Background: Phospholemman regulates the plasmalemmal sodium pump in excitable tissues.
Results: In cardiac muscle, a subpopulation of phospholemman with a unique phosphorylation signature associates with other phospholemman molecules but not with the pump.
Conclusion: Phospholemman oligomers exist in cardiac muscle.
Significance: Much like phospholamban regulation of SERCA, phospholemman exists as both a sodium pump inhibiting monomer and an unassociated oligomer.
Keywords: Caveolae; Heart; Na,K-ATPase; PP2A; Protein Palmitoylation; Protein Phosphatase; Protein Phosphorylation; Serine-Threonine Protein Phosphatase; FXYD Proteins; Phospholemman
Abstract
Phospholemman (PLM), the principal quantitative sarcolemmal substrate for protein kinases A and C in the heart, regulates the cardiac sodium pump. Much like phospholamban, which regulates the related ATPase SERCA, PLM is reported to oligomerize. We investigated subpopulations of PLM in adult rat ventricular myocytes based on phosphorylation status. Co-immunoprecipitation identified two pools of PLM: one not associated with the sodium pump phosphorylated at Ser63 and one associated with the pump, both phosphorylated at Ser68 and unphosphorylated. Phosphorylation of PLM at Ser63 following activation of PKC did not abrogate association of PLM with the pump, so its failure to associate with the pump was not due to phosphorylation at this site. All pools of PLM co-localized to cell surface caveolin-enriched microdomains with sodium pump α subunits, despite the lack of caveolin-binding motif in PLM. Mass spectrometry analysis of phosphospecific immunoprecipitation reactions revealed no unique protein interactions for Ser63-phosphorylated PLM, and cross-linking reagents also failed to identify any partner proteins for this pool. In lysates from hearts of heterozygous transgenic animals expressing wild type and unphosphorylatable PLM, Ser63-phosphorylated PLM co-immunoprecipitated unphosphorylatable PLM, confirming the existence of PLM multimers. Dephosphorylation of the PLM multimer does not change sodium pump activity. Hence like phospholamban, PLM exists as a pump-inhibiting monomer and an unassociated oligomer. The distribution of different PLM phosphorylation states to different pools may be explained by their differential proximity to protein phosphatases rather than a direct effect of phosphorylation on PLM association with the pump.
Introduction
The FXYD family of type 1 membrane proteins (1) regulates the sodium pump (Na pump)2 by modifying its substrate affinities and maximum transport rate (2). In cardiac myocytes, the Na pump associates with FXYD1 (phospholemman (PLM)) (3, 4). Phosphoregulation of the cardiac Na pump by the adrenergic system is through phosphorylation of PLM (4–10), which is unique in the FXYD family in having multiple phosphorylation sites on its intracellular carboxyl terminus (5, 11). Unphosphorylated PLM inhibits the cardiac Na pump, and phosphorylation of PLM by PKA (on serine 68) or PKC (on serines 63 and 68 and threonine 69) relieves this inhibition (4–10, 12). Activation of the cardiac sodium pump via phosphorylation of PLM is necessary to protect against catecholamine-induced sodium and calcium overload, which leads to cardiac arrhythmias in vivo (13).
Parallels exist between regulation of the Na pump by PLM and regulation of the closely related P-type ATPase sarco/endoplasmic reticulum Ca-ATPase (SERCA 2a) by phospholamban (PLB). Unphosphorylated PLB inhibits SERCA, and PLB phosphorylation partially relieves this inhibition (14). It is proposed that PLB exists as a homo-pentamer in the sarcoplasmic reticulum membrane, but that the active species that inhibits SERCA 2a is a PLB monomer in equilibrium with this homopentamer (15, 16). Phosphorylation of PLB promotes pentamer formation (17), although the precise molecular details of the PLB-SERCA relationship remain incomplete. SERCA either dissociates from PLB in the calcium-bound E1 state (18, 19) or calcium binding reorganizes PLB relative to SERCA (20, 21), a phenomenon that has not been reported for PLM (although ouabain, which stabilizes the E2 conformation of the Na pump, abolishes FRET but not co-immunoprecipitation between PLM and the pump α subunit (7)). Like PLB, PLM is reported to homo-oligomerize: it was originally proposed to form an ion channel in its own right (22), and although a role in regulation of cell volume remains unproven (23), stable tetramers of the PLM transmembrane domain have been observed in perfluoro-octaneoate polyacrylamide gels (24). Fusion proteins of PLM-YFP and PLM-CFP exhibit significant intermolecular FRET in HEK cells, which is enhanced upon PLM phosphorylation (25), suggesting that oligomerization is promoted by both transmembrane and intracellular regions of the protein. Most recently, FRET measurements of phosphomimetic PLM fluorescent protein fusions expressed in HEK cells have suggested that the PLM oligomer is a tetramer (26).
The existence of PLM oligomers has not been reported in cardiac muscle, raising the possibility that PLM oligomerization is an artifact caused by its overexpression or fusion to a fluorescent protein. The aim of this investigation was to characterize a pool of PLM in cardiac muscle that we find is not associated with the Na pump. We report the existence of a subpopulation of PLM that interacts only with other PLM molecules and not with the Na pump, with a unique phosphorylation status driven by differential proximity of protein phosphatases.
EXPERIMENTAL PROCEDURES
Drugs, Antibodies, and Chemicals
Antibodies to the PLM amino terminus (4) and phosphorylation sites (5) were as previously described. Antibody C2 (specific for unphosphorylated PLM) was kindly provided by Dr. J. Y. Cheung (Temple University). The monoclonal antibodies α6F and α5 raised against the sodium pump α1 subunit and all sodium pump α subunits, respectively, by Douglas M. Fambrough were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD, National Institutes of Health and maintained by the University of Iowa Department of Biology (Iowa City, IA). The anti-sodium pump α2 subunit was from Millipore. The anti-clathrin heavy chain and anti-PP2A catalytic subunit were from BD Biosciences. All other antibodies were as previously described (27). Unless indicated otherwise, all reagents were obtained from Sigma and were of the highest grade available.
Adult Rat Ventricular Myocytes
Calcium-tolerant adult rat ventricular myocytes (ARVMs) were isolated by retrograde perfusion of collagenase in the Langendorff mode. The myocytes were left to recover for 2 h at 35 °C before experiments. All of the drugs were applied at 35 °C.
Quantitative Western Immunoblotting
Chemiluminescent images were obtained using the Bio-Rad ChemiDoc XRS imaging system, and band density was quantified using the Quantity One software package (Bio-Rad).
Immunoprecipitation
Immunoprecipitation was largely carried out as described previously (5). Briefly, for co-immunoprecipitation cells, myocytes or sonicated (six times for 20 s, 5-μm amplitude) whole ventricular homogenates were lysed in 2 mg/ml C12E10 or 1% Nonidet P-40 in PBS supplemented with protease (Calbiochem) and phosphatase (Sigma) inhibitor cocktails, as well as a mixture of chemical phosphatase inhibitors: 5 mm sodium fluoride, 2 mm sodium orthovanadate, 2 mm sodium pyrophosphate, 2 mm sodium glycerophosphate. Insoluble material was removed by centrifugation, and targets were captured overnight using antibodies that had been preimmobilized on protein A- or G-Sepharose beads (GE Healthcare). The next day, the beads were extensively washed, and immunoprecipitated proteins were eluted in SDS-PAGE sample buffer and separated by SDS-PAGE. To ensure specificity, co-immunoprecipitation reactions were routinely immunoblotted for membrane proteins not co-purifying with the immunoprecipitated proteins: Plasma Membrane Ca ATPase 1, 3 ketoacyl coenzyme A thiolase α subunit, and PLB (not shown).
For immunoprecipitation, ARVMs or whole ventricular homogenates were lysed for 30 min in 1% SDS in PBS supplemented with protease and phosphatase inhibitors. SDS concentration was reduced by the addition of 10 volumes of 1% Triton in PBS, after which insoluble material was removed by centrifugation, and an identical protocol was followed as for co-immunoprecipitation.
Immunofluorescence
ARVMs were plated onto laminin-coated coverslips, washed with PBS, and fixed with 0.5% formaldehyde in PBS for 30 min at room temperature. The cells were permeabilized with 0.15% Triton X-100 in PBS for 15 min, incubated at room temperature in 0.1 m NH4Cl for 10 min, washed with PBS, and blocked with blocking buffer (5% donkey serum, 0.01% Triton X-100 in PBS) for 3 h at room temperature. All of the antibodies were diluted in blocking buffer; phosphospecific antibodies were preincubated with 1 μg/ml unphosphorylated blocking peptide for 30 min prior to use. Fixed ARVMs were immunolabeled with 5 μg/ml anti-PLM Ser63 or 5 μg/ml anti-PLM Ser68 and detected with Alexa Fluor 594-conjugated anti-sheep (1:400, Invitrogen). Fluorescent images were obtained using a Leica TCS LS inverted confocal microscope and analyzed using Leica LAS AF software.
Sucrose Gradient Fractionation of Caveolin-enriched Membranes
Caveolin-enriched buoyant membranes were prepared from ARVMs homogenized (five 6-s bursts Ultra-Turrax with 20-s intervals) and sonicated (six 20-s bursts, 5-μm amplitude; Soniprep) in 500 mm sodium carbonate, pH 11, supplemented with 1 mm DTT, 1 mm EDTA, protease, and phosphatase inhibitors. The lysates were immediately adjusted to 45% sucrose by the addition of an equal volume of 90% (w/v) sucrose in (25 mm MES, 150 mm NaCl, pH 6.5), and 4 ml was transferred to an ultracentrifuge tube and overlaid with 4 ml of 35% sucrose and 4 ml of 5% sucrose (dissolved in the same combination of salts). Following overnight centrifugation at 270,000 × g, the uppermost three 1-ml fractions were discarded, the next nine fractions (fractions 4–12) were collected, and equal volumes of each were separated by SDS-PAGE and analyzed by immunoblotting. Caveolin 3-enriched caveolar membranes were concentrated in fractions 4 and 5. The purity of caveolar fractions was routinely assessed by immunoblotting for mitochondrial (3 ketoacyl coenzyme A thiolase α subunit; not shown) and bulk sarcolemmal marker proteins (clathrin heavy chain). Caveolae were considered pure if ≥99% mitochondrial and bulk sarcolemmal marker proteins were excluded from gradient fraction 4.
Acyl Resin-assisted Capture
We adapted a recently published method for purification of acylated proteins by resin-assisted capture (28). ARVMs were lysed in 2.5% SDS, 100 mm HEPES, 1 mm EDTA, pH 7.4, and free cysteines were alkylated by addition of 1% methyl methanethiosulfonate and incubation at 40 °C for 4 h. Excess unreacted methyl methanethiosulfonate was removed by acetone precipitation, and protein pellets were extensively washed with 70% acetone, dried, and then resolubilized in 1% SDS, 100 mm HEPES, 1 mm EDTA, pH 7.4 (binding buffer). Acylated proteins were captured on pre-equilibrated thiopropyl-Sepharose (GE Life Sciences) in the presence of 200 mm hydroxylamine (pH 7.4) for 2.5 h at room temperature. An identical reaction in which hydroxylamine was replaced with 200 mm NaCl served as a negative control. Following capture of acylated proteins, the beads were extensively washed in binding buffer, and proteins were eluted by heating for 10 min at 60 °C in SDS-PAGE loading buffer supplemented with 100 mm DTT. Unfractionated (after methyl methanethiosulfonate block), unbound (not captured by thioproyl-Sepharose in the presence of hydroxylamine), and acylated (captured by thiopropyl-Sepharose beads) fractions were routinely analyzed to assess depletion of proteins from the unbound fraction and their enrichment in the acylated fraction.
