Abstract
The carbonic anhydrase (Cpb) from Clostridium perfringens strain 13, the only carbonic anhydrase encoded in the genome, was characterized both biochemically and physiologically. Heterologously produced and purified Cpb was shown to belong to the type I subclass of the β class, the first β class enzyme investigated from a strictly anaerobic species of the domain Bacteria. Kinetic analyses revealed a two-step, ping-pong, zinc-hydroxide mechanism of catalysis with Km and kcat/Km values of 3.1 mM CO2 and 4.8 × 106 s−1 M−1, respectively. Analyses of a cpb deletion mutant of C. perfringens strain HN13 showed that Cpb is strictly required for growth when cultured in semidefined medium and an atmosphere without CO2. The growth of the mutant was the same as that of the parent wild-type strain when cultured in nutrient-rich media with or without CO2 in the atmosphere, although elimination of glucose resulted in decreased production of acetate, propionate, and butyrate. The results suggest a role for Cpb in anaplerotic CO2 fixation reactions by supplying bicarbonate to carboxylases. Potential roles in competitive fitness are discussed.
INTRODUCTION
Carbonic anhydrase (CA) is a metalloenzyme catalyzing the reversible interconversion of CO2 and bicarbonate (CO2 + H2O = HCO3− + H+). To date, five independently evolved classes (αβγζδ) are known (1). The β class is abundant in higher plants and photosynthetic algae of the domain Eukarya (2). Several β class CAs from the domain Bacteria have been described, including those from photosynthetic (3–5) and nonphotosynthetic species of which several are pathogenic (6–10). The β class CAs of photosynthetic species function to concentrate atmospheric CO2, whereas it is proposed that nonphotosynthetic species utilize β class CAs to retain intracellular CO2 by conversion to bicarbonate for anaplerotic carboxylation reactions (11–14). The only characterized β class CA from the domain Archaea (Cab) is that from Methanothermobacter thermoautotrophicus (formerly Methanobacterium thermoautotrophicum strain ΔH) (15–18), a microbe that obtains energy for growth by reducing CO2 to methane. It is proposed that Cab functions to concentrate CO2 for both methanogenesis and anaplerotic carboxylation reactions.
CAs are important for survival and growth of several pathogens and, therefore, a potential target for development of novel antimicrobial agents (19–22). Clostridium perfringens strains are ubiquitous, strictly anaerobic pathogens that inhabit many environmental niches, including soil, sediments, and the intestinal tracts of mammals (23). In humans, different strains of C. perfringens cause gas gangrene (myonecrosis), acute food poisoning, and necrotic enteritis. The CPE0413 gene of strain 13 is annotated as encoding a probable CA (Comprehensive Microbial Resource [http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi]) that has yet to be investigated for enzymatic activity or physiological function. Here, we report the results of a phylogenetic, biochemical, and physiological characterization of the protein encoded by CPE0413. The results show that the protein is a type I β class CA, the first biochemical and physiological investigation of a CA from the genus Clostridium. The results further reveal that Cpb is essential for growth in semidefined media with no added CO2, suggesting that Cpb functions in nature to retain intracellular levels of CO2 and supply bicarbonate for anaplerotic reactions.
MATERIALS AND METHODS
Bacterial strains, growth conditions.
Bacterial strains, plasmids, and primers used in this study are listed in Table 1. Escherichia coli was grown in Luria-Bertani (LB) medium supplemented with antibiotics as needed: 400 μg/ml erythromycin, 100 μg/ml ampicillin, or 20 μg/ml chloramphenicol. Clostridium perfringens was grown in a Coy anaerobic chamber at 37°C. Four different media were used for growth of C. perfringens: brain heart infusion (BHI) medium (Difco), TY medium (3% tryptone, 2% yeast extract, 0.1% sodium thioglycolate), PGY medium (3% proteose peptone 3, 2% glucose, 1% yeast extract, 0.1% sodium thioglycolate) and semidefined media (0.1% yeast extract, 50 mM glucose, 50 mM NaKPO4 [pH 7.0], 19.79 mg/liter MnCl2 · 4H2O, 122 mg/liter MgCl2 · 6H2O, 29.4 mg/liter CaCl2 · 2H2O, 11.5 mg/liter ZnSO4 · 7H2O, 0.025 mg/liter CuSO4 · 5H2O). Strains were first inoculated onto plates containing BHI medium (with chloramphenicol for growth of the complemented strain) incubated in a 100% nitrogen atmosphere and then inoculated onto two identical agar plates containing semidefined media, one incubated in the anaerobic chamber (10% CO2) and the other in an anaerobic jar containing 100% nitrogen.
Table 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant characteristic(s) or sequence (5′ to 3′) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH10B | F− mcrAΔ mrr-hsdRMS mcrBC ϕ80dlacZΔM15 lacX74 deoR recA1 araD139Δ ara leu7697 galU galK ΔrpsL endA1 nupG | Gibco/BRL |
| C. perfringens | ||
| HN13 | Strain 13; ΔgalKT | 24 |
| WH1 | Strain HN13 with deletion of the region from bp 18 to 555 in cpb | This study |
| Plasmids | ||
| pGEM-T Easy | PCR cloning vector; ampicillin resistance | Promega |
| pKRAH1 | Contains bgaR-PbgaL; chloramphenicol resistance | 25 |
| pCM-GALK | Contains a Clostridium beijerinckii galK gene under the control of a ferredoxin promoter from C. perfringens | 24 |
| pET22b | Protein production vector | Novagen |
| pCPE0413 | cpb gene in pET22b | This study |
| pWH5 | pGEM-T Easy with overlapping PCR product of DNA flanking the cpb gene | This study |
| pWH6 | pCM-GALK with cpb deletion fragment | This study |
| pWH7 | pGEM-T Easy with cpb gene from HN13 | This study |
| pWH8 | pKRAH1 with cpb gene from HN13 | This study |
| Primers | ||
| CA forward | TTTTTTCATATGGAAAAGAATGTGAGAAGACTAGAAG | |
| CA reverse | TTTTTTAAGGTTATTCGTATCCATTTACTATAACCTTAAGTTCTCC | |
| OWH9 | GTCGACTTCACAAATCAATAGAAGAAATTGAAG | |
| OWH10 | GTTTTTTAATTTTTATTTTATTCGTATCCATTCACATTCTTTTCCATACTAAATACC | |
| OWH11 | GGTATTTAGTATGGAAAAGAATGTGAATGGATACGAATAAAATAAAAATTAAAAAAC | |
| OWH12 | GGATCCCTTTTTGTTTATTTTCAATTCCAAATATAC | |
| OWH13 | AATGAAAAAGGAGATAAAACTCCAG | |
| OWH14 | CCATATACAGGACTAATTGGTCC | |
| OWH15 | GTCGACGTGGGTTTAAAGTAAAGACTTTAAATC | |
| OWH24 |
In-frame deletion of gene CPE0413 (cpb).
