complex with ASPP2, regulates cell-cell adhesion in Drosophila | 51 |
| RHG12 | Rho GTPase-activating protein 12 | Positive regulator of GTPase activity | 48 |
| RHG23 | Rho GTPase-activating protein 23 | Positive regulator of GTPase activity | 43 |
| SCRIB | Protein scribble homolog | Polarity protein/interacts with VANGL2, LPP | 55 |
| SDCB1 | Syntenin-1 | Adherens junction protein, colocalizes with e-cadherin, scaffolding protein | 94 |
| SH3K1 | SH3 domain-containing kinase-binding protein 2, CIN85 | Adaptor protein involved in many diverse signaling pathways | 95 |
| SRBS2 | Sorbin and SH3 domain-containing protein 2 | Adaptor protein involved in signaling pathways | 49 |
| US6NL | USP6 N-terminal-like protein, RN-tre | GTPase-activating protein for RAB5A. Involved in receptor trafficking | 50 |
| VANG1/2 | Vang-like protein 1, Stabismus 2 | Membrane protein, polarity protein, co-localizes with e-cadherin in intestinal epithelial cells | 62 |
| WIPF2 | WAS/WASL-interacting protein family member 2 | Cytoskeleton, may cooperate with WASP and WASL in actin reorganization interacts w/Tuba | 96 |
a Proteins present at approximately equal levels in biotin ligase alone and ZO-1 fusion protein samples but were included because they were part of the function network described here; all other proteins were enriched at least 3-fold (as determined by the average of normalized PSMs from three samples) in ZO-1 samples compared with biotin ligase alone or were not present in biotin ligase alone samples.
Direct comparison of proteins biotinylated in cells expressing either BL-ZO-1 or ZO-1-BL revealed that the majority of proteins were common to both sets. However, some proteins were uniquely tagged by either BL-ZO-1 or ZO-1-BL or much more abundant in one set or the other (supplemental File S5). Analysis of functional categories of these proteins revealed striking differences between the two groups (Fig. 3, bottom). Of the tight and adherens junction proteins tagged in BL-ZO-1 cells, 15 are unique to or much more heavily tagged in (>3-fold, average normalized PSMs) cells expressing this construct. Similarly, eight polarity proteins and 15 kinase/phosphatases are uniquely or more heavily tagged in the BL-ZO-1 cells (Fig. 3, bottom left). In contrast, only one tight or adherens junction protein (JCAD), one polarity protein (SCRIB), and one kinase/phosphatase (PP1G) are uniquely tagged in the ZO-1-BL-expressing cells (Fig. 3, bottom right). In contrast, a slightly larger number of cytoskeletal proteins (28 compared with 24) are tagged by ZO-1-BL. The preferential tagging by the N-terminally fused ZO-1 with junction proteins compared with the C-terminally fused ZO-1 is generally consistent with what is known about the roles of different protein interaction domains in ZO-1. However, the differential enrichment in kinases, trafficking, and polarity proteins between the two ends was not expected. These results suggest that there are further functional distinctions between the N and C terminus of ZO-1 yet to be explored.
Despite the large differences in localization, there were only minor differences between the functional categories of proteins tagged in the BL-ZO-1 cells cultured with and without calcium (not shown). It seems likely that even in low calcium, the proximity of ZO-1 to many neighboring proteins is maintained.
Specific Proteins Are Differentially Tagged in Cells Expressing ZO-1 with Biotin Ligase Fused to the N or C Terminus
Beyond the differences in functional categories of proteins tagged when BL is at the N or C terminus of ZO-1, there were clearly differences in the tagging of specific proteins (Fig. 4) as detected by mass spectrometry. Consistent with the ability of the biotin ligase fusion protein to self-biotinylate with high efficiency, ZO-1 had the highest spectral counts in all mass spectrometry experiments, and recovery was equivalent when BL was fused to the N or C terminus or cells were grown in low calcium medium (Fig. 4, top row, left). In contrast, the recovery of tagged ZO-2 is higher in samples from the N-terminal BL-ZO-1 fusion protein compared with C-terminal fused ZO-1-BL protein (Fig. 4, top row, middle), consistent with its known interaction with PDZ2, which is closer to the N terminus (17). This same pattern is observed for another tight junction scaffolding protein, MAGI3 (Fig. 4, top row, right). More dramatic are the findings for the integral membrane proteins of the tight junction; the Coxsackie and adenovirus receptor (CXAR) is very poorly tagged by ZO-1-BL (Fig. 4, second row, left), and neither occludin (OCLN; second row, middle) nor any of the claudins (claudin-2 (CLDN2) is shown here; second row, right) are present in any of the samples from cells expressing biotin ligase positioned at the C-terminal end of ZO-1. As described previously, the C-terminal PDZ-binding motif on claudin-2 interacts with the first PDZ domain of ZO-1 (16), whereas occludin interacts with the GUK domain (20). CXAR has been shown to co-immunoprecipitate with ZO-1 (32) and ends in a C-terminal PDZ binding motif that is likely to interact with one of the three N-terminal PDZ domains in ZO-1.
Not just transmembrane proteins were enriched in the BL-ZO-1 samples compared with ZO-1-BL samples. Two kinases were markedly better recovered in the BL-ZO-1 samples: MARK2 (Fig. 4, third row, left), which is involved in microtubule dynamics, Wnt signaling, and cell polarization (33) and casein kinase 1 α (KC1A; third row middle), which partially colocalizes with E-cadherin in MDCK cells (34). Phosphorylation of E-cadherin by KC1A inhibits its localization to cell contacts, negatively regulating cell adhesion (34). In addition, KC1A can phosphorylate occludin in vitro, but the functional significance of this is unknown (35). ASPP2 (apoptosis stimulating of 53 protein 2) (Fig. 4, third row, right), which interacts with the tight junction protein MAGI1 and polarity protein PAR3 and has been implicated both in cell polarity and in tight junction function (36, 37), and the F-BAR domain-containing protein, TOCA-1 (FBP1L) (Fig. 4, fourth row, left) are also enriched in samples from BL-ZO-1- compared with ZO-1-BL-expressing cells. TOCA-1 binds to and remodels lipid bilayers and localizes to cell junctions in Caenorhabditis elegans (38) and to some extent in A431 cells (39).
