Mutations in genes encoding the spliceosome machinery recently have been described in hematological malignancies,1, 2 particularly in myelodysplastic syndrome (MDS) and chronic lymphocytic leukemia (CLL). These mutations can occur in genes encoding different components of the spliceosome.3 However, the most frequent of such mutations observed in CLL occurs in the gene-encoding splicing factor 3 subunit 1 (SF3B1), a core component of the RNA splicing machinery,1, 4, 5, 6 and is found in 5–17% of CLL patients.5, 6, 7 SF3B1 mutations have been associated with a relatively poor prognosis in CLL,6, 7, 8 but appear associated with a relatively good prognosis in MDS.2, 9, 10, 11 Although initially proposed to represent a driver mutation in CLL,4 a recent study noted two cases in which the CLL cells appeared to have acquired SF3B1 mutations during clonal evolution.7
In this report, we investigated the progression of SF3B1 mutations in the CLL B cells over time in an attempt to elucidate whether there exists subclonal evolution involving SF3B1 mutations in CLL. Accumulation of CLL cells harboring mutations in SF3B1 suggests that such subclones have some competitive advantage, which might account for accelerated progression of the disease in some patients over time. Alternatively, subclones of CLL cells might be selected during therapy, similar to what has been observed in mutations involving TP53 in CLL cells of patients treated with standard chemotherapy.
To address these questions, we examined for SF3B1 mutations in the CLL cells of 545 patients. The CLL samples of 448 patients were acquired before any therapy, and 97 following treatment for CLL. We investigated for changes in the proportionate representation of subclones harboring SF3B1 mutations over time, with or without therapeutic intervention.
Thirty-six cases (6.6%) had CLL cells with detectable mutations in the 545 cases examined. In all, 20 (56%) of these 36 cases had K700E and 6 had K666E/Q/N/T (6/36, 17%). As noted in another study,7 the proportion of cases that had SF3B1 mutations was significantly higher in the samples of patients who had received prior therapy (12.4%, 12/97) than in samples from patients with no prior therapy (5.4%, 24/448, P=0.02) (Supplementary Figure 1A). Cases found to have SF3B1 mutations more commonly were found to have unfavorable prognostic features than did cases without SF3B1 mutations, such as expression of unmutated immunoglobulin heavy chain variable region genes (75%, 27/36, P=0.0002), as reported previously.5 Furthermore, a higher proportion of cases with SF3B1 mutations also expressed ZAP-70 (75%, 27/36, P<0.0001) or CD38 (58%, 21/36, P=0.0007) than did cases without SF3B1 mutations (Supplementary Figure 1B). Patients with CLL cells harboring SF3B1 mutations were found to have a significantly shorter median treatment-free survival (3.0 versus 6.0 years, P<0.0001) and overall survival (OS, 11.4 versus 21.0 years, P=0.0021) than did patients with CLL cells lacking detectable SF3B1 mutations (Supplementary Figures 1C and D).
Fluorescence in situ hybridization (FISH) data were available for 362 (71%) out of 509 CLL patients with leukemic cells without SF3B1 mutations and 33 (92%) out of 36 CLL patients with leukemic cells with SF3B1 mutations. A higher proportion of the SF3B1-mutated cases lacked detectable chromosomal abnormalities than did those cases without SF3B1 mutations (15/33, 45% versus 88/362, 24%, P=0.012). Regarding the genetic abnormalities detected by FISH (del17p, del11q, +12 and del13q), we did not observe any one genetic abnormality associated with cases that did or did not have SF3B1 mutations (Supplementary Figures 2A and B).
