Abstract
Salmonella virulence is largely mediated by two type III secretion systems (T3SS) that deliver effector proteins from the bacterium to a host cell; however, the secretion signal is poorly defined. Effector N termini are thought to contain the signal, but they lack homology, possess no identifiable motif, and adopt intrinsically disordered structures. Alternative studies suggest that RNA-encoded signals may also be recognized and that they can be located in the 5′ untranslated leader sequence. We began our study by establishing the minimum sequence required for reporter translocation. Untranslated leader sequences predicted from 42 different Salmonella effector proteins were fused to the adenylate cyclase reporter (CyaA′), and each of them was tested for protein injection into J774 macrophages. RNA sequences derived from five effectors, gtgA, cigR, gogB, sseL, and steD, were sufficient for CyaA′ translocation into host cells. To determine the mechanism of signal recognition, we identified proteins that bound specifically to the gtgA RNA. One of the unique proteins identified was Hfq. Hfq had no effect upon the translocation of full-length CigR and SteD, but injection of intact GtgA, GogB, and SseL was abolished in an hfq mutant, confirming the importance of Hfq. Our results demonstrated that the Salmonella pathogenicity island 2 (SPI-2) T3SS assembled into a functional apparatus independently of Hfq. Since particular effectors required Hfq for translocation, Hfq-RNA complexes may participate in signal recognition.
INTRODUCTION
Type III secretion systems (T3SS) are employed by numerous Gram-negative pathogens to facilitate infection and are widely considered to be a potential target for antimicrobial drug development (1). Because of their ability to secrete proteins, they have also been studied for therapeutic and industrial applications, such as delivering protective antigens and protein purification (2, 3). A better understanding of the secretion signal could prove useful in these endeavors. Salmonella enterica serovar Typhimurium is an excellent model because it is an important intracellular pathogen with an extensive body of literature describing its T3SS and effector repertoire. S. Typhimurium encodes two T3SS on Salmonella pathogenicity islands 1 and 2 (SPI-1 and SPI-2, respectively). In a mouse model of infection, the SPI-1 T3SS is required for the invasion of nonphagocytic cells and dissemination from the intestine, whereas the SPI-2 T3SS promotes intracellular replication and is essential for systemic disease. Effector activities upon host cell targets mediate these processes (4, 5).
The 30 N-terminal amino acids of an effector are generally sufficient for secretion. However, effector N termini lack an obvious consensus sequence and are intrinsically disordered based on structural studies (6, 7). Intrinsic disorder has been proposed to function as the signal (1), but there are countervailing arguments. It is estimated that approximately 40% of all soluble proteins encoded by enteric bacteria possess an intrinsically disordered N terminus (8, 9), yet only a small subset of proteins have been identified as type III substrates (4, 5). Alternatively, effector-chaperone complexes have been proposed to function as the signal. Chaperones maintain effectors in an unfolded state prior to translocation, but unlike effectors, chaperones remain in the bacterial cytoplasm (10). While intrinsic disorder and chaperone interaction describe properties of the signal, RNA sequence may be another component.
Several effectors encode RNA signals. Elegant experiments using the Yersinia effectors YopE, YopN, and YopQ (YopENQ) demonstrated that N-terminal frameshifts, which dramatically altered the amino acid sequence, had little effect upon secretion. Furthermore, silent mutations in the codon wobble positions of yopN and yopQ that altered the RNA, but not the amino acid sequence, blocked secretion (11–15). However, the YopENQ experiments evaluated secretion into media as opposed to translocation into cells. In the cases of YopE and YopQ, the RNA signal is located within the 15 N-terminal amino acids and is sufficient for secretion into media (11, 16), but chaperone binding to a downstream domain is required for injection into animal cells (15–17). Only one Salmonella effector has been tested for the presence of an RNA signal, SopE, a guanine nucleotide exchange factor that induces membrane ruffling and actin rearrangements (18). Using an experimental strategy similar to that used in the YopENQ analyses mentioned above, SopE secretion required an N-terminal amino acid sequence (19).
Flagella are evolutionarily related to T3SS and may also utilize RNA signals. Unlike with the RNA signals encoded within Yersinia effectors, 173 bp of sequence upstream of the Escherichia coli fliC start codon was sufficient for flagellar secretion of two proteins into media: E. coli enolase and PebI from Campylobacter jejuni (2). The fliC gene encodes the flagellum filament protein (20), enolase is a glycolysis enzyme (21), and PebI is a major cell adherence molecule utilized by C. jejuni during infection (22).
Given the conflicting evidence on the nature of the T3S signal, we hypothesized that T3SS may recognize multiple signal types. To approximate one type of signal, we focused on untranslated RNA (UTR). As with the fliC RNA signal, we found five unique 25-bp RNA sequences sufficient for reporter translocation into animal cells by S. Typhimurium. This is the first report of RNA facilitating protein injection, and our data provide insight into how RNA may direct particular effector transcripts to the secretory apparatus.
MATERIALS AND METHODS
Culture conditions, strains, and plasmids.
All bacteria were grown in Luria Bertani (LB) broth at 37°C on a shaker set to 300 rpm. Carbenicillin and chloramphenicol were used at 100 and 30 μg/ml, respectively. J774 cells were cultured at 37°C in 5% CO2 using Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, sodium pyruvate, sodium bicarbonate, and nonessential amino acids. The strains and plasmids used in this study are listed in Table S1 in the supplemental material. Primers are listed in Table S2. For inserts of ≤50 bp, complementary oligonucleotides were duplexed in a thermocycler set to 95°C for 1 min and cooled thereafter by 1°C per minute to room temperature. Duplexing reaction mixtures contained 1 nmol of primers in 10 mM Tris-HCl, 1 mM EDTA, 50 mM NaCl, pH 8.0. Flanking 5′ XbaI and 3′ PvuII restriction sites enabled directional cloning into pMJW1753 (23) cut with XbaI and SmaI. Point mutants were generated with the QuikChange site-directed mutagenesis kit (Stratagene). Fusions were verified by automated sequencing according to LT2 (20) or 14028 (24) annotations and transformed into any of four isogenic backgrounds: wild-type (WT) 14028 and the ssaK::cat, invA::cat, and Δhfq mutants.