Mutagenesis of C. perfringens HN13, a derivative of C. perfringens strain 13, was performed as previously described (24). Briefly, overlap extension PCR using primers OWH9, OWH10, OWH11, and OWH12 was used to amplify ∼900-bp regions flanking CPE0413 (cpb). The resulting DNA fragment was cloned into pGEM-T Easy (pWH5) and then pCM-GALK (24), yielding pWH6. In-frame deletion mutagenesis was then performed using this construct (24), resulting in a deletion of the region from 18 to 555 bp out of the 570-bp cpb gene reading frame; this strain was named WH1. The mutation was confirmed using PCR with flanking primers OWH13 and OWH14.
Complementation.
To complement the cpb mutation in strain WH1, a plasmid was constructed by amplifying the cpb gene plus 134 bp upstream with primers OWH15 and OWH24, cloning the cpb gene into plasmid pGEM-T Easy (pWH7) and then into pKRAH1 (pWH8), which placed the gene under the control of the lactose-inducible promoter, PbgaL (25). Plasmids were introduced into strain WH1 by electroporation (26).
Cloning and heterologous production of Cpb.
The CPE0413 gene encoding Cpb was amplified by PCR using Clostridium perfringens strain 13 genomic DNA as a template. The forward primer (CA forward) partially corresponds to nucleotides encoding amino acids 1 to 9, and the reverse primer (CA reverse) corresponds to nucleotides 536 to 572 downstream of the beginning of the gene (Table 1). NdeI and HindIII restriction sites were introduced in the forward and reverse primers, respectively. The PCR product was digested with NdeI and HindIII and cloned into the digested pET22b(+) vector (Novagen) to yield pCPE0413. This constructed plasmid was transformed into E. coli strain Rosetta (DE3) pLacI (Novagen) which was used to inoculate Luria-Bertani broth containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol. The cells were grown at 37°C to an A600 of 0.6 to 0.8 and then induced with 1 mM isopropyl thiogalactopyranoside (IPTG) and 500 μM ZnSO4 to overproduce Cpb. Induced cultures were allowed to continue growth for 4 to 5 h. The cells were harvested by centrifugation and stored frozen at −80°C until lysis.
Enzyme purification.
Cell paste (5 g [wet weight]) was thawed in 30 ml of buffer A (50 mM potassium phosphate [pH 7.0]) containing 0.1 mM zinc and passed twice through a chilled French pressure cell at 138 MPa. The cell debris was removed by centrifuging the cell lysate at 29,000 × g for 30 min. After centrifugation, the supernatant was passed through a 0.45-μm filter before loading onto a Q-Sepharose (GE Healthcare) column equilibrated with buffer A. The column was developed with the same buffer to elute unbound protein. Bound proteins were then eluted with a linear gradient (0 to 1 M) of NaCl in buffer A. The fractions with CA activity were concentrated and chromatographed on a Sephadex G-150 superfine gel filtration column (GE Healthcare) using buffer B (50 mM potassium phosphate [pH 7.0]) containing 0.1 M NaCl. Fractions with CA activity were pooled and stored at −20°C. CA activity was measured at room temperature by using the electrometric method described previously (27).
Enzyme activity.
Steady-state CO2 hydration activity was measured by stopped-flow spectroscopy using the changing pH indicator method described previously (28). Saturated solutions of CO2 (32.9 mM) were prepared by bubbling CO2 into distilled, deionized water at 25°C. The CO2 concentration was varied from 5 to 27 mM by mixing with an appropriate volume of N2-saturated water. The buffer/indicator pairs (the wavelengths used shown in parentheses) were morpholineethanesulfonic acid (MES)/chlorophenol red (574 nm) at pH 5.5 to 6.8, morpholinepropanesulfonic acid (MOPS)/p-nitrophenol (400 nm) at pH 6.8 to 7.5, HEPES/phenol red (557 nm) at pH 7.5 to 8.0, and N-Tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid (TAPS)/m-cresol purple (578 nm) at pH 8.0 to 9.0. Buffer concentrations were 50 mM. The total ionic strength was adjusted to 150 mM with Na2SO4. The final pH indicator concentration was 50 μM. The initial 5 to 10% of the total absorbance change was used to calculate the initial steady-state rate with the mean of 5 to 10 reaction traces per experiment. The steady-state parameters kcat and kcat/Km and their standard errors were determined by fitting the observed initial rates (corrected for the uncatalyzed reaction) to the Michaelis-Menten equation using the KaleidaGraph data analysis and graphing software. The pH-independent values of kcat and pKa for the CO2 hydration reaction were determined by fitting the experimental pH-dependent Michaelis-Menten parameters to equations 1 and 2 where kcatobs is the observed kcat and Kmobs is the observed Km.
| (1) |
| (2) |
Esterase activity, using p-nitrophenylacetate as a substrate, was determined as previously described (29).
Protein, metal, and volatile fatty acid (VFA) analyses.
Protein concentrations were estimated by measuring the absorbance of solutions (A280), using a theoretical monomer extinction coefficient of 11,710 cm−1 M−1 and a calculated monomer molecular mass of 21,351 Da. For analysis of metals, protein concentrations were determined by the biuret method (30) using bovine serum albumin as the standard.
The homogeneity of the enzyme was determined by 12% SDS-PAGE (31). The gels were stained for protein with Coomassie brilliant blue. Markers in the molecular mass range from 20 to 250 kDa were used to estimate the molecular mass. The native molecular mass was determined by gel filtration chromatography using a Sephadex G-150 gel filtration column (GE Healthcare) equilibrated with buffer B and calibrated with lysozyme (14.7 kDa), bovine erythrocyte CA (29 kDa), bovine serum albumin (monomer, 66 kDa; dimer, 132 kDa), the β class CA (Cab) from M. thermautotrophicus (90 kDa), and urease (trimer, 272 kDa; hexamer, 545 kDa). The column was developed with a flow rate of 0.2 ml/min using buffer B.
A comprehensive metal analysis (20 elements) was conducted using inductively coupled plasma (ICP) atomic emission spectroscopy at the Chemical Analysis Laboratory, University of Georgia, Athens. All solutions were made metal free by treating with Chelex 100 (Sigma). Prior to metal ion analysis, samples were dialyzed for about 24 h at 4°C with 4 to 6 changes against 20 mM potassium phosphate (pH 7.0).