In general, the proteins isolated from cells incubated in low calcium were not qualitatively different from cells expressing the same construct (BL-ZO-1) in normal calcium. There tended to be lower recoveries of neighboring proteins but a similar pattern. One of the few proteins consistently more heavily tagged in BL-ZO-1-expressing cells grown in low calcium than in normal calcium is YAP1 (Yorkie Homolog, Yes-associated protein; Fig. 4, fourth row, middle). YAP1 has been reported to play a key role in cell proliferation in response to cell contact (40) and has been reported to interact with angiomotin at tight junctions and ZO-2 in the nucleus (41, 42). It seems possible that the loss of tight and adherens junctions in low calcium might thus trigger changes in the proximity to ZO-1/2 and YAP1.
Finally, a number of proteins are more heavily tagged in cells expressing ZO-1-BL, including uncharacterized protein Rho GTPase-activating protein 23 (RHG23; Fig. 4, fourth row, right) (43), cingulin (fifth row, left), Lim domain only protein 7 (LMO7) (fifth row, middle), and cytoplasmic actin (ACTB) (fifth row, right). Consistent with this differential tagging, cingulin (22) and actin (21) are known to interact with the C terminus of ZO-1. LMO7 is known to interact at cell junctions with nectin and afadin and to shuttle to the nucleus, where it functions as a transcriptional regulator (44).
Immunoblot analysis was used to verify selected proteomic findings (Fig. 5). Lysates (Fig. 5, left) and streptavidin-purified samples (right) from BL, BL-ZO-1, ZO-1-BL, and BL-ZO-1 cells grown in low calcium were probed for the Myc tag on the fusion proteins (top). Myc-tagged constructs in streptavidin-eluted samples show some degradation, but it appears that degradation happens during the purification on or elution from the beads, because the lysate samples are mostly intact. The same samples were probed for ZO-2 (second panel), claudin-2 (third panel), occludin (fourth panel), KC1A (fifth panel), and cingulin (bottom panel). ZO-2, claudin-2, occludin, and casein kinase 1 α are all present in markedly lower amounts in samples from cells expressing ZO-1-BL, and cingulin is present in higher amounts than in samples from cells expressing BL-ZO-1. Culture of cells in low calcium results in decreased levels of most of the proteins, but the differences are less dramatic than is seen between the N- and C-terminal fusion proteins. Although limited examples, these data strongly support the differential tagging observed by mass spectrometry in Fig. 4.
FIGURE 5.
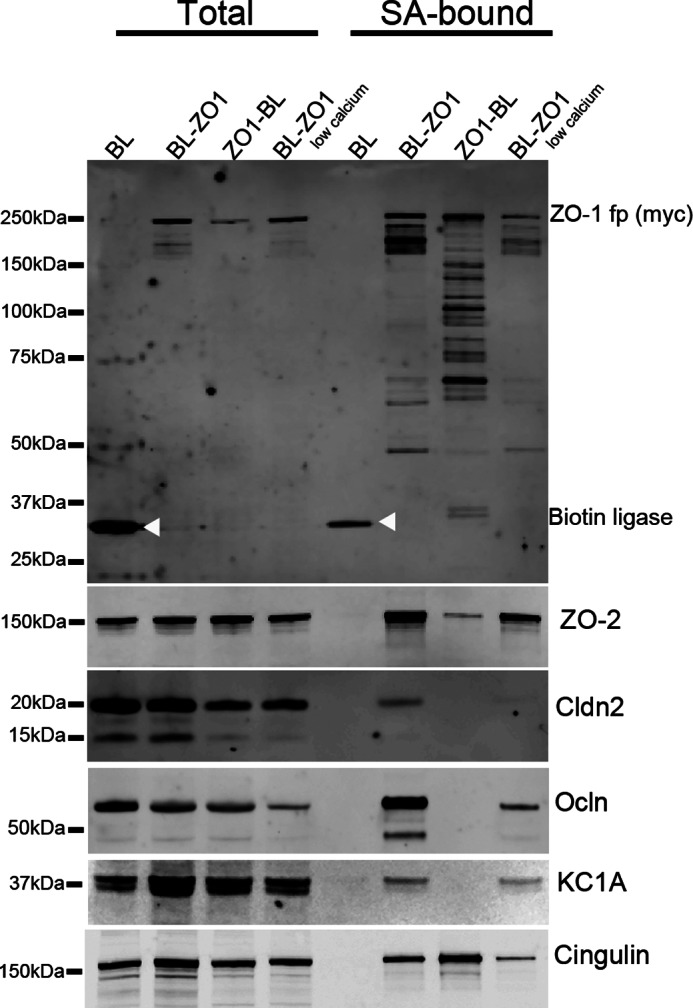
Immunoblot of selected proteins purified on streptavidin resin from MDCK cells expressing biotin ligase alone (BL), biotin ligase fused to the N-terminal (BL-ZO1) or C-terminal end of ZO-1 (ZO1-BL), or BL-ZO-1-expressing cells incubated in low calcium. Shown are samples from cell lysate before (left side, total) and after purification on streptavidin resin (right side, SA-bound). Cells were probed for the fusion protein with an anti-Myc antibody (top panel; white arrowheads mark the location of the biotin ligase alone fusion protein); the same samples were also probed for ZO-2 (second panel), claudin-2 (third panel), occludin (Ocln; fourth panel), CK1A (fifth panel), and cingulin (bottom panel).
