Background: Vertebrate APC collaborates with Dia through its Basic domain to assemble actin filaments.
Results: Despite limited sequence homology between the vertebrate and Drosophila APC Basic domains, Drosophila APC1 collaborates with Dia to stimulate actin assembly in vitro.
Conclusion: The mechanism of actin assembly is highly conserved over evolution.
Significance: APC-Dia collaborations may be crucial in a wide range of animal cells.
Keywords: Actin, Fluorescence, Formin, Protein Purification, Protein-Protein Interactions, APC1B, Drosophila, TIRF Microscopy
Abstract
Adenomatous polyposis coli (APC) is a large multidomain protein that regulates the cytoskeleton. Recently, it was shown that vertebrate APC through its Basic domain directly collaborates with the formin mDia1 to stimulate actin filament assembly in the presence of nucleation barriers. However, it has been unclear whether these activities extend to homologues of APC and Dia in other organisms. Drosophila APC and Dia are each required to promote actin furrow formation in the syncytial embryo, suggesting a potential collaboration in actin assembly, but low sequence homology between the Basic domains of Drosophila and vertebrate APC has left their functional and mechanistic parallels uncertain. To address this question, we purified Drosophila APC1 and Dia and determined their individual and combined effects on actin assembly using both bulk fluorescence assays and total internal reflection fluorescence microscopy. Our data show that APC1, similar to its vertebrate homologue, bound to actin monomers and nucleated and bundled filaments. Further, Drosophila Dia nucleated actin assembly and protected growing filament barbed ends from capping protein. Drosophila APC1 and Dia directly interacted and collaborated to promote actin assembly in the combined presence of profilin and capping protein. Thus, despite limited sequence homology, Drosophila and vertebrate APCs exhibit highly related activities and mechanisms and directly collaborate with formins. These results suggest that APC-Dia interactions in actin assembly are conserved and may underlie important in vivo functions in a broad range of animal phyla.
Introduction
Actin filament assembly is highly regulated to ensure the production of temporal and spatial patterns of filaments that are essential for dynamic processes such as cytokinesis, cell motility, membrane trafficking, and morphogenesis (1). The cellular environment generally suppresses new filament assembly through the activity of proteins such as profilin and thymosin β4 which sequester actin monomers, and capping protein which blocks filament elongation (2). To overcome these barriers, the cell deploys actin nucleation and elongation factors such as the Arp2/3 complex, formins, and the group of tandem monomer binders including Spire, Cobl, and adenomatous polyposis coli (APC)3 (1).
Formins are unique among this group of actin assembly factors in their ability both to nucleate and elongate actin filaments (for review, see Refs. 2–4). Diaphanous (Dia)-related formins are characterized by C-terminal FH1, FH2, and DAD domains that form a “tripartite nucleation machine” to promote filament nucleation (5). The FH2 domain forms a doughnut-shaped dimer that remains associated with the growing barbed end of the filament and protects it from capping protein. The FH2 domain also coordinates with the profilin-actin monomer binding FH1 domain to accelerate filament elongation (2). Recent in vitro and in vivo studies suggest that formins, rather than acting alone, often join forces with formin-binding nucleation-promoting factors (NPFs) to co-stimulate actin assembly. These NPF-formin pairs include: Spire-Capu/FMN, Bud6-Bni1, Flightless-DAAM, and APC-mDia1 (7–10). Among them, only the functions of Spire-Capu/FMN have been examined in an organismal context in metazoans (Drosophila melanogaster (8, 11) and mouse oocytes (12)). Less is known about the APC-Dia pair, but Drosophila offers a powerful model system in which to address their functions (13).
Vertebrate APC (vAPC) is a large (∼350 kDa) multifunctional protein with roles in the negative regulation of Wnt signal transduction and in the regulation and organization of the microtubule and actin cytoskeletons (14–17). Disruption of vAPC initiates up to 80% of both inherited and sporadic colorectal cancers, due in part to misregulation of Wnt signaling. In addition, it has been suggested that vAPC cytoskeletal functions may contribute to cancer initiation or progression (15). vAPC promotes microtubule stability together with EB1 and Dia (18) and can affect the actin cytoskeleton through its direct interactions with actin (19), Asef (a RacGEF) and IQGAP (20, 21). In particular, the C-terminal Basic domain of vAPC directly nucleates actin filament assembly in vitro by recruiting multiple actin monomers and stimulates excess actin assembly when introduced into NIH 3T3 cells (10). Further, vAPC and mDia1 collaborate to form a potent actin filament nucleation and elongation machine, capable of overcoming the dual barrier of profilin and capping protein in vitro (10, 23). This is achieved via a direct interaction between the Basic domain of vAPC and the formin tail region, which includes its DAD domain. Using triple-color total internal reflection fluorescence (TIRF) microscopy, it was shown that vAPC and mDia1 first interact with each other and actin monomers to form a ternary nucleation complex, then vAPC and mDia1 separate as the filament elongates, with vAPC remaining bound at the nucleation site and mDia1 riding away on the fast growing barbed end (23).
Like vertebrates, Drosophila has two separate APC genes/proteins, APC1 and APC2 (24). Drosophila APC1 shares a Basic domain with vAPC (24). However, there is limited sequence conservation between the Basic domain of vAPC (which harbors actin nucleation and formin-binding activities) and the C-terminal Basic domain of Drosophila APC1. Although we previously showed that the Basic domain of Drosophila APC1 binds the C-terminal half of Drosophila Dia (13), it is unclear whether invertebrate APC1 has related actin assembly activities to vAPC and/or collaborates with formins in a related manner.
Here, we addressed these open questions by purifying Drosophila APC1 and Dia and characterizing their individual and combined effects on actin assembly kinetics using bulk fluorescence assays and TIRF microscopy. We show that Drosophila APC1 and Dia each nucleates actin assembly and that they directly collaborate to assemble actin filaments in the combined presence of profilin and capping protein through mechanisms that appear to be remarkably conserved with their vertebrate counterparts.
EXPERIMENTAL PROCEDURES
Molecular Cloning
The C-terminal portion of Drosophila Dia (amino acids 512–1091; see Fig. 2A) was expressed from a modified pQE80 (Qiagen) vector as a fusion protein with a ZZ tag at the N terminus, and a His6 tag at the C terminus, graciously provided by Jörg Grosshans (25). The FH1 + FH2 fragment (amino acids 512–1002; see Fig. 2A) and the DAD-containing fragment (amino acids 1003–1091; see Fig. 2A) were PCR-amplified from this plasmid and cloned into the modified pQE80 vector. The Basic domain of Drosophila APC1 (APC1B; amino acids 2136–2417; see Fig. 1A) was PCR-amplified from a Drosophila APC1 cDNA (kindly provided by David Roberts, Franklin and Marshall College), cloned into pGEM-T-Easy (Promega), and subcloned into the BglII and EcoRI sites of pLM1 (26) (a modified pGEX-2T vector) for the GST fusion, or the NdeI and XhoI sites of pET15b (EMD Millipore) for the His6 fusion. The Basic domain contains a polymorphism Gln2347 compared with the Flybase record of His2347.
