Abstract
We investigated phenotypes of mutants of Methylotenera mobilis JLW8 with lesions in genes predicted to encode functions of the denitrification pathway, as well as mutants with mutations in methanol dehydrogenase-like structural genes xoxF1 and xoxF2, in order to obtain insights into denitrification and methanol metabolism by this bacterium. By monitoring the accumulation of nitrous oxide, we demonstrate that a periplasmic nitrate reductase, NAD(P)-linked and copper-containing nitrite reductases, and a nitric oxide reductase are involved in the denitrification pathway and that the pathway must be operational in aerobic conditions. However, only the assimilatory branch of the denitrification pathway was essential for growth on methanol in nitrate-supplemented medium. Mutants with mutations in each of the two xoxF genes maintained their ability to grow on methanol, but not the double XoxF mutant, suggesting that XoxF proteins act as methanol dehydrogenase enzymes in M. mobilis JLW8. Reduced levels of nitrous oxide accumulated by the XoxF mutants compared to the wild type suggest that these enzymes must be capable of donating electrons for denitrification.
INTRODUCTION
Methanol-enhanced denitrification as part of sewage treatment is a well-described phenomenon (1), and a number of methylotrophs have been implicated in this process, carried out by mixed cultures (2–5). However, the role of denitrification and its connection to methylotrophy in natural systems is not well understood, and data from studies involving pure cultures of methylotrophs are sparse. One of the most well-studied denitrifiers is Paracoccus denitrificans (6). Over 3 decades ago, it has been proposed that anaerobic growth of this bacterium on methanol with nitrate was made possible by reduction of nitrate to nitrite using electrons derived from the pyridine nucleotide-linked dehydrogenations of formaldehyde and formate and that the nitrite so produced then served as an electron acceptor for methanol dehydrogenase via cytochrome c and nitrite reductase (7). However, in none of the later studies employing this organism was denitrification further linked to methylotrophy (6). Likewise, studies of denitrification in other methylotroph species, including classic denitrifiers, such as Hyphomicrobium spp., were inconclusive in terms of whether denitrification is directly linked to methylotrophy (8, 9). Recently, methanol-dependent denitrification by Methyloversatilis has been investigated using proteomics (10). This study demonstrated that Methyloversatilis reduces nitrate to nitrite in anoxic conditions, by expressing a classic membrane-bound nitrate reductase and all the typical methylotrophy pathways that operate during aerobic growth (10). However, the protein components proposed to be involved in either denitrification or methanol oxidation have not been investigated via mutant analysis.
Methylotenera species represent a recently described novel genus within the family Methylophilaceae (11). Apart from genomic divergence (12), one of the main properties that distinguishes the described species of Methylotenera from species representing three other genera within this family (Methylophilus, Methylobacillus, and Methylovorus) is extremely poor growth on methanol and the lack of genes for the classic (MxaFI) methanol dehydrogenase (11). However, Methylotenera appears to thrive in a variety of environments (12), and these organisms appear to be one of the major species that consume methanol in situ (13). The growth of some of the Methylotenera species is stimulated by nitrate, and it has been proposed that the metabolism of methanol by Methylotenera may be linked to denitrification (13). Denitrification by M. mobilis JLW8 has been previously demonstrated, and putative genes for denitrification have been identified in the genome (13, 14). The denitrification pathway is incomplete, due to the lack of nitrous oxide reductase, thus resulting in accumulation of nitrous oxide (13, 14). XoxF enzymes that are homologs of the large subunit of methanol dehydrogenase have been implicated in playing a role in nitrate-dependent growth on methanol (13, 14). The two goals of the present study were the confirmation of the functions of the genes proposed to be involved in denitrification and investigation of the role of XoxF that is a homolog of the large subunit of methanol dehydrogenase in methanol metabolism and denitrification.
MATERIALS AND METHODS
Cultivation.
M. mobilis strain JLW8 was routinely cultivated in nitrate-containing MM2 medium supplemented by methylamine, as previously described (13, 14). Growth on methanol was tested on plates using solidified MM2 medium or using solidified Hypho (ammonia-containing) medium, or in the respective liquid media, as previously described (14). Methanol and formaldehyde were added to plates as vapors in a closed system (plates sealed with parafilm) at a final concentration of ∼1 mM, with periodic additions. Methanol was added to liquid cultures at 1 mM, with periodic additions.
