Abstract
The plant-pathogenic bacterium Dickeya dadantii produces several pectinolytic enzymes that play a major role in the soft-rot disease. Eight characterized endopectate lyases are secreted in the extracellular medium by the type II secretion system, Out. They cleave internal glycosidic bonds of pectin, leading to plant tissue maceration. The D. dadantii pectate lyases belong to different families, namely, PL1, PL2, PL3, and PL9. Analysis of the D. dadantii 3937 genome revealed a gene encoding a new protein of the PL9 family, which already includes the secreted endopectate lyase PelL and the periplasmic exopectate lyase PelX. We demonstrated that PelN is an additional extracellular protein secreted by the Out system. However, PelN has some unusual characteristics. Although most pectate lyases require a very alkaline pH and Ca2+ for their activity, the PelN activity is optimal at pH 7.4 and in the presence of Fe2+ as a cofactor. PelN is only weakly affected by the degree of pectin methyl esterification. The PelN structural model, constructed on the basis of the PelL structure, suggests that the PelL global topology and its catalytic amino acids are conserved in PelN. Notable differences concern the presence of additional loops at the PelN surface, and the replacement of PelL charged residues, involved in substrate binding, by aromatic residues in PelN. The pelN expression is affected by different environmental conditions, such as pH, osmolarity, and temperature. It is controlled by the repressors KdgR and PecS and by the activator GacA, three regulators of D. dadantii pectinase genes. Since a pelN mutant had reduced virulence on chicory leaves, the PelN enzyme plays a role in plant infection, despite its low specific activity and its unusual cofactor requirement.
INTRODUCTION
The plant cell wall consists of a complex network of polysaccharides that can be degraded by depolymerizing enzymes produced by phytopathogenic, or plant-associated, microorganisms. The variety of enzymes involved in their degradation reflects the complexity of the plant cell wall polysaccharides. Dickeya dadantii (formerly Erwinia chrysanthemi) produces a large set of enzymes and isoenzymes to disassemble the plant cell wall (1). It is responsible for soft-rot diseases in a wide range of plants. The combination of different enzymatic activities allows D. dadantii to efficiently degrade pectin and to use the liberated pectic oligomers as carbon sources for growth. Maceration of the plant tissue mainly results from the action of endocleaving pectate lyases (2). Endopectate lyases are the major macerating enzymes produced by D. dadantii, and they are secreted by the type II secretion system, Out (3–5). In the D. dadantii strain 3937, eight secreted endopectate lyases have already been characterized: PelA, PelB, PelC, PelD, PelE, PelI, PelL, and PelZ (1, 6, 7). In addition, D. dadantii 3937 produces two exocleaving pectate lyases, PelW and PelX, which are located in the cytoplasm and in the periplasm, respectively (8, 9). Pectate lyases cleave the α1,4-glycosidic linkages of polygalacturonate by a β-elimination mechanism, generating oligogalacturonides with C4-C5 unsaturation at the nonreducing end. Pectate lyases are usually specific for the nonmethylated polysaccharide or for pectins with a low degree of methyl esterification (pectate). The D. dadantii 3937 pectate lyases differ in terms of specific activity, optimum pH, substrate preference, and length of products (7, 10, 11, 12). Their cleavage mechanism is cation dependent. Ca2+ is the most frequent cofactor of characterized pectate lyases (11), but other cations, such as Mn2+, Co2+, or Ni2+, are cofactors of certain intracellular pectate lyases (8, 9, 13).
Pectate lyases are classified into different families of polysaccharide lyases (PL) according to their primary amino acid sequences (14–16; http://www.cazy.org/). D. dadantii pectate lyases belong to four families: PL1, PL2, PL3, and PL9. The PL1 family comprises most known pectin or pectate lyases of plant, fungal or bacterial origin, including PelA, PelB, PelC, PelD, PelE, and PelZ of D. dadantii (17, 18). The cytoplasmic exopectate lyase PelW of D. dadantii is a member of the PL2 family (8, 19), and the D. dadantii endopectate lyase PelI belongs to the PL3 family (7, 20). The endopectate lyase PelL and the exopectate lyase PelX of D. dadantii are both members of the PL9 family (9, 10, 21). The study of the three-dimensional structure of D. dadantii PelC led to the discovery of a novel protein structure, a right-handed parallel β-helix (22), that is conserved in the PL1 members and also in pectate lyases of the PL3 and PL9 families (20, 21).
Analysis of the D. dadantii genome, to search for any genes encoding other potential pectate lyases, revealed the presence of pelN, a gene predicted to encode a PL9 family protein. We report here the characterization of the PelN protein's enzymatic activity, its secretion by the Out system of D. dadantii, a modeling of the PelN structure, and the regulation of pelN expression, in different growth conditions, involving previously characterized pectinase gene regulators.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains of D. dadantii or Escherichia coli, the plasmids, and the oligonucleotides used in the present study are listed in Table 1. The phi-EC2 generalized transducing phage was used for transduction (23).
Table 1.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Genotype, phenotype, and/or sequence (5′–3′)a | Source or reference |
|---|---|---|
| Strains | ||
| D. dadantii | ||
| 3937 | Wild type | Laboratory collection |
| A1524 | rafR ganB pecS::Mu-Cmr | 45 |
| A3953 | pecS::Mu-Cmr | 45 |
| A4239 | gacA::Cmr | 40 |
| A4114 | kdgR::Smr | Laboratory collection |
| A4415 | pelD::uidA-Kmr | 36 |
| A5051 | pelN::uidA-Kmr | This study |
| A5185 | rafR ganB pecS::Mu-Cmr pelN::uidA-Kmr | This study |
| A5186 | rafR ganB pecS::Mu-Cmr pelL::uidA-Kmr | This study |
| A5189 | rafR ganB ΔoutD pecS::Mu-Cmr pelN::uidA-Kmr | This study |
| A5190 | rafR ganB ΔoutD pecS::Mu-Cmr pelL::uidA-Kmr | This study |
| A5194 | gacA::Cmr pelN::uidA-Kmr | This study |
| A5218 | pecS::Mu-Cmr pelN::uidA-Kmr | This study |
| A5293 | gacA::Cmr pelL::uidA-Kmr | This study |
| A5298 | pelL::uidA-Kmr | 10 |
| A5299 | kdgR::Smr pelN::uidA-Kmr | This study |
| E. coli | ||
| NM522 | Δ(lac-proAB) Δ(mcrB-hsdSM)5 supE thi [F′ proAB lacIq30 lacZΔM15] | Laboratory collection |
| BL21(DE3) | E. coli B, F− dcm ompT hsdS gal λ(DE3), T7 polymerase gene under the lacUV5 promoter | 29 |
| Plasmids | ||
| pGEM-T | lacZ; Apr | Promega |
| pBS Ap | pBluescript SK(+); Apr | Stratagene |
| pT7-5 | T7 phi10; Apr | 28 |
| pI2805 | pBS Ap; pelN+ | This study |
| pI2812 | pBS Ap; pelN::uidA-Kmr | This study |
| pI3372 | pGEM-T; pelN+ | This study |
| pNA13 | pT7-5 derivative with a 1.4-kb BamHI-XbaI fragment from pI3372, pelN+ | This study |
| Oligonucleotides | ||
| PelNG | GCGGATCCCGGTAAATGCTTTGGGTTTG | This study |
| PelND | CGTCTAGAGTTTCGCCGCTTATTTCAGTTC | This study |
Cmr, chloramphenicol resistance; Apr, ampicillin resistance; Smr, streptomycin resistance; Kmr, kanamycin resistance.
Media and growth conditions.