CyaA′ expression.
Adenylate cyclase reporter (CyaA′) fusions were expressed from the lac promoter encoded by pMJW1753 (23), which is constitutively expressed in Salmonella. To evaluate protein expression from LB broth, cultures were pelleted, resuspended in phosphate-buffered saline (PBS), normalized to an optical density at 600 nm (OD600), and lysed in Laemmli sample buffer, and a volume corresponding to ∼105 bacteria was resolved by SDS-PAGE. To assess expression from J774 macrophages, cells were infected under SPI-2 conditions and lysed in a buffered solution containing 50 mM HEPES, pH 7.4, 1% Triton X-100, 10% glycerol, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, and protease inhibitor cocktail (Roche). Lysates were precipitated with trichloroacetic acid (TCA), suspended in Laemmli sample buffer, and resolved by SDS-PAGE. Expression was evaluated by Western blotting using anti-CyaA′ antibody (Santa Cruz; 1:1,000), and loading was confirmed by probing for DnaK (Assay Designs; 1:10,000). To calculate relative expression levels, band density was measured with Fiji (25), values were normalized to that for DnaK, and the ratios between isogenic backgrounds were compared. Statistical significance was calculated using both the Student t test and analysis of variance (ANOVA).
CyaA′ translocation assays.
CyaA′ translocation assays were performed as previously described (23). To test for SPI-1 translocation, overnight cultures were diluted 1:33 in LB broth and incubated at 37°C for 3 h on a shaker to obtain late-log-phase cultures. J774 cells were infected at a multiplicity of infection (MOI) of 50 for 1 h. For SPI-2 translocation, bacteria were grown in LB broth overnight to late stationary phase, and J774 cells were infected at an MOI of 250 for 6 to 8 h. A structural mutant, i.e., the invA::cat mutant for SPI-1 and the ssaK::cat mutant for SPI-2, was tested under each of these conditions to verify translocation through the appropriate apparatus (23). Translocation was measured by cyclic AMP (cAMP) enzyme-linked immunosorbent assay (ELISA) (Assay Designs). An effector was deemed secreted by the SPI-1 or SPI-2 T3SS if the cAMP responses between the WT and the corresponding structural mutant were approximately 5-fold different. Error bars represent standard deviations.
RNA aptamer affinity chromatography.
Twenty-five milliliters of overnight LB broth cultures were resuspended in 5 ml lysis buffer containing 200 mM NaCl, 1% Triton X-100, 10 mM MgCl2, 10 mM HEPES, pH 7, and Roche protease inhibitor cocktail. Bacteria were lysed in a French press and centrifuged at 13,000 × g for 15 min at 4°C, and the soluble supernatant was transferred to fresh tubes. Protein concentration was measured by determining the A280, and lysates were blocked with 10 μg egg white avidin (EMD Chemicals) and 10 mg Saccharomyces cerevisiae RNA (Sigma) per mg of soluble protein for 20 min at 4°C. Lysates were centrifuged again, and clarified supernatants were used for affinity chromatography. RNA was synthesized in a runoff in vitro transcription reaction (Epicentre) using a PCR template encoding a 5′ T7 promoter, the desired UTR, and the RNA aptamer (see Table S6 in the supplemental material). RNA was purified by ammonium acetate precipitation, diluted in 10 mM MgCl2, 10 mM HEPES, pH 7, heated to 65°C for 5 min in a thermocycler, and renatured by slowly cooling the mixture to room temperature. For each sample, a 50-μl bed volume of streptavidin beads (Pierce) was washed twice in lysis buffer, tethered to 50 μg of renatured RNA for 20 min at 4°C, and washed two times with lysis buffer to remove unbound RNA. Affinity chromatography was performed in the presence of SUPERas-In (Ambion) using 8 mg of soluble protein for 1.5 h at 4°C. Beads were washed five times with lysis buffer, and bound proteins were eluted with Laemmli sample buffer.
MS.
Proteins were resolved by SDS-PAGE and stained using a Pierce silver stain kit (Thermo Scientific). Stained regions were excised and destained according to the manufacturer's protocol with a modified wash solution containing 40% acetonitrile, 200 mM ammonium bicarbonate. Trypsin-digested gel slices were separated by liquid chromatography followed by tandem mass spectrometry (MS/MS) using an LTQ ion trap mass spectrometer (Thermo) (26). With SEQUEST, peptides were identified by referencing the tandem mass spectra against the S. Typhimurium 14028 FASTA file. Results were filtered using the MS-GeneratingFunction (MS-GF), a software tool that assigns P values to peptide identifications (27). The number of peptide identifications from each protein (spectral count) was used to measure relative abundance. To increase stringency, the analysis was limited to proteins with at least two unique peptides.
qRT-PCR.
J774 cells were infected with late-stationary-phase bacteria as described for the CyaA′ translocation assays. Cells were stabilized with RNAprotect bacterial reagent (Qiagen), and RNA was isolated with a miniprep kit (Qiagen) and by on-column DNase digestion (Qiagen). Total RNA was reverse transcribed with iScript (Bio-Rad), and quantitative reverse transcription-PCR (qRT-PCR) was performed using SYBR green reagent (Applied Biosystems) and a StepOnePlus real-time PCR system (Applied Biosystems). Comparative threshold cycle (CT) values from cyaA′ were normalized to gyrB values, and expression levels between the wild-type and Δhfq backgrounds were compared [ΔΔCT = ΔCT hfq mutant − ΔCT WT = (CT hfq cyaA′ mutant − CT hfq gyrB mutant) − (CT WT cyaA′ strain − CT WT gyrB strain); relative quantity was equal to 2−ΔΔCT]. Statistical significance was calculated using both the Student t test and ANOVA.
