Abstract
The methylotroph Methylobacterium extorquens AM1 oxidizes methanol and methylamine to formaldehyde and subsequently to formate, an intermediate that serves as the branch point between assimilation (formation of biomass) and dissimilation (oxidation to CO2). The oxidation of formaldehyde to formate is dephosphotetrahydromethanopterin (dH4MPT) dependent, while the assimilation of carbon into biomass is tetrahydrofolate (H4F) dependent. This bacterium contains two different enzymes, MtdA and MtdB, both of which are dehydrogenases able to use methylene-dH4MPT, an intermediate in the oxidation of formaldehyde to formate. Unique to MtdA is a second enzymatic activity with methylene-H4F. Since methylene-H4F is the entry point into the biomass pathways, MtdA plays a key role in assimilatory metabolism. However, its role in oxidative metabolism via the dH4MPT-dependent pathway and its apparent inability to replace MtdB in vivo on methanol growth are not understood. Here, we have shown that an mtdB mutant is able to grow on methylamine, providing a system to study the role of MtdA. We demonstrate that the absence of MtdB results in the accumulation of methenyl-dH4MPT. Methenyl-dH4MPT is shown to be a competitive inhibitor of the reduction of methenyl-H4F to methylene-H4F catalyzed by MtdA, with an estimated Ki of 10 μM. Thus, methenyl-dH4MPT accumulation inhibits H4F-dependent assimilation. Overexpression of mch in the mtdB mutant strain, predicted to reduce methenyl-dH4MPT accumulation, enhances growth on methylamine. Our model proposes that MtdA regulates carbon flux due to differences in its kinetic properties for methylene-dH4MPT and for methenyl-H4F during growth on single-carbon compounds.
INTRODUCTION
Methylotrophy is the ability of microorganisms to utilize reduced compounds with no carbon-carbon bonds as a sole source of energy and carbon, a metabolism that has been studied in detail for over 50 years (1, 2). Examples of these carbon sources include methanol, methylamine, and methane, placing methylotrophs as key players in global cycling of carbon and nitrogen (3, 4, 5, 6). From a physiological and biochemical perspective, methylotrophy is an intriguing model of study, since methylotrophs must accommodate high flux through toxic metabolites such as formaldehyde and glyoxylate. The metabolism of one-carbon compounds in the facultative methylotroph Methylobacterium extorquens AM1 (Fig. 1) (7) involves the oxidation of methanol and methylamine to formaldehyde via methanol dehydrogenase or methylamine dehydrogenase (MaDH), respectively. Formaldehyde is incorporated into the cytoplasm and coupled with the carbon carrier dephosphotetrahydromethanopterin (dH4MPT) via Fae (formaldehyde-activating enzyme) to generate methylene-dH4MPT. MtdA (methylene-H4MPT/H4F dehydrogenase) and MtdB (methylene-H4MPT dehydrogenase) catalyze the oxidation of methylene-dH4MPT to methenyl-dH4MPT with NAD(P)+ as a cosubstrate. Mch (methenyl-H4MPT cyclohydrolase) catalyzes the conversion of methenyl-dH4MPT to formyl-dH4MPT. Fhc (formyltransferase/hydrolase complex) catalyzes the conversion of formyl-dH4MPT to formate via a methanofuran derivative. Together, these reactions constitute the dH4MPT-dependent oxidative pathway. The partitioning of carbon between assimilatory and oxidative metabolism occurs at formate (8). Oxidative metabolism involves CO2 production via formate dehydrogenases (Fdh). Four different formate dehydrogenases are known, two NAD linked (Fdh1 and Fdh2) and two non-NAD linked, for which the in vivo electron acceptors are not known (Fdh3 and Fdh4) (9). Assimilatory metabolism starts with the tetrahydrofolate (H4F) pathway. FtfL (formate-H4F ligase) catalyzes the conversion of formate and H4F to generate formyl-H4F. Fch (methenyl-H4F cyclohydrolase) catalyzes the reversible dehydration to methenyl-H4F. MtdA (methylene-H4MPT/H4F dehydrogenase) catalyzes the reversible reduction to methylene-H4F, the intermediate that is incorporated into the assimilatory cycles (Fig. 1). The interlinked assimilatory cycles comprise 22 enzymes (10). An alternative pathway for methylamine oxidation, the N-methyl glutamate (NMG) pathway, has been described for several microorganisms (11). Although biochemical details of the pathway are not well understood, it is known that three enzymes are necessary for the conversion of methylamine to presumably methylene-H4F (11): an NMG synthase, a gamma-glutamylmethylamide synthetase (GMA synthetase), and an NMG dehydrogenase (NMGDH). Low NMG dehydrogenase activity was detected in cell extracts of M. extorquens, suggesting that M. extorquens has the capacity to oxidize methylamine via the indirect N-methyl glutamate pathway (12). However, a mau mutant (lacking MaDH) is unable to grow on methylamine (12), questioning the functionality of the pathway.
Fig 1.
