Abstract
Advances in imaging techniques and high-throughput technologies are providing scientists with unprecedented possibilities to visualize internal structures of cells, organs and organisms and to collect systematic image data characterizing genes and proteins on a large scale. To make the best use of these increasingly complex and large image data resources, the scientific community must be provided with methods to query, analyze and crosslink these resources to give an intuitive visual representation of the data. This review gives an overview of existing methods and tools for this purpose and highlights some of their limitations and challenges.
By their very nature, microscopy and magnetic resonance imaging (MRI) (Fig. 1 and Boxes 1 and 2) are dependent on data visualization. Whereas in the past it was considered sufficient to show images (photographs or digitized images) in the printed version of an article to illustrate an experimental result, the presentation of image data has become more challenging for three reasons. First, new imaging techniques allow the generation of massive datasets that cannot be adequately presented on paper nor be browsed and looked at with older software tools. MRI, which is mostly used to acquire three-dimensional (3D) imagery, has faced some of these problems for many years. Second, the availability of high-throughput techniques enables experiments on a large scale, generating large sets of image data, and even though the readout of each single experiment may be easily visualized, this is no longer true for whole screens consisting of thousands of such experiments. Third, microscopy and MRI are increasingly part of a broader analytical context that may include quantitative measurement, statistical analysis, mathematical modeling and simulation and/or automated reasoning over multiple datasets reflecting different properties and possibly resulting from different acquisition techniques at different scales of resolution, often generated at different institutions. This review describes how the visualization challenges in these three areas are addressed for a range of imaging modalities.
Figure 1.
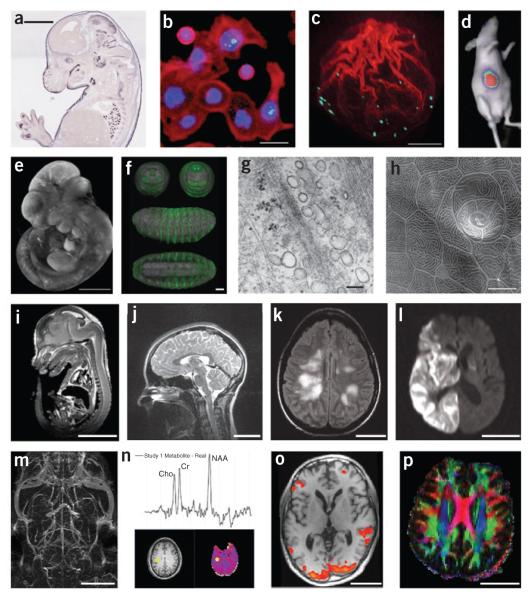
Imaging techniques. (a) Brightfield microscopy: mouse embryo, in situ expression pattern of Irx1, Eurexpress; scale bar, 2 mm. (b) Fluorescence microscopy: HT29 cells stained for DNA (blue), actin (red) and phospho-histone H3 (green)75; scale bar, 20 μm. (c) Confocal microscopy: actin polymerization along the breaking nuclear envelope during meiotic maturation of a starfish oocyte. Actin filaments, red (rhodamine-phalloidin stain); chromosomes, cyan (Hoechst 33342 stain). Projection of confocal sections, (image courtesy P. Lénárt); scale bar, 20 μm. (d) Bioluminescence imaging: in vivo bioluminescence imaging of mice after implantation of Gli36-Gluc cells76, (figure courtesy B.A. Tannous). (e) Optical projection tomography: mouse embryo, EMAP33,66; scale bar, 1 mm. (f) Single/selective plane illumination microscopy: late-stage Drosophila embryo probed with anti-GFP antibody and DRAQ5 nuclear marker: frontal, caudal, lateral and ventral views of the same embryo77; scale bar, 50 μm. (g) Transmission electron microscopy: human fibroblast, glancing section close to surface (image courtesy R. Parton and M. Floetenmeyer); scale bar, 100 nm. (h) Scanning electron microscopy: zebrafish peridermal skin cells (courtesy R. Parton and M. Floetenmeyer); scale bar, 10 μm. (i) microMRI: mouse embryo (source: http://mouseatlas.caltech.edu/); scale bar, 5 mm. (j) T2-weighted MRI: human cervical spine (source: http://www.radswiki.net/); scale bar, 5 cm. (k) Fluid attenuation inversion recover (FLAIR) image of a human brain with acute disseminated encephalomyelitis. Bright areas indicate demyelination and possibly some edema (image courtesy N. Salamon); scale bar, 5 cm. (l) Diffusion-weighted image of a human brain after a stroke. Bright areas indicate areas of restricted diffusion (image courtesy N. Salamon); scale bar, 5 cm. (m) Maximum intensity projection image of a magnetic resonance angiogram of a C57BL/6J mouse brain acquired in vivo using blood pool contrast78 (image courtesy G. Howles); scale bar, 5 mm. (n) 3D proton magnetic resonance spectroscopic imaging study of normal human brain. Graph shows proton spectrum for the brain location identified by yellow markers on the T1-weighted MRI (lower left) and N-acetylaspartate (NAA; lower right) images. Data acquired using the MIDAS/EPSI methodology79 (image courtesy J. Alger); scale bar, 5 cm. (o) Functional MRI activation map overlaid on a T1-weighted MRI: human brain (image courtesy L. Foland-Ross); scale bar, 5 cm. (p) Direction-encoded color map computed from DTI. Red, left–right directionality; green, anterior–posterior; blue, superior–inferior; scale bar, 5 cm.
BOX 1. MICROSCOPY TECHNIQUES.
Brightfield microscopy with colorimetric stains is the primary technique for capturing tissue and whole organism morphology (Fig. 1a). For high-throughput capture of in situ expression patterns, automated bright-field microscopy has been used for whole-genome projects such as the Allen Brain Atlas.
Widefield fluorescence microscopy is the most widely used imaging technique in biology (Fig. 1b). Fluorescent markers make it possible to see particular structures with high contrast, either in fixed samples using immunostaining or in living cells with expressed GFP-tagged proteins83. The resolution is limited by diffraction to about 200 nm.
Confocal scanning microscopy generates optical sections through a specimen by pointwise scanning of different focal planes and thereby reduces both scattered light from the focal plane and out-of-focus light84. The image quality of twodimensional images is therefore improved, and 3D images can be taken (axial resolution is typically 2–3 times lower than lateral resolution; see Fig. 1c). The method is also applicable to live cell imaging. There are variants of this method increasing axial resolution (for example, 4Pi microscopy)85.
Computational optical sectioning microscopy (COSM) achieves optical sectioning by taking a series of two-dimensional images with a widefield microscope focusing in different planes of the specimen84. Out-of-focus light is then removed computationally.
Structured illumination microscopy acquires several widefield images at different focal planes using spatial illumination patterns84. As the out-of-focus light is less dependent on the spatial illumination pattern than the in-focus light, combinations of different images at the same focal plane under laterally shifted illumination patterns allow computational attenuation of out-of-focus light.
Two-photon microscopy is similar to confocal scanning microscopy but uses nonlinear excitation involving two-photon (or multiphoton) absorption86. This allows the use of longer excitation wavelengths, permitting deeper penetration into the tissue and—owing to the nonlinearity—confines emission to the perifocal region, leading to substantial reduction of scattering.
Super-resolution fluorescence microscopy groups several recently developed methods in light microscopy capable of significantly increasing resolution and visualizing details at the nanometer scale. In stimulated emission depletion (STED) microscopy85, the focal spot is ‘narrowed’ by overlapping it with a doughnut-shaped spot that prevents the surrounding fluorophores from fluorescing and thereby contributing to the collected light. In PALM (photo-activated localization microscopy)87 and STORM (stochastic optical reconstruction microscopy)88, subsets of the fluorophores present are activated and localized. Iterating this process and combining the acquired raw images yields a high-resolution image.
Bioluminescence imaging (Fig. 1d) is based on the detection of light produced by luciferase-mediated oxidation of a substrate in living organisms. Transfected cells expressing luciferase can be injected into animals, or transgenic animals can be created that express luciferase as a reporter gene. When such animals are injected with a luciferase substrate, light is produced by the luciferase-expressing cells in the presence of oxygen. The bioluminescence image is often superimposed on a white-light image to show localization of the light-producing cells.
Optical projection tomography captures object projections in different directions as line integrals of the transmitted light89 (Fig. 1e). From these projections (corresponding to the ‘shadow’ of the object), a volumetric model can be calculated by means of back-projection algorithms.
Light sheet–based fluorescence microscopy uses a thin sheet of laser light for optical sectioning and a perpendicularly oriented objective with a CCD camera for detection of the fluorescent signal. Single- or selective plane illumination microscopy (SPIM)90 (Fig. 1f) adds sample rotation that enables acquisition of large samples from multiple angles. Low phototoxicity, high acquisition speed and ability to cover large samples make it particularly suitable for in toto time-lapse imaging of developing biological specimens, such as model organism embryos, with cellular resolution.
Transmission electron microscopy (TEM) (Fig. 1g) uses accelerated electrons instead of visible light for imaging. As a result, the achievable resolution (typically 2 nm) is much higher than in light microscopy. The method is not applicable to live cell imaging, and the specimen preparation is technically very complex. In electron tomography, the specimen is physically sectioned and 3D images are obtained by imaging each section at progressive angles of rotation, followed by computational reassembly to yield a tomogram. Resolution ranges from 20–30 nm to 5 nm or less.
Scanning electron microscopy (SEM) (Fig. 1h) produces an image of the 3D structure of the surface of the specimen by collecting the scattered electrons (rather than the transmitted electrons as in TEM). The resolution is typically lower than for TEM.
BOX 2. MAGNETIC RESONANCE IMAGING TECHNIQUES.
Magnetic resonance imaging uses the intrinsic nuclear magnetization of materials to probe their general physical and chemical structure. A sample to be imaged is first placed in a strong static magnetic field. Gradients in the static field force the Larmor frequency (resonance frequency) of the sample’s atomic nuclei to be a function of their spatial position within the sample space. The sample is then excited by a carefully crafted radio frequency electromagnetic pulse that deflects the magnetic moments of the sample’s nuclei away from their steady-state orientation. The relaxation of the magnetic moments back to their steady state creates a radio frequency echo that is detected by an acquisition system. The composition of the material, the spatially dependent Larmor frequency and the magnetic pulse itself determine the characteristics of that echo. Variations in the power, orientation and duration of the radio frequency pulse allow different tissue properties to be probed while retaining some details of differentiation (different composition) and position. Paramagnetic T1 contrast agents, such as gadolinium, may be injected into the subject. The agent alters the relaxation characteristic of water, and the image appears hyperintense in areas of contrast agent concentration; applications include vascular imaging (Fig. 1m) and detection of active tumors or lesions. Some of the widely used acquisition methods are described below.
Clinical MRI devices typically use static field strengths in the range of 1.5–3T and have resolutions on the order of 1 mm. Small-animal scanners apply the same principles but use stronger field strengths (typically in the range 7–11T) and are capable of resolutions on the order of tens of microns.
T1 applies a short excitation time and a short relaxation time; fat appears bright, water appears dark. In brain images, white matter appears bright, gray matter slightly darker and cerebrospinal fluid very dark (Fig. 1o).