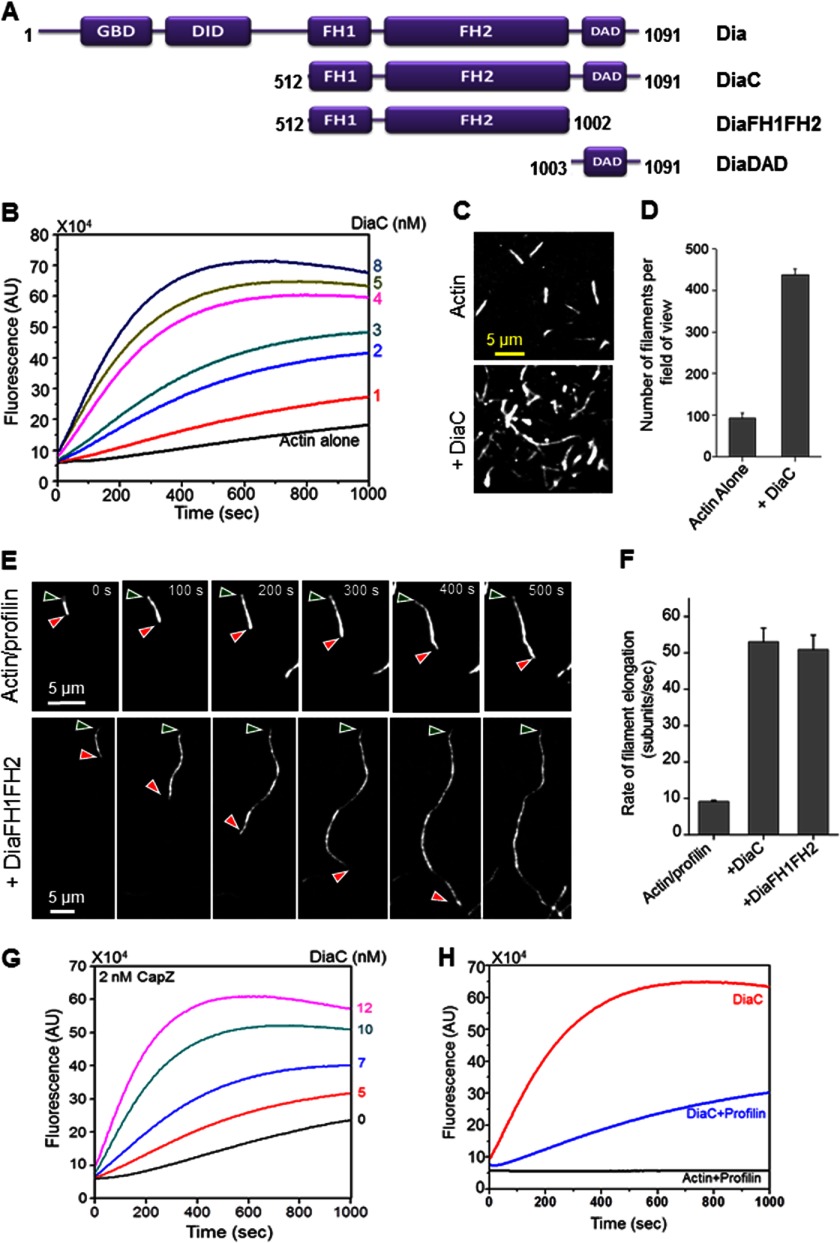
FIGURE 2.
Drosophila Dia nucleates actin assembly, accelerates filament elongation in the presence of profilin, and protects growing barbed ends from capping protein/CapZ. A, schematic of full-length Drosophila Dia and the constructs used in this paper. B, effects of DiaC (0–8 nm) on the assembly of 2 μm actin monomers (5% pyrene-labeled). C, TIRF microscopy images of actin filaments assembled from 1 μm monomeric actin (10% Oregon Green-labeled) with and without 2 nm DiaC. Images were taken 200 s after the initiation of actin assembly. D, quantification of number of filaments per TIRF microscopy field of view (135 × 135 μm) from reactions in C (n = 3 fields of view from each of three separate reactions). Error bars represent S.D. E, TIRF microscopy time-lapse images of filaments assembled from 1 μm actin monomers (10% Oregon Green-labeled) and 3 μm human profilin in the presence or absence of 2 nm DiaFH1FH2. Red and green arrowheads mark the barbed and pointed ends of filaments, respectively. Time is indicated in seconds. F, quantification of filament elongation rates from TIRF microscopy time lapse images as in E. Error bars represent S.E. (n >10 filaments). G, concentration-dependent effects of DiaC (0–12 nm) on the assembly of 2 μm actin monomers (5% pyrene-labeled) in the presence of 2 nm CapZ. H, comparison of rates of actin assembly induced by 2 nm DiaC in the presence and absence of 3 μm human profilin.
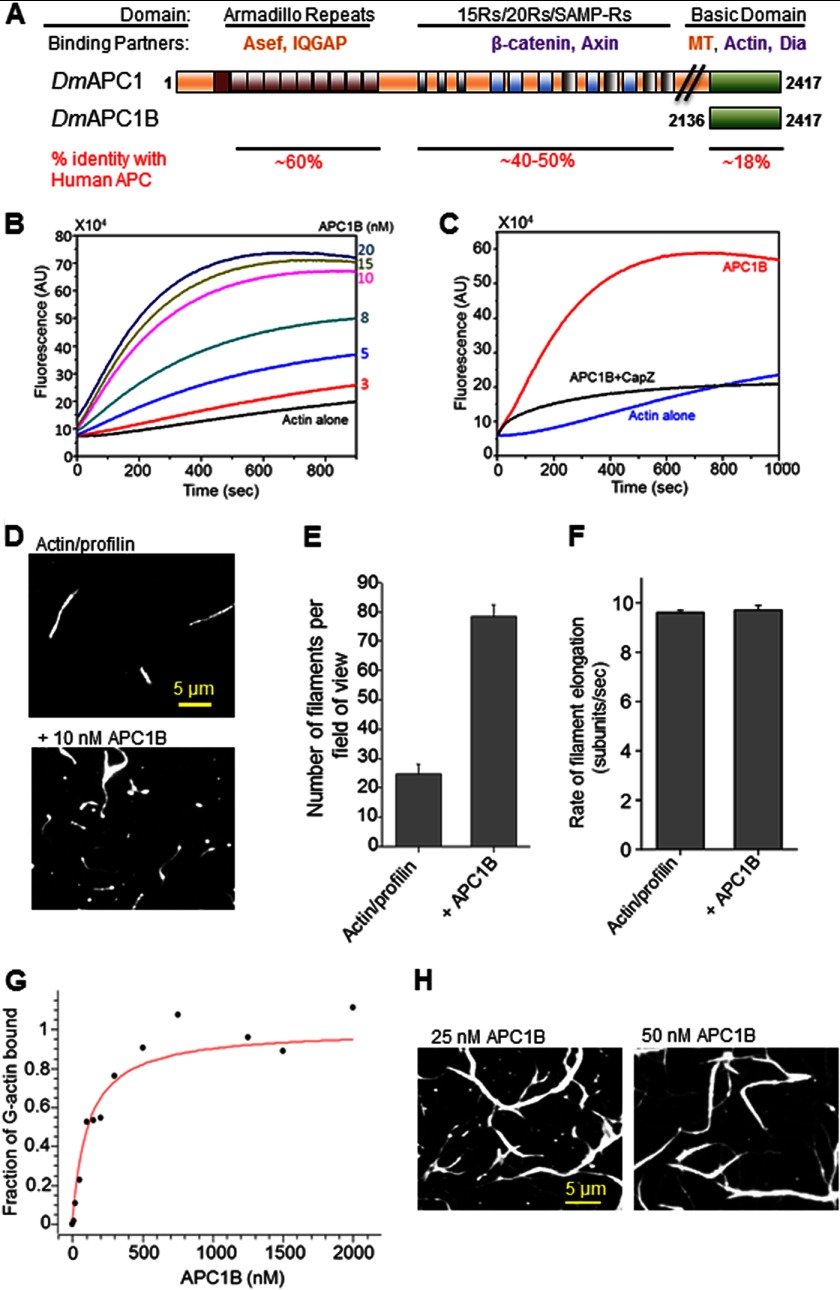
FIGURE 1.