Lactate and formate were determined using commercial kits (Megazymes, Ireland). Cells were grown for ∼15 h to stationary phase and then removed by centrifugation, and the supernatant was filtered through a 0.22-μm filter and stored at −80°C until analyzed.
Acetate, butyrate, and propionate were determined by liquid chromatography-mass spectrometry (LC-MS). Samples were analyzed using an Agilent 6890 N gas chromatograph (GC) with a Waters GCT classic mass spectrometer operated in electron ionization (EI) mode. Aliquots (2 μl) were injected using a split/splitless injector at 250°C with a 5:1 split onto a VF5 (5% diphenyl–95% dimethyl polysiloxane) capillary column (Varian) (20-m column with 150-μm internal diameter and 0.15-μm film thickness) using helium as the carrier gas at a flow rate of 1.4 ml/min. The column was held at an initial temperature of 50°C for 1 min and then heated at 10°C/min to 100°C where it was held for 2 min before heating at 30°C/min to a final temperature of 230°C. Following a 6-min solvent delay, EI mass spectra were acquired over the mass range of 45 to 400 kDa at 1 scan/second. Under these conditions, the pentafluorobenzyl (PFB) derivatives of acetic acid, propionic acid-2,2-d2, propionic acid (internal standard), and butyric acid had retention times of 7.05, 8.74, 8.78, and 9.69 min, respectively. The reconstructed ion chromatograms (RIC) for m/z 240, 256, 254, and 268 were generated, and the integrated areas were used for quantitation. A standard curve was prepared from a mixture of the three VFAs at a concentration of 240, 120, 60, 30, 15, 7.5, and 3.75 μg/ml containing 8 μg/ml of the internal standard. The amounts of each of the VFAs in the samples were calculated from the integrated area of the RIC, using the response factor for each VFA and normalized for the internal standard (IS) response.
RESULTS AND DISCUSSION
Purification and characterization of Cpb encoded by CPE0413.
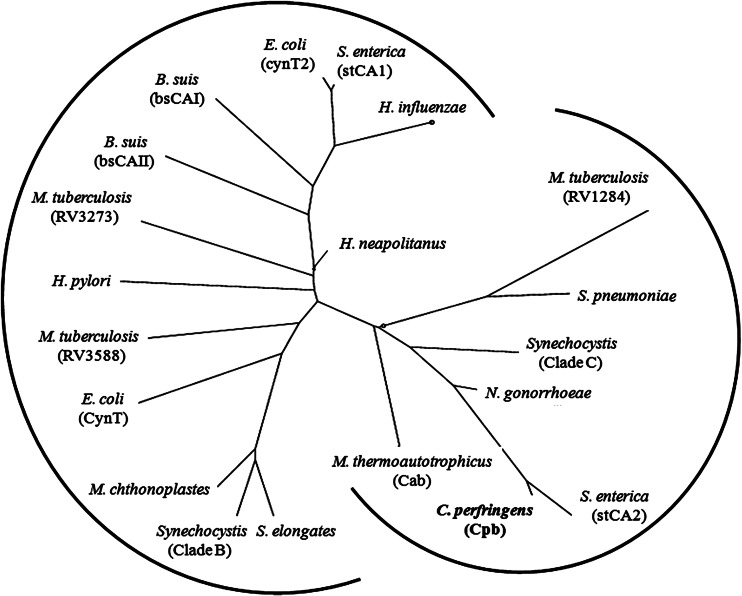
The protein encoded by the CPE0413 locus of the C. perfringens genome is annotated as a probable CA (32). The gene was amplified by PCR from C. perfringens genomic DNA using primers shown in Table 1. The PCR product was cloned into pET22b(+) to yield the expression vector pCPE0413 that was transformed into E. coli strain Rosetta (DE3) pLacI for overproduction of the encoded protein. The protein was purified to homogeneity (Fig. 1) from cell extract of IPTG-induced cells by ion-exchange and gel filtration chromatography. CA activity was monitored using the electrometric method (27). The purified protein showed CO2 hydration activity comparable to the activities of kinetically characterized β class CAs (Table 2) validating the annotation; therefore, the gene is designated cpb (Clostridium perfringens beta class carbonic anhydrase). SDS-PAGE (Fig. 1) revealed a molecular mass of 23 kDa consistent with 21.3 kDa calculated from the deduced amino acid composition. The molecular mass of ∼88 kDa, determined by gel filtration chromatography (data not shown), indicated that the native enzyme is a tetramer. A comprehensive metal analysis (20 elements) performed by plasma emission spectroscopy, using two independent enzyme preparations with similar specific activities, revealed 0.89 and 0.93 zinc per subunit, indicating that the enzyme contains one active site zinc per subunit. The kinetic constants and subunit composition of Cpb (Table 2) most resemble those of the β class CA (Cab) from M. thermautotrophicus of the domain Archaea (15–17). Both Cpb and Cab have lower Km values for CO2 compared to all other β class CAs (Table 2), a result consistent with a role for Cpb and Cab in capturing environmental CO2 and retaining intracellular levels for anaplerotic CO2 fixation reactions. Cpb and Cab belong to one of two clades identified in the phylogenetic analysis of prokaryotic β class CAs (Fig. 2) from species other than Clostridium species. A search of the nonredundant databases revealed 23 closely related homologs of C. perfringens strain 13 cpb belonging to the genus Clostridium represented by pathogenic species and nonpathogenic species of environmental and biotechnological importance (see Fig. S1 in the supplemental material).
Fig 1.
