Abstract
SMC and MukB complexes consist of a central SMC dimer and two essential binding partners, ScpA and ScpB (MukE and MukF), and are crucial for correct chromosome compaction and segregation. The complexes form two bipolar assemblies on the chromosome, one in each cell half. Using fluorescence recovery after photobleaching (FRAP), we provide evidence that the SMC complex has high exchange rates. This depends to a considerable degree on de novo protein synthesis, revealing that the bacterial SMC complex has high on and off rates for binding to the chromosome. A mutation in SMC that affects ATPase activity and results in exaggerated DNA binding in vitro causes a strong segregation defect in vivo and affects the localization of the entire SMC complex, which localizes to many more sites in the cell than under normal conditions. These data indicate that ATP turnover is important for the function of Bacillus subtilis SMC. In contrast, the centromere protein Spo0J and DNA gyrase showed much less exchange between distinct binding sites on the chromosome than that seen with SMC. Binding of Spo0J to the origin regions was rather static and remained partially conserved until the next cell cycle. Our experiments reveal that the SMC complex has a high, condensin-like turnover rate and that an alteration of the ATPase cycle affects SMC function in vivo, while several nucleoid-associated proteins feature limited or slow exchange between different sites on the nucleoid, which may be the basis for epigenetic-like phenomena observed in bacteria.
INTRODUCTION
The bacterial chromosome is highly compacted and has a quite-defined arrangement in the cell in those bacteria that have been investigated (1). Compaction, replication, and transcription occur in parallel and are achieved by a multitude of DNA-binding proteins and proteins indirectly associated with the nucleoid. The condensation of the bacterial chromosome and the segregation of duplicated chromosome regions depend on several DNA-binding proteins. In Escherichia coli, the histone-like proteins HU, H-NS, FIS, and IHF play overlapping functions in chromosome compaction and bind to DNA with a preference for AT-rich sequences (2). Bacillus subtilis contains a similar protein, HBsu, which is essential for viability and also binds with low selectivity throughout the chromosome (3–5). These small and basic DNA-binding proteins are thought to compact DNA through tight bending via dimeric structures, but they bear no sequence similarity to histones.
Key proteins in several aspects of chromosome dynamics are SMC proteins (6), which are conserved essential proteins throughout all domains of life. Eukaryotes contain several SMC complexes, among which cohesin bridges duplicated sister chromosomes, while condensin leads to chromosome compaction during prophase (7), in an ATP-dependent manner (8). SMC proteins are the central components of such complexes and have a characteristic organization: a Walker A motif at the N terminus and a Walker B motif at the C terminus. These globular domains are connected by a long coiled-coil region which contains a central hinge domain. Through antiparallel interaction, the coiled-coil region folds back onto itself. Thereby, the Walker A and Walker B motifs interact and form a functional ABC transporter-type ATPase, i.e., the head domain (9). This overall structure is common in eukaryotic and bacterial SMC complexes, but B. subtilis SMC and E. coli MukB share only 30% sequence homology within the head domains. Cohesin, condensin, B. subtilis SMC, and MukB complexes have different biochemical properties, e.g., in terms of ATPase activity (10–12). Nevertheless, both eukaryotic and prokaryotic SMC and MukB complexes interact with two (or more) small regulatory proteins. Deletion of either of the subunits leads to severe chromosome decondensation and segregation defects, revealing common schemes of activity for SMC proteins (7, 12).
The bacterial SMC complex (MukBEF complex in E. coli) consists of the SMC (MukB) protein, ScpA (MukE), and ScpB (MukF) (12). While SMC forms a dimer, ScpA and ScpB form a 2:4 subcomplex that binds to the SMC head domains (13) and appears to regulate ATPase activity and, thereby, DNA binding of the SMC dimer (14). MukE and -F can bridge MukB heads (15), and likewise, the kleisin subunit (homologous to ScpA) bridges cohesion heads (16). ATP disrupts the MukEF-MukB interaction, while kleisin is cleaved to release condensin from DNA (15, 16). The bacterial protein complexes form two (and sometimes four) subcellular centers, one (or two) within each cell half, which are essential for the proper arrangement as well as segregation of the chromosome (17, 18). It is unclear how these condensation centers are established and how the SMC complex binds to the chromosome. DNA binding appears to involve the closing of the SMC dimer into a ring-like structure (13), and it depends on proper supercoiling of the chromosome (19). Higher-order structures of SMC have been inferred from atomic force microscopy (AFM) and biochemical experiments, and the MukB complex was shown to be able to bridge different DNA molecules (19, 20). Multimeric SMC-MukB complexes could be the basis of the observed condensation centers.
Another protein required for chromosome compaction and segregation is DNA gyrase, which sets up overall negative supercoiling of the chromosome and is also essential for DNA replication (21). Gyrase is present throughout the chromosome, but it accumulates at and dissipates from the centrally located DNA replication machinery on a time scale of a few minutes (22). A further protein involved in chromosome segregation is Spo0J, which sets up a centromere-like structure around the origin of replication by binding to at least 8 conserved parS sequences, which are spaced in the origin-proximal region, comprising about 20% of the chromosome (23). Spo0J belongs to the class of ParB proteins and has been shown to affect chromosome replication and segregation, in concert with Soj, the corresponding ParA protein (24, 25). SMC is enriched at regions surrounding the origin of replication, according to chromatin immunoprecipitation with microarray technology (ChIP-chip), and Spo0J assists the loading of SMC at this region, possibly through a direct interaction (26–28). ParB proteins have been shown to bind to specific DNA sequences and also to spread along DNA from their binding sites (29, 30).
We wished to investigate the dynamics of the SMC complex and of other nucleoid-associated proteins in live bacterial cells. We therefore employed fluorescence recovery after photobleaching (FRAP) and investigated the B. subtilis SMC complex, Spo0J, and DNA gyrase. We found that the SMC complex shows dynamic exchange kinetics within the condensation centers, revealing that these structures turn over many times during the cell cycle. A transition-state mutation in SMC that leads to exaggerated DNA binding in vitro has a dominant-negative phenotype, indicating that dynamic binding is important for the function of SMC. In contrast, all other DNA-binding proteins showed only limited diffusion through the nucleoid, revealing that several nucleoid-associated proteins feature constrained diffusion through the chromosome.
