Abstract
The activities of DNA methyltransferases are important for a variety of cellular functions in bacteria. In this study, we developed a modified high-throughput technique called methyl homopolymer tail mediated sequencing (methyl HTM-seq) to identify the undermethylated sites in the Vibrio cholerae genome for the two DNA methyltransferases, Dam, an adenine methyltransferase, and VchM, a cytosine methyltransferase, during growth in rich medium in vitro. Many of the undermethylated sites occurred in intergenic regions, and for most of these sites, we identified the transcription factors responsible for undermethylation. This confirmed the presence of previously hypothesized DNA-protein interactions for these transcription factors and provided insight into the biological state of these cells during growth in vitro. DNA adenine methylation has previously been shown to mediate heritable epigenetic switches in gene regulation. However, none of the undermethylated Dam sites tested showed evidence of regulation by this mechanism. This study is the first to identify undermethylated adenines and cytosines genomewide in a bacterium using second-generation sequencing technology.
INTRODUCTION
Toxigenic strains of Vibrio cholerae are the causative agent of cholera and, according to the World Health Organization, are annually responsible for 3 to 5 million infections worldwide. During its life cycle, this pathogen transits between the human host and aquatic environments, and as a result, it must respond to shifts in temperature, osmolarity, and concentrations of organic and inorganic nutrients (1). It is, therefore, important for V. cholerae to sense its environment and specifically regulate the genes that are required for survival in each niche.
One mechanism of gene regulation occurs via DNA methylation. One of the best-characterized DNA methyltransferases (MTases) is the DNA-adenine MTase (Dam) that is commonly expressed among enteric bacteria, including V. cholerae. Dam methylates the A residue on both strands of the palindromic sequence GATC. It has been shown to be involved in timing of chromosome replication, control of transposition, mismatch repair, gene regulation, and phase variation (2, 3). In a given cell, >99% of all Dam sites will be fully methylated, and thus, the presence of stably unmethylated sites is atypical and defines genomic loci that are potentially sites of tightly bound transcriptional regulators (4).
Binding of some transcription factors has been shown to inhibit methylation of DNA sites. This has been characterized in depth in Escherichia coli, and this activity has been exploited to identify transcription factor binding sites across an entire genome (5). In some instances, methylated adenine but not methylated cytosine has been shown to alter the ability of transcription factors to bind DNA. If transcription factor binding inhibits DNA methylation and methylation inhibits transcription factor binding, there is the potential for the DNA methylation status of a promoter to define the transcriptional activity of that locus.
A classic approach to study gene regulation by MTases is to generate a mutant strain lacking the MTase and determine what genes are differentially expressed at the transcriptional level. In V. cholerae, however, this type of analysis is not possible for the major MTase Dam because its activity is essential for the replication of both chromosomes and, therefore, survival (6). In addition to Dam, V. cholerae also encodes an orphan cytosine MTase, VchM, which methylates the proximal C on both strands of the palindromic sequence RCCGGY (7).
To gain further insight into the role of methylation in V. cholerae, we sought to define the undermethylated sites for both Dam and VchM in the O1 serogroup, El Tor biotype clinical isolate E7946. Since undermethylation can be caused by transcription factor binding, this analysis would potentially define novel transcription factor binding sites that overlap Dam and VchM methylation sites in the genome. Additionally, since methyladenine can regulate gene expression, this analysis may also identify regions in the genome that are regulated by Dam methylation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The V. cholerae strain used in this study was a streptomycin-resistant variant of the clinical isolate E7946. Cells were usually grown at 37°C in Luria Bertani (LB) broth and on LB agar. Where indicated, cells were also grown at 37°C in M9 minimal medium supplemented with a carbon source as indicated below at a final concentration of 0.4%. When appropriate, cultures were supplemented with streptomycin (100 μg/ml) and/or kanamycin (100 μg/ml) as appropriate. To study lacZ fusions on solid medium, plates were supplemented with 40 μg/ml 5-bromo-4-chloro-3-indolyl-d-galactoside (X-Gal). The PVCA0063 lacZ fusion strain produced dark-blue colonies on LB plates, so in order to reduce the amount of product to enable increases in LacZ expression to be observed, 0.3 mM PETG (phenyl-ethyl-β-d-thiogalactoside, a competitive inhibitor of X-Gal) was added to the plates, which was the minimum amount required to see light-blue colonies. This lacZ fusion strain was light blue on LB plates containing 200 μM FeSO4, so PETG was not added to this medium.
Methyl homopolymer tail mediated-seq (methyl HTM-seq).
Genomic DNA (gDNA) was isolated using the DNeasy blood and tissue kit (Qiagen) according to the manufacturer's instructions. After isolation, between 1 and 2 μg of gDNA was sheared to ∼400 bp (range 200 to 600 bp) using a prechilled Branson high-intensity cup horn sonifier (Branson) for 2 min at 50% intensity with a 5-s-on/5-s-off duty cycle. Homopolymer tails of cytosine were added to the 3′ ends of all sheared molecules using terminal deoxynucleotidyl transferase (TdT) according to the manufacturer's instructions (Promega). For these reactions, a 20:1 mixture of dCTP and ddCTP (a chain terminator) was used to generate C-tails of approximately 20 cytosines. After C-tailing, reaction mixtures were run through a Performa spin column (Edge Biosystems) according to the manufacturer's instructions to remove excess nucleotides and to desalt reaction mixtures. The eluate was digested using the appropriate methylation-sensitive restriction enzyme (MeSR). For Dam, the MeSR used was MboI, while for VchM, the MeSR used was BsrFI (New England BioLabs). After digestion, samples were heat inactivated when appropriate and run through a Performa spin column to desalt reactions. The eluate was then ligated to the tIL1 adaptor (final concentration of 1 μM) using the Quick Ligase kit (New England BioLabs), in a ligation mixture volume of 60 μl, according to the manufacturer's instructions. The tIL1 adaptor was generated by annealing the oligonucleotides ABD013 and ABD013D for Dam and ABD013 and ABD013B for VchM (see Table S1 in the supplemental material). Samples were run through a Performa spin column to desalt reaction mixtures. Then, 6 μl of the eluate was used as the template in PCRs to amplify the samples, using OLJ 131 as the forward primer and either OLJ 573 (Dam) or BC33G (VchM) as the reverse primer (see Table 1 in the supplemental material). These forward and reverse primers contain the sequences specific for capture and sequencing on the Illumina HiSeq2000 platform (Illumina). Also, the reverse primers used provide a unique barcode index sequence that can be used to multiplex samples onto a single lane in the Illumina flow cell. After PCR, the DNA concentrations of samples were determined on a Nanodrop 2000 spectrophotometer (Nanodrop) and submitted for sequencing at the Tufts University Core Facility via single-end 50-bp reads on the Illumina HiSeq2000.
Table 1.