Drosophila APC1B directly promotes actin filament nucleation and bundling. A, schematic of full-length Drosophila APC1 protein and the C-terminal APC1B fragment used in this study. Dm, Drosophila melanogaster; MT, microtubules. Binding partners indicated in purple have been identified for both vertebrate and Drosophila APC, whereas those in orange have been demonstrated for vAPC only. B, concentration-dependent effects of APC1B (0–20 nm) on the assembly of monomeric actin (2 μm, 5% pyrene-labeled) into filaments. C, effects of 2 nm CapZ on actin assembly (as above) induced by 20 nm APC1B. D, TIRF microscopy images showing the effects of 10 nm APC1B on the assembly of 1 μm actin (10% Oregon Green-labeled) in the presence of 3 μm human profilin. Images were taken 400 s after the initiation of actin assembly. E, quantification of number of filaments per field of view (135 × 135 μm) from TIRF microscopy reactions in D (n = 3 fields of view from each of three separate reactions). Error bars represent S.D. F, quantification of filament elongation rates from TIRF microscopy reactions in D. Error bars represent S.E. (n >10 filaments). G, concentration-dependent binding of APC1B (0–2 μm) to 100 nm G-actin (100% pyrene-labeled) in the presence of latrunculin B, measured in a fluorescence-based assay. H, TIRF microscopy images showing actin filament bundles assembled by 25 or 50 nm APC1B and 1 μm actin (10% Oregon Green-labeled).
Protein Purification
Rabbit skeletal muscle actin (RMA) was purified as described (27) and gel-filtered. Chicken capping protein (CapZ) and human profilin were purified as described (28, 29). For fluorometric assays, RMA was labeled with pyrenyl iodoacetamide using a standard protocol (30). For TIRF experiments, RMA was labeled with Oregon Green-actin as described (31). Vertebrate mDiaC (FH1-FH2-C) was expressed and purified from yeast as an N-terminal His6 fusion protein as described (5, 32). Drosophila DiaC, DiaFH1FH2, DiaDAD, and APC1B were expressed in BL21 DE3 Rosetta-2 cells (EMD Millipore), induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside, harvested, and snap frozen in liquid nitrogen. Following a quick thaw, cells were suspended in ice-cold lysis buffer (20 mm sodium phosphate, pH 8.0, 20 mm imidazole, pH 8.0, 500 mm NaCl, 14 mm β-mercaptoethanol, 0.1% Triton X-100, 5% glycerol, 200 μg/ml lysozyme, 0.2 μm PMSF, and 2 mm DTT for GST fusions) and sonicated. The lysate was cleared by centrifugation at 10,000 rpm in a Sorvall SA600 rotor for 10 min. The supernatant was incubated with 0.4 ml of either nickel-nitrilotriacetic acid or GST-agarose and eluted with 250 mm imidazole or 15 mm glutathione, respectively, in elution buffer (20 mm sodium phosphate, pH 8.0, 150 mm NaCl, 0.1% Triton X-100, 5% glycerol, 14 mm β-mercaptoethanol, or 1 mm DTT for GST fusion). Eluates were pooled and dialyzed in Spectra/Por 6000–8000 MWCO membrane tubing (Spectrum labs132 650) against at least a 250-fold excess of HEKG5 buffer (20 mm HEPES, pH 7.4, 1 mm EDTA, 50 mm KCl, 5% glycerol, 1 mm DTT) at 4 °C overnight, and against fresh buffer for 6 more h. Concentration of full-length protein was determined on SDS-polyacrylamide gels by comparison with known quantities of BSA. Proteins were stored at 4 °C for short term use or snap frozen and stored at −80 °C for long term use.
GST Pulldown Assays
GST-APC1B or GST alone was immobilized on glutathione-agarose beads (Sigma) in binding buffer (20 mm HEPES, pH 7.4, 50 mm KCl, 1 mm DTT). The beads were washed two times with a 20-fold volume of binding buffer and then resuspended in a 20-fold bead bed volume of binding buffer. Beads were mixed with soluble DiaC or Dia-DAD proteins (His6-tagged) and incubated together for 1 h at 4 °C. All proteins were used at a final concentration of 0.5 μm in each reaction. Beads were gently centrifuged at 2000 × g for 3 min, and the supernatant was saved for analysis on SDS-PAGE. Beads were washed by gently resuspending in a 50-fold bed volume of binding buffer followed by four washes with a 50-fold bed volume of binding buffer. Beads were then resuspended in 30 μl of 1× SDS sample buffer. Samples were loaded and fractionated on SDS-polyacrylamide gels, blotted, and probed with HRP-conjugated anti-His6 antibodies (Bethyl Laboratories, Montgomery, TX) at 1:10,000.
Pyrene-Actin Assembly Assays
Ca2+-ATP actin monomers (2 μm, 5% pyrene-labeled) in G-buffer (10 mm Tris-Cl−, 0.1 mm CaCl2, 0.2 mm DTT, and 0.2 mm ATP, pH 7.5) were converted to Mg2+-ATP actin before each reaction (32). The actin was mixed with other proteins or control buffer, and polymerization was initiated by addition of 3 μl of 20× initiation mix (40 mm MgCl2, 10 mm ATP, 1 m KCl). Pyrene fluorescence was monitored over time at excitation 365 nm and emission 407 nm at 25 °C in a fluorometer (Photon Technology International, Lawrenceville, NJ).
G-actin Binding Assays
For measuring the affinity of APC1B for G-actin (see Fig. 1G), 100 nm Ca-ATP-G-actin (100% pyrene-labeled) was preincubated with 500 nm latrunculin-B for 5 min in G-buffer. Variable concentrations of APC1B (0–2 μm) in HEKG5 buffer (20 mm HEPES-KOH, pH 7.5, 1 mm EDTA, 50 mm KCl, and 5% glycerol) were added to each reaction and incubated for 45 min at room temperature. Fluorescence intensities of the reactions were recorded after they reached steady state. Nonlinear fitting of the data was executed using SciDavis (free software application for Scientific Data Analysis and Visualization).