Nitric oxide measurements.
N2O was measured in cells grown on methylamine as described above. Cells from 100 ml of culture were pelleted, resuspended in fresh MM2 medium, and supplemented with either 25 mM methylamine or 25 mM methanol. Cultures were placed in 150-ml vials sealed with rubber stoppers and incubated with shaking at 30°C. The headspace atmosphere was sampled at different time intervals using a syringe. N2O was detected by gas chromatography on an SRI 8610C gas chromatograph with an electron capture detector (ECD). Samples (0.1-ml volume) were injected manually, and gases were separated on a 2-m HyaSepD column at 103°C using 95% argon–5% methane as carrier gas. N2O was detectable after ∼1 h of incubation with methylamine and after ∼4 h of incubation with methanol, with maximum accumulation observed at approximately 6 to 8 h and 18 to 20 h of incubation, respectively.
Enzyme assays.
Enzyme activities were measured in cells grown on methylamine as described above and in cells induced with methanol as follows. Cells were grown on methylamine, pelleted, washed, resuspended in a fresh medium, supplemented with 10 mM methanol, and incubated overnight with shaking at room temperature. Nitrate reductase was measured at room temperature in a reaction mixture containing (per a total volume of 1 ml) 50 mM potassium phosphate buffer (pH 7.5), 20 mM NaNO3−, 1 mM methyl viologen, and cell extract. Reaction was started by the addition of 50 μl of a freshly made solution of 100 mM dithionite in 0.1 M NaHCO3. The reaction was stopped by rapid oxidation of the viologen achieved by vigorous mixing. Aliquots of 0.2 ml were assayed for nitrite (15). Copper nitrite reductase was measured as described above, except 10 mM NaNO2−was substituted for NaNO3−. Nitrite disappearance was monitored with a reaction mixture diluted 100-fold (15). NAD(P)H-dependent nitrite reductase was measured as previously described (16). Methanol dehydrogenase was assayed and in-gel staining was carried out as described previously (17).
Mutant generation.
Mutants were generated as previously described (18), using the suicide vector pCM184. Briefly, 3′ and 5′ fragments of each gene to be mutated were PCR amplified and inserted into pCM184, respectively, upstream and downstream of the kanamycin (Km) resistance gene. The native gene was then replaced by the deletion/insertion version via homologous recombination. The following primers were used to PCR amplify respective gene fragments, introducing a specific cloning site on each end of the fragment, and these are indicated in the name of each primer: for nap deletion, ATGAATTCGGTGATCCGAACCATCCGG (1648Eco) and AAGGTACCGCAGGTTAATCAGCGCTGCAT (1648Kpn) (the upstream fragment) and AAGGGCCCGCGATGCTAGTGGTAAGGC (1648Apa) and ATGAGCTCGCAGTCGCCAGGGTCTCAC (1648Sac) (the downstream fragment); for nirB deletion, ATGAATTCGTACAGTGGAAGAGTTG (1652Eco) and AAGGTACCGCTACGCACTCGCCCAC (1652Kpn) (the upstream fragment) and AAGGGCCCTCAGTGATTCCACGTATG (1652Apa) and ATGAGCTCGGTTGTTCACGCGCTCAGC (1652Sac) (the downstream fragment); for nirK deletion, ATGAATTCGATACCGCTTTTAGTACTGG (1061Eco) and AAGGTACCACAAGAATCAGCCCATAC (1061Kpn) (the upstream fragment) and AAGGGCCCATCTACTCAGGAAAAATG (1061Apa) and ATGAGCTCTCGACATGTTTTACAGGTG (1061Sac) (the downstream fragment); for norB deletion, ATGAATTCAATATCAATCTCAGGCAG (1051Eco) and AAGGTACCCAACACCAGCACTAGAC (1051Kpn) (the upstream fragment) and AAGGGCCCCTTTGCGTTCAATATGGTG (1051Apa2) and ATGAGCTCGATGAAAAAGCTAGTGATG (1051Sac2) (the downstream fragment); for nod deletion, ATGAATTCGTTATCAGCAGCAGCAAAACC (1063Eco) and AAGGTACCACATCATCGGATTGATGC (1063Kpn) (the upstream fragment) and AAGGGCCCTATCCATGTGGCAACTTTAC (1063Apa) and ATGAGCTCATGATATGGTTCAGGCTTTC (1063Sac) (the downstream fragment); for xoxF1 deletion, ATGAATTCAACACTAATGAAGGATG (1770Eco) and TTGGTACCGATATAGCAGCGTACGCCG (1770Kpn) (the upstream fragment) and TTGGGCCCATGTTCTACGTACCGC (1770Apa) and TAGAGCTCGCCACCGCCAGGTGCAGCG (1770Sac) (the downstream fragment); and for xoxF2 deletion, GATCGAATTCCTACCGGGCCGTCTACCTC (2048Eco) and CTAGGGTACCATGCAGACCAATATTTAC (2048Kpn) (the upstream fragment) and GATCGGGCCCGATGGCAACTTAGTAGC (2048Apa) and CTAGGAGCTCATGTGTAAGCGAAGCCG (2048Sac) (the downstream fragment).