RESULTS
Screen for RNA signals.
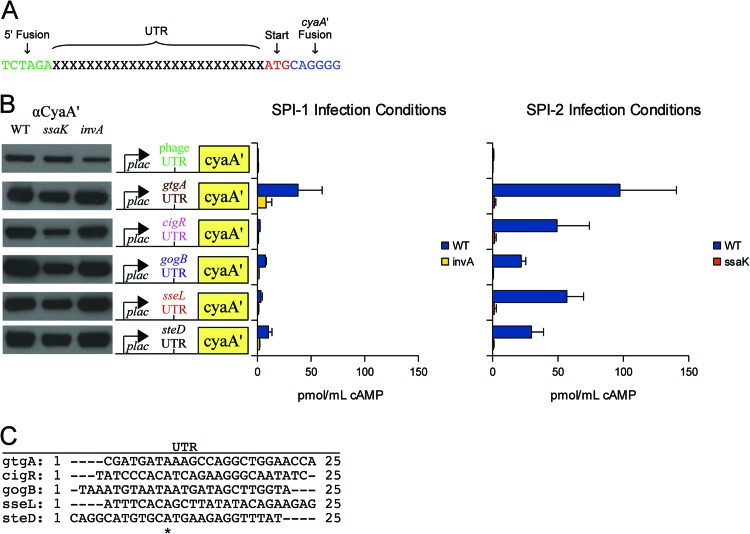
Flagellar T3S signals were recently shown to reside within untranslated RNA (2), and this evidence prompted us to screen for similar signals in S. Typhimurium. We employed a minimalist approach to identify UTR sequences that mediated reporter translocation. Twenty-five-base-pair UTRs from 42 Salmonella type III effectors were fused directly to cyaA′ (Fig. 1A). Sequences of this length were chosen to ensure that the ribosome-binding site (Shine-Dalgarno sequence) was present. Constructs were constructed with the low-copy-number plasmid pMJW1753 (23) and were designed to omit any effector amino acid sequence except for the start codon (Fig. 1A). Expression was confirmed by Western blotting using an antibody against CyaA′ (Fig. 1B), and translocation was evaluated by measuring cAMP levels from infected J774 macrophages (Fig. 1B). For each construct, a type III structural mutant was tested concurrently to verify translocation through the appropriate apparatus: the invA::cat mutant for SPI-1 and the ssaK::cat mutant for SPI-2 (23). We found that the leader sequences of gtgA, cigR, gogB, sseL, and steD were sufficient for CyaA′ translocation into J774 macrophages under SPI-2 infection conditions (Fig. 1B). We also screened each of these fusions under SPI-1 infection conditions, but only the gtgA fusion was injected (Fig. 1B). Levels of CyaA′ expression were similar in all backgrounds tested (Fig. 1B), and results from the 37 nontranslocated fusions are listed in Table S3 in the supplemental material. The reported functions, activities, and cellular targets of intact SseL, GtgA, CigR, GogB, and SteD are summarized in Table S4. Previous studies demonstrated that full-length SseL, GtgA, CigR, GogB, and SteD were translocated by SPI-2 (26, 28, 29), and the secretion profiles of the UTR fusions were generally consistent with SPI-2 utilization. We also analyzed bacteria expressing the UTR fusions by mass spectrometry to confirm that the predicted translational start sites were correct. No UTR-encoded peptides were observed (Table S5), indicating that the start sites were accurate. Taken together, these results demonstrated that RNA signals mediated protein translocation.
Fig 1.
RNA leader sequences sufficient for CyaA′ translocation. (A) Construction of UTR::cyaA′ fusions. UTR fusions were designed to express CyaA′ without any amino acid sequence corresponding to a type III effector. Each UTR encodes a Shine-Dalgarno sequence required for ribosome binding and translation. (B) Translocated UTR::cyaA′ fusions. Forty-two effector UTRs were fused directly to cyaA′ and screened for injection into J774 macrophages. Five were found to be sufficient for CyaA′ translocation. The data are summarized here, and the complete data set is provided in Table S1 in the supplemental material. (Left) Western blots showing CyaA′ expression from LB cultures. Samples were normalized to an OD600, and ∼105 bacteria were loaded into each lane. (Middle) Construct map showing the plac promoter, which is constitutive in Salmonella, the UTR to initiate translation, and the CyaA′ reporter. (Right) Bacteria were induced for SPI-1 and SPI-2 expression and used to infect J774 macrophages. Translocation was evaluated by cAMP ELISA. The invA and ssaK mutants are functional SPI-1 and SPI-2 mutants, respectively (23). (C) ClustalX alignment of UTRs sufficient for CyaA′ translocation. *, aligned residues.
General utility.
As shown in Fig. 1B, the gtgA fusion gave the highest cAMP response from infected macrophages. It was therefore used to characterize the RNA signals that we identified. To our knowledge, the only other published example of a UTR-encoded secretion signal is E. coli fliC, which encodes the flagellin protein. In this case, an upstream sequence of 173 bp mediated secretion of both PebI and enolase (2). Although the gtgA leader facilitated CyaA′ translocation (Fig. 1B), an enolase fusion was not injected into host cells (see Fig. S1 in the supplemental material). Thus, UTRs that mediate the translocation of one protein may not necessarily target a different one, indicating that sequence downstream of the start codon affects RNA signal functionality. To gain additional insight, we analyzed the gtgA RNA in further detail.
Dissection of the gtgA RNA.