TIRF Microscopy
All procedures, including preparation of coverslips, assembly of flow cells, image acquisition, and image processing, were as described previously (23). All imaging was done on a Nikon-Ti2000 inverted microscope equipped with a cooled, back-illuminated EMCCD camera (Andor Ixon, Belfast, Northern Ireland), a 150-mW Ar+ laser (emission 488 nm, from Mellot Griot, Carlsbad, CA; operated at maximum power in all experiments), and a CFI Apo TIRF 60× H objective (Nikon Instruments Inc., New York, NY). Before each reaction, 4 μg/ml streptavidin in 20 mm Tris-Cl, pH 8.0, 1 mm DTT, 100 mm KCl was flowed in for 15 s, followed by washing with 50 μl of PBS + 1% BSA. The flow cell was then equilibrated with TIRF buffer (10 mm imidazole, pH 7.4, 50 mm KCl, 1 mm MgCl2, 1 mm EGTA, 0.2 mm ATP, 10 mm DTT, 15 mm glucose, 20 μg/ml catalase, 100 μg/ml glucose oxidase, and 0.5% methylcellulose (viscosity 4000 cP) (Sigma). For monitoring actin filament polymerization, actin regulatory proteins or control buffer was mixed with 1 μm actin (10% Oregon Green-labeled and 0.2% biotinylated) and introduced into the flow cell, which was then mounted on the microscope stage for imaging. Filament elongation rates were determined by measuring change in filament length over time (10 min) as described (23). Image acquisition was controlled by NIS-Elements software (Nikon Instruments Inc.).
RESULTS
The Basic Domain of Drosophila APC1 Promotes Actin Assembly in Vitro
The Basic domains of Drosophila APC1 and vAPC have limited sequence conservation (18% identity and 28% similarity; Fig. 1A and supplemental Fig. 1), which has left it uncertain whether the actin assembly-promoting properties of vertebrate APC extend to Drosophila APC1. To address this, we purified the APC1 Basic domain (APC1B) and asked whether it has actin nucleation activity. Like vertebrate APC-B (10), Drosophila APC1B was sufficient to promote actin assembly strongly at low nanomolar concentrations in a concentration-dependent manner (Fig. 1B). Next, we investigated whether APC1B nucleates filaments that have free barbed ends and therefore are vulnerable to the inhibitory effects of capping protein (CapZ). Addition of capping protein to reactions containing APC1B strongly suppressed the assembly of pyrene actin (Fig. 1C), suggesting that Drosophila APC1B, like vertebrate APC-B, does not protect barbed ends from capping protein (10).
Because bulk assays do not distinguish between the assembly of branched versus unbranched filaments, or between effects on actin filament nucleation versus elongation, we next performed TIRF microscopy to visualize the assembly of single filaments. TIRF analysis revealed that APC1B promotes the assembly of unbranched filaments (Fig. 1D) and increases the number of filaments per field of view (Fig. 1E) without affecting the rate of filament elongation (Fig. 1F). This suggests that APC1B primarily regulates the nucleation step of actin filament assembly rather than the elongation step.
vAPC is a tandem actin monomer binder that nucleates actin assembly by efficiently seeding polymer formation (10). Consistent with this mechanism, we found that Drosophila APC1B binds to G-actin strongly with a dissociation constant of ∼110 nm (Fig. 1G). Further, Drosophila APC1B induced bundling of actin filaments at higher concentrations (Fig. 1H), similar to the bundling properties of vAPC (19, 23).
Actin Assembly Properties of Drosophila Dia
We next expressed and purified a C-terminal construct of Drosophila Diaphanous that includes the FH1, FH2, and DAD domains (DiaC) (Fig. 2A) and characterized its effects on actin assembly. DiaC, like the equivalent C-terminal fragment of mDia1, induced actin assembly at low nanomolar concentrations (1–8 nm) (Fig. 2B). In addition, TIRF microscopy was performed to distinguish the effects of DiaC on actin nucleation versus elongation. We observed an increase in the number of filaments per field of view in the presence of DiaC (Fig. 2C, quantified in Fig. 2D), demonstrating that DiaC promotes nucleation. We also monitored increase in filament length over time in the presence and absence of DiaC, as well as a shorter Dia construct consisting of only the FH1 and FH2 domains (Fig. 2A; DiaFH1FH2) (Fig. 2E). Quantification of the rates of filament elongation (Fig. 2F) revealed that both DiaC and DiaFH1FH2 accelerated elongation by ∼5-fold compared with the rate of elongation in profilin-actin control reactions. This suggests a conserved mechanism for Dia-induced actin assembly and elongation in the presence of profilin. Similar to the behavior of mammalian mDia1C with profilin (23, 31), we observed that filaments assembled by Drosophila DiaFH1FH2 in the presence of profilin were also dimmer compared with filaments assembled in the absence of the formin (Fig. 2E); the same was observed for DiaC.4 This observation is explained by the reduced binding affinity of profilin for labeled actin monomers, which results in a decrease in the incorporation of labeled compared with unlabeled subunits into filaments elongated by formins (31).
Previously, many other formins have been shown to move processively on the growing barbed ends of filaments and protect them from the inhibitory effects of capping proteins (10, 23, 33). To determine whether Drosophila DiaC shares this mechanistic feature, we added increasing concentrations of DiaC to reactions containing a concentration of CapZ that inhibits assembly in the absence of formins. We observed a DiaC concentration-dependent increase in the rate of actin assembly in the presence of CapZ, indicating that DiaC protects growing barbed ends from capping protein (Fig. 2G). Importantly, this activity was not observed for APC1B (Fig. 1C).
Although formins collaborate with profilin in promoting filament elongation, profilin conversely suppresses formin-mediated actin nucleation by sequestering actin monomers (2). Therefore, we examined the effects of profilin on DiaC-induced actin assembly and found that, similar to its effect on vertebrate mDiaC, profilin suppressed the actin assembly activity of DiaC (Fig. 2H).
Drosophila APC1B and DiaC Collaborate to Assemble Actin Filaments
Given the conserved mechanisms of actin assembly displayed by Drosophila APC1 and DiaC, we next asked whether they could also collaborate to stimulate actin assembly, like their vertebrate counterparts, by overcoming a dual barrier to filament assembly imposed by profilin and capping protein (10). In the combined presence of profilin and CapZ, Drosophila APC1B and DiaC individually failed to induce actin assembly (Fig. 3A). However, when APC1B and DiaC were combined they overcame this barrier and promoted robust actin assembly. Under these conditions, APC1B and DiaC together produced ∼10-fold higher activity than for either protein alone and ∼4-fold higher activity compared with the sum of their individual effects (Fig. 3B). These findings were corroborated using TIRF microscopy, which revealed that APC1B and DiaC synergistically increased the average number of filaments per field of view (Fig. 3, C and D). We also noticed an accumulation of immobilized fluorescent actin spots in reactions containing DiaC and to a lesser extent in reactions containing DiaFH1FH2. In addition, a subset of brighter spots was visible in reactions containing APC1B (Fig. 3C). These spots could represent short actin filaments initiated by APC1B and/or Dia that become capped by CapZ early in the reactions, or alternatively, they could represent immobilized APC1B and/or Dia molecules that retain the ability to recruit G-actin but are not capable of polymerizing a filament.
