Abstract
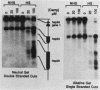
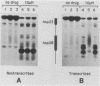
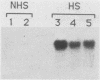
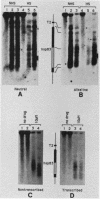
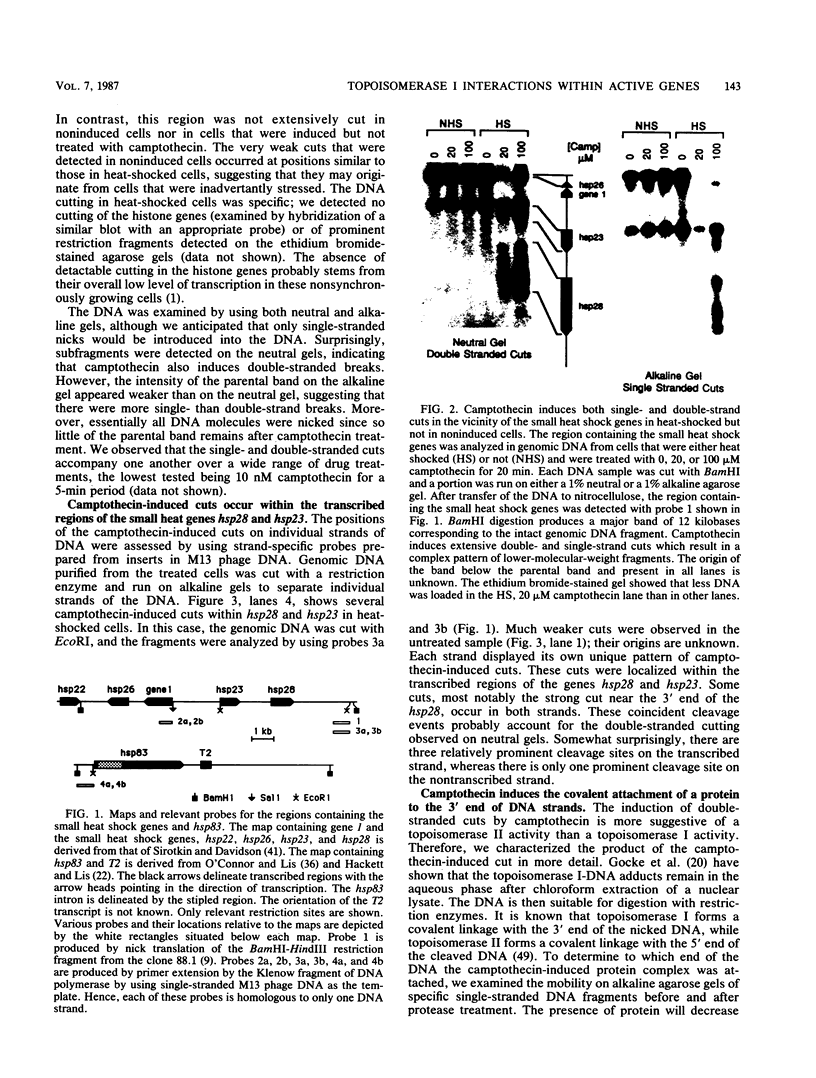
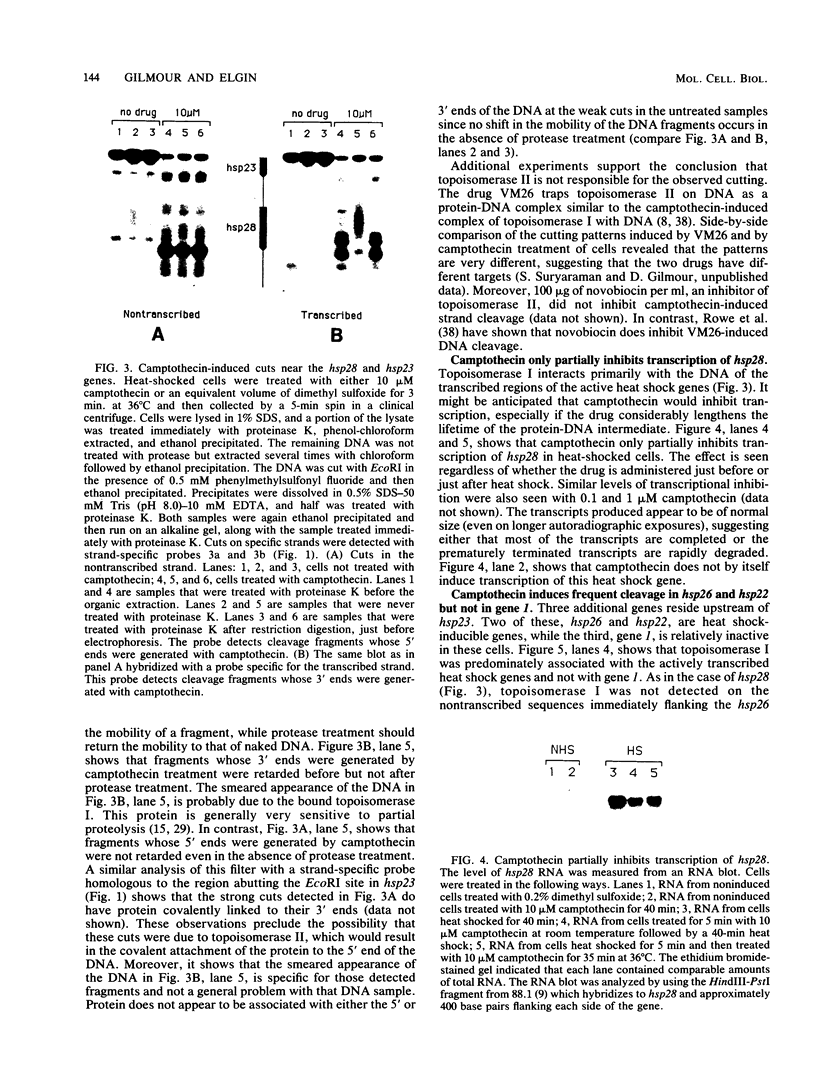
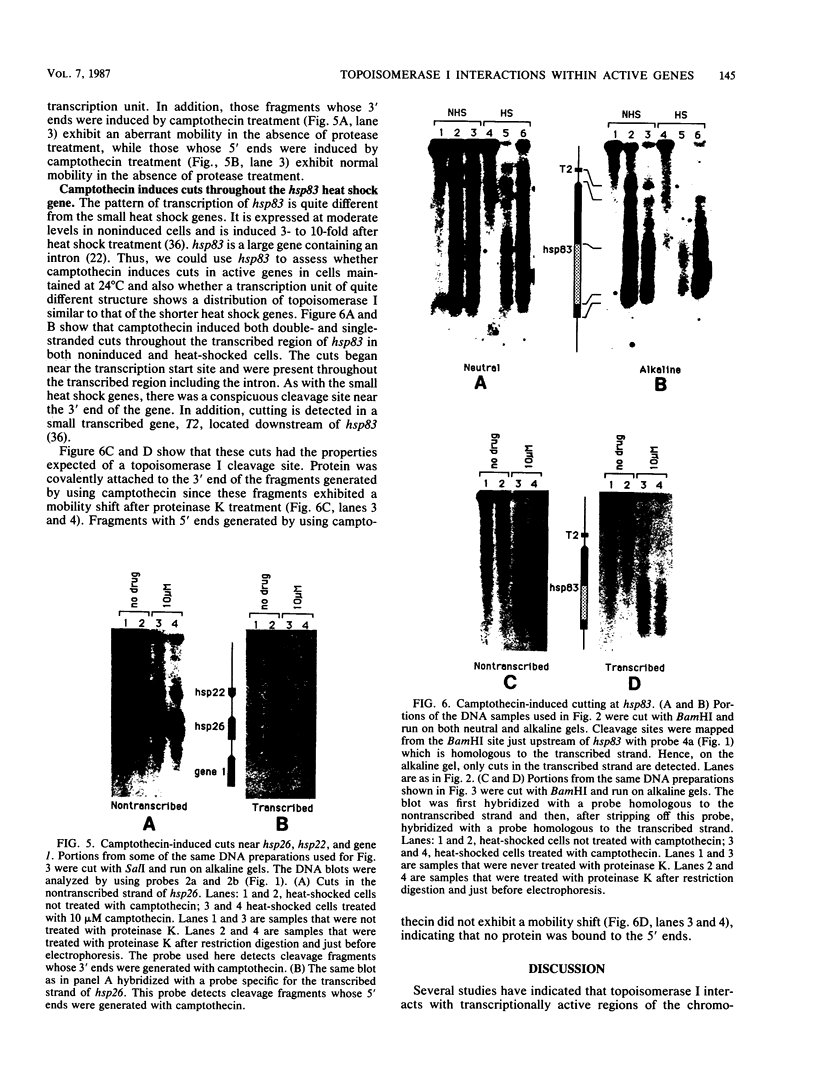
Camptothecin stabilizes the topoisomerase I-DNA covalent intermediate that forms during the relaxation of torsionally strained DNA. By mapping the position of the resultant DNA nicks, we analyzed the distribution of the covalent intermediates formed on heat shock genes in cultured Drosophila melanogaster cells. Topoisomerase I was found to interact with the transcriptionally active genes hsp22, hsp23, hsp26, and hsp28 after heat shock but not with the inactive genes before heat shock. The interaction occurred predominantly within the transcribed region, with specific sites occurring on both the transcribed and nontranscribed strands of the DNA. Little interaction was seen with nontranscribed flanking sequences. Camptothecin only partially inhibited transcription of the hsp28 gene during heat shock, causing a reduced level of transcripts which were nonetheless full length. Topoisomerase I also interacted with the DNA throughout the transcriptionally active hsp83 gene, including an intron, in both heat-shocked and non-heat-shocked cells. The results point to a dynamic set of interactions at the active locus.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baase W. A., Wang J. C. An omega protein from Drosophila melanogaster. Biochemistry. 1974 Oct 8;13(21):4299–4303. doi: 10.1021/bi00718a009. [DOI] [PubMed] [Google Scholar]
- Berrios M., Osheroff N., Fisher P. A. In situ localization of DNA topoisomerase II, a major polypeptide component of the Drosophila nuclear matrix fraction. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4142–4146. doi: 10.1073/pnas.82.12.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonven B. J., Gocke E., Westergaard O. A high affinity topoisomerase I binding sequence is clustered at DNAase I hypersensitive sites in Tetrahymena R-chromatin. Cell. 1985 Jun;41(2):541–551. doi: 10.1016/s0092-8674(85)80027-1. [DOI] [PubMed] [Google Scholar]