T2 typically uses a long excitation time and a long relaxation time; fluid (for example, cerebrospinal fluid) appears bright in these scans, and fat is less bright (Fig. 1j).
T2* (usually pronounced “T2-star”) is observed in long-excitation-time gradient echo images; contrast is sensitive to local magnetic field inhomogeneities produced, for example, by iron oxide T2 contrast agents and air-tissue interfaces.
Proton density information is obtained from scans with a short excitation time and a long relaxation time, or by extrapolating relaxation-weighted datasets back to zero time.
Fluid attenuation inversion recovery (FLaIr) pulse sequences suppress the fluid signal, which allows otherwise hidden fluid-covered lesions to be observed (Fig. 1k).
magnetic resonance angiography uses the water proton signal to produce millimeterscale images of arteries and veins without the addition of contrast agents.
magnetic resonance spectroscopy acquires localized spectra from a defined region within the sample, with spectral peaks indicating the presence of various metabolites or biomolecules such as lactate, creatine, phosphocreatine and glutamate (Fig. 1l).
Functional MRI (fMRI) measures the signal change that occurs when blood is deoxygenated; neuronal activity relates to increased oxygen demand, allowing maps of activation to be made by examining the blood oxygenation level–dependent (BOLD) signal (Fig. 1o).
Diffusion MRI uses the reduction in the detected MR signal produced by diffusion of water molecules along the magnetic gradient. Areas with lower diffusion are affected less than areas with high diffusion, producing brighter signals (Fig. 1l). Performing multiple acquisitions with different gradients and field strengths allows models of the directionality of the local diffusion properties to be resolved in the form of diffusion tensors (DTI) or more complicated patterns (Q-Ball and DSI). The diffusion properties are governed by local physical structures in the material. (Fig. 1p).
To be useful to the immediate research group and more broadly to the scientific community, massive datasets must be presented in a way that enables them to be browsed, analyzed, queried and compared with other resources—not only other images but also molecular sequences, structures, pathways and regulatory networks, tissue physiology and micromorphology. In addition, intuitive and efficient visualization is important at all intermediate steps in such projects: proper visualization tools are indispensable for quality control (for example, identification of dead cells, ‘misbehaving’ markers or image acquisition artifacts), the sharing of generated resources among a network of collaborators or the setup and validation of an automated analysis pipeline.
The first section of this review briefly describes issues related to digital images. The second section deals with visualization techniques for complex multidimensional image datasets at relatively low throughput. Next, we discuss typical visualization problems arising with an increase in scale: here, the challenges are to provide tools allowing the user to navigate through large image-derived datasets at different levels of abstraction and to develop meaningful profiles and clustering methods. The last section deals with how images can be shared with collaborators or with the community. Finally, we conclude with the need for integration and linking of different image-based source data (including computational models) into a comprehensive view of biological entities.
Accessing the images
Digital representation of images
The use of digital images as a convenient replacement for photographic film has paved the way for the increase in the volume of images produced. While we expect a digital image to carry the same amount of visual information as its analog counterpart, it is amenable to faster and more complex processing, and the task of viewing an image is complicated by the lack of standard image representation. Whereas photographic film used to provide a common format for image representation, digital images have different formats with respect to the number of bits per pixel or whether the encoded values are signed or unsigned.
Although most image-handling software programs support unsigned 8-bit images (values between 0 and 255) and unsigned 16-bit images (values between 0 and 65,535), care must be taken with more ‘exotic’ formats, such as unsigned 12-bit images (values between 0 and 4,095) or signed 16-bit images (values between −32,768 and 32,767), which are routinely produced by modern imaging equipment. If, for instance, an unsigned 12-bit image is simply interpreted as an unsigned 16-bit image, only ~6% of the dynamic range will be used and the images may appear ‘dark’. If the image is rescaled to cover the maximal dynamic range (as it is the default behavior of many image viewers), the absolute intensity information is lost, which makes any comparison between different images impossible. Signed values are also often misinterpreted by the image-handling software (for example, negative values may be ignored). Although the above may be trivial issues for imaging experts, they are pitfalls routinely encountered by biologists.
Image file formats
The fields of microscopy and MRI both face significant challenges in the sharing and processing of data owing to the variety of digital file formats that are used.
For microscopy images, no format has been adopted as a universal standard. Faced with a choice, many new users are unaware that image quality is degraded when using a file format that relies on a lossy compression algorithm (for example, JPEG). Image files can also hold further information about the image. Instrument manufacturers use either a proprietary format or a customized version of a pre-existing extensible format (for example, TIFF) to include metadata such as the time the image was acquired within the image file itself. These embedded metadata usually do not survive conversion to another format. To address this important issue, the BioFormats project has been working to create translators for a variety of image formats and has accomplished this task for over 70 image file formats so far (http://www.loci.wisc.edu/software/bio-formats).
However, most high-dimensional and high-throughput projects require devising a system to store and query further metadata about the images. For example, interpreting a time-lapse experiment requires understanding which images represent which time points for which samples, and there is no standard way of organizing the images to reflect this information (typically, a time-lapse experiment is stored as a stack of images, where the time information is encoded in the file names). Hence, researchers must often rely on their notes to determine what each image represents, which becomes an issue, particularly when the data are to be shared between collaborators. The most common practice is to duplicate images and share metadata in spreadsheets, although a suitable laboratory information management system (LIMS) informatics platform could be used for managing the metadata in a reliable and convenient way. An attempt to overcome these issues is the OMERO platform from the Open Microscopy Environment (OME), which provides a client-server system for managing images and their associated metadata through a common interface1–3.
Commercial microscopy and image analysis software companies often engage in format ‘wars’, whereas open-source solutions struggle to bridge the gaps among the many proprietary formats. A movement toward universally adopted standards, with a degree of data integration like that which has been achieved for genome sequences (for example, GenBank) and microarray data (for example, MIAME), must become a common goal of industry and academia.
MRI is an inherently digital medium and similarly faces problems with file formats. Acquisition systems from different scanners often use proprietary file formats. Though clinical scanners support the DICOM standard managed by NEMA, the Association of Electrical and Medical Imaging Equipment Manufacturers, writing a validly formatted DICOM image file is neither practical nor required for many academic imaging projects. In addition, emerging imaging techniques are often not fully standardized within DICOM, and implementation of the standard varies by scanner vendor. As a result, investigators often rely on file formats that are both simpler and better able to capture the parameters required for their particular domains. The Analyze 7.5 file format (Analyze Direct) has been widely used in many software packages, but its interpretation often differs among these. As a result, ambiguities arise regarding the orientation of the stored data, and great care must be taken to ensure that the right and left sides of the image volume are interpreted correctly. Furthermore, the Analyze format is not designed to store much of the metadata that is contained in DICOM or other proprietary formats.
The NIfTI file format4 (http://nifti.nimh.nih.gov/nifti-1/) was recently developed to address many of these problems and is rapidly becoming the standard in the neuroimaging community. It is supported by many of the popular image analysis suites and provides unambiguous information about image orientation, additional codes that describe aspects of the image including its intent and a standardized method for adding extensions to the format. Although standardized formats address many interoperability issues, significant challenges remain in digitally describing the full experimental paradigm used to collect the data. For example, functional MRI stimulus paradigms must typically be hand coded into an application-specific proprietary format for statistical analysis. Similar issues appear in the analysis of dynamic contrast enhanced images, diffusion images and other new scanning techniques.
Visualization of high-dimensional image data
As technology develops, images are carrying more and more information in the form of additional dimensions. Typically, these dimensions correspond to space (3D imaging techniques; Fig. 1c,e–f,i–p), time (for example, live cell imaging, functional MRI; Fig. 1o) and channels (for example, different fluorescent markers, multispectral imaging; Fig. 1b,c,f). Emerging microscopy techniques, such as single plane illumination microscopy (SPIM; Fig. 1f) or high-throughput, time-lapse live cell imaging, combine all these dimensional expansions and generate massive 3D, time-lapse, multichannel acquisitions. High-dimensional visualization is not limited to raw image data; it can also be useful for understanding features derived from the image data, such as segmentations of cells and annotations of subcompartments or tissues. Care must be taken in the interpretation of visualized samples, analysis results and derived measurements, as each acquisition method has its own resolution limitation, and therefore not all biological structures might be imaged at sufficient resolution to show relevant detail. Last but not least, intuitive visualization using simulated behavior of biological entities can aid understanding of scientific methods, models and hypotheses not only for scientists themselves but also for the general public. Visualization and analysis of many complex datasets are beyond the capabilities of existing software packages and rely on cuttingedge research in computer graphics and computer vision fields.
In most biological experiments, visualization means displaying the variations in several channels over the spatiotemporal dimensions. As standard computer monitors can only display two spatial dimensions directly, some sort of data reduction must be applied to visualize multidimensional images. The simplest solution is to display only selected dimensions from the multidimensional dataset at a time—for instance, one two-dimensional image—and allow the user to interactively change the remaining dimensions. Because computer memory becomes limiting for large datasets, multidimensional image browsers must ensure that only data that are being viewed are loaded into memory. Proper memory management is particularly important for online browsing applications that must minimize the amount of image data transferred between the client and the server5.
3D visualization techniques
Multidimensional images can typically be observed as a collection of separate slice planes, but often dimensions are combined using various projection methods to form a single display object (Fig. 2). For two-dimensional display, one spatial dimension can be collapsed by an orthographic projection (for example, maximum intensity projection), creating a partially flattened image (Fig. 2a,b). The projection can also be applied along any other axis, such as time (creating a kymogram) or joint display of color-coded channels. A more advanced technique, the perspective projection, preserves the 3D appearance of the object in the two-dimensional projection image (Fig. 2c). In perspective projection, the geometry of the image is modified to have the x and y coordinates of objects in the image converge toward vanishing points, whereas in the so-called isometric projection, the original sizes of the objects are preserved. Perspective views look more realistic, but isometric views are useful if the image is to be used for distance measurements.
Figure 2.
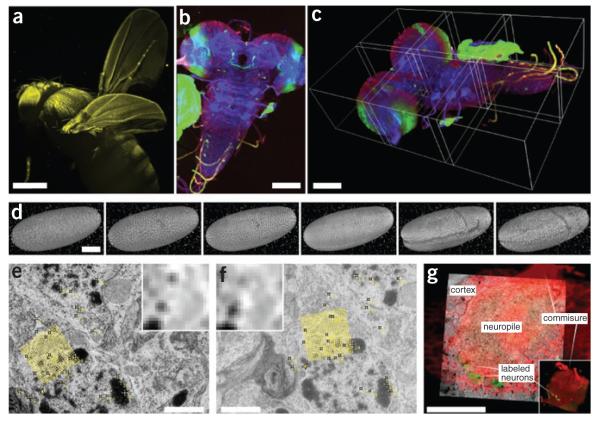
Visualization of high–dimensional image data. (a) SPIM scan of autofluorescent adult Drosophila female gives an impression of 3D rendering in maximum intensity projection (image courtesy D.J. White); scale bar, 100 μm. (b) Maximum-intensity projection of tiled 3D multichannel acquisition of Drosophila larval nervous system; scale bar, 400 μm. (c) The corresponding 3D rendering in Fiji 3D viewer; borders of the tiles are highlighted; scale bar, 100 μm. (d) Visualization of gastrulation in Drosophila expressing His-YFP in all cells by time-lapse SPIM microscopy. The images show six reconstructed time points covering early Drosophila embryonic development rendered in Fiji 3D viewer. Fluorescent beads visible around sample were used as fiduciary markers for registration of multi-angle SPIM acquisition; scale bar, 100 μm. (e,f) Two consecutive slices from serial section transmission electron microscopy dataset of first-instar larval brain. Yellow marks, corresponding SIFT features that can be used for registration; yellow grid, position and orientation of one of the SIFT descriptors; inset, corresponding pixel intensities in the area covered by the descriptor; scale bar, 1 μm. (g) Multimodal acquisition of Drosophila first-instar larval brain by confocal (red, green) and electron microscopy (underlying gray). The two separate specimens were registered using manually extracted corresponding landmarks (not shown). Main anatomical landmarks of the brain correspond in the two modalities after registration (white labels). (Electron microscopy images courtesy A. Cardona; confocal image courtesy V. Hartenstein). Scale bar, 20 μm.
