Abstract
Three evolutionarily distinct families of replicative DNA polymerases, designated polymerase B (Pol B), Pol C, and Pol D, have been identified. Members of the Pol B family are present in all three domains of life, whereas Pol C exists only in Bacteria and Pol D exists only in Archaea. Pol B enzymes replicate eukaryotic chromosomal DNA, and as members of the Pol B family are present in all Archaea, it has been assumed that Pol B enzymes also replicate archaeal genomes. Here we report the construction of Thermococcus kodakarensis strains with mutations that delete or inactivate key functions of Pol B. T. kodakarensis strains lacking Pol B had no detectable loss in viability and no growth defects or changes in spontaneous mutation frequency but had increased sensitivity to UV irradiation. In contrast, we were unable to introduce mutations that inactivated either of the genes encoding the two subunits of Pol D. The results reported establish that Pol D is sufficient for viability and genome replication in T. kodakarensis and argue that Pol D rather than Pol B is likely the replicative DNA polymerase in this archaeon. The majority of Archaea contain Pol D, and, as discussed, if Pol D is the predominant replicative polymerase in Archaea, this profoundly impacts hypotheses for the origin(s), evolution, and distribution of the different DNA replication enzymes and systems now employed in the three domains of life.
INTRODUCTION
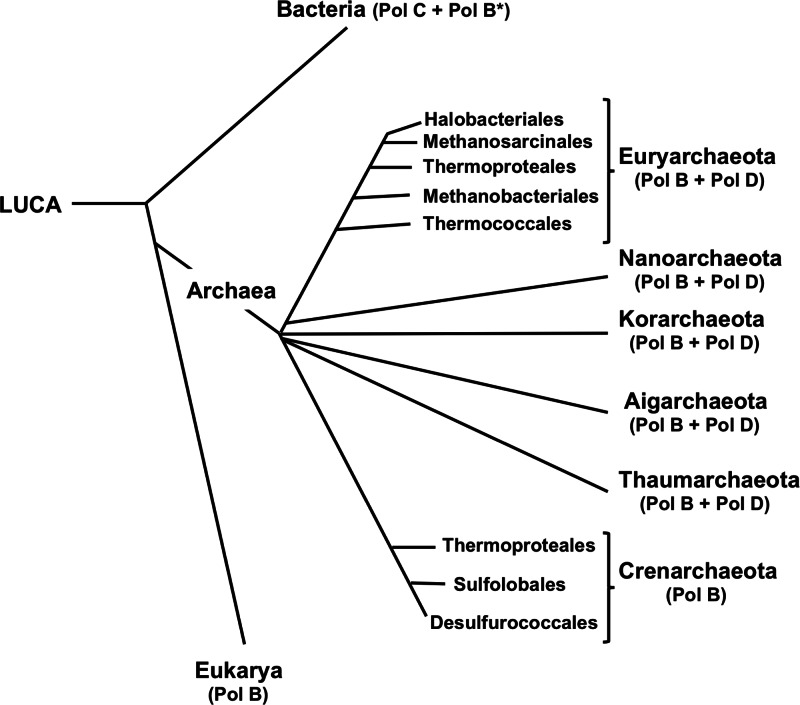
DNA replication, an essential event for all cellular life, is catalyzed by protein complexes designated replisomes, in which individual activities are tightly regulated and coordinated. DNA polymerases are the functional center of the replisome, but structurally distinct DNA polymerases, designated family C (Pol C) and family B (Pol B) polymerases, catalyze genome replication in Bacteria and eukaryotes, respectively (1–3). This difference has led to much debate, most fundamentally regarding whether DNA replication has evolved more than once, possibly independently in different biological lineages (1, 4–10). All known archaeal genomes encode at least one member of the Pol B family, and given that Archaea are evolutionarily closer to eukaryotes than are Bacteria (11, 12), it has been tacitly assumed, but challenged (13, 14), that Pol B enzymes must also replicate archaeal genomes. Presumably, this must be the case for the Crenarchaeota, as their genomes appear to encode only Pol B enzymes. This is, however, only an assumption for all members of the Euryarchaeota, Thaumarchaeota, Korarchaeota, Aigarchaeota, and Nanoarchaeota lineages, as their genomes encode not only Pol B enzymes but also members of an archaeon-specific DNA polymerase family designated Pol D (Fig. 1) (11, 13–15).
Fig 1.
Phylogenetic distribution of Pol B, Pol C, and Pol D family DNA polymerases. The distribution of Pol B and Pol D in the archaeal lineages is based on all available genome sequences. Pol B (*) occurs only infrequently in Bacteria and is not employed for DNA replication. LUCA, last universal common ancestor.
The Thermococcales are hyperthermophilic Euryarchaea, and given the commercial value of thermostable processive DNA polymerases, Pol B polymerases from this genus have received extensive characterization (13, 16, 17). Within these single polypeptide enzymes, the regions and residues directly responsible for nucleotide polymerization, 3′→5′ exonuclease proofreading (18–20), deaminated base sensing (21, 22), and interactions with the processivity factors are well defined (23, 24). In contrast, Pol D enzymes have received much less attention. The enzyme is composed of a small subunit (DP1) with 3′→5′ exonuclease activity and a large subunit (DP2) that contains the polymerase activity. Individually, the subunits have low activity, but when assembled into a heterodimer, or further into a heterotetramer, both activities are substantially increased (14, 15, 25–27). In vitro, Pol D behaves as a template-dependent DNA polymerase, has proofreading activity, and is capable of strand displacement DNA synthesis (13, 28). Pol D also interacts strongly with PCNA, conferring the ability to extend rapidly along long stretches of template DNA (28–30). These properties and fast, accurate, and processive DNA synthesis together with evidence that Pol D assembles in vivo into complexes that contain other replisome components (13, 31, 32) provide consistent support for the hypothesis that Pol D may replicate archaeal genomic DNA.