- Bullock P., Champoux J. J., Botchan M. Association of crossover points with topoisomerase I cleavage sites: a model for nonhomologous recombination. Science. 1985 Nov 22;230(4728):954–958. doi: 10.1126/science.2997924. [DOI] [PubMed] [Google Scholar]
- Cartwright I. L., Elgin S. C. Nucleosomal instability and induction of new upstream protein-DNA associations accompany activation of four small heat shock protein genes in Drosophila melanogaster. Mol Cell Biol. 1986 Mar;6(3):779–791. doi: 10.1128/mcb.6.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J., Dulbecco R. An activity from mammalian cells that untwists superhelical DNA--a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay). Proc Natl Acad Sci U S A. 1972 Jan;69(1):143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. L., Yang L., Rowe T. C., Halligan B. D., Tewey K. M., Liu L. F. Nonintercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J Biol Chem. 1984 Nov 10;259(21):13560–13566. [PubMed] [Google Scholar]
- Corces V., Holmgren R., Freund R., Morimoto R., Meselson M. Four heat shock proteins of Drosophila melanogaster coded within a 12-kilobase region in chromosome subdivision 67B. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5390–5393. doi: 10.1073/pnas.77.9.5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costlow N., Lis J. T. High-resolution mapping of DNase I-hypersensitive sites of Drosophila heat shock genes in Drosophila melanogaster and Saccharomyces cerevisiae. Mol Cell Biol. 1984 Sep;4(9):1853–1863. doi: 10.1128/mcb.4.9.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig E. A., McCarthy B. J. Four Drosophila heat shock genes at 67B: characterization of recombinant plasmids. Nucleic Acids Res. 1980 Oct 10;8(19):4441–4457. doi: 10.1093/nar/8.19.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S., Voelkel K., Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc Natl Acad Sci U S A. 1984 May;81(9):2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Halligan B., Cooke C. A., Heck M. M., Liu L. F. Topoisomerase II is a structural component of mitotic chromosome scaffolds. J Cell Biol. 1985 May;100(5):1706–1715. doi: 10.1083/jcb.100.5.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher L. M. DNA supercoiling and gene expression. Nature. 1984 Feb 23;307(5953):686–687. doi: 10.1038/307686a0. [DOI] [PubMed] [Google Scholar]
