Abstract
A subset of non-O1/non-O139 serogroup strains of Vibrio cholerae cause disease using type 3 secretion system (T3SS)-mediated mechanisms. An ∼50-kb genomic island carries genes encoding the T3SS structural apparatus, effector proteins, and two transmembrane transcriptional regulators, VttRA and VttRB, which are ToxR homologues. Previous experiments demonstrated that VttRA and VttRB are necessary for colonization in vivo and promote bile-dependent T3SS gene expression in vitro. To better understand the scope of genes that are potential targets of VttRA and VttRB regulation, we performed deep RNA sequencing using O39 serogroup strain AM-19226 and derivatives carrying deletions in vttRA and vttRB grown in bile. Comparison of the transcript profiles from ΔvttRA and ΔvttRB mutant strains to the isogenic parent strain confirmed that VttRA and VttRB regulate expression of some T3SS island genes and provided additional information about relative expression levels and operon organization. Interestingly, the data also suggested that additional genes, located outside the T3SS island and encoding functions involved in motility, chemotaxis, type 6 secretion, transcriptional regulation, and stress responses, may also by regulated by VttRA and VttRB. We verified transcript levels for selected genes by quantitative reverse transcription (RT)-PCR and then focused additional studies on motility and biofilm formation. The results suggest that VttRA and VttRB act as part of a complex transcriptional network that coordinates virulence gene expression with multiple cellular phenotypes. VttRA and VttRB therefore represent horizontally acquired transcriptional regulators with the ability to influence global gene expression in addition to modulating gene expression within the T3SS genomic island.
INTRODUCTION
In general, water- and food-borne enteric pathogens must recognize multiple host niches encountered during infection and correctly regulate gene expression to promote virulence. Persistence in an environmental reservoir adds the additional challenge of integrating a broader set of external signals to facilitate environmental adaptation as well as recognition of a host niche. Regulation of virulence gene expression is therefore a critical parameter affecting the success of a pathogen. As an environmental organism that is also an important cause of infectious diarrhea, Vibrio cholerae has served as a paradigm for understanding coordinated virulence gene regulation and adaptation to the human host (1, 2).
V. cholerae strains vary in their genetic composition but are classified into serogroups based on O-antigen structure (3). Strains that cause epidemic cholera are restricted to O1 and O139 serogroups, having clonal origins and conserved virulence mechanisms (4, 5). The major virulence factors of epidemic O1 and O139 serogroup strains are the toxin-coregulated pilus (TCP), required for colonization, and cholera toxin (CT), whose enzymatic activity results in secretory diarrhea (6). Genes encoding both factors reside on horizontally transferred sequences; TCP is encoded on a genomic island having several characteristics of mobile elements, and the cholera toxin genes are acquired via lysogenic conversion by the filamentous CTX phage (7, 8). Although dedicated transcriptional regulators are often encoded within the mobile elements that carry virulence genes, expression of TCP and CT is governed by both a core genome-encoded transcriptional regulator, ToxR, and a regulator encoded within the TCP island, ToxT (1). ToxR is a transmembrane transcriptional regulator that modulates porin gene expression and multiple metabolic pathways in all V. cholerae strains, in addition to regulating virulence gene expression in pathogenic strains (9). ToxR can directly bind promoters under specific conditions, but it can also indirectly regulate TCP and CT expression by activating expression of toxT, which encodes a cytoplasmic AraC family transcriptional regulator (10, 11). toxT expression also requires the activity of another membrane-localized transcriptional regulator, TcpP, encoded within the TCP genomic island. The transcription of tcpP itself is activated by two chromosomal regulators, AphA and AphB (1). The ToxR-ToxT transcriptional hierarchy is therefore an important component of V. cholerae virulence gene expression, which also involves accessory proteins (10, 12, 13) and an integrated response to multiple external signals, such as bile salts, pH, and temperature (14).
Strains that cause nonepidemic disease belong to other serogroups and are collectively referred to as non-O1/non-O139 serogroup strains. Whereas epidemic disease is typically restricted to a biennial pattern in Southeast Asia, Africa, South America, and areas experiencing disrupted civil infrastructure, non-O1/non-O139 strains cause sporadic disease throughout the year and in diverse geographic locations worldwide (15–17). The clinical characteristics of cholera caused by nonepidemic strains are similar to epidemic disease, although non-O1/non-O139 strains do not typically encode TCP and CT. All strains, however, encode ToxR (16). Non-O1/non-O139 strains can carry a diverse array of virulence factors, but a subset of pathogenic non-O1/non-O139 serogroup strains possess an ∼50-kb, horizontally acquired, genomic island that carries genes encoding a type 3 secretion system (T3SS) (15, 18, 19). The T3SS virulence mechanism is widespread among Gram-negative pathogenic bacteria, including Salmonella, Shigella, Yersinia, and notably Vibrio parahaemolyticus, where the T3SS2 shares sequence similarity to the V. cholerae T3SS (15, 20–22). Our laboratory uses a clinically isolated O39 serogroup strain named AM-19226 to study T3SS-mediated V. cholerae pathogenesis, and we have identified 12 effector proteins encoded within the T3SS genomic island that presumably function in colonization and disease (23). We have also identified two ToxR-like transcriptional regulators, VttRA and VttRB, which are encoded within the T3SS genomic island. In vitro, VttRA, VttRB, and ToxR can regulate expression of the T3SS structural genes and some genes encoding effector proteins, in a bile-dependent manner (23, 24). The data also suggest that autoregulation plays a role in VttRA and VttRB expression and that under some conditions, VttRA may control expression of vttRB (24, 25).
In general, however, we have a limited understanding of how horizontally acquired virulence regulators influence global gene expression. We were therefore interested in exploring whether the regulatory activities of VttRA and VttRB extend beyond the T3SS genomic island or were restricted to genes within the T3SS island. We used deep RNA sequencing (RNA-seq) to compare the expression profiles of wild-type (WT) and ΔvttRA and ΔvttRB mutant strains grown in bile, with the intention of discovering genes or gene classes that might be part of the VttRA and VttRB transcriptomes. We found that strains deleted for VttRA and VttRB had altered expression levels of T3SS island genes, as predicted. Interestingly, however, the results suggest that VttRA and VttRB can also modulate the expression of core chromosomal genes that regulate multiple phenotypes and encode proteins necessary for motility and type 6 secretion. The results support the hypothesis that when virulence gene expression is controlled by horizontally acquired regulators, those regulators must also participate in coordinately regulating other phenotypes important for infection in order to promote successful colonization of the host niche and subsequent disease.
MATERIALS AND METHODS
Strains, RNA isolation, and sample preparation.
V. cholerae R− M+ strain AM-19226 (MD992) was used as the parental strain for experiments reported in this study. MD992 has been previously described and contains a deletion in the type II restriction endonuclease (R−) but is methyltransferase positive (M+) (24). MD992 and ΔvttRA (AAC228) and ΔvttRB (AAC40) (24) mutant derivatives were grown in Luria-Bertani (LB) broth supplemented with 0.4% bile (B-3883; Sigma) on a roller drum at 37°C for approximately 15 h to an optical density at 600 nm (OD600) ranging from 1.2 to 2.3. At least two independent RNA isolations were performed for each strain, as previously described (26) with slight modifications. Briefly, 1 ml of bacterial cells was harvested by centrifugation, the pellet was resuspended in 10 ml TRIzol (Invitrogen) and incubated at 4°C for 1 h followed by addition of 2 ml chloroform. The suspension was centrifuged at 6,804 × g (JA-20; Beckman) for 45 min at 4°C, and the aqueous phase was removed. A 1/10 volume of 3 M sodium acetate (pH 5.9) was added to the aqueous phase, and samples were precipitated by the addition of isopropanol and centrifuged at 6,804 × g (JA-20; Beckman) for 30 min at 4°C. The pellet was resuspended in 0.1% (vol/vol) diethyl pyrocarbonate (DEPC)-treated water, and the RNA was further purified using an RNeasy minikit (Qiagen) according to the manufacturer's instructions. On-column DNase digestion was performed using the Qiagen RNase-free DNase set, and the absence of contaminating DNA was tested by PCR using primers for a housekeeping gene (A33_0788). For RNA-seq, mRNA enrichment was performed twice using MICROBExpress (Ambion) according to the manufacturer's protocol. Enriched mRNA integrity was evaluated using a Bioanalyzer (Agilent) at University of Rochester's Functional Genomics Center (UR-FGC).
Transcriptome analysis.
Deep RNA-seq was performed at UR-FGC using the Applied Biosystems SOLiD 3 Plus (replicate 1) and Applied Biosystems SOLiD 4 Plus (replicate 2) platforms. All procedures were carried out as recommended by the manufacturer and as described by Isabella and Clark (27). Briefly, mRNA was fragmented using RNase III, ligated to adapters, and reverse transcribed to generate cDNA, which was then clonally amplified on magnetic beads and covalently affixed to glass slides. Sequencing of monoclonal cDNA molecules was performed by multiple rounds of ligation of fluorescently labeled oligonucleotides to derive color sequences that were then mapped to the annotated AM-19226 genome (National Center for Biotechnology Information [NCBI]) using BioScope v.1.2 and BioScope v.1.3 software (Applied Biosystems) for replicates 1 and 2, respectively. Contig DS265226, containing the AM-19226 T3SS genomic island, was remapped to obtain RPKM (reads per kilobase of coding sequence per million unique mapped reads) values for open reading frames (ORFs) that likely have alternative start sites compared to the NCBI annotation (23). We also mapped and calculated RPKM values for five unannotated genes, to which we have assigned the following locus tags: acfA, A33_1660a; ORFs 67a and 65 encoding conserved hypothetical proteins (CHPs), A33_1662a and A33_1664a, respectively; vcsC2, A33_1670; and ORF58 encoding a hypothetical protein (HP), A33_1704a. The total number of ORFs used for mapping is thus 3,712.