MATERIALS AND METHODS
Cell growth and preparation.
Bacillus subtilis strains were grown in S750 defined medium (31) complemented with 0.002% Casamino Acids. The doubling time was 93 min under our experimental conditions, i.e., slightly overlapping rounds of replication (32). For induction of the xylose promoter, glucose was exchanged for 0.5% fructose, and xylose was added to 0.5%. For induction of the hyperspank promoter, the culture medium was supplemented with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The antibiotics chloramphenicol (5 μg/ml) and spectinomycin (50 μg/ml) were used when necessary. Overnight cultures were grown at 25°C. Daily cultures were prepared by diluting the overnight culture 1:100. These cultures were grown in a Unitron rotary shaker (Infors AG) at 200 rpm and 25°C until the exponential growth phase. For inhibition of translation, 150 μg/ml puromycin was added, and images were acquired 1 h later.
Strain construction.
The E1118Q mutation in SMC was generated by PCR-based site-directed mutagenesis of the plasmid pSG1729, carrying an smc gene. The following primers were used: GCCGTTTTGCGTCCTTGACCAAGTAGAGGCTGCGCTCGAC (E1118Qup) and GTCGAGCGCAGCCTCTACTTGGTCAAGGACGCAAAACGGC (E1118Qdw). After amplification, the template plasmid was degraded by DpnI at 37°C for 1 h, and the reaction mixture was transformed into E. coli DH5α and selected with ampicillin. The presence of the mutation was confirmed by sequencing, and the plasmid was transformed into either wild-type PY79 or strain PG92, carrying scpA-YFP. The SMC-GFP clpX mutant strain was constructed by transforming the SMC-GFP strain (PG72) with genomic DNA of a clpX::kan strain (33).
Image acquisition.
Fluorescence microscopy was performed on a Zeiss Axioimager microscope, using 100× TIRF objectives with an aperture of 1.45. Images were acquired with a digital charge-coupled device (CCD) camera (Photometrix Cool Snap HQ). For staining of DNA, 4′,6-diamidino-2-phenylindole (DAPI) was applied to a final concentration of 0.2 ng ml−1. Membranes were stained with FM4-64 (2.5 mg ml−1). Cells were mounted on agarose gel pads (1% agarose in S750 minimal medium) on object slides.
FRAP analysis.
For FRAP studies, a 50-mW argon laser (488 nm) was mirrored onto the specimen from above the filter plane, using the side port of the Zeiss microscope. The size of the laser beam (50 μm) was generated by a pinhole inserted into the laser beam within a module that incorporated the optical wire into the side port (custom made by A&S, Munich, Germany). Only cells in which only a single focus or cell half was bleached, which continued to grow (cell extension during the experiment), and in which foci did not move out of the plane during the time of the experiment were analyzed.
Image analysis.
Images of a FRAP sequence were recorded by using Metamorph 6.3 software and were subsequently analyzed by using ImageJ software, version 1.43m (W. Rasband, National Institutes of Health, Bethesda, MD [http://rsb.info.nih.gov/ij/]). In order to align a stack of images, the StackReg plug-in was used. With this tool, each slice is used as the template with respect to which the next slice is aligned, such that the alignment proceeds by propagation (34). The gradual bleaching of the image during acquisition was compensated by normalizing the fluorescence of the bleached region to the integral fluorescence of the entire cell in the same image (35). Background fluorescence was subtracted. To facilitate comparisons of multiple experiments with different bleaching depths and cluster intensities, the relative fluorescence intensity of the region of interest (ROI) of the image sequence was normalized again, using the relative ROI intensity before bleaching. Fits to the FRAP data points were determined using a nonlinear regression algorithm from Wolfram Mathematica 9 software, which calculates parameter estimates and their standard errors (SE). As a model, we used the monoexponential decay function f(t) = A[1 − exp(−t/t0)], with the recovery time constant t0 and the maximum recovery amplitude A.
RESULTS
To determine the dynamics of the SMC condensation centers in live cells, we performed FRAP experiments. FRAP is a powerful technique because it is not invasive and detects time-resolved localization in live cells. A fraction of fluorescently labeled protein is bleached, which leaves the protein functional and only destroys the chromophore. Recovery of fluorescence reveals the exchange of bleached protein fusions for proteins from the nonbleached area of the cell and therefore determines exchange rates within a given subcellular space. A recovery of SMC condensation centers could be based on true exchange of bound proteins or on mere addition of newly synthesized molecules to the assemblies. The latter might be prevalent if condensation centers have a flexible number of assembled proteins, which should be reflected in a highly variable number of components in the centers within and between the cells. To determine whether the number of SMC molecules within the centers varies greatly or is rather uniform, we measured the relative fluorescence intensities of SMC-GFP foci. From 60 cells captured in 3 acquisitions, an average intensity (above background) of 41.2 relative units (RU) (standard deviation [SD] = 10.8; SE = 2.0) was observed, with 25 units being the lowest value and 55 units being the highest value (data not shown). Thus, condensation centers, on average, vary in size by 25%, revealing a relatively constant number of bound molecules. For a continuously growing number of molecules in the centers, fluorescence would be expected to recover gradually with the progressing cell cycle, whereas recovery faster than the time of the cell cycle would imply continuous exchange dynamics. Cells were grown in a medium in which the cell cycle takes about 93 min (overlapping rounds of replication) (32); the cell cycle slowed down under the microscope (due to oxygen limitation), but cells continued to grow for several doubling times, showing that bleaching and image acquisition did not disturb the physiology of the cells. All fluorescent protein (FP) fusions employed functionally complemented the wild-type proteins (ScpA-YFP [17, 18], SMC-GFP [13, 18], Spo0J-GFP [36, 37], and GyrA-YFP [22]) and were expressed under the native promoter (except for GFP-Mbl), as the sole source of the protein. We generally bleached half a cell, using a 488-nm argon laser, and monitored the recovery of fluorescence in the bleached cell half. For FtsZ-CFP and GFP-Mbl, we bleached a spot of only about 1 μm, with the center of the spot being outside the cell; this way, only half an FtsZ ring or one side of an Mbl filament, and much less than one cell half, was bleached.