- Fleischmann G., Pflugfelder G., Steiner E. K., Javaherian K., Howard G. C., Wang J. C., Elgin S. C. Drosophila DNA topoisomerase I is associated with transcriptionally active regions of the genome. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6958–6962. doi: 10.1073/pnas.81.22.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Rouvière-Yaniv J., Yaniv M., Brutlag D. Nicking-closing enzyme assembles nucleosome-like structures in vitro. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3779–3783. doi: 10.1073/pnas.76.8.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour D. S., Lis J. T. In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster. Mol Cell Biol. 1985 Aug;5(8):2009–2018. doi: 10.1128/mcb.5.8.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour D. S., Pflugfelder G., Wang J. C., Lis J. T. Topoisomerase I interacts with transcribed regions in Drosophila cells. Cell. 1986 Feb 14;44(3):401–407. doi: 10.1016/0092-8674(86)90461-7. [DOI] [PubMed] [Google Scholar]
- Gocke E., Bonven B. J., Westergaard O. A site and strand specific nuclease activity with analogies to topoisomerase I frames the rRNA gene of Tetrahymena. Nucleic Acids Res. 1983 Nov 25;11(22):7661–7678. doi: 10.1093/nar/11.22.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T., Wang J. C. Cloning of yeast TOP1, the gene encoding DNA topoisomerase I, and construction of mutants defective in both DNA topoisomerase I and DNA topoisomerase II. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7178–7182. doi: 10.1073/pnas.82.21.7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett R. W., Lis J. T. Localization of the hsp83 transcript within a 3292 nucleotide sequence from the 63B heat shock locus of D. melanogaster. Nucleic Acids Res. 1983 Oct 25;11(20):7011–7030. doi: 10.1093/nar/11.20.7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Udvardy A., Schedl P. Novobiocin blocks the Drosophila heat shock response. J Mol Biol. 1985 May 5;183(1):13–29. doi: 10.1016/0022-2836(85)90277-3. [DOI] [PubMed] [Google Scholar]