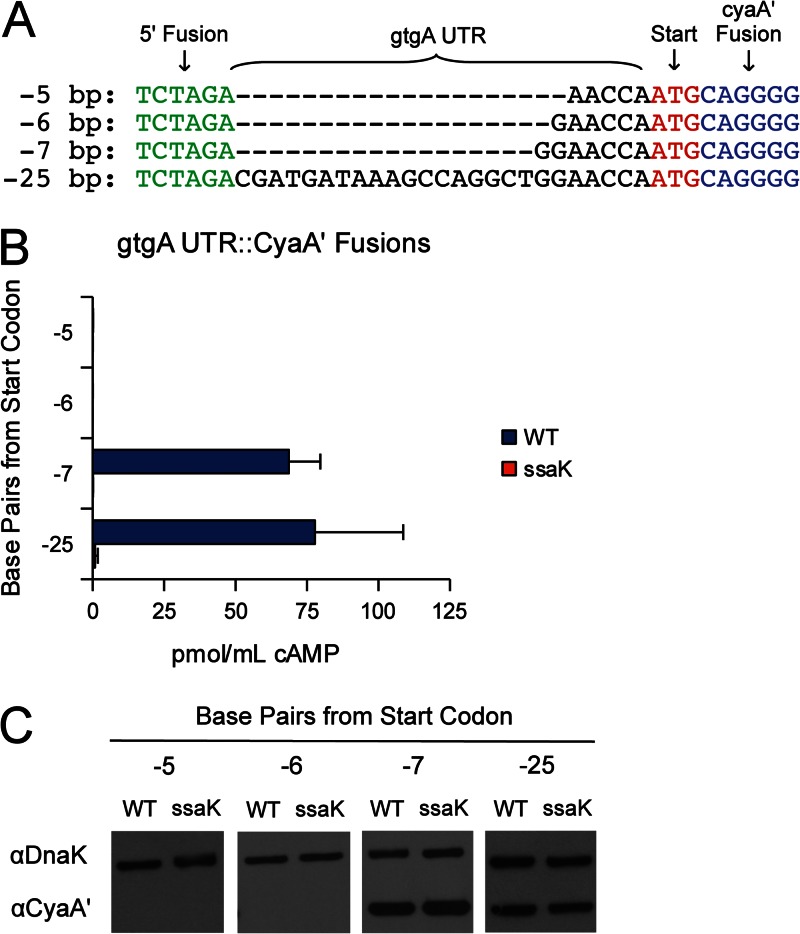
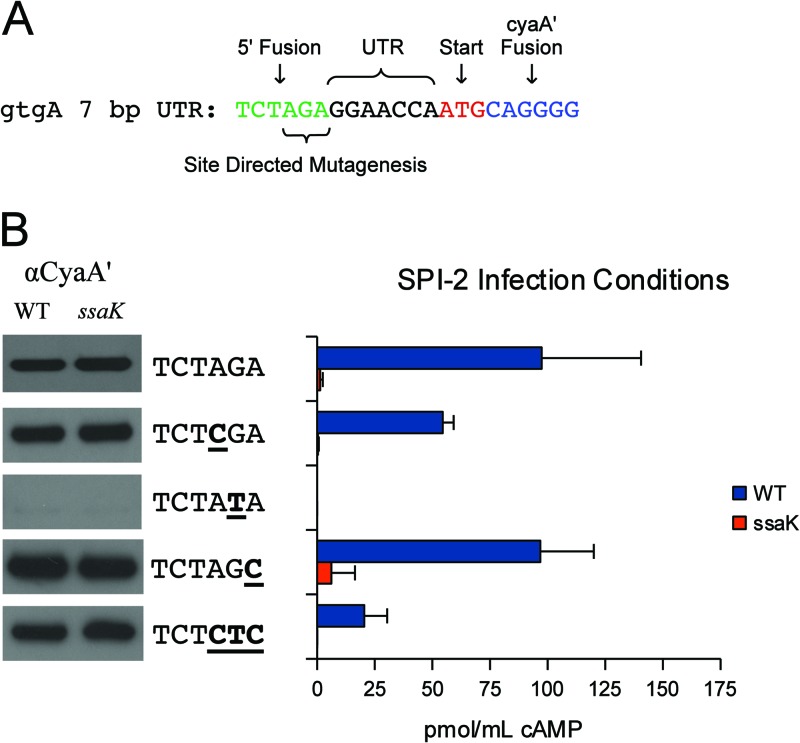
Starting with the original gtgA construct, containing 25 bp of upstream sequence, truncations were fused to cyaA′ for analysis (Fig. 2A). Fusions containing less than 6 bp of upstream sequence were not expressed, so no conclusion could be drawn from these constructs (Fig. 2B). However, a 7-bp sequence was sufficient for both the expression and translocation of CyaA′ (Fig. 2B). To simplify construction, all fragments were cloned with the same restriction sites. To determine if the 5′ XbaI site (5′-TCTAGA-3′) introduced an essential sequence, strains with single nucleotide transversions were constructed and tested (Fig. 3A). All of these point mutants were expressed and secreted into J774 macrophages, with the exception of a mutant with a G-to-T transversion at position 5, resulting in 5′-TCTATA-3′. Data from this mutant could not be interpreted because no expression was observed (Fig. 3B). Nevertheless, the collective data indicate that the 5′ cloning site had a nominal impact upon secretion and that as little as 7 bp of untranslated RNA imparted CyaA′ translocation. We next attempted to identify the molecular mechanisms by which RNA signals are recognized.
Fig 2.
Minimal gtgA UTR sequence required for CyaA′ translocation. (A) ClustalX alignment of gtgA UTR truncations. (B) Translocation of truncated gtgA UTR::CyaA′ fusions. Bacteria were induced for SPI-2 expression and used to infect J774 macrophages. Translocation was evaluated by cAMP ELISA. The ssaK mutant is a functional SPI-2 mutant. (C) Western blots showing CyaA′ expression from LB cultures. Samples were normalized to an OD600, and ∼105 bacteria were loaded into each lane.
Fig 3.