Projections can be combined with other techniques from computer graphics, such as wire frame models, shading, reflection and illumination, to create a realistic 3D rendering of the biological object. When only the outer shape of the 3D object needs to be realistically visualized, surface rendering of the manually or automatically extracted outlines of organelles, cells or tissue can help in assessing their topological arrangement within the 3D volume of the imaged specimen. In contrast, when the interior of 3D objects is of interest, ‘volume rendering’ coupled with transparency manipulations or orthogonal sectioning is required.
In direct volume rendering, viewing rays are projected through the data6. Data points in the volume are sampled along these rays, and their visual representation is accumulated using a transfer function that maps the data values to opacity and color values (Fig. 3a). The transfer functions can be adjusted to emphasize different structures or features and may introduce color or opacity changes as a function of the local intensity gradient. Similarly, the intensity gradient vector can be used to emulate the effect of external light sources interacting with tissue boundaries. Although direct volume rendering can be computationally expensive, the advent of high-powered graphical processing units has allowed many software tools (for example, OsiriX, ImageVis3D in SCIRun, 3D Slicer7,8, VTK (Tables 1 and 2)) to provide these capabilities interactively on personal computers.
Figure 3.
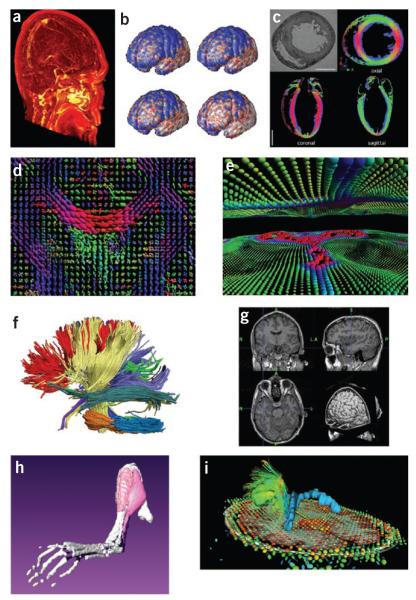
Visualization of anatomical features in MRI. (a) Volume rendering of a difference image computed from a pre- and post-gadolinium contrast scan. Brighter areas indicate a concentration of gadolinium, emphasizing the vasculature. (b) Time-lapse imaging of a subpopulation with Alzheimer’s disease showing loss of cortical gray matter density at 0, 6, 12 and 18 months80. Blue, no significant difference in cortical thickness from elderly control subjects; red and white, significant differences in cortical thickness (image courtesy P. Thompson). (c) Cardiac MRI analysis using anatomical scans and DTI of an ex vivo rat heart81. Color encoding of the DTI indicates the direction of the primary eigenvector: x direction, green; y, red; z, blue. (d) Visualization of a human brain DTI field during a fluid deformation process for image registration82. Orientation and shape of each ellipsoid indicate the pattern of diffusion at that location. Color encoding: low diffusion, green, to high diffusion, red. (e) Interactive visualization of high angular resolution diffusion imaging (HARDI) data using spherical harmonics27. Each shape represents the orientation distribution function measured at that point, which indicates the probability of diffusion in each angular direction. Colors indicate direction of maximum probability: red, lateral; blue, inferior–superior; green, anterior–posterior. Visible in this frame are portions of corpus callosum (central red area) and corticospinal tracts (blue vertical areas near edges). (f) White matter tracts computed from diffusion spectral imaging (DSI) data using Diffusion Toolkit (http://www.trackvis.org/dtk/). The tracts were then clustered automatically into bundles based on shape similarity measures and finally rendered using BrainSuite27. Each color indicates a different bundle. (g) 3D orthogonal views of an MRI volume, displayed with an automatically extracted surface mesh model of the surface of the cerebral cortex (BrainSuite27). (h) 3D surface reconstructions (Amira) from micro-MRI data: left hindlimb of a mouse with peroneal muscular atrophy. (i) Surgical planning visualization for assessment of white matter integrity: tumor model (green mass), ventricles (blue), local diffusion for one slice plane (ellipsoid scale and orientation indicating local diffusion tensor: red, low anisotropy; blue, high) and white matter fiber tracts shaded red to blue with increasing local anisotropy (thin lines, peri-tumoral; thick lines, corticospinal tracts). 3D Slicer: http://wiki.na-mic.org/Wiki/index.php/IGT:ToolKit/Neurosurgical-Planning.
Table 1.
A selective list of image visualization tools
| Name | Cost | OS | Description | URL |
|---|---|---|---|---|
|
Stand-alone
| ||||
| Amira* | $ | Win, Mac, Linux | Multichannel 4D images, image processing, extensible (in C++), scripting (in Tcl) |
http://www.amiravis.com/ |
| Arivis | $ | Win | Multichannel 4D images, image acquisition, processing, collaborative annotation and browsing, extensible (MATLAB and Python) |
http://www.arivis.com/ |
| Axiovision | $ | Win | Multichannel 4D images, image acquisition, image processing | http://tiny.cc/cZUbB |
| BioImageXD | Free | Win, Mac, Linux | 3D image analysis and visualization, in Python using VTK library | http://www.bioimagexd.net/ |
| Blender | Free | Win, Mac, Linux | 3D content creation suite, the open source Maya | http://www.blender.org/ |
| Fiji* | Free | Win, Mac, Linux | ImageJ distribution focused on registration and analysis of confocal and electron microscopy data. Six scripting languages, extensive wiki documentation, video tutorials |
http://pacific.mpi-cbg.de/ |
| Imaris* | $ | Win, Mac | Multichannel 4D images, image processing | http://www.bitplane.com/ |
| IMOD | Free | Win, Mac, Linux | Monochannel 4D images, extensible (in C/C++) | http://tiny.cc/kfLgQ |
| Huygens | $ | Win, Mac, Linux | Multichannel 4D images, image processing, scripting (in Tcl), web interface for batch processing |
http://www.svi.nl/ |
| Image-Pro | $ | Win Multichannel | 4D images, image acquisition, image processing | http://www.mediacy.com/ |
| ImageJ* | Free | Win, Mac, Linux | Image processing, extensible (in Java) | http://rsbweb.nih.gov/ij/ |
| LSM image browser |
Free | Win | Multichannel 4D images | http://tiny.cc/WMHsE |
| MetaMorph | $ | Win | Multichannel 4D images, image acquisition, image processing, extensible (in Visual Basic), scripting (with macros) |
http://tiny.cc/YrCK3 |
| POV Ray | Free | Win, Mac, Linux | High-quality tool for creating impressive 3D graphics | http://www.povray.org/ |
| Priism/IVE | Free | Mac, Linux | Multichannel 4D images, image processing, extensible (in C and Fortran) | http://tiny.cc/SIAMF |
| V3D, VANO and Cell Explorer |
Free | Win, Mac, Linux | 3D Image visualization, analysis and annotation | http://tiny.cc/JWdFb |
| VisBio | Free | Win, Mac, Linux | Multichannel 3D images, image processing (with ImageJ), connection to an OMERO server |
http://tiny.cc/TOZad/ |
| Volocity | $ | Win, Mac | Multichannel 4D images, image acquisition, image processing | http://www.improvision.com/ |
| VOXX | Free | Win, Mac, Linux | Real-time rendering of large multichannel 3D and 4D microscopy datasets | http://tiny.cc/b0KRt |
| VTK* | Free | Win, Mac, Linux | Library of C++ code for 3D computer graphics, image processing and visualization |
http://www.vtk.org/ |
|
Web-based
| ||||
| Brain Maps | Free | Win, Mac, Linux | Interactive multiresolution next-generation brain atlas | http://brainmaps.org/ |
| CATMAID | Free | Win, Mac, Linux | Collaborative Annotation Toolkit for Massive Amounts of Image Data: distributed architecture, modeled after Google Maps |
http://fly.mpi-cbg.de/~saalfeld/catmaid/ |
Recommended and popular tools. Free means the tool is free for academic use; $ means there is a cost. OS, operating system: Win, Microsoft Windows; Mac, Macintosh OS X. Tools running on Linux usually also run on other versions of Unix. 4D, four-dimensional.
Table 2.