Data analysis.
.wig files containing raw data for RNA sequencing runs from each strain were visualized using the Integrative Genomics Viewer (IGV) (28). The IGV is a computer-based tool that allows visualization of high-throughput sequencing data, such as those generated by RNA-seq. It also facilitates simultaneous filtering, grouping, and sorting of data generated from multiple samples and data sets. Data from each sample are viewed as an individual track, which can be displayed as a heat map, as shown here. In this study, data were aligned with 154 concatenated contigs from the AM-19226 genome sequencing project (NCBI project accession no. NZ_AATY00000000.1, WGS AATY01000001:AATY01000154) and were also mapped against the revised contig annotated as described above. Reads that mapped uniquely to the AM-19226 sequenced genome were used to compute the RPKM values for each ORF. ORFs with an RPKM value of <10 in all three strain backgrounds were excluded from further analysis. ORFs whose expression was positively influenced by VttRA or VttRB were identified based on two criteria: an RPKM value of ≥10 in the parental strain and an RPKM value at least 2.5-fold lower in the ΔvttRA or ΔvttRB strain(s). Conversely, for ORFs whose expression was decreased in the presence of VttRA/B, we required an RPKM value of ≥10 in the deletion strains and a 2.5-fold-level change compared to expression in the parental strain. RPKM values obtained for each of the 3,712 ORFs were compared across strains, and ORFs with ≥2.5-fold differences in expression were identified for subsequent analyses.
For quantitative reverse transcription-PCR (qRT-PCR), motility, and biofilm assays, all data are presented as means ± standard deviations (SD). Statistical analyses were conducted using Microsoft Excel and/or GraphPad, and Student's t test was performed to determine statistical significance. P values of <0.05 were considered statistically significant.
qRT-PCR.
qRT-PCR was performed using independently isolated samples of RNA, in addition to using RNA from the second RNA-seq run. cDNA was generated using 500 ng of RNA and qScript cDNA SuperMix (Quanta BioSciences, Inc.) according to the manufacturer's instructions. Gene-specific primers (sequences available upon request) were used to amplify short fragments of the gene of interest in qRT-PCRs using an Applied Biosystems StepOne Plus real-time PCR system and Power SYBR green PCR master mix (Applied Biosystems). The cycling conditions were as follows: one 10-min incubation at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Non-reverse-transcribed RNA was used as a template for negative controls in the qRT-PCRs. Transcript levels for individual genes were calculated based on absolute copy numbers obtained using genomic DNA standard curves, as previously described (29). Samples from each RNA source were assayed in triplicate.
Swarming motility assay.
Single colonies of wild-type and ΔvttRA, ΔvttRB, ΔvttRA ΔvttRB (MD1069), ΔtoxR (AD10), and ΔvcsN2 (MD1018) mutant strains (23, 24) were used to inoculate LB plates containing 0.3% agar with or without 0.4% bile. Zones of swarming were measured after incubation for 8 h at 37°C. The assay was performed three times using three to five colonies for each strain.
Static biofilm assay.
Wild-type and ΔvttRA, ΔvttRB, and ΔtoxR mutant strains were grown overnight in LB with 0.4% bile. Cells were centrifuged, resuspended in fresh medium, and seeded in a 96-well microtiter plate at an OD600 of 0.1. The plate was incubated at 30°C for 24 h and washed twice with phosphate-buffered saline (PBS), and the remaining biofilm-associated cells were fixed using 95% ethanol at 4°C for 2 h and stained at room temperature using 0.1% crystal violet solution. The wells were then washed once with PBS, and the remaining biofilm-associated stain was solubilized using 95% ethanol. Total biofilm biomass was determined by measuring absorbance at 570 nm using a Bio-TEK Power Wave XS plate reader. The experiment was conducted using eight individual colonies per strain and repeated three times. For statistical analyses, the highest and the lowest values were excluded and the total biofilm mass was calculated based on 6 colonies per strain.
RESULTS
Gene expression profiles for the AM-19226 parent and isogenic ΔvttRA and ΔvttRB strains.
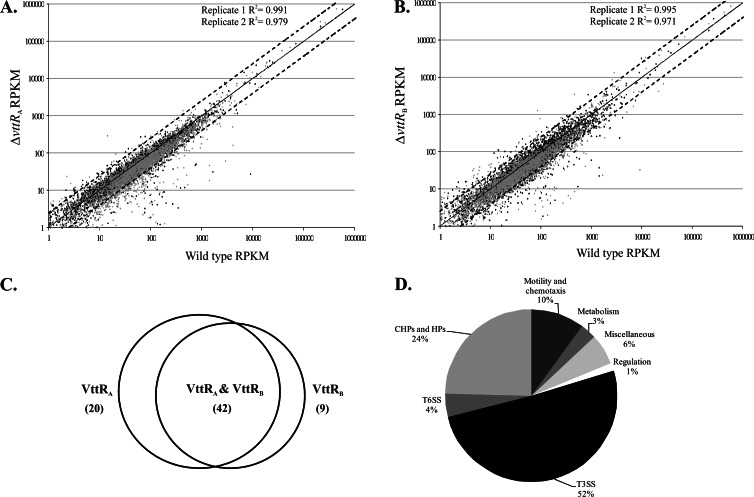
We performed RNA-seq on RNA extracted from an AM-19226 ΔvttRA strain, a ΔvttRB strain, and the isogenic parent strain (carrying wild-type copies of the vttRA and vttRB genes) grown to late exponential-early stationary phase in LB broth supplemented with 0.4% bile. The growth conditions were based on previous results demonstrating that maximal expression of vttRA, vttRB, and T3SS genes encoding structural components of the apparatus occurred under such conditions (24). Two independently isolated RNA samples for each strain were processed for transcriptome analysis. SOLiD sequencing generated 50-bp reads that were mapped to the AM-19226 sequenced genome, resulting in 58% uniquely mapped reads to locus tags for the parental strain (vttRA and vttRB wild type) in replicate 1 and 71% uniquely mapped reads to locus tags for replicate 2. The percentages of uniquely mapped reads to locus tags were 56% for the ΔvttRA strain for replicate 1 and 65% for replicate 2, and 59% for the ΔvttRB strain for replicate 1 and 69% for replicate 2 (see Table S1 in the supplemental material). Each of the 3,712 locus tags (representing ORFs) in the AM-19226 genome was assigned a normalized RPKM (reads per kilobase of coding sequence per million unique mapped reads) value, providing an estimate of the expression level of each ORF/gene. We observed similar trends for gene expression between the two RNA-seq experiments and, for simplicity, list fold changes derived from RPKM values obtained from replicate 1 only in Tables 1 and 2. The complete RPKM data set for each run is provided in Tables S2 and S3 in the supplemental material.
Table 1.
Genes outside the AM-19226 T3SS with ≥2.5-fold-decreased expression in the absence of VttRA or VttRBa
| Gene class and locus tag | Gene product | Fold difference in expression between WT and: |
|
|---|---|---|---|
| ΔvttRA mutant | ΔvttRB mutant | ||
| Motility and chemotaxis | |||
| A33_2046 | FlaD, flagellin | 3.1 | 3.0 |
| A33_2047 | FlaE, flagellin | 2.2 | 2.7 |
| A33_2100 | FlaC, flagellin | 3.0 | 2.1 |
| A33_2149 | FlaA, flagellin | 3.5 | 2.6 |
| A33_1181 | Methyl-accepting chemotaxis protein | 3.2 | 2.0 |
| A33_A0275 | CheA, chemotaxis protein | 3.8 | 2.6 |
| A33_A0320 | Methyl-accepting chemotaxis protein | 3.6 | 1.8 |
| T6SS | |||
| A33_1343 | Hcp-2, hemolysin-coregulated protein | 5.1 | 2.7 |
| A33_1344 | VgrG1, valine-glycine repeat protein | 3.5 | 0.7 |
| A33_A0307 | Hcp-1, hemolysin-coregulated protein | 4.3 | 2.7 |
| Regulation | |||
| A33_1014 | Two-component response regulator | 2.9 | 1.3 |
| Metabolism | |||
| A33_A0971 | Acetoacteyl coenzyme A reductase | 1.8 | 3.1 |
| A33_A0977 | GlgC2, glucose-1-phosphate adenylyltransferase | 3.1 | 3.1 |
| Miscellaneous | |||
| A33_0283 | Etp, Low-mol-wt protein-tyrosine phosphatase | 3.0 | 0.9 |
| A33_0572 | DksA, dnaK suppressor protein | 2.5 | 2.3 |
| A33_A0309 | MurE, UDP-N-acetylmuramyl tripeptide synthetase | 2.5 | 0.9 |
| A33_A0497 | EmrD-2, multidrug resistance protein D | 3.6 | 0.3 |
| CHPs and HPsb | |||
| A33_0127 | CHP | 1.4 | 2.9 |
| A33_0281 | HP | 3.3 | 0.8 |
| A33_1022 | CHP | 2.6 | 1.1 |
| A33_1302 | CHP | 2.6 | 2.9 |
| A33_1345 | CHP | 3.0 | 0.5 |
| A33_1879 | HP | 1.3 | 3.4 |
| A33_1915 | HP | 2.5 | 2.3 |
| A33_2162 | HP | 2.7 | 2.7 |
| A33_2284 | CHP | 3.5 | 3.9 |
| A33_A0004 | HP | 4.1 | 5.2 |
| A33_A0071 | HP | 2.7 | 3.1 |
| A33_A0072 | HP | 2.6 | 3.2 |
| A33_A0073 | HP | 2.7 | 7.4 |
| A33_A0121 | HP | 4.2 | 4.3 |
| A33_A0164 | HP | 5.1 | 3.1 |
| A33_A0247 | CHP | 2.2 | 3.2 |
| A33_A0321 | HP | 4.6 | 2.8 |
Based on RPKM values.