Point mutant analysis. (A) Description of the 7-bp gtgA UTR fusion. The 5′ XbaI site was mutated by site-directed mutagenesis to incorporate a series of transversions. (B) Expression and translocation of UTR point mutants. (Left) Western blots showing CyaA′ expression from LB cultures. Samples were normalized to an OD600, and ∼105 bacteria were loaded into each lane. (Middle) Transversions incorporated into the XbaI site are underlined. (Right) Bacteria were induced for SPI-2 expression and used to infect J774 macrophages. Translocation was evaluated by cAMP ELISA. The ssaK mutant is a functional SPI-2 mutant.
Sequence motifs and RNA secondary structure.
Sequence comparison of the five leader sequences by ClustalX alignment did not identify an underlying motif (Fig. 1C). We also evaluated secondary structure because it has been proposed as an alternative means of RNA signal identification (11). However, RNAfold structural analysis (30) was equivocal because a variety of stem-loop structures were present in both signals and nonsignals (see Fig. S2 in the supplemental material). Thus, neither sequence homology nor secondary structure clearly distinguished the RNA signals that we identified. These characteristics indicated that an RNA-binding protein might be involved.
Identification of RNA-binding proteins.
RNA-binding proteins are difficult to identify because RNA is a flexible and unstable molecule to which many proteins adhere nonspecifically. However, RNA aptamers that specifically bind to a solid matrix have been used to purify interacting proteins from crude lysates. In our case, we used a tRNA aptamer that binds to streptavidin (31). We fused the 25-bp gtgA leader to the aptamer and, as a control, the spvD UTR, which did not translocate its CyaA′ fusion (see Table S1 in the supplemental material). RNA was synthesized in vitro, immobilized to streptavidin beads, and suspended in a crude S. Typhimurium lysate to which avidin, yeast RNA, and RNase inhibitors were added to minimize nonspecific interactions and degradation. Bound proteins were washed extensively and identified by MS. Protein identifications unique to the gtgA RNA are summarized in Table 1, and the full data set is catalogued in Table S6 in the supplemental material. MS analysis identified only two nonribosomal proteins specific to the gtgA RNA: TruD and Hfq (Table 1). TruD was likely a nonspecific interaction because it is a pseudouridine synthase, the product of which is a modified nucleoside and constituent of tRNA and rRNA (32). On the other hand, Hfq is an abundant RNA chaperone that regulates numerous physiological processes and at least 20% of the genes borne by S. Typhimurium (33, 34). Since Hfq is a global regulator and because we identified it in complex with the gtgA RNA, we tested an hfq mutant to determine if it altered the secretion and expression of the five UTR::CyaA′ fusions that we identified.
Table 1.
MS identification of gtgA RNA-binding proteinsa
| LT2 locus | Gene | Mass (kDa) | Protein description | No. of unique peptides | Total no. of peptides |
|---|---|---|---|---|---|
| STM4361 | hfq | 11.13 | RNA-binding protein | 3 | 4 |
| STM2928 | truD | 39.33 | tRNA pseudouridine synthase | 2 | 2 |
Proteins unique to the gtgA RNA were affinity purified and analyzed by liquid chromatography-MS/MS. Values are spectral counts, i.e., the numbers of times a peptide corresponding to a protein was identified.
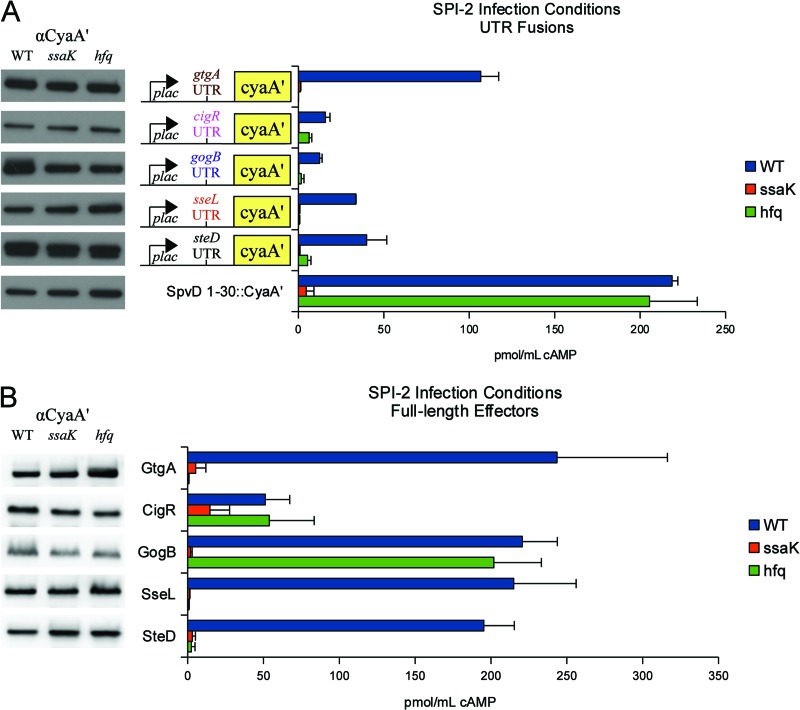
Regulation of RNA T3S signals by Hfq.
An hfq mutant blocked the translocation of the gtgA, cigR, gogB, sseL, and steD fusions (Fig. 4A). In S. Typhimurium, hfq mutants display pleiotropic phenotypes, such as the outer membrane stress responses, motility defects, and greatly attenuated virulence (35, 36). Thus, we also tested for a functional secretion apparatus using SpvD containing amino acids 1 to 30 fused to CyaA′ (SpvD1–30::CyaA′), for which the leader sequence was insufficient for CyaA′ translocation (see Table S3 in the supplemental material). As shown in Fig. 4A, an hfq mutant secreted SpvD1–30::CyaA′ into J774 macrophages, confirming a functional SPI-2 T3SS. Consistent with these results, Hfq was also required for translocation of the 7-bp gtgA UTR::cyaA′ fusion shown in Fig. 2B (and see Fig. S3).