Bacteria were grown in Luria-Bertani (LB) medium or in M63 minimal medium (24). When required, the media were solidified with 15 g of agar liter−1. D. dadantii cells were usually incubated at 30°C, and E. coli cells at 37°C. Carbon sources were added at 2 g liter−1. When required, antibiotics were added at the following concentrations: ampicillin (Ap), 50 μg ml−1; kanamycin (Km), 20 μg ml−1; and chloramphenicol (Cm), 20 μg ml−1.
Enzyme assays.
The β-glucuronidase activity was measured by monitoring the degradation of p-nitrophenyl-β-d-glucuronide into p-nitrophenol at 405 nm (25). Specific activity is expressed as nmol of product liberated per min per mg of bacterial dry weight (nmol min−1 mg−1).
Pectate lyase activity was determined by monitoring, spectrophotometrically, the formation of unsaturated products from pectic substrates, at 230 nm. The mixture usually used to assay the D. dadantii pectate lyases contains 100 mM Tris-HCl buffer (pH 8.5), polygalacturonate at 0.5 g liter−1, and 0.1 mM Ca2+.
The assay mixture adapted to PelN activity consisted of 100 mM TES (N-tris[hydroxymethyl] methyl-2-aminoethane-sulfonic acid) buffer (pH 7.4), polygalacturonate at 0.5 g liter−1, 0.1 mM Fe2+, and the enzyme, in a total volume of 1 ml. The appearance of products was monitored for 3 min, at 37°C. Specific activity is expressed as nmol of unsaturated products liberated per min, either per mg of bacterial dry weight or per mg of proteins (nmol min−1 mg−1). The Vmax and Km of PelN were determined in the standard conditions using polygalacturonate concentrations from 0.1 to 0.75 g liter−1. The optimum pH was determined using 100 mM MES or TES buffer from pH 5.5 to 8. The influence of divalent cations was assessed in the presence of the chloride salt of Ca2+, Cu2+, Co2+, Ba2+, Fe2+, Mg2+, Co2+, or Mn2+ at a final concentration of 0.1 or 1 mM. To complex the cations, EDTA was added at a final concentration of 1 to 10 mM. Either polygalacturonate or pectins, with different degrees of methylation (9.7 to 91% from Copenhagen pectin), were used as substrates at a final concentration of 0.5 g liter−1.
Recombinant DNA techniques.
Preparation of plasmid DNA, restriction digestions, ligations, DNA electrophoresis, and transformations were all carried out as previously described (26). The pelN gene was amplified by PCR and cloned in the pGEM-T vector (Promega). The nucleotide sequence of the 1,392 nucleotide (nt) cloned fragment was verified.
After subcloning in the vector Bluescript KS(+) Apr (Stratagene), a genetic fusion was constructed by insertion of the uidA-Km cassette (25) into the EcoRI site situated inside pelN. The resulting plasmid was introduced into D. dadantii cells, and the insertion was integrated into the chromosome by marker-exchange recombination, after successive cultures in low phosphate medium in the presence of the appropriate antibiotic (27). The correct recombination of the insertion was verified by PCR.
PelN overproduction.
The protein PelN was overproduced using the T7 promoter-T7 RNA polymerase system (28). The 1.4-kb pelN fragment was inserted into the pT7-5 vector under the control of the T7 promoter. The resulting plasmid, pNA13, was introduced into the E. coli strain BL21(DE3), which contains a chromosomal copy of the T7 RNA polymerase gene under the control of a lacUV5 promoter (29). The BL21(DE3)/pNA13 cells were grown, at 30°C, in LB medium with 200 μg of ampicillin ml−1. When the optical density at 600 nm reached 0.4 to 0.6, the synthesis of T7 RNA polymerase was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) at a final concentration of 2 mM, and cells were grown for an additional 2 h. A culture without IPTG induction was performed as a control.
Cellular fractionation.
Different fractions were prepared from E. coli BL21(DE3)/pNA13-induced cells. The bacterial cells were recovered by centrifugation for 2 min at 8,000 rpm. The pellet was suspended in 0.7 ml of 80 mM Tris-HCl (pH 8.0)–0.1% Triton X-100, and the cells were broken by sonication. After centrifugation for 2 min at 10 000 rpm, the supernatant containing the soluble proteins (fraction S) was recovered. The pellet, corresponding to insoluble proteins and cell debris, was suspended in 0.7 ml of 80 mM Tris-HCl buffer (pH 8; fraction I), and the periplasmic fraction (P) was obtained by osmotic shock (30). A variation of this method was also used. After centrifugation of the culture, a mixture of 33 mM Tris-HCl buffer (pH 8), 1 mM EDTA, and 40% sucrose was added to the cell pellet. After a 5-min incubation on ice, the supernatant, containing periplasmic proteins (Ps), was recovered by centrifugation for 4 min at 12,000 rpm.
Analytical procedures.
The protein concentrations were determined by the Bradford method using a commercial protein assay kit (Bio-Rad) and bovine serum albumin (BSA) as a standard. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on slab gels (4% stacking gel and 12% separating gel) using the mini-Protean II system (Bio-Rad). The proteins were revealed by staining with Coomassie blue G250.
Western blot analysis.
The D. dadantii cultures were centrifuged for 4 min at 12,000 rpm to separate the cells (fraction C) and the culture supernatant (fraction Sp). The proteins of the supernatant were concentrated by acetone precipitation (31). After separation by SDS-PAGE, proteins were transferred onto a nitrocellulose membrane and analyzed by immunoblotting with PelL antibody (21) at a dilution of 1:1,500.
Pathogenicity test.
Plant infections were performed as previously described (32). Chicory leaves were infected using 106 bacteria per inoculation site. After incubation in a dew chamber for 24 h at 30°C, the length of rotted tissue was measured to estimate the symptom severity. Potato tubers were infected using 107 bacteria per inoculation site. After incubation in a dew chamber for 48 h at 30°C, the weight of rotted tissue was measured to estimate the symptom severity. After infection, the plant macerated tissue was recovered and used to perform bacterial cell numeration by dilution plating and the β-glucuronidase assay. The specific activity was calculated in nmol of products formed per min per 109 bacteria. In each experiment, the wild-type strain and the pelD mutant were used as controls. The wild-type strain shows no detectable β-glucuronidase activity. The pelD mutant gives reduced symptoms and the expression of the pelD::uidA fusion is induced during infection.
Molecular modeling.
The structure of the predicted mature form of PelN (residues 26 to 439) was modeled using the homology molecular modeling program MODELLER 9v10 (33). The software identified a unique crystal structure as the template, providing a good level of confidence in the modeling: PDB ID 1RU4 (Erwinia chrysanthemi pectate lyase Pel9A, designated in the present study by D. dadantii PelL). Twenty distinct models were generated and their geometry was assessed by a Ramachandran plot, calculated with the program PROCHECK (34). The most satisfactory model was then retained. It has 92.4% of nonproline and nonglycine residues in the most favored regions, 7.6% in the additionally allowed regions, and none in the disallowed regions.