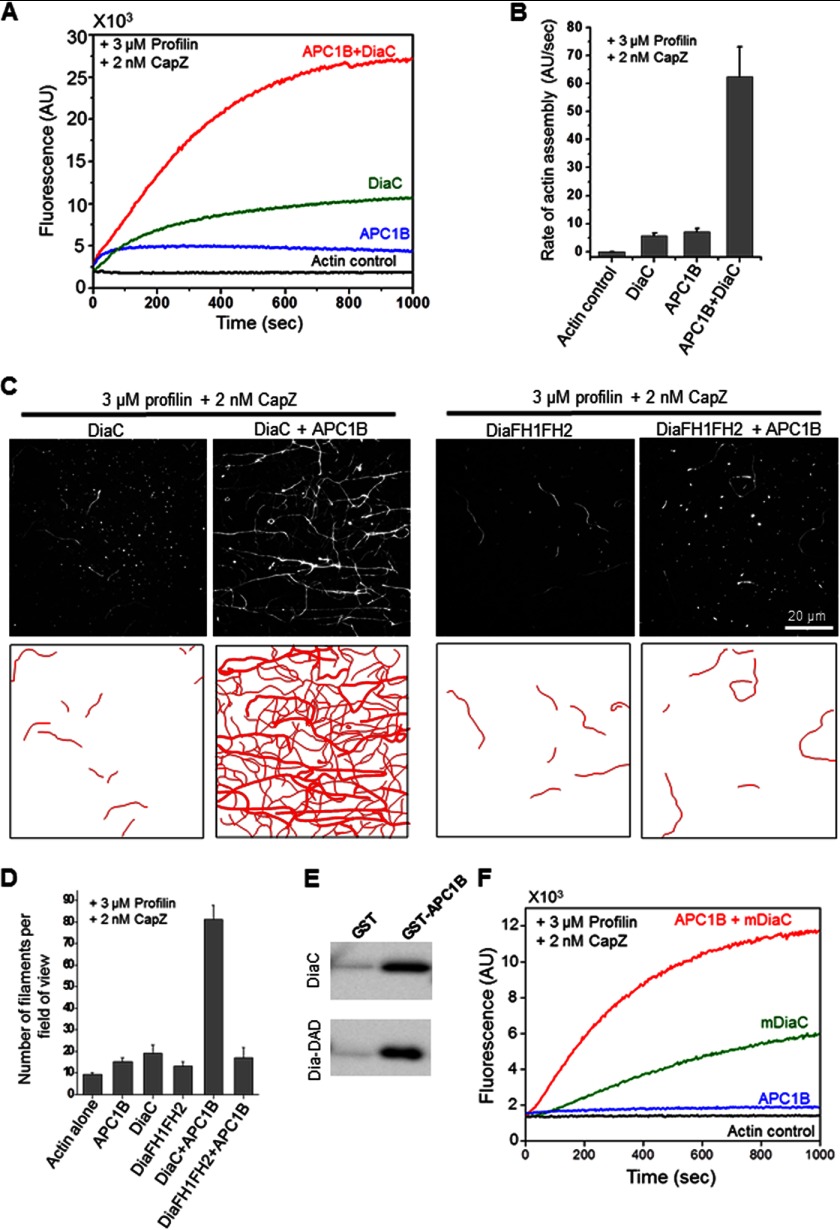
FIGURE 3.
APC1B and DiaC directly interact and collaborate to stimulate actin assembly in the combined presence of profilin and capping protein/CapZ. A, effects of 20 nm APC1B and 2 nm DiaC, individually and combined, on the assembly of 2 μm actin monomers (5% pyrene-labeled) in the presence of 3 μm human profilin and 2 nm CapZ. B, quantification of actin assembly rates determined from the slopes of curves from reactions as in A. Error bars represent S.D. (n = 2). C, upper panels, TIRF microscopy images (top panels) comparing the effects of 2 nm DiaC or DiaFH1FH2 with and without 5 nm APC1B. All reactions contained 1 μm actin monomers (10% Oregon Green-labeled), 3 μm human profilin, and 2 nm CapZ. Images were captured 400 s after initiation of actin assembly. C, lower panels, corresponding filament traces. D, quantification of number of filaments per field of view for reactions as in C (n = 3 fields of view from each of three separate reactions). Error bars represent S.D. E, Western blot probed with antibody against His6 tag showing levels of soluble His6-DiaC and His6-DiaDAD that bound to GST-APC1B or GST on beads. F, cross-species effects of 20 nm Drosophila APC1B with and without 2 nm vertebrate mDiaC on the assembly of 2 μm actin monomers (5% pyrene-labeled) in the presence of 3 μm human profilin and 2 nm CapZ.
Previous studies have shown that the vertebrate APC-Dia collaboration in actin assembly depends on a direct physical interaction between the C-terminal DAD domain-containing region of mDia1 and the APC Basic domain (23). Therefore, we tested whether Drosophila APC1B and DiaC similarly interact directly. In bead binding assays, soluble DiaC specifically bound to immobilized GST-APC1B, but not GST (Fig. 3E). Further, a much shorter C-terminal tail fragment of Dia that contains the DAD domain (DiaDAD; Fig. 2A) was sufficient to bind GST-APC1B (Fig. 3E). Finally, we asked whether DiaFH1FH2, which lacks the APC1B binding C-terminal tail region, but alone has similar effects on actin assembly compared with DiaC (Fig. 2F), is capable of collaborating with APC1B. In stark contrast to DiaC, DiaFH1FH2 failed to collaborate with APC1B in overcoming inhibition by profilin and capping protein (Fig. 3, C and D). We also tested whether Drosophila APC1B was capable of collaborating with vertebrate mDia1 (mDiaC) in promoting actin assembly. Remarkably, this cross-species invertebrate and vertebrate NPF-formin pair exhibited collaborative effects in actin filament assembly in the combined presence of CapZ and profilin (Fig. 3F).
Taken together, these results demonstrate that Drosophila APC1B and DiaC collaborate by an evolutionarily conserved mechanism to promote actin assembly and that this collaboration requires their direct interaction. Further, our results suggest that the collaborative effects are derived in part from an ability of APC1B and DiaC to nucleate filaments more effectively together than alone and in part from the ability of DiaC to accelerate filament elongation. All of these findings are highly consistent with the activities of the vertebrate counterparts (10, 23).