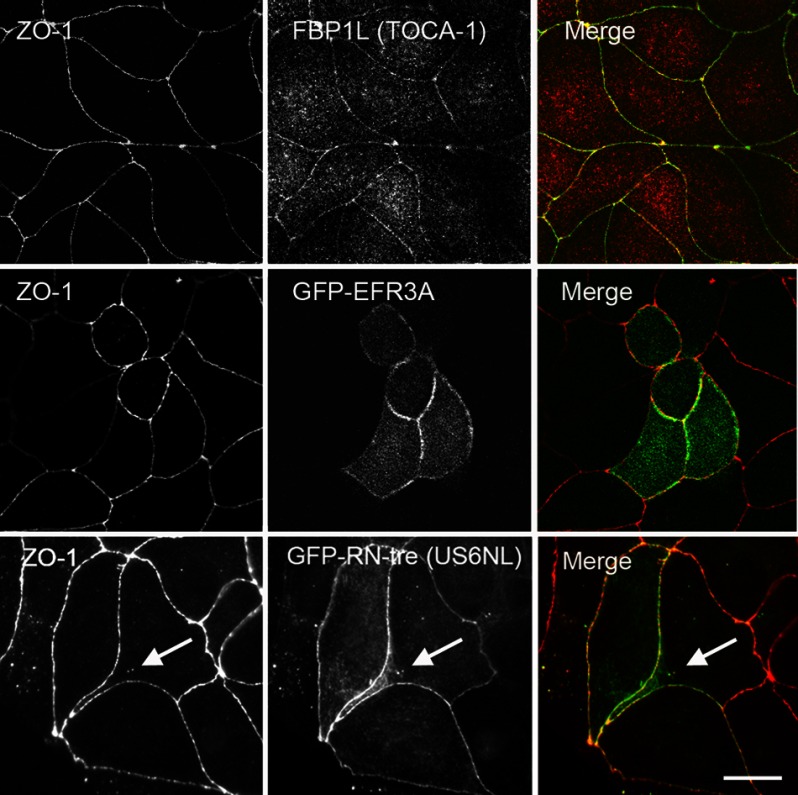
To test if some of the unexpected proteins identified by MS were partially associated with cell junctions, we analyzed the localization of FBP1L (TOCA-1), EFR3A, and US6NL (RN-tre) (Fig. 6). FBP1L and US6NL were chosen because they are both identified more in BL-ZO-1 than in ZO-1-BL samples and were recovered at relatively high levels. EFR3A was detected at lower levels, but a literature search suggested that it might play an important role in membrane identity (45). The availability of an antibody that could detect canine FBP1L allowed us to assess localization of endogenous FBP1L (Fig. 6, top middle panel). Although the FBP1L signal is localized throughout the cells, there is some clear colocalization with ZO-1 (Fig. 6, top left); this colocalization is evident in the yellow signal in the merged images (top right). In contrast, we were unable to identify antibodies to either EFR3A or US6NL that gave us clear signals on immunoblots from MDCK cells, so in these cases, we investigated the localization of GFP-tagged proteins. Transient transfection with a GFP-EFR3A-encoding plasmid (Fig. 6, second row, middle panel) showed that GFP-EFR3A clearly partially overlapped with the ZO-1 signal (second row, left panel); this can be seen in the merged signals (right panel), although the GFP-EFR3A is also distributed somewhat along the lateral membrane. Similarly, the distribution of GFP-US6NL (RN-tre) (Fig. 6, bottom row, middle panel) is also partially colocalized with ZO-1 (left panel, merge, right panel). In this case, both proteins are also present in occasional intracellular vesicles (white arrows).
FIGURE 6.
Confocal microscopy reveals that several proteins identified by biotinylation as ZO-1 neighbors partially colocalize with ZO-1 in MDCK cells. Top panels, endogenous FBP1L (TOCA-1; middle panel) is localized both intracellularly and at cell contacts. ZO-1 immunofluorescence (left panel) demarcates the tight junction; FBP1L staining overlaps with ZO-1 signal at the tight junction (right panel, Merge; green, ZO-1; red, FBP1L). Middle panels, similarly, ZO-1 (left panel) and GFP-EFR3A (middle panel) are partially colocalized (Merge, right panel; red, ZO-1; green, GFP), although GFP-EFR3A is also found inside cells and somewhat on the lateral membrane. Bottom panels, ZO-1 (left panel) also partially colocalizes with GFP-RN-tre (US6NL (middle panel) and Merge (left panel); red, ZO-1; green, GFP). In this case, colocalization can be also seen in several intracellular vesicles (white arrows). Bar, 10 μm.
DISCUSSION
Our findings identify a large set of proteins as ZO-1 neighbors, confirming known proximities and providing a rich set of proteins for further study of tight junction biology. In addition, fusing biotin ligase separately to the N- and C-terminal ends of ZO-1 allowed unexpected discrimination in identification of both shared and distinct proximal proteins at each end of ZO-1. This reveals the existence of distinct functional compartments within the junction and supports the notion that this labeling method is specific and can provide high spatial resolution. As expected, we identified many known tight junction proteins, but we also found many other unexpected, but in most cases easily rationalized, proteins that are likely to play a role in the regulation of tight junction structure and function. Others have used proteomics approaches to enrich the list of tight junction proteins, notably Yamazaki et al. (46). By first isolating junction-enriched membranes and then stripping peripheral proteins, they identified several novel transmembrane proteins that do not appear on our list presumably because they are not close to ZO-1 or do not have accessible lysines. The different outcome between our studies highlights the need to employ a variety of technically distinct approaches to identify the entire tight junction proteome.
Some methodological issues require that the proteins identified using biotin ligase fusion proteins be interpreted with a degree of caution. First, although biotinylated proteins appear to be concentrated at the tight junction (Fig. 1), the extended period (15 h) of exposure to biotin required for maximal labeling means the biotin ligase is also active during protein synthesis, trafficking to the tight junction, and at least the initial phases of degradation. Proteins proximal to ZO-1 during these processes will thus also be labeled with biotin. For example, a large number of ribosomal proteins were identified as proximal to ZO-1 (see “Experimental Procedures” and supplemental File S6). Because many of the same ribosomal proteins were tagged by the biotin ligase fusion protein alone, we assume that their identification may represent co-translational tagging; similarly, trafficking or other proteins may interact with ZO-1 in a non-junctional setting. In order to study dynamic as opposed to steady state proximities, this method would need to be refined to allow detectable labeling to occur during a much shorter time interval. Second, although MDCK cells are canine in origin, for reasons described above (see “Experimental Procedures”), we used the better curated and annotated SwissProt human database to identify peptides. The use of this human database meant that not all identified proteins were included in these analyses, but comparison of the human and dog results confirmed the identity and relative abundance of the proteins discussed above. Using the human database, we did not find some expected proteins (e.g. apical polarity proteins, E-cadherin, and β-catenin), but the absence of these proteins was verified in the protein lists from the RefSeq dog database. Third, detection of a neighbor requires that the neighbor have primary amines and that these primary amines be accessible to the ligase. In addition, the level of tagging cannot be assumed to be directly proportional to proximity because value is also influenced both by the cellular protein level and the number of lysines in the target.