Undermethylated intergenic sites identified by methyl HTM-seq
| Chromosome | Positiona | Intergenic locusa | Frequencyb | Fold overrepresentionc |
|---|---|---|---|---|
| Dam data | ||||
| chrI | 1306007 | VC1231-VC1232 | 6.96E−04 | 26.2 |
| chrI | 1306010 | VC1231-VC1232 | 5.74E−04 | 21.6 |
| chrI | 1933145 | VC1783-VC1784 | 3.28E−04 | 12.4 |
| chrI | 1933148 | VC1783-VC1784 | 4.60E−04 | 17.3 |
| chrI | 2364224 | K139p05-K139p04 | 2.02E−04 | 7.6 |
| chrI | 2364255 | K139p05-K139p04 | 1.22E−04 | 4.6 |
| chrII | 68810 | VCA0062-VCA0063 | 3.11E−04 | 11.7 |
| chrII | 68813 | VCA0062-VCA0063 | 1.07E−03 | 40.3 |
| VchM data | ||||
| chrI | 296268 | VC0286-VC0287 | 1.10E−02 | 47.0 |
| chrI | 296271 | VC0286-VC0287 | 9.66E−03 | 41.4 |
| chrI | 1356851 | VC1280-VC1281 | 5.55E−03 | 23.8 |
| chrI | 1356854 | VC1280-VC1281 | 2.72E−03 | 11.6 |
| chrI | 1670727 | VC1558-VC1559 | 5.27E−03 | 22.6 |
| chrI | 1670730 | VC1558-VC1559 | 3.80E−03 | 16.3 |
The position and locus are based on the annotated N16961 genome containing the K139 prophage genome in chromosome I. The unmethylated site is located in the intergenic region between the indicated loci.
Indicates the frequency of reads mapping to the indicated site relative to the total number of reads mapped.
Indicates the degree of overrepresentation of the reads at the indicated site relative to the mean number of reads obtained for all sites.
After sequencing and demultiplexing, reads were further filtered to identify sequences that represent true Dam and VchM sites. For Dam data, reads were filtered to obtain sequences that started with GATC, while for the VchM data, reads were filtered to obtain sequences that started with either CCGGC or CCGGT. The reads were then trimmed to a length of 21 bp and mapped to the O1 El Tor N16961 genome containing the sequence for the K139 prophage inserted into chromosome I (since this prophage sequence is absent from N16961 but present in the E7946 strain used in this study [8]), using the program Bowtie, and no mismatches were allowed during mapping (9). Finally, the total numbers of forward and reverse reads mapping to methylation sites were determined.
MeSR digestion and qPCR for characterization of undermethylated sites.
Assays were performed essentially as previously described (10). Briefly, between 10 and 100 ng of gDNA was digested using a MeSR in a final reaction mixture volume of 20 μl. For Dam, the MeSR used was MboI, while for VchM, the MeSR used was MspI. After digestion, reaction mixtures were heat inactivated when appropriate. Then, 2 μl of this digestion (1 to 10 ng) was used as the template for quantitative PCR (qPCR) using primers that span a methylation site of interest. All primers used for MeSR digestion and qPCR are listed in Table S1 in the supplemental material. An uncut control reaction mixture (no-enzyme control) was run for every sample. The abundance of DNA in all samples was determined relative to a standard curve generated using dilutions of purified gDNA. Reaction mixtures were run on an Mv3005P qPCR instrument (Stratagene) using the dye incorporation method (SYBR green) and analyzed using MxPro qPCR software (Stratagene).
Generation of mutant strains.
Deletion mutants were generated using the natural competence of V. cholerae, and PCR products generated by splicing overlap extension (SOE) PCR essentially as previously described (11, 12). The oligonucleotides used for SOE PCRs are shown in Table S1 in the supplemental material. For all SOE products, PCRs were performed to generate an up arm (primers F1/R1), a down arm (primers F2/R2), and a kanamycin resistance cassette (primers FRT-Kan-F/FRT-Kan-R). The PCR products for the up arm, down arm, and kanamycin resistance cassette were then mixed 1:1:1 (∼50 ng each) and used as the template in a PCR mixture using the F1 and R2 primers to generate the full-length SOE product. This product was then purified using QIAquick PCR cleanup columns (Qiagen), and 1- to 5-μg amounts were used to transform V. cholerae via natural competence as previously described (11). The lacZ fusion strain was generated by a similar method. A SOE product was generated to delete the endogenous lacZ (VC2338) as described above, and a second SOE product was generated to transcriptionally fuse the VCA0063 promoter to the V. cholerae lacZ gene at the VCA0063 locus. Both constructs were simultaneously transformed into V. cholerae via natural transformation. All mutant strains were confirmed by PCR and/or sequencing.
Purification of Fur and electrophoretic mobility shift assays (EMSAs).
The gene encoding the V. cholerae Fur protein was amplified using primers ABD084 and ABD085 and cloned into the NdeI and BamHI sites of pET15b-His-Tev to generate a Fur protein containing a His tag that can be removed by cleavage with the Tev protease as previously described (see Table S1 in the supplemental material) (13). This expression vector was transformed into E. coli BL21(DE3), a single colony was picked, and the sequence of the insertion was confirmed by PCR and sequencing. To induce expression, cells were grown to an optical density at 600 nm (OD600) of ∼0.8 in LB medium containing 100 μg/ml ampicillin at 37°C with shaking (250 rpm), and then isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture to a final concentration of 1 mM and the culture incubated for 16 h at 18°C shaking (250 rpm). Protein was then purified from these cells essentially as previously described using a Ni-nitrilotriacetic acid (NTA) column (13). The His tag was then cleaved from Fur by incubating purified protein with Tev protease at 4°C overnight as previously described (13). Untagged Fur protein was then stored in 10 mM Tris, pH 8.0, 250 mM NaCl, 5 mM β-mercaptoethanol, and 50% glycerol at −20°C for short-term storage (up to 1 week) or at −80°C for long-term storage (tested up to 2 months).
The primers used to generate probes are listed in Table S1 in the supplemental material. For the probe of PVCA0063, oligonucleotides end labeled with Cy5 were ordered (IDT) and were annealed in vitro, while for PSodA and PAphA, Cy5 was incorporated into probes during PCR using Cy5-labeled dCTP (GE Healthcare). For the latter, we specifically determined the titer of the ratio of unlabeled dCTP to Cy5-labeled dCTP added to PCRs to result in the incorporation of only 1 or 2 labeled C residues, as we found this to be sufficient to result in a robustly labeled EMSA probe. Where indicated, EMSA probes were in vitro methylated with Dam methylase according to the manufacturer's instruction (NEB). After methylation, reaction mixtures were run through a Performa spin column to remove excess S-adenosylmethionine and to desalt the mixtures.
The binding reaction was done in a 20-μl mixture that contained probe (2 nM), Fur protein (as indicated below), 10 mM Tris-borate buffer, 40 mM KCl, 1 mM MgCl2, 100 μM MnCl2, 2 mM dithiothreitol (DTT), 100 μg/ml bovine serum albumin (BSA), 5 μg sheared calf thymus DNA, and 10% glycerol. The reaction mixtures were incubated for 30 min at room temperature prior to loading onto prerun 6% native polyacrylamide gels. Gels were made in 0.5× Tris-borate buffer (5.4 g/liter Tris base and 2.25 g/liter boric acid) and prerun for 1 h in 0.5× Tris-borate buffer containing 100 μM MnCl2. Gels were imaged using the Cy5 setting on the FLA-9000IR instrument (GE Healthcare Life Sciences).
Transcript abundance by qRT-PCR.