Biotinylation and Purification of Cell Surface Proteins
Cell surface proteins were prepared from ARVMs that had been plated on laminin-coated multiwell dishes using a protocol identical to that previously described for immortalized cell lines (27).
Mass Spectrometry
Phosphospecific immunoprecipitation reactions were carried out using antibodies specific for unphosphorylated, Ser63-phosphorylated, and Ser68-phosphorylated PLM that had been cross-linked to protein A- or G-Sepharose beads using dimethyl pimelimidate. After overnight immunoprecipitation and extensive washes, immunoprecipitated proteins were eluted with 1 m glycine, pH 3. A sample was retained for analysis by SDS-PAGE, and the remainder was reductively alkylated by addition of 2 volumes of 2 m urea, 200 mm ammonium bicarbonate, 20 mm DTT (then incubated for 60 min at room temperature), followed by a further 2 volumes of 50 mm iodoacetamide in water (then incubated for 30 min at room temperature in the dark). Protein mixtures were concentrated and buffer-exchanged into 0.8 m urea, 20 mm ammonium bicarbonate, 2 mm DTT using 3-kDa cutoff centrifugal concentrators (Millipore), then digested with trypsin (Promega), acidified with 0.1% TFA, and loaded directly onto a C18 liquid chromatography column connected to a LTQ Orbitrap Velos Pro (Thermo). The peptides were matched and assigned using Mascot with a minimum of two peptides with an ion score of >34 required for identification.
For analysis, rat IPI identifiers supplied by Mascot were mapped to human Uniprot identifiers using a custom-built IPI to Uniprot dictionary. No proteins were excluded from the initial analysis of overlap using the VENNY Venn diagram tool (Ref. 29 and supplemental Table S1); however, likely contaminants based on mitochondrial/sarcoplasmic reticulum subcellular localization are annotated in supplemental Table S1. For the analysis presented in Fig. 4 and Table 1, only keratin contaminants were removed from the protein lists.
FIGURE 4.
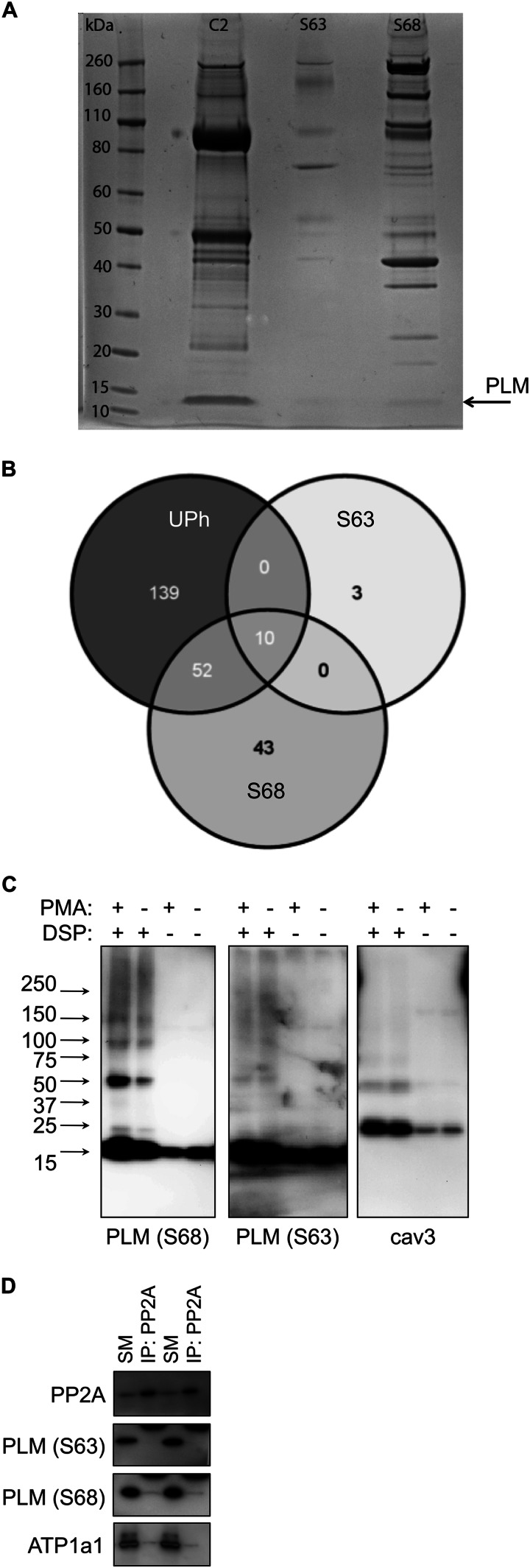
Protein interactions of Ser63-phosphorylated PLM. A, co-immunoprecipitation reactions using antibodies specific for unphosphorylated (C2), Ser63-phosphorylated (S63), and Ser68-phosphorylated (S68) PLM. Despite purifying similar quantities of PLM, the number and abundance of co-purifying proteins are substantially lower in the Ser63 immunoprecipitation reaction compared with the Ser68 immunoprecipitation reaction. B, co-immunoprecipitation reactions shown in A were analyzed by LC-MS/MS to identify all proteins. Venn diagram showing overlap in the proteins identified from each reaction. Protein identifications are shown in Table 1 and supplemental Table S1. UPh, unphosphorylated. C, control and PMA-treated ARVMs (300 nm, 10 min) were treated with the homobifunctional cross-linker DSP (1 mm, 60 min at 4 °C) and immunoblotted as indicated. No unique interactions for Ser63-phosphorylated PLM were revealed. cav3, caveolin 3. D, PP2A catalytic subunit was immunoprecipitated from ARVMs, and co-purifying sodium pump α1 and PLM phosphorylation states were immunoblotted. Ser63-phosphorylated PLM does not co-purify with PP2A. Duplicate immunoprecipitations are shown. SM, immunoprecipitation starting material; IP, immunoprecipitation.
TABLE 1.
Proteins identified by LC-MS/MS co-immunoprecipitated with unphosphorylated, Ser63-phosphorylated, and Ser68-phosphorylated PLM and proteins uniquely co-immunoprecipitated with Ser63-phosphorylated PLM
| Uniprot id | Entry name | Protein name |
|---|---|---|
| Common proteins in unphosphorylated and Ser63- and Ser68-phosphorylated immunoprecipitations | ||
| P05023 | ATP1A1 | Na pump subunit α1 |
| P50993 | ATP1A2 | Na pump subunit α2 |
| P05026 | ATP1B1 | Na pump subunit β1 |
| P13533 | MYH6 | Myosin-6 |
| Q562R1 | ACTBL | β-Actin-like protein 2 |
| P08590 | MYL3 | Myosin light chain 3 |
| P38646 | GRP75 | Stress-70 protein, mitochondrial |
| P07477 | TRY1 | Trypsin-1 |
| P02585 | TNNC2 | Troponin C, skeletal muscle |
| P09493 | TPM1 | Tropomyosin α1 chain |
| Proteins only in Ser63 immunoprecipitations | ||
| Q9UKX2 | MYH2 | Myosin heavy chain 2 |
| P05976 | MYL1 | Myosin light chain 1/3, skeletal muscle isoform |
| O14983 | ATP2A1 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 |
Cross-linking
Intact ARVMs were exposed to the amine reactive, cell-permeable cross-linking reagent dithiobis[succinimidyl propionate] (DSP, 1 mm; Pierce) in PBS for 60 min at 4 °C. Cross-linking reactions were quenched with nonreducing SDS-PAGE sample buffer. Formaldehyde was applied to intact ARVMs in PBS for 20 min at 35 °C, after which myocytes were recovered by centrifugation, washed three times with PBS, and solubilized with SDS-PAGE loading buffer. Unless indicated, the samples were heated at 60 °C for 10 min prior to electrophoresis.
Unphosphorylatable PLM Knockin Transgenic Mouse Line
This animal model is described in detail in manuscripts under preparation. All of the animals were age-matched and were either littermates or cousins. Following terminal anesthesia (intraperitoneal injection of sodium pentobarbital (200 mg/kg) in combination with sodium heparin (200 IU/kg)), the hearts were removed and briefly perfused in the Langendorff mode to remove contaminating blood before being snap frozen prior to homogenization and sonication.
Phos-Tag PAGE
PLM phosphorylation states were resolved using Phos-Tag PAGE (30). Proteins were resolved using standard SDS-PAGE in which the resolving gel was supplemented with 62.5 μm Phos-Tag reagent (Wako Chemical GmBH), and 125 μm MnCl2. Prior to transfer, the gels were washed for 20 min with standard transfer buffer supplemented with 2 mm EDTA and then washed twice more for 10 min with standard transfer buffer.
Whole Cell Pump Currents in ARVMs
Pump currents were measured with 50 mm pipette sodium as described previously (5). Briefly, ARVMs were perfused with 1 μm bisindolylmaleimide or 0.1% (v/v) Me2SO (vehicle) at 35 °C. Whole cell patch clamp recordings were established between 10 and 20 min after drug application, and sodium/potassium pump current was measured as the potassium-dependent current inhibited on switching from 5 to 0 mm potassium solution at 0 mV.
Modeling
We used our existing model of PLM with the Na pump α1 subunit (10, 27, 31). Palmitates were built onto the cysteines of PLM using PyMOL and extended in the direction lipid tails would likely adopt in the phospholipid membrane.
Statistical Analysis
The quantitative data are presented as the means ± S.E. The differences between experimental groups were analyzed by one-way analysis of variance, followed by post hoc t tests. The differences were considered statistically significant when p < 0.05.
RESULTS
PLM Phosphorylated at Ser63 Is Not Associated with the Sodium Pump α Subunit
PLM is substantially phosphorylated at Ser63 and Ser68 but not Thr69 in unstimulated ARVMs (5). We immunoprecipitated phosphorylated and unphosphorylated PLM from freshly isolated ARVMs under conditions designed to favor co-purification of sodium pump subunits. Representative immunoblots are shown in Fig. 1A. Note that immunoprecipitation of Thr69-phosphorylated PLM purifies very little PLM, because this site is largely unphosphorylated in these cells. The antibody raised to unphosphorylated PLM shows some cross-reactivity with Ser68-phosphorylated PLM but essentially none with Ser63-phosphorylated PLM. Most strikingly, although Ser63-phosphorylated PLM is abundant in ARVMs (5) and the Ser63-specific antibody purifies a substantial quantity of PLM, very little sodium pump α1 subunit is co-immunoprecipitated (Fig. 1A). This is in stark contrast to Ser68-specific immunoprecipitations, which purify a similar quantity of PLM but co-purify considerably more sodium pump α1 subunit.
FIGURE 1.