FRAP kinetics were determined by measuring the fluorescence intensities of the bleached ROI over time. Due to the general bleaching of the images caused by image acquisition, normalization of the fluorescence intensities was required. Therefore, the gradual bleaching of the image during acquisition was compensated by normalizing the fluorescence of the bleached region to the integral fluorescence of the entire cell in the same image. Because the foci/chromosome regions had different fluorescence intensities before bleaching and the bleaching depth changed within every experiment, the relative fluorescence intensity of the ROI of the image sequence was normalized again, using the relative ROI intensity before bleaching. To compare the dynamics of different proteins, the means of the fluorescence intensities of the single experiments (5 to 17 experiments) were plotted against time. Only those experiments in which the cells unequivocally grew (extended during the experiment), where only a single focus or cell half was bleached, and where no out-of-focus signals were present during the recovery were finally analyzed.
For FtsZ, we found a rapid recovery of the Z ring (see Fig. S1A and B in the supplemental material), with a recovery half time of 9.7 s, which is close to the previously determined value of 8 s (38). Recovery was determined from a fit to the raw data (Table 1; see Fig. S1B). The cytoskeletal elements MreB and Mbl have also been shown to feature rapid recovery of their membrane-associated structures (39, 40). For GFP-Mbl, we obtained a recovery half time of 4.79 min (see Fig. S2), which is somewhat shorter than the value obtained in an earlier study (8.5 min) (39). These experiments verify that the Z ring remodels within a few seconds, and Mbl structures within a time frame of a few minutes, and validate our experimental FRAP setup.
Table 1.
FRAP data
| Protein(s) | t1/2 fit (min) | SE of fit (min) | Saturation level (%) | SE of saturation level (%) | No. of expts |
|---|---|---|---|---|---|
| ScpA-YFP | 4.68 | 0.61 | 65.82 | 0.6 | 10 |
| ScpA-YFPa | 6.29 | 2.14 | 60.23 | 13.8 | 3 |
| SMC-GFP | 2.66 | 0.56 | 67.88 | 3.8 | 9 |
| ScpA-YFP plus SMC E1118Q | 5.95 | 1.09 | 92.35 | 9.2 | 10 |
| Spo0J-GFP | 23.43 | 18.50 | 66.12 | 38.3 | 17 |
| GyrA-GFP | 5.71 | 0.85 | 35.91 | 1.7 | 12 |
| GFP-Mbl | 4.79 | 0.59 | 48.72 | 1.9 | 10 |
| FtsZ-CFP | 0.16 | 0.02 | 28.69 | 1.6 | 5 |
Whole-cell FRAP.
Exchange of the SMC complex within condensation centers during the cell cycle.
We used SMC-GFP and ScpA-YFP to gain insight into the in vivo binding to the chromosome of two components of the SMC complex. When a single ScpA-YFP focus was bleached in one cell half, it quickly regained fluorescence, providing evidence for high exchange rates within the condensation centers during the course of a cell cycle (Fig. 1A and B; see Fig. S3D in the supplemental material). Please note that foci show movement within a confined region on a time scale of minutes (17, 18). For fluorescence intensity profile plots of the panels, see Fig. S3A and B. Interestingly, recovery reached levels of up to 66% (SD of fit = 0.6), although only 50% would be expected upon bleaching of a cell half (Fig. 1C). These data indicate that during the experimental time of 30 min, substantial de novo protein synthesis may occur. From a fit to the experimental data, the recovery half time was determined to be 4.7 ± 0.6 min (Fig. 1C; Table 1), i.e., a small fraction of the cell cycle of about 93 min. When an entire cell was bleached on purpose, fluorescence of ScpA-YFP foci recovered within a few minutes (Fig. 1D; Table 1), although more slowly than that in half-bleached cells. This experiment supports the idea that the exchange of ScpA molecules within the centers may be based, at least in part, on de novo protein synthesis, and not solely on the exchange between assembled and freely diffusing molecules. It should be noted that due to the maturation of green fluorescent protein (GFP) or yellow fluorescent protein (YFP) (7 to 30 min) (41, 42), recovery based on new protein synthesis is seen for proteins synthesized much earlier than during the actual experiment. For ScpB, it was more difficult to obtain accurate FRAP data, which showed large variations due to low signal intensities. In principle, ScpB also showed exchange rates in the range of those of ScpA (data not shown). Thus, at least the ScpA subunit of the SMC complex, and likely also ScpB, shows dynamic on and off rates within the condensation centers, in a time frame of a few minutes.
Fig 1.
Sample FRAP measurements of ScpA-YFP. (A and B) FRAP sequences for an ScpA-YFP fusion expressed in exponentially growing Bacillus subtilis PY79. White lines indicate cell ends. A prebleach image was acquired, and an ROI was bleached using a 488-nm laser beam. The bleached region is indicated by a white dashed circle. Filled white triangles indicate ScpA-YFP foci that reappeared within the time of the experiment. (C) Plot of relative fluorescence intensity as a function of time (minutes), corrected for background brightness and total YFP fading over time. The prebleach intensity was normalized to 100%, and the postbleach intensity was normalized to 0%. Error bars represent standard errors of the means (SEM; n = 10). (D) FRAP experiment with a cell in which the fluorescence was fully bleached. Fluorescence recovered, as indicated by the arrowheads.