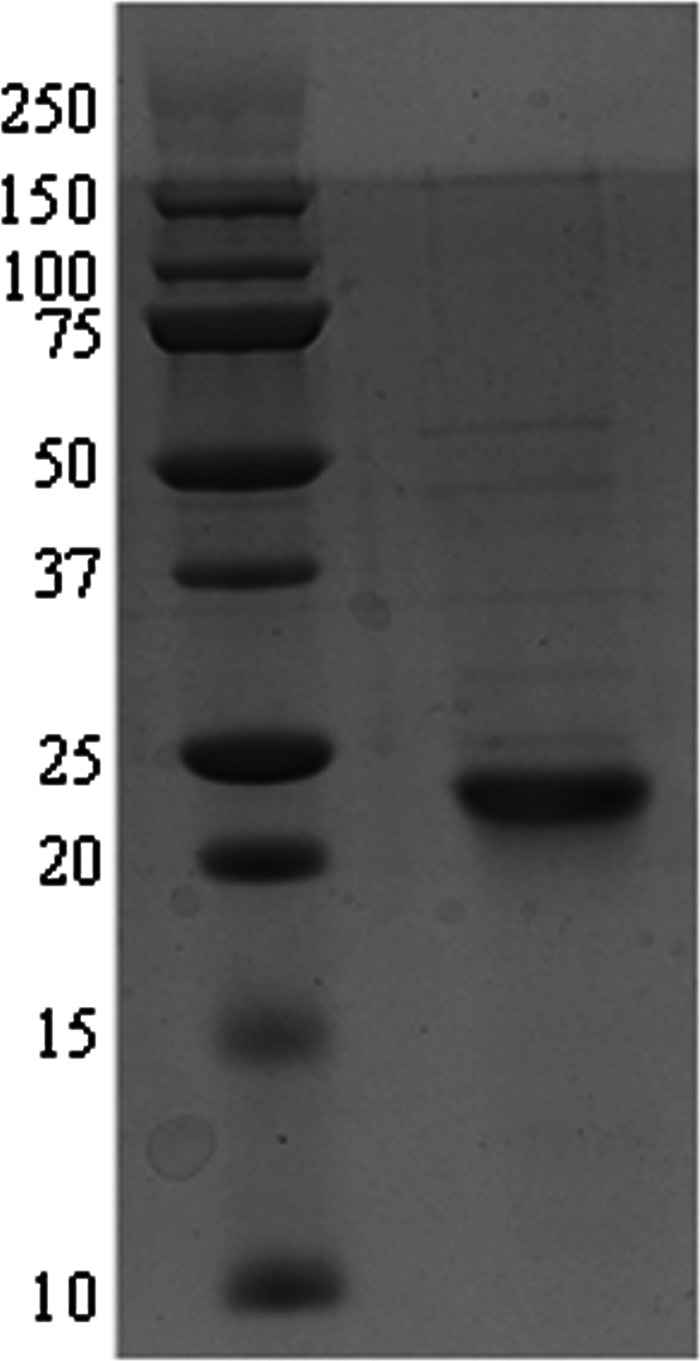
Denaturing polyacrylamide gel electrophoresis of Cpb. The right lane contains 10 μg of purified protein. The left lane contains molecular mass standards. The positions of molecular mass standards (in kilodaltons) are shown to the left of the gel. The gel was stained for protein with Coomassie blue R-250.
Table 2.
Subunit composition and kinetic parameters of characterized prokaryotic β class carbonic anhydrases
| Enzyme source | Compositiona |
Kinetic parametersa |
Reference(s) | |||
|---|---|---|---|---|---|---|
| Subunit mass (kDa) | Holoenzyme | kcat (s−1) | Km (mM) | kcat/Km (s−1 M−1) | ||
| Clostridium perfringens strain 13 | 21.3 | Tetramer | 1.5 × 104 | 3.1 | 4.8 × 106 | This study |
| Methanothermobacter thermautotrophicus | 18.9 | Tetramer | 1.7 × 104 | 2.9 | 5.9 × 106 | 16 |
| Mycobacterium tuberculosis (Rv1284) | 18.2 | Tetramer | 3.9 × 105 | 10.5 | 3.7 × 107 | 9, 33 |
| Mycobacterium tuberculosis (Rv3588c) | 21.8 | Dimer | NA | NA | NA | 9 |
| Brucella suis (bsCAI) | 25 | Dimer | 6.4 × 105 | 16.4 | 3.9 × 107 | 6 |
| Brucella suis (bsCAII) | 25 | NA | 1.1 × 106 | 12.3 | 8.9 × 107 | 20 |
| Helicobacter pylori | 25.8 | NA | 7.1 × 105 | 14.7 | 4.8 × 107 | 7 |
| Streptococcus pneumoniae | 21.1 | NA | 7.4 × 105 | 11.3 | 6.5 × 107 | 34 |
| Salmonella enterica (stCAI) | 24.8 | NA | 1.0 × 106 | 12 | 8.3 × 107 | 35 |
| Salmonella enterica (stCAII) | 26.6 | NA | 7.9 × 105 | 15.2 | 5.2 × 107 | 35 |
| Halothiobacillus neapolitanus | 57.3 | NA | 8.9 × 105 | 3.2 | 2.8 × 107 | 36 |
NA, not available.
Fig 2.
Phylogenetic analysis of prokaryotic β class carbonic anhydrases. The sequences were from the following species (gene identification numbers shown in parentheses or brackets): Clostridium perfringens strain 13 (gi|18309395), Methanothermobacter thermautotrophicus strain ΔH (gi|15679577), Escherichia coli (cynT [gi|386607709] and cynT2 [gi|14277938]), Brucella suis (bsCAI [gi|260567947] and bsCAII [gi|260568899]), Helicobacter pylori (gi|188526816), Mycobacterium tuberculosis (strain RV1284 [gi|298524787], RV3588C [gi|15610724], and RV3273 [gi|15610409]), Salmonella enterica serovar Typhimurium (stCA2 [gi|378987335] and stCA1 [gi|16763561]), Neisseria gonorrhoeae (gi|59802380), Streptococcus pneumoniae (gi|307066706), Haemophilus influenzae (gi|99031788), Synechocystis (clade C [gi|1001130] and clade B [gi|1653251]), Synechococcus elongates (gi|56685078), Microcoleus chthonoplastes (gi|78057950), and Halothiobacillus neapolitanus (gi|88193005). The tree was constructed using the phylogeny/evolution analysis tools obtained at ExPASy (http://expasy.org/phylogeny_evolution). A distance-based method was used for the construction.
CAs characterized thus far employ a two-step, ping-pong, metal-hydroxide mechanism of catalysis (equations 3 and 4) where E represents enzyme residues, M is metal, and B is buffer.