DISCUSSION
APC is a central regulator of cell polarity, cytoskeleton remodeling, and cell differentiation that is broadly conserved from humans and other vertebrates to echinoderms and insects. The C-terminal Basic domain of vertebrate APC binds directly to actin and microtubules (19, 34, 35), strongly nucleates actin assembly alone and in collaboration with formins (10, 23), bundles filaments (19), and helps localize APC to cytoskeleton-rich regions of cells (17, 36–40). However, it was unclear until now whether these cytoskeletal activities of vertebrate APC are conserved in the invertebrate homologues given that there is limited sequence conservation within the Basic domain. Here we investigated this issue by purifying the Basic domain of Drosophila APC1 and defining its effects on the kinetics of actin assembly in vitro. Similar to the Basic domain of vertebrate APC, Drosophila APC1B bound to actin monomers, directly and potently stimulated actin nucleation, did not affect the rate of filament elongation, and assembled filaments with free barbed ends that were strongly inhibited by capping protein (Fig. 1C). Further, at higher concentrations APC1B promoted actin filament bundling, as observed for vertebrate APC-B (Fig. 1D and Ref. 19). Thus, despite the lack of sequence conservation, Drosophila APC1B shows strikingly conserved activities with vertebrate APC-B in regulating actin dynamics.
In addition, because vertebrate APC-B collaborates with the Diaphanous formin mDia1 in stimulating actin assembly (10, 23), we investigated the potential for a Drosophila APC-Dia collaboration. We started by purifying the C-terminal half of the Drosophila formin Diaphanous, which contains the conserved FH1, FH2, and DAD domains (DiaC) and characterizing its effects on actin assembly. Similar to its mammalian formin counterparts (mDia1 and mDia2), Drosophila DiaC nucleated actin assembly, accelerated the rate of filament elongation in a profilin-dependent manner, and protected the growing ends of filaments from capping protein (Fig. 2). Further, Drosophila APC1B and Dia directly interacted and collaborated to stimulate filament assembly, overcoming the dual barrier to assembly imposed by profilin and capping protein (Fig. 3). These properties are remarkably indistinguishable from collaborations between the vertebrate homologues (10, 23). Further, Drosophila APC1B was capable of collaborating with vertebrate mDia1 to stimulate actin assembly (Fig. 3F). These data, together with our biochemical characterization of APC1B, demonstrate that despite the low sequence homology shared between the Basic regions of fly and vertebrate APC, the interactions of this domain with both actin and formins are highly conserved.
These findings contribute to an emerging body of work on the relationship between formins and their interacting co-factors or NPFs, which together provide an enhanced platform for filament formation in vitro. Identification of these factors has helped to address the longstanding question of how formins nucleate new filaments in vitro and in cells in the presence of multiple inhibitory factors. It is now clear that formins utilize a variety of domains and partners to accomplish nucleation, including the monomer binding DAD domain (5, 41) and direct collaboration with actin monomer-binding NPFs including Bud6, APC, and Spire (9, 10, 23, 42–48). These NPF activities can be mediated by a variety of domains, including WASp homology 2 (WH2) or WH2-like domains (WASp, Bud6, and Spire), the Basic domain (APC), and Gelsolin repeats (Fli-I) (7, 9, 10, 23, 42–48). Despite these structural differences, a number of common mechanistic features are shared among most of these NPFs, such as binding to the formin C-terminal tail, binding to actin monomers, and interacting with microtubules.
It is also interesting to note that collaborator pairs are not monogamous. Bud6 directly collaborates with two different formins in yeast, Bni1 and Bnr1, which is achieved through distinct mechanisms that have different regulatory requirements5 (9). Fli-I can collaborate in vitro and in cultured cells with two different formins, Daam1 and mDia1 (7). APC functions in pathways with both Daam1 and mDia1 (10, 13, 18, 23, 49, 50). Thus, it appears that a given NPF can have more than one collaborator, and a given formin may associate with more than one NPF. This mix-and-match system provides a broader repertoire of related mechanisms for formin regulation that could play important physiological roles in complex multicellular organisms with the need to control actin assembly in spatial, temporal, and tissue-specific patterns.
One question that this growing appreciation for formin cofactors raises is whether this is an obligate relationship in vivo for most or only some formins. On a fundamental level, the physiological roles of formin collaborations are not yet well understood. The interactions between Bud6 and the formin Bni1, and Spire and the formin Capu/Fmn2, have clear physiological significance in yeast, and in Drosophila and mammals, respectively (8, 9, 12, 51). Bud6 and Bnr1 are required for the assembly of actin cables that direct polarized cell growth in Saccharomyces cerevisiae (9). Loss of Spire in the Drosophila ovary leads to loss of an actin meshwork essential for normal oogenesis (8, 51), and loss of both Spire 1 and 2 in mouse oocytes results in a reduced cytoplasmic actin network that is necessary for asymmetric spindle positioning and oocyte maturation (12). Whereas overexpression of the human APC basic domain in cultured fibroblasts leads to an mDia1-dependent increase in actin assembly (10), the physiological role of the vertebrate APC-Dia collaboration has not yet been established in a true in vivo context. Drosophila presents an opportunity to fill this gap in our understanding.
In Drosophila, genetic analysis of APC1 has defined clear functional roles in Wnt signaling (52–54) as well as in non-Wnt-mediated processes, including photoreceptor morphogenesis, centrosome orientation, cadherin-based adhesion, and maintenance of muscle-tendon junctions (55–58). Some of these functions may be attributed to the potent actin nucleation activity of APC1, revealed here, as well as its interactions with microtubules (53). There is also a clear role for Drosophila Diaphanous in regulating actin-based processes including cytokinesis and coordinating adhesion and contractility of the actomyosin ring that underlies the adherens junction during dorsal closure, an embryonic epithelial closure event analogous to wound healing (6, 13, 22, 59). In addition, Drosophila APC2 and Dia together promote actin furrow formation in the syncytial embryo (13), suggesting that APC-Dia collaborations play a physiological role in Drosophila development. The strong in vitro collaboration between fly APC1 and Dia that we observed, together with that of vertebrate APC and mDia1, suggests that APC contributes directly to actin-based cellular processes via its Basic domain and points to a conserved physiological importance for APC in directly regulating actin dynamics. The remarkable conservation in functional activities between the vertebrate and Drosophila APC Basic domains, and between the vertebrate and Drosophila APC-Dia collaboration, strongly suggests that what we learn about the in vivo role of APC-Dia mediated actin assembly in Drosophila will illuminate our understanding of the human proteins, their roles in normal actin assembly, and the consequence of disruption to human disease. The current study lays the foundation for that future in vivo work.
Acknowledgments
We thank Dennis Breitsprecher for preparing TIRF reagents; Jörg Grosshans, David Roberts, the Gordon Rule laboratory, and the Adam Linstedt laboratory for providing useful reagents and equipment; and Dennis Breitsprecher and Avital Rodal for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants GM083137 and GM098143 (to B. L. G.) and GM073891 (to B. M. M.).

This article contains supplemental Fig. 1.
R. Jaiswal, unpublished observations.
B. Graziano and B. L. Goode, unpublished observations.
- APC
- adenomatous polyposis coli
- APC1B
- APC1 Basic domain
- Dia
- Diaphanous
- DAD
- Diaphanous autoregulatory domain
- FH
- formin homology
- NPF
- nucleation-promoting factor
- TIRF
- total internal reflection fluorescence
- vAPC
- vertebrate APC.