To test this hypothesis, we took advantage of the genetic technologies now available for Thermococcus kodakarensis (33) to manipulate the in vivo structure and expression of Pol B (encoded by TK0001 [34]) (Fig. 2) and Pol D (encoded by TK1902 [DP1] and TK1903 [DP2]). The results reveal that, although abundant in cells during the exponential growth phase, Pol B is not essential for growth but does provide resistance to UV irradiation. Targeted inactivation of the Pol B proofreading 3′→5′ exonuclease and uracil-sensing functions (18–20) had no detectable deleterious effects. In contrast, all attempts to generate strains with Pol D inactivated were unsuccessful. The discovery that Pol B is not necessary and that Pol D is sufficient for DNA replication in this model archaeon adds a fundamentally important new feature to the questions of when, where, and how often DNA replication has evolved in the three domains of life (5–10).
Fig 2.
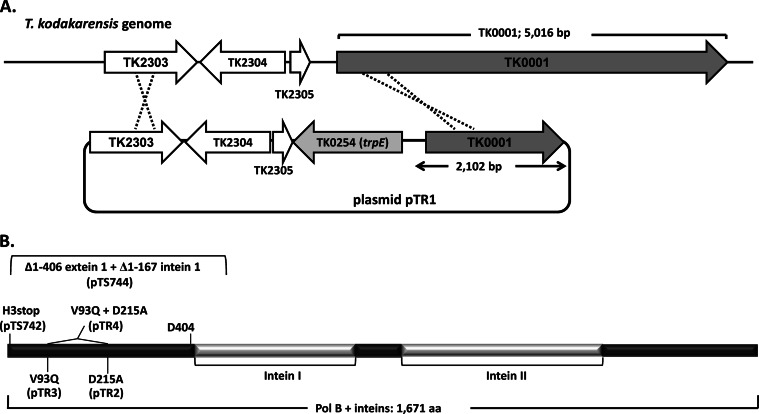
Structures of pTR1, Pol B, and Pol B variants. (A) Organization of genes adjacent to TK0001 in the T. kodakarensis genome (34) above the structure of plasmid pTR1. Transformation of T. kodakarensis KW128 (ΔpyrF ΔtrpE::pyrF) with pTR1 and homologous recombination, indicated by the dotted lines, generated the transformant T. kodakarensis TR1 (Table 1), which grows in the absence of tryptophan as a consequence of TK0254 (trpE) expression. (B) TK0001 encodes a polypeptide with 1,671 amino acid (aa) residues that includes two inteins, which are excised to produce mature Pol B. Site-directed mutagenesis of pTR1 generated plasmids pTR2, pTR3, pTR4, and pTS742, which encode the Pol B variants shown (H3stop indicates a nonsense codon at position 3). Plasmid pTS744 was generated by deleting the 5′-terminal sequence of TK0001 that encodes the first 406 amino acids of Pol B, including a universally conserved aspartic acid (D404) required for deoxynucleoside triphosphate polymerization (19, 20), the extein-1–intein-1 junction, and the first 167 amino acids of intein-1. These mutations in TK0001 in plasmids pTR2, pTR3, pTR4, pTS742, and pTS744 were transferred into the T. kodakarensis genome, generating T. kodakarensis TR2, TR3, TR4, TS742, and TS744, respectively (Table 1), by transformation, cointegration, and selection for TK0254 expression.
MATERIALS AND METHODS
T. kodakarensis strain construction and confirmation of genome structures.
T. kodakarensis strains were grown in artificial seawater (ASW) with 5 g/liter each of yeast extract and tryptone (YT) and 2 g/liter sulfur (S0) at 85°C, with the growth of cultures measured by an increase in the optical density at 600 nm (OD600), as previously described (33). Standard molecular biology techniques were used to construct plasmids pTR1, pTR2, pTR3, pTR4, pTS742, and pTS744, which were maintained and amplified in Escherichia coli. The sequences of the oligonucleotides employed are available upon request. T. kodakarensis strains TR1, TR2, TR3, TR4, TS742, and TS744 were constructed by transformation of T. kodakarensis KW128 (35) with transformants selected by growth in the absence of tryptophan, as previously described (36). Construction of T. kodakarensis TS742 introduced an additional DdeI cleavage site; DdeI digestion of the appropriate amplicon from T. kodakarensis TS742 genomic DNA confirmed the presence of this site, and sequencing confirmed the presence of the nonsense mutation introduced at codon 3 of TK0001. Approximately 1.7 kbp was deleted from the 5′ terminus of TK0001 in T. kodakarensis TS744, and this was confirmed by the sizes of the amplicons generated using primers that flanked the locus and by failure to generate amplicons using primers that would have hybridized within the deleted region. The inability to generate amplicons from within TK0001 also confirmed that this coding region was not present elsewhere in the T. kodakarensis TS744 genome. Additional confirmation of the deletion in TK0001 in T. kodakarensis TS744 was provided by Southern blotting of PsiI-digested genomic DNA. The PsiI restriction fragments that hybridized to a digoxigenin (DIG)-labeled amplicon probe, generated by PCR from within TK0001, were identified by using anti-DIG antibodies coupled to alkaline phosphatase, as previously described (37).
Western blots with anti-Pol B and anti-Pol D polyclonal antibodies.
Cells were concentrated from exponentially growing cultures of T. kodakarensis KW128, TS742, and TS744 by centrifugation; resuspended in a solution containing 20 mM Tris-HCl (pH 7.5), 10% (vol/vol) glycerol, and 100 mM NaCl; and lysed by repeated freezing and thawing. After centrifugation, the resulting clarified lysates were quantified via Bradford assays (38), and the polypeptides present in 50-μg protein aliquots were separated by electrophoresis through denaturing 10% polyacrylamide gels and electroblotted onto a nitrocellulose membrane (Protran BA83; Whatman Inc.). Western blots were generated by using a combination of guinea pig polyclonal antibodies (Cocalico Biologicals Inc.) raised against recombinant Pol B and the small subunit of Pol D and rabbit anti-guinea pig antibodies coupled to horseradish peroxidase (Sigma-Aldrich). The blots were developed by using enhanced chemiluminescence (ECL; GE Healthcare).