| (3a) |
| (3b) |
| (4a) |
| (4b) |
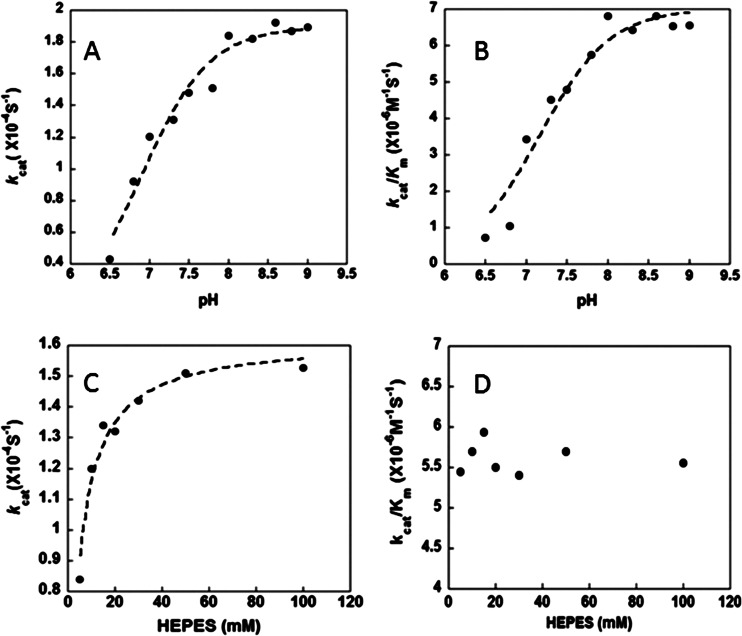
In the first step, a pair of electrons residing on the metal-bound hydroxide attack the carbon of CO2 (equation 3a), producing metal-bound HCO3− that is next displaced by water (equation 3b). In the next step, a proton is extracted from the metal-bound water (equation 4a) and transferred to bulk solvent or buffer (equation 4b), regenerating the metal-bound hydroxide (37). The steady-state parameters of kcat and kcat/Km for Cpb increased with increasing pH (Fig. 3A and B), suggesting that an unprotonated form is required for activity consistent with a metal-hydroxide mechanism of catalysis. Single ionizations with pKa values of 6.9 ± 0.15 and 7.1 ± 0.2 were observed for kcat and kcat/Km (Fig. 3A and B). The dependence of CO2 hydration on the concentration of HEPES buffer was determined at pH 7.5. The kcat was dependent on the concentration of buffer in a saturable manner with an apparent Km of 4 mM for HEPES (Fig. 3C). This result indicates that buffer acts as the second substrate in a ping-pong mechanism accepting a proton from the enzyme during CO2 hydration. The Km parameter also increased with increasing buffer concentration, resulting in a kcat/Km that was independent of the buffer concentration (Fig. 3D). These results are consistent with the metal-hydroxide mechanism in which the interconversion of CO2 and HCO3− (equation 3) reflected by kcat/Km is separate from the intermolecular proton transfer reflected by kcat (equation 4).
Fig 3.
pH and buffer dependence of CO2 hydration catalyzed by Cpb. (A) pH versus kcat; (B) pH versus kcat/Km; (C) HEPES concentration versus kcat; (D) HEPES concentration versus kcat/Km.
There are two distinct subclasses of the β class (2). The type 1 subclass is characterized by ionizable water serving as a fourth ligand to zinc and pH rate profiles for CO2 hydration featuring only one ionization (pKa). In contrast, the type II subclass is characterized by Asp serving as the fourth ligand to zinc and a cooperative pH rate profile with multiple ionizations and only slight activity below pH 8 compared to type I. The type II subclass is further distinguished by a trio of residues (Trp, Tyr, and Arg) in a putative bicarbonate-binding allosteric site represented by the enzyme from Haemophilus influenzae (38). The Cpb CA does not contain the cognate Trp39 or Arg64 of the H. influenzae bicarbonate-binding allosteric site, and phylogenetic analyses place Cpb farthest from the H. influenzae enzyme (Fig. 2). In contrast, Cpb from C. perfringens is in the same clade (Fig. 2) and has substantial sequence identity with Cab from M. thermautotrophicus (Fig. 4), which is representative of the type I subclass. Furthermore, the pH profiles of Cpb revealed only one pKa each with activity extending well below pH 8 (Fig. 3A and B). A distinctive feature of all β class CAs is ligation of the active site zinc with sulfur atoms of two Cys residues and a nitrogen atom of His (39). Comparison of sequences in Fig. 4 indicates that zinc ligands in type I Cab are conserved in Cpb, in addition to the Asp/Arg pair essential for ensuring that water is the fourth ligand to zinc and playing a role in the proton transfer step in the mechanism of Cab (15, 18). Thus, the kinetic parameters, phylogeny, and sequence comparisons establish that Cpb of C. perfringens belongs to the type I subclass of β class CAs.
Fig 4.
Alignment of the sequences for Cpb from Clostridium perfringens and Cab from Methanothermobacter thermautotrophicus. The deduced sequences were aligned by using ClustalX (50). Abbreviations and gene identification numbers: C.p., C. perfringens, gi|110799007; M.t., M. thermautotrophicus, gi|1272331. The numbering corresponds to the M. thermautotrophicus sequence. Residues 32, 87, and 90 are ligands to zinc. Residues 34 and 36 are the Asp/Arg pair important for the proton transfer step of catalysis.
Although commercially available bovine α class CA showed an esterase activity of ∼37 mol of p-nitrophenylacetate hydrolyzed min−1 mol−1 of enzyme, the C. perfringens CA (Cpb) had no detectable esterase activity (<0.05 mol of p-nitrophenylacetate hydrolyzed min−1 mol−1 of tetramer), also a property of Cab from M. thermautotrophicus (16, 17).
Physiological role of Cpb.
Annotation of the genome of C. perfringens strain 13 indicated that a single CA-encoding gene, CPE0413 (cpb), was present (40). To confirm that no other CAs are present, the deduced cpb gene product was entered into the BLASTP program and compared to the strain 13 genome sequence. No significant homology was seen to any other proteins, suggesting there is only a single CA in this strain. The same analysis was done with the other 8 C. perfringens strains with sequenced genomes, and only a single CA gene was detected in each, with 100 to 94% identity to the CA from strain 13 (blast.ncbi.nlm.nih.gov). The cpb gene of strain 13 is predicted to be monocistronic by the operon prediction software available in MicrobesOnline (http://www.microbesonline.org/cgi-bin/fetchLocus.cgi?locus=184876&disp=1). A rho-independent transcription terminator is predicted to lie in the 141-bp intergenic region between cpb and CPE0414 using the ARNold software (http://rna.igmors.u-psud.fr/toolbox/arnold/index.php).