Finally, the activity radius of the active BioAMP is unknown. Roux et al. (12) conservatively estimated that roughly 50% of the detected proteins were likely to reside within 20–30 nm of lamin A, their biotin ligase fusion protein. Our results suggest that the activity radius of BioAMP may be smaller, because purified ZO-1 has a Stokes radius of 8–9 nm (47) and yet we see differential and even exclusive recovery of proteins from ZO-1 tagged at the N or C terminus. However, definitive studies to define the active radius of the BioAMP have not yet been performed.
Despite these caveats, the use of ZO-1 biotin ligase fusion proteins appears to be an extremely useful method to identify proteins proximal to ZO-1 at the tight junction. The finding that a large number of tight and adherens junction proteins are tagged lends support to the overall specificity of this method. Previously recognized scaffolding, integral membrane, and cytoskeletal proteins were all found (Table 1). In addition, we confirmed reports for several proteins that had been localized to the tight or adherens junction on the basis of a single publication (e.g. LMO7 (44), KC1A (34), RHG12 (48), and SRBS2 (49)). Other proteins have not previously been specifically associated with tight junctions but were heavily tagged by ZO-1 (e.g. the F-bar-containing protein, FBP1L (38), US6NL (50), and the ASPP2-interacting protein, RASF8 (51)), suggesting that these proteins may be functionally important at this site. Partial colocalization of two of these proteins, FBP1L and US6NL, with ZO-1 was demonstrated by immunofluorescent confocal microscopy, as was colocalization of EFR3A, a palmitoylated protein that targets PI4KIIa to the plasma membrane (45). Finally, a number of uncharacterized proteins were tagged, including some transmembrane proteins with unknown functions (e.g. CL023, IGS11 (immunoglobulin superfamily member 11), and SLIT4 (Slit and Trk-like protein 4)); further investigation of these proteins may identify a contribution to tight junction structure or function.
In addition to single tagged proteins, we also find tagged proteins to cluster into groups of proteins with reported interactions that suggest they are components of local functional networks. One example of such a network is the subset of polarity and/or Wnt-signaling proteins tagged by ZO-1. Unexpectedly, none of the polarity proteins typically reported as associated with tight junctions and apical-basal polarity (reviewed in Ref. 52) was tagged by either ZO-1 fusion protein; this absence included Crb1 to -3, PALS1, and the PAR6-atypical protein kinase C-PAR3 complex. Instead, more laterally distributed polarity and associated proteins were identified, including MARK2 (full names and annotations appear in Table 2) and several MARK2 substrates, including the IBAR protein BAIP2 (53) and DVL1 and DVL3 (54); these latter proteins are members of the Wnt-signaling and planar cell polarity pathways. In addition, SCRIB, which co-immunoprecipitates with ZO-1 (55, 56), the SCRIB-interacting protein, LPP (57), and the planar cell polarity proteins VANG1 and VANG2 were identified. The only isoform of PAR3 tagged to a significant extent was PAR3L, which, unlike some other PAR3 splice forms, does not bind atypical PKC (58) but is recruited to tight junctions via an interaction with JAM; PAR3L recruits PP1A, which was also identified. Finally, ASPP2, which partially localized to tight junctions and interacts with MAGI1, PAR3, and RASF8 (51) was tagged; ASPP2 has been implicated both in cell polarity and in tight junction function (36, 37). Many of these proteins interact and also have effects on tight junction organization (53, 55, 56, 59–64).
Although tight junctions demarcate the apical and basolateral domains of polarized epithelia, they are not required for setting up these domains (65), and the interplay between tight junctions and polarity complexes is incompletely understood. However, it seems likely that at least in stable monolayers, the interactions with the basolaterally positioned polarity complex proteins are quantitatively more important than interactions with apical members. In addition, based on the localization of the streptavidin signal in Fig. 1D, ZO-1 fusion proteins have access to basolateral membrane domains but not the apical domain, which probably reveals dynamic interactions with adherens junction proteins (66). The role of the laterally distributed polarity proteins that we did find is less well defined than in those of the apical complex. However, recent data suggest that along with a minor function in apical/basolateral polarity (52), a number of the identified proteins, notably SCRIB, VANG, and DVL, are implicated in control of planar cell polarity and directed cell migration (reviewed in Ref. 67).
Finally, our findings that the two ends of ZO-1 are associated with different although overlapping neighborhoods of proximal proteins confirms the hypothesis that ZO-1 functions as a scaffold protein linking the transmembrane proteins of the tight junction to cytoskeletal proteins. In addition, the biased association of other types of proteins with the N and C terminus of ZO-1 suggests that these domains may also be differentially involved in subjunctional networks for signaling, polarity, and trafficking. For example, we found not only that claudins and occludin were enriched in samples tagged by the ligase fused to N-terminal end of ZO-1 but also that many of the above mentioned polarity proteins were also enriched in these same samples. Conversely, the proteins preferentially tagged by ZO-1-BL support the idea that this end is positioned within the cortical cytoskeleton. Even within “signaling” proteins, we find distinctions with most kinases and phosphatases near the N terminus and the erbin adaptor LAP2 and RhoGAP23 near the C terminus. Together these spatial distinctions point to a significant level of functional compartmentalization within the small dimensions of the tight junction. We are currently performing similar tagging methods with other known tight junction proteins with the goal of visualizing other functional subcompartments.
The biotin ligase tagging has provided a large and relatively unbiased list of ZO-1-proximal proteins. Many proteins on this list invite further investigation regarding their putative roles at the junction. As proof of principle, we have verified that three new proteins colocalize with ZO-1 and deserve further study of their biologic role at the junction. Overall, the use of this method will allow us to ask more informed questions about junction structure and regulation based on a more complete definition of the tight junction proteome.
Acknowledgments
We thank Sandra Citi (University of Geneva) for the generous gift of cingulin antibody. We thank Daniela Malide and Christian Combs (Light Microscopy Core Facility, NHLBI, National Institutes of Health) and Mark A. Knepper and Jason Hoffert (Systems Biology Center, NHLBI, National Institutes of Health) for invaluable help. We also thank Alan Fanning (University of North Carolina) for thoughtful discussions.
This work was supported, in whole or in part, by the National Institutes of Health Division of Intramural Research.

This article contains supplemental Files S1–S7.