- Harland R. M., Weintraub H., McKnight S. L. Transcription of DNA injected into Xenopus oocytes is influenced by template topology. Nature. 1983 Mar 3;302(5903):38–43. doi: 10.1038/302038a0. [DOI] [PubMed] [Google Scholar]
- Heller R. A., Shelton E. R., Dietrich V., Elgin S. C., Brutlag D. L. Multiple forms and cellular localization of Drosophila DNA topoisomerase II. J Biol Chem. 1986 Jun 15;261(17):8063–8069. [PubMed] [Google Scholar]
- Holm C., Goto T., Wang J. C., Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985 Jun;41(2):553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- Hsiang Y. H., Hertzberg R., Hecht S., Liu L. F. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985 Nov 25;260(27):14873–14878. [PubMed] [Google Scholar]
- Jackson D. A., Cook P. R. Transcription occurs at a nucleoskeleton. EMBO J. 1985 Apr;4(4):919–925. doi: 10.1002/j.1460-2075.1985.tb03719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javaherian K., Tse Y. C., Vega J. Drosophila topoisomerase I: isolation, purification and characterization. Nucleic Acids Res. 1982 Nov 11;10(21):6945–6955. doi: 10.1093/nar/10.21.6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec E. B., Ryoji M., Worcel A. Gyration is required for 5S RNA transcription from a chromatin template. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1305–1309. doi: 10.1073/pnas.83.5.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchnik A. N., Bakayev V. V., Zbarsky I. B., Georgiev G. P. Elastic torsional strain in DNA within a fraction of SV40 minichromosomes: relation to transcriptionally active chromatin. EMBO J. 1982;1(11):1353–1358. doi: 10.1002/j.1460-2075.1982.tb01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedospasov S. A., Georgiev G. P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980 Jan 29;92(2):532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- O'Connor D., Lis J. T. Two closely linked transcription units within the 63B heat shock puff locus of D. melanogaster display strikingly different regulation. Nucleic Acids Res. 1981 Oct 10;9(19):5075–5092. doi: 10.1093/nar/9.19.5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S. I., Nelkin B. D., Vogelstein B. The ovalbumin gene is associated with the nuclear matrix of chicken oviduct cells. Cell. 1982 Jan;28(1):99–106. doi: 10.1016/0092-8674(82)90379-8. [DOI] [PubMed] [Google Scholar]