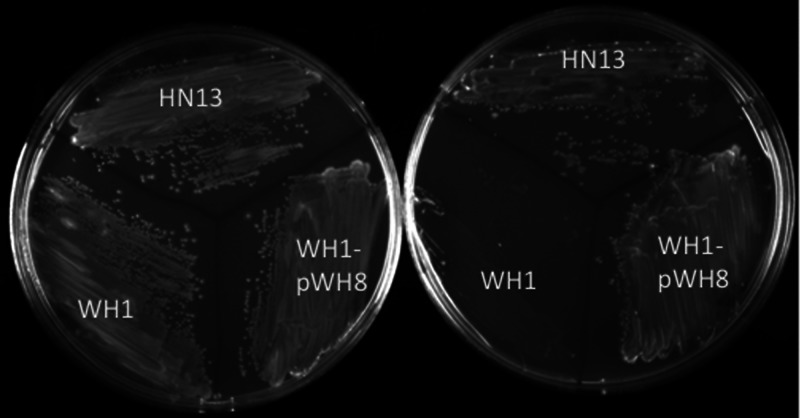
CAs have been shown to mediate a wide variety of physiological effects on species from the domains Bacteria and Eukarya (11, 13, 21, 22, 41–47). Therefore, an in-frame deletion of the cpb gene was constructed in wild-type strain HN13, a derivative of C. perfringens strain 13 that has a deletion of the galT-galK genes (24). Introducing a heterologous galK gene into the chromosome by insertion of a nonreplicating plasmid (pWH6) at the cpb locus established a counterselectable marker strategy where the addition of galactose is toxic to the cell due to the accumulation of galactose-1-phosphate (Gal-1-P) (24). Cells that have undergone a second recombination event that leads to deletion of the cpb gene are resistant to galactose treatment. This strategy was used to produce mutant strain WH1 for evaluating the physiological role of Cpb. There were no differences in the growth rate or final A600 of the mutant strain and wild-type strain when cultured in nutrient-rich PY, BHI, or PGY liquid medium (not shown). However, E. coli and other prokaryotes lacking a functional CA have shown deficiencies in the ability to grow in an atmosphere largely devoid of CO2 required for anaplerotic carboxylation reactions (11, 13, 21, 46). Therefore, we tested the ability of the cpb deletion mutant WH1 to grow on solid media in the absence of CO2 by replacing the headspace gas in anaerobic jars with 100% N2. As shown in Fig. 5, the cpb mutant strain WH1 was unable to grow in the absence of CO2 on a semidefined medium comprised of 0.1% yeast extract, 50 mM glucose, and mineral salts. However, growth in semidefined medium was restored when a wild-type copy of the cpb gene was provided to strain WH1, yielding complemented strain WH1-pWH8 (Fig. 5). In contrast, growth was comparable for wild-type strain HN13 and mutant strain WH1 when cultured on solid nutrient-rich medium (BHI, TY, or PGY) in an atmosphere of either 100% N2 or 80% N2, 10% H2, and 10% CO2 (not shown). These results demonstrate a strict requirement for Cpb during growth of C. perfringens in semidefined medium.
Fig 5.
Growth of C. perfringens strains on solid semidefined medium. Wild-type (HN13), cpb deletion mutant (WH1), and complemented mutant (WH1-pWH8) strains were cultivated on solid agar containing semidefined medium with 10% CO2 (left) and without CO2 (right) added to the atmosphere (see Materials and Methods).
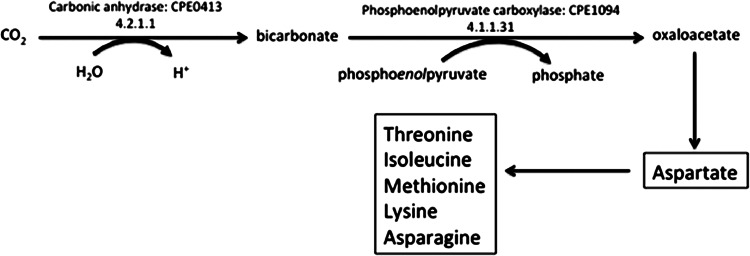
The absence of growth for the cpb deletion mutant strain WH1 when cultured in a semidefined medium and an atmosphere devoid of CO2 is likely due to low activity of anaplerotic CO2-fixing reactions dependent on Cpb to supply HCO3− to carboxylases. One candidate anaplerotic enzyme is the phosphoenolpyruvate (PEP) carboxylase from C. perfringens (48) that produces oxaloacetate (Fig. 6). Oxaloacetate is the first intermediate in the reverse tricarboxylic acid (TCA) cycle that C. perfringens strains use to synthesize the carbon backbones of amino acids (KEGG database [http://www.genome.jp/kegg/kegg2.html]) that are most likely present in nutrient-rich media although in low concentrations or absent in semidefined media. The PEP carboxylase from C. perfringens utilizes HCO3− and is homologous in sequence (49) (Fig. 2) and similar in structure (48) to the PEP carboxylase of M. thermautotrophicus that also utilizes HCO3− (49). The C. perfringens archaeal-type PEP carboxylase is only one of three known in the domain Bacteria (49). The finding that Cpb has a relatively low Km for CO2 (Table 2) is, at least, consistent with a role for Cpb to supply HCO3− for anaplerotic reactions similar to that catalyzed by PEP carboxylase (Fig. 6).
Fig 6.
Proposed involvement of C. perfringens Cpb CA and PEP carboxylase leading to oxaloacetate and amino acid biosynthetic pathways. Carbonic anhydrase is necessary for bicarbonate formation, which is the substrate for the anaplerotic enzyme, PEP carboxylase, leading to oxaloacetate formation and amino acid biosynthesis (boxes).
There was no statistically significant difference in the wild-type strain (HN13) and the cpb deletion mutant strain (WH1) in l-lactate, d-lactate, or formate production grown in either low-carbohydrate (PY) medium or high-carbohydrate (PGY) medium containing 111 mM glucose (data not shown). There was also no statistically significant difference in either the total or ratio of the acetate, butyrate, and propionate VFAs produced when the strains were cultured in PGY medium (data not shown). In contrast, mutant strain WH1 was deficient in the amount of total VFAs produced in PY medium compared to wild-type strain HN13, although the ratios of VFAs and final A600 were unchanged (Table 3). Addition of the wild-type cpb gene under the control of the lactose-inducible promoter PbgaL to mutant strain WH1 restored VFA production in the complemented strain (WH1-pWH8) to wild-type levels (Table 3). While C. perfringens can ferment lactose, the amount of lactose added for induction had no significant effects on VFA production by the wild-type strain HN13 cultured in PGY or PY medium (data not shown). Finally, there was no significant difference in the growth rate or final A600 between wild-type strain HN13 and the cpb deletion mutant strain WH1 (data not shown). These results establish that maximum production of VFAs in a low-carbohydrate medium is dependent on Cpb.
Table 3.
Volatile fatty acid production in three strains of C. perfringens cultured in PY medium
| Trial | Total Ac/Pr/Bua produced (μg/ml) [ratio] in strain: |
||
|---|---|---|---|
| HN13b | WH1c | WH1-pWH8d | |
| 1 | 1,310/460/280 [2.8/1/0.6] | 300/110/63 [2.7/1/0.6] | 970/415/240 [2.3/1/0.6] |
| 2 | 1,230/430/280 [2.9/1/0.7] | 311/110/62 [.8/1/0.6] | 1,030/420/580 [2.5/1/1.4] |
Ac/Pr/Bu, acetate/propionate/butyrate.