CHP, conserved hypothetical protein; HP, hypothetical protein.
Table 2.
Genes outside the AM-19226 T3SS with ≥2.5-fold-increased expression in the absence of VttRA or VttRBa
| Locus tag | Gene product/pBLAST top hit | Fold difference in expression between WT and: |
|
|---|---|---|---|
| ΔvttRA mutant | ΔvttRB mutant | ||
| A33_0677 | NTPase | 3.0 | 4.8 |
| A33_1527 | Agglutination protein | 3.4 | 2.7 |
| A33_2142 | K139p07 phage protein | 1.0 | 2.7 |
| A33_2143 | K139p06 phage protein (transcriptional regulator) | 0.6 | 3.2 |
| A33_A0028 | HPb | 2.0 | 3.0 |
| A33_A0125 | Sensory box/GGDEF family protein | 3.6 | 2.9 |
| A33_A0330 | HP | 3.6 | 5.0 |
Based on RPKM values.
HP, hypothetical protein.
Candidate genes comprising the VttRA/VttRB transcriptomes.
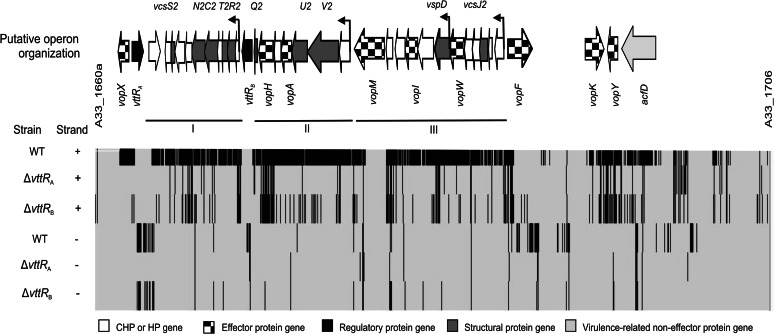
The RPKM values representing the expression of each gene in the ΔvttRA and ΔvttRB strains and the RPKM value for that gene in the parental strain are plotted in Fig. 1A and B. More than 98% of the genes exhibit similar transcriptional expression levels in the different strains. However, we identified a total of 71 genes (∼1.9% of the genome) with a ≥2.5-fold reduction in mRNA levels in the ΔvttRA or ΔvttRB backgrounds compared to the wild-type strain (Fig. 1C). Although we did not conduct extensive statistical analyses of the data sets from both sequencing runs, a 2.5-fold change in expression was consistent with the expression patterns observed for genes within predicted operons and correlated with data obtained from lacZ-transcriptional reporter fusion studies (24). We therefore viewed genes as potentially differentially expressed based on the ≥2.5-fold change and classified them according to predicted functions (Fig. 1D). Of the 71 genes expressed at lower levels in the absence of VttRA and/or VttRB, 37 were located within the AM-19226 T3SS island (see Table S4 in the supplemental material), consistent with the lacZ transcriptional fusion data from our laboratory showing that the T3SS structural gene operons (vcsRTCNS2, vcsJ2, vcsVUQ2, and vspD) require VttRA and VttRB for bile-induced expression (24). Combined with the RNA-seq data, the results suggest that genes encoding the structural apparatus and several effector proteins are expressed as three major operons: operon I (A33_1674 to A33_1665) containing vcsRTCNS2 and proteins of unknown function, operon II (A33_1683 to A33_1676) containing vcsVUQ2, and operon III (A33_1695 to A33_1684) containing vcsJ2 and vspD (Fig. 2) (24) (RT-PCR data not shown). The collective data also predict an internal promoter that lies within operon III, between A33_1690 (vopW) and A33_1689 (vspD) (Fig. 2). Unique effectors such as VopG and VopK that lie in the 3′ region flanking the “core” region that encodes structural proteins do not appear to be transcribed as an operon, based on lacZ transcriptional fusion data and sequence analysis (data not shown).
Fig 1.
VttRA and VttRB transcriptome. (A and B) Scatter plot depicting RPKM values for wild-type and ΔvttRA strains (A) and wild-type and ΔvttRB strains (B) across two independent samples (black and gray). Each data point represents an RPKM value of ≥1 for a specific locus tag/ORF. Solid lines represent x = y, and the dotted lines above and below depict y = 2.5x and x = 2.5y, respectively. R is the coefficient of correlation. (C) Venn diagram depicting genes with reduced expression in the ΔvttRA and ΔvttRB strains. A total of 71 genes are differentially expressed with ≥2.5-fold reduction in expression levels compared to the wild-type strain. (D) Differentially expressed genes in ΔvttRA and ΔvttRB strains potentially categorized according to function.
Fig 2.
Putative operon organization of AM-19226 T3SS genomic island. Heat map representation of the AM-19226 T3SS genomic island spanning from A33_1660a (AcfA) to A33_1706 (CHP) as viewed in the IGV. The lines above the map indicate three major operons. The operons encode genes for structural proteins (vcsRTCNS2, vcsVUQ2, vcsJ2, and vspD) and some effector proteins (vopA, vopH, vopI, vopM, and vopW). Additional effector genes in the 5′ and 3′ flanking regions, as well as acfD, encoding a putative accessory colonization factor, are also indicated. All genes are shown aligned to the IGV heat map with the following settings: positive strand, base count 0 to 100 in gray, 100 to maximum in black; negative strand, 0 to 50 in gray, 50 to maximum in black.
We previously identified 12 genes within the AM-19226 T3SS island that encode translocated effector proteins (23). Nine of the 12 effector genes have lower expression levels in the ΔvttRA and ΔvttRB strains compared to the parent strain carrying intact vttRA and vttRB loci: vopA, vopF, vopG, vopH, vopI, vopM, vopW, vopX, and vopY. For example, the RPKM values for vopX were 346, 3, and 7 for the wild-type and ΔvttRA and ΔvttRB mutant strains, respectively (see Table S4 in the supplemental material). These observations are consistent with results obtained using a vopX-lacZ transcriptional reporter fusion (23). The acfD gene, encoding a predicted accessory virulence factor, and several uncharacterized genes within the T3SS island, such as A33_1665, A33_1668, and A33_1703, also showed reduced expression in the ΔvttRA and ΔvttRB strains.
Interestingly, 34 of the 71 ORFs that were expressed at lower levels in the absence of VttRA or VttRB lie outside the T3SS genomic island. These include genes encoding the FlaA, FlaC, FlaD, and FlaE flagellins, the CheA chemotaxis protein, type 6 secretion system (T6SS) proteins Hcp-1, Hcp-2, and VgrG-1, a protein involved in the stringent response (DksA), and a two-component response regulator (A33_1014) (Table 1). A33_1014 appears to lie within a large operon that also includes three other genes predicted to encode response regulators (A33_1015, A33_1019, and A33_1020), two genes encoding sensory histidine kinases (A33_1018 and A33_1021), a gene predicted to encode a sensor box sensory histidine kinase (A33_1017), and a gene encoding a hypothetical protein (A33_1016). The operon is conserved in the epidemic O1 biovar El Tor strain N16961, which does not encode VttRA or VttRB, and lies in a similar genomic context. We also identified glgC2 (A33_A0971), encoding a protein important for glycogen synthesis in V. cholerae (30) and A33_A0971, involved in the formation of polyhydroxylbutyrates (31–33), as potentially regulated in the deletion strains. Finally, 17 genes predicted to encode hypothetical or conserved hypothetical proteins showed ≥2.5-fold differential expression in the ΔvttRA and/or ΔvttRB strains compared to the wild-type strain (Table 1), although we did not confirm their expression patterns by other methods and thus regard them as potential members of the VttRA and VttRB transcriptomes. We also identified seven genes with expression patterns that suggest they are targets of VttRA and VttRB repression (Table 2). Two genes, A33_A0028 (identical to VCA0834 in strain N16961) and A33_A0330 (predicted to encode a 38-amino-acid protein), encode hypothetical proteins based on NCBI annotation and our own BLAST analyses. The combination of annotation and BLAST analyses was used to identify the function of other encoded products listed in Table 2, based on high degrees of amino acid similarity, or predicted roles based on more limited similarity as described below. A33_0677 encodes a protein identical to VC0702 in O1 serogroup strain N16961. VC0702 is predicted to encode an NTPase and lies within the mba operon that has been shown to play a role in biofilm formation and the maintenance of biofilm architecture (34). A33_1527 is similar to the N16961 FrhC protein (VC1621), a flagellum-regulated hemagglutination protein with a TolC (type I secretion outer membrane protein) domain (35). A33_2142 and A33_2143 are >98% identical to K139 phage proteins K139p07 (unknown function) and K139p06 (putative regulator), which are present in numerous V. cholerae strains, although A33_2143 appears to be differentially regulated by VttRB only. A33_A0125 is predicted to encode a member of the sensory box/GGDEF family of proteins, which are abundant in V. cholerae strains and have roles in numerous cellular processes and responses to environmental stimuli (36).
VttRA and VttRB positively influence expression of flagellins.