A selective list of MRI visualization tools
| Name | Cost | OS | Description | URL |
|---|---|---|---|---|
| 3D Slicer* | Free | Win, Mac, Linux | Tools for visualization, registration, segmentation and quantification of medical data; extensible; uses VTK and ITK |
http://www.slicer.org/ |
| Amira | $ | Win, Mac, Linux | Allows 2D slices to be viewed from any angle; provides image segmentation, 3D mesh generation; surface rendering; data overlay and quantitative measurements |
http://www.amiravis.com/ |
| Analyze | $ | Win, Mac, Linux | Many processing and visualization features for many types of medical imaging data | http://tiny.cc/gXO76 |
| Anatomist | Free | Win, Mac, Linux | Visualization software that works in concert with BrainVisa; can map data onto 3D renderings of the brain; provides manual drawing tools |
http://brainvisa.info/ |
| AVS | $ | Win, Mac, Linux | General purpose data visualization package | http://www.avs.com/ |
| BioImage Suite* |
Free | Win, Mac, Linux | Tools for biomedical image analysis; includes preprocessing, voxel-based classification; image registration; diffusion image analysis; cardiac image analysis; fMRI activation detection |
http://bioimagesuite.org/ |
| BrainSuite | Free | Win, Mac, Linux | Automated cortical surface extraction from MRI; orthogonal image viewer; automated and interactive segmentation and labeling; surface visualization |
http://tiny.cc/Qxv6x |
| BrainVisa | Free | Win, Mac, Linux | Toolbox for segmentation of T1-weighted images; performs classification and mesh generation on brain images; automated sulcal labeling |
http://brainvisa.info/ |
| BrainVoyager | $ | Win, Mac, Linux | Analysis and visualization of MRI and fMRI data and for EEG and MEG distributed source imaging |
http://tiny.cc/kFKhv |
| Cardiac Image Modeller |
$ | Irix | Visualization and functional analysis, in 3D space and through time of cardiac cine data | http://tiny.cc/4KY6E |
| DTIStudio | Free | Win | Tools for tensor calculation, color mapping, fiber tracking and 3D visualization | http://tiny.cc/pvU9B |
| FreeSurfer | Free | Mac, Linux | Automated tools for reconstruction of the brain’s cortical surface from structural MRI data and overlay of functional MRI data onto the reconstructed surface |
http://tiny.cc/H3uG5 |
| FSL* | Free | Win, Mac, Linux | Comprehensive library of analysis tools for fMRI, MRI and DTI brain imaging data; includes widely used registration and segmentation tools |
http://tiny.cc/NFPHO |
| ImageJ | Free | Win, Mac, Linux | Image processing, extensible (in Java), large user community | http://rsb.info.nih.gov/ij/ |
| ImagePro | $ | Win | 2D and 3D image processing and enhancement software | http://www.mediacy.com/ |
| ITK | Free | Win, Mac, Linux | Extensive suite of software tools for image analysis | http://www.itk.org/ |
| Jim | $ | Win, Mac, Linux | Calculates T1 and T2 relaxation times, magnetization transfer, diffusion maps from MRI data | http://www.xinapse.com/ |
| MBAT | Free | Win, Mac, Linux | Workflow environment bringing together online resources, a user’s image data and biological atlases in a unified workspace; extensible via plug-ins |
http://tiny.cc/W2Tx2 |
| MedINRIA | Free | Win, Mac, Linux | Many algorithms dedicated to medical image processing and visualization; provides many modules, including DTI and HARDI viewing and analysis |
http://tiny.cc/RCptw |
| MIPAV | Free | Win, Mac, Linux | Quantitative analysis and visualization of medical images of numerous modalities such as PET, MRI, CT or microscopy |
http://mipav.cit.nih.gov/ |
| OpenDX | Free/$ | Win, Mac, Linux | General-purpose data visualization package | http://www.opendx.org/ |
| OsiriX* | Free | Mac | Image processing and viewing tool for DICOM images, provides 2D viewers, 3D planar reconstruction, surface and volume rendering, export to QuickTime |
http://tiny.cc/kOTzy |
| SCIRun | Free | Win, Mac, Linux | Environment for modeling, simulation and visualization of scientific problems; includes many biomedical analysis components, such as BioTensor, BioFEM and BioImage |
http://tiny.cc/eLufx |
| SPM | Free | Win, Mac, Linux | Analysis of brain imaging data sequences; applies statistical parametric mapping methods to sequences of images; widely used in fMRI; provides segmentation and registration |
http://tiny.cc/dVc7v |
| TrackVis | Free | Win, Mac, Linux | Tools to visualize and analyze fiber track data from diffusion MRI (DTI, DSI, HARDI, Q-Ball) tractography |
http://trackvis.org/ |
| TractoR | Free | Linux | Tools to segment comparable tracts in group studies using FSL tractography | http://tiny.cc/OsBH9 |
| VTK* | Free | Win, Mac, Linux | Library of C++ code that implements many state-of-the-art visualization techniques with a consistent developer interface |
http://www.vtk.org/ |
Recommended and popular tools. Free means the tool is free for academic use; $ means there is a cost; free/$ means free for Windows and Linux, at a cost for Mac OS X. OS, operating system: Win, Microsoft Windows; Mac, Macintosh OS X. Tools running on Linux usually also run on other versions of Unix. Irix is SGI’s Unix operating system. 2D, two-dimensional; CT, computed tomography; PET, positron emission tomography; HARDI, high angular resolution diffusion imaging.
Direct volume rendering has the advantage of requiring little preprocessing to produce high-quality renderings of multidimensional data and is best suited for data in which the structures of interest are readily differentiated by the pixel intensity. When this is not the case, further analytical techniques are required to clearly visualize these structures. It is also possible to view all three spatial dimensions at once. In stereoscopic views, an image is presented to the right eye and the same image rotated by a small angle is presented to the left eye. This can be achieved by presenting the two images in the two halves of the monitor or by superimposing the two images with a small relative shift. The final frontier in this area is volume visualization of biological image data that combines various visualization approaches and couples them to virtual reality environments to allow not only seamless navigation through the data but also intuitive interaction with the visualized biological entities.
Treatment of the time dimension
The changes along the time axis in dynamically changing biological specimens are best visualized by assembling a static gallery of images from different time points (for printed media, see Fig. 2d) or presenting a movie (for the web). The biological processes are often too slow to be shown in real time, and time-lapse techniques, where the frames are replayed faster, may reveal surprising details. Nevertheless, movies are significant simplifications of the acquired multidimensional data (for example, they do not allow rigorous time point comparisons and typically discard too much of the captured data) and researchers should always have the possibility of browsing through the raw data along arbitrary dimensions. Movies of 3D volume renderings of biological data tend to be particularly impressive but require substantial computational power. Alternatively, signal changes over time can be visualized as heat maps overlaid on the other dimensions of the image9, on normalized reference templates or on surface models of cellular or anatomical structures. For example, functional MRI acquires many images in the span of several minutes during the application of some study paradigm. These are then processed using statistical methods to produce maps of activation, using tools such as SPM10 or FSL11. These maps may then be aligned to higher-spatial-resolution structural MRIs to provide anatomical context for the functional activation information (Fig. 1o).
Analysis and visualization of temporal information also depend on the time scale of the studies. Typical studies in molecular biology cover relatively short time intervals, ranging from several minutes to several hours and sometimes to several days. Often these studies require reliable tracking algorithms12,13 that enable researchers to follow the same object (for example, single molecules or cells) over time and to extract and visualize trajectories and other measurements (for example, color-coded speed of cells in a developing embryo14). In other cases, imaging is only a means to derive various parameters, whose kinetics are then visualized. For instance, variations in fluorescence intensity over time can be used to measure diffusion coefficients or concentrations. Another example is high temporal resolution MRI (‘cine-MRI’) of the heart, where epi- and endocardial borders can be traced in the images to obtain global cardiac functional parameters such as ventricular volumes or wall thickening. Alternatively, displacement or velocities of the ventricular wall can be measured temporally to quantify transmural wall motion and to assess cardiac function regionally.
In the medical sciences, there is interest in long-term studies—often measuring effects over time periods of several months or years. For example, in neurology, the ability to accurately measure the local thickness of the cerebral cortex provides an important measure of pathological changes associated with Alzheimer’s disease and other types of cognitive decline. Since these studies rely upon separate acquisitions and compare multiple subjects, the data must be registered spatially (see Registration below) for the time series to be analyzed. These dynamic effects are often analyzed using many subjects; this adds the requirement that all subject data be spatially resampled to bring them into anatomical correspondence. The FreeSurfer package, for instance, accomplishes this using a cortical surface matching technique that aligns brains on the basis of their cortical folding patterns and is widely used for detecting these brain changes, for populations and for single subjects over time15–17. Once such a spatial normalization has been performed, statistical maps may be computed to examine changes in various biomarkers, such as cortical thickness, and these measures may then be mapped onto images or surface models for visualization in the form of renderings or time-lapse animations (Fig. 3b).
Extra dimensions
Image data can have more dimensions than space and time in two ways. Either extra channels can be recorded or each voxel can be associated with a dataset encoding various properties. Although different channels could be browsed as extra dimensions, they are usually color coded and jointly displayed. For more than three channels, however, the combinations of channel values do not result in unique colors. Dimensionality reduction techniques can be applied to map meaningful channel combinations to unique colors. This, however, only partially alleviates the problem as the number of combinations to display could easily exceed the number of available colors. Color coding becomes useless when tens or hundreds of channels must be visualized simultaneously (for example, in multi-epitope-ligand cartography18). To solve this problem, some authors have even considered converting data to sound (‘data sonification’ (T. Hermann, T. Nattkemper, H. Ritter and W. Schubert. Proc. Mathematical and Engineering Techniques in Medical and Biological Sciences, 745–750, 2000) to take advantage of people’s ability to distinguish subtle variations in sound patterns, which shows that in this challenging field, there is still room for new, sophisticated visualization tools.
The advent of diffusion MRI has increased the number of dimensions of MRI images. In their most basic form, these images present a scalar value at each voxel indicating a measure of the local water molecules diffusion properties of the imaged sample along a particular direction19. These water diffusion properties indicate the local structure along that direction in the image and can be used to examine, for example, the architecture of white matter in the brain. When multiple images are acquired using different gradient directions, a more complete spatial approximation of the diffusion can be formed. In the case of diffusion tensor imaging (DTI), a rank-2 diffusion tensor is estimated at each voxel20. These images may be visualized by using color to represent the principal direction of diffusion (Figs. 1p and 3c). They may also be visualized as fields of glyphs representing the two-dimensional tensor as an ellipsoid or other shapes that indicate the pattern of water diffusion and thus provide an indication of the structure in the image (Fig. 3d). As the number of angular samples increases (for example, in Q-Ball imaging21), to resolve multiple white matter fiber populations in each voxel, the orientation distribution function, which describes the probability of diffusion in a given angular direction, becomes more complicated and can be represented using higher-order functions, such as spherical harmonic series (Fig. 3e). For both DTI and Q-ball, the data is represented as two-dimensional surfaces at each point in a 3D volume. Diffusion spectral imaging (DSI)22 further increases the dimensionality with multiple acquisitions at different magnetic gradient strengths yielding a 3D dataset at each voxel in the 3D volume.
The challenge of processing and visualizing these data is to convert the raw data into the tensor or glyph representations and display them in a meaningful way to the user. This may include additional processing to reduce the data dimensionality into scalar measures (for example, fractional anisotropy) or to extract features from the tensor, Q-ball or DSI data that indicate structure in the sample. A prime example of this is white matter tractography in the brain. Identification and visualization of white matter tracts (see, for example, Fig. 3f) in diffusion imaging are provided by several of the software packages listed in Table 2 (for example, DTIStudio, TrakVis, ITK, 3D Slicer, MedINRIA, TractoR and FSL).
Although in microscopy some tools have been developed aimed at visualizing directional data such as diffusion properties and mapping them onto two- or 3D datasets (for example, spatio-temporal image correlation spectroscopy (STICS)23), the microscopy application field seems to be less advanced than in MRI. It will therefore be interesting to see whether the tools developed for visualization of diffusion-weighted MRI data will be adopted for microscopy applications.
Segmentation
Whether analyzing scalar images, vector volumes or more complicated data types, a frequent task in processing and visualizing 3D data is the segmentation of cellular or anatomical structures to define the boundaries of target structures. Once the boundaries of a structure have been defined, a surface mesh model can then be generated to represent that structure. These models are often generated using isosurface approaches such as the marching cubes algorithm24. The meshes may then be rendered rapidly using accelerated 3D graphics hardware that is optimized for drawing triangles. The VTK software library provides a widely used implementation of these techniques and is incorporated in, for example, OsiriX, BioImage Suite and 3D Slicer. Surface mesh techniques can visualize anatomy, produce 3D digital reconstructions and make volumetric measurements (Fig. 3g,h). Extra related data, such as statistical maps of tissue changes, can also be represented on these surfaces for display purposes (Fig. 3b).