Mutants were selected on solid media supplemented with methylamine as a carbon source and containing Km. Double-crossover mutants were identified by diagnostic PCR tests. In order to generate the double xoxF mutant, the Km resistance gene in the original pCM184-based construct for mutating gene xoxF1 (mmol_1770) was replaced by a gentamicin (Gm) resistance gene, which was amplified from the vector pDdxRGent, and this construct was used for generating a mutation in the Km resistant mutant with lesion in the xoxF2 gene (mmol_2048). Mutants were selected on methylamine in the presence of Km and Gm, and double-crossover mutants were identified via diagnostic PCR tests.
RESULTS
Mutant growth phenotypes.
Null mutants in the following genes predicted to encode catalytic functions in nitrate reduction pathways were generated: the single subunit Nap-type periplasmic nitrate reductase (Nap; mmol_1648; latter gene designations as per the published genome of M. mobilis JLW8 [12]), the single subunit copper-containing (dissimilatory) nitrite reductase (NirK; mmol_1061), the large subunit of the NAD(P)-linked (assimilatory) nitrite reductase (NirB; mmol_1652), and the large subunit of nitric oxide reductase (NorB; mmol_1051). We also mutated a gene predicted to encode a nitric oxide dioxygenase (Nod; mmol_1063) and each of two xoxF genes (mmol_1770 and mmol_2048) that encode homologs of the large subunit of methanol dehydrogenase (11, 14, 19, 20). In addition, a double mutant lacking both XoxF proteins was generated. Growth phenotypes of the mutants were investigated on methanol plates using either nitrate or ammonia as nitrogen sources. Although M. mobilis shows very poor growth on methanol (13), especially when ammonia is used as the nitrogen source, we demonstrated that all of the mutants with the exception of the double xoxF mutant were positive for methanol growth. When nitrate was used as the nitrogen source, mutants in nitrate reductase and the assimilatory nitrite reductase showed growth defects, as well as the double xoxF mutant (Table 1). These data were supported by experiments with liquid cultures. However, we were unable to generate growth curves or measure growth rates, as cultures took months to produce turbidity, over which time multiple additions of methanol were required due to the necessity of keeping methanol concentration at a submillimolar level (13). Although both XoxF mutants were capable of growth on methanol, they revealed phenotypes distinguishing them from each other and from the wild type. The mmol_1770 mutant grew poorly on methanol compared to wild type, while the mmol_2048 mutant grew better than the wild type, forming significantly larger colonies. No differences from wild type could be detected for any of the mutants when grown on methylamine. All mutants also grew on formaldehyde with a wild-type rate, based on plate tests (Table 1).
Table 1.