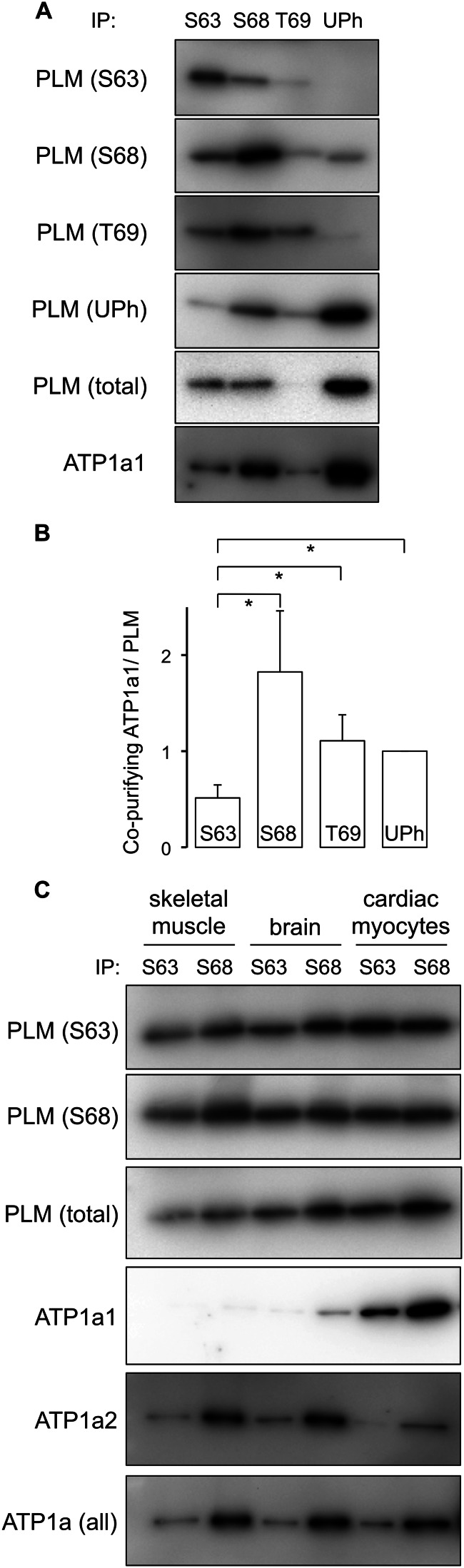
PLM phosphorylated at Ser63 co-immunoprecipitates poorly with the Na pump α subunit. A, PLM was immunoprecipitated (IP) from ARVMs using antibodies specific for Ser63 (S63), Ser68 (S68), and Thr69 (T69) phosphorylated and unphosphorylated (UPh) forms. In the representative immunoblots shown, although the PLM-Ser63 antibody precipitates a substantial amount of PLM, co-precipitation of sodium pump α1 subunit is relatively poor, particularly compared with the adjacent Ser68 immunoprecipitation, which precipitates the same amount of PLM but a considerably larger amount of sodium pump α1 subunit. B, quantitative analysis of Western blot data presented in A. For comparison purposes, the quantity of sodium pump α1 subunit co-precipitated by each antibody was normalized to the quantity of PLM precipitated in the same reaction. To allow comparison of multiple experiments, this ratio was arbitrarily defined as equal to 1 for the immunoprecipitation reactions using antibodies specific for the unphosphorylated form of PLM. *, p < 0.05 (n = 7). C, co-immunoprecipitation reactions from homogenates of whole brain, skeletal muscle (quadriceps), and ARVMs using antibodies specific for PLM phosphorylated at Ser63 and Ser68. Na pump α subunits from both brain and skeletal muscle exhibit the same preferential enrichment of the pump α subunit in PLM-Ser68 immunoprecipitation reactions only.
Quantitative analysis of phosphospecific immunoprecipitations is presented in Fig. 1B. For comparison purposes, the quantity of sodium pump α1 subunit co-precipitated by each antibody was normalized to the quantity of PLM precipitated in the same reaction. For comparison of multiple experiments, this ratio was arbitrarily defined as equal to 1 for immunoprecipitation reactions using antibodies raised to the unphosphorylated form of PLM. Fig. 1B highlights that although quantitatively similar amounts of sodium pump α1 subunit are co-purified with antibodies specific for Ser68-phosphorylated, Thr69-phosphorylated, and unphosphorylated PLM, very little pump α1 subunit co-purifies with Ser63-phosphorylated PLM.
The poor co-purification of pump α1 subunit with Ser63-phosphorylated PLM may reflect a Na pump subunit-specific association of this phosphorylation state in ARVMs. The α1 subunit is the dominant isoform in ARVMs (70% of total pumping capacity (32)). Na pump α2 subunit has been proposed to regulate sarcoplasmic reticulum calcium content through its functional coupling to NCX1 (33, 34) and is specifically functionally coupled to PLM phosphorylated at Ser63 (10). Fig. 1C indicates that we found essentially no pump α2 subunit associated with Ser63-phosphorylated PLM in ARVMs, whereas pump α2 could be readily detected co-purifying with Ser68-phosphorylated PLM.
We next investigated whether the relative inability of Ser63-phosphorylated PLM to co-purify Na pump α subunit is a cardiac-specific phenomenon by immunoprecipitating PLM phosphorylation states from other tissues expressing PLM. Fig. 1C indicates that an almost identical phenomenon occurs when PLM is immunoprecipitated from skeletal muscle (quadriceps). The dominant pump isoform in this tissue is the α2 subunit, which co-purifies extremely poorly with Ser63-phosphorylated PLM but very well with Ser68-phosphorylated PLM. When Ser63- and Ser68-phosphorylated PLM were immunoprecipitated from whole brain, an antibody that reacts with all forms of pump α subunit shows the same preference for co-purification of α with Ser68-phosphorylated rather than Ser63-phosphorylated PLM (this is predominantly of pump α3 subunit, the dominant isoform in the brain). Hence the failure of Ser63-phosphorylated PLM to co-purify pump subunits is common to at least three tissues in which PLM is expressed and occurs irrespective of the dominant isoform of the α subunit expressed.
Phosphorylation of PLM at Ser63 Does Not Abrogate Association of PLM with Na Pump α Subunit
The physical interaction between PLM and Na pump α subunits is well established (5, 7), so we considered the unlikely possibility that the failure of our antibody specific for Ser63-phosphorylated PLM to co-immunoprecipitate the pump α1 subunit in ARVMs is an experimental artifact. Experiments depicted in Fig. 2 indicate that this is not the case. First, we increased the availability of Ser63-phosphorylated PLM in ARVM lysates by treating with PMA prior to lysis (Fig. 2A). Increasing the quantity of Ser63-phosphorylated PLM by activating PKC does not increase the quantity of PLM immunoprecipitated by the Ser63-specific antibody (because the amount of antibody in the immunoprecipitation experiment is limiting), but it does increase the quantity of sodium pump α1 subunit co-precipitated. This suggests that two pools of PLM are available to the PLM-Ser63 antibody: one not associated with the pump (in untreated cells) and one associated with the pump (in PMA-treated cells only).
FIGURE 2.
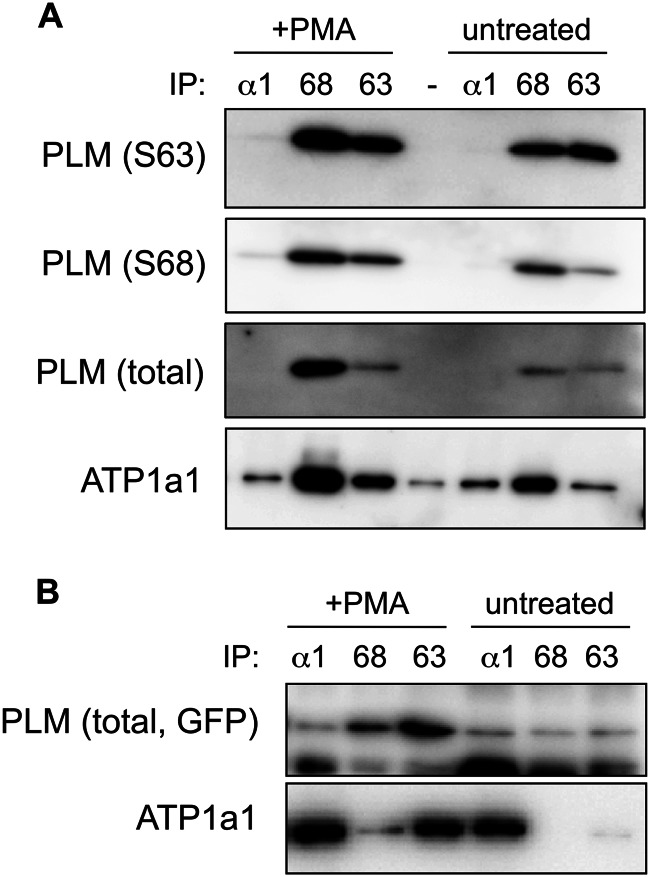
PLM-Ser63 antibody performance in co-immunoprecipitation experiments. A, immunoprecipitation (IP) reactions from untreated and PMA-treated (300 nm, 10 min) ARVMs. In untreated cells, sodium pump α1 subunit co-precipitated poorly with PLM-Ser63. Increasing the quantity of Ser63-phosphorylated PLM by activating PKC increased the quantity of sodium pump α1 subunit co-precipitated. B, immunoprecipitation reactions from FT-293 cells stably expressing PLM-YFP. In the absence of PMA, PLM is largely unphosphorylated in these cells and therefore not immunoprecipitated by Ser63 (S63) and Ser68 (S68) antibodies. Following PMA treatment, sodium pump α1 subunit is co-precipitated considerably more efficiently with Ser63-phosphorylated than Ser68-phosphorylated PLM.
In a second series of experiments, we immunoprecipitated Ser63- and Ser68-phosphorylated PLM from FT-293 cells stably expressing tet-inducible PLM-YFP. PLM is essentially unphosphorylated in this cell line, so co-precipitation of sodium pump α1 subunit from unstimulated cells was very poor with both Ser63- and Ser68-specific antibodies. Upon activation of PKC with PMA, we found that the antibody directed against Ser63-phosphorylated PLM co-precipitated Na pump α1 subunit far more efficiently than the Ser68-specific antibody. Hence in this cell type, Ser63-phosphorylated PLM is better associated with the Na pump α1 subunit than Ser68-phosphorylated PLM. The Ser63-specific antibody is therefore undoubtedly capable of co-precipitating Na pump α subunit.
Subcellular Localization of Ser63-phosphorylated PLM
We investigated the subcellular distribution of Ser63-phosphorylated PLM using a variety of techniques, all of which point to Ser63- and Ser68-phosphorylated PLM residing in the same cellular compartments (Fig. 3). Fig. 3A shows immunofluorescent localization of the two phosphorylation states in ARVMs. Both are localized to surface sarcolemma, t-tubules, and intercalated disks; we found neither qualitative nor quantitative differences in their distribution in stained myocytes. We also prepared cell surface proteins using a membrane-impermeable amine reactive biotinylation reagent (Fig. 3B). The enrichment of Ser63-phosphorylated and Ser68-phosphorylated PLM in cell surface fractions relative to unfractionated starting material were indistinguishable from each other and also from the enrichment of total PLM. Notably, sodium pump α1 subunit was generally more enriched in cell surface membrane preparations than PLM, but this probably reflects the relative abundance of primary amines in the extracellular region of rat sodium pump α1 subunit compared with rat PLM (the only primary amine available being the extracellular amino terminus of PLM). Cell surface proteins were prepared using conditions that promote the dissociation of the Na pump-PLM complex (5).
FIGURE 3.