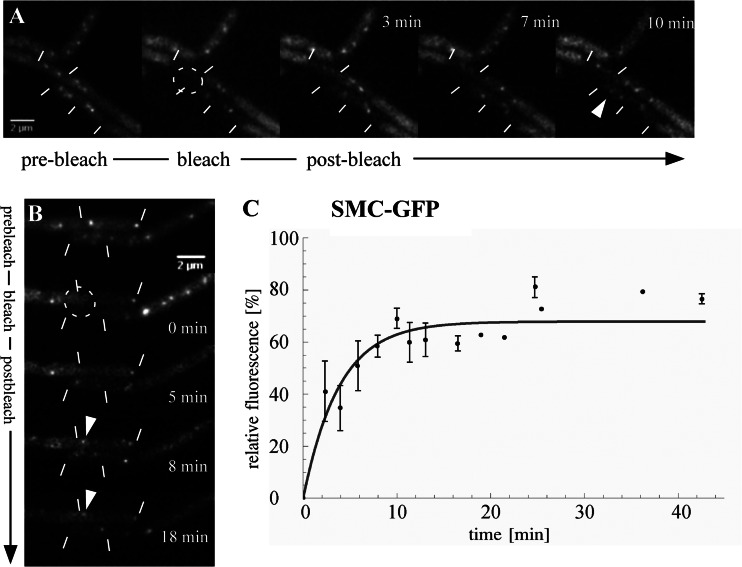
When SMC-GFP was bleached, fluorescence of the bleached focus also recovered rapidly (Fig. 2A and B), with an average recovery half time of only 2.7 ± 0.6 min (Fig. 2C; Table 1). Based on the fact that condensation centers do not show a high degree of variability in size, but deviate by only 25%, on average, these data show that recovery is based on the exchange of bound molecules for nonbound molecules from the surrounding area. Like the case for ScpA, the final fluorescence recovery was 67.9% ± 3.8%, indicating a significant contribution of de novo protein synthesis. We have found that fluorescence of SMC-GFP in anucleate cells is similar to background fluorescence (13), suggesting that most SMC molecules are present on and bound to the nucleoid. Thus, the recovery half time of 2.7 min for one condensation center indicates a high and constant turnover rate, i.e., association with and dissociation from DNA. Thus, the central SMC component, as well as the assumed regulatory ScpA and ScpB subunits of the SMC complex, shows dynamic binding to the condensation center, with SMC having considerably higher exchange rates than those of ScpA.
Fig 2.
(A and B) FRAP sequences for an SMC-GFP fusion expressed in exponentially growing B. subtilis PY79. The white lines represent the cell length. A prebleach image was acquired, and an ROI was bleached using a 488-nm laser beam. The dashed circle indicates the bleached ROI. The white arrowhead indicates the reappearance of a bleached focus. (C) Plot of relative fluorescence intensity as a function of time (minutes), corrected for background brightness and total GFP fading over time. Error bars represent SEM (n = 9). Recovery of SMC reached up to 68% of the initial fluorescence, with a recovery half time of 2.7 ± 0.6 min.
These experiments argue against a previous model proposing that SMC molecules are loaded onto newly duplicated chromosome regions at the replication machinery and are transported to the condensation centers (19). For this scenario, we would have expected a much more gradual and slow regaining of fluorescence, based on a continuous addition of SMC transported along with the newly duplicated DNA. The cell cycle of B. subtilis under our experimental conditions takes about 93 min, whereas SMC centers showed several rounds of turnover within this period. Based on epifluorescence experiments, cells contain very few freely diffusing SMC or ScpA molecules (13, 19), suggesting that the SMC and ScpA pools within the centers are largely replenished from newly synthesized molecules. However, freely diffusing molecules are detectable only through high-speed acquisitions, and for MukB, such a mobile fraction indeed exists (43). Thus, a mobile SMC fraction may exchange with the condensation centers. It is possible that SMC is replenished through Spo0J-guided loading at origin regions, where it is enriched, based on CHIP-chip experiments (26, 27). The bacterial SMC complex thus clearly has condensin-like properties (condensin shows high FRAP rates, with a 2-min recovery half time [44]) in terms of its continuous association with and dissociation from the chromosome, rather than the behavior of cohesin, which is loaded onto chromosomes during S phase and is released only during anaphase, and thus shows a much more long-lived association with the chromosome (45).
The number of condensation centers is affected through a clpX mutation.
SMC has been shown to be partially proteolyzed within each cell cycle (19), and the above data suggest that protein turnover may play a role in the exchange dynamics of SMC protein within the condensation centers. ClpX has been shown to affect the stability of SMC during entry into stationary phase (19), so we wished to investigate if a mutation in the major protease in B. subtilis would have an effect on the nature of condensation centers. Therefore, we tested the localization of the SMC complex in a clpX-deficient strain. Note that a clpX deletion mutant has several pleiotrophic phenotypes (46), and the deletion of an increasing number of proteases leads to additive effects (47). Thus, as a caveat to the analysis, the effect of the absence of ClpX could be indirect. Interestingly, the SMC complex showed abnormal localization in clpX mutant cells (Fig. 3A), in that cells contained, on average, 3.2-fold more SMC-GFP foci on the nucleoids than the case in wild-type cells (three to eight foci versus one to four foci in wild-type cells; 220 cells were analyzed), indicating that reduced proteolysis affects the localization of the SMC complex. To rule out an effect of the clpX mutation on cell division, we measured the lengths of 105 cells and found an average length of 3.2 μm (SD = 0.8; SE = 0.08), while wild-type cells were 3.1 μm long (SD = 0.6; SE = 0.08029). A second interesting phenotype of clpX mutant cells was that a major proportion of cells contained decondensed chromosomes (>80%; 250 cells were analyzed) (Fig. 3A), but no anucleate cells were observed. Thus, a mutation in ClpX affects chromosome condensation and the number of SMC condensation centers.
Fig 3.
Fluorescence microscopy of exponentially growing B. subtilis cells. DNA is stained in green, and the membrane is stained in red. (A) The first panel shows ΔclpX cells expressing SMC-GFP, showing abnormal localization and decondensed chromosomes. The second panel shows DAPI-stained DNA, the third panel shows FM4-64 membrane staining, and the fourth panel shows an overlay of the membrane (red) and SMC-GFP (green) staining. (B) Wild-type cells. (C) Wild-type cells expressing ScpA-YFP from the endogenous gene locus. Ends of cells are indicated by white lines. (D to G) Cells ectopically expressing SMC E1118Q, leading to hypercompacted nucleoids (D and F), generation of anucleate cells (indicated by a white triangle) (E), and altered cell shape (D and G). (H to J) Cells ectopically expressing SMC E1118Q and expressing ScpA-YFP. In panel H, nucleoids lacking a clear ScpA-YFP signal are indicated by white triangles. Panel J shows multiple ScpA-YFP foci. Bars, 2 μm. (K) FRAP measurement of exponentially growing B. subtilis PY79 cells expressing SMC E1118Q (ectopically) and ScpA-YFP. The white lines represent the cell length. A prebleach image was acquired, and an ROI was bleached using a 488-nm laser beam. The circle indicates the bleached ROI. The white arrows indicate bleached foci and their reappearance. (L) Plot of relative fluorescence intensity as a function of time (minutes), corrected for background brightness and total GFP fading over time. Error bars represent SEM (n = 10).