Total cellular RNA was purified from V. cholerae using the RNeasy minikit (Qiagen) and then treated with DNase from the TURBO DNA-free kit (Ambion) according to the manufacturer's instructions. Next, cDNA was generated from 1 μg of treated RNA using random priming with the iScript cDNA synthesis kit in a volume of 20 μl according to the manufacturers' instructions (Bio-Rad). Two microliters of each sample was loaded in quadruplicate onto a qPCR plate. Two wells were used to assess the transcript abundance of the query amplicon (genes VCA0063 or VC1784), while the other two wells were used to assess the transcript abundance of the housekeeping gene rpoB. The primers used for all quantitative reverse transcriptase PCR (qRT-PCR) experiments are listed in Table S1 in the supplemental material, and qPCR was performed as described above. The relative abundance of transcripts was determined using the standard curve method: for each reaction plate, two standard curves of gDNA were included and tested with the amplicon-specific primers and the primers for rpoB. The expression of each amplicon was determined relative to the expression of rpoB. To compare between different growth conditions, expression is shown relative to one of the conditions (usually relative to growth in LB). For all samples, a control reaction mixture lacking reverse transcriptase was run to confirm that RNA samples were not contaminated with gDNA.
Statistical comparisons and DNA analysis.
All data were plotted using GraphPad Prism version 5.0 (GraphPad), and statistical comparisons were made as indicated in figure legends. Analysis of DNA was performed using CLC Main Workbench 6 (CLC Bio).
RESULTS
Identification of undermethylated sites in the V. cholerae genome by a modified high-throughput approach.
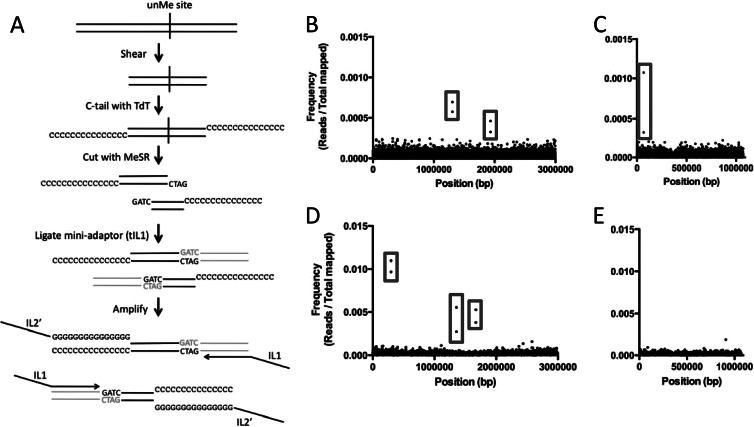
To identify the undermethylated sites in both chromosomes of the V. cholerae O1 El Tor strain E7946, we undertook a high-throughput approach using methylation-sensitive restriction enzymes (MeSR) and next-generation sequencing (NGS). This approach is similar to the methyl-seq technique used to study methylation profiles in eukaryotes (14). Methyl-seq uses MeSRs to cut undermethylated sites in the genome and then ligates the ends generated to adaptors that allow for NGS. Thus, a single unmethylated site should be represented by two independent sequences (reads). The methyl-seq strategy is limited, however, since areas where unmethylated sites are rare and thus widely spaced in the genome will not be represented in the output due to the constraints of size selection for sequencing on NGS platforms like Illumina (14, 15). This can result in these sites being completely absent in the output or can result in the loss of one of the two paired sequences that define an unmethylated site (15). To circumvent this problem, we utilized a modification of the recently described homopolymer tail-mediated ligation PCR (HTML-PCR) method for generating Illumina genomic libraries to probe for unmethylated sites in the genome (16). In this approach, DNA is first sheared to ∼200 to 600 bp, and then homopolymer C-tails are added to the 3′ ends of all molecules using terminal deoxynucleotidyl transferase (TdT) (Fig. 1A). Next, a MeSR is used to cut all unmethylated sites, and the cut ends are ligated to an adaptor (tIL1) (Fig. 1A). Once ligated, libraries are amplified using oligonucleotide primers containing the sequences necessary for sequencing on the Illumina platform (IL-1 and IL-2) (Fig. 1A). Therefore, we are calling this modified technique methyl homopolymer tail mediated sequencing (methyl HTM-seq). One potential issue that can arise during methyl HTM-seq is that unmethylated sites that are in close proximity to one another (<100 bp apart) may lose one of the sequences that forms a pair for each unmethylated site, since the likelihood of shearing the genomic DNA (gDNA) between these two sites is relatively low. The outside reads that span the two pairs, however, should still be represented and constitute a pseudopair that can be used to infer the unmethylated status of closely spaced methylation sites.
Fig 1.
Identification of undermethylated Dam and VchM sites in V. cholerae. (A) Using an unmethylated Dam site, we diagram the methyl HTM-seq protocol used to identify undermethylated sites in the genome. Genomic DNA is shown as black lines, while the adaptor (tIL1) is shown in gray. (B to E) Visual representation of Dam (B and C) and VchM (D and E) methyl HTM-seq data. Frequencies of mapped Illumina reads (y axis) are plotted against the V. cholerae genome (x axis) for chromosome I (B and D) and chromosome II (C and E). Examples of paired reads that make up a single undermethylated Dam site are highlighted by gray boxes.
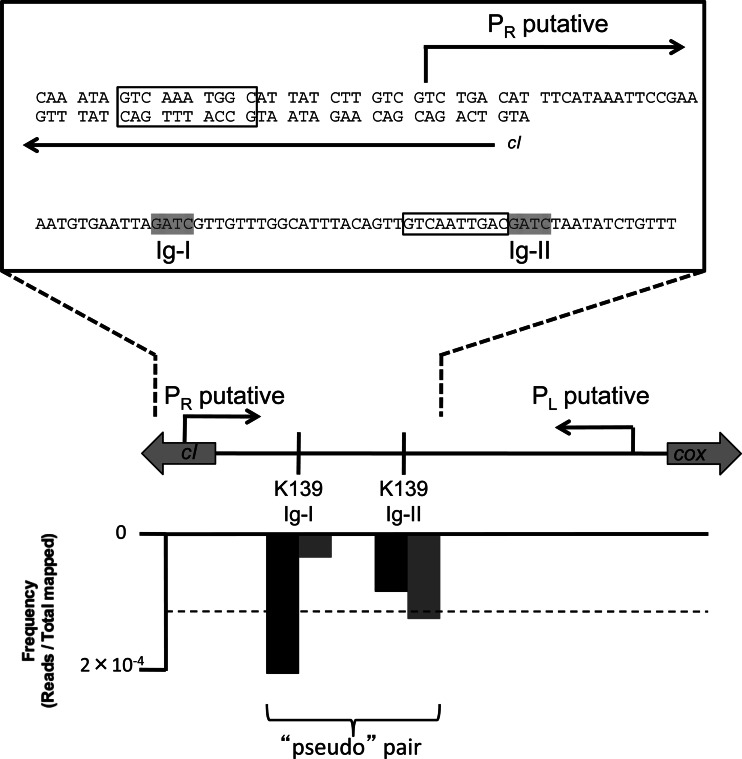
To assess undermethylation of Dam and VchM sites in V. cholerae, we used the MeSRs MboI and BsrFI, respectively, which perfectly overlap the recognition sites of these two MTases. We assessed the undermethylated profile for these two MTases in cultures grown to late exponential phase in the rich medium lysogeny broth (LB). The late exponential phase represents a phase of growth when nutrient depletion is sensed and major changes in bacterial gene regulation occur. These data sets were mapped to the V. cholerae genome. Assessing the graphed frequency of reads mapped along the V. cholerae genome, we find pairs of reads that constitute undermethylated sites (Fig. 1B to E). As a stringent measurement of undermethylated sites, we determined the paired sites (separated by <100 bp) that are above the 99% confidence interval for the mean of the data. Strikingly, there are very few Dam and VchM sites that are undermethylated in the V. cholerae genome (see Table S2 in the supplemental material). Many of the sites that are undermethylated, however, are in intergenic regions, indicating that these are likely due to transcription factor binding (Table 1). Specifically for the undermethylated intergenic sites identified in the K139 prophage, we find that the paired reads are separated by 31 bp, indicating that this site may represent two unmethylated sites in close proximity, which generates a pseudopair. Indeed, upon closer examination of this region, we found that the outside reads spanning the two Dam sites are overrepresented compared to the inside reads between the two Dam sites, consistent with a model wherein these two sites are undermethylated (Fig. 2).