In many biomedical imaging applications, structures to be segmented are identified in the data either through manual delineation or through automated and semiautomated computational approaches. Image analysis tools, such as 3D Slicer25, MIPAV26, BrainSuite27, MedINRIA and Amira (Visage Imaging), can be used to display and manually delineate 3D volumetric data, which are then turned into 3D surface models. Although manual delineation is often the gold standard for identifying structure, many computational approaches have been developed. Extensible tool suites such as ITK, SCIRun, MIPAV and ImageJ provide collections of automated approaches to the general problem of segmentation for two- and 3D images, as well as tools designed to extract specific anatomical structures. Several tools have been developed for the task of extracting, analyzing and visualizing models of the brain from MRI (for example, FreeSurfer28, BrainSuite27, BrainVoyager29, MedINRIA and BrainVisa30), as have tools specific to cardiac image processing and analysis (for example, Cardiac Image Modeller). Many of these display tools provide facilities for image, volume or surface registration, allowing 3D surface-rendered data acquired during different experiments to be overlaid and displayed. These capabilities allow 3D anatomy to be digitally reconstructed and enable comparison of various samples. Combining organ- and tissue-specific analysis techniques with calibrated MRI acquisition sequences can support accurate in vivo measurement of anatomical structures.
Registration
The dimensional expansion of biological image data goes beyond individual datasets. Gene activity, to take one exemplar of the properties of biological systems, may be imaged and visualized one gene at a time but systematically for many genes in different specimens31–36. Furthermore, different imaging techniques offering different resolutions may be used to visualize different aspects of gene activity. The gene identity becomes yet another dimension in the data, and to quantitatively compare across this dimension the datasets must be properly registered. Many tools have been developed for image registration36–39. In its simplest form, registration is achieved by designating one acquisition as the reference and registering all other acquisitions to it, but this approach introduces reference-specific bias to the data. The computationally most elegant solution is to register all datasets to one another simultaneously in an empty output image space, but such an approach is also the most computationally expensive. Therefore, the most commonly used technique is atlas registration, whereby individual acquisitions are registered to an idealized expert-defined atlas based on prior knowledge of the imaged system.
Registration algorithms can take advantage of the actual pixel intensities in the 3D datasets and iteratively minimize some cost function that reflects the overall image content similarity. Given the size of typical 3D image data, such intensity-based approaches are often slow or unfeasible. Therefore, the image content is typically reduced to some relatively small set of salient features (Fig. 2e,f) and correspondence analysis is used to match the features in different 3D acquisitions and iteratively minimize their displacement. The features may be extracted from the images fully automatically, as in the popular ‘scale invariant feature transform’ (SIFT40), or an expert can define them manually. The manual definition of the corresponding landmarks is at present the only option when registering multimodal data of vastly different scales, such as from confocal and electron microscopy (Fig. 2g). When technically possible, it is beneficial to uncouple the registration problem from the image intensities by using fiduciary markers such as fluorescent beads41 or gold particles. Regardless of the image content representation, the algorithms used for registration use some form of iterative optimization of an appropriately chosen cost function.
An interesting idea for multimodal image registration is to establish a reference output space where the different modalities are registered to each other once, and subsequently new instances of one modality are mapped onto the already registered example. This process can also be iterative, increasing the registration precision with each new incoming dataset. The visualization of registered multimodal image data of different scales presents a new set of challenges (Fig. 2g). Proper down-sampling techniques based on Gaussian convolution must be used when changing the scale dimension of the multiresolution data42.
Multimodal image registration is also an important issue for MRI data. Multiple scanning technologies are often applied to the same subject to provide an integrated assessment. For example, the integration of structural, functional and diffusion MRI has been productively applied to the analysis of and treatment planning for deep brain tumors, in which functional MRI is used to indicate the ‘eloquent cortex’ (involved in tasks to be preserved during surgery) while diffusion imaging indicates the white matter fiber bundles and how they are invaded and/or displaced by the tumor43. Interactive visualization techniques allow clinicians to superimpose the 3D renderings of the various image modalities to better understand the clinical situation and evaluate treatment options (Fig. 3i). Creating an integrated visualization is complicated by patient motion between scans and by inherent geometric distortion associated with different scan techniques—for example, eddy current–induced distortions in diffusion MRI. Automated registration and distortion correction techniques can be used to compensate for these effects when creating the integrated view.
Multimodal MRI visualization can be also used together with real-time data to guide therapeutic procedures such as neurosurgery. Current neurosurgical practice is often augmented with so-called ‘navigation’ systems consisting of surgical tools whose position and orientation are digitally tracked. This information is used to provide a reference between the preoperative image data and the live patient with submillimeter accuracy. In this context, it is possible to support the procedure with visualizations of MRI data collected preoperatively. For many interventions, nonlinear deformation of the image data are required owing to significant changes between pre- and intraoperative patient anatomy. The VectorVision (BrainLabAG) System is an example of a state-of-the-art MRI surgical navigation system, while BioImage Suite, 3D Slicer and other open source software tools are available for researchers looking to provide enhanced functionality.
Implementation issues
The main commercial software tools providing methods for viewing primary image data in cell biology are MetaMorph, Imaris, Volocity, Amira and LSM Image Browser. There is also a wide range of open source tools, such as various plug-ins to the image analysis suite ImageJ and Fiji, its distribution specialized in 3D registration and visualization; BioImageXD, based on the state-of-the-art VTK library; and the V3D toolkit, emerging from systematic 3D imaging efforts in neurobiology at Janelia Farm in Ashburn, Virginia, USA (see Table 1). Because visualization strategies are very diverse, no single software fits all needs, and the availability of either the program’s source code or a functional application programming interface (API) is a must for the programming required to realize complex visualizations.
Most of these software tools require that the image volumes of interest be read into the computer memory before they can provide efficient visualization and reasonable interactive response. Recent commercial and open-source tools have started to use graphics hardware to accelerate 3D visualization. Wider adoption of these approaches is prevented by the small spectrum of graphical processing units (GPUs) accessible for parallel programming using standard programming languages and the relatively small size of GPU memory. Image datasets produced nowadays are often so large that it is impossible to load them even into the CPU memory except on systems configured expressly (and expensively) for the purpose. Open-source software suites such as ImageJ and Fiji provide practical solutions for managing memory and displaying massive datasets without unrealistic hardware requirements. But although these tools are very user friendly and popular, they lack generic multidimensional data structures like those developed by the ITK/VTK project that enable programmatic abstraction of the access to arbitrary dimensions in the image data residing on the hard drive. Approaches that rely on random data access from the hard drive at multiple predefined resolutions are now available for very large-scale two-dimensional images in a client-server mode and can be accessed over the Internet, which greatly enhances their visibility and the possibilities for collaborative annotation. Examples include Google Maps, Zoomify and CATMAID. Browser-based visualization of 3D image data are so far limited to slice-by-slice browsing of the z dimensions (CATMAID, BrainMaps). Web viewers for 3D data enabling section browsing at arbitrary angles and scales are just now emerging44,45.
Visualization of high-throughput microscopy data
In recent years, systems for performing high-throughput microscopy-based experiments have become available and are often used to test the effects of chemical or genetic perturbations on cells46,47, to determine the subcellular localization of proteins48,49 or to study gene expression patterns in development50. These screens produce huge amounts of image data (sometimes tens of terabytes and millions of images) that must be managed, quality controlled, browsed, annotated and interpreted. As a consequence, tools for visualization and analysis are key at virtually all levels of such projects (see Fig. 4).
Figure 4.
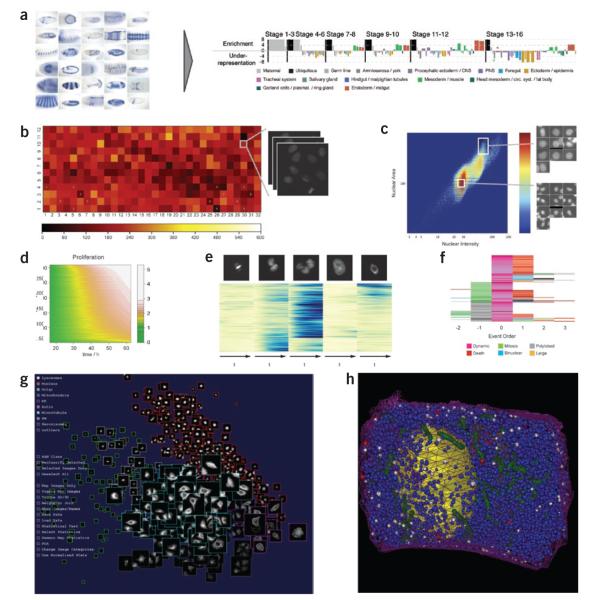
Visualization of high–throughput data. (a) By analogy with the ‘eisengram’ for microarray data, the discrete spatial gene expression (left) annotation data can be summarized by a so-called ‘anatogram’ (right), wherein anatomical structures are color coded, grouped temporally (vertical black lines) and ordered consistently within the temporal groups. Over- or under-representation of anatomical term in a group of genes is expressed by height of the color-coded bar; width of the bar is proportional to the frequency of the anatomical term in the annotation dataset. (b) Typical visualizing and browsing of high-throughput data at experiment level: color-coded cell density on a 384-well plate with link to raw data. (c) Typical visualizing and browsing of high-throughput data at the level of exploratory analysis: density plots of nuclear features (area and intensity), linked to the single segmented nuclei. (d) Joint visualization of 2,600 time-lapse experiments with one-dimensional readout (here proliferation curves): values are color-coded; each row corresponds to one experiment. (e) Time-resolved heat map for multidimensional read-out (here percentages of nuclei in the different morphological classes shown at the top): values are color-coded; each row corresponds to one RNAi experiment. Rows are arranged according to trajectory clustering64. (f) Event order map visualizing the relative order of phenotypic events in cell populations: events are color-coded and centered around one phenotype (here dynamic). (g) Visualizing high-throughput subcellular localization data (iCluster): images of ten subcellular localizations (indicated by outline color) spatially arranged by statistical similarity to identify outliers and representative images. (h) Visualization of spatially mapped simulation results (The Visible Cell): simulation of insulin secretion within a beta cell based on electron microscope tomography data (resolution, 15 nm). Blue granules are primed for insulin release, white are docked into the membrane (releasing insulin) and red are returning to the cytoplasm after having been docked.
Some large-scale experiments involving particularly complex read-outs have been annotated manually using controlled vocabularies51, in some cases—as for high-content analysis of gene expression during development—eased by the use of custom-built annotation tools34,52,53. Several visualization aids have been developed to succinctly summarize the complex, multidimensional annotations and organize them using clustering methods borrowed from the microarray data analysis field. The main challenge of representing qualitative rather than quantitative annotation data was addressed by introducing discrete color coding for the controlled vocabulary terms collapsed to the most informative level in the annotation ontology. In that way, the analog microarray ‘eisengram’ evolved into the digital ‘anatogram’ capable of visually summarizing the gene expression properties of arbitrary groups of genes (Fig. 4a). Once large, expert-annotated image sets became available, computational approaches were successfully used to automate the annotation process (for example, automatic annotation of subcellular protein localization54 and gene expression patterns55,56). In most cases, large image datasets are automatically processed to extract a wide range of attributes from the images.