Fig 4.
Regulation of RNA T3S signals by Hfq. (A) Hfq-dependent translocation of the UTR::cyaA′ fusions. SpvD1–30::CyaA′ was a positive control to verify the functionality of the SPI-2 T3SS. (Left) Western blots showing CyaA′ expression from LB cultures. Samples were normalized to an OD600, and ∼105 bacteria were loaded into each lane. (Middle) UTR::cyaA′ fusions. (Right) Bacteria were induced for SPI-2 expression and used to infect J774 macrophages. Translocation was evaluated by cAMP ELISA. The ssaK mutant is a functional SPI-2 mutant. (B) Hfq-dependent translocation of intact S. Typhimurium 14028 effectors. Constructs were evaluated for expression and secretion into J774 macrophages as described above.
Hfq regulation typically occurs by interaction with small regulatory RNAs, by modulation of transcript stability, or by repression of translation (34, 37). However, Hfq did not significantly affect protein expression in LB broth cultures (Fig. 4A) or in infected J774 cells (Table 2). To determine if Hfq regulated transcript stability, we also evaluated RNA levels from infected J774 cells. Only the gogB fusion had increased RNA levels (∼2-fold) as a result of hfq mutation (Table 2). Since Hfq did not affect protein expression and because only one construct was slightly regulated at the RNA level, we next evaluated RNA signal utilization by intact effectors.
Table 2.
Relative CyaA′ expression from infected J774 cellsa
| Effector locus | Δhfq mutant vs WT |
|||
|---|---|---|---|---|
|
cyaA′ transcript level |
CyaA′ protein level |
|||
| UTR fusion | Intact effector | UTR fusion | Intact effector | |
| gtgA | 1.4 ± 1.8 | 1.4 ± 1.2 | 0.8 ± 0.3 | 0.2 ± 0.1* |
| cigR | 1.7 ± 1.1 | 1.2 ± 1.0 | 4.5 ± 6.3 | 0.6 ± 0.1 |
| gogB | 2.1 ± 0.6* | 0.9 ± 0.4 | 1.5 ± 0.2 | 0.7 ± 0.2 |
| sseL | 1.9 ± 0.9 | 0.5 ± 0.3 | 3.1 ± 3.6 | 0.5 ± 0.1 |
| steD | 1.7 ± 1.1 | 0.4 ± 0.3* | 0.9 ± 0.1 | 0.5 ± 0.1 |
For cyaA′ transcript levels, RNA was collected from infected J774 macrophages and analyzed by RT-PCR to determine cyaA′ RNA levels in the hfq mutant relative to WT bacteria. For CyaA′ protein levels, lysates from infected J774 cells were evaluated by Western blotting for CyaA′ and DnaK. Representative blots are shown in Fig. S4 in the supplemental material. Protein levels in the hfq mutant are relative to levels in WT bacteria. Standard deviation denotes the error of results from at least three independent experiments. Both ANOVA and the Student t test were used to calculate significant differences.
indicates a P of ≤0.05.
Hfq-dependent translocation of intact effectors.
To determine if RNA signals directed translocation of full-length effectors, intact GtgA, CigR, GogB, SseL, and SteD were fused to CyaA′ and tested for translocation. As with the UTR fusions, Hfq was required for the injection of GtgA, SseL, and SteD (Fig. 4B). Hfq had no impact upon protein expression from LB broth cultures (Fig. 4B), but some differences were observed from infected J774 cells. GtgA protein levels were approximately 5-fold lower (Table 2 and see Fig. S4 in the supplemental material), but no effect was observed with intact SseL and SteD. Overall, Hfq modestly regulated RNA stability and the translation of full-length proteins (Table 2 and Fig. 4B), and these findings were consistent with the actions of the UTR fusions described in Table 2 and Fig. 4A. Since Hfq was required for translocation of certain effectors but had only modest effects upon RNA stability and protein expression, the data suggest that Hfq-RNA complexes may play a role in signal recognition.
DISCUSSION
The type III secretion signal has been surprisingly difficult to identify, but it is widely assumed to reside within the effector's N terminus (1). However, we and other labs have found that RNA can play a critical role in directing T3S. In an effort to define the minimal signal required for T3S, we developed a simple, well-defined system in which levels of expression and translocations could be directly compared. Using this approach, we found that untranslated RNA could be fused directly to the cyaA′ reporter to facilitate injection into host cells. We identified RNA sequences corresponding to five Salmonella effectors that were sufficient for CyaA′ translocation: gtgA, cigR, gogB, sseL, and steD.
Mechanism of Hfq regulation.
The mechanism facilitating signal recognition may be dependent upon the RNA chaperone Hfq. Hfq binding sites are short AU-rich sequences often interrupted by cytidines and guanosines and are influenced by flanking stem-loop structures (37). These attributes do in fact describe all five of the RNA sequences that we identified: no discernible motif (Fig. 1C), an AT-rich motif (61.5% AT [on average]), and flanking stem-loop structures (see Fig. S2 in the supplemental material). Hfq was purified from crude extracts using the gtgA RNA (Table 1), and Hfq was required for the translocation of all five UTR fusions that we characterized (Fig. 4A), suggesting an Hfq-RNA complex.