Sensitivity to UV light irradiation, MMS, and mitomycin C.
Aliquots (5 μl) of serial dilutions of cells sampled from growing cultures of T. kodakarensis KW128, TR1, TS742, and TS744 were plated, under anaerobic conditions, on ASW-S0 medium that contained all 20 amino acids (33) with or without 0.1% methylmethane sulfonate (MMS) or 100 μg mitomycin C/ml. The colonies present on each plate after 60 h of incubation at 85°C were counted. Similarly, aliquots of dilutions of T. kodakarensis KW128, TR1, TS742, and TS744 cultures were spotted onto ASW-S0 plates containing all 20 amino acids and were then exposed to UV irradiation (model UVH-46550; International Biotechnologies Inc., New Haven, CT) for 1 min before the plates were incubated anaerobically at 85°C for 60 h to allow colony formation.
RESULTS
Cloning and mutagenesis of the gene (TK0001) encoding Pol B.
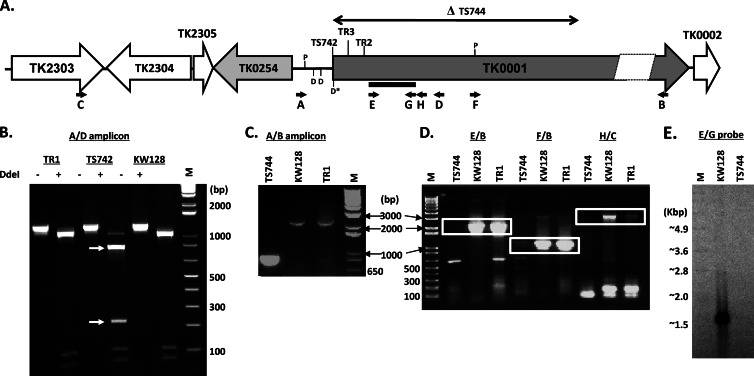
Pol B is encoded by TK0001 (14, 34), a >5-kbp open reading frame that also encodes two inteins (Fig. 2B). By using standard molecular biology techniques, chromosomal regions flanking and containing TK0001 were PCR amplified from T. kodakarensis genomic DNA, cloned, sequenced, and replicated within plasmids in E. coli. Subcloning and site-specific mutagenesis generated plasmids pTR1 (Fig. 2A), pTR2, pTR3, pTR4, pTS742, and pTS744 (Fig. 2B), which were used as donor DNAs to transform T. kodakarensis KW128 (Table 1) with transformants selected by the expression of trpE (TK0254). Sequencing of genomic DNA amplified from the transformants studied further in detail, designated T. kodakarensis TR1, TR2, TR3, TR4, TS742, and TS744 (Table 1), confirmed that they differed only by the mutation(s) introduced into TK0001 and that there were no changes to the promoter or ribosome binding site of TK0001. In T. kodakarensis TR2 and TR3, the mutations in TK0001 result in D215A and V93Q substitutions, which inactivate 3′→5′ exonuclease proofreading (18–20) and deaminated base recognition activities of the encoded Pol B, respectively (21, 22). Both mutations were introduced into TK0001 in T. kodakarensis strain TR4, resulting in a Pol B variant that lacked both accuracy-conferring features (Fig. 2B and Table 1). Initially, two strains were constructed with mutations designed to eliminate Pol B synthesis. In T. kodakarensis TS742, the TK0001 reading frame was terminated by a nonsense mutation at codon 3, and in T. kodakarensis TS744, ∼1.7 kbp was deleted from the 5′ terminus of TK0001, removing the first extein, the extein-1–intein-1 boundary, and part of the first intein (Fig. 2B). Surprisingly, none of the mutations introduced into TK0001 had any detectable negative effects on growth or viability (see below). Given this result, and to confirm that there was no essential function encoded anywhere within the TK0001 sequence, T. kodakarensis TS745 was constructed with TK0001 completely deleted (see below; see also the supplemental material). The presence of each mutation and all genome structures was unequivocally verified by multiple diagnostic PCRs, restriction enzyme digestions, sequencing of amplicons, and Southern blotting (Fig. 3B to E; see also Fig. S2 in the supplemental material).
Table 1.
T. kodakarensis strains constructed and used in this study
| Strain | Genotype or description | Reference |
|---|---|---|
| KW128 | ΔpyrF ΔtrpE::pyrF | 35 |
| TR1 | KW128 with trpE (TK0254) integrated upstream of TK0001 | This work |
| TR2 | TR1 with mutation in TK0001 encoding Pol B (D215A) | This work |
| TR3 | TR1 with mutation in TK0001 encoding Pol B (V93Q) | This work |
| TR4 | TR1 with mutations in TK0001 encoding Pol B (V93Q + D215A) | This work |
| TS742 | TR1 with nonsense mutation in codon 3 of TK0001 | This work |
| TS744 | TR1 with 1.7-kbp deletion of the 5′ region of TK0001 | This work |
| TS745 | KW128 with TK0001 deleted and replaced by trpE (TK0254) | This work |
Fig 3.