RESULTS
D. dadantii encodes an additional protein of the PL9 family.
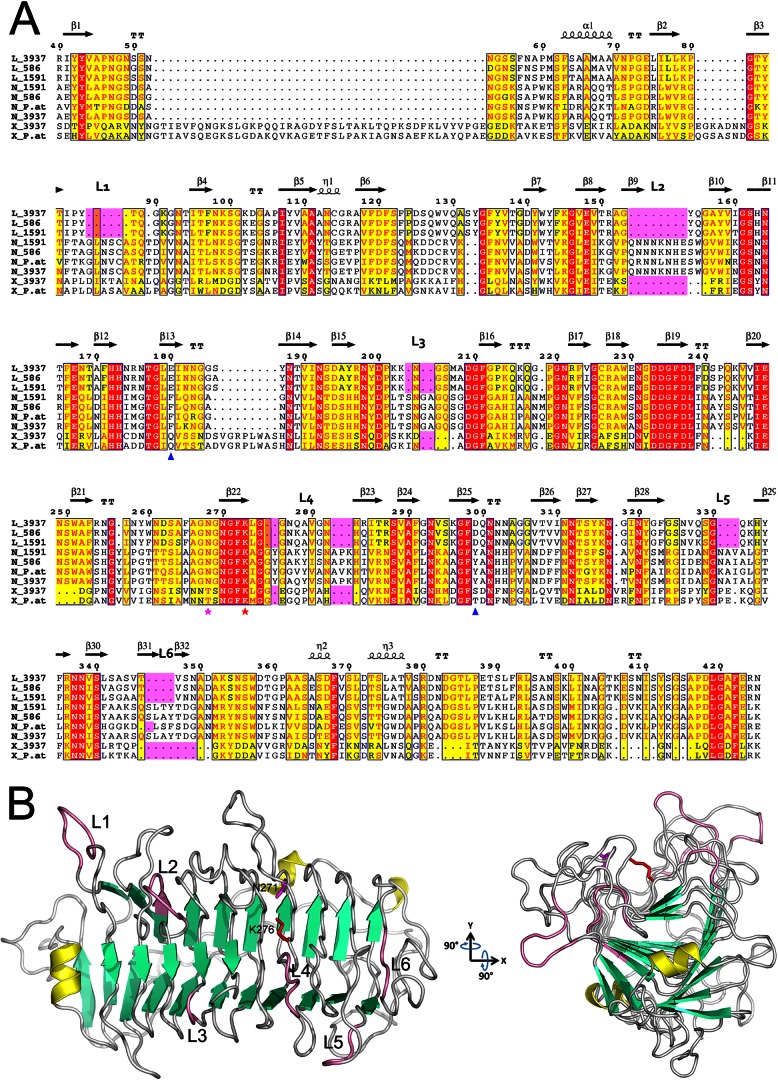
The D. dadantii 3937 genome (35; GenBank accession number CP002038.1) encodes three proteins of the PL9 family, including the two previously characterized enzymes PelL and PelX. The third gene (ID19391) was named pelN. The predicted pelN product is a 439-amino-acid protein homologous to the endopectate lyase PelL (38% identity) and to the exopectate lyase PelX (26% identity) of D. dadantii (Fig. 1A). The main difference between PelX and PelN is the presence of a large extension in PelX (320 residues), typical of exocleaving enzymes (9). The N-terminal sequences of PelL and PelN show the characteristics of classical signal sequences of proteins exported by the Sec system. Since the mature PelL protein has been crystallized, the PelL signal sequence cleavage site is already known (21). From a comparison with PelL, the PelN cleavage site is predicted to be between the two alanine residues at positions 25 and 26.
Fig 1.
Alignment of the PL9 family pectate lyases and the PelN structural model. (A) Alignment of representative examples from the PL9 family, namely, PelL, PelN, and PelX, of Dickeya strains 3937, Ech586, and Ech1591, and of Pectobacterium atrosepticum SCRI1043 (UniProt/NCBI accession codes PLYL_DICD3, D2BWX2_DICD5, C6CF33_DICZE, C6CJG7_DICZE, D2C125_DICD5, CAG75452.1, E0SAR9_DICD3, Q9Z5P8_ERWCH, and YP_052593.1). The residue numbering is for D. dadantii 3937 PelL. Secondary structure elements identified in the crystal structure of D. dadantii PelL (PDB 1RU4) are indicated at the top. The additional loops (L1 to L6), specific for PelN, are also indicated at the top and highlighted in pink. Residues shown with red or yellow backgrounds are identical or similar, respectively, throughout the sequences. The catalytic asparagine and lysine involved in PelL activity are shown by magenta and red asterisks, respectively. Asp299 and Glu180, presumed to form calcium-mediated interactions with the substrate in PelL, are indicated with blue triangles. Alignment was generated using the ESPript web server (44). (B) The PelN model (residues 26 to 439) was generated using the program MODELLER 9v10, with the D. dadantii PelL structure (PDB 1RU4) as a template. The secondary structure elements are shown in yellow for α-helices, in green for β-strands, and in pink for specific PelN loops. Catalytic asparagine and lysine are shown in magenta and red, respectively, using the PelN numbering (N271 and K276). To highlight the presumed substrate-binding cleft and the surrounding extra loops, the structure was rotated around the x and y axes by 90°. The image in panel B was generated using PYMOL.
The pelN gene is not situated in a cluster encoding other pectinases. On its 5′ side, the pelN gene is separated by a 299-nt intergenic space from the preceding gene, kdsB, which encodes an enzyme involved in lipopolysaccharide biosynthesis. The kdsB gene is followed by a sequence typical of a rho-independent termination site, situated 29 nt after its TGA stop codon. On its 3′ side, pelN is separated by 264 nt from the gene ycbJ, of unknown function but conserved in Enterobacteriaceae. The pelN gene is followed by a typical rho-independent transcription termination site, situated 61 nt after its TAA stop codon. Thus, on the basis of this genetic organization, pelN is thought to constitute an independent transcriptional unit.
PelN is exported and secreted by the Out system.
The pelN gene was inserted in the pT7-5 vector and the resulting plasmid, pNA13, was used to overproduce the PelN protein in E. coli BL21(DE3) (Fig. 2). Analysis of different cellular fractions revealed the overproduction of a protein of ∼43 kDa, mainly located in the periplasmic fraction (Fig. 2). This size is close to that of the 44,498 Da estimated for the PelN mature form from the pelN nucleotide sequence. The periplasmic localization of the overproduced protein confirmed the functionality, in E. coli, of the PelN signal sequence.
Fig 2.
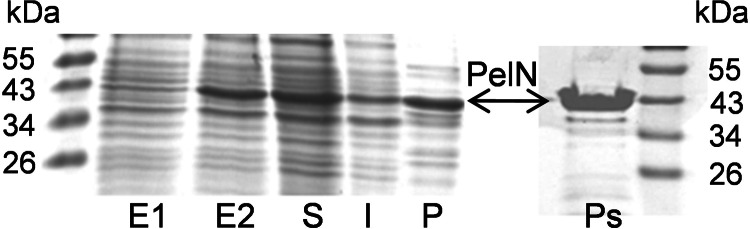
Overproduction and cellular localization of the PelN protein in E. coli. Whole-cell lysates of BL21(DE3)/pNA13 before induction (E1) and after induction with IPTG (E2). Fractions extracted from IPTG-induced cells: soluble (S), insoluble (I), and periplasmic fractions obtained by osmotic shock (P) or by a variation of this method (Ps). The proteins were separated by SDS-PAGE and stained with Coomassie blue. The positions of the molecular mass standards are indicated.
Since PelN and PelL share 38% identity and 51% similarity, we tested whether polyclonal antibodies raised against PelL (21) could also detect PelN. Analysis of periplasmic extracts of the PelN overproducing E. coli strain demonstrated a cross-reaction of the PelL antibodies with PelN (Fig. 3A). Consequently, the PelL antibodies could be used to detect PelN in the different cellular fractions of a D. dadantii mutant in which pelL is inactivated (Fig. 3B). The detection of PelN in culture supernatants indicated that it is an extracellular protein. We next used a pelL outD mutant to test whether PelN is secreted by the Out system. Although PelN was found to be secreted in the culture supernatant when the Out system was functional, PelN was recovered in the cell fraction when outD was inactivated (Fig. 3B). Since PelL is known to be secreted by the Out system, a parallel analysis of the pelN and pelN-outD mutants was performed as a control, giving similar results (Fig. 3C). These data demonstrate that PelN is an additional protein secreted by the Out system and that the Out system is involved in the secretion of all of the characterized D. dadantii extracellular pectinases.