HN13 is the wild-type strain.
WH1 is the cpb deletion mutant strain.
WH1-pWH8 is the cpb-complemented mutant cultured with lactose present in PY medium.
The finding that the cpb deletion mutant (WH1) showed no discernible defect in growth phenotype when cultured in nutrient-rich media diminishes the possibility that Cpb plays an important role in gas gangrene infections where C. perfringens is exposed to a surplus of nutrients by breaking down tissue and releasing soluble components. However, it remains to be determined whether Cpb has a more direct role in pathogenicity. Furthermore, although the results indicate that Cpb is necessary for maximum VFA production when carbohydrates are limiting, the effects of reduced VFA levels on pathogenicity have yet to be determined. Nonetheless, it appears likely that Cpb is valuable in soil environments where there is greater competition for nutrients and CO2 levels are limiting. The results reported here encourage further research to determine whether Cpb homologs identified from other Clostridium species, isolated from diverse anaerobic environments, play important roles in the physiology and ecology of these organisms.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy through grant DE-FG02-95ER20198 MOD16 to J.G.F. and National Institutes of Health grants R21 AI088298 and NSF grant 1057871 to S.B.M.
Footnotes
Published ahead of print 8 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02288-12.
REFERENCES
- 1. Tripp BC, Smith K, Ferry JG. 2001. Carbonic anhydrase: new insights for an ancient enzyme. J. Biol. Chem. 276:48615–48618 [DOI] [PubMed] [Google Scholar]
- 2. Rowlett RS. 2010. Structure and catalytic mechanism of the beta-carbonic anhydrases. Biochim. Biophys. Acta 1804:362–373 [DOI] [PubMed] [Google Scholar]
- 3. So AKC, Espie GS. 1998. Cloning, characterization and expression of carbonic anhydrase from the cyanobacterium Synechocystis PCC6803. Plant Mol. Biol. 37:205–215 [DOI] [PubMed] [Google Scholar]
- 4. Sawaya MR, Cannon GC, Heinhorst S, Tanaka S, Williams EB, Yeates TO, Kerfeld CA. 2006. The structure of β-carbonic anhydrase from the carboxysomal shell reveals a distinct subclass with one active site for the price of two. J. Biol. Chem. 281:7546–7555 [DOI] [PubMed] [Google Scholar]
- 5. Kupriyanova EV, Sinetova MA, Markelova AG, Allakhverdiev SI, Los DA, Pronina NA. 2011. Extracellular beta-class carbonic anhydrase of the alkaliphilic cyanobacterium Microcoleus chthonoplastes. J. Photochem. Photobiol. B 103:78–86 [DOI] [PubMed] [Google Scholar]
- 6. Joseph P, Turtaut F, Ouahrani-Bettache S, Montero JL, Nishimori I, Minakuchi T, Vullo D, Scozzafava A, Kohler S, Winum JY, Supuran CT. 2010. Cloning, characterization, and inhibition studies of a beta-carbonic anhydrase from Brucella suis. J. Med. Chem. 53:2277–2285 [DOI] [PubMed] [Google Scholar]
- 7. Nishimori I, Minakuchi T, Kohsaki T, Onishi S, Takeuchi H, Vullo D, Scozzafava A, Supuran CT. 2007. Carbonic anhydrase inhibitors: the beta-carbonic anhydrase from Helicobacter pylori is a new target for sulfonamide and sulfamate inhibitors. Bioorg. Med. Chem. Lett. 17:3585–3594 [DOI] [PubMed] [Google Scholar]
- 8. Cronk JD, O'Neill JW, Cronk MR, Endrizzi JA, Zhang KY. 2000. Cloning, crystallization and preliminary characterization of a beta-carbonic anhydrase from Escherichia coli. Acta Crystallogr. D Biol. Crystallogr. 56:1176–1179 [DOI] [PubMed] [Google Scholar]
- 9. Suarez Covarrubias A, Larsson AM, Hogbom M, Lindberg J, Bergfors T, Bjorkelid C, Mowbray SL, Unge T, Jones TA. 2005. Structure and function of carbonic anhydrases from Mycobacterium tuberculosis. J. Biol. Chem. 280:18782–18789 [DOI] [PubMed] [Google Scholar]
- 10. Vullo D, Nishimori I, Minakuchi T, Scozzafava A, Supuran CT. 2011. Inhibition studies with anions and small molecules of two novel beta-carbonic anhydrases from the bacterial pathogen Salmonella enterica serovar Typhimurium. Bioorg. Med. Chem. Lett. 21:3591–3595 [DOI] [PubMed] [Google Scholar]
- 11. Merlin C, Masters M, McAteer S, Coulson A. 2003. Why is carbonic anhydrase essential to Escherichia coli? J. Bacteriol. 185:6415–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Amoroso G, Morell-Avrahov L, Muller D, Klug K, Sultemeyer D. 2005. The gene NCE103 (YNL036w) from Saccharomyces cerevisiae encodes a functional carbonic anhydrase and its transcription is regulated by the concentration of inorganic carbon in the medium. Mol. Microbiol. 56:549–558 [DOI] [PubMed] [Google Scholar]
- 13. Mitsuhashi S, Ohnishi J, Hayashi M, Ikeda M. 2004. A gene homologous to β-type carbonic anhydrase is essential for growth of Corynebacterium glutamicum under atmospheric conditions. Appl. Microbiol. Biotechnol. 63:592–601 [DOI] [PubMed] [Google Scholar]
- 14. Guilloton MB, Lamblin AF, Kozliak EI, Geraminejad M, Tu C, Silverman D, Anderson PM, Fuchs JA. 1993. A physiological role for cyanate-induced carbonic anhydrase in Escherichia coli. J. Bacteriol. 175:1443–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Strop P, Smith KS, Iverson TM, Ferry JG, Rees DC. 2001. Crystal structure of the “cab”-type β-class carbonic anhydrase from the archaeon Methanobacterium thermoautotrophicum. J. Biol. Chem. 276:10299–10305 [DOI] [PubMed] [Google Scholar]
- 16. Smith KS, Ferry JG. 1999. A plant-type (beta-class) carbonic anhydrase in the thermophilic methanoarchaeon Methanobacterium thermoautotrophicum. J. Bacteriol. 181:6247–6253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smith KS, Cosper NJ, Stalhandske C, Scott RA, Ferry JG. 2000. Structural and kinetic characterization of an archaeal β-class carbonic anhydrase. J. Bacteriol. 182:6605–6613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smith KS, Ingram-Smith C, Ferry JG. 2002. Roles of the conserved aspartate and arginine in the catalytic mechanism of an archaeal β-class carbonic anhydrase. J. Bacteriol. 184:4240–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Supuran CT. 2011. Bacterial carbonic anhydrases as drug targets: toward novel antibiotics? Front. Pharmacol. 2:34 doi:10.3389/fphar.2011.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Joseph P, Ouahrani-Bettache S, Montero JL, Nishimori I, Minakuchi T, Vullo D, Scozzafava A, Winum JY, Kohler S, Supuran CT. 2011. A new beta-carbonic anhydrase from Brucella suis, its cloning, characterization, and inhibition with sulfonamides and sulfamates, leading to impaired pathogen growth. Bioorg. Med. Chem. Lett. 19:1172–1178 [DOI] [PubMed] [Google Scholar]