- SH3
- Src homology 3
- PSM
- peptide spectrum match
- MDCK
- Madin-Darby canine kidney
- CXAR
- Coxsackie and adenovirus receptor
- OCLN
- occludin
- KC1A
- casein kinase 1 α.
REFERENCES
- 1. Anderson J. M., Van Itallie C. M. (2009) Physiology and function of the tight junction. Cold Spring Harb. Perspect. Biol. 1, a002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shen L., Weber C. R., Raleigh D. R., Yu D., Turner J. R. (2011) Tight junction pore and leak pathways. A dynamic duo. Annu. Rev. Physiol. 73, 283–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cao X., Surma M. A., Simons K. (2012) Polarized sorting and trafficking in epithelial cells. Cell Res. 22, 793–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rodgers L. S., Fanning A. S. (2011) Regulation of epithelial permeability by the actin cytoskeleton. Cytoskeleton 68, 653–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Furuse M. (2010) Molecular basis of the core structure of tight junctions. Cold Spring Harb. Perspect. Biol. 2, a002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stevenson B. R., Siliciano J. D., Mooseker M. S., Goodenough D. A. (1986) Identification of ZO-1. A high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J. Cell Biol. 103, 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Furuse M., Hirase T., Itoh M., Nagafuchi A., Yonemura S., Tsukita S. (1993) Occludin. A novel integral membrane protein localizing at tight junctions. J. Cell Biol. 123, 1777–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Furuse M., Sasaki H., Fujimoto K., Tsukita S. (1998) A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell Biol. 143, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Morita K., Furuse M., Fujimoto K., Tsukita S. (1999) Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc. Natl. Acad. Sci. U.S.A. 96, 511–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jesaitis L. A., Goodenough D. A. (1994) Molecular characterization and tissue distribution of ZO-2, a tight junction protein homologous to ZO-1 and the Drosophila discs-large tumor suppressor protein. J. Cell Biol. 124, 949–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haskins J., Gu L., Wittchen E. S., Hibbard J., Stevenson B. R. (1998) ZO-3, a novel member of the MAGUK protein family found at the tight junction, interacts with ZO-1 and occludin. J. Cell Biol. 141, 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roux K. J., Kim D. I., Raida M., Burke B. (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ikenouchi J., Umeda K., Tsukita S., Furuse M. (2007) Requirement of ZO-1 for the formation of belt-like adherens junctions during epithelial cell polarization. J. Cell Biol. 176, 779–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fanning A. S., Anderson J. M. (2009) Zonula occludens-1 and -2 are cytosolic scaffolds that regulate the assembly of cellular junctions. Ann. N.Y. Acad. Sci. 1165, 113–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Willott E., Balda M. S., Fanning A. S., Jameson B., Van Itallie C., Anderson J. M. (1993) The tight junction protein ZO-1 is homologous to the Drosophila discs-large tumor suppressor protein of septate junctions. Proc. Natl. Acad. Sci. U.S.A. 90, 7834–7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Itoh M., Furuse M., Morita K., Kubota K., Saitou M., Tsukita S. (1999) Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J. Cell Biol. 147, 1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Utepbergenov D. I., Fanning A. S., Anderson J. M. (2006) Dimerization of the scaffolding protein ZO-1 through the second PDZ domain. J. Biol. Chem. 281, 24671–24677 [DOI] [PubMed] [Google Scholar]
- 18. Ebnet K., Schulz C. U., Meyer Zu Brickwedde M. K., Pendl G. G., Vestweber D. (2000) Junctional adhesion molecule interacts with the PDZ domain-containing proteins AF-6 and ZO-1. J. Biol. Chem. 275, 27979–27988 [DOI] [PubMed] [Google Scholar]
- 19. Müller S. L., Portwich M., Schmidt A., Utepbergenov D. I., Huber O., Blasig I. E., Krause G. (2005) The tight junction protein occludin and the adherens junction protein α-catenin share a common interaction mechanism with ZO-1. J. Biol. Chem. 280, 3747–3756 [DOI] [PubMed] [Google Scholar]
- 20. Fanning A. S., Ma T. Y., Anderson J. M. (2002) Isolation and functional characterization of the actin binding region in the tight junction protein ZO-1. FASEB J. 16, 1835–1837 [DOI] [PubMed] [Google Scholar]
- 21. Fanning A. S., Jameson B. J., Jesaitis L. A., Anderson J. M. (1998) The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J. Biol. Chem. 273, 29745–29753 [DOI] [PubMed] [Google Scholar]
- 22. Citi S., Pulimeno P., Paschoud S. (2012) Cingulin, paracingulin, and PLEKHA7. Signaling and cytoskeletal adaptors at the apical junctional complex. Ann. N.Y. Acad. Sci. 1257, 125–132 [DOI] [PubMed] [Google Scholar]
- 23. Van Itallie C. M., Fanning A. S., Bridges A., Anderson J. M. (2009) ZO-1 stabilizes the tight junction solute barrier through coupling to the perijunctional cytoskeleton. Mol. Biol. Cell 20, 3930–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Van Itallie C. M., Fanning A. S., Anderson J. M. (2003) Reversal of charge selectivity in cation or anion-selective epithelial lines by expression of different claudins. Am. J. Physiol. Renal Physiol. 285, F1078–F1084 [DOI] [PubMed] [Google Scholar]
- 25. Akashi M., Higashi T., Masuda S., Komori T., Furuse M. (2011) A coronary artery disease-associated gene product, JCAD/KIAA1462, is a novel component of endothelial cell-cell junctions. Biochem. Biophys. Res. Commun. 413, 224–229 [DOI] [PubMed] [Google Scholar]
- 26. Gonzalez-Mariscal L., Chávez de Ramirez B., Cereijido M. (1985) Tight junction formation in cultured epithelial cells (MDCK). J. Membr. Biol. 86, 113–125 [DOI] [PubMed] [Google Scholar]
