Abstract
We linked the cDNA coding region for the bean storage protein phaseolin to the promoter and regulatory region of the Saccharomyces cerevisiae repressible acid phosphatase gene (PHO5) in multicopy expression plasmids. Yeast transformants containing these plasmids expressed phaseolin at levels up to 3% of the total soluble cellular protein. Phaseolin polypeptides in S. cerevisiae were glycosylated, and their molecular weights suggested that the signal peptide had been processed. We also constructed a series of plasmids in which the phaseolin signal-peptide-coding region was either removed or replaced with increasing amounts of the amino-terminal coding region for acid phosphatase. Phaseolin polypeptides with no signal peptide were not posttranslationally modified in S. cerevisiae. Partial or complete substitution of the phaseolin signal peptide with that from acid phosphatase dramatically inhibited both signal peptide processing and glycosylation, suggesting that some specific feature of the phaseolin signal amino acid sequence was required for these modifications to occur. Larger hybrid proteins that included approximately one-half of the acid phosphatase sequence linked to the amino terminus of the mature phaseolin polypeptide did undergo proteolytic processing and glycosylation. However, these polypeptides were cleaved at several sites that are not normally used in the unaltered acid phosphatase protein.
Full text
PDF
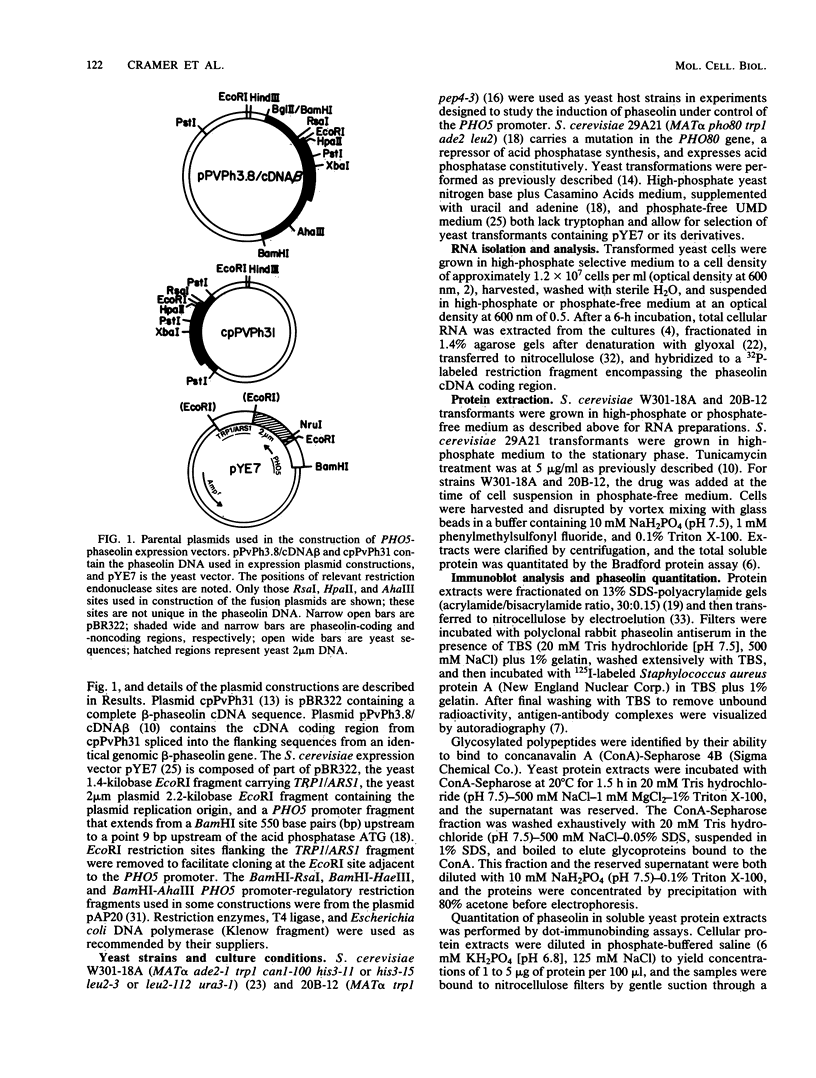

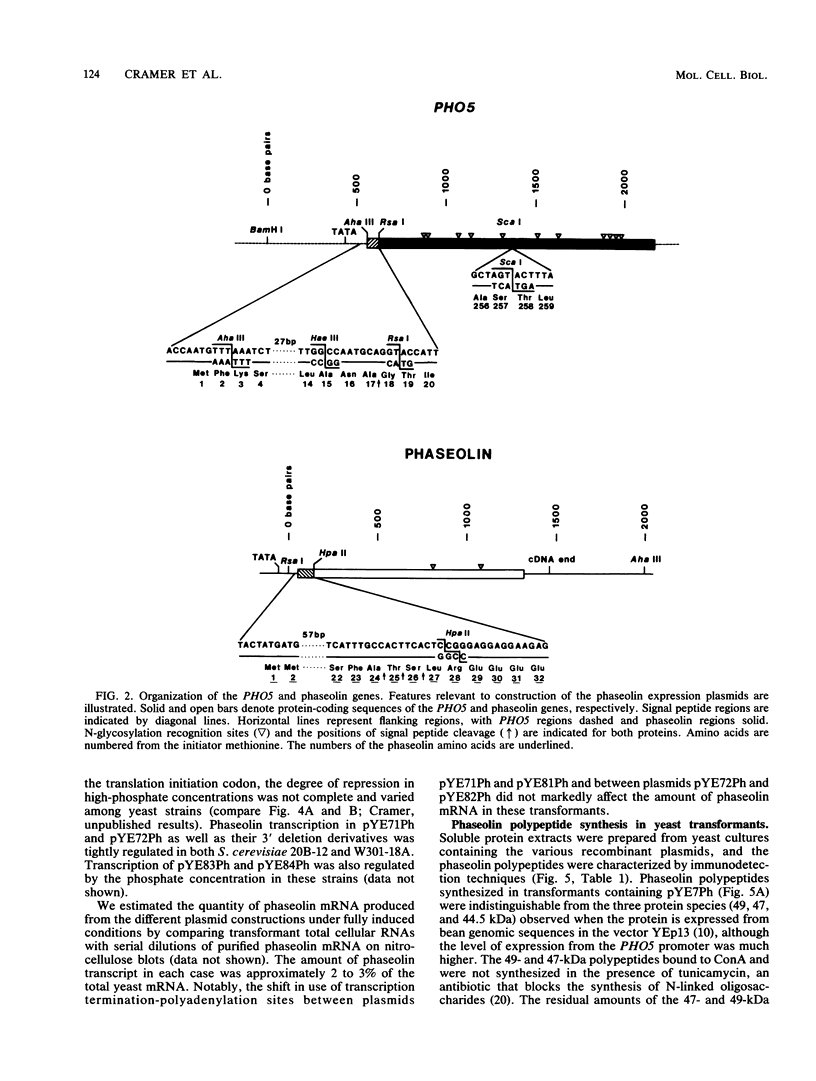
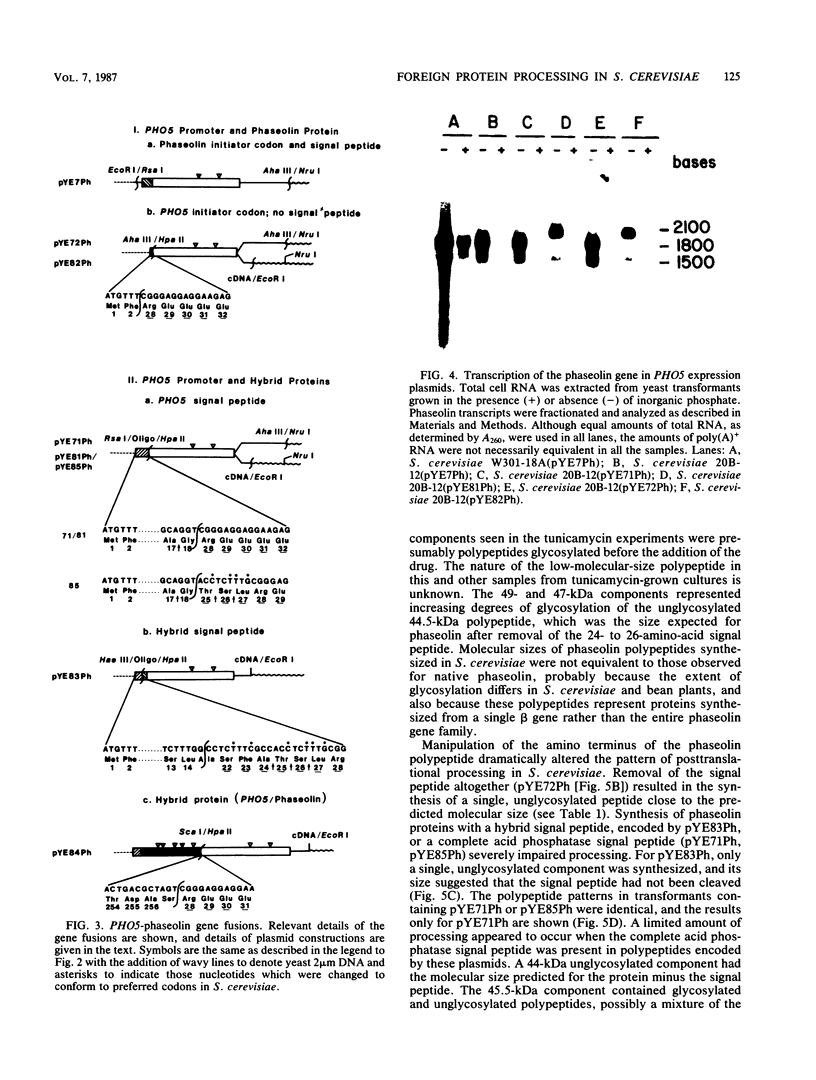


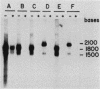
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersen N., Thill G. P., Kramer R. A. RNA and homology mapping of two DNA fragments with repressible acid phosphatase genes from Saccharomyces cerevisiae. Mol Cell Biol. 1983 Apr;3(4):562–569. doi: 10.1128/mcb.3.4.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arima K., Oshima T., Kubota I., Nakamura N., Mizunaga T., Toh-e A. The nucleotide sequence of the yeast PHO5 gene: a putative precursor of repressible acid phosphatase contains a signal peptide. Nucleic Acids Res. 1983 Mar 25;11(6):1657–1672. doi: 10.1093/nar/11.6.