Growth phenotypes and N2O production by mutants compared to wild type
| Protein (derivation) | Growtha on: |
Mean productionb in mM ± SEM (%) of: |
||||
|---|---|---|---|---|---|---|
| Methanol (NO3−) | Methanol (NH4+) | Formaldehyde (NH4+) | N2O− (NO3−*, methylamine†) | N2O (NO3−*, methanol†) | N2O (NO2−*, methanol†) | |
| Wild type | + | + | + | 8.1 ± 0.8 (100) | 5.3 ± 0.6 (100) | 6.8 ± 0.9 (100) |
| Nap (nitrate reductase, mmol_1648) | – | + | + | 0.04 ± 0.006 (0.5) | 0.03 ± 0.005 (0.6) | 6.0 ± 0.7 (88.2) |
| NirK (nitrite reductase, mmol_1061) | + | + | + | 0.1 ± 0.01 (1.2) | 0.1 ± 0.02 (1.9) | 0.2 ± 0.02 (2.9) |
| NirB (nitrite reductase, mmol_1652) | – | + | + | 0.1 ± 0.06 (1.2) | 0.3 ± 0.02 (5.7) | 0.2 ± 0.07 (2.9) |
| NorB (nitric oxide reductase, mmol_1051) | + | + | + | 0.2 ± 0.06 (2.4) | 0.3 ± 0.04 (5.7) | 0.2 ± 0.01 (2.9) |
| Nod (nitric oxide dioxygenase, mmol_1063) | + | + | + | 6.4 ± 0.08 (78.0) | 5.7 ± 0.1 (107.5) | 8.3 ± 0.6 (122.1) |
| XoxF1 (mmol_1770) | +/– | +/– | + | 4.3 ± 0.06 (53.1) | 0.1 ± 0.03 (1.9) | 0.2 ± 0.05 (2.9) |
| XoxF2 (mmol_2048) | ++ | ++ | + | 3.4 ± 0.2 (42.0) | 0.07 ± 0.02 (1.3) | 0.1 ± 0.08 (1.5) |
| XoxF12 (mmol_1770 mmol_2048) | – | – | + | 2.1 ± 0.08 (26.0) | 0.04 ± 0.07 (0.8) | 0.2 ± 0.03 (2.9) |
+, Wild-type growth; –, no growth; +/–, poor growth compared to wild type; ++, enhanced growth compared to wild type. The nitrogen source is indicated in parentheses.
*, Electron acceptor; †, electron donor. Cells for all N2O measurements were grown on methylamine, washed, and incubated with the indicated electron donors/acceptors as described in Materials and Methods. Cultures incubated with methylamine were assayed after 8 h, and cultures incubated with methanol were assayed after 20 h.
Metabolite analysis.
We have previously demonstrated that cultures of M. mobilis accumulate nitrous oxide when supplemented with nitrate (13). We used this metabolite as a proxy for testing the capability of denitrification. Both methylamine and methanol were used as electron donors. As alternative electron donors, pyruvate, succinate, and glucose were tested, but no nitrous oxide accumulation was observed with these substrates (data not shown). Decreased levels of N2O accumulation were demonstrated for all mutants with the exception of the nitric oxide dioxygenase mutant, which behaved like wild type (Table 1). The drop in N2O production was most severe for mutants defective in the denitrification functions, including the assimilatory nitrite reductase, in all of which N2O was only measured at background levels when either methylamine or methanol was supplied as an electron donor. The xoxF mutants behaved differently. The single mutants accumulated approximately half of the wild-type level of N2O, and the double mutant accumulated approximately a quarter of the wild-type level when methylamine was used as electron donor. However, with methanol as an electron donor, N2O was at a background level. We also measured N2O production from nitrite using methanol as an electron donor. The results from these experiments agreed well with the results from the experiments described above, except that N2O production in the nitrate reductase mutant was at a wild-type level, as expected (Table 1).
Enzyme activities.
The identities of some of the mutants were also confirmed by measurements of the predicted enzyme activities. The nitrate reductase mutant was negative for nitrate reductase activity, while the rest of the mutants had wild-type levels of this enzyme (1.6 ± 0.2 nmol min−1 mg of protein−1) in both methylamine-grown and methanol-induced cells. Nitrite reductase activity using methyl viologen as an artificial electron donor was detected in all mutants (2.5 ± 0.5 nmol min−1 mg of protein−1) with the exception of the dissimilatory nitrite reductase mutant, as expected. Assays for nitrite reductase activity using NAD(P)H were negative in all mutants, as well as in the wild-type M. mobilis, suggesting that the activity of this enzyme was under detection limit of the enzyme system used (∼1 nmol min−1 mg of protein−1). The denitrification enzyme activities were also assayed in cells transferred into anaerobic conditions by incubating them in an atmosphere of nitrogen overnight, but assays were negative in these cells, suggesting that the enzymes of interest are expressed in aerobic conditions. The aerobic nature of the denitrification enzymes and low enzyme activities agree with prior data on relatively low expression of the respective genes/proteins (14). Assays for methanol dehydrogenase were also negative, as previously reported for wild-type M. mobilis JLW8 (20, 21), in each of the xoxF mutants, including the mutant with enhanced growth on methanol.