REFERENCES
- 1. Campellone K. G., Welch M. D. (2010) A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11, 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chesarone M. A., DuPage A. G., Goode B. L. (2010) Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat. Rev. Mol. Cell Biol. 11, 62–74 [DOI] [PubMed] [Google Scholar]
- 3. Faix J., Grosse R. (2006) Staying in shape with formins. Dev. Cell 10, 693–706 [DOI] [PubMed] [Google Scholar]
- 4. Goode B. L., Eck M. J. (2007) Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76, 593–627 [DOI] [PubMed] [Google Scholar]
- 5. Gould C. J., Maiti S., Michelot A., Graziano B. R., Blanchoin L., Goode B. L. (2011) The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr. Biol. 21, 384–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Castrillon D. H., Wasserman S. A. (1994) Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development 120, 3367–3377 [DOI] [PubMed] [Google Scholar]
- 7. Higashi T., Ikeda T., Murakami T., Shirakawa R., Kawato M., Okawa K., Furuse M., Kimura T., Kita T., Horiuchi H. (2010) Flightless-I (Fli-I) regulates the actin assembly activity of diaphanous-related formins (DRFs) Daam1 and mDia1 in cooperation with active Rho GTPase. J. Biol. Chem. 285, 16231–16238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dahlgaard K., Raposo A. A., Niccoli T., St Johnston D. (2007) Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev. Cell 13, 539–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Graziano B. R., DuPage A. G., Michelot A., Breitsprecher D., Moseley J. B., Sagot I., Blanchoin L., Goode B. L. (2011) Mechanism and cellular function of Bud6 as an actin nucleation-promoting factor. Mol. Biol. Cell 22, 4016–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Okada K., Bartolini F., Deaconescu A. M., Moseley J. B., Dogic Z., Grigorieff N., Gundersen G. G., Goode B. L. (2010) Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J. Cell Biol. 189, 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rosales-Nieves A. E., Johndrow J. E., Keller L. C., Magie C. R., Pinto-Santini D. M., Parkhurst S. M. (2006) Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nat. Cell Biol. 8, 367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pfender S., Kuznetsov V., Pleiser S., Kerkhoff E., Schuh M. (2011) Spire-type actin nucleators cooperate with Formin-2 to drive asymmetric oocyte division. Curr. Biol. 21, 955–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Webb R. L., Zhou M. N., McCartney B. M. (2009) A novel role for an APC2-Diaphanous complex in regulating actin organization in Drosophila. Development 136, 1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McCartney B. M., Näthke I. S. (2008) Cell regulation by the APC protein APC as master regulator of epithelia. Curr. Opin. Cell Biol. 20, 186–193 [DOI] [PubMed] [Google Scholar]
- 15. Näthke I. (2006) Cytoskeleton out of the cupboard: colon cancer and cytoskeletal changes induced by loss of APC. Nat. Rev. Cancer 6, 967–974 [DOI] [PubMed] [Google Scholar]
- 16. Näthke I. S. (2004) The adenomatous polyposis coli protein: the Achilles heel of the gut epithelium. Annu. Rev. Cell Dev. Biol. 20, 337–366 [DOI] [PubMed] [Google Scholar]
- 17. Rosin-Arbesfeld R., Ihrke G., Bienz M. (2001) Actin-dependent membrane association of the APC tumour suppressor in polarized mammalian epithelial cells. EMBO J. 20, 5929–5939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wen Y., Eng C. H., Schmoranzer J., Cabrera-Poch N., Morris E. J., Chen M., Wallar B. J., Alberts A. S., Gundersen G. G. (2004) EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 6, 820–830 [DOI] [PubMed] [Google Scholar]
- 19. Moseley J. B., Bartolini F., Okada K., Wen Y., Gundersen G. G., Goode B. L. (2007) Regulated binding of adenomatous polyposis coli protein to actin. J. Biol. Chem. 282, 12661–12668 [DOI] [PubMed] [Google Scholar]
- 20. Kawasaki Y., Senda T., Ishidate T., Koyama R., Morishita T., Iwayama Y., Higuchi O., Akiyama T. (2000) Asef, a link between the tumor suppressor APC and G-protein signaling. Science 289, 1194–1197 [DOI] [PubMed] [Google Scholar]
- 21. Watanabe T., Wang S., Noritake J., Sato K., Fukata M., Takefuji M., Nakagawa M., Izumi N., Akiyama T., Kaibuchi K. (2004) Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev. Cell 7, 871–883 [DOI] [PubMed] [Google Scholar]
- 22. Homem C. C., Peifer M. (2008) Diaphanous regulates myosin and adherens junctions to control cell contractility and protrusive behavior during morphogenesis. Development 135, 1005–1018 [DOI] [PubMed] [Google Scholar]
- 23. Breitsprecher D., Jaiswal R., Bombardier J. P., Gould C. J., Gelles J., Goode B. L. (2012) Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science 336, 1164–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McCartney B. M., Price M. H., Webb R. L., Hayden M. A., Holot L. M., Zhou M., Bejsovec A., Peifer M. (2006) Testing hypotheses for the functions of APC family proteins using null and truncation alleles in Drosophila. Development 133, 2407–2418 [DOI] [PubMed] [Google Scholar]
- 25. Grosshans J., Wenzl C., Herz H. M., Bartoszewski S., Schnorrer F., Vogt N., Schwarz H., Müller H. A. (2005) RhoGEF2 and the formin Dia control the formation of the furrow canal by directed actin assembly during Drosophila cellularisation. Development 132, 1009–1020 [DOI] [PubMed] [Google Scholar]
- 26. Pai L. M., Kirkpatrick C., Blanton J., Oda H., Takeichi M., Peifer M. (1996) Drosophila α-catenin and E-cadherin bind to distinct regions of Drosophila armadillo. J. Biol. Chem. 271, 32411–32420 [DOI] [PubMed] [Google Scholar]
- 27. Spudich J. A., Watt S. (1971) The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 28. Moseley J. B., Sagot I., Manning A. L., Xu Y., Eck M. J., Pellman D., Goode B. L. (2004) A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell 15, 896–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soeno Y., Abe H., Kimura S., Maruyama K., Obinata T. (1998) Generation of functional β-actinin (CapZ) in an E. coli expression system. J. Muscle Res. Cell Motil. 19, 639–646 [DOI] [PubMed] [Google Scholar]
- 30. Cooper J. A., Walker S. B., Pollard T. D. (1983) Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J. Muscle Res. Cell Motil. 4, 253–262 [DOI] [PubMed] [Google Scholar]
- 31. Kovar D. R., Harris E. S., Mahaffy R., Higgs H. N., Pollard T. D. (2006) Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124, 423–435 [DOI] [PubMed] [Google Scholar]
- 32. Moseley J. B., Maiti S., Goode B. L. (2006) Formin proteins: purification and measurement of effects on actin assembly. Methods Enzymol. 406, 215–234 [DOI] [PubMed] [Google Scholar]