Fig 3.
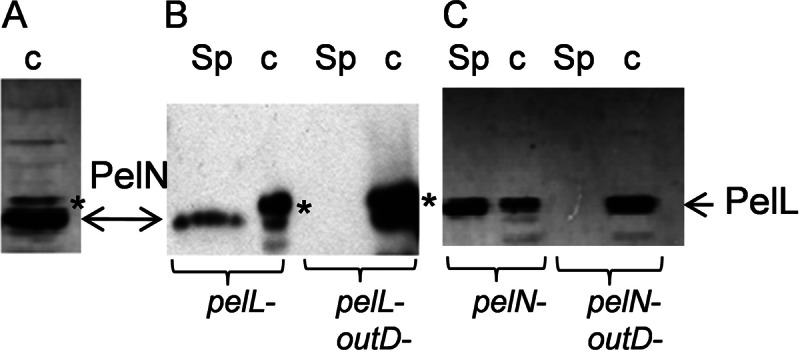
Secretion of the PelN protein by the Out system. After separation of the proteins by SDS-PAGE, PelL antibodies were used to detect the proteins PelN and PelL by Western blotting. (A) The periplasmic fraction was prepared from IPTG-induced cells of E. coli BL21(DE3)/PNA13 overproducing PelN. Cultures of various D. dadantii mutants were used to prepare culture supernatants (Sp) and cell lysates (c). (B and C) PelN was detected in the pelL and pelL-outD mutants (B), and PelL was detected in the pelN and pelN-outD mutants (C). In each D. dadantii strain, the pecS gene was also inactivated in order to increase PelL and PelN production. Although the development usually lasted 1 min to detect PelL, it was increased to 10 min for PelN detection in D. dadantii. A nonspecific band is indicated with an asterisk.
Biochemical characterization of the PelN activity.
The PelN protein, recovered in the periplasmic fraction of E. coli BL21(DE3)/pNA13 cells (Fig. 2), was used for biochemical characterization. Enzymatic assays indicated that PelN has a pectate lyase activity and that it requires very particular conditions to cleave its substrates, be they polygalacturonate or pectins. Notably, PelN shows optimum activity at pH 7.4 (Fig. 4A), a value which is clearly lower than the optimal pH of other D. dadantii extracellular pectate lyases. The activity of most known pectate lyases requires Ca2+ or, for some intracellular enzymes, Mn2+, Co2+, or Ni2+ as cofactor(s). PelN had no enzymatic activity in the presence of EDTA (Fig. 4B), confirming the need for a cation. No increase in enzymatic activity was observed after the addition of Ca2+, Co2+, Cu2+, Mg2+, Mn2+, Ni2+, Zn2+, or Ba2+, but a clear stimulation of the pectate lyase activity was detected in the presence of Fe2+ (Fig. 4B and data not shown). Optimal activity was observed at Fe2+ concentrations ranging from 0.1 to 0.3 mM (data not shown). PelN is the first example of a pectate lyase which requires Fe2+ as a cofactor.
Fig 4.
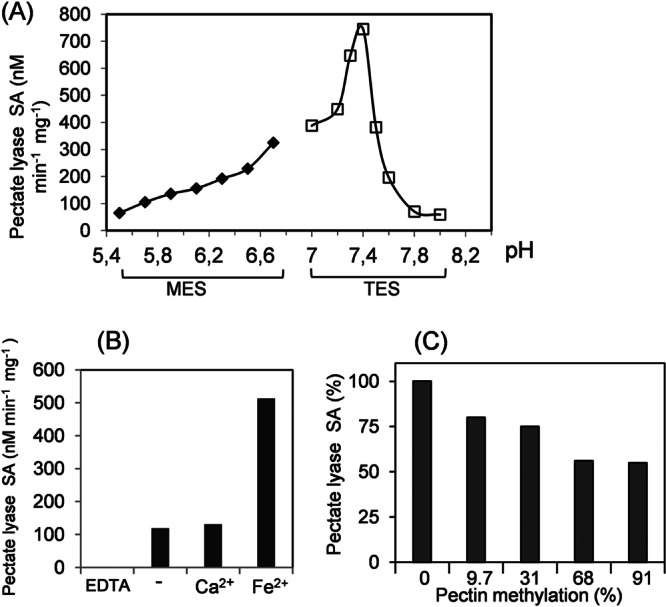
Influence of cations, pH, and substrate methylation on PelN activity. (A) The pectate lyase specific activity (SA) was assayed at different pHs, from 5.5 to 8, in 100 mM MES or TES buffer, in the presence of 0.1 mM Fe2+ and 0.5 g of polygalacturonate liter−1. (B) The pectate lyase specific activity (SA) was assayed in 100 mM TES buffer (pH 7.4), 0.5 g of polygalacturonate liter−1 (−) and after the addition of 1 mM EDTA, 0.1 mM Ca2+, or 0.1 mM Fe2+. (C) The PelN specific activity (SA) was tested using pectins with different degrees of methylation. The assay was performed in the presence of 0.5 g of pectin liter−1 in 100 mM TES buffer (pH 7.4) and 0.1 mM Fe2+. These results are the average of at least three independent assays, with standard deviations of <10%.
Consequently, the standard conditions used for the PelN assay were 0.1 M TES buffer (pH 7.4) with 0.1 mM Fe2+. The initial velocity of the enzyme was determined using different polygalacturonate concentrations. The Km and Vmax values for PelN were 2.5 g liter−1 and 170 nmol min−1 mg−1, respectively. The influence of substrate methylation on PelN activity was tested using polygalacturonate and pectins with various degrees of methyl esterification (Fig. 4C). Although PelN showed maximal activity on polygalacturonate, it also exhibited activity on all of the substrates tested. PelN retained >50% of the maximal activity on pectin with 91% methyl esterification. The PelN enzymatic activity was not affected by the addition of different monomeric monosaccharides: galacturonate, galactose, arabinose, or rhamnose (data not shown).
The PelN thermal stability was tested at 50°C in the presence of different compounds (Fig. 5). PelN retained >50% of its activity after 90 min at 50°C. Its stability was not at all, or only weakly, increased by the addition of Fe2+ or polygalacturonate. In contrast, with EDTA 1 mM, PelN lost 50% of its activity after 30 min at 50°C. Thus, PelN stability is probably enhanced in the presence of divalent cations, which may serve to stabilize the protein structure.
Fig 5.
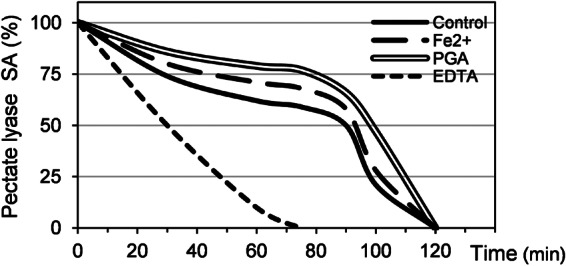
PelN thermal stability. The pectate lyase specific activity (SA) of PelN was assayed after incubation at 50°C for different time intervals of up to 120 min. The compounds were added at the following concentrations: 0.1 mM Fe2+, 0.5 g of polygalacturonate (PGA) liter−1, and 1 mM EDTA. After incubation, the assay was performed using 100 mM TES buffer (pH 7.4), 0.1 mM Fe2+, 0.5 g of polygalacturonate liter−1. These results are the averages of at least three independent experiments, with standard deviations between 7 to 10% for each value.