Confirmation of the genome structures of T. kodakarensis strains. (A) Diagram illustrating the positions of the mutations in TK0001 (Fig. 2B). The locations of the oligonucleotides used as primers in diagnostic PCRs are indicated by the small black arrows labeled A through H. The positions of PsiI (P) and DdeI (D) cleavage sites are shown, with D* indicating an additional DdeI site introduced into the genome of T. kodakarensis TS742. The solid black bar indicates the amplicon generated by using primers E and G that was DIG labeled and used as a probe in Southern blot experiments (see panel E). (B) Electrophoretic separation of the amplicons generated by using primers A and D from T. kodakarensis TR1, TS742, and KW128 genomic DNAs with (+) or without (−) DdeI digestion. Changing codon 3 to a stop codon introduced a DdeI cleavage site (D* in panel A) in DNA from T. kodakarensis TS742, which resulted in the DdeI fragments identified by white arrows. Molecular size markers, noted in base pairs, were separated in lane M. (C) Electrophoretic separation of amplicons generated by using primers A and B from genomic DNA isolated from T. kodakarensis TS744, KW128, and TR1. Deletion of the 5′ region of TK0001 in T. kodakarensis TS744 resulted in an ∼700-bp amplicon rather than the ∼2,400-bp amplicon generated from T. kodakarensis KW128 and TR1 DNAs. Molecular size markers (in base pairs) were separated in lane M. (D) Electrophoretic separation of the amplicons generated with primer pairs E and B, F and B, and H and C. The boxes identify the amplicons from genomes that contained a full-length TK0001. Molecular size markers (in base pairs) were separated in lane M. (E) Southern blot of PsiI-digested genomic DNA from T. kodakarensis KW128 and TS744 probed with a DIG-labeled amplicon generated by using primers E and G from T. kodakarensis KW128 genomic DNA. This region of TK0001 is not present in the genome of T. kodakarensis TS744. DIG-labeled molecular size markers (in kilobase pairs) were separated in lane M.
In contrast to the ready construction of T. kodakarensis strains with mutations in TK0001, despite using the same procedures and selections with many different plasmid donor DNAs, we were unable to generate a T. kodakarensis strain with an inactivating mutation in TK1902 and/or TK1903, encoding DP1 and DP2, respectively.
T. kodakarensis lacking Pol B grows normally.
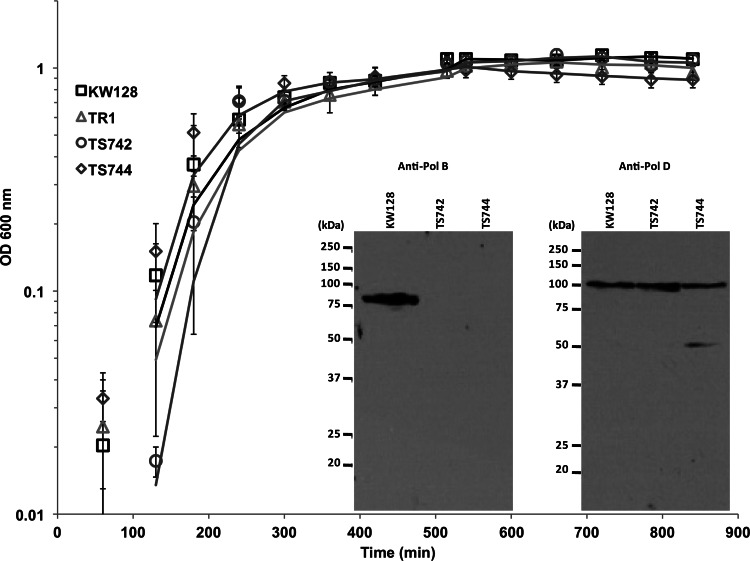
Cultures of T. kodakarensis TS742, TS744, and TS745 grew at the same rate and reached the same final cell densities as cultures of T. kodakarensis KW128 and TR1 (Fig. 4; see also Fig. S3 in the supplemental material). These strains also had indistinguishable plating efficiencies and formed colonies with the same sizes on several different Gelrite-solidified media. Western blotting confirmed that Pol B was present in lysates of T. kodakarensis KW128 cells taken from cultures during exponential growth, but identical processing and probing failed to detect any full-length or truncated Pol B in lysates of T. kodakarensis TS742 and TS744 (Fig. 4). In contrast, Western blotting using antiserum raised against DP1 confirmed the presence of Pol D in lysates of all three strains (Fig. 4). In the absence of Pol B, there was no detectable increase in the abundance of Pol D in T. kodakarensis TS742 and TS744 (Fig. 4).
Fig 4.
Growth curves and Western blots. Average values for the growth of at least three cultures of T. kodakarensis KW128, TR1, TS742, and TS744 in ASW-YT-S0 medium at 85°C are shown, with errors and with moving averages provided as trend lines. The growth of T. kodakarensis TR2, TR3, and TR4 cultures was indistinguishable from the growth of the strains shown. To generate the Western blots, all soluble proteins in clarified lysates of cells harvested from exponentially growing cultures of T. kodakarensis KW128, TS742, and TS744 were separated by electrophoresis, transferred onto membranes, and probed by incubation with polyclonal antisera raised against T. kodakarensis Pol B (anti-PolB) or Pol D (anti-PolD).