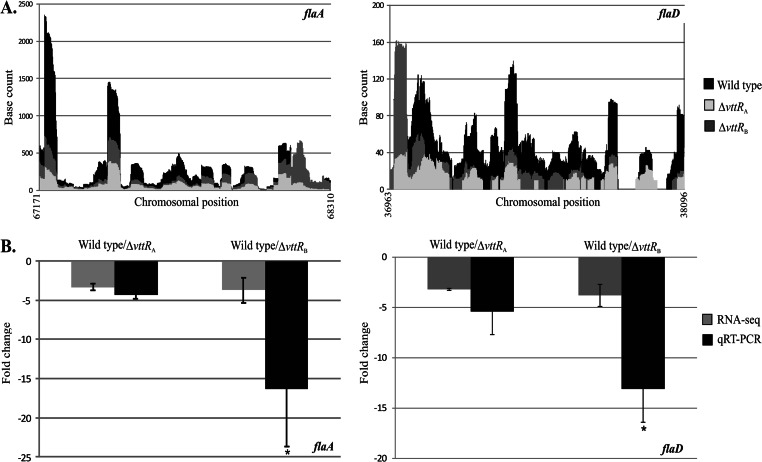
V. cholerae strains, including AM-19226, encode five flagellins whose genes are found within two operons: one that encodes the FlaA essential flagellin and the minor flagellin FlaC, and the other encoding the three minor flagellins FlaE, FlaD, and FlaB (35, 37). flaA transcription is dependent on σ54 and requires an additional regulator, FlrC, which must be phosphorylated as part of a two-component system (FlrBC) in order for the σ54-dependent transcription of flaA to proceed. The genes encoding the four minor flagellins are under the transcriptional control of σ28 (encoded by fliA), whose activity is controlled by the anti-σ28 factor FlgM (35). The RPKM values of four of the five genes encoding flagellins (flaACDE) were at least 2.5-fold lower in the ΔvttRA and/or ΔvttRB strains compared to the wild-type strain (Table 1). The RPKM values of flaB (A33_2045) were reduced as follows in replicates 1 and 2, respectively: 2.1- and 1.4-fold in the ΔvttRA strain and 2.7- and 2.2-fold in the ΔvttRB strain compared to the wild-type strain (see Tables S2 and S3 in the supplemental material) and therefore did not meet the 2.5-fold cutoff for differentially expressed genes. The raw RNA-seq data for flaA and flaD are shown in Fig. 3, demonstrating that across the entire open reading frame, fewer transcripts were mapped from the deletion strains (light gray and medium gray) compared to the parent strain (black). The qRT-PCR results indicated that the levels of flaA expression in the ΔvttRA and ΔvttRB strains were 4.3- and 16.4-fold lower, respectively, than that in the parental, VttRA VttRB wild-type strain (Fig. 3B). Similarly, flaD expression was reduced by 5.4- and 13.1-fold in the ΔvttRA and ΔvttRB strains compared to that in the parental strain when quantified by qRT-PCR (Fig. 3B).
Fig 3.
VttRA and VttRB influence transcription of flaA and flaD. (A) Raw RNA-seq data obtained from .wig files indicating the number of times each base was mapped for flaA and flaD in the wild-type and deletion backgrounds. (B) qRT-PCR analysis of flaA and flaD transcript levels in the wild-type and ΔvttRA and ΔvttRB strains. Mean fold change values obtained from qRT-PCR are shown adjacent to the mean fold change obtained from RNA-seq. qRT-PCR was performed four times on RNA used for RNA-seq and independently isolated RNA. Error bars represent standard deviations. *, P < 0.05 compared to the wild-type strain.
VttRA and VttRB influence the expression of genes encoding regulators of the flagellar biosynthesis pathway.
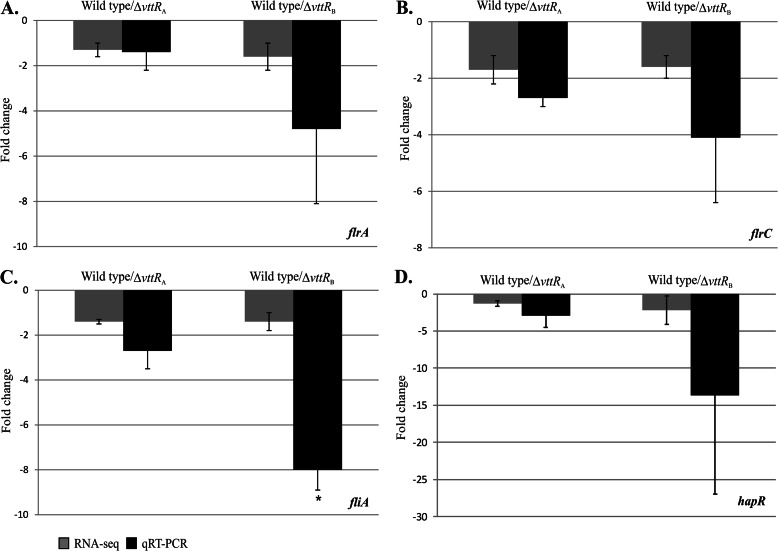
The RNA-seq results indicated modest reductions (<2-fold) in the expression levels of flrA, flrC, and fliA in the ΔvttRB strain compared to the wild type (see Table S3 in the supplemental material). Since FlrA, FlrC, and FliA regulate the expression of class II, class III, and class IV flagellar genes, respectively, we explored whether their transcript levels were decreased in independently isolated RNA samples using qRT-PCR (35). We found that flrA expression was decreased 4.8-fold in the ΔvttRB strain, although transcript levels were similar to wild-type levels in the ΔvttRA strain. On the other hand, flrC transcript levels were decreased 2.7-fold in the ΔvttRA strain and 4.1-fold in the ΔvttRB strain. We observed similar trends for fliA transcript levels, with 2.7- and 8-fold reductions in the ΔvttRA and ΔvttRB strains, respectively, compared to the parental strain (Fig. 4A to C).
Fig 4.
VttRA and VttRB influence expression of other transcriptional regulators. Fold change representations (±SD) for ΔvttRA and ΔvttRB strains compared to the wild-type strain for the flagellar biosynthesis regulatory genes flrA (A), flrC (B), and fliA (C) and the QS master regulatory gene, hapR (D). qRT-PCR was performed four times on three technical replicates for flrA and hapR and three times for flrC and fliA using cDNA generated from RNA isolated independently and for RNA-seq. *, P < 0.005 compared to the wild-type strain.
ΔvttRA and ΔtoxR strains display altered swarming motility.
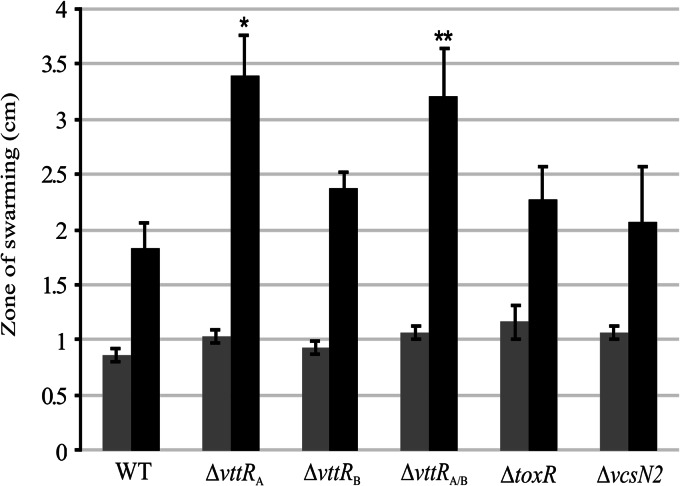
The decreased expression of flagellin and motility transcriptional regulatory genes in the ΔvttRA and ΔvttRB strains led us to predict that we might be able to detect motility defects in the deletion strains using standard methods. We therefore examined the swarming ability of the following strains on semisolid media with and without bile: the parental, ΔvttRA, and ΔvttRB strains, a ΔvttRA ΔvttRB double deletion strain, a ΔtoxR strain, and a T3SS-deficient (ΔvcsN2) strain. toxR mutant strains have been previously described as hypermotile (38), and we therefore included the ΔtoxR strain both as a control and to determine its swarming pattern in the presence of bile. The ΔvcsN2 strain was included so that we could evaluate any effect of the T3SS on motility, since the vcsN2 mutant strain does not produce a functional T3SS apparatus (23, 39). The results are shown in Fig. 5. We observed that the ΔtoxR strain had a larger zone of swarming (mean, 1.2 cm) than the parental strain carrying a wild-type toxR allele (mean, 0.9 cm) on standard LB motility plates without bile, as previously reported (P < 0.05) (Fig. 5, gray bars). The other deletion strains displayed similar zones of swarming, as shown in Fig. 5. Surprisingly, however, the ΔvttRA strain showed a robust increase in the swarming diameter (3.4 cm) compared to the parental strain (1.8 cm) on motility plates that included bile, whereas the ΔvttRB strain displayed a less dramatic increase in swarming diameter (2.4 cm) compared to the parental strain under the same conditions (Fig. 5, black bars). A double deletion (ΔvttRA ΔvttRB) strain also displayed an increased ability to swarm (3.2 cm) compared to the parental strain. The ΔtoxR and ΔvcsN2 strains exhibited a slightly larger zone of swarming compared to the wild-type strain, but the differences in their swarming patterns were not statistically significant.
Fig 5.
Swarming diameters of AM-19226 wild-type and regulator deletion strains. The zones of swarming for individual strains were measured as diameters across the zone for the wild-type and ΔvttRA, ΔvttRB, ΔvttRAΔvttRB, ΔtoxR, and ΔvcsN2 mutant strains. Single colonies were inoculated onto plates containing LB broth only (gray bars) or LB broth supplemented with 0.4% bile (black bars), and zones were measured after 8 h at 37°C. n = 3. P values denoted by asterisks correspond to comparisons of the deletion strains to the wild type. *, P < 0.005; and **, P < 0.01.
VttRA and VttRB influence hapR expression.
HapR positively regulates the expression of the hapA gene, which encodes a hemagglutinin (HA) called HA/protease that can cleave a variety of substrates, such as lactoferrin and mucin (40). HapR is also an important regulator of biofilm formation in V. cholerae and is a LuxR homolog, functioning at high cell density to represses the expression of virulence-associated genes such as aphA and polysaccharide synthesis genes (vps genes), leading to reduced biofilm formation (36). Because of its role as a transcriptional regulator influencing multiple phenotypes, we further examined hapR transcript levels in both ΔvttRA and ΔvttRB strains by qRT-PCR since the RNA-seq data suggested that hapR (A33_0561) expression is reduced in the ΔvttRB strain (see Table S2 in the supplemental material). Data obtained from two individual qRT-PCR experiments showed at least 2.8-fold reduction in hapR transcript in the absence of VttRB. We also observed an ∼2- to 4-fold reduction in hapR expression (Fig. 4D) in the absence of VttRA, suggesting that VttRA and VttRB may act indirectly through HapR in some cases to influence gene expression. The results are consistent with those demonstrating that effects on motility may also be mediated indirectly through other transcriptional regulators (as reported above).