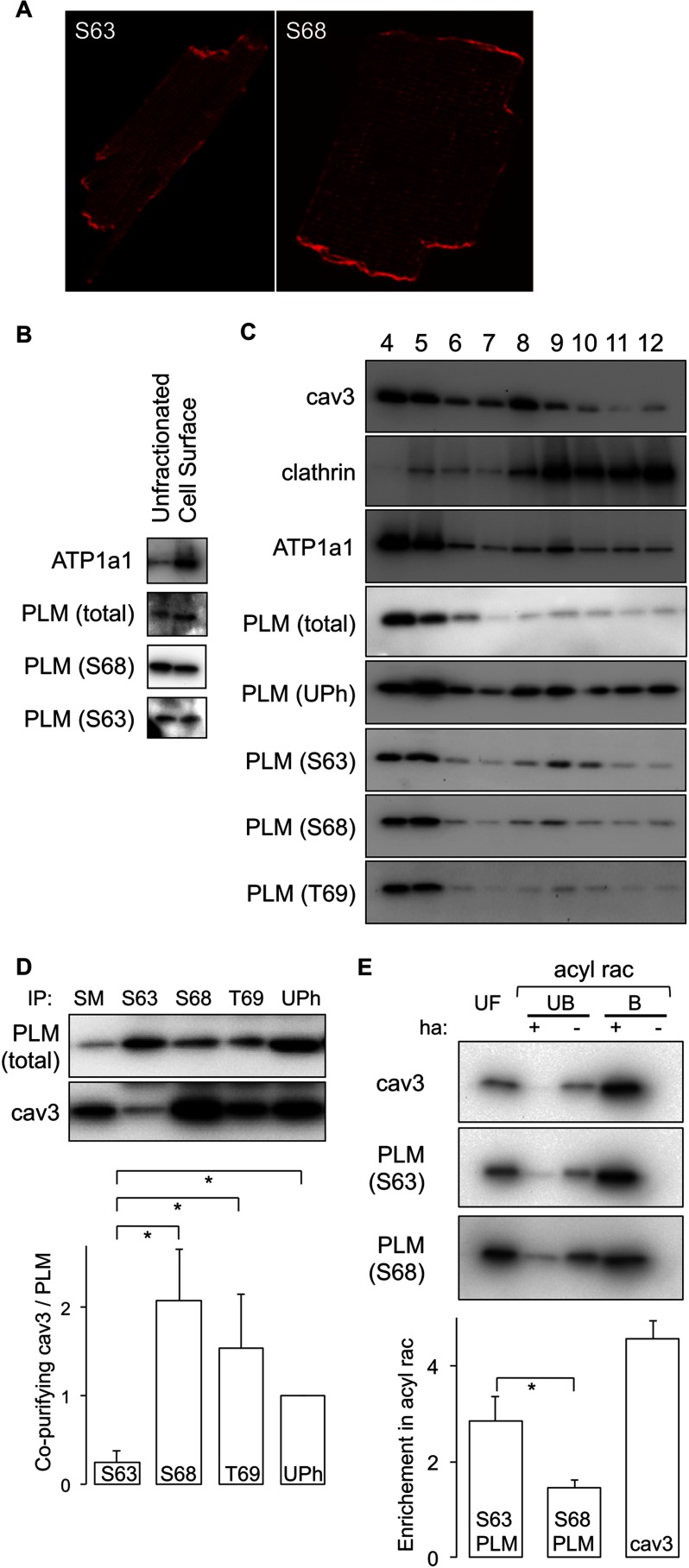
Subcellular localization of Ser63-phosphorylated PLM. A, immunofluorescent localization of Ser63 (S63) and Ser68 (S68) phosphorylated PLM in ARVMs. Both forms are distributed grossly identically in myocyte surface membranes in t-tubules, surface sarcolemma, and intercalated disks. B, cell surface fractions were prepared from ARVMs using cell-impermeable biotinylation reagents and analyzed alongside the unfractionated cell lysate from which they were derived. Ser63- and Ser68-phosphorylated forms are equally enriched in cell surface membranes compared with unfractionated starting material. C, sucrose gradient fractionation of ARVMs to purify a caveolar/lipid raft component. All phosphorylation states and unphosphorylated (UPh) PLM are localized in buoyant membranes from sucrose gradients (fractions 4 and 5), which are also enriched in the caveolar marker protein caveolin 3 (cav3) but not the bulk sarcolemma marker clathrin. T69, Thr69. D, phosphospecific immunoprecipitations (IP) of PLM indicate very poor co-purification of caveolin 3 with Ser63-phosphorylated PLM despite its localization to the caveolar membrane compartment. Quantitative analysis of the relative co-purification of caveolin 3 with PLM in phosphospecific co-immunoprecipitation reactions is shown (for an explanation of the calculation see legend to Fig. 1). SM, immunoprecipitation starting material. *, p < 0.05 (n = 7). E, palmitoylated proteins were purified by acyl rac in the presence (+) and absence (−) of hydroxylamine. UF lanes, unfractionated starting material; UB lanes, proteins not purified by acyl rac; B lanes, proteins purified by acyl rac. Ser63-phosphorylated PLM is purified to a considerably greater extent relative to the unfractionated starting material than Ser68-phosphorylated PLM, indicating that it is considerably more palmitoylated. *, p < 0.05 (n = 5).
In cardiac muscle, the sodium pump is reported to localize to buoyant caveolin-enriched microdomains (35), which we prepared from homogenized and sonicated myocyte lysates by floatation on a discontinuous sucrose gradient. Fig. 3C confirms that the Na pump α1 subunit is indeed localized to cardiomyocyte buoyant caveolin-enriched membranes, as are all PLM phosphorylation states and most unphosphorylated PLM. Hence, although Ser63-phosphorylated PLM is not physically associated with the pump, it does co-localize to the same membrane microdomain, despite the lack of caveolin-binding motif in PLM (ΦXΦXXXXΦ, ΦXXXXΦXXΦ, or ΦXΦXXXXΦXXΦ, where Φ = F, W, or Y (36), although the requirement for a caveolar binding motif to target proteins to caveolae has recently been challenged (37)). We therefore investigated co-immunoprecipitation of caveolin 3 with PLM phosphorylation states. Much like the sodium pump α subunit, Ser63-phosphorylated PLM co-immunoprecipitated caveolin 3 very poorly compared with all other PLM phosphorylation states (Fig. 3D). We do not propose a direct interaction between PLM and caveolin 3, rather an interaction bridged by the Na pump α subunit, which has two consensus caveolin-binding motifs (38). Hence the failure of Ser63-phosphorylated PLM to co-precipitate caveolin 3 is likely due to its lack of association with the pump.
We investigated why Ser63-phosphorylated PLM is localized to cardiomyocyte caveolae. For certain proteins, fatty acid anchors such as palmitate are proposed to mediate localization to lipid rafts/caveolae (39). Because PLM may be palmitoylated (27), we investigated whether Ser63-phosphorylated PLM is palmitoylated in ARVMs. We purified palmitoylated proteins from ARVMs by resin-assisted capture (28) and investigated the relative enrichment of PLM phosphorylation states and caveolin 3 (Fig. 3E). Constitutively palmitoylated caveolin 3 is enriched ∼5-fold in these experiments, and Ser63-phosphorylated PLM is enriched substantially more than Ser68-phosphorylated PLM, suggesting that its caveolar localization may be achieved through fatty acylation.
Protein Interactions of Ser63-phosphorylated PLM
Because Ser63-phosphorylated PLM interacts with neither Na pump nor caveolin 3, we sought to identify unique partner proteins for Ser63 using large scale immunoprecipitations from ARVMs followed by mass spectrometry to identify binding partners. Fig. 4A shows proteins co-immunoprecipitated with unphosphorylated, Ser63-phosphorylated, and Ser68-phosphorylated PLM separated by SDS-PAGE and stained with Coomassie Blue. Despite purifying similar quantities of PLM, the number and abundance of co-purifying proteins is substantially fewer in the Ser63 immunoprecipitation reaction compared with the Ser68 immunoprecipitation reaction. In a separate experiment, the proteins were eluted from immunoprecipitation reactions, tryptically digested, and identified by LC-MS/MS. For analysis purposes, the rat proteins identified were mapped to human homologues. The overlap between the three immunoprecipitation reactions is shown in Fig. 4B, with the 10 proteins common to all reactions listed in Table 1, and further information is shown in supplemental Table S1. Apart from Na pump subunits, the only proteins common to all three immunoprecipitation reactions are abundant cardiac contractile and mitochondrial proteins, which are common contaminants of these experiments (not shown). The three proteins unique to the Ser63-specific immunoprecipitation are myosin heavy and light chains and SERCA, which in our experience are also common contaminants of mass spectrometry experiments from cardiac tissue. Hence we could identify no specific interactors for Ser63-phosphorylated PLM using this approach.
We next investigated Ser63-phosphorylated PLM partner proteins using heterobifunctional cross-linking reagents. An example using the cell-permeable cross-linker DSP (reactive toward primary amines; distance between reactive groups, 12.0 Å) is shown in Fig. 4C. We investigated cross-linking of control and PMA-treated myocytes with DSP. The most abundant cross-linked species identified cross-linked to Ser68-phosphorylated PLM migrates at 50 kDa (consistent with formation of a PLM tetramer), and lower abundance species were identified at 100 kDa (consistent with Na pump α subunit with PLM) and 150 kDa (consistent with Na pump α and β subunit with PLM). No unique interactions for Ser63-phosphorylated PLM were revealed in these experiments.
PP2A is reported to interact directly with the Na pump α subunit (40, 41). Because PP2A dephosphorylates PLM at Ser63 but not Ser68 and Thr69 (42), we investigated co-purification of sodium pump α1 subunit and PLM phosphorylation states with PP2A. Fig. 4D shows that Ser68-phosphorylated PLM and Na pump α1 subunit but not Ser63-phosphorylated PLM co-immunoprecipitate with PP2A in ARVM lysates.