To navigate efficiently through this sea of data, users need visualization software that can display informative summaries at different levels: in the acquisition and quality-control phase, multiwell plate and similar visualizations (Fig. 4b) that show image-based data values with a raw data link or thumbnails of images that can be enlarged for careful examination are very helpful. Tools are also needed to show relevant image-based data to biologists in an intuitive manner and enable them to identify meaningful characteristics and explore potential correlations and relationships between data and to point them toward the most interesting samples in their experiment. For this exploratory data analysis, data-enhanced scatter and density plots (Fig. 4c) and histograms of image-derived data can be used, in which the user can select subsets of data, view examples of the raw images producing those data points and filter data points for further analysis. Browsing these graphical representations linked to the raw data allows biologists to identify interesting subsets (for example, the morphological classes present in a cell-based screen or training sets for subsequent supervised machine learning) in an interactive and intuitive manner. Linking to the original data are particularly important: first, because users must frequently locate relevant images to manually confirm the automated, quantitative results and, second, because there is often no obvious a priori link between quantitative image descriptors and biological meaning. Further analysis of these attributes and eventually identified subsets leads to image annotations (for example, phenotypes) and/or classifications (for example, as a ‘hit’), often by means of supervised learning methods.
When putting the experiment into the context of existing biological knowledge, researchers are concerned about how images and their derived data relate to known biological entities. For example, one may want to browse all images related to a given gene, gene ontology term or chemical treatment. This requires integration with other sources of information, usually external databases. The visualization methods suited for this are also commonly used in systems biology: heat maps and projections in two-dimensional maps57.
However, many of these goals remain unaddressed by existing software tools. Gracefully and intuitively presenting rich image data representing possibly hundreds of attributes extracted from billions of cells is a demanding task for a visual analysis tool. Still, some recent developments have begun to ease aspects of these visualization challenges for high-throughput experiments. Several software tools offered by screening-oriented microscope companies enable certain data visualizations (Table 3), as does third-party software such as Cellenger and the open-source CellProfiler project58,59 (Table 3). These packages integrate image processing algorithms with statistical analysis and graphical representations of the data and also offer machine learning methods that capitalize on the multiple attributes measured in the images. In workflow management software (for example, HCDC; Table 3), where modules communicate through defined inputs and outputs, user-defined visualization modules can be integrated into a data acquisition and processing workflow; this increase in flexibility and history tracking typically comes with a loss in user-interactivity and browsing capabilities.
Table 3.
A selective list of high-throughput visualization tools
| Name | Cost | OS | Description | URL |
|---|---|---|---|---|
| BD Pathway | $ | Win | Automated image acquisition, analysis, data mining and visualization | http://tiny.cc/093OJ |
| Cellenger | $ | Win | Automated image analysis | http://tiny.cc/rARky |
| CellHTS Bioconductor (R) | Free | Win, Mac, Linux | Analysis of cell-based screens, visualization of screening data, statistical analysis, links to bioinformatics resources |
http://www.bioconductor.org/ |
| CellProfiler CP-Analyst* | Free | Win, Mac, Linux | Automated image analysis, classification, interactive data browsing, data mining and visualization; extensible, supports distributed processing |
http://www.cellprofiler.org/ |
| CompuCyte | $ | Win | Automated image acquisition, analysis, data mining and visualization | http://tiny.cc/jHsAm |
| GE IN Cell Investigator, Miner | $ | Win | Automated image acquisition, analysis, data mining and visualization | http://tiny.cc/9rFoh |
| Genedata Screener* | $ | Win, Mac, Linux | Data analysis and visualization | http://tiny.cc/HBfpY |
| Evotec Columbus, Acapella | $ | Win, Linux | Automated image analysis, distributed processing, data management (OME compatible), data mining and visualization |
http://tiny.cc/yvyek |
| HCDC | Free | Win, Mac, Linux | Workflow management, data mining, statistical analysis, visualization, based on KNIME (http://www.knime.org/) |
http://hcdc.ethz.ch/ |
| iCluster | Free | Win, Mac, Linux | Spatial layout of imaging by statistical similarity, statistical testing for difference |
http://icluster.imb.uq.edu.au/ |
| MetaMorph MetaXpress, AcuityXpress* |
$ | Win, Mac, Linux | Automated image acquisition, analysis, data mining and visualization | http://tiny.cc/OU9sf |
| Olympus Scan^R | $ | Win | Image acquisition, automated image analysis, extensible (with LabView) | http://tiny.cc/NtEhH |
| Pathfinder Morphoscan | $ | Win | Automated image analysis, cell and nuclear analysis, karyotyping, histoand cytopathology, high-content screening |
http://www.imstar.fr/ |
| Pipeline Pilot | $ | Win, Linux | Workflow management, image processing, data mining and visualization | http://tiny.cc/uY4ZO |
| Spotfire* | $ | Win, Mac, Linux | Data analysis and visualization | http://tiny.cc/rtVeL |
| Thermo Scientific Cellomics | $ | Win | Automated image analysis, analysis, data mining and visualization | http://tiny.cc/Bt7Ov |
| TMALab | $ | Win | Automated image acquisition, storage, analysis, scoring, remote sharing and annotation; mainly for clinical pathology |
Recommended and popular tools. Free means the tool is free for academic use; $ means there is a cost. OS, operating system: Win, Microsoft Windows; Mac, Macintosh OS X. Tools running on Linux usually also run on other versions of Unix.
Although presentation, representation and querying of primary visual and quantitative data are a significant problem, an associated difficulty is that the dimensionality of data derived from or associated with each image or object is rapidly growing. The problem is to visualize such high-dimensional data in a concise way so that it may be explored to identify patterns and trends at the image level. A common strategy linearly projects high-dimensional data into low dimensions for visualization using various forms of multidimensional scaling60 (for example, principal component analysis, Sammon mapping61). Multidimensional scaling aims to map high-dimension vectors into low dimensions in such a way as to preserve some measure of distance between the vectors. Once such an embedding or mapping into two or three dimensions has been accomplished, the data can be visualized and any relationships observed. One approach to visualizing and interacting with high-dimensional data and microscopy imaging is the iCluster system62, developed in association with the Visible Cell63. Here, large image sets from single or multiple fluorescence microscopy experiments may be visualized in three dimensions (Fig. 4g). Spatial placement in three dimensions can be automatically generated by Sammon mapping using high-dimensional texture measures or through user-supplied statistics associated with each image. Thus, sets of images that are statistically and visually similar are presented as spatially proximate, whereas dissimilar images are distant. This allows outliers and unusual images to be detected easily, while differences between classes (for example, treatment versus control) or multiple classes within an experiment can be seen as spatial separation. Visualization of relationships and correlations among the data allow the user to find and define the unusual, the representative and broad patterns in the data.
Most of the above visualization schemes apply to cellular-level measurements of populations of cells, but none of these methods takes into consideration time-resolved data. Although the temporal evolution of one or several cellular or population features from a single experiment can easily be plotted over time, this approach is impractical when relationships between hundreds or thousands of experiments must be visualized. In this case, the time series can be ordered according to some similarity criterion and visualized as a color-coded matrix (Fig. 4d). Similarly, heat maps can be extended to represent multidimensional time series (Fig. 4e); the time series corresponding to different dimensions can be concatenated. Here, the most difficult part is to define an appropriate distance function for multidimensional time series according to whether absolute or relative temporal information is important64.
Often, the time itself is less informative than the relative order in which events occur. In this case, it is also possible to estimate a representative order of events from the time-lapse experiments (for example, phenotypic events on single-cell level). This event order can be used for characterizing, grouping and visualizing experimental conditions, creating an event order map (Fig. 4f)64.
Dissemination of image datasets
High-throughput microscopy techniques have led to an exponential increase in visual biological data. Although every high-throughput imaging project strives to perform a comprehensive analysis of its image data, the sheer volume of the images and the inadequacy of the computational tools make such efforts incomplete. It is likely that distributed, research community–driven and competitive analysis of these datasets will lead to new discoveries, as it has for publicly released genome sequences. Although standalone applications are catching up with the immediate needs of primary data visualization, solutions for distributing image data to the community or in collaborative environments are lagging behind. Traditional paper publication or publication as online supplementary materials is clearly inadequate, and the Journal of Cell Biology and Journal of the Optical Society of America have attempted to address this by implementing software systems to link original image data to articles. The former’s DataViewer (http://jcb-dataviewer.rupress.org/) is based on OMERO and provides web-based interactivity, whereas the latter’s ISP software uses VTK and requires readers of ISP-enabled articles to download and install the ISP software on their machine.
The first attempts at distributing large image datasets to the biology community have come from atlases of gene expression in model organisms31–35,65 (Table 4). Two projects have now completed respectively a transcriptome atlas for the adult mouse brain35 (Allen Mouse Brain Atlas) and for the mouse embryo (Eurexpress). In these projects images of tissue sections are captured at about 0.5 μm pixel resolution, resulting in images with pixel dimensions of about 4,000 × 4,000. With sampling through the brain or embryo at about 150 μm and for most (~20,000) expressed genes, this results in an archive of millions of images. These have been manually and automatically annotated; in the case of the brain data, this was done using 3D registration.
Table 4.
Selected image repositories
| Name | Description | URL |
|---|---|---|
| 4DXpress | Cross-species gene expression database | http://tiny.cc/DT6lh |
| ADNI | Imaging and genetics data from elderly controls and subjects with mild cognitive impairment and Alzheimer’s disease |
http://www.loni.ucla.edu/ADNI/ |
| Allen Brain Atlas | Interactive, genome-wide image database of gene expression |
http://www.brain-map.org/ |
| APOGEE | Atlas of patterns of gene expression in Drosophila embryogenesis |
http://tiny.cc/ZASKo |
| BIRN | Lists of tools and datasets, mostly from the neuroimaging community |
http://www.birncommunity.org/ |
| Bisque | Exchange and exploration of biological images |
http://www.bioimage.ucsb.edu/ |
| Cell Centered Database | Database for high-resolution 2D, 3D and 4D data from light and electron microscopy, including correlated imaging |
http://ccdb.ucsd.edu/index.shtm |
| Edinburgh Mouse Atlas | Atlas of mouse embryonic development (emap) and gene expression patterns (EMAGE), 2D, 3D spatially annotated data |
http://www.emouseatlas.org/ |
| Fly-FISH | Atlas of patterns of RNA localization in Drosophila embryogenesis |
http://fly-fish.ccbr.utoronto.ca/ |
| fMRIDC | Public repository of peer-reviewed functional MRI studies and underlying data |
http://www.fmridc.org/ |
| ICBM | Web-based query interface for selecting subject data from the ICBM archive |
http://tiny.cc/0JDXE |
| Mitocheck | Microscopy-based RNAi screening data | http://www.mitocheck.org/ |
| OASIS | Cross-sectional MRI data in young, middle-aged, undemented and demented older adults; longitudinal MRI data in undemented and demented older adults |
http://www.oasis-brains.org/ |
| ZFIN | The zebrafish model organism database | http://tiny.cc/uitMn |
2D, 3D and 4D: two-, three- and four-dimensional, respectively.
With the exception of Phenobank (http://worm.mpi-cbg.de/phenobank2/cgi-bin/ProjectInfoPage.py), which provides data for a genome-wide time-lapse screen in Caenorhabditis elegans, image data from high-throughput, image-based RNAi screens have not yet arrived in the public domain, although several projects (Mitocheck, GenomeRNAi) aim to make their images available. The logistics and storage requirements are formidable, and perhaps a publicly funded centralized repository similar to GenBank or ArrayExpress should be established. Success of such a repository would depend on the willingness of data producers to share their images through a central system. Alternatively, a distributed infrastructure could be considered. Querying these resources relies on textual annotations of all images. Although already useful, text-based queries are limited by the lack of ontologies for many descriptive attributes. For example, how can the user retrieve all images of mitotic phenotypes when some annotations are free text and use wording such as “chromosome segregation defect”? However, ontologies will not solve all problems in image retrieval, as many images will not have been annotated with the required level of detail. For example, in a screen, most images are just annotated as ‘not a hit’ for a given phenotype.