Several documented pathways have been described for Hfq regulation: interaction with regulatory small RNA to facilitate interaction with their targets, modulation of RNA decay, and inhibition of translation (34, 37). However, Hfq had modest effects upon translation (Fig. 4, Table 2, and Fig. S4 in the supplemental material) and a slight impact on RNA stability (Table 2), and the secretion block was not a consequence of a defective apparatus (Fig. 4). An alternative mechanism by which Hfq directs some transcripts to the cell periphery is supported by Hfq interactions with other proteins and ribosomes and localization to the bacterial inner membrane (34, 37). To investigate this possibility, we examined deep sequencing data from a previously published Hfq-RNA study (38). Sequences corresponding to gtgA, cigR, gogB, steD, and sseL were not identified in complex with Hfq. However, this analysis was limited to bacteria grown to early stationary phase. GtgA, CigR, GogB, SteD, and SseL are restricted to the SPI-2 T3SS (Table S2), which is induced by late-stationary-phase growth. Differences in the growth conditions may account for the disparity. Additional experiments are required to determine if Hfq directly associates with the transcripts that we identified and if Hfq interacts with the secretory apparatus.
Multiple secretion signals.
Intact GtgA, SseL, and SteD required Hfq for translocation (Fig. 4B), suggesting that RNA signals may be utilized during infection. However, the full-length effectors yielded higher cAMP responses than their respective UTR::CyaA′ fusions (Fig. 4). In general, the differences ranged between 2- and 6-fold and were functionally comparable (Fig. 4). GogB was a notable exception. The UTR and full-length fusions had an approximate 12-fold difference in cAMP responses (Fig. 4). Overall, these observations indicate that RNA is not the only signal recognized. Other signals, such as protein sequence or chaperones, likely contribute. As mentioned previously, Yersinia YopE and YopQ contain at least two signals: an initial RNA sequence that permits secretion into media and a subsequent, downstream signal for translocation into cells (11–15). One important distinction between this work and the Yop experiments is that the Yersinia studies focused upon the sequence encoding the N-terminal amino acid. Our data indicate that RNA signals may extend into the untranslated leader. Signal complexity is further complicated by other attributes, such as chaperones. For example, when the chaperone-binding domains of the Salmonella effectors SptP and SopE were deleted, flagellar secretion resulted, indicating that the chaperones provided specificity for one apparatus versus another (39). Similarly, several of the RNA signals that we tested were restricted for SPI-2 translocation (Fig. 2B), suggesting that RNA signals also convey apparatus specificity.
Signal classification and evolution.
The variety of secretion signals documented in the literature indicates that different effectors encode different signals, which may include RNA, amino acid sequence, and chaperone interactions. Multiple signals may improve translocation efficiency or, as Galán and Wolf-Watz proposed, may be important in the timing of T3S so that effector activities are coordinated (1). Conversely, some signals may exert dominance over others. Since identification of a single, defined motif may be impossible, it may be useful to subdivide effectors into groups based upon their signal types. How and why these assorted signals evolved is unknown. T3SS and flagella are ancient, and both presumably preceded the evolution of multicellular eukaryotes hundreds of millions of years ago (40). Like many other evolutionary changes, multiple secretion signals may simply reflect what worked in different organisms at different times in the distant past. Alternatively, signal flexibility may provide a platform by which a gene encoding a weak signal may evolve into an effector with a more complex or efficient signal.
Supplementary Material
ACKNOWLEDGMENTS
Support for this work was provided by the National Institute of Allergy and Infectious Diseases, NIH/DHHS, through interagency agreement Y1-A1-8401-01, by NIH/NIAID grant A1022933-22A1 to F.H., and the National Institute of General Medical Sciences (grant GM094623). This work used instrumentation and capabilities developed with support from the National Center for Research Resources (grant RR 018522 to R.D.S.) and the U.S. Department of Energy's Office of Biological and Environmental Research (DOE/BER).
Proteomic analyses were performed in the Environmental Molecular Sciences Laboratory, a DOE/BER national scientific user facility on the Pacific Northwest National Laboratory (PNNL) campus in Richland, WA. PNNL is a multiprogram national laboratory operated by Battelle for the DOE under contract DE-AC05-76RL01830. Mass spectrometry results are available via sysbep.org and omics.pnl.gov.
Footnotes
Published ahead of print 8 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00024-13.
REFERENCES
- 1. Galan JE, Wolf-Watz H. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444:567–573 [DOI] [PubMed] [Google Scholar]
- 2. Majander K, Anton L, Antikainen J, Lang H, Brummer M, Korhonen TK, Westerlund-Wikstrom B. 2005. Extracellular secretion of polypeptides using a modified Escherichia coli flagellar secretion apparatus. Nat. Biotechnol. 23:475–481 [DOI] [PubMed] [Google Scholar]
- 3. Russmann H. 2003. Bacterial type III translocation: a unique mechanism for cytosolic display of heterologous antigens by attenuated Salmonella. Int. J. Med. Microbiol. 293:107–112 [DOI] [PubMed] [Google Scholar]
- 4. Heffron F, Niemann G, Yoon H, Kidwai AS, Brown RN, McDermott JE, Smith RD, Adkins JN. 2011. Salmonella-secreted virulence factors, p 187–223 In Porwallik S. (ed), Salmonella: from genome to function. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 5. McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V. 2009. Salmonella takes control: effector-driven manipulation of the host. Curr. Opin. Microbiol. 12:117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buchko GW, Niemann G, Baker ES, Belov ME, Smith RD, Heffron F, Adkins JN, McDermott JE. 2010. A multi-pronged search for a common structural motif in the secretion signal of Salmonella enterica serovar Typhimurium type III effector proteins. Mol. Biosyst. 6:2448–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gazi AD, Charova SN, Panopoulos NJ, Kokkinidis M. 2009. Coiled-coils in type III secretion systems: structural flexibility, disorder and biological implications. Cell. Microbiol. 11:719–729 [DOI] [PubMed] [Google Scholar]