Partial Formaldehyde Fixation to Identify Multiprotein Complexes
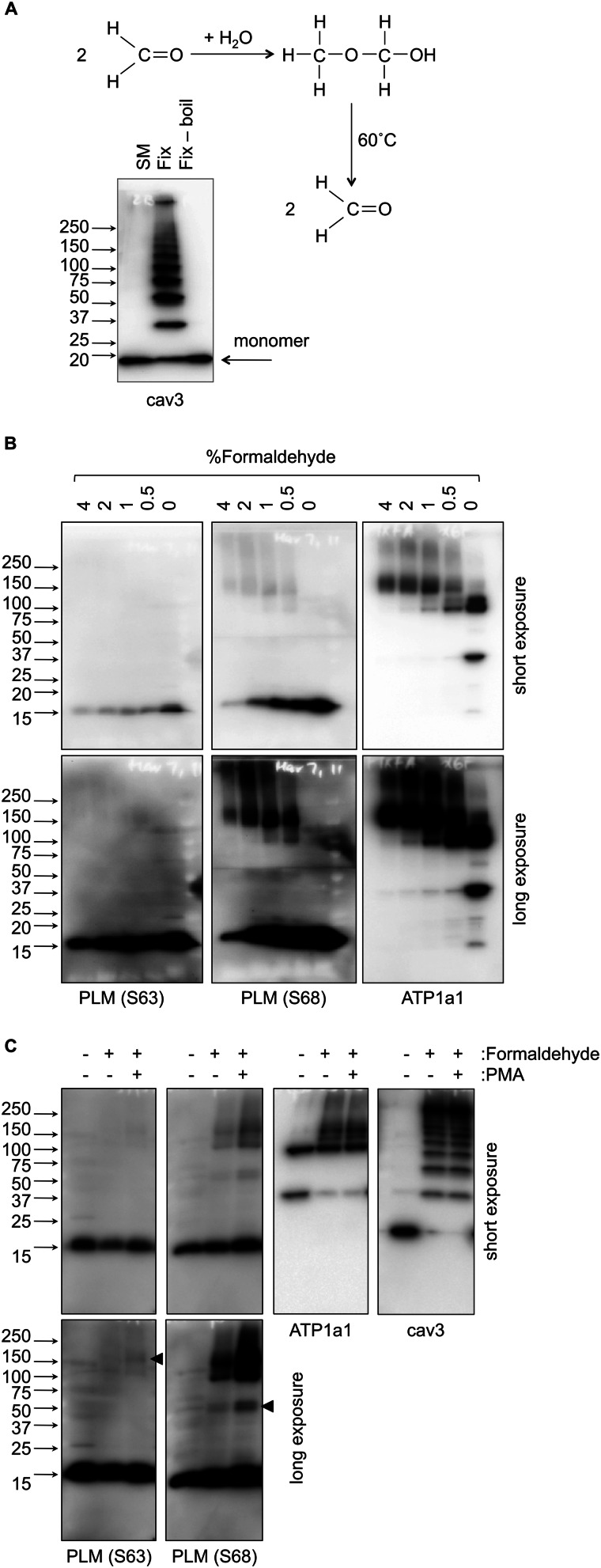
Formaldehyde cross-linking, although commonly used in chromatin immunoprecipitation and as a fixative prior to immunofluoresence, is relatively neglected in the study of protein-protein interactions (43). Although the distance between reactive groups of formaldehyde is very short (2 Å), its ability to reversibly polymerize (Fig. 5A) means it may cross-link moieties separated by any multiple of ∼2 Å. Additionally, its broad reactivity (as well as free amino termini, the side chains of lysine, histidine, arginine, glutamine, asparagine, tryptophan, tyrosine, and cysteine may all be modified (43)), means a whole spectrum of protein-protein interactions may be visualized using formaldehyde that would usually be unavailable to inflexible cross-linkers with fixed chemistries and distances between their reactive groups (such as DSP).
FIGURE 5.
Partial formaldehyde fixation to identify multiprotein complexes. A, reaction scheme for formaldehyde monomer (left) in the presence of water to form formaldehyde polymer (right), which is hydrolyzed back to the monomer on heating/boiling. The primary amine reactive formaldehyde is therefore capable of cross-linking primary amines any multiple of 2 Å apart. A representative caveolin 3 immunoblot shows the result of partially fixing ARVMs with formaldehyde. The partially fixed caveolin 3 (cav3) multimer is resolved up to a nonamer, which may be quantitatively retrieved as a monomer upon hydrolysis of the formaldehyde polymer. SM, immunoprecipitation starting material. B, ARVMs were treated with the concentration of formaldehyde indicated above each lane, and immunoblotted as shown. Formaldehyde fixation reveals no interactions for Ser63-phosphorylated PLM (S63). Ser68-phosphorylated PLM (S68) is observed at 100 kDa (likely PLM interacting with the sodium pump α subunit) and 150 kDa (likely PLM-α-β complexes). Short and long exposures of the same immunoblots are shown. C, ARVMs were treated with PMA (300 nm, 10 min), fixed with 2% formaldehyde in the continued presence of PMA, and immunoblotted as shown without heating prior to electrophoresis. Ser63-phosphorylated PLM may be detected at 150 kDa following formaldehyde fixation in the presence of PMA (arrowhead). A species migrating at 50 kDa is immunoreactive with the Ser68-phosphorylated PLM antibody, consistent with the formation of a PLM tetramer (arrowhead).
Fig. 5A shows an example of direct cross-linking of intact ARVMs with formaldehyde, immunoblotted for the structural protein caveolin 3. Caveolins homo-oligomerize in multimers of 14–16 units (44). In the experiment depicted in Fig. 5A, all oligmerization states from monomer to nonamer are readily visualized on a single minigel. We therefore used partial formaldehyde cross-linking to investigate interactions of Ser63- and Ser68-phosphorylated PLM. Representative immunoblots are presented in Fig. 5B. Partial cross-linking reveals adducts immunoreactive with Ser68-specific antibody migrating at 100 and 150 kDa that are identical in mobility to adducts immunoreactive with Na pump α1 subunit antibody. In contrast, no unique interactions for Ser63-phosphorylated PLM were identified.
In an attempt to identify readily hydrolyzed formaldehyde-cross-linked adducts, we refined our formaldehyde cross-linking protocol by eliminating all heating steps prior to electrophoresis. Fig. 5C shows representative immunoblots of control and PMA-treated ARVMs partially fixed with formaldehyde. Following activation of PKC, a small amount of Ser63-phosphorylated PLM is visible in cross-linked adducts at 150 kDa. Additional species visible to the Ser68-specific antibody only at 50 kDa are consistent with the formation of a PLM tetramer.
PLM Multimers Identified by Co-immunoprecipitation
We investigated the hypothesis that the Ser63-phosphorylated pool of PLM not associated with the Na pump is a homo-oligomer using co-immunoprecipitation. The existence of PLM homo-oligomers has been reported in transfected cells (25, 26), artificial lipid bilayers (22), and perfluoro-octaneoate polyacrylamide gels (24) but never for the endogenous protein in cardiac muscle, raising the possibility that previous reports are artifacts caused by overexpression/fusion to a fluorescent protein. We addressed these shortcomings using a transgenic knockin (KI) mouse in which wild type PLM has been replaced with unphosphorylatable PLM (S63A/S68A/S69A (3SA PLM)) by targeted mutation of the endogenous PLM locus. These animals express normal quantities of PLM, and heterozygous KI animals express both wild type and 3SA PLM, which can be distinguished biochemically both according to their reactivity toward PLM phospho-specific antibodies and by their mobility on Phos-Tag PAGE (in which the electrophoretic mobility of phosphorylated forms of a protein is reduced by their interaction with the Phos-Tag reagent within the polyacrylamide gel (30)).
Fig. 6A shows characterization of wild type and homozygous KI cardiac lysates on standard SDS-PAGE and Phos-Tag PAGE. Singly and doubly phosphorylated states are clearly resolved on Phos-Tag PAGE, which are immunoreactive with Ser63 and Ser68 (not shown) phosphospecific antibodies, but not with a Ser69 phosphospecific antibody (not shown). 3SA PLM is detectable with antibodies raised against the PLM amino terminus but is not detected by phosphospecific antibodies in immunoblots and migrates entirely as unphosphorylated in Phos-Tag gels.
FIGURE 6.
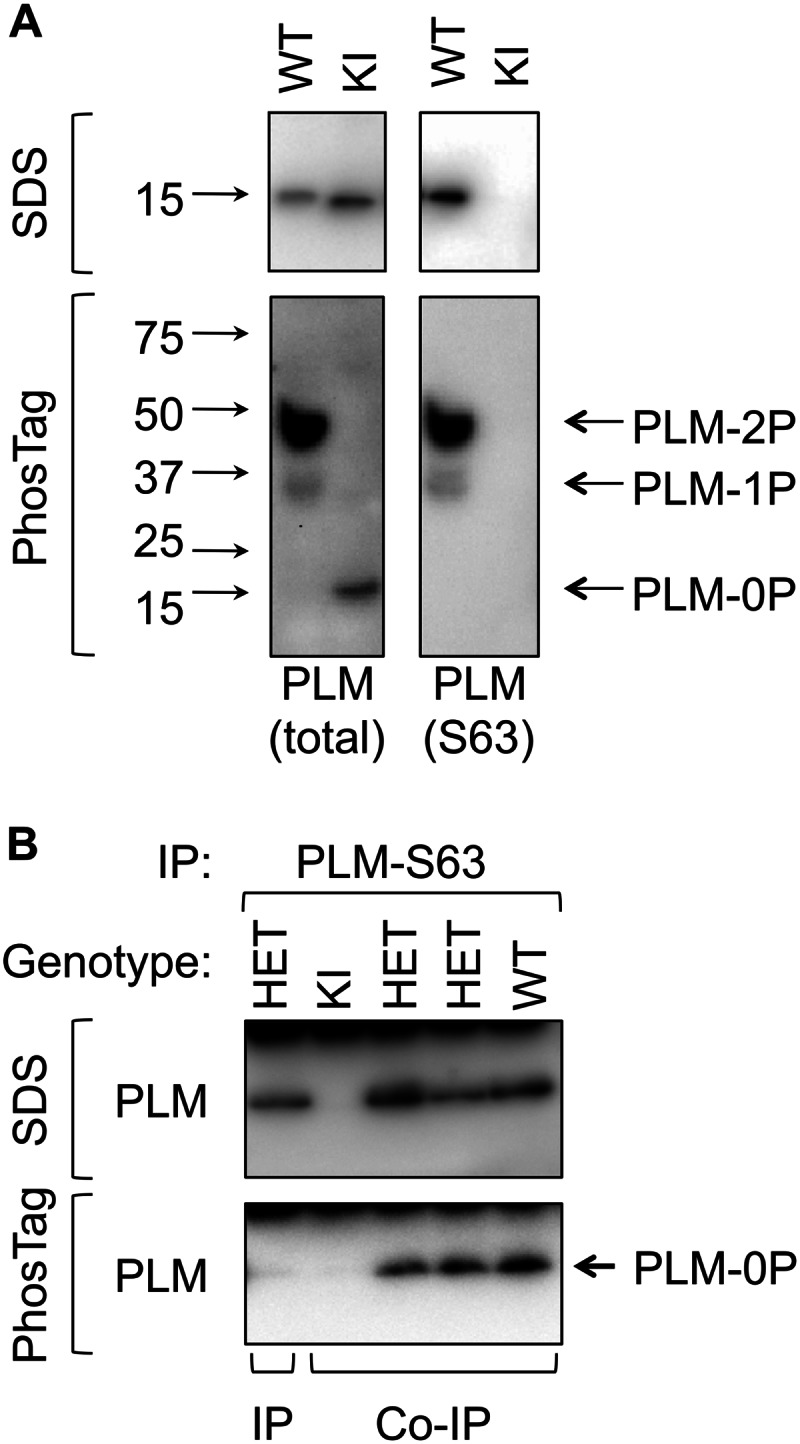
Phospholemman multimers in cardiac muscle. A, characterization of PLM from WT and KI animals on standard and Phos-Tag SDS-PAGE. The mobility of molecular mass standards is indicated alongside the immunoblots. Unphosphorylated (0P, immunoreactive with total PLM antibody only) and singly (1P) and doubly (2P) phosphorylated states (immunoreactive with Ser63 (S63) phosphospecific antibody) are resolved on Phos-Tag PAGE. 3SA PLM migrates entirely as unphosphorylated in Phos-Tag gels. B, Ser63-phosphorylated PLM was immunoprecipitated from WT, heterozygous KI (HET), and homozygous KI hearts under co-immunoprecipitation (Co-IP) and immunoprecipitation (IP) conditions. Purification of unphosphorylated PLM (0P) in co-immunoprecipitation reactions confirms the existence of interactions between WT and 3SA/unphosphorylated PLM.