Another challenge is to allow browsing without significant download time. In the Edinburgh Mouse Atlas33,66 (Table 4), the data range from medium-resolution (0.5 μm) tissue section images captured using light microscopy to full 3D images captured typically with optical projection tomography (see Box 1). The data are mapped onto a standard mouse embryo model to allow direct comparison and analysis in spatial terms. Mapping the spatial patterns of gene expression provides some powerful options for query and analysis and avoids the partiality, ambiguity and resolution problems of text annotation. For example, the EMAGE gene expression database67, which has spatially mapped patterns, allows query on the basis of spatial location and pattern similarity. Using a straightforward Jaccard index measure, the spatial search is sorted according to similarity with the search area, which can either be manually drawn or can be the pattern resulting from an analysis of an input image—that is, “find one like this.” The same similarity measures can be used to cluster the data and enable varieties of automated data mining. Another example comes from neurobiology: the anatomic gene expression atlas (AGEA)38 of the mouse brain allows users to interactively explore spatial patterns of gene expression through correlation maps, to apply hierarchical, multiscale partitioning of the image volumes according to spatial gene expression similarity and to identify genes with localized enrichment in a chosen region of interest.
Systematic efforts are underway in neurobiology to map the anatomy and connectivity of entire nervous systems. Various imaging modalities are used in tiling mode across serially sectioned tissues, resulting in massive layered image canvases. Several projects use a Google Maps–like user interface to provide web access to the data. In BrainMaps (Table 1), images of serial sections of both primate and nonprimate brains scanned with submicrometer resolution are presented in a Google Maps–like viewer with possibilities for controlled vocabulary annotation by registered users. CATMAID also uses the same navigation principle to allow collaborative manual annotation of serial-section transmission electron microscopy images of the Drosophila larval brain imaged at a resolution of 4 nm per pixel5. CATMAID implements synchronized browsing and annotation of linked multimodal image data (such as confocal and electron microscopy). CATMAID does not require the use of a central data repository but instead allows the images to be distributed across several laboratories and thus avoids duplication of massive datasets. The use of a lightweight web client that is ‘aware’ of various datasets around the world through a collection of expert-submitted annotations in a centralized database seems to be a good approach to start mapping large biological image collections using a community-driven effort. The alternative possibility would be the creation of well-structured repositories. Such repositories should ideally provide three levels of access to the data. The first level represents the raw data, and the second level comprises the data analysis results and/or annotations, with relevant external information. The third level consists in integrating different data sources such that data from multiple datasets can be simultaneously queried.
These efforts at making biological image data accessible and usable by the scientific community are paralleled by similar efforts in the MRI community. But whereas clinical scanners often rely upon specialized servers dedicated to the storage and distribution of images (so-called picture archiving and communication systems, or PACS), most research facilities rely upon local systems and protocols for data storage and organization. Large-scale research databases do exist that provide access for the neuroimaging community. Both the Alzheimer’s Disease Neuroimaging Initiative (ADNI68) and the International Consortium for Brain Mapping (ICBM69) make use of the Laboratory of Neuro Imaging’s Image Data Archive (IDA) to store data for thousands of subjects, including MRI, positron emission tomography, magnetic resonance angiography and DTI, as well as related meta-data70. The system provides investigators with fine-grained control of data access rights, ranging from allowing data to be made fully public to restricting it to collaborators. The Functional MRI Data Center (fMRIDC71) provides investigators with a repository for peerreviewed fMRI studies and underlying data, which may then be requested by visitors to the fMRIDC website. The Open Access Series of Imaging Studies (OASIS72) also provides collections of hundreds of brain MRI volumes to the scientific community at no charge. The Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC; http://nitrc.org/) provides a central repository through which neuroimaging resources, such as software tools or data, may be described, disseminated and discussed. The development of large, publicly available collections of data provides opportunities for large-scale neuroimaging studies, which introduces new challenges for information visualization.
Perspectives on image data integration
Although some tools are available for browsing and querying a complex large-scale dataset, a systems understanding will require more-comprehensive queries and views. Just as the sequencing revolution led to tools such as BLAST73 that allow users to find, compare, sort and make inferences from vast numbers of sequences, a corresponding set of tools must be developed for imaging to fully exploit the flood of data becoming available from modern imaging techniques and provide a foundation from which to build a spatially aware systems-biology model of the cell. At their best, “graphics are instruments for reasoning about quantitative information”74. In this context, visualization extends beyond presentation of image data per se to enable, facilitate and integrate statistical tests, mathematical modeling and simulation and automated reasoning over multiple data types. Primary image data are combined with instrument and experimental meta-data, data derived from analysis of the image series and information from diverse external resources. In this way, visualization can help biologists and modelers address broader questions: for example, how specific pathways or functions are organized spatially within a cell and how they change with cell type, disease state or treatment. Already environments such as Virtual Cell and BioSPICE embed analyses (for example, biomolecular interaction networks, or simulations of metabolite flow through pathways) in an abstract two- or 3D space. The Visible Cell environment Illoura63 (Fig. 4h) aims to visualize such analysis in empirical two- or 3D microscopy data. Within the Visible Cell, first primary microscopy data are segmented and the resulting objects are marked up ontologically and stored in a way that supports further annotation. Key elements are database federation to enable multiuser access; data storage in Semantic Web format Resource Description Framework (RDF) to enable complex semantic queries across multiple data types; and visualization components to view and interact with two- and 3D spatial data and results of queries.
It is said that a picture is worth a thousand words. It is difficult to imagine how many words the enormous image space of biological data would be worth. Many primary data in modern biology are in the form of images. These images are a rich source of qualitative as well as quantitative information about the biological system in many dimensions and across many scales. Biologists and computer scientists put substantial efforts into untangling this vast image space and giving meaning to these visual data. Its visualization provides the scientific community with tools to gain systematic and unprecedented insights at many levels of biological scale.
ACKNOWLEDGMENTS
P.T. thanks Carl Zeiss Microimaging for SPIM prototype access. T.W. was supported by a grant to J.E. (within the Mitocheck European Integrated Project LSHG-CT-2004-503464). D.W.S. was partially supported by US National Institutes of Health (NIH) grant P41 RR013642. M.E.B. was partly supported by NIH grant R01 EB004155-03. S.P. was partially supported by NIH grant P41 RR13218. S.D. was supported by the Wellcome Trust. J.-K.H. was supported by the ENFIN European Network of Excellence (contract LSHG-CT-2005-518254) awarded to J.E. A.E.C. and A.F. were supported by NIH grant 5 RL1 CA133834-03. J.E.S. was supported by the British Heart Foundation (grant BS/06/001) and the BBSRC (grant E003443). This work was funded in part through the NIH Roadmap for Medical Research grants U54 RR021813 (D.W.S.) and U54 EB005149 (S.P.). Information on the US National Centers for Biomedical Computing can be obtained from http://nihroadmap.nih.gov/bioinformatics/.
Footnotes
COMPETING INTERESTS STATEMENT The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Goldberg IG, et al. The Open Microscopy Environment (OME) Data Model and XML file: open tools for informatics and quantitative analysis in biological imaging. Genome Biol. 2005;6:R47. doi: 10.1186/gb-2005-6-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore J, et al. Open tools for storage and management of quantitative image data. Methods Cell Biol. 2008;85:555–570. doi: 10.1016/S0091-679X(08)85024-8. [DOI] [PubMed] [Google Scholar]
- 3.Swedlow JR, Goldberg IG, Eliceiri KW, the OME Consortium Bioimage informatics for experimental biology. Annu. Rev. Biophys. 2009;38:327–346. doi: 10.1146/annurev.biophys.050708.133641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox R, et al. A (sort of) new image data format standard: NifTI-1. Neuroimage. 2004;22:99. [Google Scholar]
- 5.Saalfeld S, Cardona A, Hartenstein V, Tomancák P. CATMAID: Collaborative Annotation Toolkit for Massive Amounts of Image Data. Bioinformatics. 2009;25:1984–1986. doi: 10.1093/bioinformatics/btp266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levoy M. Display of surfaces from volume data. IEEE Comput. Graph. Appl. 1988;8:29–37. This paper is a seminal work on the rendering of volumetric data by directly shading each voxel value and projecting it onto the viewing plane. the method provides realistic volumetric rendering without the need to model the data with geometric primitives.
- 7.Pieper SD, Halle M, Kikinis R. 3D Slicer. IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 2004:632–635. [Google Scholar]
- 8.Pieper S, Lorensen B, Schroeder W, Kikinis R. The na-mic kit: Itk, vtk, pipelines, grids and 3d slicer as an open platform for the medical image computing community. IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 2006:698–701. [Google Scholar]
- 9.Gordon JL, Buguliskis JS, Buske PJ, Sibley LD. Actin-like protein 1 (ALP1) is a component of dynamic, high molecular weight complexes in Toxoplasma gondii. Cell Motil. Cytoskeleton. 2009;67:23–31. doi: 10.1002/cm.20414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friston KJ, et al. Statistical parametric maps in functional imaging: a general linear approach. Hum. Brain Mapp. 1994;2:189–210. [Google Scholar]
- 11.Smith SM, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(suppl. 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 12.Dufour A, et al. Segmenting and tracking fluorescent cells in dynamic 3-D microscopy with coupled active surfaces. IEEE Trans. Image Process. 2005;14:1396–1410. doi: 10.1109/tip.2005.852790. [DOI] [PubMed] [Google Scholar]
- 13.Jaqaman K, et al. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EH. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322:1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- 15.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc. Natl. Acad. Sci. USA. 2000;97:11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischl B, et al. Sequence-independent segmentation of magnetic resonance images. Neuroimage. 2004;23(suppl. 1):S69–S84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 17.Salat DH, et al. Age-associated alterations in cortical gray and white matter signal intensity and gray to white matter contrast. Neuroimage. 2009;48:21–28. doi: 10.1016/j.neuroimage.2009.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schubert W, et al. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat. Biotechnol. 2006;24:1270–1278. doi: 10.1038/nbt1250. [DOI] [PubMed] [Google Scholar]
- 19.Le Bihan D, et al. MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology. 1986;161:401–407. doi: 10.1148/radiology.161.2.3763909. [DOI] [PubMed] [Google Scholar]
- 20.Basser PJ, Mattiello J, LeBihan D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J. Magn. Reson. B. 1994;103:247–254. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- 21.Tuch DS, et al. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn. Reson. Med. 2002;48:577–582. doi: 10.1002/mrm.10268. [DOI] [PubMed] [Google Scholar]
- 22.Wedeen VJ, Hagmann P, Tseng W-YI, Reese TG, Weisskoff RM. Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magn. Reson. Med. 2005;54:1377–1386. doi: 10.1002/mrm.20642. [DOI] [PubMed] [Google Scholar]
- 23.Hebert B, Costantino S, Wiseman PW. Spatiotemporal image correlation spectroscopy (STICS) theory, verification, and application to protein velocity mapping in living CHO cells. Biophys. J. 2005;88:3601–3614. doi: 10.1529/biophysj.104.054874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorensen WE, Cline HE. Marching cubes: a high resolution 3D surface construction algorithm. SIGGRAPH ‘87: Proc. 14th Ann. Conf. Computer Graphics and Interactive Techniques.1987. pp. 163–169. this paper presented a fast algorithm for computing a triangular mesh corresponding to an isosurface in a 3d data volume.