DISCUSSION
In this study, mutant phenotype and metabolite analyses were used to confirm functions of the genes that have been predicted to encode denitrification pathways in M. mobilis JLW8 (Fig. 1). The single subunit nitrate reductase (Nap), Mmol_1648, appears to be involved in both the assimilatory and the dissimilatory denitrification pathways. The role in the former is supported by the methanol growth deficiency of the mutant when nitrate is used as a nitrogen source, and the role in the latter is supported by the lack of accumulation of N2O in the mutant. The predicted roles of the dissimilatory nitrite reductase (NirK; Mmol_1061) and the nitric oxide reductase (NorB; Mmol_1051) are supported by the defect in N2O accumulation. Growth phenotypes of these mutants do not suggest that dissimilatory nitrite reductase or nitric oxide reductase is essential for metabolism of methanol, in aerobic conditions. Indeed, other characterized Methylophilaceae that grow on methanol do not contain genes that encode these enzymes (12). The assimilatory nitrite reductase (NirB; Mmol_1652), however, is essential, confirming its role in generating ammonium for cell biosynthesis. There is no obvious explanation for why the mutant in the putative NAD(P)-linked (assimilatory) nitrite reductase gene (mmol_1652) is defective for accumulation of N2O. No activity could be detected for this enzyme using standard assay. This phenotype may suggest an unusual requirement for measuring in vitro enzyme activity, or a regulatory mechanism that interconnects the functions of the two nitrite reductases, for example, through sharing a reductant with the dissimilatory nitrite reductase, but such a mechanism has not been previously described for known homologs. All of the components of the denitrification pathway are functional in aerobic conditions.
Fig 1.
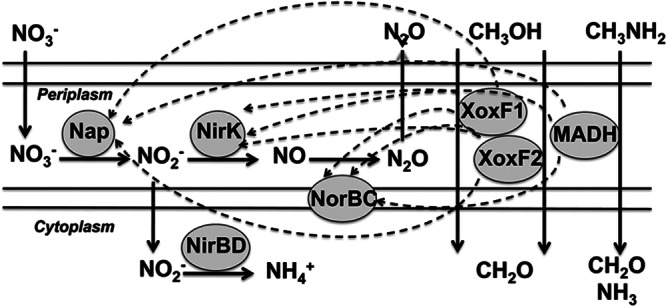
Proposed denitrification pathways in M. mobilis JLW8, and their connection with methylotrophy pathways. Dashed arrows show potential electron transport chains, specific components (cytochromes and/or other redox proteins) of which remain unknown. Cytochromes c encoded by genes in close proximity to the genes for XoxF proteins and methylamine dehydrogenase functions (12, 14) are likely involved. For simplicity, electron transfers from MADH and XoxF to oxygen are not shown.
The phenotypes of the xoxF mutants strongly suggest their involvement in methanol metabolism, identifying them as likely methanol dehydrogenases, with redundant functions. Such a role has been previously proposed for XoxF proteins (22, 23), and weak methanol dehydrogenase activity has been measured for a purified XoxF protein from Methylobacterium extorquens AM1 (24). It is assumed that, like the MxaFI methanol dehydrogenase, the immediate electron acceptor is a cytochrome c (12). However, there has been no previous evidence from analysis of mutants which suggested that this enzyme may be an alternative methanol dehydrogenase. Although we were unable to measure methanol dehydrogenase activity in either wild-type or mutant cells, in accordance with previous observations (21), the fact that either XoxF mutant retains its ability to grow on methanol but the double XoxF mutant is unable to grow on methanol strongly suggests that both XoxF proteins are able to carry out a methanol dehydrogenase reaction. The positive formaldehyde growth phenotype for all xoxF mutants supports this hypothesis.