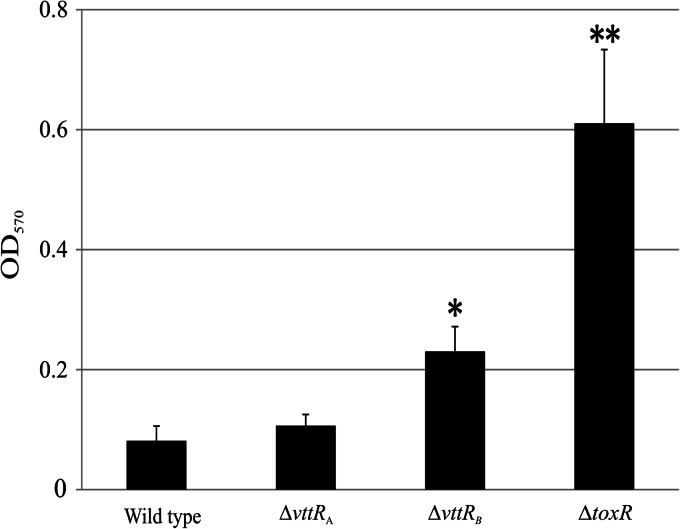
VttRB and ToxR influence biofilm formation in AM-19226.
Based on the decreased expression levels of hapR in the ΔvttRA and ΔvttRB strains, we predicted that biofilm formation would be altered in the deletion strains. We performed static biofilm assays on the ΔvttRA and ΔvttRB strains and also chose to include a ΔtoxR strain because of the documented role of ToxR in multiple phenotypes (41–43). Strains were grown in LB broth plus 0.4% bile and allowed to form biofilms during static growth for 24 h at 30°C. The ΔvttRA strain formed biofilms of similar mass to the wild-type strain, whereas both ΔvttRB and ΔtoxR strains formed more robust biofilms than the wild-type strain. The total biofilm mass for the ΔvttRB strain was approximately 2-fold greater than that of the wild type (P < 0.0016) and was approximately 6-fold greater for the ΔtoxR strain (P < 0.0001) (Fig. 6). These data suggest that under the conditions tested, VttRA has little impact on biofilm formation, whereas VttRB and ToxR appear to negatively regulate biofilm formation in the presence of bile.
Fig 6.
VttRB and ToxR negatively impact biofilm formation. Static biofilms were measured after incubation at 30°C for 24 h. Total biofilm biomass was determined by measuring absorbance at 570 nm. *, P < 0.0016, and **, P < 0.0001 compared to the wild-type strain. A representative of three independent experiments with n = 6 for each strain is shown.
VttRA and VttRB influence the transcription of T6SS substrates.
T6SS in V. cholerae has been associated with cytotoxicity toward the social amoeba Dictyostelium and antimicrobial properties toward enterohemorrhagic Escherichia coli (EHEC), enteropathogenic E. coli (EPEC), and Salmonella enterica serovar Typhimurium (44, 45). The T6SS was first described in V. cholerae strain V52 (44), which encodes five T6SS proteins that have homologs in strain AM-19226: two hemolysin-coregulated proteins, Hcp-1 and Hcp-2 and three valine glycine repeat proteins, VgrG-1 to VgrG-3. Hcp-1 and Hcp-2 are nearly identical proteins that differ in only one amino acid. On the other hand, the VgrG proteins show slight sequence divergence. VgrG-1 (A33_1343; 1,163 amino acids [aa]) shares homology with the N16961 homolog VC1416 (99% identity). VgrG-2 (A33_A0308; 698 aa) also shares homology with its N16961 homolog, VCA0018 (94% identity). VgrG-3 (A33_A0406) is annotated to be a 108-aa protein according to NCBI, but its N16961 homolog (VCA0123) is 1,017 aa in length. Sequence analysis of the intergenic region between A33_A0404 and A33_A0406 revealed the presence of a pseudogene annotated by NCBI. ClustalW2 alignments (EMBL-EBI) (46) of the 685-aa AM-19226 pseudogene, the 108-aa A33_A0406, and the 1,017-aa VCA0123 from strain N16961 suggest that the start codon of the pseudogene is likely the start codon for the AM-19226 VgrG-3, with the ORF continuing through the annotated A33_0406 ORF. However, AM-19226 appears to encode a truncated version of VgrG3, which is missing 224 C-terminal residues (data not shown).
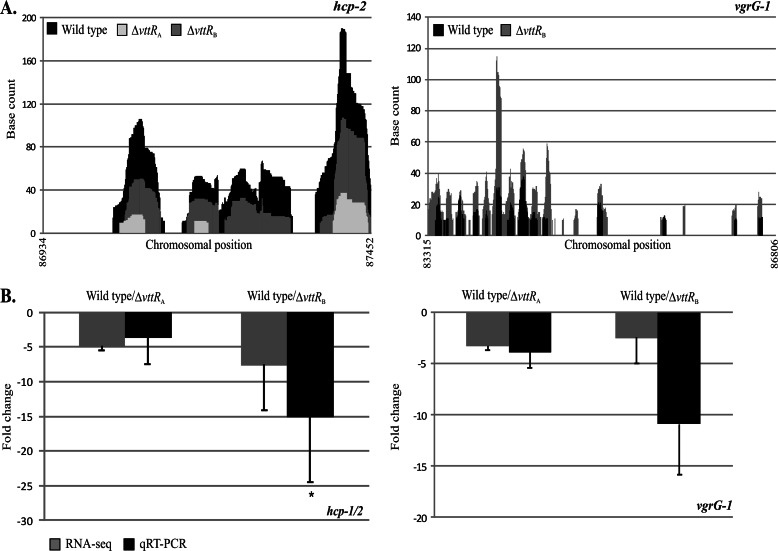
Recent reports suggest that Hcp and VgrG proteins are both structural and effector components of T6SSs (47, 48). RPKM values for hcp-1 and hcp-2 were at least 2.5-fold lower in the ΔvttRA and ΔvttRB strains than in the parental strain. In the case of vgrG-1, the RPKM values in the ΔvttRA strain were at least 2.5-fold lower than in the parental strain in both replicates (Table 1; see Tables S2 and S3 in the supplemental material), although the RPKM value obtained for the ΔvttRB strain was decreased compared to the parental strain in replicate 2 only (Table 1 and Fig. 7A; see Tables S2 and S3 in the supplemental material). We therefore further examined the expression levels of hcp-2 and vgrG-1 using qRT-PCR. qRT-PCR estimated 3.7- and 15.1-fold reductions in hcp-2 transcript levels in the ΔvttRA and ΔvttRB strains, respectively, compared to the parental strain. Similarly, vgrG-1 transcript levels were decreased ∼4-fold in the ΔvttRA strain and ∼11-fold in the ΔvttRB strains (Fig. 7).
Fig 7.
T6SS genes are expressed at lower levels in the absence of VttRA and/or VttRB. (A) Raw RNA-seq data obtained from .wig files indicating the number of times each base was mapped for hcp-2 and vgrG-1 in the wild-type and ΔvttRA and ΔvttRB mutant strains. For vgrG-1, base counts obtained for the ΔvttRA strain were zero throughout most of the ORF and therefore are not represented. (B) qRT-PCR verification of hcp-1/2 and vgrG-1 transcript levels in the wild-type and ΔvttRA and ΔvttRB strains. Mean fold change values (±SD) obtained from qRT-PCR are shown adjacent to mean fold change values obtained from RNA-seq runs. qRT-PCR was performed at least three times in triplicate using cDNA generated from two independently isolated RNA samples. *, P < 0.05 compared to the wild-type strain.
DISCUSSION
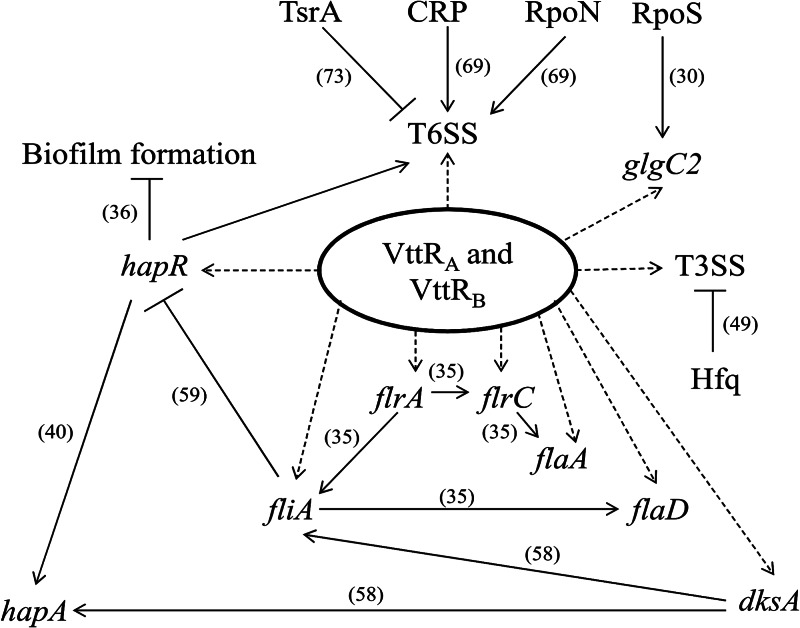
Our results indicate that VttRA and VttRB can modulate the expression of genes belonging to diverse functional classes and may influence expression of ≈2% of the AM-19226 genome. A model summarizing the results of our experiments (dotted arrows) combined with previously published results from numerous laboratories (solid lines with references) is presented in Fig. 8. Based on a ≥2.5-fold change in expression values in the deletion strains compared to the strain carrying wild-type copies of the VttRA and VttRB transcriptional regulators, we identified a total of 78 genes that are candidates for differential expression. Of the candidate genes, 60 genes are carried on the large chromosome and 18 are found on the small chromosome. As expected, genes within the T3SS genomic island were identified as part of the VttRA/VttRB transcriptomes, consistent with previous results using lacZ-transcriptional reporter fusions that identified VttRA and VttRB as transcriptional regulators of T3SS genes (24). Remapping of the contig containing the T3SS genomic island allowed us to obtain RPKM values for five genes that were not originally annotated (see Materials and Methods) by NCBI. Not all genes within the T3SS island were identified as differentially regulated by VttRA/VttRB. Presumably, VttRA and VttRB may not regulate all genes within the T3SS island; some genes may be expressed at low levels, and others may require different growth conditions for expression. For example, genes encoding the VopE, VopZ, and VopK effector proteins have very low levels of expression in the parent strain grown in bile (RPKM values of 2 for vopE and 6 for vopK and vopZ). Other regulators of effector gene expression have been reported, such as the influence of Hfq on VopF expression (Fig. 8) (49).