- 33. Kovar D. R., Wu J. Q., Pollard T. D. (2005) Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast. Mol. Biol. Cell 16, 2313–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deka J., Kuhlmann J., Müller O. (1998) A domain within the tumor suppressor protein APC shows very similar biochemical properties as the microtubule-associated protein tau. Eur. J. Biochem. 253, 591–597 [DOI] [PubMed] [Google Scholar]
- 35. Munemitsu S., Souza B., Müller O., Albert I., Rubinfeld B., Polakis P. (1994) The APC gene product associates with microtubules in vivo and promotes their assembly in vitro. Cancer Res. 54, 3676–3681 [PubMed] [Google Scholar]
- 36. Jarrett C. R., Blancato J., Cao T., Bressette D. S., Cepeda M., Young P. E., King C. R., Byers S. W. (2001) Human APC2 localization and allelic imbalance. Cancer Res. 61, 7978–7984 [PubMed] [Google Scholar]
- 37. Langford K. J., Askham J. M., Lee T., Adams M., Morrison E. E. (2006) Examination of actin- and microtubule-dependent APC localisations in living mammalian cells. BMC Cell Biol. 7, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Langford K. J., Lee T., Askham J. M., Morrison E. E. (2006) Adenomatous polyposis coli localization is both cell type- and cell context-dependent. Cell Motil. Cytoskeleton 63, 483–492 [DOI] [PubMed] [Google Scholar]
- 39. Miyashiro I., Senda T., Matsumine A., Baeg G. H., Kuroda T., Shimano T., Miura S., Noda T., Kobayashi S., Monden M. (1995) Subcellular localization of the APC protein: immunoelectron microscopic study of the association of the APC protein with catenin. Oncogene 11, 89–96 [PubMed] [Google Scholar]
- 40. Näthke I. S., Adams C. L., Polakis P., Sellin J. H., Nelson W. J. (1996) The adenomatous polyposis coli tumor suppressor protein localizes to plasma membrane sites involved in active cell migration. J. Cell Biol. 134, 165–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Heimsath E. G., Jr., Higgs H. N. (2012) The C terminus of formin FMNL3 accelerates actin polymerization and contains a WH2 domain-like sequence that binds both monomers and filament barbed ends. J. Biol. Chem. 287, 3087–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bosch M., Le K. H., Bugyi B., Correia J. J., Renault L., Carlier M. F. (2007) Analysis of the function of Spire in actin assembly and its synergy with formin and profilin. Mol. Cell 28, 555–568 [DOI] [PubMed] [Google Scholar]
- 43. Pechlivanis M., Samol A., Kerkhoff E. (2009) Identification of a short Spir interaction sequence at the C-terminal end of formin subgroup proteins. J. Biol. Chem. 284, 25324–25333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Quinlan M. E., Hilgert S., Bedrossian A., Mullins R. D., Kerkhoff E. (2007) Regulatory interactions between two actin nucleators, Spire and Cappuccino. J. Cell Biol. 179, 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sitar T., Gallinger J., Ducka A. M., Ikonen T. P., Wohlhoefler M., Schmoller K. M., Bausch A. R., Joel P., Trybus K. M., Noegel A. A., Schleicher M., Huber R., Holak T. A. (2011) Molecular architecture of the Spire-actin nucleus and its implication for actin filament assembly. Proc. Natl. Acad. Sci. U.S.A. 108, 19575–19580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tu D., Graziano B. R., Park E., Zheng W., Li Y., Goode B. L., Eck M. J. (2012) Structure of the formin-interaction domain of the actin nucleation-promoting factor Bud6. Proc. Natl. Acad. Sci. U.S.A. 109, E3424–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vizcarra C. L., Kreutz B., Rodal A. A., Toms A. V., Lu J., Zheng W., Quinlan M. E., Eck M. J. (2011) Structure and function of the interacting domains of Spire and Fmn family formins. Proc. Natl. Acad. Sci. U.S.A. 108, 11884–11889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zeth K., Pechlivanis M., Samol A., Pleiser S., Vonrhein C., Kerkhoff E. (2011) Molecular basis of actin nucleation factor cooperativity: crystal structure of the Spir-1 kinase non-catalytic C-lobe domain (KIND)*formin-2 formin SPIR interaction motif (FSI) complex. J. Biol. Chem. 286, 30732–30739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Habas R., Kato Y., He X. (2001) Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell 107, 843–854 [DOI] [PubMed] [Google Scholar]
- 50. Yamana N., Arakawa Y., Nishino T., Kurokawa K., Tanji M., Itoh R. E., Monypenny J., Ishizaki T., Bito H., Nozaki K., Hashimoto N., Matsuda M., Narumiya S. (2006) The Rho-mDia1 pathway regulates cell polarity and focal adhesion turnover in migrating cells through mobilizing APC and c-Src. Mol. Cell. Biol. 26, 6844–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Manseau L. J., Schüpbach T. (1989) Cappuccino and Spire: two unique maternal-effect loci required for both the anteroposterior and dorsoventral patterns of the Drosophila embryo. Genes Dev. 3, 1437–1452 [DOI] [PubMed] [Google Scholar]
- 52. Ahmed Y., Nouri A., Wieschaus E. (2002) Drosophila APC1 and APC2 regulate Wingless transduction throughout development. Development 129, 1751–1762 [DOI] [PubMed] [Google Scholar]
- 53. Akong K., Grevengoed E. E., Price M. H., McCartney B. M., Hayden M. A., DeNofrio J. C., Peifer M. (2002) Drosophila APC2 and APC1 play overlapping roles in Wingless signaling in the embryo and imaginal discs. Dev. Biol. 250, 91–100 [DOI] [PubMed] [Google Scholar]
- 54. Hayashi S., Rubinfeld B., Souza B., Polakis P., Wieschaus E., Levine A. J. (1997) A Drosophila homolog of the tumor suppressor gene adenomatous polyposis coli down-regulates β-catenin but its zygotic expression is not essential for the regulation of Armadillo. Proc. Natl. Acad. Sci. U.S.A. 94, 242–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ahmed Y., Hayashi S., Levine A., Wieschaus E. (1998) Regulation of Armadillo by a Drosophila APC inhibits neuronal apoptosis during retinal development. Cell 93, 1171–1182 [DOI] [PubMed] [Google Scholar]
- 56. De Graeve F. M., Van de Bor V., Ghiglione C., Cerezo D., Jouandin P., Ueda R., Shashidhara L. S., Noselli S. (2012) Drosophila APC regulates delamination of invasive epithelial clusters. Dev. Biol. 368, 76–85 [DOI] [PubMed] [Google Scholar]
- 57. Subramanian A., Prokop A., Yamamoto M., Sugimura K., Uemura T., Betschinger J., Knoblich J. A., Volk T. (2003) Shortstop recruits EB1/APC1 and promotes microtubule assembly at the muscle-tendon junction. Curr. Biol. 13, 1086–1095 [DOI] [PubMed] [Google Scholar]
- 58. Yamashita Y. M., Jones D. L., Fuller M. T. (2003) Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301, 1547–1550 [DOI] [PubMed] [Google Scholar]
- 59. Afshar K., Stuart B., Wasserman S. A. (2000) Functional analysis of the Drosophila diaphanous FH protein in early embryonic development. Development 127, 1887–1897 [DOI] [PubMed] [Google Scholar]