A hyper-DNA-binding mutant of SMC (“transition-state mutant”) shows a dominant-negative segregation defect.
Because the SMC complex showed high on and off rates on the nucleoids, we wondered if exaggerated DNA binding of SMC may cause a problem during the cell cycle. Hirano and Hirano generated an E1118Q mutant version of SMC, termed a “transition-state mutant,” which shows strongly reduced ATPase activity but a much higher DNA-binding capability in vitro than that of wild-type SMC (14). It has been speculated that the transition-state mutation locks head domains in a dimeric state (which it does for ATP-binding cassettes of ABC transporters), which would strongly reduce the off rate for DNA release and thus lead to exaggerated DNA binding. We expressed the fusion from the amylase locus under the control of the xylose-inducible promoter. Expression of mutant SMC for 2 h led to a strong growth defect and to numerous different nucleoid morphology defects. Induction of SMC E1118Q led to the formation of nucleoids that were hypercompacted (in 20% of the cells) (Fig. 3D and F), stretched and decondensed (60%), cut (15%), or misplaced (5%) in the cells (Fig. 3D, E, and G), in contrast with wild-type nucleoids (Fig. 3B). Different nucleoid morphologies did not correlate with cell size, and thus with a particular state of the cell cycle. Growth of cells ceased after 4 to 5 doubling times (data not shown), revealing that extended expression of SMC E1118Q was toxic for the cells. Even in the absence of induction, a subset of the cells (20 to 30%) displayed all phenotypes described above, but in a less pronounced fashion. Contrarily, overproduction of wild-type SMC does not slow down the growth rate and leads to general hypercompaction of nucleoids (13), but not to any other of the observed changes due to expression of mutant SMC. Therefore, the transition-state mutation is dominant and blocks chromosome segregation in B. subtilis. Strikingly, the single point mutation also led to a severe alteration of cell morphology. Concomitant with increasing changes in nucleoid structure, cells became irregularly curved and bent (Fig. 3D and G), and often had an irregular width. These data indicate that the shape of the nucleoid may have an effect on the shape of the cell, with the caveat that alterations in cell morphology may be based on misregulation of gene expression through the SMC E1118Q mutation.
Despite many attempts, we have not been able to generate a transition-state SMC-GFP version for study of its localization. However, to visualize the formation of the bipolar condensation centers, we monitored ScpA-YFP as a marker for the SMC complex. In contrast to the bipolar wild-type pattern of localization (Fig. 3C), ScpA-YFP formed more foci (between 4 and 9), exclusively on the nucleoids, upon the induction of transition-state SMC in a majority of the cells (Fig. 3I and J), or a single, less discrete focus (Fig. 3H), showing that the induction of mutant SMC affects the localization of the entire SMC complex, which localizes to many more sites in the cell than under normal conditions. To test for the exchange dynamics of the irregular ScpA foci, we performed FRAP experiments, again bleaching a cell half. Figure 3K shows that bleached ScpA-YFP foci reappeared, with a somewhat longer half time of 6 ± 1.1 min than the 4.7 ± 0.6 min in wild-type cells (Table 1; Fig. 3L), showing that the defect in activity caused by transition-state SMC affects exchange rates of its complex partner ScpA.
Spo0J shows slow exchange around origin regions on the chromosome.
Spo0J binds to at least 8 specific sites on the chromosome, surrounding the origin region (comprising about 20% of the chromosome), and is involved in the initiation of replication and ensuing segregation of chromosomes (23), as well as in loading of SMC onto DNA at the origin region (26, 27). Due to the compaction of the chromosome (and the limit of resolution of light microscopy), all binding sites can be seen as a single focus within growing cells. Figure 4A shows an example of a cell containing four Spo0J-GFP foci (corresponding to four origin regions), which is at a late stage in the cell cycle and soon to divide. Two origin regions in the right side of the cell are in close proximity and were bleached; they regained fluorescence within several minutes and separated from each other 12 to 13 min after the bleach. In Fig. 4B, the single bleached Spo0J-GFP focus also regains fluorescence, but much slower than the SMC complex. In Fig. S3E in the supplemental material, in the cell adjacent to the partially bleached cell, a single Spo0J-GFP focus can be seen to duplicate between 22 and 29 min after the bleach, showing that the cell cycle continued under the experimental conditions. Figure 4C reveals that Spo0J-GFP fluorescence also recovered exponentially, but much more slowly than that of the SMC complex: after 10 min, only 20% of the Spo0J molecules were exchanged. The recovery half time was determined to be 23.4 min (Table 1). Slow exchange may be based on a mixture of two presumed fractions of Spo0J: one dynamically exchanging fraction, which may consist of Spo0J spreading between the parS sites, and a second, much more static fraction, composed of those molecules that tightly bind to parS sites (23). In any event, exchange of Spo0J proceeds slowly, such that the origin-proximal site on the chromosome is a rather stable structure in this regard.
Fig 4.
FRAP measurement of Spo0J-GFP. (A) The upper image shows an overlay of FM4-64 membrane staining (red), revealing the borders of cells, and staining of Spo0J-GFP fusion proteins (green). White lines indicate cell length, and the circle indicates the bleached ROI. White arrowheads indicate bleached foci and the slow reappearance of the foci, which separated from each other during the course of the experiment. (B) The upper image shows a DNA stain, the circle the bleached ROI, and the arrowheads the reappearance of foci. Bar, 2 μm. (C) Plot of relative fluorescence intensity as a function of time (minutes), corrected for background brightness and total GFP fading over time. Error bars represent SEM (n = 17).
DNA gyrase shows limited diffusion throughout the nucleoid.