Since the PelN optimal conditions appeared very different from that defined for previously characterized pectate lyases, we reexamined the enzymatic activities of D. dadantii 3937 pectate lyases (Table 2). In contrast to PelL and PelX, two other enzymes of the PL9 family, PelN showed no detectable activity in the usual pectate lyase assay conditions (0.1 M Tris-HCl buffer [pH 8.5], 0.1 mM Ca2+). When measured in the PelN optimal assay conditions (0.1 M TES buffer [pH 7.4], 0.1 mM Fe2+), PelL and PelX retained 70 and 85% of their activity, respectively (Table 2). The major D. dadantii pectate lyases, PelA, PelB, PelC, PelD, and PelE, were also tested in these two conditions (Table 2). When their activity was compared, in the PelN buffer relative to the usual buffer, PelC and PelE retained a low level of activity (0.3 to 0.4%), PelB at 7%, and PelD at 58%, and PelA showed similar activity in both conditions.
Table 2.
Pectate lyase activity of PelN and other D. dadantii pectate lyases
| Enzyme(s)a | Enzyme activity (nM min−1)b in: |
|
|---|---|---|
| Optimal PelN conditions | Usual pectate lyase conditionsc | |
| PelN | 5.98 | ND |
| PelL | 5.63 | 7.98 |
| PelL + PelN | 9.52 | – |
| PelX | 8.19 | 9.69 |
| PelX + PelN | 14.40 | – |
| PelA | 6.46 | 6.89 |
| PelA + PelN | 9.72 | – |
| PelB | 6.63 | 90.2 |
| PelB + PelN | 11.92 | – |
| PelC | 9.33 | 2,446 |
| PelC + PelN | 16.13 | – |
| PelD | 9.52 | 16.35 |
| PelD + PelN | 16.06 | – |
| PelE | 5.25 | 1,752 |
| PelE + PelN | 7.48 | – |
PelN and other pectate lyases (PelA, PelB, PelC, PelD, PelL, and PelX) were assayed independently or concomitantly.
The quantity of each enzyme was chosen to give an activity between 5 and 10 nM min−1 when the assay was performed using the optimal PelN conditions: 100 mM TES buffer (pH 7.4) and 0.1 mM Fe2+. The activity of each enzyme was also determined in the usual pectate lyase conditions: 100 mM Tris-HCl buffer (pH 8.5) and 0.1 mM Ca2+.
ND, not detected; –, not tested.
Natural pectins are complex substrates that include various minor sugars and multiple modifications. Consequently, different pectate lyase isoenzymes can act in synergy, i.e., product of one isoenzyme can be a better substrate for another isoenzyme. In this way, the combined activity of two isoenzymes would be higher than a simple sum of them. Therefore, combinations of different isoenzymes could disclose synergism between them. Using the PelN optimal assay conditions, we analyzed whether PelN activity could be enhanced by the presence of another D. dadantii pectate lyase: PelL, PelX, PelA, PelB, PelC, PelD, or PelE (Table 2). The activity of each enzyme was determined independently, and concomitantly, with PelN. A simple additive effect was observed after the concomitant addition of PelN with either PelX, PelB, PelC, or PelD. However, in the case of PelL, PelA, and PelE, the activity observed in the presence of the two enzymes was even lower than the sum of the two independent activities. Thus, no synergistic effect was observed between PelN and any of the other D. dadantii pectate lyases.
Identification of PelN orthologs and molecular modeling of the PelN structure.
A PelN ortholog is encoded by all of the sequenced genomes of the Dickeya and Pectobacterium species, such as D. zeae Ech586, D. chrysanthemi Ech1591, D. paradisiaca Ech703, P. carotovorum PC1 and WPP14, P. wasabiae WWPP163, P. brasiliensis 1692, and P. atrosepticum SCRI1043 (72 to 93% identity) (Fig. 1A and data not shown). In contrast, PelL orthologs are observed only in Dickeya species (79 to 93% identity between PelL of strains 3937, Ech586, Ech703, and Ech1591) but not in the Pectobacterium species. In addition to the Dickeya and Pectobacterium genera, PelN homologues are also found in certain pathovars of Pseudomonas syringae pv. syringae and pv. phaseolicola (62 to 64% identity), which are also pectinase-producing plant pathogens.
Since most pectate lyases share a particular three-dimensional structure, consisting of a parallel β-helix, we wanted to try to model the PelN structure. The structural data of D. dadantii PelL (21), sharing 38% identity with PelN, seemed to provide a good level of confidence for the PelN modeling (Fig. 1B). This analysis suggests that PelL and PelN share a highly similar global topology. The predicted PelN structure consists of a 10-coil parallel β-helix, including stabilizing intramolecular bonds, such as an asparagine ladder and aromatic stacks. The Asn and Lys residues involved in the PelL catalytic site are conserved in PelN. However, two aromatic chains replace two carboxylic chains of PelL, both involved in Ca-mediated substrate binding. Moreover, six additional loops, located around the potential substrate-binding groove, are predicted at the PelN surface (Fig. 1B). These structural differences could be responsible for the unusual enzymatic characteristics of PelN.
Virulence of the pelN mutant.
The extent of soft-rot caused by the pelN mutant was tested on chicory leaves and potato tubers and compared to the symptoms caused by the wild-type strain 3937 (Fig. 6 and data not shown). Although no significant difference was detected after infection of potato tubers, we observed a weak decrease in the degree of maceration of chicory leaves caused by the pelN mutant, compared to the wild-type strain. This decrease was similar to that observed with the pelD mutant in which a major pectate lyase gene is inactivated. The expression of the pelN fusion in the macerated tissue was compared to that of pelD which is induced in planta (36). The pelN fusion was transcribed in planta at a level similar to that of pelD, after infection of either chicory leaves or potato tubers (Fig. 6 and data not shown), confirming that pelN is expressed during plant infection.
Fig 6.
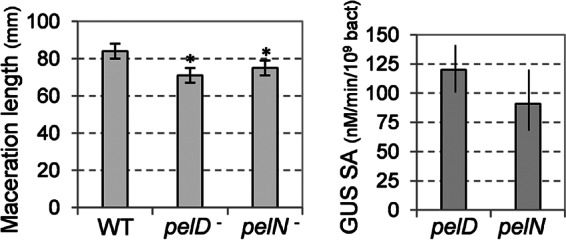
Infection of chicory leaves with the pelN mutant. (A) The length of macerated tissue was measured after a 24 h infection with the mutant and with the wild-type strain 3937 (WT). (B) The β-glucuronidase (GUS) assay and bacterial enumeration were performed on the macerated tissue in order to estimate the expression of the uidA fusions in pelD and pelN. The mean value and the standard deviations reported correspond to 10 infections with each strain. Asterisks indicate statistically significant differences in the degree of maceration of the mutants, compared to the wild-type strain (P < 0.05 [Student t test]).
Effect of different conditions and regulators on pelN expression.
We analyzed the potential effect of different sugars and plant compounds on pelN transcription. The expression of the pelN::uidA fusion increased, by ∼2-fold, in the presence of polygalacturonate (Table 3). In contrast, the pelN expression was not significantly affected by the addition of plant extracts, ferulic acid, glucose, xylose, galactose, galacturonate, glucuronate, or rhamnose (Table 3 and data not shown). We observed that pelN expression was influenced by variations in the growth conditions, such as temperature, pH, and medium osmolarity. Since pelL is also affected by these conditions, we compared their effect on pelN and pelL expression (Fig. 7). Both the pelN and pelL fusions were expressed at 25 or 30°C but repressed at 37°C. Similarly, both pelN and pelL fusions were expressed at neutral or alkaline pH but repressed at acidic pH, although pelN appeared to be more sensitive than pelL to the acidity of the external medium. In contrast, the pelN and pelL fusions responded in opposite ways to variations in the osmolarity of the medium. Although the pelN expression increased with increasing osmolarity, the pelL expression was enhanced at low osmolarity (Fig. 7).
Table 3.