T. kodakarensis lacking Pol B retains a wild-type mutation profile.
T. kodakarensis incorporates exogenous purines, and this scavenging activity results in sensitivity to the analogue 6-methylpurine (6MP) (33, 39). Sensitivity is conferred by the activity of a hypoxanthine/guanine phosphoribosyltransferase encoded by TK0664. Virtually all mutants isolated as spontaneously resistant to 6MP (6MPr) have mutations in TK0664, and this was used as an assay to measure the frequency and determine the patterns of spontaneous mutations that occurred in T. kodakarensis strains KW128, TR1, TR2, TR3, TR4, and TS744. Dilutions of cultures were plated onto media with or without 100 μM 6MP, and comparisons of the numbers of colonies formed, in at least three separate experiments, revealed that mutations conferring 6MPr occurred spontaneously at statistically the same frequency (1 per 9.4 × 107 ± 7 × 107 plated cells) in the wild-type, Pol B-defective, and Pol B-deleted strains. To determine if these mutations were qualitatively different, the TK0664 locus was PCR amplified from >100 6MPr mutants from each strain and sequenced. Given that only inactivating mutations were likely to result in 6MPr, as expected, the majority of mutations in all strains were deletions or insertions, and many mutations changed residues predicted to be components of the enzyme's active site (see Fig. S1 in the supplemental material). In total, 163 different transitions and transversions were identified, most resulting in translation-terminating nonsense mutations, and transitions outnumbered transversions 2-fold in all strains (see Table S1 in the supplemental material). Consistent with a hyperthermophilic life-style increasing cytosine deamination (40, 41), C-to-T transitions occurred three times more often than T-to-C transitions, although the overall percentage of C-to-T transitions remained relatively constant, ranging from 22% to 28% in all strains. There were no striking differences in the profiles of spontaneous mutations that conferred 6MPr in the strains that contained versus those that lacked Pol B, although deletions and insertions of >3 bp occurred more often in the absence of Pol B (see Table S1 in the supplemental material).
T. kodakarensis lacking Pol B has increased sensitivity to UV irradiation.
Direct comparisons of the sensitivity of T. kodakarensis KW128, TS742, and TS744 to methyl methanesulfonate (MMS), mitomycin C, and UV light irradiation were made. All three treatments were toxic, causing substantial cell death, but whereas there were no differences in the sensitivities of these strains to MMS and mitomycin C, T. kodakarensis TS742 and TS744 were more sensitive to UV irradiation than T. kodakarensis KW128 (Fig. 5).
Fig 5.
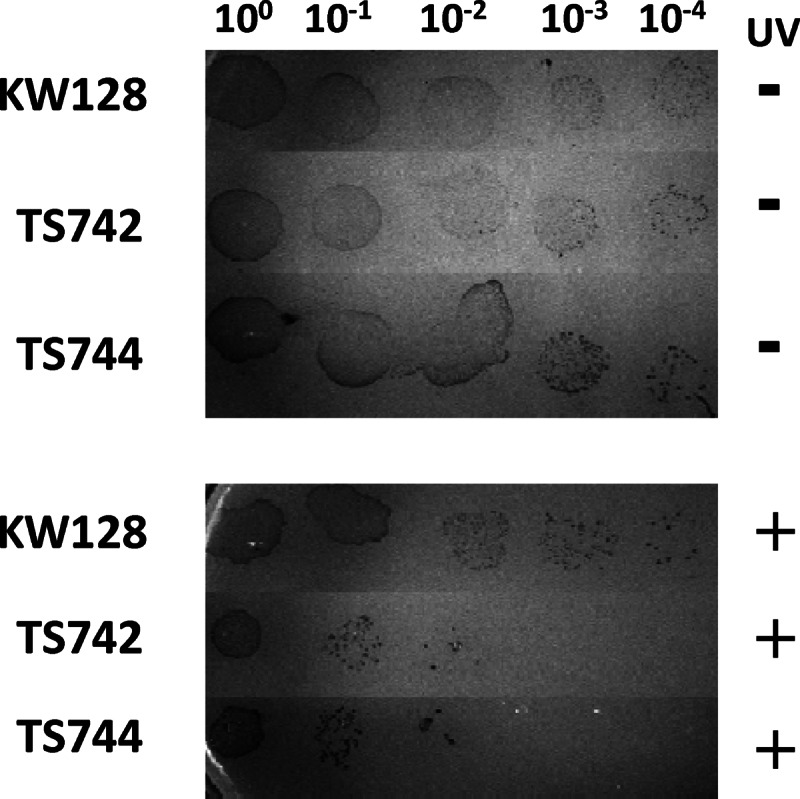
T. kodakarensis strains lacking Pol B are UV sensitive. An aliquot (5 μl) of cells (100) and of 10-fold serial dilutions (10−1 to 10−4) of these cells taken from exponentially growing cultures of T. kodakarensis KW128, TS742, and TS744 were spotted onto duplicate ASW-YT-S0 plates. The plates were (+) or were not (−) exposed to UV irradiation for 1 min before being incubated at 85°C for ∼60 h.
The entire 5,016-bp sequence of TK0001 is nonessential.
A review of the original submission recommended that we construct a strain with the entire TK0001 sequence deleted to categorically establish that no TK0001-encoded function remained. As described in the supplemental material, T. kodakarensis TS745 (ΔTK0001) was therefore constructed (see Fig. S2A in the supplemental material), and the complete and precise deletion of TK0001 was confirmed by multiple diagnostic PCR amplifications and Southern blotting (see Fig. S2B to S2F in the supplemental material). T. kodakarensis TS745 grows at the same rate and to the same final cell density as the parental strain T. kodakarensis KW128 in liquid media (see Fig. S3 in the supplemental material) and has the same plating efficiency on solidified media. As documented for T. kodakarensis TS742 and TS744 (Fig. 5), T. kodakarensis TS745 also has increased sensitivity to UV irradiation.
DISCUSSION
Based on the results obtained, Pol B is not required for viability or growth at wild-type rates in the euryarchaeon T. kodakarensis. The absence of Pol B does not increase the rate of spontaneous mutation, and the only pronounced phenotype detected so far is increased sensitivity to UV irradiation. In contrast, exhaustive attempts to delete either of the genes (TK1902 and TK1903) that encode the two subunits of Pol D, the second DNA polymerase present in T. kodakarensis (13, 34), were unsuccessful. We conclude that Pol D can, and most likely does, function normally as the genome-replicating DNA polymerase in T. kodakarensis. This conclusion, coupled with the nonessentiality of Pol B, raises some intriguing questions, addressed below.
Is Pol D the replicative DNA polymerase?