Fig 2.
The genetic architecture of undermethylated Dam sites in the K139 prophage. Shown is the structure of the intergenic region between the outward-facing genes encoding cI and Cox. Putative PL and PR sites were annotated by Nesper et al. (32). PL drives the expression of cI, and PR drives the expression of cox. Above the gene layout is a zoomed inset showing the sequence of putative cI operator sites (boxed) and the undermethylated Dam sites Ig-I and Ig-II (highlighted in gray). Below the gene layout is a graph plotting the frequency of mapped reads (y axis) against the genome (x axis) for the two Dam sites in this region from the methyl HTM-seq data set. Two bars are shown for each Dam site, representing the forward (gray) and reverse (black) reads that result from cleavage of each site. The dotted line on this graph represents the 99% cutoff value used as a measure of significant undermethylation in this experiment.
Confirmation of undermethylated sites by MeSR digestion and qPCR.
To validate and further characterize undermethylated sites, we used a previously described technique that combines MeSR digestion and qPCR (10). The gDNA is first digested with a MeSR and, optionally, also with a methylation dependent restriction enzyme (MeDR). Digestion at a site reduces the number of intact template molecules spanning the region. Samples are heat inactivated and then used as the template in a qPCR using oligonucleotide primers that span the methylation site of interest. The relative abundance of DNA in experimental samples is then determined against a standard curve, and the percentage of undermethylation is determined relative to the undigested control. To characterize the undermethylation of Dam sites, we used the MeSR MboI and the MeDR DpnI, and to characterize the undermethylation of VchM sites, we used the MeSR MspI.
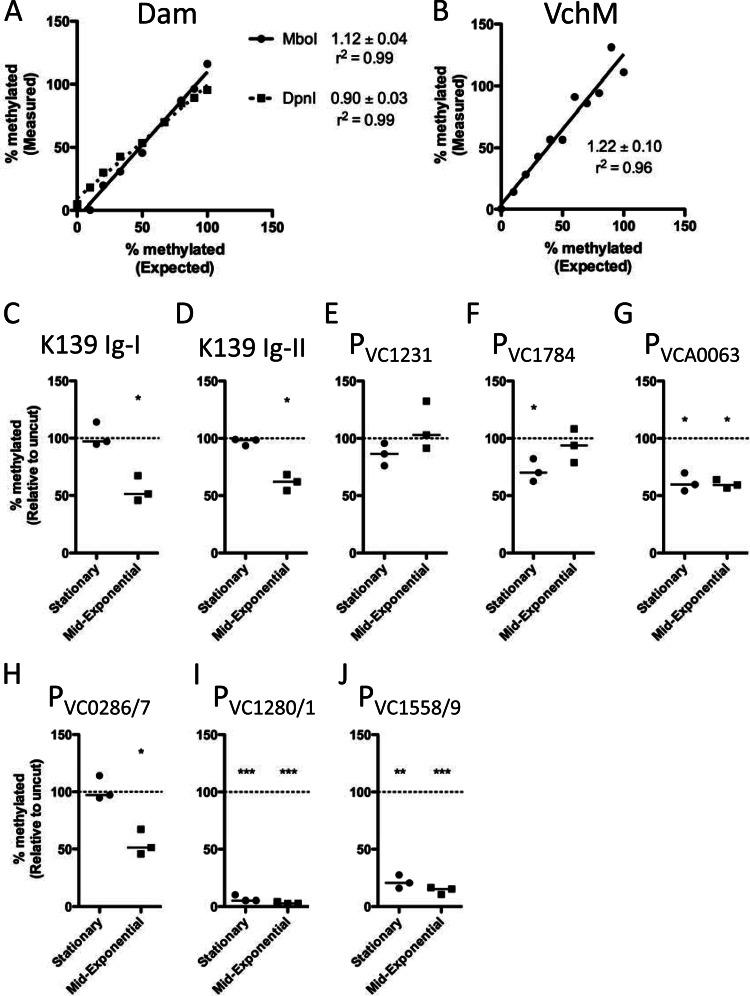
To confirm that this method yields a quantitative assessment of methylation status, we performed two control experiments. E. coli Dam+ and Dam− DNA (KEIO collection strains [17]) were mixed at different ratios, ranging from 100% to 0% Dam+ in 10% increments, and subjected to the method described above. The experimentally derived values (measured) for percentage of unmethylated DNA were compared to the actual values (expected), and, as expected, both the MboI and DpnI data showed linearity over this range, with slopes near 1 (Fig. 3). Thus, subsequent data for Dam undermethylation were assessed by MboI digestion alone. The same analysis was performed using mixtures of wild-type E7946 and ΔvchM mutant strain gDNA to validate the assay using the MeSR MspI. Again, as expected, the data show linearity over the range of ratios tested, with a slope near 1 (Fig. 3).
Fig 3.
Characterization of intergenic undermethylated sites by MeSR digestion and qPCR. (A and B) Assays were assessed for reliable detection of undermethylation using gDNA from methylase-expressing and methylase-null mutants. (A) For MboI (solid line) and DpnI (dotted line), gDNA from Dam+ and Dam− E. coli strains were mixed in 10% increments from 100% to 0% Dam+ and the experimental values obtained (Measured) were compared to actual mixed ratios (Expected). (B) For MspI, gDNA from wild-type E7946 and a ΔvchM mutant strain were mixed and analyzed as described for panel A. Data are shown as percent methylated relative to an uncut control reaction mixture. The slope of the linear regression ± standard error is indicated. Data are representative results from one of at least two independent experiments. (C to J) Characterization of Dam (C to G) and VchM (H to J) undermethylated intergenic sites using MeSR digestion and qPCR of gDNA from cultures grown to mid-exponential (optical density at 600 nm [OD600] of ≈0.5) or stationary (OD600, ≈3.0) phase. Data are shown as percent methylated relative to an uncut control reaction mixture, and significance was determined by one-sampled Student's t test to determine if means were significantly different from 1. Each data point in panels C to J represents an independent biological replicate, and a horizontal line represents the median of each sample. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Using this method, we sought to extend the analysis of the undermethylated sites we identified as described above to cells grown to either mid-exponential or stationary phases. All sites assessed were significantly undermethylated in either mid-exponential, stationary, or both phases, except for PVC1231, which trended toward undermethylation in stationary phase but not to a statistically significant degree (Fig. 3).
Identification of transcription factors responsible for undermethylation in V. cholerae.