Expression of the pelN transcriptional fusion
| Fusion, mutation | Potential inducera | Mean GUS sp act ± SDb |
|---|---|---|
| pelN::uidA | None | 69 ± 9 |
| Polygalacturonate | 171 ± 20 | |
| Plant extract | 82 ± 8 | |
| Ferulic acid | 70 ± 17 | |
| pelN::uidA, kdgR | None | 219 ± 36 |
| Polygalacturonate | 232 ± 18 | |
| pelN::uidA, pecS | None | 202 ± 22 |
| Polygalacturonate | 235 ± 26 | |
| pelN::uidA, gacA | None | 34 ± 5 |
| Polygalacturonate | 68 ± 7 |
Potential inducers were added at the following concentrations: polygalacturonate at 2 g liter−1 or 0.25 mM ferulic acid. Plant extract (10 g liter−1) was juice obtained after the crushing and filtration of chicory leaves.
The β-glucuronidase (GUS) specific activity (nmol min−1 mg−1) is the average of at least three independent experiments.
Fig 7.
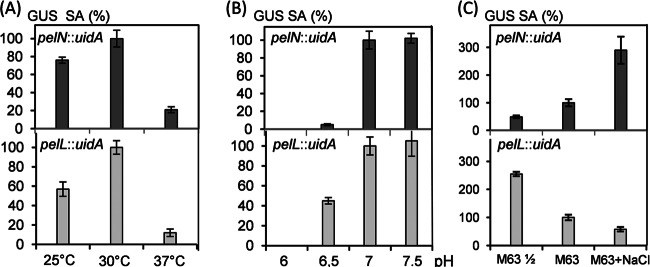
Expression of the pelN and pelL transcriptional fusions in different conditions. (A) The effect of temperature on pelN and pelL expression was tested by growing the bacteria in M63 medium at 25, 30, or 37°C. (B) The effect of the external pH on pelN and pelL expression was tested by growing the bacteria in M63 medium buffered at pH 6, 6.5, 7, or 7.5. (C) The effect of osmolarity on pelN and pelL expression was tested by growing the bacteria in 2-fold-diluted M63 (M63 ½), M63, or M63 + 0.3 M NaCl. In each case, the medium was supplemented with 2 g of glycerol liter−1 as the sole carbon source. The β-glucuronidase (GUS) specific activity (SA) was determined to estimate the expression of the pelN::uidA and pelL::uidA fusions. The values reported are the averages of at least three independent experiments, and the standard deviations are indicated.
The pelN fusion was introduced into different regulatory mutants of D. dadantii (Table 3). In each mutant, a gene encoding a transcriptional regulator of pectinase transcription (1) is inactivated. The pelN transcription was not significantly affected by inactivation of CRP or PecT (data not shown). On the other hand, expression of the pelN fusion increased in the kdgR or pecS mutants, by a factor of ∼3 (Table 3). In accordance with these results, a previous transcriptomic analysis of the pecS mutant indicated that PecS represses both pelL and pelN transcription (37). When gacA was inactivated, the expression of the pelN fusion decreased 2-fold, in a similar way to pelL (Table 3 and data not shown). These results demonstrate that KdgR and PecS negatively control, and GacA positively controls, pelN transcription.
DISCUSSION
We report here the characterization of a novel pectate lyase of D. dadantii 3937. Digestion of polygalacturonate by the PelN enzyme yielded products with an absorbance peak at 230 nm, characteristic of unsaturated oligogalacturonides, and indicative of lyase activity. According to its primary amino acid sequence, PelN belongs to the PL9 family of polysaccharide lyases (http://www.cazy.org/), which already includes two characterized D. dadantii enzymes, the extracellular endopectate lyase, PelL, and the periplasmic exopectate lyase, PelX. A search for PelN homologues in the protein databases showed that D. dadantii PelL is the protein presenting the highest degree of identity with PelN (Fig. 1). Analysis of the known bacterial genomes indicated that the presence of pelN is a feature common to all Dickeya and Pectobacterium species. It is also found in certain strains of another plant pathogenic species, Pseudomonas syringae. Although PelX is widely found, even in non-plant-pathogenic genera such as Yersinia and Klebsiella, PelL orthologs are restricted to the genus Dickeya.
Classical pectate lyases require an alkaline pH and bivalent cations, often Ca2+, for their activity. The PelN optimal pH of 7.4 is only weakly alkaline and lower than that of other extracellular D. dadantii pectate lyases. For instance, the optimal pH of PelL and PelX is ∼8.5 (9, 10). Moreover, PelN is the first example of a pectate lyase which uses Fe2+ as a cofactor for its enzymatic activity (Fig. 4B). The preferential cofactor of the two PL9 enzymes, PelL and PelX, is Ca2+ and Mn2+, respectively (9, 10). PelN also differs from classical pectate lyases in its ability to cleave pectins with various degrees of methylation (Fig. 4C).
The presence of a putative signal sequence, at the N-terminal end of the PelN deduced protein, is consistent with the recovery of the protein in the periplasmic fraction of E. coli expressing the pelN gene (Fig. 2). In D. dadantii, PelN is secreted to the extracellular medium by the Out system (Fig. 3B), which is involved in the secretion of all of the extracellular pectinases (5).
Pectate lyases, belonging to the PL1, PL3, and PL9 families, share a right-handed parallel β-helix topology (15, 16, 20, 21, 22). Each coil of this helical architecture is composed of three consecutive strand-turn motifs. These proteins contain from 8 to 14 helical coils, creating three parallel β-sheets extending along the helix axis. The stability of the cylindrical core is ensured by inner stacks of aliphatic and aromatic side chains. In PL1 and PL9 enzymes, a characteristic “asparagine ladder” is formed by the internal side chains of asparagine residues (21, 22). These intramolecular bonds generate a very stable protein fold. The main differences between pectate lyases are found in the two extremities of the β-helix, where N-terminal and C-terminal extensions form diverse structures (20, 21, 22). The strands and turns involved in the substrate-binding site, and contributing to the active site, also show great variability that could allow each enzyme to adapt to its preferred substrate(s). The catalytic reaction is initiated by an essential basic amino acid abstracting the proton from the substrate carbon, C5. This residue is an arginine in the PL1 family (22) and a lysine in the PL3 and PL9 families (20, 21). The pectate lyase activity is cation dependent, with Ca2+ being the most frequent cofactor. Ca2+ is also involved in substrate interactions, by binding the enzyme and anionic charges of the substrate.
Modeling of the PelN structure (Fig. 1B) suggests that PelN and PelL share a highly similar overall folding. Both proteins are made up of a 10-coil parallel β-helix domain. An asparagine ladder and aromatic stacks, characteristics of the PelL superhelical structure, are well conserved in PelN. Moreover, the positions of two presumed catalytic residues, the catalytic base Lys276 and Asp271 involved in the acidification of C5, are also conserved in PelN (Fig. 1A). However, the two PelL carboxylates, Glu180 and Asp299, which are thought to instigate Ca2+-mediated interactions between PelL and the substrate (21), are replaced, in PelN, by the aromatics Phe179 and Tyr307, respectively (Fig. 1A). Such a difference is consistent with the fact that PelN tolerates methylation of the substrate (Fig. 4). Indeed, part of the carboxyl groups in the C6 position of pectin are methylated and hydrophobic interactions with the aromatic side chains could enhance stabilization of the methylated substrates in the catalytic site. Another notable difference between the structure of PelL and PelN is the presence of several extra loops in PelN. These loops (L1 to L6) are located around the presumed substrate-binding groove (Fig. 1B) and, hence, could affect the substrate binding and/or the catalytic properties of PelN. Notably, loop L2 is extremely rich in Asn and other potential metal coordinating amino acids, namely, Lys, Glu, and His (Fig. 1A). Additional studies are needed to explain the potential link between this loop and the unexpected metal requirement of PelN.