In the absence of Pol B, Pol D must be the polymerase responsible for synthesizing both the leading and lagging DNA strands of the T. kodakarensis genome. All of the biochemical properties established for Pol D in vitro are consistent with a replication function (13–15, 25–30), and Pol D exists in vivo in a stable complex(es) that also contains many other components predicted for the T. kodakarensis replisome, including MCM1, the GINS subcomplex, DNA ligase, Cdc6, PCNA1, PCNA2, and GAN (31, 32). In contrast, when similarly isolated from cell lysates, Pol B was not present in a stable complex with these replisome components. The location of the genes (TK1902 and TK1903) that encode Pol D, directly adjacent to the gene (TK1901) that encodes Cdc6 and next to the presumed origin of chromosomal replication in T. kodakarensis, also hints strongly at a replication role (4, 34).
What does Pol B do?
Pol B is abundant in T. kodakarensis and is encoded in all archaeal genomes (4, 6, 8, 13); however, given that its absence does not compromise the viability or growth of T. kodakarensis, the simplest argument is that Pol B does not play a critical role in genome replication in this archaeon. It is, however, important to note that DNA polymerase ϵ (DNAP-ϵ) has been shown conclusively to replicate the leading strand of chromosomal DNA in eukaryotes (2, 42, 43), and despite this, deletions of the N-terminal DNA-polymerizing catalytic domain of DNAP-ϵ are not lethal in yeasts (44–46). These deletion mutations do result in substantial growth defects, whereas point mutations in the same gene, which inactivate only the catalytic activity of DNAP-ϵ, are lethal (47). The explanation appears to be that catalytically active DNAP-ϵ is essential when full-length DNAP-ϵ is assembled into the replisome, but replisomes that support viability are formed with DNAP-δ when full-length DNAP-ϵ is not available for incorporation (2, 42, 47). Given this precedent, it could be argued that Pol B does normally catalyze genome replication in T. kodakarensis but that this function can be fully replaced by Pol D, as unlike the growth defects of yeast strains that lack the N terminus of DNAP-ϵ, T. kodakarensis strains lacking Pol B grow normally (Fig. 4; see also Fig. S3 in the supplemental material). Arguing against Pol B normally catalyzing genome replication in T. kodakarensis, the introduction of mutations that resulted in Pol B variants that lack 3′→5′ exonuclease proofreading and uracil-sensing mechanisms had no detectable effects on the frequency or profile of spontaneous mutation (see Table S1 in the supplemental material).
If Pol B is not primarily involved in replication, likely alternatives would be a role(s) in DNA repair and/or recombination, and there are precedents for the participation of Pol B family members in repair. For example, bacterial Pol II is involved in the restart of collapsed replication forks (48), and after a repair polymerase inserts a normal base opposite a damaged template base, eukaryotic Pol ζ extends the resulting mismatch (49). The increased UV sensitivity of T. kodakarensis strains lacking Pol B argues for a role in resolving DNA damage, but it is then difficult to explain why there were no similar increases in sensitivity to MMS and mitomycin C. It is nevertheless well established that the Thermococcales are unusually resistant to ionizing radiation (50–52), and this could reflect the constitutive and abundant presence of Pol B facilitating the reassembly of a radiation-fragmented genome.
DNA replication systems in the three domains of life.
The existence of analogous but nonorthologous replicative DNA replication proteins, including many DNA polymerases of the B family, in different biological lineages has led to the hypothesis that multiple replicative systems were established in the last universal common ancestor (LUCA) (Fig. 1) (8). One or more systems and replicative polymerases were then subsequently retained and/or lost in the lineages that now constitute the three domains of life. An alternative hypothesis posits that the different DNA replicative systems evolved successively over time, with transfer and replacement of the existing system in some lineages, likely facilitated by viral infections given that many viral genomes encode DNA polymerases (7). Providing experimental support for these hypotheses, establishing an evolutionary sequence, and determining the consequences of a nonorthologous DNA replication system replacement are clearly major research challenges. Currently, the literature arguments focus on the dichotomy of Pol C in Bacteria versus Pol B in Archaea, with the presence of Pol B in the eukaryotes being a consequence and consistent with Archaea and eukaryotes more recently sharing a common ancestor (11, 12). The discovery that Pol D is likely the replicative enzyme in T. kodakarensis, and, by extrapolation, perhaps in many archaeal lineages, adds a further complication. Most archaeal research to date has investigated members of only two lineages, the Euryarchaea, which have Pol D, and the Crenarchaea, which do not have Pol D. However, now with the rapid increases in genome and metagenome sequencing, it has become clear that there are many other early-branching archaeal lineages and that Pol D is also present in the Korarchaeota, Aigarchaeota, Nanoarchaeota, and Thaumarchaeota (Fig. 1) (13). Based on parsimony, possibly both Pol B and Pol D were present in the ancestral archaeal lineage and Pol D was lost in a branch that led to the Crenarchaea and eukaryotic replication systems. Alternatively, Pol D could have evolved in the archaeal lineage after the divergence of this putative crenarchaeal-eukaryotic lineage. The distribution of histones would, in contrast, argue against this lineage and hypothesis, as the Pol D-containing Archaea, in common with all eukaryotes, have histones, whereas the Crenarchaea do not (37, 53). There are clearly exceptions, but as a current generalization, we now propose that genomic replication can also be viewed meaningfully as conforming to the biological domain trichotomy (11, 12): Archaea employ Pol D, Bacteria employ Pol C, and eukaryotes employ Pol B.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Heslop for technical assistance.
This work was supported by National Institutes of Health grant GM098176 (to J.N.R. and T.J.S.) and National Science Foundation grant MCB-0815646 (to Z.K.). T.R. was a UK-BBSRC-supported Ph.D. student.
Footnotes
Published ahead of print 15 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02037-12.