- 27. Huling J. C., Pisitkun T., Song J. H., Yu M. J., Hoffert J. D., Knepper M. A. (2012) Gene expression databases for kidney epithelial cells. Am. J. Physiol. Renal Physiol. 302, F401–F407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) DAVID. Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 29. UniProt Consortium (2012) Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 40, D71–D75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ivanov A. I., Bachar M., Babbin B. A., Adelstein R. S., Nusrat A., Parkos C. A. (2007) A unique role for nonmuscle myosin heavy chain IIA in regulation of epithelial apical junctions. PLoS One 2, e658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Katsube T., Takahisa M., Ueda R., Hashimoto N., Kobayashi M., Togashi S. (1998) Cortactin associates with the cell-cell junction protein ZO-1 in both Drosophila and mouse. J. Biol. Chem. 273, 29672–29677 [DOI] [PubMed] [Google Scholar]
- 32. Cohen C. J., Shieh J. T., Pickles R. J., Okegawa T., Hsieh J. T., Bergelson J. M. (2001) The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc. Natl. Acad. Sci. U.S.A. 98, 15191–15196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marx A., Nugoor C., Panneerselvam S., Mandelkow E. (2010) Structure and function of polarity-inducing kinase family MARK/Par-1 within the branch of AMPK/Snf1-related kinases. FASEB J. 24, 1637–1648 [DOI] [PubMed] [Google Scholar]
- 34. Dupre-Crochet S., Figueroa A., Hogan C., Ferber E. C., Bialucha C. U., Adams J., Richardson E. C., Fujita Y. (2007) Casein kinase 1 is a novel negative regulator of E-cadherin-based cell-cell contacts. Mol. Cell. Biol. 27, 3804–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dörfel M. J., Westphal J. K., Huber O. (2009) Differential phosphorylation of occludin and tricellulin by CK2 and CK1. Ann. N.Y. Acad. Sci. 1165, 69–73 [DOI] [PubMed] [Google Scholar]
- 36. Cong W., Hirose T., Harita Y., Yamashita A., Mizuno K., Hirano H., Ohno S. (2010) ASPP2 regulates epithelial cell polarity through the PAR complex. Curr. Biol. 20, 1408–1414 [DOI] [PubMed] [Google Scholar]
- 37. Sottocornola R., Royer C., Vives V., Tordella L., Zhong S., Wang Y., Ratnayaka I., Shipman M., Cheung A., Gaston-Massuet C., Ferretti P., Molnár Z., Lu X. (2010) ASPP2 binds Par-3 and controls the polarity and proliferation of neural progenitors during CNS development. Dev. Cell 19, 126–137 [DOI] [PubMed] [Google Scholar]
- 38. Giuliani C., Troglio F., Bai Z., Patel F. B., Zucconi A., Malabarba M. G., Disanza A., Stradal T. B., Cassata G., Confalonieri S., Hardin J. D., Soto M. C., Grant B. D., Scita G. (2009) Requirements for F-BAR proteins TOCA-1 and TOCA-2 in actin dynamics and membrane trafficking during Caenorhabditis elegans oocyte growth and embryonic epidermal morphogenesis. PLoS Genet. 5, e1000675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hu J., Mukhopadhyay A., Craig A. W. (2011) Transducer of Cdc42-dependent actin assembly promotes epidermal growth factor-induced cell motility and invasiveness. J. Biol. Chem. 286, 2261–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., Zheng P., Ye K., Chinnaiyan A., Halder G., Lai Z. C., Guan K. L. (2007) Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oka T., Schmitt A. P., Sudol M. (2012) Opposing roles of angiomotin-like-1 and zona occludens-2 on pro-apoptotic function of YAP. Oncogene 31, 128–134 [DOI] [PubMed] [Google Scholar]
- 42. Gonzalez-Mariscal L., Bautista P., Lechuga S., Quiros M. (2012) ZO-2, a tight junction scaffold protein involved in the regulation of cell proliferation and apoptosis. Ann. N.Y. Acad. Sci. 1257, 133–141 [DOI] [PubMed] [Google Scholar]
- 43. Katoh M. (2004) Identification and characterization of human ARHGAP23 gene in silico. Int. J. Oncol. 25, 535–540 [PubMed] [Google Scholar]
- 44. Ooshio T., Irie K., Morimoto K., Fukuhara A., Imai T., Takai Y. (2004) Involvement of LMO7 in the association of two cell-cell adhesion molecules, nectin and E-cadherin, through afadin and α-actinin in epithelial cells. J. Biol. Chem. 279, 31365–31373 [DOI] [PubMed] [Google Scholar]
- 45. Nakatsu F., Baskin J. M., Chung J., Tanner L. B., Shui G., Lee S. Y., Pirruccello M., Hao M., Ingolia N. T., Wenk M. R., De Camilli P. (2012) PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J. Cell Biol. 199, 1003–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yamazaki Y., Okawa K., Yano T., Tsukita S. (2008) Optimized proteomic analysis on gels of cell-cell adhering junctional membrane proteins. Biochemistry 47, 5378–5386 [DOI] [PubMed] [Google Scholar]
- 47. Anderson J. M., Stevenson B. R., Jesaitis L. A., Goodenough D. A., Mooseker M. S. (1988) Characterization of ZO-1, a protein component of the tight junction from mouse liver and Madin-Darby canine kidney cells. J. Cell Biol. 106, 1141–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Matsuda M., Kobayashi Y., Masuda S., Adachi M., Watanabe T., Yamashita J. K., Nishi E., Tsukita S., Furuse M. (2008) Identification of adherens junction-associated GTPase activating proteins by the fluorescence localization-based expression cloning. Exp. Cell Res. 314, 939–949 [DOI] [PubMed] [Google Scholar]
- 49. Murase K., Ito H., Kanoh H., Sudo K., Iwamoto I., Morishita R., Soubeyran P., Seishima M., Nagata K. (2012) Cell biological characterization of a multidomain adaptor protein, ArgBP2, in epithelial NMuMG cells, and identification of a novel short isoform. Med. Mol. Morphol. 45, 22–28 [DOI] [PubMed] [Google Scholar]
- 50. Lanzetti L., Rybin V., Malabarba M. G., Christoforidis S., Scita G., Zerial M., Di Fiore P. P. (2000) The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature 408, 374–377 [DOI] [PubMed] [Google Scholar]