We also compared pelN expression to that of other D. dadantii pectate lyase genes, particularly pelL. Analysis of regulatory mutants indicated that pelN expression is moderately controlled by the KdgR, PecS, and GacA regulators, by a factor of 2- to 3-fold (Table 3). The pelL and pelN genes respond in a similar way to PecS and GacA, but pelL is not regulated by KdgR (10). KdgR is a regulator responding to the presence of pectin or its derivatives, and this repressor mostly controls the genes involved in pectin degradation and catabolism (38). The absence of a potential KdgR binding site in the promoter region of pelN suggests that it is controlled by an indirect mechanism. PecS is a negative regulator controlling a large set of genes involved in the symptomatic phase of infection (37, 39). GacA is a positive regulator involved in the control of virulence genes during plant infection (40, 41). In culture media, the pelN expression was weakly induced in the presence of polygalacturonate (Table 3), but it was not affected by the addition of other molecular inducers of pectate lyase synthesis, such as plant extracts or ferulic acid (1, 42). On the other hand, the pelN expression was sensitive to several external physicochemical conditions. It was repressed at a high temperature, at acidic pH and in a low osmolarity medium (Fig. 7). Although the effects of pH and temperature were similar on pelN and pelL expression, these genes showed opposite responses to osmolarity, with an increase for pelN and a decrease for pelL in high osmolarity conditions (Fig. 7). Opposite effects of osmolarity, such as these, were previously observed for pelD and pelE expression, with an increase for pelE and a decrease for pelD at a high osmolarity level (36). Thus, osmolarity is a factor that seems to have different effects on the production of pectate lyases of the same family. The pelN expression during plant infection (Fig. 6) probably results from favorable environmental conditions rather than from induction by plant compounds. Like the PelN enzymatic properties, pelN expression shows specific characteristics allowing this pectate lyase isozyme to be a complementary element of the enzymatic complex which confers to D. dadantii its remarkable pectinolytic activity.
There may be several reasons for the evolution and retention of multiple isoforms of pectate lyases in soft-rot bacteria. The pectate lyases secreted by D. dadantii play multiple roles in pathogenesis. During the infection process, these enzymes directly damage the host tissue, they facilitate the bacterial invasion in surrounding tissues, and they give rise to carbon sources for the bacterial growth. They have probably also a nutritional role outside the host, when bacteria are present in ecological habitats containing plant debris. In attempting to understand the functional basis for the multiplicity of pectate lyase isozymes, it is important to keep in mind that significant differences have been reported for the action patterns of each isozyme. In the case of D. dadantii 3937, their optimal pH varies from nearly neutral to highly alkaline (7.4 for PelN to 9.3 for PelB). They tolerate low to high ranges of pectin methylation (0 to 25% for PelD to 0 to 91% for PelN). Considering its enzymatic properties, PelN is of particular interest because some of its traits are complementary to those of other known D. dadantii secreted pectate lyases. Variations among isozymes with respect to catalytic properties and gene regulation could allow the bacterium to attack a variety of host plants under diverse conditions. These conditions could vary depending on the plant but also throughout the plant infection. For instance pH varies from about 5 at the early stage of infection to nearly 7 in the macerated tissue (43). In contrast to other D. dadantii 3937 pectate lyases, which are mainly induced in the presence of pectin degradation products and plant extracts, PelN regulation is mostly connected with environmental conditions, probably under the control of global regulators. The multiple pectate lyases secreted by soft-rot bacteria could play complementary roles in pathogenesis, increasing the bacterial adaptability and host range. The combined traits of the complete set of D. dadantii secreted enzymes define a large spectrum of potential plant cell wall degradation activity.
ACKNOWLEDGMENTS
We thank Guy Condemine and Valerie James for reading the manuscript. We thank our Lyon coworkers for helpful discussions and the members of the International Erwinia Consortium for the exchange of unpublished data concerning the D. dadantii 3937 genome.
This study was supported by a Ph.D. grant from the Syrian government to S.H. and by grants from the Centre National de la Recherche Scientifique, from the Ministère de l'Enseignement Supérieur et de la Recherche, and from the Région Rhône-Alpes (cluster 9).
Footnotes
Published ahead of print 8 March 2013
REFERENCES
- 1. Hugouvieux-Cotte-Pattat N, Condemine G, Nasser W, Reverchon S. 1996. Regulation of pectinolysis in Erwinia chrysanthemi. Annu. Rev. Microbiol. 50:213–257 [DOI] [PubMed] [Google Scholar]
- 2. Garibaldi A, Bateman DF. 1971. Pectic enzymes produced by Erwinia chrysanthemi and their effects on plant tissue. Physiol. Plant Pathol. 1:25–40 [Google Scholar]
- 3. He SY, Lindeberg M, Chatterjee AK, Collmer A. 1991. Cloned Erwinia chrysanthemi out genes enable Escherichia coli to selectively secrete a diverse family of heterologous proteins to its milieu. Proc. Natl. Acad. Sci. U. S. A. 88:1079–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Condemine G, Dorel C, Hugouvieux-Cotte-Pattat N, Robert-Baudouy J. 1992. Some of the out genes involved in the secretion of pectate lyases in Erwinia chrysanthemi are regulated by KdgR. Mol. Microbiol. 6:3199–3211 [DOI] [PubMed] [Google Scholar]
- 5. Bouley J, Condemine G, Shevchik VE. 2001. The PDZ somain of OutC and the N-terminal region of OutD determine the secretion specificity of the type II Out pathway of Erwinia chrysanthemi. J. Mol. Biol. 308:205–219 [DOI] [PubMed] [Google Scholar]
- 6. Pissavin C, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. 1998. Biochemical characterization of the pectate lyase PelZ of Erwinia chrysanthemi 3937. Biochim. Biophys. Acta 1383:188–196 [DOI] [PubMed] [Google Scholar]
- 7. Shevchik VE, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. 1997. The pectate lyase PelI of Erwinia chrysanthemi belongs to a new family. J. Bacteriol. 179:7321–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shevchik VE, Condemine G, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. 1999. The exopolygalacturonate lyase PelW and the oligogalacturonate lyase Ogl, two cytoplasmic enzymes of pectin catabolism in Erwinia chrysanthemi 3937. J. Bacteriol. 181:3912–3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shevchik VE, Kester HCM, Benen JAE, Visser J, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. 1999. Characterisation of exopolygalacturonate lyase PelX of Erwinia chrysanthemi 3937. J. Bacteriol. 181:1652–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lojkowska E, Masclaux C, Boccara M, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. 1995. Characterization of the pelL gene encoding novel pectate lyase of Erwinia chrysanthemi 3937. Mol. Microbiol. 16:1183–1195 [DOI] [PubMed] [Google Scholar]