REFERENCES
- 1. Braithwaite D, Ito J. 1993. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 21:787–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burgers PMJ. 2009. Polymerase dynamics at the eukaryotic DNA replication fork. J. Biol. Chem. 284:4041–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kelman Z, O'Donnell M. 1995. DNA polymerase holoenzyme—structure and function of a chromosomal replicating machine. Annu. Rev. Biochem. 64:171–200 [DOI] [PubMed] [Google Scholar]
- 4. Berthon J, Cortez D, Forterre P. 2008. Genomic context analysis in Archaea suggests previously unrecognized links between DNA replication and translation. Genome Biol. 9:R71 doi:10.1186/gb-2008-9-4-r71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Edgell DR, Doolittle WF. 1997. Archaea and the origin(s) of DNA replication proteins. Cell 89:995–998 [DOI] [PubMed] [Google Scholar]
- 6. Filee J, Forterre P, Laurent J. 2002. Evolution of DNA polymerase families: evidence for multiple gene exchange between cellular and viral proteins. J. Mol. Evol. 54:763–773 [DOI] [PubMed] [Google Scholar]
- 7. Forterre P. 1999. Displacement of cellular proteins by functional analogues from plasmids or viruses could explain puzzling phylogenies of many DNA informational proteins. Mol. Microbiol. 33:457–465 [DOI] [PubMed] [Google Scholar]
- 8. Koonin EV. 2006. Temporal order of evolution of DNA replication systems inferred by comparison of cellular and viral DNA polymerases. Biol. Direct 1:39 doi:10.1186/1745-6150-1-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leipe DD, Aravind L, Koonin EV. 1999. Did DNA replication evolve twice independently? Nucleic Acids Res. 27:3389–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tahirov TH, Makarova KS, Rogozin IB, Pavlov YI, Koonin EV. 2009. Evolution of DNA polymerases: an inactivated polymerase-exonuclease module of Pol E and a chimeric origin of eukaryotic polymerases from two classes of archaeal ancestors. Biol. Direct 4:11 doi:10.1186/1745-6150-4-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brochier-Armanet C, Forterre P, Gribaldo S. 2011. Phylogeny and evolution of the Archaea: one hundred genomes later. Curr. Opin. Microbiol. 14:274–281 [DOI] [PubMed] [Google Scholar]
- 12. Woese CR, Fox GE. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. U. S. A. 74:5088–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ishino Y, Ishino S. 2011. Rapid progress of DNA replication studies in Archaea, the third domain of life. China Life Sci. 55:386–403 [DOI] [PubMed] [Google Scholar]
- 14. Ishino Y, Komori K, Cann IK, Koga Y. 1998. A novel DNA polymerase family found in Archaea. J. Bacteriol. 180:2232–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cann IK, Komori K, Toh H, Kanai S, Ishino Y. 1998. A heterodimeric DNA polymerase: evidence that members of the Euryarchaeota possess a distinct DNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 95:14250–14255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perler FB, Kumar S, Kong H. 1996. Thermostable DNA polymerases. Adv. Protein Chem. 48:377–435 [DOI] [PubMed] [Google Scholar]
- 17. Southworth MW, Kong H, Kucera RB, Ware J, Jannasch HW, Perler FB. 1996. Cloning of thermostable DNA polymerases from hyperthermophilic marine Archaea with emphasis on Thermococcus sp. 9 degrees N-7 and mutations affecting 3′-5′ exonuclease activity. Proc. Natl. Acad. Sci. U. S. A. 93:5281–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chapin Rodrigues A, Park H-W, Mao C, Beese LS. 2000. Crystal structure of a Pol α family DNA polymerase from the hyperthermophilic archaeon Thermococcus sp. 9°N-7. J. Mol. Biol. 299:447–462 [DOI] [PubMed] [Google Scholar]
- 19. Hashimoto H, Nishioka M, Fujiwara S, Takagi M, Imanaka T, Inoue T, Kai Y. 2001. Crystal structure of DNA polymerase from the hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. J. Mol. Biol. 306:469–477 [DOI] [PubMed] [Google Scholar]
- 20. Hopfner KP, Eichinger A, Engh RA, Laue F, Ankenbauer W, Huber R, Angerer B. 1999. Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius. Proc. Natl. Acad. Sci. U. S. A. 96:3600–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Firbank SJ, Wardle J, Heslop P, Lewis RJ, Connolly BA. 2008. Uracil recognition in archaeal DNA polymerases captured by X-ray crystallography. J. Mol. Biol. 381:529–539 [DOI] [PubMed] [Google Scholar]
- 22. Killelea T, Ghosh S, Tan SS, Heslop P, Firbank SJ, Kool ET, Connolly BA. 2010. Probing the interaction of archaeal DNA polymerases with deaminated bases using X-ray crystallography and non-hydrogen bonding isosteric base analogues. Biochemistry 49:5772–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mayanagi K, Kiyonari S, Nishida H, Saito M, Kohda D, Ishino Y, Shirai T, Morikawa K. 2011. Architecture of the DNA polymerase B-proliferating cell nuclear antigen (PCNA)-DNA ternary complex. Proc. Natl. Acad. Sci. U. S. A. 108:1845–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vivona JB, Kelman Z. 2003. The diverse spectrum of sliding clamp interacting proteins. FEBS Lett. 546:167–172 [DOI] [PubMed] [Google Scholar]
- 25. Gueguen Y, Rolland JL, Lecompte O, Azam P, Le Romancer G, Flament D, Raffin JP, Dietrich J. 2001. Characterization of two DNA polymerases from the hyperthermophilic euryarchaeon Pyrococcus abyssi. Eur. J. Biochem. 268:5961–5969 [DOI] [PubMed] [Google Scholar]
- 26. Shen Y, Musti K, Hiramoto M, Kikuchi H, Kawarabayashi Y, Matsui I. 2001. Invariant Asp-1122 and Asp-1124 are essential residues for polymerization catalysis of family D DNA polymerase from Pyrococcus horikoshii. J. Biol. Chem. 276:27376–27383 [DOI] [PubMed] [Google Scholar]