- 8. Goulding CW, Perry LJ. 2003. Protein production in Escherichia coli for structural studies by X-ray crystallography. J. Struct. Biol. 142:133–143 [DOI] [PubMed] [Google Scholar]
- 9. Noordewier M, Brown J. 2002. Unfolding the secrets of the Salmonella genome to aid drug development. Trends Pharmacol. Sci. 23:397–399 [DOI] [PubMed] [Google Scholar]
- 10. Stebbins CE, Galan JE. 2001. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 414:77–81 [DOI] [PubMed] [Google Scholar]
- 11. Anderson DM, Schneewind O. 1997. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278:1140–1143 [DOI] [PubMed] [Google Scholar]
- 12. Anderson DM, Schneewind O. 1999. Yersinia enterocolitica type III secretion: an mRNA signal that couples translation and secretion of YopQ. Mol. Microbiol. 31:1139–1148 [DOI] [PubMed] [Google Scholar]
- 13. Ramamurthi KS, Schneewind O. 2002. Yersinia enterocolitica type III secretion: mutational analysis of the yopQ secretion signal. J. Bacteriol. 184:3321–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schesser K, Frithz-Lindsten E, Wolf-Watz H. 1996. Delineation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic cellular membranes. J. Bacteriol. 178:7227–7233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sory MP, Boland A, Lambermont I, Cornelis GR. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. U. S. A. 92:11998–12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheng LW, Anderson DM, Schneewind O. 1997. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol. Microbiol. 24:757–765 [DOI] [PubMed] [Google Scholar]
- 17. Jackson MW, Day JB, Plano GV. 1998. YscB of Yersinia pestis functions as a specific chaperone for YopN. J. Bacteriol. 180:4912–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rudolph MG, Weise C, Mirold S, Hillenbrand B, Bader B, Wittinghofer A, Hardt WD. 1999. Biochemical analysis of SopE from Salmonella typhimurium, a highly efficient guanosine nucleotide exchange factor for RhoGTPases. J. Biol. Chem. 274:30501–30509 [DOI] [PubMed] [Google Scholar]
- 19. Karavolos MH, Roe AJ, Wilson M, Henderson J, Lee JJ, Gally DL, Khan CM. 2005. Type III secretion of the Salmonella effector protein SopE is mediated via an N-terminal amino acid signal and not an mRNA sequence. J. Bacteriol. 187:1559–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856 [DOI] [PubMed] [Google Scholar]
- 21. Lima T, Auchincloss AH, Coudert E, Keller G, Michoud K, Rivoire C, Bulliard V, de Castro E, Lachaize C, Baratin D, Phan I, Bougueleret L, Bairoch A. 2009. HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res. 37:D471–D478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pei Z, Burucoa C, Grignon B, Baqar S, Huang XZ, Kopecko DJ, Bourgeois AL, Fauchere JL, Blaser MJ. 1998. Mutation in the peb1A locus of Campylobacter jejuni reduces interactions with epithelial cells and intestinal colonization of mice. Infect. Immun. 66:938–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Geddes K, Worley M, Niemann G, Heffron F. 2005. Identification of new secreted effectors in Salmonella enterica serovar Typhimurium. Infect. Immun. 73:6260–6271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jarvik T, Smillie C, Groisman EA, Ochman H. 2010. Short-term signatures of evolutionary change in the Salmonella enterica serovar Typhimurium 14028 genome. J. Bacteriol. 192:560–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez Y-V, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9:676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Niemann GS, Brown RN, Gustin JK, Stufkens A, Shaikh-Kidwai AS, Li J, McDermott JE, Brewer HM, Schepmoes A, Smith RD, Adkins JN, Heffron F. 2011. Discovery of novel secreted virulence factors from Salmonella enterica serovar Typhimurium by proteomic analysis of culture supernatants. Infect. Immun. 79:33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim S, Gupta N, Pevzner PA. 2008. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J. Proteome Res. 7:3354–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coombes BK, Lowden MJ, Bishop JL, Wickham ME, Brown NF, Duong N, Osborne S, Gal-Mor O, Finlay BB. 2007. SseL is a Salmonella-specific translocated effector integrated into the SsrB-controlled Salmonella pathogenicity island 2 type III secretion system. Infect. Immun. 75:574–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rytkonen A, Poh J, Garmendia J, Boyle C, Thompson A, Liu M, Freemont P, Hinton JC, Holden DW. 2007. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc. Natl. Acad. Sci. U. S. A. 104:3502–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Capriotti E, Marti-Renom MA. 2008. RNA structure alignment by a unit-vector approach. Bioinformatics 24:i112–8 doi:10.1093/bioinformatics/btn288 [DOI] [PubMed] [Google Scholar]
- 31. Iioka H, Loiselle D, Haystead TA, Macara IG. 2011. Efficient detection of RNA-protein interactions using tethered RNAs. Nucleic Acids Res. 39:e53 doi:10.1093/nar/gkq1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Charette M, Gray MW. 2000. Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49:341–351 [DOI] [PubMed] [Google Scholar]
- 33. Ansong C, Yoon H, Porwollik S, Mottaz-Brewer H, Petritis BO, Jaitly N, Adkins JN, McClelland M, Heffron F, Smith RD. 2009. Global systems-level analysis of Hfq and SmpB deletion mutants in Salmonella: implications for virulence and global protein translation. PLoS One 4:e4809 doi:10.1371/journal.pone.0004809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vogel J, Luisi BF. 2011. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9:578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sittka A, Pfeiffer V, Tedin K, Vogel J. 2007. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 63:193–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yoon H, McDermott JE, Porwollik S, McClelland M, Heffron F. 2009. Coordinated regulation of virulence during systemic infection of Salmonella enterica serovar Typhimurium. PLoS Pathog. 5:e1000306 doi:10.1371/journal.ppat.1000306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brennan RG, Link TM. 2007. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 10:125–133 [DOI] [PubMed] [Google Scholar]
- 38. Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. 2008. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 4:e1000163 doi:10.1371/journal.pgen.1000163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee SH, Galan JE. 2004. Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol. Microbiol. 51:483–495 [DOI] [PubMed] [Google Scholar]
- 40. Gophna U, Ron EZ, Graur D. 2003. Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events. Gene 312:151–163 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.