For all undermethylated intergenic sites, we attempted to identify a transcription factor that caused the observed methylation pattern. For the K139 prophage, we found two undermethylated sites between the outward-facing genes encoding the cI repressor and Cox (Fig. 2). In the homologous bacteriophage HP1, the cI repressor is responsible for repressing PR, which is the promoter that drives the expression of cox and the downstream genes involved in lytic growth, while Cox is involved in repressing PL, which is the promoter that drives the expression of cI and the downstream genes involved in the maintenance of lysogeny (18). Since these sites are undermethylated in E7946, a K139 lysogen, we predict that the repressor responsible for the observed methylation profile is the cI repressor; however, this is difficult to confirm experimentally since a mutant strain lacking this repressor would be nonviable. To determine whether we could identify putative operator sites for the cI repressor based on the location of the undermethylated sites in the K139 prophage, we visually inspected the genomic region between PR and PL for palindromic sequences or regions of dyad symmetry. This analysis identified two 10-bp sites that overlap or are in close proximity to the two undermethylated Dam sites and, thus, may constitute operator sites for the cI repressor (Fig. 2). The putative cI operator site located between the two undermethylated Dam sites (Ig-I and Ig-II) is a perfect palindrome with the sequence GTCAATTGAC, while the site located upstream from PR deviates from this palindrome at 2 sites (GTCAAaTGgC) (Fig. 2).
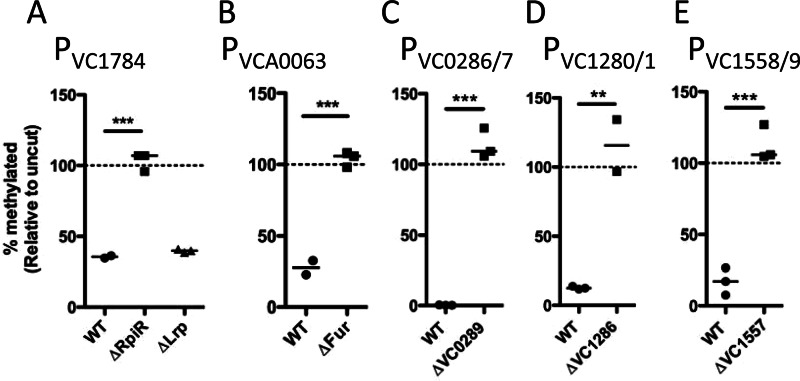
Based on the literature and the promoter prediction software BPROM, there are two potential repressors that may promote undermethylation of PVC1784; these are RpiR and Lrp (19). To test this hypothesis, we generated mutant strains and found that RpiR is fully responsible for undermethylation at the Dam site in this promoter (Fig. 4). It was previously shown that VCA0063 is repressed by Fur, and we have determined by using a Δfur mutant strain that this repressor is responsible for undermethylation of the Dam site in this promoter (Fig. 4) (20). The three undermethylated VchM sites are located within the intergenic space between divergent genes VC0286-VC0287, VC1280-VC1281, and VC1558-VC1559, respectively, and each has a gene encoding a putative repressor protein in close proximity in the genome. Analysis of mutant strains lacking these repressors confirmed that they were fully responsible for undermethylation at these VchM sites (Fig. 4).
Fig 4.
Identification of transcription factors responsible for undermethylation of intergenic sites. Putative transcription factors were deleted and replaced with a kanamycin resistance cassette, and the effect on undermethylation of the indicated site was determined by MeSR digestion and qPCR. Data are shown as percent methylated relative to an uncut control reaction mixture, and significance was determined by two-tailed Student's t test. (A) Strains were grown to mid-exponential phase in M9 medium with glucose as the sole carbon source, since this showed the greatest level of undermethylation at the Dam site in PVC1784, as shown by the results in Figure 6. (B) Strains were grown to mid-exponential phase in iron-replete medium (LB plus 200 μM FeSO4), since this showed the greatest level of undermethylation at the Dam site in PVCA0063, as shown by the results in Figure 5. (C to E) Strains were grown to mid-exponential phase in LB medium, as this resulted in severe undermethylation of the VchM sites in the indicated promoters, as shown by the results in Figure 3. Each data point in these plots represents an independent biological replicate, and a horizontal line represents the median of each sample. WT, wild type; **, P < 0.01; ***, P < 0.001.
Overlapping Fur binding and Dam methylation do not mediate an epigenetic switch.
The promoter upstream from VCA0063 controls the expression of ptrB and hutR, two genes involved in heme utilization in V. cholerae (20). This region contains an undermethylated Dam site that overlaps with the Fur binding site and the −10 consensus sequence of PVCA0063 (Fig. 5A). An almost identical regulatory arrangement is seen in the promoter of the sci1 gene cluster, which controls the expression of the type VI secretion system in E. coli. Recently, it was shown for the sci1 promoter that Fur binding results in undermethylation of the Dam site that overlaps the Fur consensus sequence and that Dam methylation reduced the affinity for Fur binding at this site (21). Analogous regulatory arrangements have been shown to promote a heritable epigenetic switch in gene regulation in other systems (2, 22). The best-described example is the control of Pap pilus expression in uropathogenic E. coli. In this system, the global response regulator Lrp acts as a repressor and competes with Dam to prevent methylation at a Dam site proximal to the promoter driving expression of the pilus. At a low, stochastic frequency within a population of cells, Dam will gain entry to this site and methylate it. This, in turn, inhibits Lrp from binding to the region proximal to the Pap promoter and allows for expression of the pilus. This phenotype is heritable because, upon cell division, DNA becomes hemimethylated, which still results in poor binding of Lrp, resulting in the Dam site becoming fully methylated in the daughter cells. Conversely, at a low, stochastic frequency within a population of cells, Lrp will bind to hemimethylated DNA following cell division, which will sequester this site from Dam. After a second round of DNA replication and cell division, this site can become fully unmethylated and Lrp will be stably bound to switch pilus expression into the OFF phase. This system, therefore, defines a heritable epigenetic switch for gene regulation.
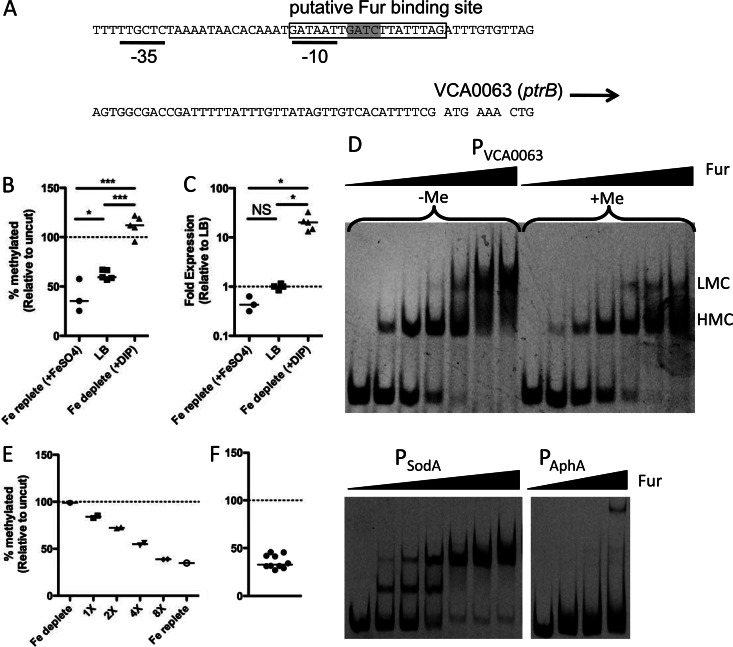
Fig 5.