- 27. Uemori T, Sato Y, Kato I, Doi H, Ishino Y. 1997. A novel DNA polymerase in the hyperthermophilic archaeon, Pyrococcus furiosus: gene cloning, expression, and characterization. Genes Cells 2:499–512 [DOI] [PubMed] [Google Scholar]
- 28. Henneke G, Flament D, Hübscher U, Querellou J, Raffin J-P. 2005. The hyperthermophilic euryarchaeote Pyrococcus abyssi likely requires the two DNA polymerases D and B for DNA replication. J. Mol. Biol. 350:53–64 [DOI] [PubMed] [Google Scholar]
- 29. Castrec B, Rouillon C, Henneke G, Flament D, Querellou J, Raffin JP. 2009. Binding to PCNA in euryarchaeal DNA replication requires two PIP motifs for DNA polymerase D and one PIP motif for DNA polymerase B. J. Mol. Biol. 394:209–218 [DOI] [PubMed] [Google Scholar]
- 30. Tori K, Kimizu M, Ishino S, Ishino Y. 2007. DNA polymerases B and D from the hyperthermophilic archaeon Pyrococcus furiosus both bind to proliferating cell nuclear antigen with their C-terminal PIP-box motifs. J. Bacteriol. 189:5652–5657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Z, Santangelo TJ, Cubonova L, Reeve JN, Kelman Z. 2010. Affinity purification of an archaeal DNA replication protein network. mBio 1(5):e00221–10 doi:10.1128/mBio.00221-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Z, Pan M, Santangelo TJ, Chemnitz W, Yuan W, Edwards JL, Hurwitz J, Reeve JN, Kelman Z. 2011. A novel DNA nuclease is stimulated by association with the GINS complex. Nucleic Acids Res. 39:6114–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hileman TH, Santangelo TJ. 2012. Genetic techniques for Thermococcus kodakarensis. Front. Microbiol. 3:195 doi:10.3389/fmicb.2012.00195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fukui T, Atomi H, Kanai T, Matsumi R, Fujiwara S, Imanaka T. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15:352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sato T, Fukui T, Atomi H, Imanaka T. 2005. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl. Environ. Microbiol. 71:3889–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Santangelo TJ, Cubonova L, James CL, Reeve JN. 2007. TFB1 or TFB2 is sufficient for Thermococcus kodakaraensis viability and for basal transcription in vitro. J. Mol. Biol. 367:344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cubonova L, Katano M, Kanai T, Atomi H, Reeve JN, Santangelo TJ. 2012. An archaeal histone is required for transformation of Thermococcus kodakarensis. J. Bacteriol. 194:6864–6874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 39. Santangelo TJ, Cubonova L, Reeve JN. 2010. Thermococcus kodakarensis genetics: TK1827-encoded beta-glycosidase, new positive-selection protocol, and targeted and repetitive deletion technology. Appl. Environ. Microbiol. 76:1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lindahl T, Nyberg B. 1974. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry 13:3405–3410 [DOI] [PubMed] [Google Scholar]
- 41. Schroeder GK, Wolfenden R. 2007. Rates of spontaneous disintegration of DNA and the rate enhancements produced by DNA glycosylases and deaminases. Biochemistry 46:13638–13647 [DOI] [PubMed] [Google Scholar]
- 42. Kunkel TA, Burgers PM. 2008. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 18:521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. 2007. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science 317:127–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Feng W, D'Urso G. 2001. Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase epsilon are viable but require the DNA damage checkpoint control. Mol. Cell. Biol. 21:4495–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kesti T, Flick K, Keränen S, Syväoja JE, Wittenberg C. 1999. DNA polymerase ϵ catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell 3:679–685 [DOI] [PubMed] [Google Scholar]
- 46. Ohya T, Kawasaki Y, Hiraga S-I, Kanbara S, Nakajo K, Nakashima N, Suzuki A, Sugino A. 2002. The DNA polymerase domain of pol ϵ is required for rapid, efficient, and highly accurate chromosomal DNA replication, telomere length maintenance, and normal cell senescence in Saccharomyces cerevisiae. J. Biol. Chem. 277:28099–28108 [DOI] [PubMed] [Google Scholar]
- 47. Dua R, Levy DL, Campbell JL. 1999. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol ϵ and its unexpected ability to support growth in the absence of the DNA polymerase domain. J. Biol. Chem. 274:22283–22288 [DOI] [PubMed] [Google Scholar]
- 48. Rangarajan S, Woodgate R, Goodman MF. 1999. A phenotype for enigmatic DNA polymerase II: a pivotal role for pol II in replication restart in UV-irradiated Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 96:9224–9229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lawrence CW. 2004. Cellular functions of DNA polymerase ζ and Rev 1 protein. Adv. Protein Chem. 69:167–203 [DOI] [PubMed] [Google Scholar]
- 50. Confalonieri F, Sommer S. 2011. Bacterial and archaeal resistance to ionizing radiation. J. Physics Conf. Ser. 261:012005. doi:1088/1742-6596 [Google Scholar]
- 51. Jolivet E, L'Haridon S, Corre E, Forterre P, Prieur D. 2003. Thermococcus gammatolerans sp. nov., a hyperthermophilic archaeon from a deep-sea hydrothermal vent that resists ionizing radiation. Int. J. Syst. Evol. Microbiol. 53:847–851 [DOI] [PubMed] [Google Scholar]
- 52. Jolivet E, Matsunaga F, Ishino Y, Forterre P, Prieur D, Myllykallio H. 2003. Physiological responses of the hyperthermophilic archaeon Pyrococcus abyssi to DNA damage caused by ionizing radiation. J. Bacteriol. 185:3958–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Samson R, Reeve JN. 2007. DNA binding proteins and chromatin, p 110–119 In Cavicchioli R. (ed), Archaea. Molecular and cellular biology. ASM Press, Washington, DC [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.