Fig 8.
Model for coordinated regulation of multiple phenotypes in AM-19226. VttRA and VttRB modulate the expression of horizontally acquired T3SS and T6SS genes and core chromosomal genes involved in motility (flrAC, flaAD, fliA), stringent response (dksA), and biofilm formation (hapR), as indicated by dotted lines. Evidence exists for cross talk between regulators encoded by the flrA, fliA, dksA, and hapR and genes involved in motility, stringent response, and biofilm formation, based on reports in the literature and indicated by solid lines and references. Other known regulators for T6SS and T3SS genes are indicated by solid lines. The metabolic genes of the VttRA/VttRB regulon (glgC2 shown here) might also play a role in reconfiguring regulatory networks in response to different environmental stimuli.
Interestingly, RNA-seq results led us to further explore whether genes located outside the T3SS genomic island are part of the VttRA and VttRB transcriptomes. The data suggest that VttRA and VttRB transcriptional activity is not restricted to genes within the T3SS genomic island. Rather, the activity of these horizontally acquired regulators likely extends to influence the expression of core chromosomal genes. Non-T3SS genes that showed differential expression in the absence of VttRA and/or VttRB belong to several gene classes based on functions such as motility and chemotaxis, type 6 secretion, and metabolism, suggesting that VttRA and VttRB have important roles in coordinating global gene regulation during the infectious process. The ability of a horizontally acquired transcriptional regulator to influence global gene expression has been described for S. enterica (50), but is not as common as core genome-encoded regulators, such as ToxR, controlling the expression of both ancestral genes (ompU, ompT, etc.) and horizontally acquired virulence factors (TCP and CT). Interestingly, all of the non-T3SS island genes identified as putative targets of VttRA- and/or VttRB-positive regulation are present in the TCP/CT-positive, epidemic O1 serogoup strain N16961 (which does not encode VttRA or VttRB), except for four: A33_0283, A33_A0309, A33_0281, and A33_1879. We did not perform statistical analyses to determine the definitive VttRA and VttRB transcriptomes, but rather chose specific gene classes and phenotypes for our immediate focus.
We selected genes involved in motility for further analysis, since it is an important virulence determinant in V. cholerae (38, 51) and has been studied within the context of T3SS regulation in other pathogens. In S. enterica serovar Typhimurium, regulation of HilA, an OmpR-ToxR family transcriptional regulator of the Salmonella pathogenicity island 1 (SPI1) T3SS invasion genes, has been linked with the products encoded by the fliAZY motility operon. FliA regulates motility and chemotaxis, whereas FliZ positively influences hilA expression indirectly by activating another transcriptional activator, and collective studies indicate cross talk in regulation of expression of flagellar and SPI1 genes (52–54). In Pseudomonas aeruginosa, absence of flagella was linked to increased T3SS gene expression and cytotoxicity (55). We specifically examined the influence of VttRA and VttRB on the flagellins and T6SS substrates (described below) in order to gain a better understanding of regulation of motility and T6SS in AM-19226.
To provide supporting evidence that VttRA and VttRB positively influence flagellin gene expression, we selected one flagellin gene from each of the two flagellin operons and determined expression levels using qRT-PCR. Both flaA and flaD showed reduced expression in the ΔvttRA and ΔvttRB strains (Fig. 2). Combined with RNA-seq data for genes encoding transcriptional regulators of motility operons, we therefore predicted that the ΔvttRA and ΔvttRB strains would be less motile than the wild-type strain in the standard semisolid, motility agar swarming assay. Unexpectedly, we observed increased swarming in the absence of VttRA. Several possible explanations could account for the hypermotile phenotype of the ΔvttRA strain, including regulatory mechanisms at the posttranscriptional, translational, or posttranslational levels. While it seems likely that VttRA and VttRB influence the expression of motility genes, perhaps via positive regulation of flrA, flrC, and fliA expression, it is also likely that regulation of other genes counters positive effects such that the ΔvttRA strain is hypermotile in our assay. Although the RNA-seq data did not identify many genes whose expression was negatively influenced by VttRA or VttRB, it is possible that VttRA or VttRB might repress the expression of inhibitors of motility. Such interactions could involve the products of A33_0677 and/or A33_A0125, whose products have been implicated in motility and biofilm phenotypes in other strains (34, 36, 56). It is also likely that flagellar synthesis and motility gene regulation in AM-19226 differ between solid (motility plate assay) and liquid (RNA isolation for RNA-seq and qRT-PCR) culture conditions or that the function of the sodium salt-driven flagellum may be impacted by the bile salts in the media (57). DksA may also play a role, since RNA-seq results suggested that VttRA and VttRB positively influence dksA expression (Table 1). In V. cholerae, DksA has been implicated in modulating multiple phenotypes, including motility, in part by influencing fliA expression (58). Note also that FliA has been shown to repress HapR production (59). We therefore favor the explanation that a combination of direct and indirect effects on motility genes and, importantly, genes involved in chemotaxis (Table 1) account for the phenotype observed on swarming plates, which likely measure both motility and chemotaxis. Nonetheless, our data suggest that VttRA and VttRB can influence expression of genes involved in motility and support existing data that demonstrate a strong association between motility and virulence factor gene(s) expression in multiple species (14, 38, 51, 52, 60–62). Future experiments will address the molecular mechanisms and specific regulatory interactions that coordinate motility and disease in T3SS-positive V. cholerae strains.
Our results suggest that VttRA and VttRB not only influence the expression of flagellar biosynthesis pathway transcriptional regulators but also the quorum-sensing (QS) global regulator, HapR (Fig. 8). In epidemic O1 and O139 serogroup V. cholerae strains, HapR expression increases at high cell densities and leads to reduced biofilm formation and virulence factor production (36, 63). We found decreased hapR transcript levels in ΔvttRA and ΔvttRB strains, and data obtained from qRT-PCR showed that hapR transcript levels are diminished in ΔvttRA and ΔvttRB strains by an average of 2.6- and 11-fold, respectively. The results suggest that VttRA and VttRB might play a role in maintaining wild-type HapR levels and in turn, affect T3SS gene expression and/or quorum sensing during infection. We observed enhanced biofilm formation in the ΔtoxR and ΔvttRB strains but not the ΔvttRA strain (and observed a greater fold change in hapR transcript levels in the ΔvttRB strain compared to the ΔvttRA strain), consistent with a more prominent role for VttRB in regulating hapR expression. However, we cannot at this time, reconcile the results that VttRA and VttRB appear to positively regulate expression of flrA, fliA, and dksA, whose encoded products can repress hapR expression (and therefore result in greater biofilm formation in the wild-type strain). Again, note that fliA was shown to regulate hapR and could be integrated into the network regulating motility gene expression and the stress response through dksA (Fig. 8). One interpretation is that the activity of multiple regulators coordinates motility and chemotaxis as important phenotypes involved in biofilm formation. Recently, Valeru et al. showed that ToxR can influence biofilm formation in V. cholerae epidemic strain A1552 (43). In vivo-formed biofilms have been shown to have hyperinfective properties (64), and environmentally formed biofilms show increased resistance to stomach acids compared to planktonic cells and increased persistence in the aquatic environment (63). Furthermore, Hung et al. showed that V. cholerae cells within the biofilm are more resistant to bile acids compared to the planktonic form, and that bile acids stimulate biofilm formation in a VpsR-dependent manner (65). VpsR is a two-component transcriptional regulator that positively regulates expression of the vps genes, necessary for polysaccharide production during biofilm formation, and interestingly, vpsR expression is negatively regulated by HapR (36, 66). At this time, we cannot exclude a role for VpsR in the VttRA/VttRB network of regulation, although RNA-seq data did not indicate differential expression of vpsR in the deletion strains. Nonetheless, our observations suggest that in AM-19226, VttRB and ToxR likely regulate expression of HapR in the presence of bile and therefore are integrated with quorum-sensing signals in the human host and perhaps in the environment as well.