- 21. Burghout P, Cron LE, Gradstedt H, Quintero B, Simonetti E, Bijlsma JJ, Bootsma HJ, Hermans PW. 2010. Carbonic anhydrase is essential for Streptococcus pneumoniae growth in environmental ambient air. J. Bacteriol. 192:4054–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77–84 [DOI] [PubMed] [Google Scholar]
- 23. Rood JI, Cole ST. 1991. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol. Rev. 55:621–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nariya H, Miyata S, Suzuki M, Tamai E, Okabe A. 2011. Development and application of a method for counterselectable in-frame deletion in Clostridium perfringens. Appl. Environ. Microbiol. 77:1375–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hartman AH, Liu H, Melville SB. 2011. Construction and characterization of a lactose-inducible promoter system for controlled gene expression in Clostridium perfringens. Appl. Environ. Microbiol. 77:471–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Melville SB, Labbe R, Sonenshein AL. 1994. Expression from the Clostridium perfringens cpe promoter in C. perfringens and Bacillus subtilis. Infect. Immun. 62:5550–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wilbur KM, Anderson NG. 1948. Electrometric and colorimetric determination of carbonic anhydrase. J. Biol. Chem. 176:147–154 [PubMed] [Google Scholar]
- 28. Khalifah RG. 1971. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J. Biol. Chem. 246:2561–2573 [PubMed] [Google Scholar]
- 29. Alber BE, Ferry JG. 1996. Characterization of heterologously produced carbonic anhydrase from Methanosarcina thermophila. J. Bacteriol. 178:3270–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gornall AG, Bardawill CJ, David MM. 1949. Determination of serum proteins by means of the biuret reaction. J. Biol. Chem. 177:751–766 [PubMed] [Google Scholar]
- 31. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 32. Peterson JD, Umayam LA, Dickinson T, Hickey EK, White O. 2001. The Comprehensive Microbial Resource. Nucleic Acids Res. 29:123–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Minakuchi T, Nishimori I, Vullo D, Scozzafava A, Supuran CT. 2009. Molecular cloning, characterization, and inhibition studies of the Rv1284 beta-carbonic anhydrase from Mycobacterium tuberculosis with sulfonamides and a sulfamate. J. Med. Chem. 52:2226–2232 [DOI] [PubMed] [Google Scholar]
- 34. Burghout P, Vullo D, Scozzafava A, Hermans PW, Supuran CT. 2011. Inhibition of the beta-carbonic anhydrase from Streptococcus pneumoniae by inorganic anions and small molecules: toward innovative drug design of antiinfectives? Bioorg. Med. Chem. Lett. 19:243–248 [DOI] [PubMed] [Google Scholar]
- 35. Nishimori I, Minakuchi T, Vullo D, Scozzafava A, Supuran CT. 2011. Inhibition studies of the beta-carbonic anhydrases from the bacterial pathogen Salmonella enterica serovar Typhimurium with sulfonamides and sulfamates. Bioorg. Med. Chem. Lett. 19:5023–5030 [DOI] [PubMed] [Google Scholar]
- 36. Heinhorst S, Williams EB, Cai F, Murin CD, Shively JM, Cannon GC. 2006. Characterization of the carboxysomal carbonic anhydrase CsoSCA from Halothiobacillus neapolitanus. J. Bacteriol. 188:8087–8094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zimmerman SA, Ferry JG. 2008. The beta and gamma classes of carbonic anhydrase. Curr. Pharm. Des. 14:716–721 [DOI] [PubMed] [Google Scholar]
- 38. Rowlett RS, Tu C, Lee J, Herman AG, Chapnick DA, Shah SH, Gareiss PC. 2009. Allosteric site variants of Haemophilus influenzae beta-carbonic anhydrase. Biochemistry 48:6146–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rowlett RS, Chance MR, Wirt MD, Sidelinger DE, Royal JR, Woodroffe M, Wang YFA, Saha RP, Lam MG. 1994. Kinetic and structural characterization of spinach carbonic anhydrase. Biochemistry 33:13967–13976 [DOI] [PubMed] [Google Scholar]
- 40. Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, Ogasawara N, Hattori M, Kuhara S, Hayashi H. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. U. S. A. 99:996–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ueda K, Nishida H, Beppu T. 2012. Dispensabilities of carbonic anhydrase in proteobacteria. Int. J. Evol. Biol. 2012:324549 doi:10.1155/2012/324549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cot SS, So AK, Espie GS. 2008. A multiprotein bicarbonate dehydration complex essential to carboxysome function in cyanobacteria. J. Bacteriol. 190:936–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marcus EA, Moshfegh AP, Sachs G, Scott DR. 2005. The periplasmic alpha-carbonic anhydrase activity of Helicobacter pylori is essential for acid acclimation. J. Bacteriol. 187:729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sassetti CM, Rubin EJ. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U. S. A. 100:12989–12994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Badger MR, Price GD. 2003. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J. Exp. Bot. 54:609–622 [DOI] [PubMed] [Google Scholar]
- 46. Kusian B, Sultemeyer D, Bowien B. 2002. Carbonic anhydrase is essential for growth of Ralstonia eutropha at ambient CO2 concentrations. J. Bacteriol. 184:5018–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smith KS, Ferry JG. 2000. Procaryotic carbonic anhydrases. FEMS Microbiol. Rev. 24:335–366 [DOI] [PubMed] [Google Scholar]
- 48. Dharmarajan L, Kraszewski JL, Mukhopadhyay B, Dunten PW. 2011. Structure of an archaeal-type phosphoenolpyruvate carboxylase sensitive to inhibition by aspartate. Proteins 79:1820–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Patel HM, Kraszewski JL, Mukhopadhyay B. 2004. The phosphoenolpyruvate carboxylase from Methanothermobacter thermautotrophicus has a novel structure. J. Bacteriol. 186:5129–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.