Overlapping Fur and Dam methylation do not constitute an epigenetic switch at the VCA0063 promoter. (A) Genetic architecture of the VCA0063 promoter, showing the overlapping putative Fur binding site (boxed), −35 and −10 consensus sequences (black lines), and Dam site (highlighted in gray). (B and C) Cells were grown in iron-replete medium (LB plus 200 μM FeSO4), LB, or iron-depleted medium (LB plus 90 μM dipyridyl [DIP]), and the methylation level of PVCA0063 was determined by MeSR digestion and qPCR (B), while the expression level of VCA0063 was assessed by qRT-PCR (C). Each data point is from an independent biological replicate, and a horizontal line represents the median of each sample. Statistical significance was determined by two-tailed Student's t test (B) and Mann-Whitney (C). (D) EMSAs using probes of the indicated promoters and purified Fur protein. The final concentration of probe used in all EMSA reactions was 2 nM. The concentrations of Fur added in EMSAs with the probes PVCA0063 (in vitro methylated [+Me] and unmethylated [−Me]) and PSodA, from left to right, were 0, 7.8, 15.6, 31.25, 62.5, 125, and 250 nM. The concentrations of Fur added in EMSAs with the probe PAphA, from left to right, were 0, 125, 250, and 500 nM. The positions of higher- and lower-mobility complexes are designated by HMC and LMC, respectively. Data are representative results from one of at least two independent experiments. (E) The heritability of PVCA0063 methylation was tested by taking cells grown under iron-depleted conditions (fully methylated) and incubating them for the indicated number of generations under iron-replete conditions. (F) Heterogeneity in the methylation of the VCA0063 promoter was assessed among single-colony isolates by MeSR digestion and qPCR. Each data point represents an independent biological replicate, and the horizontal line represents the median of each sample. NS, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To determine whether PVCA0063 undergoes a similar mechanism of regulation, we first determined whether Fur-mediated undermethylation of PVCA0063 was responsive to the available iron concentration. Under iron-replete conditions when Fur should bind tightly, we find undermethylation of the Dam site in PVCA0063 (Fig. 5B). Furthermore, under iron-depleted conditions, when the affinity of Fur for its binding site is reduced, we find that the locus is fully methylated (Fig. 5B). The expression levels of VCA0063 (ptrB) were also assessed under these conditions by qRT-PCR, and the transcript levels were found to correlate directly with the methylation state of the promoter (Fig. 5C).
To determine whether Fur binding is inhibited by methylation of PVCA0063 (as observed for the sci1 and pap promoters in E. coli), we performed EMSAs. Fur can generate two independent shifts (higher- and lower-mobility complexes) in EMSAs due to the structure of the Fur box as two overlapping heptamer repeats (23). Using an unmethylated and an in vitro Dam-methylated probe of the VCA0063 promoter, we show that formation of the lower-mobility complex is inhibited when PVCA0063 is fully methylated (Fig. 5D). Full methylation of probes was confirmed by restriction digest of probes with the MeSR MboI and the MeDR DpnI (data not shown). It was previously shown that PSodA was bound by Fur, so a probe of this promoter was included in these assays as a positive control, while a probe of the aphA promoter was included as a negative control since this promoter is not predicted to interact with Fur (Fig. 5D).
Since we see that the mobility of Fur in EMSAs is altered by methylation of the VCA0063 promoter, it is possible that Fur binding and methylation by Dam constitute a heritable epigenetic switch that regulates this locus. There are no DNA demethylases in V. cholerae, so transition from the methylated state to the unmethylated state can only occur by DNA replication and cell division. If a site is fully methylated, upon one round of DNA replication and cell division, the site will become hemimethylated. This hemimethylated site can then either become fully remethylated (by the methylase) or can be sequestered by a transcription factor, which will prevent access of the methylase to its target site. If the site is sequestered in a hemimethylated state, a subsequent round of DNA replication and cell division will result in one daughter cell with a fully unmethylated site. So, if methylation allowed for a heritable epigenetic switch at the VCA0063 promoter, we would predict that a fully methylated and active promoter would retain both its activity and its fully methylated state through multiple generations under repressive conditions, since methylation should inhibit Fur binding and allow for Dam to remethylate the site upon cell division. To test this hypothesis, we took cells grown under iron-depleted conditions, where the locus is fully expressed and fully methylated, and incubated them for multiple generations in iron-replete medium, which should promote repression of the locus. When we performed this analysis, we found that after two generations under iron-replete conditions, the methylation of this site was significantly reduced (to ∼30%), which is what would be predicted if methylation of this site were not in the least heritable (Fig. 5E).
An alternative explanation for this result is that, among the clonal population under repressive conditions, there are cells where this site is stably methylated and cells where it is stably unmethylated, resulting in an average of 30% when viewed as a population (as was done in the experiment whose results are shown in Fig. 5E). To differentiate between these two possibilities, we analyzed the methylation of the VCA0063 promoter among single-colony isolates when cells were grown under inducing conditions (iron depleted) and subsequently plated and grown on repressive medium (iron replete). If there was methylation-dependent epigenetic control of the VCA0063 promoter (i.e., methylation states were stably and heritably maintained), we would predict heterogeneity in the methylated state of the VCA0063 promoter among these single-colony isolates, where ∼30% of isolates will be fully methylated and ∼70% will be fully unmethylated. When we performed this analysis, however, we found no heterogeneity in the methylated state of the VCA0063 promoter in these single-colony isolates (Fig. 5F). Furthermore, we generated a strain containing a transcriptional lacZ fusion to the VCA0063 promoter and tested this strain for heterogeneity in the expression of LacZ when grown under inducing conditions (iron depleted) and subsequently plated on repressive medium containing two different concentrations of iron (LB or LB plus 200 μM FeSO4) and a chromogenic substrate for LacZ (X-Gal). Screening >1,500 colonies under each condition, we found no heterogeneity in the expression of LacZ (data not shown). Taken together, these results indicate that the undermethylation observed in the VCA0063 promoter represents a steady-state competition for Dam and Fur binding at this site and is not the result of an epigenetic switch.
Overlapping RpiR binding and Dam methylation do not mediate an epigenetic switch.
Next, we sought to determine whether VC1784, which encodes the sialidase NanH in V. cholerae, is controlled by an epigenetic switch. It was previously shown that the growth of V. cholerae in medium containing sialic acid (Neu5Ac) as the sole carbon source induces the expression of this locus (19). To confirm this result and determine how the methylation pattern at PVC1784 changes when this locus is induced, we grew V. cholerae in M9 medium with either glucose or Neu5Ac as the sole carbon source. When grown with sialic acid, we find that VC1784 is induced transcriptionally and that the promoter becomes fully methylated (Fig. 6A and B). Since we have already determined that RpiR is responsible for the undermethylation of this promoter, this indicates that this transcription factor is a repressor of VC1784 and that Neu5Ac may limit the ability of RpiR to bind to PVC1784 (Fig. 4A).