- 11. Tardy F, Nasser W, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. 1997. Comparative analysis of the five major Erwinia chrysanthemi pectate lyases: enzyme characteristics and potential inhibitors. J. Bacteriol. 179:2503–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roy C, Kester HCM, Visser J, Shevchik VE, Hugouvieux-Cotte-Pattat N, Robert-Baudouy J, Benen JAE. 1999. Mode of action of five different endopectate lyases from Erwinia chrysanthemi 3937. J. Bacteriol. 181:3705–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abbott DW, Gilbert HJ, Boraston AB. 2010. The active site of oligogalacturonate lyase provides unique insights into cytoplasmic oligogalacturonate beta-elimination. J. Biol. Chem. 285:39029–39038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lombard V, Bernard T, Rancurel C, Brumer H, Coutinho PM. 2010. A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem. J. 432:437–444 [DOI] [PubMed] [Google Scholar]
- 16. Garron ML, Cygler M. 2010. Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology 20:1547–1573 [DOI] [PubMed] [Google Scholar]
- 17. Heffron S, Henrissat B, Yoder MD, Lietzke S, Jurnak F. 1995. Structure-based multiple alignment of extracellular pectate lyase sequences. Mol. Plant-Microbe Interact. 8:331–334 [DOI] [PubMed] [Google Scholar]
- 18. Henrissat B, Heffron SE, Yoder MD, Lietzke S, Jurnak F. 1995. Functional implications of structure-based sequence alignment of proteins in the extracellular pectate lyase superfamily. Plant Physiol. 107:963–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abbott DW, Boraston AB. 2007. A family 2 pectate lyase displays a rare fold and transition metal-assisted beta-elimination. J. Biol. Chem. 282:35328–35336 [DOI] [PubMed] [Google Scholar]
- 20. Creze C, Castang S, Derivery E, Haser R, Hugouvieux-Cotte-Pattat N, Shevchik VE, Gouet P. 2008. The crystal structure of pectate lyase PelI from soft rot pathogen Erwinia chrysanthemi in complex with its substrate. J. Biol. Chem. 283:18260–18268 [DOI] [PubMed] [Google Scholar]
- 21. Jenkins J, Shevchik VE, Hugouvieux-Cotte-Pattat N, Pickersgill RW. 2004. The crystal structure of pectate lyase Pel9A from Erwinia chrysanthemi. J. Biol. Chem. 279:9139–9145 [DOI] [PubMed] [Google Scholar]
- 22. Yoder MD, Keen NT, Jurnak F. 1993. New domain motif: the structure of pectate lyase C, a secreted plant virulence factor. Science 260:1503–1507 [DOI] [PubMed] [Google Scholar]
- 23. Resibois A, Colet M, Faelen M, Schoonejans T, Toussaint A. 1984. Phi-EC2, a new generalized transducing phage of Erwinia chrysanthemi. Virology 137:102–112 [DOI] [PubMed] [Google Scholar]
- 24. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 25. Bardonnet N, Blanco C. 1992. ′uidA antibiotic resistance cassettes for insertion mutagenesis, gene fusions and genetic constructions. FEMS Microbiol. Lett. 72:243–247 [DOI] [PubMed] [Google Scholar]
- 26. Hugouvieux-Cotte-Pattat N. 2004. The RhaS activator controls the Erwinia chrysanthemi 3937 genes rhiN, rhiT, and rhiE involved in rhamnogalacturonan catabolism. Mol. Microbiol. 51:1361–1374 [DOI] [PubMed] [Google Scholar]
- 27. Roeder DL, Collmer A. 1985. Marker-exchange mutagenesis of a pectate lyase isozyme gene in Erwinia chrysanthemi. J. Bacteriol. 164:51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tabor S, Richardson CC. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. U. S. A. 82:1074–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113–130 [DOI] [PubMed] [Google Scholar]
- 30. Copeland BR, Richter RJ, Furlong CE. 1982. Renaturation and identification of periplasmic proteins in two-dimensional gels of Escherichia coli. J. Biol. Chem. 257:15065–15071 [PubMed] [Google Scholar]
- 31. Kazemi-Pour N, Condemine G, Hugouvieux-Cotte-Pattat N. 2004. The secretome of the plant pathogenic bacterium Erwinia chrysanthemi. Proteomics 10:3177–3186 [DOI] [PubMed] [Google Scholar]
- 32. Van Gijsegem F, Woldarczyk A, Cornu A, Reverchon S, Hugouvieux-Cotte-Pattat N. 2008. Analysis of the LacI family regulators of Erwinia chrysanthemi 3937, involvement in the bacterial phytopathogenicity. Mol. Plant-Microbe Interact. 21:1471–1481 [DOI] [PubMed] [Google Scholar]
- 33. Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. 2007. Comparative protein structure using MODELLER. Curr. Protoc. Protein Sci. Chapter 2:Unit 2.9 [DOI] [PubMed] [Google Scholar]
- 34. Laskowski RA, MacArthur MW, Moss DS. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291 [Google Scholar]
- 35. Glasner JD, Yang CH, Reverchon S, Hugouvieux-Cotte-Pattat N, Condemine G, Bohin JP, Van Gijsegem F, Yang S, Franza T, Expert D, Plunkett G, III, San Francisco MJ, Charkowski AO, Py B, Bell K, Rauscher L, Rodriguez-Palenzuela P, Toussaint A, Holeva MC, He SY, Douet V, Boccara M, Blanco C, Toth I, Anderson BD, Biehl BS, Mau B, Flynn SM, Barras F, Lindeberg M, Birch PR, Tsuyumu S, Shi X, Hibbing M, Yap MN, Carpentier M, Dassa E, Umehara M, Kim JF, Rusch M, Soni P, Mayhew GF, Fouts DE, Gill SR, Blattner FR, Keen NT, Perna NT. 2011. Genome sequence of the plant-pathogenic bacterium Dickeya dadantii 3937. J. Bacteriol. 193:2076–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hugouvieux-Cotte-Pattat N, Dominguez H, Robert-Baudouy J. 1992. Environmental conditions affect the transcription of the pectinases gene of Erwinia chrysanthemi 3937. J. Bacteriol. 174:7807–7818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hommais F, Oger-Desfeux C, Van Gijsegem F, Castang S, Ligori S, Expert D, Nasser W, Reverchon S. 2008. PecS is a global regulator of the symptomatic phase in the phytopathogenic bacterium Erwinia chrysanthemi 3937. J. Bacteriol. 190:7508–7522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rodionov DA, Gelfand MS, Hugouvieux-Cotte-Pattat N. 2004. Comparative genomics of the KdgR regulon in Erwinia chrysanthemi 3937 and other Gram-negative bacteria. Microbiology 150:3571–3590 [DOI] [PubMed] [Google Scholar]
- 39. Mhedbi-Hajri N, Malfatti P, Pédron J, Gaubert S, Reverchon S, Van Gijsegem F. 2011. PecS is an important player in the regulatory network governing the coordinated expression of virulence genes during the interaction between Dickeya dadantii 3937 and plants. Environ. Microbiol. 13:2901–2914 [DOI] [PubMed] [Google Scholar]
- 40. Lebeau A, Reverchon S, Gaubert S, Kraepiel Y, Simond-Côte E, Nasser W, Van Gijsegem F. 2008. The GacA global regulator is required for the appropriate expression of Erwinia chrysanthemi 3937 pathogenicity genes during plant infection. Environ. Microbiol. 10:545–559 [DOI] [PubMed] [Google Scholar]
- 41. Yang S, Peng Q, Zhang Q, Yi X, Choi CJ, Reedy RM, Charkowski AO, Yang CH. 2008. Dynamic regulation of GacA in type III secretion, pectinase gene expression, pellicle formation, and pathogenicity of Dickeya dadantii (Erwinia chrysanthemi 3937). Mol. Plant-Microbe Interact. 21:133–142 [DOI] [PubMed] [Google Scholar]
- 42. Hassan S, Hugouvieux-Cotte-Pattat N. 2011. Identification of two feruloyl esterases in Dickeya dadantii 3937 and induction by ferulic acid of the major feruloyl esterase and of pectate lyases. J. Bacteriol. 193:963–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Effantin G, Rivasseau C, Gromova M, Bligny R, Hugouvieux-Cotte-Pattat N. 2011. Massive production of butanediol during infection by phytopathogenic bacteria of the genera Dickeya and Pectobacterium. Mol. Microbiol. 82:988–997 [DOI] [PubMed] [Google Scholar]
- 44. Gouet P, Robert X, Courcelle E. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31:3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reverchon S, Nasser W, Robert-Baudouy J. 1994. pecS, a locus controlling pectinase, cellulase, and blue pigment production in Erwinia chrysanthemi. Mol. Microbiol. 11:1127–1139 [DOI] [PubMed] [Google Scholar]