We investigated the formation of PLM multimers by immunopurifying Ser63-phosphorylated PLM from wild type, heterozygous KI, and homozygous KI cardiac lysates under conditions favoring either co-immunoprecipitation or immunoprecipitation. Fig. 6B indicates that we are able to detect PLM-PLM interactions in these experiments. No PLM was immunoprecipitated from homozygous KI hearts, indicating that the Ser63-specific antibody does not cross-react with 3SA PLM. Analysis of immunoprecipitation reactions on Phos-Tag PAGE shows that Ser63-specific co-immunoprecipitation reactions from both heterozygous KI and wild type cardiac lysates contain unphosphorylated PLM (as well as other phosphorylation states whose definitive identification is precluded by interference by the immunoprecipitating immunoglobulins).
We sought to exclude the possibility that the Ser63-specific antibody cross-reacts with wild type unphosphorylated PLM, which could account for the presence of unphosphorylated PLM in these co-immunoprecipitation reactions. Ser63-phosphorylated PLM was immunoprecipitated from lysates from heterozygous KI hearts that had been solubilized in 1% SDS to disrupt protein-protein interactions and then diluted in 1% Triton X-100. Although the Ser63-specific antibody successfully immunoprecipitates PLM under these conditions, no co-purifying unphosphorylated PLM is detected on Phos-Tag gels (Fig. 6B). Hence the unphosphorylated PLM detected in co-immunoprecipitation reactions is co-purifying in a PLM multimer/oligomer.
A Pool of PLM That Does Not Regulate the Na Pump
Although PLM phosphorylation alters FRET between the Na pump α subunit and PLM (25), it does not alter the amount physically associated in sarcolemmal membranes (4). We nevertheless investigated the possibility that phosphorylation of PLM at Ser63 prevents its interaction with the sodium pump α subunit, by immunoprecipitating α1 subunit from ARVMs that had been treated with agonists prior to lysis to induce phosphorylation of PLM (Fig. 7). Co-purifying total, Ser63-phosphorylated, and Ser68-phosphorylated PLM was normalized to the amount co-purified from vehicle-treated cells. Phosphorylation at any site is clearly without effect on the quantity of PLM found associated with the pump α1 subunit in ARVMs. Interestingly, although acute PKC inhibition with bisindolylmaleimide (bis) decreases PLM phosphorylation at Ser63 and Ser68 in unfractionated ARVMs (Fig. 7A; in agreement with our previous findings (5)), we saw no accompanying decrease in phosphorylation of PLM co-purifying with the pump (Fig. 7B), suggesting that PLM dephosphorylation following application of bis was specifically the pump-free pool of PLM. We therefore investigated whether dephosphorylation of PLM caused by acute application of bis influenced Na pump activity by measuring whole cell pump currents in ARVMs. Fig. 7C shows that bis was without effect on pump activity, confirming the existence of a pool of PLM that does not regulate the pump.
FIGURE 7.
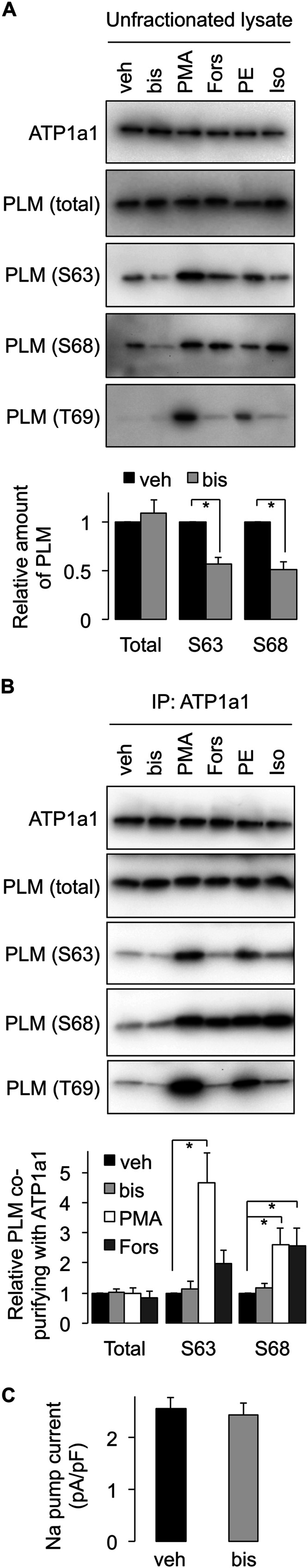
A pool of PLM that does not regulate the Na pump. A, kinase activation/inhibition causes phosphorylation/dephosphorylation of PLM in ARVMs. veh, vehicle; bis, 1 μm bisindolylmaleimide; PMA, 300 nm PMA; Fors, 10 μm forskolin; PE, 100 μm phenylephrine; Iso, 100 nm isoproterenol, all applied for 10 min at 35 °C (for clarity of presentation, only relevant treatment groups are quantitated). S63, Ser63; S68, Ser68; T69, Thr69. *, p < 0.05 (n = 8). B, kinase activation/inhibition in ARVMs leading to phosphorylation/dephosphorylation of PLM does not alter the quantity of PLM co-immunoprecipitated with the sodium pump α1 subunit. Although bis treatment causes PLM dephosphorylation in unfractionated lysates, the phosphorylation status of pump-associated PLM is not altered. *, p < 0.05 (n = 7). C, whole cell Na pump currents in ARVMs. Although bis treatment causes PLM dephosphorylation in unfractionated lysates, pump currents are not altered because the phosphorylation status of pump-associated PLM is not changed (six cells from three animals for vehicle and seven cells from five animals for bis). IP, immunoprecipitation.
DISCUSSION
A Pool of PLM Not Associated with the Na Pump
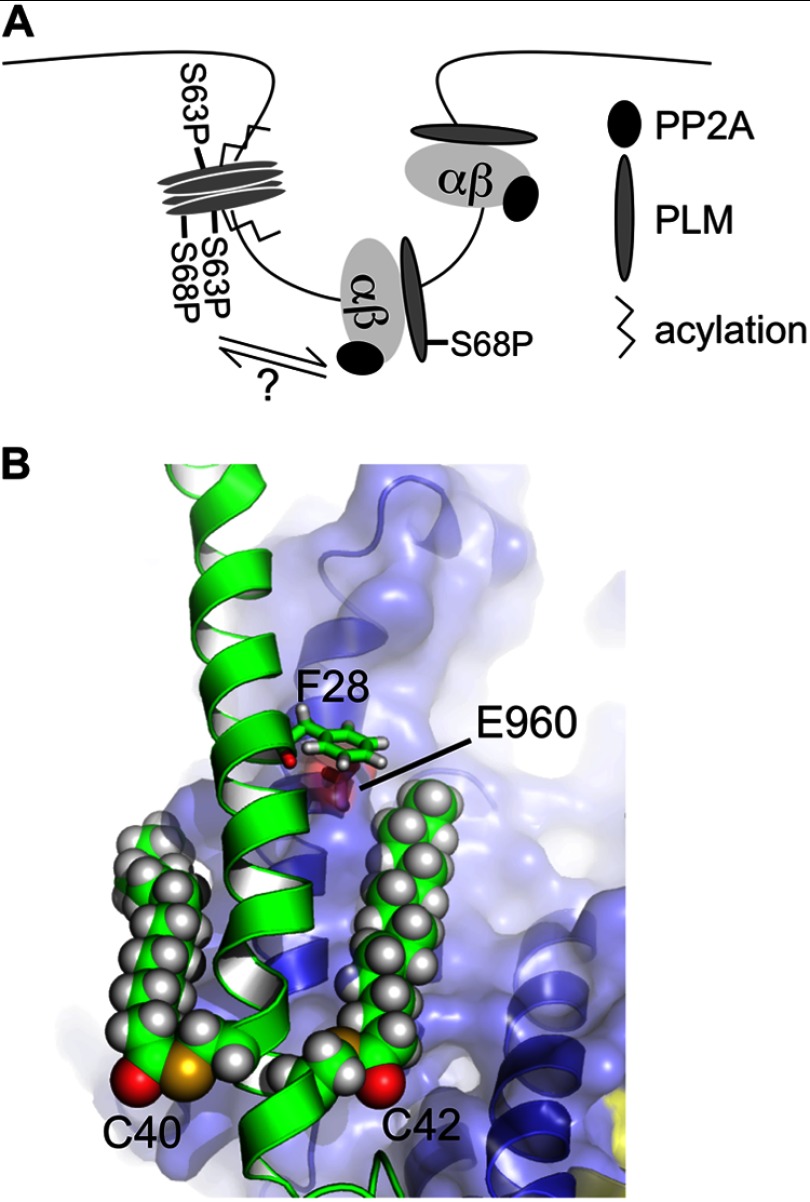
The present study describes a pool of PLM in ventricular myocytes phosphorylated at Ser63 that is not associated with any ion transporters but does co-immunoprecipitate with other PLM molecules. We would emphasize that although for simplicity we refer to this pool of PLM not associated with the pump as Ser63-phosphorylated, this should not imply that it is not also phosphorylated at other sites. Indeed, we do detect Ser68-phosphorylated PLM in Ser63-specific immunoprecipitations (Fig. 1, A and C). Rather, we find that the pool of Ser63-phosphorylated PLM is specifically not associated with the pump, not that it is not phosphorylated at other sites. The phenomenon appears to be conserved in all tissues investigated: in both brain and skeletal muscle, we find that Ser63-phosphorylated PLM co-purifies the Na pump poorly. The relationships between the proteins investigated in this study are shown in Fig. 8A: acylated, Ser63-phosphorylated PLM forms multimers in the same membrane compartment as the pump. The presence of PP2A ensures that pump-associated PLM remains unphosphorylated at Ser63.
FIGURE 8.
Modeling. A, cartoon depicting the relationships between PLM, Na pump, and PP2A established in this investigation. B, palmitates were added to the NMR structure of PLM in positions Cys40 and Cys42 (Protein Data Bank code 2JO1, shown in green) modeled with pump α subunit (Protein Data Bank code 3B8E, shown in blue) according to Ref. 27. Palmitate at Cys42 of PLM may impinge on PLM F28 (side chain shown in green)/α subunit Glu960 (side chain shown in red) and therefore alter the association of PLM with the pump.
The failure to identify this pool of PLM in myocytes before now is most likely the result of experimental design. When immunoprecipitating pump subunits, we are able to detect some co-purifying Ser63-phosphorylated PLM (for example, Ref. 5 and Fig. 7B). However, no studies to date have quantitatively assessed the amount of Na pump α subunit co-purifying with different PLM phosphorylation states. Although we find a small amount of Ser63-phosphorylated PLM associated with the pump (∼4-fold less than Ser68; Fig. 1B), the vast majority of this phosphorylation state is not. Given that >50% of PLM is phosphorylated at Ser63 in ARVMs (5), the proportion of PLM engaged in this multimer and therefore not associated with the pump is large.
The presence of both multimerized and pump-associated PLM in the same surface membrane compartment might appear to facilitate dynamic exchange between the two pools, but it remains to be seen whether this is actually the case. They may never exchange with each other, and our observations offer no mechanistic basis for such an exchange. Like PLB and SERCA, phosphorylation of PLM reduces FRET between PLM and the Na pump α subunit and increases PLM-PLM FRET (7, 25). Despite this, phosphorylated PLM remains physically associated with the α subunit (4–6, 46). Indeed the fact that we detect no quantitative changes in the total quantity of PLM associated with the pump no matter how or at which site PLM is phosphorylated (Fig. 7B) would argue against phosphorylation of PLM driving it from being pump-associated toward forming a multimer. (What drives the differential phosphorylation status of PLM in these two pools is discussed below.)