The dramatically reduced levels of N2O from nitrate or nitrite when methanol is used as an electron donor in XoxF mutants also agree with these enzymes being methanol dehydrogenases and the electron donors to the denitrification pathway (Fig. 1), likely involving cytochrome c proteins as intermediary electron transfer proteins (6). Interestingly, mutation in each of the XoxF enzymes resulted in dramatically reduced levels of accumulated N2O, suggesting some type of a cross talk between the two enzymes, such as a common cytochrome or other electron transfer protein. Moreover, the reduced levels of N2O production with methylamine as an electron donor suggest that XoxF enzymes or their dedicated electron transfer proteins may be involved in methylamine metabolism as well. Although the exact nature of this involvement remains unknown, methylamine dehydrogenases have been shown to be promiscuous in terms of electron transfer specificity (25).
In conclusion, mutant evidence presented here confirms the predicted functions of the components of assimilatory and dissimilatory denitrification pathways and links them to C1 metabolic pathways through XoxF enzymes and likely through methylamine dehydrogenase and respective electron transfer proteins.
ACKNOWLEDGMENTS
This research was supported by the National Science Foundation (grant MCB-0950183).
We are grateful to W. Martens-Habbena, A. Vorobev, and U. Setboonsarng for technical assistance and to D. M. Downs for the generous gift of pDdxRGent.
Footnotes
Published ahead of print 8 March 2013
REFERENCES
- 1. Park JY, Yoo YJ. 2009. Biological nitrate removal in industrial wastewater treatment: which electron donor we can choose. Appl. Microbiol. Biotechnol. 82:415–429 [DOI] [PubMed] [Google Scholar]
- 2. Neef A, Zaglauer A, Meier H, Amann R, Lemmer H, Scheifer KH. 1996. Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl. Environ. Microbiol. 62:4329–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Labbé N, Laurin V, Juteau P, Parnet S, Villemur R. 2007. Microbiological community structure of the biofilm of a methanol-fed, marine denitrification system, and identification of the methanol-utilizing microorganisms. Microb. Ecol. 53:621–630 [DOI] [PubMed] [Google Scholar]
- 4. Ginige MP, Hugenholtz P, Daims H, Wagner M, Keller J, Blackall LL. 2004. Use of stable-isotope probing, full-cycle rRNA analysis, and fluorescence in situ hybridization-microautoradiography to study a methanol-fed denitrifying microbial community. Appl. Environ. Microbiol. 70:588–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baytshtok V, Lu H, Park H, Kim SYR, Chandran KI. 2009. Impact of varying electron donors on the molecular microbial ecology and biokinetics of methylotrophic denitrifying bacteria. Biotechnol. Bioeng. 102:1527–1536 [DOI] [PubMed] [Google Scholar]
- 6. Baker SC, Ferguson SJ, Ludwig B, Page MD, Richter OM, van Spanning RJ. 1998. Molecular genetics of the genus Paracoccus: metabolically versatile bacteria with bioenergetic flexibility. Microbiol. Mol. Biol. Rev. 62:1046–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bamforth CW, Quayle JR. 1978. Aerobic and anaerobic growth of Paracoccus denitrificans on methanol. Arch. Microbiol. 119:91–97 [DOI] [PubMed] [Google Scholar]
- 8. Sperl GT, Hoare DS. 1971. Denitrification with methanol: a selective enrichment for Hyphomicrobium species. J. Bacteriol. 108:733–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Green SJ, Prakash O, Gihring TM, Akob DM, Jasrotia P, Jardine PM, Watson DB, Brown SD, Palumbo AV, Kostka JE. 2010. Denitrifying bacteria isolated from terrestrial subsurface sediments exposed to mixed-waste contamination. Appl. Environ. Microbiol. 76:3244–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huijie L, Kalyuzhnaya M, Chandral K. 2012. Comparative proteomic analysis reveals insights into anoxic growth of Methyloversatilis universalis FAM5 on methanol and methanol. Environ. Microbiol. 14:2935–2945 [DOI] [PubMed] [Google Scholar]