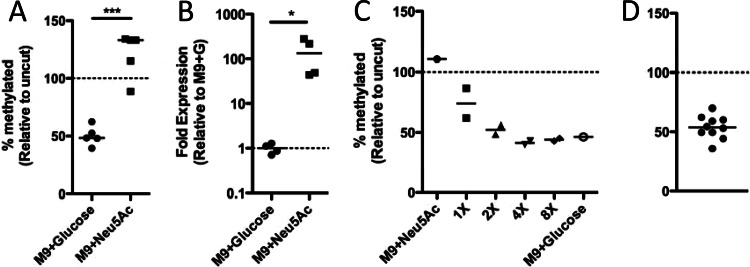
Fig 6.
PVC1784 is not controlled by an epigenetic switch. (A and B) Cells were grown in M9 medium with glucose or sialic acid (Neu5Ac) as the sole carbon source, and the methylation level of PVC1784 was determined by MeSR digestion and qPCR (A), while the expression level of VC1784 was determined by qRT-PCR (B). Significance was determined by two-tailed Student's t test (A) and Mann-Whitney (B). (C) The heritability of PVC1784 methylation was tested by taking cells grown in M9 plus Neu5Ac (fully methylated) and incubating them for the indicated number of generations in M9 medium with glucose as the sole carbon source. (D) Heterogeneity in the methylation of the VC1784 promoter was assessed among single-colony isolates by MeSR digestion and qPCR. Each data point represents an independent biological replicate, and a horizontal line represents the median of each sample. *, P < 0.05; ***, P < 0.001.
Since we have defined a condition when the locus is fully expressed and methylated, as well as a condition when the locus is repressed and undermethylated, we can determine whether a heritable epigenetic switch controls the expression of this locus, as was done for the VCA0063 promoter. To that end, we took cells grown under inducing conditions (M9 plus Neu5Ac), which fully methylates the promoter, and subsequently grew them for multiple generations in medium with glucose as the sole carbon source, which should repress this locus, to determine whether the methylation state of this promoter is heritable. When we performed this analysis, we found that the promoter quickly became undermethylated following growth under repressive conditions (∼4 generations) (Fig. 6C). Furthermore, we did not see heterogeneity in the methylation of this site among single-colony isolates when cells were grown under inducing conditions and subsequently plated and grown in repressive medium (Fig. 6D). Thus, this indicates that, similar to overlapping Fur and Dam sites, overlapping RpiR and Dam methylation do not constitute an epigenetic switch at this site (Fig. 6C and D).
DISCUSSION
In this study, we have described a modified high-throughput method for identifying unmethylated sites. To our knowledge, this is the first study to use second-generation sequencing technology to identify genome-wide undermethylation in a bacterium. Using this approach, we identified the undermethylated sites for Dam and VchM, the two known MTases in V. cholerae.
Many of these undermethylated sites occurred in intergenic regions, and we have identified the transcription factors responsible for the observed methylation pattern for five of these sites. All of these transcription factors were predicted to regulate the identified undermethylated promoters; however, none had previously been confirmed. Binding of transcription factors at these sites prevents the MTase from accessing its target site, and thus, our study provides the first direct evidence for a DNA-protein interaction between these DNA-binding proteins and the promoters they regulate (5). Furthermore, the location of the undermethylated site indicates the location where the transcription factor is bound. This is especially interesting for the K139 prophage, which has two undermethylated Dam sites between PL and PR, which are likely the result of cI repressor binding. Using the location of the undermethylated sites, we identified putative cI operator sites for the K139 prophage. It is tempting to speculate that Dam methylation of these sites affects lytic induction of the phage; however, this hypothesis remains to be tested.
Interestingly, four of the undermethylated intergenic sites are located in operons involved in uptake/catabolism of carbon sources. PVC1784 is in a sialic acid utilization operon, PVC0286/7 is in a gluconate utilization operon, PVC1280/1 is in a chitobiose utilization operon, and PVC1558/9 is upstream from 6-phospho-beta-glucosidase (19, 24). A fifth site (PVCA0063) is upstream from a heme utilization operon, indicating that five undermethylated sites are in regions of the genome involved in scavenging of essential nutrients (iron and carbon) (20). For two of these sites (PVCA0063 and PVC1784), we show that undermethylation correlates with reduced transcription of the locus, suggesting that under normal growth in rich medium, these operons are repressed. It is likely that the other three transcription factors that promote undermethylation are also repressors, because two of these, VC1286 and VC1557, are homologs of the LacI repressor, while VC0289 is a homolog of the gluconate transcriptional repressor from E. coli. Thus, these undermethylated sites have revealed that the iron- and carbon-scavenging loci they reside in are likely tightly repressed during growth in LB. This is consistent with a report indicating that the primary carbon source during growth in LB is amino acids (25). Additionally, it is known that LB contains ∼17 μM iron, making it an iron-rich growth medium (26).
Dam methylation has previously been shown to control gene expression via an epigenetic switch in other bacteria (2). Thus, we determined whether two of the undermethylated Dam sites we identified by methyl HTM-seq promoted a Dam-mediated epigenetic switch. We found that in PVCA0063, methylation reduced the affinity for Fur binding at this site. However, we found no evidence that the methylation state of cells was heritable, indicating that methylation likely did not constitute an epigenetic switch at this site. A similar analysis of PVC1784 also showed that this promoter was not subject to regulation by an epigenetic switch.
It was recently suggested that overlapping Dam and Fur mediate an epigenetic switch that controls the expression of a type VI secretion system in a pathogenic strain of E. coli (21). In that study, Fur binding was shown to prevent methylation of a Dam site in the sci1 promoter both in vitro and in vivo. Additionally, methylation of this Dam site was shown to reduce the affinity of Fur for this promoter in EMSAs. This was used to suggest that this regulatory arrangement mediates an epigenetic switch for the expression of the type VI secretion apparatus. What was not analyzed in that study, however, was a heritability of the methylation state following cell division to prove that regulation of this site truly occurred via an epigenetic switch (21). In our study, we found a very similar regulatory arrangement at PVCA0063. While we found a difference in the mobility of Fur in EMSAs for the methylated and unmethylated VCA0063 promoter, we did not find evidence for the methylation state of cells being heritable, indicating that this arrangement likely does not constitute an epigenetic switch. Thus, it may be pertinent to further test the role of overlapping Fur and Dam in mediating an epigenetic switch for the type VI secretion system in pathogenic E. coli.
In E. coli, the expression of Pap pili is controlled by an epigenetic switch, and phase OFF cells are selected for during growth in rich medium in vitro, likely due to the cost of production of this pilus, while phase ON cells are selected for during infection, likely due to the importance of this pilus for virulence (27–29). The E7946 V. cholerae strain used in this study is a clinical isolate that has been subsequently passaged an estimated five times by colony purification on LB agar and expansion in LB broth, which represents ∼170 generations in vitro. During this extensive growth in vitro, we would expect any analogous phase ON virulence loci that impose a fitness cost, and whose expression requires methylation at their promoters, to shift to the OFF phase (undermethylation). Since we did not identify any such loci, we conclude that there is likely not a virulence locus regulated by a Dam-dependent epigenetic switch in V. cholerae.