We also further examined the role of VttRA and VttRB in T6SS gene expression, since T6SSs have been shown to function in multiple roles, including virulence (V. cholerae), symbiosis (Helicobacter hepaticus), and interbacterial competition (P. aeruginosa) (67). qRT-PCR confirmed the RNA-seq results, indicating that the transcript levels of hcp-2 and vgrG-1 were reduced in the absence of VttRA and VttRB. The hcp-1 and hcp-2 genes differ by only eight nucleotides, and qRT-PCR therefore measured the total transcripts from both hcp loci, since amplification primed from highly conserved regions of the hcp genes. RNA-seq is more likely to measure transcripts specific to either hcp-1 or hcp-2, depending on where sequencing reads were mapped (Table 1 and Fig. 7). On the other hand, the vgrG-1 primers for RT-PCR were unique and measured transcript levels only from the vgrG-1 locus. The fold change values from both RNA-seq and qRT-PCR were comparable (3.3- and 3.9-fold mean reductions, respectively) in the ΔvttRA strain. The ΔvttRB strain, however, showed a more drastic decrease in vgrG-1 levels by qRT-PCR in comparison to RNA-seq (10.9-fold versus 2.5-fold). One possible explanation is that fewer unique reads were aligned to the vgrG-1 locus in the parental strain during the mapping stage of RNA-seq, since reads that are not unique (i.e., can align equally well to more than one genomic locus) are discarded. This seems plausible since the vgrG genes in AM-19226 exhibit considerable nucleotide sequence similarity (data not shown). qRT-PCRs were also conducted on multiple, independently isolated RNA samples, which could account for variations in transcript levels observed among experiments and samples.
Complex regulatory networks have been proposed to mediate coregulation of T6SS genes with other virulence determinants in several organisms. For example, in P. aeruginosa QS represses different T6SS gene clusters at the transcriptional level, the Gac/Rsm pathway exerts its influence at the posttranscriptional level, whereas threonine phosphorylation regulates the T6SS posttranslationally (68, 69). In A. hydrophila, there is evidence to suggest cross talk between polar and lateral flagellar systems, a T3SS, and a T6SS that is mediated by the T3SS regulator AxsA, the QS regulator AhyR, and σ54 (encoded by rpoN) (70). A recent report by Dong and Mekalanos reveals that in V. cholerae strain V52, RpoN differentially regulates expression of motility and T6SS genes (Fig. 8) (71). In S. enterica serovar Typhimurium, T6SS-associated genes have been implicated in the coordination of T6SS gene expression with the virulence cascade during host infection (72). It is clear that in various organisms, different regulatory networks have been integrated to control T6SS gene expression at multiple levels.
Similar complexity has been documented in V. cholerae. In O1 El Tor V. cholerae strains A1552 and C6706, HapR can directly bind the hcp-1 and hcp-2 promoters, and along with RpoN and VasH (an activator of RpoN) can activate T6SS gene expression (69, 73, 74). Our RNA-seq data suggest that hapR expression is not affected by VttRA but is reduced in the ΔVttRB strain, as described earlier. We were unable to definitively determine whether expression of tsrA (whose protein product A33_0095 is known to repress T6SS gene expression in V. cholerae strain C6706 [73]) is controlled by VttRA and VttRB because of modest reductions in transcript levels as assayed by RNA-seq and technical difficulties with tsrA specific qRT-PCR. Our observations do suggest, however, that VttRA and VttRB might influence T6SS gene expression indirectly through HapR, indicating cross talk between the T3SS and the T6SS genes in the presence of bile.
Recent genetic analysis of the T6SS pathogenicity island genes of V. cholerae strain V52 suggests that structural proteins (e.g., VipA, VipB, VasH, VasK, and ClpV) and secreted proteins Hcp-1, Hcp-2, VgrG-1, and VgrG-2 are required for Hcp secretion (75, 76). Our experiments were conducted in the presence of bile, and only the genes encoding substrate Hcp proteins and VgrG-1 consistently showed decreased expression in the absence of VttRA and VttRB (data not shown) (Table 1). However, we cannot rule out a role for VttRA and VttRB in modulating T6SS gene expression under parameters that reflect the environment outside the human host; the T6SS is not expressed in El Tor epidemic cholera strains under laboratory conditions (73). In AM-19226, if the reduced expression of Hcps and VgrGs is bile dependent, it is possible that VttRA and VttRB coordinate the expression of T3SS and T6SS proteins in response to cues in the small intestine. Evidence supporting this hypothesis has been reported for the O37 serogroup V. cholerae strain V52, where the actin cross-linking domain of VgrG-1 was shown to be important for T6SS-dependent fluid accumulation and subsequent intestinal inflammation in the infant mouse model (77). It is interesting to note that in vivo transcriptome studies indicate that T6SS gene expression is induced in the infant mouse model but not in a rabbit model of infection, further underscoring that we do not fully understand the signals required for T6SS gene expression (78). We speculate that as for other strains, T6SS assembly and function are coordinated by multiple regulators in AM-19226, including HapR, RpoN, VttRA, and VttRB. Whether T6SS gene expression is induced in response to environmental conditions and cell density signals in the human intestine in order to optimize virulence remains to be determined.
For most organisms, a simple model of coordinated virulence gene expression has been expanded to encompass complex networks and hierarchies that modulate gene expression in response to environmental stimuli and compensatory adaptive responses, with temporal and spatial sensitivity. Our results, which address the question of whether horizontally acquired transcriptional regulators are restricted to control only virulence gene expression, offer the ability to explore a relatively understudied aspect of coordinated regulation. The data indicate that VttRA and VttRB can indeed exert transcriptional control outside the T3SS island to influence the expression of core chromosomal genes, and we predict that both direct and indirect regulation serve to modulate gene expression. It is possible that VttRA and VttRB compete to bind to the promoter regions of their target genes, and evidence from our laboratory suggests that precise concentrations of VttRA and VttRB are critical for gene activation or repression in vitro, which might in turn be influenced by various environmental conditions, such as the bile gradient in the intestine or different niches during infection (25). In addition, hierarchical activation of vttRB expression by VttRA and subsequent activation of different sets of downstream target genes by both VttRA and VttRB might be required for a successful infection and/or survival in the aquatic habitat (24, 25). Our system, therefore, reflects a unique opportunity to dissect the molecular mechanism(s) of both direct and indirect gene regulation by two transcriptional regulators as it relates to coordinating multiple phenotypes related to pathogenesis and provides a platform to further study the properties of an expanding family of transcriptional regulatory proteins with novel transmembrane features.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chin-Yi Chu, Anthony Durkin, Steve Gill, Vincent Isabella, Jason Myers, and Steve Welle for support with transcriptome sequencing and data analysis, the J. Lemos and R. Quivey laboratories for advice and use of the qRT-PCR equipment, and Scott Butler, Steve Gill, and the Dziejman laboratory for helpful discussion and critical reading of the manuscript.
This work was supported by NIH/NIAID AI073785.
Footnotes
Published ahead of print 22 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02151-12.
REFERENCES
- 1. Matson JS, Withey JH, DiRita VJ. 2007. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect. Immun. 75:5542–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Faruque SM, Nair GB, Mekalanos JJ. 2004. Genetics of stress adaptation and virulence in toxigenic Vibrio cholerae. DNA Cell Biol. 23:723–741 [DOI] [PubMed] [Google Scholar]
- 3. Chatterjee SN, Chaudhuri K. 2003. Lipopolysaccharides of Vibrio cholerae. I. Physical and chemical characterization. Biochim. Biophys. Acta 1639:65–79 [DOI] [PubMed] [Google Scholar]
- 4. Harris JB, LaRocque RC, Qadri F, Ryan ET, Calderwood SB. 2012. Cholera. Lancet 379:2466–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wachsmuth IK, Olsvik Ø, Evins GM, Popovic T. 1994. Molecular epidemiology of cholera, p 357–370 In Wachsmuth IK, Blake PA, Olsvik Ø. (ed), Vibrio cholerae and cholera: molecular to global perspectives. American Society for Microbiology, Washington, DC [Google Scholar]
- 6. Faruque SM, Albert MJ, Mekalanos JJ. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Faruque SM, Mekalanos JJ. 2003. Pathogenicity islands and phages in Vibrio cholerae evolution. Trends Microbiol. 11:505–510 [DOI] [PubMed] [Google Scholar]
- 8. Waldor MK, Mekalanos JJ. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910–1914 [DOI] [PubMed] [Google Scholar]
- 9. Bina J, Zhu J, Dziejman M, Faruque S, Calderwood S, Mekalanos J. 2003. ToxR regulon of Vibrio cholerae and its expression in vibrios shed by cholera patients. Proc. Natl. Acad. Sci. U. S. A. 100:2801–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Higgins DE, DiRita VJ. 1996. Genetic analysis of the interaction between Vibrio cholerae transcription activator ToxR and toxT promoter DNA. J. Bacteriol. 178:1080–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Higgins DE, Nazareno E, DiRita VJ. 1992. The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators. J. Bacteriol. 174:6974–6980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Krukonis ES, Yu RR, Dirita VJ. 2000. The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol. Microbiol. 38:67–84 [DOI] [PubMed] [Google Scholar]
- 13. Li CC, Crawford JA, DiRita VJ, Kaper JB. 2000. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol. Microbiol. 35:189–203 [DOI] [PubMed] [Google Scholar]
- 14. Lee SH, Hava DL, Waldor MK, Camilli A. 1999. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99:625–634 [DOI] [PubMed] [Google Scholar]
- 15. Dziejman M, Serruto D, Tam VC, Sturtevant D, Diraphat P, Faruque SM, Rahman MH, Heidelberg JF, Decker J, Li L, Montgomery KT, Grills G, Kucherlapati R, Mekalanos JJ. 2005. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc. Natl. Acad. Sci. U. S. A. 102:3465–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Faruque SM, Chowdhury N, Kamruzzaman M, Dziejman M, Rahman MH, Sack DA, Nair GB, Mekalanos JJ. 2004. Genetic diversity and virulence potential of environmental Vibrio cholerae population in a cholera-endemic area. Proc. Natl. Acad. Sci. U. S. A. 101:2123–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Connolly MA, Gayer M, Ryan MJ, Salama P, Spiegel P, Heymann DL. 2004. Communicable diseases in complex emergencies: impact and challenges. Lancet 364:1974–1983 [DOI] [PubMed] [Google Scholar]