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bostian K. A., Hopper J. E., Rogers D. T., Tipper D. J. Translational analysis of the killer-associated virus-like particle dsRNA genome of S. cerevisiae: M dsRNA encodes toxin. Cell. 1980 Feb;19(2):403–414. doi: 10.1016/0092-8674(80)90514-0. [DOI] [PubMed] [Google Scholar]
- Bostian K. A., Lemire J. M., Cannon L. E., Halvorson H. O. In vitro synthesis of repressible yeast acid phosphatase: identification of multiple mRNAs and products. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4504–4508. doi: 10.1073/pnas.77.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Chang C. N., Matteucci M., Perry L. J., Wulf J. J., Chen C. Y., Hitzeman R. A. Saccharomyces cerevisiae secretes and correctly processes human interferon hybrid proteins containing yeast invertase signal peptides. Mol Cell Biol. 1986 May;6(5):1812–1819. doi: 10.1128/mcb.6.5.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer J. H., Lea K., Slightom J. L. Expression of phaseolin cDNA genes in yeast under control of natural plant DNA sequences. Proc Natl Acad Sci U S A. 1985 Jan;82(2):334–338. doi: 10.1073/pnas.82.2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edens L., Bom I., Ledeboer A. M., Maat J., Toonen M. Y., Visser C., Verrips C. T. Synthesis and processing of the plant protein thaumatin in yeast. Cell. 1984 Jun;37(2):629–633. doi: 10.1016/0092-8674(84)90394-5. [DOI] [PubMed] [Google Scholar]
- Haguenauer-Tsapis R., Hinnen A. A deletion that includes the signal peptidase cleavage site impairs processing, glycosylation, and secretion of cell surface yeast acid phosphatase. Mol Cell Biol. 1984 Dec;4(12):2668–2675. doi: 10.1128/mcb.4.12.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopp T. P., Woods K. R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. W. Proteinase mutants of Saccharomyces cerevisiae. Genetics. 1977 Jan;85(1):23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer R. A., Andersen N. Isolation of yeast genes with mRNA levels controlled by phosphate concentration. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6541–6545. doi: 10.1073/pnas.77.11.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer R. A., DeChiara T. M., Schaber M. D., Hilliker S. Regulated expression of a human interferon gene in yeast: control by phosphate concentration or temperature. Proc Natl Acad Sci U S A. 1984 Jan;81(2):367–370. doi: 10.1073/pnas.81.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lehle L., Tanner W. The specific site of tunicamycin inhibition in the formation of dolichol-bound N-acetylglucosamine derivatives. FEBS Lett. 1976 Nov 15;72(1):167–170. doi: 10.1016/0014-5793(76)80922-2. [DOI] [PubMed] [Google Scholar]
- Linnemans W. A., Boer P., Elbers P. F. Localization of acid phosphatase in Saccharomyces cerevisiae: a clue to cell wall formation. J Bacteriol. 1977 Aug;131(2):638–644. doi: 10.1128/jb.131.2.638-644.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Schaber M. D., DeChiara T. M., Kramer R. A. Yeast vectors for production of interferon. Methods Enzymol. 1986;119:416–423. doi: 10.1016/0076-6879(86)19061-6. [DOI] [PubMed] [Google Scholar]
- Slightom J. L., Drong R. F., Klassy R. C., Hoffman L. M. Nucleotide sequences from phaseolin cDNA clones: the major storage proteins from Phaseolus vulgaris are encoded by two unique gene families. Nucleic Acids Res. 1985 Sep 25;13(18):6483–6498. doi: 10.1093/nar/13.18.6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slightom J. L., Sun S. M., Hall T. C. Complete nucleotide sequence of a French bean storage protein gene: Phaseolin. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1897–1901. doi: 10.1073/pnas.80.7.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. A., Duncan M. J., Moir D. T. Heterologous protein secretion from yeast. Science. 1985 Sep 20;229(4719):1219–1224. doi: 10.1126/science.3939723. [DOI] [PubMed] [Google Scholar]
- Thill G. P., Kramer R. A., Turner K. J., Bostian K. A. Comparative analysis of the 5'-end regions of two repressible acid phosphatase genes in Saccharomyces cerevisiae. Mol Cell Biol. 1983 Apr;3(4):570–579. doi: 10.1128/mcb.3.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]