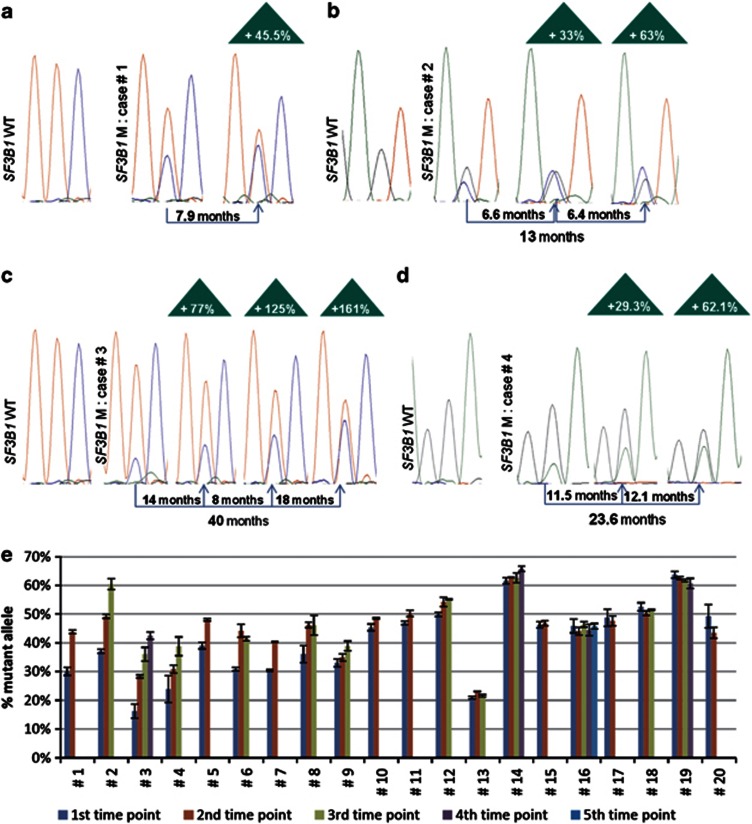
We then examined serial samples from the patients identified as having SF3B1 mutations. Of 36 cases identified to have SF3B1 mutations, 20 cases were included in these analyses (Table 1) with time intervals between sample collections ranging from 6 to 61.5 months (median 23.7 months). We measured the height of the peak corresponding to the wild-type or mutant SF3B1 and calculated the proportionate representation of the height for the mutant allele of samples collected at different times (Figure 1). For some of the samples, this value exceeded 50% (for example, cases 2,12, 14 and 19). None of these samples had the most common SF3B1 mutation (for example, K700E), making it unlikely that these samples had the same mutation on both alleles. Rather it is more likely that each of these samples had cells harboring a mutation in SF3B1 and a deletion of the wild-type allele. Prior studies had identified uncommon cases of CLL that had deletions in 2q (the chromosomal location for SF3B1), which were not detected by conventional cytogenetics.12 For seven cases (no. 1–7), we noted significant increases in the proportionate representation of the SF3B1-mutant allele over time (Table 1). For three out of the seven cases (cases 2, 3 and 4), all serial samples were collected before therapy; for the other four cases, all serial samples were collected after therapy (Supplementary Table 1). These data indicate that the proportion of subclones harboring SF3B1 mutations can increase spontaneously over time independent of any therapeutic intervention.
Table 1. Twenty cases with SF3B1 mutations examined serially.
| Patient number | Prior Tx | SF3B1 mutation | IGHV status | ZAP-70 status | FISH abnormality | Initial ratio (%)a | Increase over time (%) | First to last SC (months) | Increase/month (%) | P-value |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Yes | K700E | UM | Pos | del(17p) | 30.1 | 45.5 | 7.9 | 5.8 | 0.0091 |
| 2 | No | E622D | MU | Neg | del(13q) | 37.0 | 63.1 | 12.9 | 4.9 | 0.0095 |
| 3 | No | K700E | MU | Neg | None | 16.3 | 161.2 | 40.2 | 4.0 | 0.0022 |
| 4 | No | G740E | UM | Pos | None | 23.9 | 62.1 | 23.6 | 2.6 | 0.0096 |
| 5 | Yes | G740E | UM | Pos | None | 39.0 | 23.1 | 9.8 | 2.4 | 0.0111 |
| 6 | Yes | R625C | UM | Pos | None | 30.8 | 34.2 | 16.6 | 2.1 | 0.0464 |
| 7 | Yes | K700E | MU | Pos | None | 30.5 | 32.3 | 27.3 | 1.2 | 0.0017 |
| 8 | No | K700E | UM | Pos | del(13q) | 36.1 | 27.7 | 24.1 | 1.1 | 0.1472 |
| 9 | No | K700E | UM | Pos | None | 33.0 | 17.8 | 16.1 | 1.1 | 0.2104 |
| 10 | No | K666E | UM | Neg | None | 45.4 | 6.8 | 6.3 | 1.1 | 0.0726 |
| 11 | No | K666E | UM | Pos | del(11q) | 46.9 | 7.3 | 11.1 | 0.7 | 0.1641 |
| 12 | No | K666N | UM | Neg | trisomy 12 | 49.9 | 10.4 | 23.8 | 0.4 | 0.1643 |
| 13 | Yes | K666Q | UM | Pos | del(11q) | 20.9 | 3.0 | 11.0 | 0.3 | 0.0750 |
| 14 | No | E622D | UM | Pos | del(11q) | 61.6 | 6.8 | 36.1 | 0.2 | 0.1413 |
| 15 | Yes | K700E | MU | Neg | del(13q) | 46.3 | 1.2 | 10.3 | 0.1 | 0.7482 |
| 16 | Yes | K700E | UM | Pos | None | 45.9 | −0.3 | 35.7 | 0.0 | 0.7762 |
| 17 | Yes | K700E | UM | Pos | del(11q) | 48.7 | −2.3 | 61.5 | 0.0 | 0.7643 |
| 18 | No | N626Y | UM | Pos | del(13q) | 52.5 | −1.9 | 23.8 | −0.1 | 0.4690 |
| 19 | Yes | K741T | MU | Neg | del(13q) | 63.7 | −4.7 | 52.0 | −0.1 | 0.1531 |
| 20 | Yes | K700E | MU | Neg | del(13q) | 49.2 | −11.7 | 26.7 | −0.4 | 0.2838 |
Abbreviations: FISH, fluorescence in situ hybridization; IGHV, immunoglobulin heavy chain variable; MU, mutated; neg, negative; pos, positive; SC, sample collection ; UM, unmutated.