DNA gyrase localizes throughout the nucleoid in B. subtilis cells but is also concentrated at the center of the nucleoid, i.e., close to the replication machinery. Gyrase constantly accumulates at and dissipates from the replication forks, in a time frame of a few minutes; therefore, GyrA-GFP (one of the two subunits of the holoenzyme is fused to GFP) foci are apparent only in a subset of the cells or nucleoids. In Fig. 5A, a large part of a nucleoid (in a cell having two separate nucleoids) is bleached and continuously regains fluorescence. In the experiment shown in Fig. 5B, an entire nucleoid was bleached in a cell having two separate nucleoids. On average, fluorescence recovered to 50% within 5.7 min, with saturation reaching only 36% (Fig. 5C; Table 1). Because we wanted to minimize the extent of fluorescence excitation, we did not monitor the occurrence of cell division (through the observation of a membrane stain) with every time interval and therefore cannot determine if and when cell division occurred in the larger cells. Because of this caveat, we can make only limited statements about the loss of saturation levels, and our data may be small underestimates of this value. Initially, fluorescence recovery on a nucleoid or part of a nucleoid occurred at the expense of the other nucleoid. This can be seen in the fluorescence loss in photobleaching (FLIP) graph, which was deduced from the fluorescence intensity of nonbleached nucleoids (corrected for the gradual loss of fluorescence in the field due to image acquisition) in bleached cells (Fig. 5C). Thus, DNA gyrase showed exchange between different sites on the nucleoids in a range of several minutes rather than seconds, as might have been expected for an enzyme that needs seconds or less for its activity at a given site on the DNA. This apparent limited exchange between distant parts of the chromosome could be due in part to a tight association of gyrase with the replication machinery, as seen in previous time-lapse experiments (22).
Fig 5.
FRAP measurements of GyrA-GFP. (A and B) White triangles indicate bleached nucleoids (or parts thereof) and the reappearance of the foci. (A) A large part of a nucleoid was bleached, and fluorescence was recovered at the expense of the nonbleached part. (B) The first picture shows DNA stained with DAPI. The entire nucleoid was bleached in a cell with two separate nucleoids. Fluorescence recovered slowly, initially at the expense of the other nucleoid. (C) Plot of relative fluorescence intensity as a function of time (minutes), corrected for background brightness and total GFP fading over time. Error bars represent SEM (n = 12).
DISCUSSION
Our experiments reveal important novel insights into the mode of interaction of the SMC complex with the nucleoid, as well as distinct binding dynamics of major nucleoid-associated proteins in exponentially growing B. subtilis cells. The SMC condensation complex shows exchange rates of a few minutes within the condensation centers, one or two of which are present in each cell half. This finding has important implications for the mode of DNA binding of the complex. The SMC complex binds to DNA as a ring-like complex and can bridge distinct DNA molecules. It condenses the entire chromosome and constrains supercoils in vivo (6). Based on the high-level exchange dynamics, we conclude that the SMC complex does not bind statically to the chromosome and capture DNA strands but shuttles on and off its substrate, capturing and releasing DNA strands in a time frame of a few minutes. Thus, the B. subtilis SMC complex, with a recovery half time of 3 to 4 min, is much more similar to the dynamic condensin complex (2-min recovery half time) (44) than to the more stably bound cohesin complex, which tethers together sister chromosomes (9- to 25-min recovery half time) (45, 48), and this may be true for many bacteria. Interestingly, the SMC dimer subunit exchanged almost twice as fast as the ScpA complex partner (2.7-min versus 4.7-min recovery half time) within the centers. Any complex is in equilibrium with its nonbound constituents, and ScpA and ScpB have been shown to form a stable subcomplex in vitro, while free and ScpAB-bound SMC fractions exist in cell extracts (19). Our in vivo findings suggest that there is a larger pool of free SMC than free ScpA in the cell, such that upon dissociation of the complex, SMC is exchanged more rapidly between condensation centers and the surrounding space than the ScpAB subcomplex. It was recently shown for MukB that besides the static pool in the condensation centers, a mobile fraction outside the centers exists in vivo (43), in agreement with the above idea underlying the different dynamics of the SMC subunits. It remains to be investigated if less free ScpAB indeed exists in cells, which would set up the asymmetric exchange. Based on the fact that a mobile fraction of MukB (and of SMC [our unpublished results]) exists outside the condensation centers, it is possible that the mode of DNA binding is different between condensation centers and nonspecific sites on the chromosome.
An additional novel concept is our finding that de novo protein synthesis plays a role in the exchange dynamics of SMC and ScpA within the condensation centers, suggesting that the released components of the SMC complex are degraded to some extent and are exchanged for newly synthesized molecules. This is unusual for B. subtilis cells, because more than 95% of all soluble proteins expressed during exponential phase are rather stable (49). We found that in clpX mutant cells, the number of SMC condensation centers increased, and that chromosomes were generally more decondensed than in wild-type cells, indicating that reduced degradation of the complex may affect the formation of the bipolar condensation centers, although the effect of the loss of ClpX activity may be indirect.
A transition-state mutant of SMC has exaggerated DNA-binding abilities and reduced ATPase activity compared with wild-type SMC (14, 50). The corresponding mutation in the homologous ATP-binding cassette of ABC transporters locks the domains in a dimeric state, and it has been suggested that transition-state mutant SMC does not release bound DNA due to a much reduced opening of the head domains. In Caulobacter crescentus, the mutation is dominant negative, and it has been proposed that it leads to a failure of chromosome cohesion (50). We found that the induction of transition-state mutant SMC leads to chromosome hypercompaction and missegregation and blocks cell growth. Thus, exaggerated DNA binding blocks proper chromosome segregation in B. subtilis. In contrast to the case in C. crescentus, we take our observations to support the idea that SMC acts as a condensin and that a block in the ATPase cycle is counterproductive for the activity of the SMC complex. We have not been able to test if transition-state mutant SMC has lower exchange rates in vivo, but exchange rates of ScpA are indeed lowered. Thus, induction of mutant SMC leads to the mislocalization of ScpA, and thus of the SMC complex, and to different exchange dynamics, showing that the mutation interferes with the proper functioning of the entire SMC complex, in a dominant manner.