- Rowe T. C., Wang J. C., Liu L. F. In vivo localization of DNA topoisomerase II cleavage sites on Drosophila heat shock chromatin. Mol Cell Biol. 1986 Apr;6(4):985–992. doi: 10.1128/mcb.6.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoji M., Worcel A. Chromatin assembly in Xenopus oocytes: in vivo studies. Cell. 1984 May;37(1):21–32. doi: 10.1016/0092-8674(84)90297-6. [DOI] [PubMed] [Google Scholar]
- Saavedra R. A., Huberman J. A. Both DNA topoisomerases I and II relax 2 micron plasmid DNA in living yeast cells. Cell. 1986 Apr 11;45(1):65–70. doi: 10.1016/0092-8674(86)90538-6. [DOI] [PubMed] [Google Scholar]
- Sirotkin K., Davidson N. Developmentally regulated transcription from Drosophila melanogaster chromosomal site 67B. Dev Biol. 1982 Jan;89(1):196–210. doi: 10.1016/0012-1606(82)90307-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steiner E. K., Eissenberg J. C., Elgin S. C. A cytological approach to the ordering of events in gene activation using the Sgs-4 locus of Drosophila melanogaster. J Cell Biol. 1984 Jul;99(1 Pt 1):233–238. doi: 10.1083/jcb.99.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Yanagida M. Isolation of type I and II DNA topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J. 1984 Aug;3(8):1737–1744. doi: 10.1002/j.1460-2075.1984.tb02040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeponteau B., Lundell M., Martinson H. Torsional stress promotes the DNAase I sensitivity of active genes. Cell. 1984 Dec;39(3 Pt 2):469–478. doi: 10.1016/0092-8674(84)90454-9. [DOI] [PubMed] [Google Scholar]
- Vosberg H. P. DNA topoisomerases: enzymes that control DNA conformation. Curr Top Microbiol Immunol. 1985;114:19–102. doi: 10.1007/978-3-642-70227-3_2. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Wu C., Wong Y. C., Elgin S. C. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979 Apr;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]