- 51. Langton P. F., Colombani J., Chan E. H., Wepf A., Gstaiger M., Tapon N. (2009) The dASPP-dRASSF8 complex regulates cell-cell adhesion during Drosophila retinal morphogenesis. Curr. Biol. 19, 1969–1978 [DOI] [PubMed] [Google Scholar]
- 52. St Johnston D., Ahringer J. (2010) Cell polarity in eggs and epithelia. Parallels and diversity. Cell 141, 757–774 [DOI] [PubMed] [Google Scholar]
- 53. Cohen D., Fernandez D., Lázaro-Diéguez F., Müsch A. (2011) The serine/threonine kinase Par1b regulates epithelial lumen polarity via IRSp53-mediated cell-ECM signaling. J. Cell Biol. 192, 525–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sun T. Q., Lu B., Feng J. J., Reinhard C., Jan Y. N., Fantl W. J., Williams L. T. (2001) PAR-1 is a Dishevelled-associated kinase and a positive regulator of Wnt signalling. Nat. Cell Biol. 3, 628–636 [DOI] [PubMed] [Google Scholar]
- 55. Ivanov A. I., Young C., Den Beste K., Capaldo C. T., Humbert P. O., Brennwald P., Parkos C. A., Nusrat A. (2010) Tumor suppressor scribble regulates assembly of tight junctions in the intestinal epithelium. Am. J. Pathol. 176, 134–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yates L. L., Schnatwinkel C., Hazelwood L., Chessum L., Paudyal A., Hilton H., Romero M. R., Wilde J., Bogani D., Sanderson J., Formstone C., Murdoch J. N., Niswander L. A., Greenfield A., Dean C. H. (2013) Scribble is required for normal epithelial cell-cell contacts and lumen morphogenesis in the mammalian lung. Dev. Biol. 373, 267–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Petit M. M., Meulemans S. M., Alen P., Ayoubi T. A., Jansen E., Van de Ven W. J. (2005) The tumor suppressor Scrib interacts with the zyxin-related protein LPP, which shuttles between cell adhesion sites and the nucleus. BMC Cell Biol. 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gao L., Macara I. G., Joberty G. (2002) Multiple splice variants of Par3 and of a novel related gene, Par3L, produce proteins with different binding properties. Gene 294, 99–107 [DOI] [PubMed] [Google Scholar]
- 59. Traweger A., Wiggin G., Taylor L., Tate S. A., Metalnikov P., Pawson T. (2008) Protein phosphatase 1 regulates the phosphorylation state of the polarity scaffold Par-3. Proc. Natl. Acad. Sci. U.S.A. 105, 10402–10407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nethe M., de Kreuk B. J., Tauriello D. V., Anthony E. C., Snoek B., Stumpel T., Salinas P. C., Maurice M. M., Geerts D., Deelder A. M., Hensbergen P. J., Hordijk P. L. (2012) Rac1 acts in conjunction with Nedd4 and dishevelled-1 to promote maturation of cell-cell contacts. J. Cell Sci. 125, 3430–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Elbert M., Cohen D., Müsch A. (2006) PAR1b promotes cell-cell adhesion and inhibits dishevelled-mediated transformation of Madin-Darby canine kidney cells. Mol. Biol. Cell 17, 3345–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Anastas J. N., Biechele T. L., Robitaille M., Muster J., Allison K. H., Angers S., Moon R. T. (2012) A protein complex of SCRIB, NOS1AP and VANGL1 regulates cell polarity and migration and is associated with breast cancer progression. Oncogene 31, 3696–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Torban E., Kor C., Gros P. (2004) Van Gogh-like2 (Strabismus) and its role in planar cell polarity and convergent extension in vertebrates. Trends Genet. 20, 570–577 [DOI] [PubMed] [Google Scholar]
- 64. Torban E., Wang H. J., Groulx N., Gros P. (2004) Independent mutations in mouse Vangl2 that cause neural tube defects in looptail mice impair interaction with members of the Dishevelled family. J. Biol. Chem. 279, 52703–52713 [DOI] [PubMed] [Google Scholar]
- 65. Umeda K., Ikenouchi J., Katahira-Tayama S., Furuse K., Sasaki H., Nakayama M., Matsui T., Tsukita S., Furuse M. (2006) ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell 126, 741–754 [DOI] [PubMed] [Google Scholar]
- 66. Matter K., Balda M. S. (2003) Signalling to and from tight junctions. Nat. Rev. Mol. Cell Biol. 4, 225–236 [DOI] [PubMed] [Google Scholar]
- 67. Muthuswamy S. K., Xue B. (2012) Cell polarity as a regulator of cancer cell behavior plasticity. Annu. Rev. Cell Dev. Biol. 28, 599–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hull B. E., Staehelin L. A. (1979) The terminal web. A reevaluation of its structure and function. J. Cell Biol. 81, 67–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mandai K., Nakanishi H., Satoh A., Obaishi H., Wada M., Nishioka H., Itoh M., Mizoguchi A., Aoki T., Fujimoto T., Matsuda Y., Tsukita S., Takai Y. (1997) Afadin. A novel actin filament-binding protein with one PDZ domain localized at cadherin-based cell-to-cell adherens junction. J. Cell Biol. 139, 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ohnishi H., Nakahara T., Furuse K., Sasaki H., Tsukita S., Furuse M. (2004) JACOP, a novel plaque protein localizing at the apical junctional complex with sequence similarity to cingulin. J. Biol. Chem. 279, 46014–46022 [DOI] [PubMed] [Google Scholar]
- 71. Citi S., Sabanay H., Jakes R., Geiger B., Kendrick-Jones J. (1988) Cingulin, a new peripheral component of tight junctions. Nature 333, 272–276 [DOI] [PubMed] [Google Scholar]
- 72. Furuse M., Fujita K., Hiiragi T., Fujimoto K., Tsukita S. (1998) Claudin-1 and -2. Novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 141, 1539–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Butz S., Kemler R. (1994) Distinct cadherin-catenin complexes in Ca2+-dependent cell-cell adhesion. FEBS Lett. 355, 195–200 [DOI] [PubMed] [Google Scholar]
- 74. Yap A. S., Mullin J. M., Stevenson B. R. (1998) Molecular analyses of tight junction physiology. Insights and paradoxes. J. Membr. Biol. 163, 159–167 [DOI] [PubMed] [Google Scholar]