In marked contrast to the SMC condensation complex, Spo0J shows slower exchange from its binding sites at the origin region. Although Spo0J does equilibrate within the time frame of a cell cycle, exchange is slow, indicating that the centromere-like structure is quite stable, which may be important for reliable control of the replication and segregation of chromosomes. Surprisingly, DNA gyrase also shows limited exchange between distinct parts on the nucleoid, with exchange half times in the range of several minutes rather than seconds, although the protein does not have any specific (albeit preferential) binding sites and must cycle on and off DNA in order to perform its function in supercoiling of the chromosome (51). This finding is interesting in light of the fact that it was recently proposed that a gradient of negative supercoiling from the origin to the terminal region on the E. coli chromosome coordinates changes in gene expression between different growth phases, e.g., exponential versus stationary phase, in conjunction with the chromosomal locations of different histone-like regulatory proteins, which affects the expression of sets of genes mostly positioned within defined chromosome domains (52). Our finding that gyrase indeed shows limited diffusion through the nucleoid shows that such a gradient of supercoiling is possible, and intriguingly, genes for DNA gyrase are most frequently positioned close to the origin region on bacterial genomes (52). Thus, limited diffusion through the nucleoid and gradient-like positioning in vivo may have to be considered for many DNA-binding proteins, providing another factor in the generation of genetic noise in a noncompartmentalized cell, in addition to low protein concentration for some regulatory factors or other factors (53). Inheritance-like phenomena have been observed in bacteria (54), and limited exchange of nucleoid-associated proteins may provide one basis for these highly intriguing phenomena.
Supplementary Material
ACKNOWLEDGMENTS
We thank Victor Sourjik and Sonja Schulmeister of Heidelberg University for help with FRAP analysis.
This work was supported by the Deutsche Forschungsgemeinschaft (IRTG1478) and the Excellence Initiative of the German Federal and State Governments (EXC 294).
Footnotes
Published ahead of print 8 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02097-12.
REFERENCES
- 1. Toro E, Shapiro L. 2010. Bacterial chromosome organization and segregation. Cold Spring Harb. Perspect. Biol. 2:a000349 doi:10.1101/cshperspect.a000349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dorman CJ, Kane KA. 2009. DNA bridging and antibridging: a role for bacterial nucleoid-associated proteins in regulating the expression of laterally acquired genes. FEMS Microbiol. Rev. 33:587–592 [DOI] [PubMed] [Google Scholar]
- 3. Groch N, Schindelin H, Scholtz AS, Hahn U, Heinemann U. 1992. Determination of DNA-binding parameters for the Bacillus subtilis histone-like HBsu protein through introduction of fluorophores by site-directed mutagenesis of a synthetic gene. Eur. J. Biochem. 207:677–685 [DOI] [PubMed] [Google Scholar]
- 4. Micka B, Groch N, Heinemann U, Marahiel MA. 1991. Molecular cloning, nucleotide sequence, and characterization of the Bacillus subtilis gene encoding the DNA-binding protein HBsu. J. Bacteriol. 173:3191–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kohler P, Marahiel MA. 1997. Association of the histone-like protein HBsu with the nucleoid of Bacillus subtilis. J. Bacteriol. 179:2060–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hirano T. 2006. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell. Biol. 7:311–322 [DOI] [PubMed] [Google Scholar]
- 7. Nasmyth K, Haering CH. 2005. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 74:595–648 [DOI] [PubMed] [Google Scholar]
- 8. Strick TR, Kawaguchi T, Hirano T. 2004. Real-time detection of single-molecule DNA compaction by condensin I. Curr. Biol. 14:874–880 [DOI] [PubMed] [Google Scholar]
- 9. Lowe J, Cordell SC, van den Ent F. 2001. Crystal structure of the SMC head domain: an ABC ATPase with 900 residues antiparallel coiled-coil inserted. J. Mol. Biol. 306:25–35 [DOI] [PubMed] [Google Scholar]
- 10. Yamazoe M, Onogi T, Sunako Y, Niki H, Yamanaka K, Ichimura T, Hiraga S. 1999. Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J. 18:5873–5884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hirano M, Hirano T. 1998. ATP-dependent aggregation of single-stranded DNA by a bacterial SMC homodimer. EMBO J. 17:7139–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Graumann PL, Knust T. 2009. Dynamics of the bacterial SMC complex and SMC-like proteins involved in DNA repair. Chromosome Res. 17:265–275 [DOI] [PubMed] [Google Scholar]
- 13. Volkov A, Mascarenhas J, Andrei-Selmer C, Ulrich HD, Graumann PL. 2003. A prokaryotic condensin/cohesin-like complex can actively compact chromosomes from a single position on the nucleoid and binds to DNA as a ring-like structure. Mol. Cell. Biol. 23:5638–5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hirano M, Hirano T. 2004. Positive and negative regulation of SMC-DNA interactions by ATP and accessory proteins. EMBO J. 23:2664–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Woo JS, Lim JH, Shin HC, Suh MK, Ku B, Lee KH, Joo K, Robinson H, Lee J, Park SY, Ha NC, Oh BH. 2009. Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell 136:85–96 [DOI] [PubMed] [Google Scholar]
- 16. Uhlmann F, Lottspeich F, Nasmyth K. 1999. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400:37–42 [DOI] [PubMed] [Google Scholar]
- 17. Mascarenhas J, Soppa J, Strunnikov AV, Graumann PL. 2002. Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. EMBO J. 21:3108–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lindow JC, Kuwano M, Moriya S, Grossman AD. 2002. Subcellular localization of the Bacillus subtilis structural maintenance of chromosomes (SMC) protein. Mol. Microbiol. 46:997–1009 [DOI] [PubMed] [Google Scholar]
- 19. Mascarenhas J, Volkov AV, Rinn C, Schiener J, Guckenberger R, Graumann PL. 2005. Dynamic assembly, localization and proteolysis of the Bacillus subtilis SMC complex. BMC Cell Biol. 6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Petrushenko ZM, Cui Y, She W, Rybenkov VV. 2010. Mechanics of DNA bridging by bacterial condensin MukBEF in vitro and in singulo. EMBO J. 29:1126–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Breier AM, Cozzarelli NR. 2004. Linear ordering and dynamic segregation of the bacterial chromosome. Proc. Natl. Acad. Sci. U. S. A. 101:9175–9176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tadesse S, Graumann PL. 2006. Differential and dynamic localization of topoisomerases in Bacillus subtilis. J. Bacteriol. 188:3002–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin DC-H, Grossman AD. 1998. Identification and characterization of a bacterial chromosome partitioning site. Cell 92:675–685 [DOI] [PubMed] [Google Scholar]