- 18. Chen Y, Johnson JA, Pusch GD, Morris JG, Jr, Stine OC. 2007. The genome of non-O1 Vibrio cholerae NRT36S demonstrates the presence of pathogenic mechanisms that are distinct from those of O1 Vibrio cholerae. Infect. Immun. 75:2645–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chatterjee S, Ghosh K, Raychoudhuri A, Chowdhury G, Bhattacharya MK, Mukhopadhyay AK, Ramamurthy T, Bhattacharya SK, Klose KE, Nandy RK. 2009. Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India. J. Clin. Microbiol. 47:1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cornelis GR, Van Gijsegem F. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735–774 [DOI] [PubMed] [Google Scholar]
- 21. Galan JE, Wolf-Watz H. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444:567–573 [DOI] [PubMed] [Google Scholar]
- 22. Park KS, Ono T, Rokuda M, Jang MH, Okada K, Iida T, Honda T. 2004. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72:6659–6665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alam A, Miller KA, Chaand M, Butler JS, Dziejman M. 2011. Identification of Vibrio cholerae type III secretion system effector proteins. Infect. Immun. 79:1728–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alam A, Tam V, Hamilton E, Dziejman M. 2010. vttRA and vttRB encode ToxR family proteins that mediate bile-induced expression of type three secretion system genes in a non-O1/non-O139 Vibrio cholerae strain. Infect. Immun. 78:2554–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miller KA, Hamilton E, Dziejman M. 2012. The Vibrio cholerae trh gene is coordinately regulated in vitro with T3SS genes by VttRA/VttRB but does not contribute to Caco2-BBE cell cytotoxicity. Infect. Immun. 80:4444–4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kodama T, Gotoh K, Hiyoshi H, Morita M, Izutsu K, Akeda Y, Park KS, Cantarelli VV, Dryselius R, Iida T, Honda T. 2010. Two regulators of Vibrio parahaemolyticus play important roles in enterotoxicity by controlling the expression of genes in the Vp-PAI region. PLoS One 5:e8678 doi:10.1371/journal.pone.0008678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Isabella VM, Clark VL. 2011. Deep sequencing-based analysis of the anaerobic stimulon in Neisseria gonorrhoeae. BMC Genomics 12:51 doi:10.1186/1471-2164-12-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yin JL, Shackel NA, Zekry A, McGuinness PH, Richards C, Putten KV, McCaughan GW, Eris JM, Bishop GA. 2001. Real-time reverse transcriptase-polymerase chain reaction (RT-PCR) for measurement of cytokine and growth factor mRNA expression with fluorogenic probes or SYBR green I. Immunol. Cell Biol. 79:213–221 [DOI] [PubMed] [Google Scholar]
- 30. Bourassa L, Camilli A. 2009. Glycogen contributes to the environmental persistence and transmission of Vibrio cholerae. Mol. Microbiol. 72:124–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miyamoto CM, Sun W, Meighen EA. 1998. The LuxR regulator protein controls synthesis of polyhydroxybutyrate in Vibrio harveyi. Biochim. Biophys. Acta 1384:356–364 [DOI] [PubMed] [Google Scholar]
- 32. Wei YH, Chen WC, Wu HS, Janarthanan OM. 2011. Biodegradable and biocompatible biomaterial, polyhydroxybutyrate, produced by an indigenous Vibrio sp. BM-1 isolated from marine environment. Mar. Drugs 9:615–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kessler B, Witholt B. 2001. Factors involved in the regulatory network of polyhydroxyalkanoate metabolism. J. Biotechnol. 86:97–104 [DOI] [PubMed] [Google Scholar]
- 34. Bomchil N, Watnick P, Kolter R. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 185:1384–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Syed KA, Beyhan S, Correa N, Queen J, Liu J, Peng F, Satchell KJ, Yildiz F, Klose KE. 2009. The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J. Bacteriol. 191:6555–6570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yildiz FH, Visick KL. 2009. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 17:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Prouty MG, Correa NE, Klose KE. 2001. The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39:1595–1609 [DOI] [PubMed] [Google Scholar]
- 38. Gardel CL, Mekalanos JJ. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64:2246–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tam VC, Serruto D, Dziejman M, Brieher W, Mekalanos JJ. 2007. A type III secretion system in Vibrio cholerae translocates a formin/spire hybrid-like actin nucleator to promote intestinal colonization. Cell Host Microbe 1:95–107 [DOI] [PubMed] [Google Scholar]
- 40. Jobling MG, Holmes RK. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26:1023–1034 [DOI] [PubMed] [Google Scholar]
- 41. Skorupski K, Taylor RK. 1997. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol. Microbiol. 25:1003–1009 [DOI] [PubMed] [Google Scholar]
- 42. Merrell DS, Bailey C, Kaper JB, Camilli A. 2001. The ToxR-mediated organic acid tolerance response of Vibrio cholerae requires OmpU. J. Bacteriol. 183:2746–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Valeru SP, Wai SN, Saeed A, Sandstrom G, Abd H. 2012. ToxR of Vibrio cholerae affects biofilm, rugosity and survival with Acanthamoeba castellanii. BMC Res. Notes 5:33 doi:10.1186/1756-0500-5-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U. S. A. 103:1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. 2010. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. U. S. A. 107:19520–19524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 47. Pukatzki S, McAuley SB, Miyata ST. 2009. The type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12:11–17 [DOI] [PubMed] [Google Scholar]
- 48. Barret M, Egan F, Fargier E, Morrissey JP, O'Gara F. 2011. Genomic analysis of the type VI secretion systems in Pseudomonas spp.: novel clusters and putative effectors uncovered. Microbiology 157:1726–1739 [DOI] [PubMed] [Google Scholar]
- 49. Shakhnovich EA, Davis BM, Waldor MK. 2009. Hfq negatively regulates type III secretion in EHEC and several other pathogens. Mol. Microbiol. 74:347–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Worley MJ, Ching KH, Heffron F. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36:749–761 [DOI] [PubMed] [Google Scholar]
- 51. Butler SM, Camilli A. 2004. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 101:5018–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Golubeva YA, Sadik AY, Ellermeier JR, Slauch JM. 2012. Integrating global regulatory input into the Salmonella pathogenicity island 1 type III secretion system. Genetics 190:79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lucas RL, Lostroh CP, DiRusso CC, Spector MP, Wanner BL, Lee CA. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:1872–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Saini S, Slauch JM, Aldridge PD, Rao CV. 2010. Role of cross talk in regulating the dynamic expression of the flagellar Salmonella pathogenicity island 1 and type 1 fimbrial genes. J. Bacteriol. 192:5767–5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Soscia C, Hachani A, Bernadac A, Filloux A, Bleves S. 2007. Cross talk between type III secretion and flagellar assembly systems in Pseudomonas aeruginosa. J. Bacteriol. 189:3124–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hammer BK, Bassler BL. 2007. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 104:11145–11149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hase CC, Mekalanos JJ. 1999. Effects of changes in membrane sodium flux on virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 96:3183–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pal RR, Bag S, Dasgupta S, Das B, Bhadra RK. 2012. Functional characterization of the stringent response regulatory gene dksA of Vibrio cholerae and its role in modulation of virulence phenotypes. J. Bacteriol. 194:5638–5648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tsou AM, Frey EM, Hsiao A, Liu Z, Zhu J. 2008. Coordinated regulation of virulence by quorum sensing and motility pathways during the initial stages of Vibrio cholerae infection. Commun. Integr. Biol. 1:42–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Guentzel MN, Berry LJ. 1975. Motility as a virulence factor for Vibrio cholerae. Infect. Immun. 11:890–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Richardson K. 1991. Roles of motility and flagellar structure in pathogenicity of Vibrio cholerae: analysis of motility mutants in three animal models. Infect. Immun. 59:2727–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mekalanos JJ. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhu J, Mekalanos JJ. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647–656 [DOI] [PubMed] [Google Scholar]
- 64. Faruque SM, Biswas K, Udden SM, Ahmad QS, Sack DA, Nair GB, Mekalanos JJ. 2006. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl. Acad. Sci. U. S. A. 103:6350–6355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hung DT, Zhu J, Sturtevant D, Mekalanos JJ. 2006. Bile acids stimulate biofilm formation in Vibrio cholerae. Mol. Microbiol. 59:193–201 [DOI] [PubMed] [Google Scholar]
- 66. Yildiz FH, Dolganov NA, Schoolnik GK. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 183:1716–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jani AJ, Cotter PA. 2010. Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe 8:2–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bernard CS, Brunet YR, Gueguen E, Cascales E. 2010. Nooks and crannies in type VI secretion regulation. J. Bacteriol. 192:3850–3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ishikawa T, Rompikuntal PK, Lindmark B, Milton DL, Wai SN. 2009. Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS One 4:e6734 doi:10.1371/journal.pone.0006734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Leung KY, Siame BA, Snowball H, Mok YK. 2011. Type VI secretion regulation: crosstalk and intracellular communication. Curr. Opin. Microbiol. 14:9–15 [DOI] [PubMed] [Google Scholar]
- 71. Dong TG, Mekalanos JJ. 2012. Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res. 40:7766–7775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mulder DT, Cooper CA, Coombes BK. 2012. Type VI secretion system-associated gene clusters contribute to pathogenesis of Salmonella enterica serovar Typhimurium. Infect. Immun. 80:1996–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zheng J, Shin OS, Cameron DE, Mekalanos JJ. 2010. Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 107:21128–21133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tsou AM, Cai T, Liu Z, Zhu J, Kulkarni RV. 2009. Regulatory targets of quorum sensing in Vibrio cholerae: evidence for two distinct HapR-binding motifs. Nucleic Acids Res. 37:2747–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zheng J, Ho B, Mekalanos JJ. 2011. Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One 6:e23876 doi:10.1371/journal.pone.0023876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ma AT, Mekalanos JJ. 2010. In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc. Natl. Acad. Sci. U. S. A. 107:4365–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mandlik A, Livney J, Robins WP, Ritchie JM, Kelalanos JJ, Waldor MK. 2011. RNA-seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 10:165–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.