- 11. Chistoserdova L. 2011. Methylotrophy in a lake: from metagenomics to single organism physiology. Appl. Environ. Microbiol. 77:4705–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lapidus A, Clum A, LaButti K, Kaluzhnaya MG, Lim S, Beck DAC, Glavina del Rio T, Nolan N, Mavromatis K, Huntemann M, Lucas S, Lidstrom ME, Ivanova N, Chistoserdova L. 2011. Genomes of three methylotrophs from a single niche uncover genetic and metabolic divergence of Methylophilaceae. J. Bacteriol. 193:3757–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kalyuhznaya MG, Martens-Habbena W, Wang T, Hackett M, Stolyar SM, Stahl DA, Lidstrom ME, Chistoserdova L. 2009. Methylophilaceae link methanol oxidation to denitrification in freshwater lake sediment as suggested by stable isotope probing and pure culture analysis. Environ. Microbiol. Rep. 1:385–392 [DOI] [PubMed] [Google Scholar]
- 14. Beck DAC, Hendrickson EL, Vorobev A, Wang T, Lim S, Kalyuzhnaya MG, Lidstrom ME, Hackett M, Chistoserdova L. 2011. An integrated proteomics/ transcriptomics approach points to oxygen as the main electron sink for methanol metabolism in Methylotenera mobilis. J. Bacteriol. 193:4758–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grasshoff K, Kremling K, Ehrhardt M. (ed). 1999. Methods of seawater analysis, 3rd ed Wiley-VCH, New York, NY [Google Scholar]
- 16. Garrett RH. 1972. The induction of nitrite reductase in Neurospora crassa. Biochim. Biophys. Acta 264:481–489 [DOI] [PubMed] [Google Scholar]
- 17. Kalyuzhnaya MG, Hristova KR, Lidstrom ME, Chistoserdova L. 2008. Characterization of a novel methanol dehydrogenase in representatives of Burkholderiales: implications for environmental detection of methylotrophy and evidence for convergent evolution. J. Bacteriol. 190:3817–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kalyuzhnaya MG, Lapidus A, Ivanova N, Copeland AC, McHardy AC, Szeto E, Salamov A, Grigoriev IV, Suciu D, Levine SR, Markowitz VM, Rigoutsos I, Tringe SG, Bruce DC, Richardson PM, Lidstrom ME, Chistoserdova L. 2008. High resolution metagenomics targets major functional types in complex microbial communities. Nat. Biotechnol. 26:1029–1034 [DOI] [PubMed] [Google Scholar]
- 19. Chistoserdova L, Kalyuzhnaya MG, Lidstrom ME. 2009. The expanding world of methylotrophic metabolism. Annu. Rev. Microbiol. 63:477–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chistoserdova L. 2011. Modularity of methylotrophy, revisited. Environ. Microbiol. 13:2603–2622 [DOI] [PubMed] [Google Scholar]
- 21. Kalyuzhnaya MG, Bowerman S, Lidstrom ME, Chistoserdova L. 2006. Methylotenera mobilis, gen. nov. sp. nov., an obligately methylamine-utilizing bacterium within the family Methylophilaceae. Int. J. Syst. Evol. Microbiol. 56:2819–2823 [DOI] [PubMed] [Google Scholar]
- 22. Halsey KH, Carter AE, Giovannoni SJ. 2012. Synergistic metabolism of a broad range of C1 compounds in the marine methylotrophic bacterium HTCC2181. Environ. Microbiol. 14:630–640 [DOI] [PubMed] [Google Scholar]
- 23. Hou S, Makarova KS, Saw JH, Senin P, Ly BV, Zhou Z, Ren Y, Wang J, Galperin MY, Omelchenko MV, Wolf YI, Yutin N, Koonin EV, Stott MB, Mountain BW, Crowe MA, Smirnova AV, Dunfield PF, Feng L, Wang L, Alam M. 2008. Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum Verrucomicrobia. Biol. Direct 3:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmidt S, Christen P, Kiefer P, Vorholt JA. 2010. Functional investigation of methanol dehydrogenase-like protein XoxF in Methylobacterium extorquens AM1. Microbiology 156:2575–2586 [DOI] [PubMed] [Google Scholar]
- 25. Meschi F, Wiertz F, Klauss L, Blok A, Ludwig B, Merli A, Heering HA, Rossi GL, Ubbink M. 2011. Efficient electron transfer in a protein network lacking specific interactions. J. Am. Chem. Soc. 133:16861–16867 [DOI] [PubMed] [Google Scholar]