PLM is palmitoylated as well as being phosphorylated, but there is no difference in co-immunoprecipitation of palmitoylated and nonpalmitoylated PLM with the pump (27), so the enhanced palmitoylation of Ser63-phosphorylated PLM probably accounts for its presence in caveolin-enriched microdomains (39), but not for its failure to associate with the pump. That said, given the recent finding that Phe28 of PLM and Glu960 of the α subunit are critical for their interaction (47), we modeled the position of the PLM palmitates relative to both resides (Fig. 8B). It is possible that palmitoylation of PLM at Cys42 (but probably not Cys40) may impinge on PLM Phe28 and α subunit Glu960 and therefore alter their interaction; however, it is important to bear in mind that palmitates are highly flexible and according to our model will only reach F28 when fully extended. Nevertheless, the different acylation profiles of the two pools of PLM would appear to implicate differential palmitoylation as either a cause or consequence of their segregation to different pools. This will be the subject of future investigation. It may also be relevant that the recent proposal that PLM protects the Na pump from oxidative inhibition through oxidation of an intracellular cysteine (48) would be of significantly greater functional consequence for the pump if a pool of nonoxidized PLM existed to exchange with recently oxidized pump-associated PLM and continue to provide the pump with “redox protection.”
PLM Phosphorylation Status Governed by Partner Protein Co-localization
It has recently been established that PP2A A (regulatory) and C (catalytic) subunits bind directly to the Na pump α subunit; the A subunit through an interaction with amino-terminal A domain of the α subunit (41) and the C subunit through an interaction with the large intracellular loop of the α subunit (40). It is suggested that recruitment of PP2A is required for α subunit dephosphorylation at Ser18 (PKC site (41)) and/or GRK sites in the large intracellular loop (40) to promote trafficking of the Na pump α subunit to the cell surface membrane. However, PP2A is also the likely phosphatase that dephosphorylates PLM Ser63 (but not Ser68 or Thr69), based on okadaic acid sensitivity (42). Here we report that Ser68-phosphorylated but not Ser63-phosphorylated PLM co-immunoprecipitates with PP2A catalytic subunit, presumably as a result of co-purification via the Na pump α subunit. Hence this study highlights an additional role for α subunit-associated PP2A: the control of the phosphorylation status of a pump regulatory protein that is anchored close to the large intracellular loop of the pump. The pool of PLM associated with the pump is exposed to PP2A and therefore not phosphorylated at Ser63, whereas the pool of PLM that is not pump-associated is not. Indeed, we also find PP2A A subunit and PP1γ catalytic subunit co-immunoprecipitate with unphosphorylated and Ser68-phosphorylated PLM (supplemental Table S1; discussed below) but not with Ser63-phosphorylated PLM. Underlying the basal phosphorylation of PLM are rapid cycles of phosphorylation and dephosphorylation (5). Therefore the phosphorylation status of Ser68 and Thr69 is governed by PP1, inhibitor-1, PKA and PKC (42), whereas the phosphorylation status of Ser63 is governed by PP2A and PKC. That the phosphorylation status of the two pools of PLM is determined by co-localization with a phosphatase rather than the phosphorylation status determining subcellular distribution agrees well with our data that changes in PLM phosphorylation do not alter the amount associated with the pump.
The PLM Proteome
Our proteomic analysis of PLM immunoprecipitations identifies a variety of well established and hitherto unidentified members of the PLM-Na pump macromolecular complex. We find α1, α2, and α3 catalytic and β1 and β3 regulatory subunits of the pump expressed and co-purifying with PLM in ventricular myocytes. The relative contribution of the β3 subunit to the cardiac pump is of interest given that it lacks the transmembrane cysteine that is reported to mediate inhibition of the pump by oxidants (49). Co-localization of other ATP-dependent transporters with GAPDH has previously been reported (50, 51) and may represent a means to link glycolytically derived ATP to pump activity (10). Among other notable members of the Na pump macromolecular complex (in addition to phosphatase subunits mentioned above) are N-myc downstream-regulated genes 2 and 4 (the former has been implicated in regulation of Na pump β subunit turnover (52)), the vesicle fusion regulator syntaxin 7, the small G-protein rap-1b-like protein, and the E3 ubiquitin ligase TRIM21.
Parallels between Phospholemman and Phospholamban
This study strengthens the analogy between regulation of the Na pump by PLM and regulation of SERCA by PLB. Although we have failed to identify the stoichiometry of the PLM multimer, cross-linking data are consistent with the formation of a tetramer with an apparent mass of ∼50 kDa. In contrast to the PLB pentamer, which is resistant even to high concentrations of SDS unless boiled, the PLM multimer is considerably less stable.
The pentameric architecture of PLB is achieved through a leucine/isoleucine zipper in the transmembrane domain (15). The α-helical transmembrane domain of PLM is enriched in leucines and isoleucines in positions 16, 18, 21, 23, 26, 27, 29, 30, 32, 33, 34, and 36, so clearly it has the potential to do the same. However, the absence of any hydrophilic side chains to line a central pore makes it highly unlikely that the PLM multimer forms an ion conductance in its own right.
One notable difference between PLM and PLB is the “tonic” phosphorylation of PLM at rest. The identification of a pool of PLM not associated with the Na pump raises the possibility that gross changes in PLM phosphorylation in cardiac muscle do not necessarily report changes in phosphorylation of pump-associated PLM. Indeed, we find that the rapid dephosphorylation of PLM following acute PKC inhibition in ARVMs occurs only in the pump-free pool of PLM, which explains the lack of effect of acute PKC inhibition on pump activity (Fig. 7C). However, that is not to say that PLM dephosphorylation never leads to pump inhibition (see, for example, Ref. 42).
Partial Formaldehyde Fixation to Identify Multiprotein Complexes
Herein we describe a new method of partial formaldehyde fixation to identify protein-protein interactions. The broad reactivity of formaldehyde compared with inflexible cross-linkers with fixed chemistries is a significant advantage to investigate protein-protein interactions. Partial formaldehyde fixation elegantly demonstrates the existence of the caveolin 3 oligomer in ventricular muscle, as well as confirming that the principal partner protein for PLM in ventricular muscle is the Na pump. Its failure to identify the PLM multimer is most likely due to its lack of reactivity toward the relatively inert leucines and isoleucines that likely mediate the formation of this multimer, although it is notable that the elimination of all heating steps prior to electrophoresis does allow us to visualize a potential PLM tetramer with mobility of 50 kDa.
Na Pump-free PLM at the Cell Surface
Our finding that a Na pump-free pool of PLM is present at the cell surface in ventricular myocytes might appear to contradict the recent report that PLM requires association with the pump to traffic through the secretory pathway in Xenopus oocytes (45), but this is not necessarily the case. If the control of PLM trafficking is the same in ventricular muscle as it is in Xenopus oocytes, then the implication of our study is that the relationship between PLM and the Na pump is indeed dynamic and that PLM can dissociate from the pump once at the cell surface. Whether the quantity of PLM at the cell surface differs from the quantity of α subunit has not been investigated in this study, although there are reports that PLM-free pump can be detected in ventricular muscle (12). If PLM requires pump α and β subunits to reach the cell surface but is then able to dissociate from the pump, then the relative quantities of PLM and αβ at the cell surface will be determined by their individual degradation rates, and hence the quantity of PLM at the cell surface could exceed the quantity of αβ. If some Na pump in the heart is PLM-free, then the PLM multimers may be a storage pool to increase the “responsiveness” of the pump to adrenergic stimulation, by as yet unidentified means.
Concluding Remarks
We report the existence of a subpopulation of PLM that interacts only with other PLM molecules and not with the Na pump, with a unique phosphorylation status driven by differential proximity of protein phosphatases.
This work was support by grants from the British Heart Foundation (RG/07/001 to W. F., and M. J. S., and PG/10/93/28650 to W. F. and S. C. C.) and the Medical Research Council (G0700903 to W. F.).

This article contains supplemental Table S1.
- Na pump
- sodium/potassium ATPase
- PLM
- phospholemman
- SERCA 2a
- sarco/endoplasmic reticulum Ca-ATPase
- PLB
- phospholamban
- ARVM
- adult rat ventricular myocyte
- ha
- hydroxylamine
- KI
- knockin
- 3SA PLM
- unphosphorylatable S63A/S68A/S69A PLM
- bis
- bisindolylmaleimide
- DSP
- dithiobis[succinimidyl propionate]
- PMA
- phorbol 12-myristate 13-acetate.