The column marked ‘Prior Tx' indicates whether the patient did (yes) or did not (no) have prior therapy for CLL. The column marked ‘SF3B1 mutation' provides the site (middle number) and amino-acid residue of the wild-type and mutant allele, using standard single-letter amino-acid nomenclature. The column marked ‘IGHV status' provides the mutation status for the expressed IGHV gene, which was considered UM when it had ⩾98% identity with the germ-line IGHV or MU when it had <98% identity. The column marked ‘ZAP-70 status' indicates whether the CLL sample was pos or neg for ZAP-70, as assessed via flow cytometry. The column marked ‘FISH abnormality' provides the abnormal FISH finding (if any) for the initial sample. The column marked ‘Initial ratio (%)' provides the proportionate representation of the SF3B1-mutant allele in this initial sample. The column labeled ‘increase over time (%)' provides the percent increase in the proportionate representation of the mutant SF3B1 allele, which was calculated with the formula 100 × [(mutant allele ratio of the last SC - mutant allele ratio of the first SC)/mutant allele ratio of the first SC]. The column marked ‘First to last SC (months)' provides the number of months between the last SC and first SC. The column labeled ‘Increase/months (%)' was calculated by dividing the value in the column marked ‘Increase over time' by the value in the column labeled ‘First to last SC'. The P-values were calculated using the Student's t-test when two time points were available or using the one-way analysis of variance when data from three or more time points from the same case were available.
Initial proportionate representation of the SF3B1-mutant allele according to the formula 100 × (M peak intensity/(M peak intensity+WT peak intensity)).
Figure 1.
Evolution of SF3B1 mutations over time. (a–d) Representation of the serial sample sequence chromatograms for patient no. 1, 2, 3 and 4, respectively. The time intervals between each time point are indicated as well as the overall time span studied. The percentage of increase over time of the SF3B1-mutant allele is indicated in the triangles and is calculated by comparison with the first time point. (e) Bar graph representing the percentage of SF3B1-mutant allele measured at each time points for all the 20 SF3B1-mutant cases included in the serial analysis. The 20 cases are ranked by their increase/months (%), according to Table 1. Each time point has been sequenced in duplicate, and the error bars represent the mean±s.d.
We studied p53 function in two out of these four cases (no. 1 and 6, Supplementary Figure 3), for which CLL cells were available at the second sample collection and after treatment. To evaluate p53 function, we monitored for expression of p53 and p21 after γ-irradiation via flow cytometry. We found γ-irradiation-induced p53 and p21 in each of these two samples, indicating functional TP53. As such, subclonal expansion of cells harboring mutations in SF3B1 can occur in cells that have not lost p53 function.
For 19 cases out of the 20 cases analyzed serially, we conducted FISH analyses on the serial samples. We did not observe any changes between the serial samples for 15 (79%) cases, including 4 that had the largest increases in the proportionate representation of the SF3B1 allele over time in association with concurrent disease progression (Supplementary Table 1). However, in four cases, we observed one or two new cytogenetic abnormalities in the second sample (for example, for case 11, there was acquisition of del13q, for cases 17 and 18, there was acquisition of del17p and for case 14, there was acquisition of del13q and del17p).
Patient no. 1 was treated with fludarabine, cyclophosphamide and rituximab in July 2004. The patient relapsed after achieving a partial response, developing progressive disease. Between sample collections, the patient's absolute lymphocyte count (ALC) increased from 13 to 102 × 109/l, which was associated with an increase in the proportionate representation of the SF3B1 allele from 30 to 44%. Similarly, between sample collections of patient no.2, the ALC increased from 67 to 250 × 109/l, which was associated with an increase in the proportionate representation of the SF3B1 allele from 37 to 60%. Patient no. 3 experienced increases in ALC from 87 to 197 × 109/l between sample collections, for which we detected an increase in the proportionate representation of the SF3B1 allele from 16 to 43%. Finally, the ALC increased in patient no. 4 from 8 to 47 × 109/l between sample collections, for which we noted an increase in the proportionate representation of the SF3B1 allele from 24 to 39%. These data imply that subclones of CLL cells harboring mutant SF3B1 had either a higher growth rate and/or lower death rate than subclones lacking mutations in SF3B1, this potentially contributing to disease progression.