- 75. Martìn-Padura I., Lostaglio S., Schneemann M., Williams L., Romano M., Fruscella P., Panzeri C., Stoppacciaro A., Ruco L., Villa A., Simmons D., Dejana E. (1998) Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol. 142, 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ide N., Hata Y., Nishioka H., Hirao K., Yao I., Deguchi M., Mizoguchi A., Nishimori H., Tokino T., Nakamura Y., Takai Y. (1999) Localization of membrane-associated guanylate kinase (MAGI)-1/BAI-associated protein (BAP) 1 at tight junctions of epithelial cells. Oncogene 18, 7810–7815 [DOI] [PubMed] [Google Scholar]
- 77. Wu Y., Dowbenko D., Spencer S., Laura R., Lee J., Gu Q., Lasky L. A. (2000) Interaction of the tumor suppressor PTEN/MMAC with a PDZ domain of MAGI3, a novel membrane-associated guanylate kinase. J. Biol. Chem. 275, 21477–21485 [DOI] [PubMed] [Google Scholar]
- 78. Takai Y., Ikeda W., Ogita H., Rikitake Y. (2008) The immunoglobulin-like cell adhesion molecule nectin and its associated protein afadin. Annu. Rev. Cell Dev. Biol. 24, 309–342 [DOI] [PubMed] [Google Scholar]
- 79. Etournay R., Zwaenepoel I., Perfettini I., Legrain P., Petit C., El-Amraoui A. (2007) Shroom2, a myosin-VIIa- and actin-binding protein, directly interacts with ZO-1 at tight junctions. J. Cell Sci. 120, 2838–2850 [DOI] [PubMed] [Google Scholar]
- 80. Patrie K. M., Drescher A. J., Welihinda A., Mundel P., Margolis B. (2002) Interaction of two actin-binding proteins, synaptopodin and α-actinin-4, with the tight junction protein MAGI-1. J. Biol. Chem. 277, 30183–30190 [DOI] [PubMed] [Google Scholar]
- 81. Lee D. B., Jamgotchian N., Allen S. G., Kan F. W., Hale I. L. (2004) Annexin A2 heterotetramer. Role in tight junction assembly. Am. J. Physiol. Renal Physiol. 287, F481–F491 [DOI] [PubMed] [Google Scholar]
- 82. Luton F., Klein S., Chauvin J. P., Le Bivic A., Bourgoin S., Franco M., Chardin P. (2004) EFA6, exchange factor for ARF6, regulates the actin cytoskeleton and associated tight junction in response to E-cadherin engagement. Mol. Biol. Cell 15, 1134–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Routhier E. L., Burn T. C., Abbaszade I., Summers M., Albright C. F., Prendergast G. C. (2001) Human BIN3 complements the F-actin localization defects caused by loss of Hob3p, the fission yeast homolog of Rvs161p. J. Biol. Chem. 276, 21670–21677 [DOI] [PubMed] [Google Scholar]
- 84. Chen X., Macara I. G. (2006) Par-3 mediates the inhibition of LIM kinase 2 to regulate cofilin phosphorylation and tight junction assembly. J. Cell Biol. 172, 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ligon L. A., Karki S., Tokito M., Holzbaur E. L. (2001) Dynein binds to β-catenin and may tether microtubules at adherens junctions. Nat. Cell Biol. 3, 913–917 [DOI] [PubMed] [Google Scholar]
- 86. Lie P. P., Mruk D. D., Lee W. M., Cheng C. Y. (2009) Epidermal growth factor receptor pathway substrate 8 (Eps8) is a novel regulator of cell adhesion and the blood-testis barrier integrity in the seminiferous epithelium. FASEB J. 23, 2555–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kim Y., Ikeda W., Nakanishi H., Tanaka Y., Takekuni K., Itoh S., Monden M., Takai Y. (2002) Association of frabin with specific actin and membrane structures. Genes Cells 7, 413–420 [DOI] [PubMed] [Google Scholar]
- 88. Laura R. P., Witt A. S., Held H. A., Gerstner R., Deshayes K., Koehler M. F., Kosik K. S., Sidhu S. S., Lasky L. A. (2002) The Erbin PDZ domain binds with high affinity and specificity to the carboxyl termini of δ-catenin and ARVCF. J. Biol. Chem. 277, 12906–12914 [DOI] [PubMed] [Google Scholar]
- 89. Cohen D., Brennwald P. J., Rodriguez-Boulan E., Müsch A. (2004) Mammalian PAR-1 determines epithelial lumen polarity by organizing the microtubule cytoskeleton. J. Cell Biol. 164, 717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sharma S., Ang S. L., Shaw M., Mackey D. A., Gécz J., McAvoy J. W., Craig J. E. (2006) Nance-Horan syndrome protein, NHS, associates with epithelial cell junctions. Hum. Mol. Genet. 15, 1972–1983 [DOI] [PubMed] [Google Scholar]
- 91. Walsh G. S., Grant P. K., Morgan J. A., Moens C. B. (2011) Planar polarity pathway and Nance-Horan syndrome-like 1b have essential cell-autonomous functions in neuronal migration. Development 138, 3033–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Fletcher S. J., Poulter N. S., Haining E. J., Rappoport J. Z. (2012) Clathrin-mediated endocytosis regulates occludin, and not focal adhesion, distribution during epithelial wound healing. Biol. Cell 104, 238–256 [DOI] [PubMed] [Google Scholar]
- 93. Stamatovic S. M., Sladojevic N., Keep R. F., Andjelkovic A. V. (2012) Relocalization of junctional adhesion molecule A during inflammatory stimulation of brain endothelial cells. Mol. Cell. Biol. 32, 3414–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zimmermann P., Tomatis D., Rosas M., Grootjans J., Leenaerts I., Degeest G., Reekmans G., Coomans C., David G. (2001) Characterization of syntenin, a syndecan-binding PDZ protein, as a component of cell adhesion sites and microfilaments. Mol. Biol. Cell 12, 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Soubeyran P., Kowanetz K., Szymkiewicz I., Langdon W. Y., Dikic I. (2002) Cbl-CIN85-endophilin complex mediates ligand-induced downregulation of EGF receptors. Nature 416, 183–187 [DOI] [PubMed] [Google Scholar]
- 96. Salazar M. A., Kwiatkowski A. V., Pellegrini L., Cestra G., Butler M. H., Rossman K. L., Serna D. M., Sondek J., Gertler F. B., De Camilli P. (2003) Tuba, a novel protein containing bin/amphiphysin/Rvs and Dbl homology domains, links dynamin to regulation of the actin cytoskeleton. J. Biol. Chem. 278, 49031–49043 [DOI] [PubMed] [Google Scholar]