- 24. Lee PS, Lin DC, Moriya S, Grossman AD. 2003. Effects of the chromosome partitioning protein Spo0J (ParB) on oriC positioning and replication initiation in Bacillus subtilis. J. Bacteriol. 185:1326–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ogura Y, Ogasawara N, Harry EJ, Moriya S. 2003. Increasing the ratio of Soj to Spo0J promotes replication initiation in Bacillus subtilis. J. Bacteriol. 185:6316–6324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gruber S, Errington J. 2009. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137:685–696 [DOI] [PubMed] [Google Scholar]
- 27. Sullivan NL, Marquis KA, Rudner DZ. 2009. Recruitment of SMC by ParB-ParS organizes the origin region and promotes efficient chromosome segregation. Cell 137:697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Minnen A, Attaiech L, Thon M, Gruber S, Veening JW. 2011. SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Mol. Microbiol. 81:676–688 [DOI] [PubMed] [Google Scholar]
- 29. Rodionov O, Yarmolinsky M. 2004. Plasmid partitioning and the spreading of P1 partition protein ParB. Mol. Microbiol. 52:1215–1223 [DOI] [PubMed] [Google Scholar]
- 30. Breier AM, Grossman AD. 2007. Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol. Microbiol. 64:703–718 [DOI] [PubMed] [Google Scholar]
- 31. Jaacks KJ, Healy J, Losick R, Grossman AD. 1989. Identification and characterization of genes controlled by the sporulation regulatory gene spo0H in Bacillus subtilis. J. Bacteriol. 171:4121–4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Soufo CD, Soufo HJ, Noirot-Gros MF, Steindorf A, Noirot P, Graumann PL. 2008. Cell-cycle-dependent spatial sequestration of the DnaA replication initiator protein in Bacillus subtilis. Dev. Cell 15:935–941 [DOI] [PubMed] [Google Scholar]
- 33. Liu J, Cosby WM, Zuber P. 1999. Role of lon and ClpX in the post-translational regulation of a sigma subunit of RNA polymerase required for cellular differentiation in Bacillus subtilis. Mol. Microbiol. 33:415–428 [DOI] [PubMed] [Google Scholar]
- 34. Thevenaz P, Ruttimann UE, Unser M. 1998. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7:27–41 [DOI] [PubMed] [Google Scholar]
- 35. Schulmeister S, Ruttorf M, Thiem S, Kentner D, Lebiedz D, Sourjik V. 2008. Protein exchange dynamics at chemoreceptor clusters in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 105:6403–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin DC-H, Levin PA, Grossman AD. 1997. Bipolar localization of a chromosome partition protein to Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 94:4721–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Glaser P, Sharpe ME, Raether B, Perego M, Ohlsen K, Errington J. 1997. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 11:1160–1168 [DOI] [PubMed] [Google Scholar]
- 38. Anderson DE, Gueiros-Filho FJ, Erickson HP. 2004. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol. 186:5775–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carballido-Lopez R, Errington J. 2003. The bacterial cytoskeleton: in vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev. Cell 4:19–28 [DOI] [PubMed] [Google Scholar]
- 40. Defeu Soufo HJ, Graumann PL. 2006. Dynamic localization and interaction with other Bacillus subtilis actin-like proteins are important for the function of MreB. Mol. Microbiol. 62:1340–1356 [DOI] [PubMed] [Google Scholar]
- 41. Megerle JA, Fritz G, Gerland U, Jung K, Radler JO. 2008. Timing and dynamics of single cell gene expression in the arabinose utilization system. Biophys. J. 95:2103–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fisher AC, DeLisa MP. 2008. Laboratory evolution of fast-folding green fluorescent protein using secretory pathway quality control. PLoS One 3:e2351 doi:10.1371/journal.pone.0002351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Badrinarayanan A, Reyes-Lamothe R, Uphoff S, Leake MC, Sherratt DJ. 2012. In vivo architecture and action of bacterial structural maintenance of chromosome proteins. Science 338:528–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oliveira RA, Heidmann S, Sunkel CE. 2007. Condensin I binds chromatin early in prophase and displays a highly dynamic association with Drosophila mitotic chromosomes. Chromosoma 116:259–274 [DOI] [PubMed] [Google Scholar]
- 45. Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J. 2006. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr. Biol. 16:1571–1588 [DOI] [PubMed] [Google Scholar]
- 46. Nakano MM, Hajarizadeh F, Zhu Y, Zuber P. 2001. Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol. Microbiol. 42:383–394 [DOI] [PubMed] [Google Scholar]
- 47. Kirstein J, Moliere N, Dougan DA, Turgay K. 2009. Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat. Rev. Microbiol. 7:589–599 [DOI] [PubMed] [Google Scholar]
- 48. Gause M, Misulovin Z, Bilyeu A, Dorsett D. 2010. Dosage-sensitive regulation of cohesin chromosome binding and dynamics by Nipped-B, Pds5, and Wapl. Mol. Cell. Biol. 30:4940–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schindler T, Graumann PL, Perl D, Ma S, Schmid FX, Marahiel MA. 1999. The family of cold shock proteins of Bacillus subtilis. Stability and dynamics in vitro and in vivo. J. Biol. Chem. 274:3407–3413 [DOI] [PubMed] [Google Scholar]
- 50. Schwartz MA, Shapiro L. 2011. An SMC ATPase mutant disrupts chromosome segregation in Caulobacter. Mol. Microbiol. 82:1359–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stone MD, Bryant Z, Crisona NJ, Smith SB, Vologodskii A, Bustamante C, Cozzarelli NR. 2003. Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases. Proc. Natl. Acad. Sci. U. S. A. 100:8654–8659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sobetzko P, Travers A, Muskhelishvili G. 2012. Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc. Natl. Acad. Sci. U. S. A. 109:E42–E50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Raser JM, O'Shea EK. 2005. Noise in gene expression: origins, consequences, and control. Science 309:2010–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Veening J-W, Smits WK, Kuipers OP. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62:193–210 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.