This genome-wide screen revealed that there were relatively few intergenic undermethylated Dam sites located in the V. cholerae genome. This is in contrast to E. coli, which has been shown to have 23 undermethylated intergenic Dam sites (5). Additionally, in pathogenic E. coli, some undermethylated Dam sites have been shown to promote an epigenetic switch in gene regulation; however, in V. cholerae, we found no evidence for a similar mechanism of regulation at the undermethylated sites identified (22, 30). Expression of the Dam methylase is essential in V. cholerae, while in E. coli, Dam mutant strains are still viable (6, 31). Thus, it is tempting to speculate that the essential nature of Dam methylase activity in V. cholerae has evolutionarily limited its use in gene regulation.
Dam methylation in promoters has also been shown to affect transcription in multiple systems independent of mediating an epigenetic switch. These include control of transposase expression in Tn10 and Tn5 and the expression of Cre in bacteriophage P1, as well as the expression of a number of genes in E. coli (2). Thus, the undermethylated Dam sites identified in this study may contribute to transcriptional control of the promoters they reside in.
In conclusion, we have identified the undermethylated sites in the genome of a clinically relevant V. cholerae strain using a modified high-throughput technique. Six of these sites were studied further, and for five, the transcription factors responsible for the observed methylation profiles were identified. The locations of the undermethylated sites in the K139 prophage were also used to identify a putative repressor-binding site for cI. Thus, this study has identified and confirmed new DNA-protein interactions, which provides insight into the regulation of the promoters they reside in. Additionally, this approach has identified methylation sites that may play a role in regulating gene expression in V. cholerae, which will be the focus of future studies.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by U.S. National Institutes of Health grant AI055058 (A.C.), and A.C. is a Howard Hughes Medical Institute investigator.
Footnotes
Published ahead of print 15 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02112-12.
REFERENCES
- 1. Nelson EJ, Chowdhury A, Flynn J, Schild S, Bourassa L, Shao Y, LaRocque RC, Calderwood SB, Qadri F, Camilli A. 2008. Transmission of Vibrio cholerae is antagonized by lytic phage and entry into the aquatic environment. PLoS Pathog. 4:e1000187 doi:10.1371/journal.ppat.1000187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Casadesus J, Low D. 2006. Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 70:830–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Low DA, Weyand NJ, Mahan MJ. 2001. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect. Immun. 69:7197–7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ringquist S, Smith CL. 1992. The Escherichia coli chromosome contains specific, unmethylated dam and dcm sites. Proc. Natl. Acad. Sci. U. S. A. 89:4539–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tavazoie S, Church GM. 1998. Quantitative whole-genome analysis of DNA-protein interactions by in vivo methylase protection in E. coli. Nat. Biotechnol. 16:566–571 [DOI] [PubMed] [Google Scholar]
- 6. Egan ES, Waldor MK. 2003. Distinct replication requirements for the two Vibrio cholerae chromosomes. Cell 114:521–530 [DOI] [PubMed] [Google Scholar]
- 7. Banerjee S, Chowdhury R. 2006. An orphan DNA (cytosine-5-)-methyltransferase in Vibrio cholerae. Microbiology 152:1055–1062 [DOI] [PubMed] [Google Scholar]
- 8. Reidl J, Mekalanos JJ. 1995. Characterization of Vibrio cholerae bacteriophage K139 and use of a novel mini-transposon to identify a phage-encoded virulence factor. Mol. Microbiol. 18:685–701 [DOI] [PubMed] [Google Scholar]
- 9. Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 doi:10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oakes CC, La Salle S, Robaire B, Trasler JM. 2006. Evaluation of a quantitative DNA methylation analysis technique using methylation-sensitive/dependent restriction enzymes and real-time PCR. Epigenetics 1:146–152 [DOI] [PubMed] [Google Scholar]
- 11. De Souza Silva O, Blokesch M. 2010. Genetic manipulation of Vibrio cholerae by combining natural transformation with FLP recombination. Plasmid 64:186–195 [DOI] [PubMed] [Google Scholar]
- 12. Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827 [DOI] [PubMed] [Google Scholar]
- 13. Pratt JT, Ismail AM, Camilli A. 2010. PhoB regulates both environmental and virulence gene expression in Vibrio cholerae. Mol. Microbiol. 77:1595–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brunner AL, Johnson DS, Kim SW, Valouev A, Reddy TE, Neff NF, Anton E, Medina C, Nguyen L, Chiao E, Oyolu CB, Schroth GP, Absher DM, Baker JC, Myers RM. 2009. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 19:1044–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu G, Yi N, Absher D, Zhi D. 2011. Statistical quantification of methylation levels by next-generation sequencing. PLoS One 6:e21034 doi:10.1371/journal.pone.0021034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lazinski DW, Camilli A. 2013. Homopolymer tail-mediated ligation PCR: a streamlined and highly efficient method for DNA cloning and library construction. Biotechniques 54:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. doi:10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Esposito D, Wilson JC, Scocca JJ. 1997. Reciprocal regulation of the early promoter region of bacteriophage HP1 by the Cox and Cl proteins. Virology 234:267–276 [DOI] [PubMed] [Google Scholar]
- 19. Almagro-Moreno S, Boyd EF. 2009. Sialic acid catabolism confers a competitive advantage to pathogenic Vibrio cholerae in the mouse intestine. Infect. Immun. 77:3807–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mey AR, Payne SM. 2001. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 42:835–849 [DOI] [PubMed] [Google Scholar]
- 21. Brunet YR, Bernard CS, Gavioli M, Lloubes R, Cascales E. 2011. An epigenetic switch involving overlapping fur and DNA methylation optimizes expression of a type VI secretion gene cluster. PLoS Genet. 7:e1002205 doi:10.1371/journal.pgen.1002205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Braaten BA, Nou X, Kaltenbach LS, Low DA. 1994. Methylation patterns in pap regulatory DNA control pyelonephritis-associated pili phase variation in E. coli. Cell 76:577–588 [DOI] [PubMed] [Google Scholar]
- 23. Baichoo N, Helmann JD. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184:5826–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berg T, Schild S, Reidl J. 2007. Regulation of the chitobiose-phosphotransferase system in Vibrio cholerae. Arch. Microbiol. 187:433–439 [DOI] [PubMed] [Google Scholar]
- 25. Sezonov G, Joseleau-Petit D, D'Ari R. 2007. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189:8746–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abdul-Tehrani H, Hudson AJ, Chang YS, Timms AR, Hawkins C, Williams JM, Harrison PM, Guest JR, Andrews SC. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 181:1415–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Braaten BA, Blyn LB, Skinner BS, Low DA. 1991. Evidence for a methylation-blocking factor (mbf) locus involved in pap pilus expression and phase variation in Escherichia coli. J. Bacteriol. 173:1789–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hagberg L, Hull R, Hull S, Falkow S, Freter R, Svanborg Eden C. 1983. Contribution of adhesion to bacterial persistence in the mouse urinary tract. Infect. Immun. 40:265–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lindberg FP, Lund B, Normark S. 1984. Genes of pyelonephritogenic E. coli required for digalactoside-specific agglutination of human cells. EMBO J. 3:1167–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaminska R, van der Woude MW. 2010. Establishing and maintaining sequestration of Dam target sites for phase variation of agn43 in Escherichia coli. J. Bacteriol. 192:1937–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marinus MG, Morris NR. 1973. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J. Bacteriol. 114:1143–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nesper J, Blass J, Fountoulakis M, Reidl J. 1999. Characterization of the major control region of Vibrio cholerae bacteriophage K139: immunity, exclusion, and integration. J. Bacteriol. 181:2902–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.