- 25.Lindig TM, et al. Spiny versus stubby: 3D reconstruction of human myenteric (type I) neurons. Histochem. Cell Biol. 2009;131:1–12. doi: 10.1007/s00418-008-0505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McAuliffe M, et al. Medical image processing, analysis and visualization in clinical research. Proc. 14th IEEE Symp. Computer-based Medical Systems (CBMS2001).2001. pp. 381–386. [Google Scholar]
- 27.Shattuck DW, Leahy RM. BrainSuite: an automated cortical surface identification tool. Med. Image Anal. 2002;6:129–142. doi: 10.1016/s1361-8415(02)00054-3. [DOI] [PubMed] [Google Scholar]
- 28.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 29.Goebel R, Esposito F, Formisano E. Analysis of functional image analysis contest (FIAC) data with Brainvoyager QX: from single-subject to cortically aligned group general linear model analysis and self-organizing group independent component analysis. Hum. Brain Mapp. 2006;27:392–401. doi: 10.1002/hbm.20249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cointepas Y, Mangin J-F, Garnero L, Poline J-B, Benali H. BrainVISA: software platform for visualization and analysis of multi-modality brain data. Neuroimage. 2001;13:S98. [Google Scholar]
- 31.Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–D556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray PA, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- 33.Christiansen JH, et al. EMAGE: a spatial database of gene expression patterns during mouse embryo development. Nucleic Acids Res. 2006;34:D637–D641. doi: 10.1093/nar/gkj006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomancak P, et al. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2007;8:R145. doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 36.Fowlkes CC, et al. A quantitative spatiotemporal atlas of gene expression in the Drosophila blastoderm. Cell. 2008;133:364–374. doi: 10.1016/j.cell.2008.01.053. [DOI] [PubMed] [Google Scholar]
- 37.Hill DLG, Batchelor PG, Holden M, Hawkes DJ. Medical image registration. Phys. Med. Biol. 2001;46:R1–R45. doi: 10.1088/0031-9155/46/3/201. [DOI] [PubMed] [Google Scholar]
- 38.Ng L, et al. Neuroinformatics for genome-wide 3D gene expression mapping in the mouse brain. IEEE/ACM Trans. Comput. Biol. Bioinform. 2007;4:382–393. doi: 10.1109/tcbb.2007.1035. [DOI] [PubMed] [Google Scholar]
- 39.Klein A, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowe D. Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 2004;60:91–110. [Google Scholar]
- 41.Preibisch S, Saalfeld S, Rohlfing T, Tomancak P. Bead-based mosaicing of single plane illumination microscopy images using geometric local descriptor matching. Proc. SPIE. 2009;7259 [Google Scholar]
- 42.Lindeberg T. Scale-space for discrete signals. IEEE Trans. Pattern Anal. Mach. Learn. 1990;12:234–254. [Google Scholar]
- 43.Tharin S, Golby A. Functional brain mapping and its applications to neurosurgery. Neurosurgery. 2007;60:185–201. doi: 10.1227/01.NEU.0000255386.95464.52. discussion 201–202. [DOI] [PubMed] [Google Scholar]
- 44.Lau C, et al. Exploration and visualization of gene expression with neuroanatomy in the adult mouse brain. BMC Bioinformatics. 2008;9:153. doi: 10.1186/1471-2105-9-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertrand L, Nissanov J. The Neuroterrain 3D mouse brain atlas. Front. Neuroinformatics. 2008;2:3. doi: 10.3389/neuro.11.003.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carpenter AE, Sabatini DM. Systematic genome-wide screens of gene function. Nat. Rev. Genet. 2004;5:11–22. doi: 10.1038/nrg1248. [DOI] [PubMed] [Google Scholar]
- 47.Pepperkok R, Ellenberg J. High-throughput fluorescence microscopy for systems biology. Nat. Rev. Mol. Cell Biol. 2006;7:690–696. doi: 10.1038/nrm1979. [DOI] [PubMed] [Google Scholar]
- 48.Glory E, Murphy RF. Automated subcellular location determination and high-throughput microscopy. Dev. Cell. 2007;12:7–16. doi: 10.1016/j.devcel.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 49.Simpson JC, Wellenreuther R, Poustka A, Pepperkok R, Wiemann S. Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 2000;1:287–292. doi: 10.1093/embo-reports/kvd058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lécuyer E, Tomancak P. Mapping the gene expression universe. Curr. Opin. Genet. Dev. 2008;18:506–512. doi: 10.1016/j.gde.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Sönnichsen B, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 52.Tomancak P, et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3:research0088.1–0088.14. doi: 10.1186/gb-2002-3-12-research0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han L, Hemert J, Baldock R, Atkinson M. Automating gene expression annotation for mouse embryo. Proceedings of the 5th International Conference on Advanced Data Mining and Applications; Beijing: Springer; 2009. pp. 469–478. [Google Scholar]
- 54.Newberg J, Murphy RF. A framework for the automated analysis of subcellular patterns in human protein atlas images. J. Proteome Res. 2008;7:2300–2308. doi: 10.1021/pr7007626. [DOI] [PubMed] [Google Scholar]
- 55.Ji S, Sun L, Jin R, Kumar S, Ye J. Automated annotation of Drosophila gene expression patterns using a controlled vocabulary. Bioinformatics. 2008;24:1881–1888. doi: 10.1093/bioinformatics/btn347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng H, et al. Automatic image analysis for gene expression patterns of fly embryos. BMC Cell Biol. 2007;8(suppl. 1):S7. doi: 10.1186/1471-2121-8-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gehlenborg N, et al. Visualization of omics data for systems biology. Nat. Methods. 2010;7:S56–S68. doi: 10.1038/nmeth.1436. [DOI] [PubMed] [Google Scholar]
- 58.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones TR, et al. CellProfiler Analyst: data exploration and analysis software for complex image-based screens. BMC Bioinformatics. 2008;9:482. doi: 10.1186/1471-2105-9-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kruskal JB, Wish M. Multidimensional Scaling. Sage Publications; Beverly Hills, California, USA and London: 1978. [Google Scholar]
- 61.Sammon JW. A nonlinear mapping for data structure analysis. IEEE Trans. Comput. 1969;C-18:401–409. [Google Scholar]
- 62.Hamilton NA, Wang JTH, Kerr MC, Teasdale RD. Statistical and visual differentiation of subcellular imaging. BMC Bioinformatics. 2009;10:94. doi: 10.1186/1471-2105-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McComb T, et al. Illoura: a software tool for analysis, visualization and semantic querying of cellular and other spatial biological data. Bioinformatics. 2009;25:1208–1210. doi: 10.1093/bioinformatics/btp125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walter T, et al. Automatic identification and clustering of chromosome phenotypes in a genome wide RNAi screen by time-lapse imaging. J. Struct. Biol. 2009 Oct 23; doi: 10.1016/j.jsb.2009.10.004. published online, doi:10.1016/j.jsb.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 65.Ringwald M, et al. A database for mouse development. Science. 1994;265:2033–2034. doi: 10.1126/science.8091224. [DOI] [PubMed] [Google Scholar]
- 66.Richardson L, et al. EMAGE mouse embryo spatial gene expression database: 2010 update. Nucleic Acids Res. 2010;38:D703–D709. doi: 10.1093/nar/gkp763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baldock RA, et al. EMAP and EMAGE: a framework for understanding spatially organized data. Neuroinformatics. 2003;1:309–325. doi: 10.1385/NI:1:4:309. [DOI] [PubMed] [Google Scholar]
- 68.Mueller SG, et al. Ways toward an early diagnosis in Alzheimer’s disease: the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mazziotta J, et al. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Phil. Trans. R. Soc. Lond. B. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toga AW. Neuroimage databases: the good, the bad and the ugly. Nat. Rev. Neurosci. 2002;3:302–309. doi: 10.1038/nrn782. [DOI] [PubMed] [Google Scholar]
- 71.Van Horn JD, et al. The Functional Magnetic Resonance Imaging Data Center (fMRIDC): the challenges and rewards of large-scale databasing of neuroimaging studies. Phil. Trans. R. Soc. Lond. B. 2001;356:1323–1339. doi: 10.1098/rstb.2001.0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marcus DS, et al. Open Access Series of Imaging Studies (OASIS): Cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J. Cogn. Neurosci. 2007;19:1498–1507. doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]
- 73.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 74.Tufte ER. The classic text on the science of data visualization. Graphics Press; Cheshire, Connecticut, USA: 2001. The Visual Display of Quantitative Information. [Google Scholar]
- 75.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 76.Wurdinger T, et al. A secreted luciferase for ex vivo monitoring of in vivo processes. Nat. Methods. 2008;5:171–173. doi: 10.1038/nmeth.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ejsmont RK, Sarov M, Winkler S, Lipinski KA, Tomancak P. A toolkit for high-throughput, cross-species gene engineering in Drosophila. Nat. Methods. 2009;6:435–437. doi: 10.1038/nmeth.1334. [DOI] [PubMed] [Google Scholar]
- 78.Howles GP, Ghaghada KB, Qi Y, Munkundan S, Johnson GA. High-resolution magnetic resonance angiography in the mouse using a nanoparticle blood-pool contrast agent. Magn. Reson. Med. 2009;62:1447–1456. doi: 10.1002/mrm.22154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maudsley AA, et al. Comprehensive processing, display and analysis for in vivo MR spectroscopic imaging. NMR Biomed. 2006;19:492–503. doi: 10.1002/nbm.1025. 10.1002/nbm.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thompson PM, et al. Dynamics of gray matter loss in Alzheimer’s disease. J. Neurosci. 2003;23:994–1005. doi: 10.1523/JNEUROSCI.23-03-00994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Plank G, et al. Generation of histo-anatomically representative models of the individual heart: tools and application. Phil. Transact. A Math. Phys. Eng. Sci. 2009;367:2257–2292. doi: 10.1098/rsta.2009.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chiang MC, et al. Fluid registration of diffusion tensor images using information theory. IEEE Trans. Med. Imaging. 2008;27:442–456. doi: 10.1109/TMI.2007.907326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lichtman JW, Conchello JA. Fluorescence microscopy. Nat. Methods. 2005;2:910–919. doi: 10.1038/nmeth817. [DOI] [PubMed] [Google Scholar]
- 84.Conchello J-A, Lichtman JW. Optical sectioning microscopy. Nat. Methods. 2005;2:920–931. doi: 10.1038/nmeth815. [DOI] [PubMed] [Google Scholar]
- 85.Hell SW. Toward fluorescence nanoscopy. Nat. Biotechnol. 2003;21:1347–1355. doi: 10.1038/nbt895. [DOI] [PubMed] [Google Scholar]
- 86.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat. Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- 87.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 88.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sharpe J, et al. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296:541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- 90.Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]