For four cases with SF3B1 mutations (cases 6, 13, 16 and 17), we examined samples before and after therapy, which resulted in >50% reduction in ALC. For each of these cases, however, we did not observe a significant change in the proportionate representation of the mutant SF3B1 allele, indicating that such treatments did not have selective activity or inactivity for subclones with SF3B1 mutations.
Taken together, this study reveals subclonal evolution involving cells with SF3B1 mutations in CLL. Our data indicate that the proportionate representation of cells harboring SF3B1 mutations can increase independent of therapy or loss of functional p53. Finally, the data presented here suggest that subclones with SF3B1 mutations do not necessarily have a selective advantage or disadvantage in the setting of effective cytoreductive therapy. Nevertheless, the prevalence of SF3B1 mutations appears higher among patients who already have undergone therapy. This might reflect the fact that treated patients more commonly have had longer disease histories, potentially providing greater time for emergence and subclonal evolution of CLL cells harboring SF3B1 mutations. Further studies with additional cases will be required to address the therapeutic implications of the SF3B1 mutations and subclonal evolution in this disease.
Acknowledgments
This study was funded in part by NIH grant PO1-CA081534 for the CLL Research Consortium (CRC) and the UC San Diego Moores Cancer Center Blood Cancer Research Fund. We thank Drs William G Wierda, Kanti R Rai, John C Byrd, Neil E Kay, Jennifer Brown and John Gribben for providing the CLL samples and the relevant clinical data, and Nyla A Heerema, Paola Dal Cin, Ayala Aviram, Chandrika Sreekantaiah and Daniel van Dyke for cytogenetic analysis of the CLL samples. We are thankful to Sylvia Shen, Monica Cook and Jennifer Piper for their excellent technical assistance.
Author contributions
MS, EG, LR and TJK designed, analyzed the data and wrote the manuscript. LZR and TJK provided patients samples and clinical data. MO performed DNA extractions and edited the paper. MLD'A examined the cytogenetics. JFF analyzed the data and edited the paper.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Wang LL, Neuberg D, Wu CJ. SF3B1 in chronic lymphocytic leukemia. New Engl J Med. 2012;366:1057–1058. doi: 10.1056/NEJMc1201040. [DOI] [PubMed] [Google Scholar]
- Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. New Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thol F, Kade S, Schlarmann C, Loffeld P, Morgan M, Krauter J, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119:3578–3584. doi: 10.1182/blood-2011-12-399337. [DOI] [PubMed] [Google Scholar]
- Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44:47–U70. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- Wang LL, Lawrence MS, Wan YZ, Stojanov P, Sougnez C, Stevenson K, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. New Engl J Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Ramsay AJ, Lopez-Otin C. Chronic lymphocytic leukemia with SF3B1 mutation. New Engl J Med. 2012;366:2530–2530. doi: 10.1056/NEJMc1204033. [DOI] [PubMed] [Google Scholar]
- Rossi D, Bruscaggin A, Spina V, Rasi S, Khiabanian H, Messina M, et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood. 2011;118:6904–6908. doi: 10.1182/blood-2011-08-373159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri L, Cahill N, Gunnarsson R, Smedby KE, Tjonnfjord E, Hjalgrim H, et al. NOTCH1 and SF3B1 mutations can be added to the hierarchical prognostic classification in chronic lymphocytic leukemia Leukemia 2012. e-pub ahead of print 6 November 2012doi: 10.1038/leu.2012.307 [DOI] [PubMed]
- Visconte V, Makishima H, Jankowska AM, Traina F, Szpurka H, Rogers HJ, et al. Association of SF3B1 with ring sideroblasts in patients, in vivo, and in vitro models of spliceosomal dysfunction. Blood. 2011;118:211–212. [Google Scholar]
- Damm F, Thol F, Kosmider O, Kade S, Loffeld P, Dreyfus F, et al. SF3B1 mutations in myelodysplastic syndromes: clinical associations and prognostic implications. Leukemia. 2012;26:1137–1140. doi: 10.1038/leu.2011.321. [DOI] [PubMed] [Google Scholar]
- Bejar R, Stevenson K, Caughey B, Abdel-Wahab O, Galili N, Garcia-Manero G, et al. Validation of a prognostic model and the impact of SF3B1, DNMT3A, and other mutations in 289 genetically characterized lower risk MDS patient samples. Blood. 2011;118:443–443. [Google Scholar]
- Novak U, Oppliger Leibundgut E, Hager J, Muhlematter D, Jotterand M, Besse C, et al. A high-resolution allelotype of B-cell chronic lymphocytic leukemia (B-CLL) Blood. 2002;100:1787–1794. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.