REFERENCES
- 1. Sweadner K. J., Rael E. (2000) The FXYD gene family of small ion transport regulators or channels. cDNA sequence, protein signature sequence, and expression. Genomics 68, 41–56 [DOI] [PubMed] [Google Scholar]
- 2. Geering K. (2006) FXYD proteins. New regulators of Na-K-ATPase. Am. J. Physiol. Renal Physiol. 290, F241–F250 [DOI] [PubMed] [Google Scholar]
- 3. Crambert G., Fuzesi M., Garty H., Karlish S., Geering K. (2002) Phospholemman (FXYD1) associates with Na,K-ATPase and regulates its transport properties. Proc. Natl. Acad. Sci. U.S.A. 99, 11476–11481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fuller W., Eaton P., Bell J. R., Shattock M. J. (2004) Ischemia-induced phosphorylation of phospholemman directly activates rat cardiac Na/K-ATPase. FASEB J. 18, 197–199 [DOI] [PubMed] [Google Scholar]
- 5. Fuller W., Howie J., McLatchie L. M., Weber R. J., Hastie C. J., Burness K., Pavlovic D., Shattock M. J. (2009) FXYD1 phosphorylation in vitro and in adult rat cardiac myocytes. Threonine 69 is a novel substrate for protein kinase C. Am. J. Physiol. Cell Physiol. 296, C1346–C1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Silverman B., Fuller W., Eaton P., Deng J., Moorman J. R., Cheung J. Y., James A. F., Shattock M. J. (2005) Serine 68 phosphorylation of phospholemman. Acute isoform-specific activation of cardiac Na/K ATPase. Cardiovasc. Res. 65, 93–103 [DOI] [PubMed] [Google Scholar]
- 7. Bossuyt J., Despa S., Han F., Hou Z., Robia S. L., Lingrel J. B., Bers D. M. (2009) Isoform specificity of the Na/K-ATPase association and regulation by phospholemman. J. Biol. Chem. 284, 26749–26757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Despa S., Bossuyt J., Han F., Ginsburg K. S., Jia L. G., Kutchai H., Tucker A. L., Bers D. M. (2005) Phospholemman-phosphorylation mediates the β-adrenergic effects on Na/K pump function in cardiac myocytes. Circ. Res. 97, 252–259 [DOI] [PubMed] [Google Scholar]
- 9. Han F., Bossuyt J., Despa S., Tucker A. L., Bers D. M. (2006) Phospholemman phosphorylation mediates the protein kinase C-dependent effects on Na+/K+ pump function in cardiac myocytes. Circ. Res. 99, 1376–1383 [DOI] [PubMed] [Google Scholar]
- 10. Fuller W., Tulloch L. B., Shattock M. J., Calaghan S. C., Howie J., Wypijewski K. J. (2013) Regulation of the cardiac sodium pump. Cell Mol. Life Sci. 70, 1357–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walaas S. I., Czernik A. J., Olstad O. K., Sletten K., Walaas O. (1994) Protein kinase C and cyclic AMP-dependent protein kinase phosphorylate phospholemman, an insulin and adrenaline-regulated membrane phosphoprotein, at specific sites in the carboxy terminal domain. Biochem. J. 304, 635–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pavlović D., Fuller W., Shattock M. J. (2007) The intracellular region of FXYD1 is sufficient to regulate cardiac Na/K ATPase. FASEB J. 21, 1539–1546 [DOI] [PubMed] [Google Scholar]
- 13. Despa S., Tucker A. L., Bers D. M. (2008) Phospholemman-mediated activation of Na/K-ATPase limits [Na]i and inotropic state during β-adrenergic stimulation in mouse ventricular myocytes. Circulation 117, 1849–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Simmerman H. K., Jones L. R. (1998) Phospholamban. Protein structure, mechanism of action, and role in cardiac function. Physiol. Rev. 78, 921–947 [DOI] [PubMed] [Google Scholar]
- 15. Simmerman H. K., Kobayashi Y. M., Autry J. M., Jones L. R. (1996) A leucine zipper stabilizes the pentameric membrane domain of phospholamban and forms a coiled-coil pore structure. J. Biol. Chem. 271, 5941–5946 [DOI] [PubMed] [Google Scholar]
- 16. Autry J. M., Jones L. R. (1997) Functional Co-expression of the canine cardiac Ca2+ pump and phospholamban in Spodoptera frugiperda (Sf21) cells reveals new insights on ATPase regulation. J. Biol. Chem. 272, 15872–15880 [DOI] [PubMed] [Google Scholar]
- 17. Hou Z., Kelly E. M., Robia S. L. (2008) Phosphomimetic mutations increase phospholamban oligomerization and alter the structure of its regulatory complex. J. Biol. Chem. 283, 28996–29003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen Z., Akin B. L., Jones L. R. (2010) Ca2+ binding to site I of the cardiac Ca2+ pump is sufficient to dissociate phospholamban. J. Biol. Chem. 285, 3253–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen Z., Akin B. L., Stokes D. L., Jones L. R. (2006) Cross-linking of C-terminal residues of phospholamban to the Ca2+ pump of cardiac sarcoplasmic reticulum to probe spatial and functional interactions within the transmembrane domain. J. Biol. Chem. 281, 14163–14172 [DOI] [PubMed] [Google Scholar]
- 20. Bidwell P., Blackwell D. J., Hou Z., Zima A. V., Robia S. L. (2011) Phospholamban binds with differential affinity to calcium pump conformers. J. Biol. Chem. 286, 35044–35050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mueller B., Karim C. B., Negrashov I. V., Kutchai H., Thomas D. D. (2004) Direct detection of phospholamban and sarcoplasmic reticulum Ca-ATPase interaction in membranes using fluorescence resonance energy transfer. Biochemistry 43, 8754–8765 [DOI] [PubMed] [Google Scholar]
- 22. Moorman J. R., Ackerman S. J., Kowdley G. C., Griffin M. P., Mounsey J. P., Chen Z., Cala S. E., O'Brian J. J., Szabo G., Jones L. R. (1995) Unitary anion currents through phospholemman channel molecules. Nature 377, 737–740 [DOI] [PubMed] [Google Scholar]
- 23. Bell J. R., Lloyd D., Curl C. L., Delbridge L. M., Shattock M. J. (2009) Cell volume control in phospholemman (PLM) knockout mice. Do cardiac myocytes demonstrate a regulatory volume decrease and is this influenced by deletion of PLM? Exp. Physiol. 94, 330–343 [DOI] [PubMed] [Google Scholar]
- 24. Beevers A. J., Kukol A. (2006) Secondary structure, orientation, and oligomerization of phospholemman, a cardiac transmembrane protein. Protein Sci. 15, 1127–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bossuyt J., Despa S., Martin J. L., Bers D. M. (2006) Phospholemman phosphorylation alters its fluorescence resonance energy transfer with the Na/K-ATPase pump. J. Biol. Chem. 281, 32765–32773 [DOI] [PubMed] [Google Scholar]
- 26. Song Q., Pallikkuth S., Bossuyt J., Bers D. M., Robia S. L. (2011) Phosphomimetic mutations enhance oligomerization of phospholemman and modulate its interaction with the Na/K-ATPase. J. Biol. Chem. 286, 9120–9126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tulloch L. B., Howie J., Wypijewski K. J., Wilson C. R., Bernard W. G., Shattock M. J., Fuller W. (2011) The inhibitory effect of phospholemman on the sodium pump requires its palmitoylation. J. Biol. Chem. 286, 36020–36031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Forrester M. T., Hess D. T., Thompson J. W., Hultman R., Moseley M. A., Stamler J. S., Casey P. J. (2011) Site-specific analysis of protein S-acylation by resin-assisted capture. J. Lipid Res. 52, 393–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oliveros J. C. (2007) VENNY. An interactive tool for comparing lists with Venn Diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html
- 30. Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T. (2006) Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell Proteomics 5, 749–757 [DOI] [PubMed] [Google Scholar]
- 31. Howie J., Tulloch L. B., Shattock M. J., Fuller W. (2013) Regulation of the cardiac Na(+) pump by palmitoylation of its catalytic and regulatory subunits. Biochem. Soc. Trans. 41, 95–100 [DOI] [PubMed] [Google Scholar]
- 32. Despa S., Bers D. M. (2007) Functional analysis of Na+/K+-ATPase isoform distribution in rat ventricular myocytes. Am. J. Physiol. Cell Physiol. 293, C321–C327 [DOI] [PubMed] [Google Scholar]
- 33. Swift F., Tovsrud N., Enger U. H., Sjaastad I., Sejersted O. M. (2007) The Na+/K+-ATPase α2-isoform regulates cardiac contractility in rat cardiomyocytes. Cardiovasc. Res. 75, 109–117 [DOI] [PubMed] [Google Scholar]
- 34. Despa S., Lingrel J. B., Bers D. M. (2012) Na+/K+-ATPase α2-isoform preferentially modulates Ca2+ transients and sarcoplasmic reticulum Ca2+ release in cardiac myocytes. Cardiovasc. Res. 95, 480–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu L., Askari A. (2006) β-Subunit of cardiac Na+-K+-ATPase dictates the concentration of the functional enzyme in caveolae. Am. J. Physiol. Cell Physiol. 291, C569–C578 [DOI] [PubMed] [Google Scholar]
- 36. Couet J., Li S., Okamoto T., Ikezu T., Lisanti M. P. (1997) Identification of peptide and protein ligands for the caveolin-scaffolding domain. Implications for the interaction of caveolin with caveolae-associated proteins. J. Biol. Chem. 272, 6525–6533 [DOI] [PubMed] [Google Scholar]
- 37. Byrne D. P., Dart C., Rigden D. J. (2012) Evaluating caveolin interactions. Do proteins interact with the caveolin scaffolding domain through a widespread aromatic residue-rich motif? PLoS One 7, e44879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang H., Haas M., Liang M., Cai T., Tian J., Li S., Xie Z. (2004) Ouabain assembles signaling cascades through the caveolar Na+/K+-ATPase. J. Biol. Chem. 279, 17250–17259 [DOI] [PubMed] [Google Scholar]
- 39. Yang W., Di Vizio D., Kirchner M., Steen H., Freeman M. R. (2010) Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol. Cell Proteomics 9, 54–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kimura T., Han W., Pagel P., Nairn A. C., Caplan M. J. (2011) Protein phosphatase 2A interacts with the Na,K-ATPase and modulates its trafficking by inhibition of its association with arrestin. PLoS One 6, e29269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lecuona E., Dada L. A., Sun H., Butti M. L., Zhou G., Chew T. L., Sznajder J. I. (2006) Na,K-ATPase α1-subunit dephosphorylation by protein phosphatase 2A is necessary for its recruitment to the plasma membrane. FASEB J. 20, 2618–2620 [DOI] [PubMed] [Google Scholar]
- 42. El-Armouche A., Wittköpper K., Fuller W., Howie J., Shattock M. J., Pavlovic D. (2011) Phospholemman-dependent regulation of the cardiac Na/K-ATPase activity is modulated by inhibitor-1 sensitive type-1 phosphatase. FASEB J. 25, 4467–4475 [DOI] [PubMed] [Google Scholar]
- 43. Sutherland B. W., Toews J., Kast J. (2008) Utility of formaldehyde cross-linking and mass spectrometry in the study of protein-protein interactions. J. Mass Spectrom. 43, 699–715 [DOI] [PubMed] [Google Scholar]
- 44. Sargiacomo M., Scherer P. E., Tang Z., Kübler E., Song K. S., Sanders M. C., Lisanti M. P. (1995) Oligomeric structure of caveolin. Implications for caveolae membrane organization. Proc. Natl. Acad. Sci. U.S.A. 92, 9407–9411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moshitzky S., Asher C., Garty H. (2012) Intracellular trafficking of FXYD1 (phospholemman) and FXYD7 proteins in Xenopus oocytes and mammalian cells. J. Biol. Chem. 287, 21130–21141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bossuyt J., Ai X., Moorman J. R., Pogwizd S. M., Bers D. M. (2005) Expression and phosphorylation of the Na-pump regulatory subunit phospholemman in heart failure. Circ. Res. 97, 558–565 [DOI] [PubMed] [Google Scholar]
- 47. Khafaga M., Bossuyt J., Mamikonian L., Li J. C., Lee L. L., Yarov-Yarovoy V., Despa S., Bers D. M. (2012) Na+/K+-ATPase E960 and phospholemman F28 are critical for their functional interaction. Proc. Natl. Acad. Sci. U.S.A. 109, 20756–20761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bibert S., Liu C. C., Figtree G. A., Garcia A., Hamilton E. J., Marassi F. M., Sweadner K. J., Cornelius F., Geering K., Rasmussen H. H. (2011) FXYD proteins reverse inhibition of the Na+-K+ pump mediated by glutathionylation of its β1 subunit. J. Biol. Chem. 286, 18562–18572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Figtree G. A., Liu C. C., Bibert S., Hamilton E. J., Garcia A., White C. N., Chia K. K., Cornelius F., Geering K., Rasmussen H. H. (2009) Reversible oxidative modification. A key mechanism of Na+-K+ pump regulation. Circ. Res. 105, 185–193 [DOI] [PubMed] [Google Scholar]
- 50. Hong M., Kefaloyianni E., Bao L., Malester B., Delaroche D., Neubert T. A., Coetzee W. A. (2011) Cardiac ATP-sensitive K+ channel associates with the glycolytic enzyme complex. FASEB J. 25, 2456–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dhar-Chowdhury P., Malester B., Rajacic P., Coetzee W. A. (2007) The regulation of ion channels and transporters by glycolytically derived ATP. Cell Mol. Life Sci. 64, 3069–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li Y., Yang J., Li S., Zhang J., Zheng J., Hou W., Zhao H., Guo Y., Liu X., Dou K., Situ Z., Yao L. (2011) N-myc downstream-regulated gene 2, a novel estrogen-targeted gene, is involved in the regulation of Na+/K+-ATPase. J. Biol. Chem. 286, 32289–32299 [DOI] [PMC free article] [PubMed] [Google Scholar]