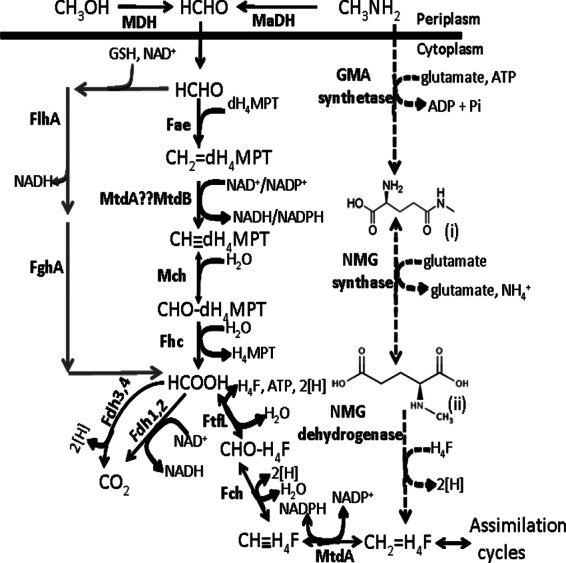
Methanol and methylamine oxidation pathway. Gene products involved in each reaction are indicated next to the arrows. Abbreviations: HCHO, formaldehyde; MDH, methanol dehydrogenase; MaDH, methylamine dehydrogenase; dH4MPT, dephosphotetrahydromethanopterin; Fae, formaldehyde-activating enzyme; CH2=dH4MPT, methylene-dH4MPT; MtdB and MtdA, methylene-tetrahydromethanopterin dehydrogenases; CH dH4MPT, methenyl-dephosphoH4MPT; Mch, methenyl-dH4MPT cyclohydrolase; CHO—dH4MPT, formyl-dH4MPT; Fhc, formyltransferase/hydrolase complex; Fdh, formate dehydrogenase; FtfL, formate-tetrahydrofolate ligase; H4F, tetrahydrofolate; CHO—H4F, to formyl-H4F; Fch, methenyl-H4F cyclohydrolase; CH
dH4MPT, methenyl-dephosphoH4MPT; Mch, methenyl-dH4MPT cyclohydrolase; CHO—dH4MPT, formyl-dH4MPT; Fhc, formyltransferase/hydrolase complex; Fdh, formate dehydrogenase; FtfL, formate-tetrahydrofolate ligase; H4F, tetrahydrofolate; CHO—H4F, to formyl-H4F; Fch, methenyl-H4F cyclohydrolase; CH H4F, methenyl-H4F; CH2—H4F, methylene-H4F; GSH, glutathione; GMA synthetase, gamma-glutamylmethylamide synthetase; NMG synthase, N-methyl glutamate synthase; NMG dehydrogenase, N-methyl glutamate dehydrogenase; FlhA, GSH/NAD-dependent formaldehyde dehydrogenase; FghA, S-formyl-GSH hydrolase. The dashed line represents the proposed NMG pathway, where “(i)” represents gamma-glutamylmethylamide and “(ii)” represents N-methyl glutamate. Gray lines represent the heterologous Paracoccus denitrificans formaldehyde detoxification pathway encoded on the plasmid pCM106 (16).
H4F, methenyl-H4F; CH2—H4F, methylene-H4F; GSH, glutathione; GMA synthetase, gamma-glutamylmethylamide synthetase; NMG synthase, N-methyl glutamate synthase; NMG dehydrogenase, N-methyl glutamate dehydrogenase; FlhA, GSH/NAD-dependent formaldehyde dehydrogenase; FghA, S-formyl-GSH hydrolase. The dashed line represents the proposed NMG pathway, where “(i)” represents gamma-glutamylmethylamide and “(ii)” represents N-methyl glutamate. Gray lines represent the heterologous Paracoccus denitrificans formaldehyde detoxification pathway encoded on the plasmid pCM106 (16).
Studies with purified MtdA have shown that it is NADP+ specific and is able to use methylene-H4MPT, methylene-dH4MPT, methenyl-H4F, and methylene-H4F as the substrates (13, 14, 15). Characterization of the second methylene-H4MPT dehydrogenase, MtdB, showed that it can use both NAD+ and NADP+ but is preferentially an NADP+-dependent enzyme (Km = 20 μM for NADP+ versus 200 μM for NAD+). Like MtdA, MtdB can use either methylene-H4MPT or methylene-dH4MPT (15). With NADP as a cosubstrate, MtdA catalyzes the dehydrogenation with an approximately 3-fold-higher catalytic efficiency (Vmax/Km) than that of MtdB (15). In addition, in cell extracts, the dH4MPT- and NADP+-dependent activity of MtdA is 10-fold higher than that of MtdB (14, 15). Therefore, it seemed likely that MtdA should be able to replace MtdB in vivo. However, an mtdB mutant strain that contained wild-type mtdA was unable to grow on either methanol or succinate in the presence of methanol. This methanol-sensitive phenotype was attributed to formaldehyde accumulation (16), suggesting that MtdA was not able to allow sufficient carbon flux from formaldehyde to formate to avoid formaldehyde toxicity. This result was inconsistent with the in vitro data regarding catalytic efficiency activity in cell extract. Further, increasing levels of MtdA relieved the formaldehyde toxicity when the mutant strain was grown with succinate plus methanol but did not allow growth in methanol liquid medium, complicating the interpretation of the role of MtdA in the oxidative step (16). The current study was undertaken to address the contradiction between the phenotypic and biochemical results and to determine the role of MtdA in methylotrophic metabolism. The results suggest that MtdA functions in the distribution of the formate pool to maintain the balance between assimilation and oxidation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All M. extorquens (17) strains and plasmids used in this study are described in Table 1. M. extorquens AM1 strains were grown at 30°C on a minimal salts medium (18) containing carbon sources at the following concentrations: 35 mM methylamine, 125 mM methanol, and 15 mM succinate. Escherichia coli strains were grown on Luria-Bertani medium. For conjugation between the helper strain E. coli S17-1 (19) and M. extorquens, Difco nutrient broth supplemented with Difco BiTek agar (1.5% [wt/vol]) was used. Antibiotics were added when needed to the following final concentrations: 100 μg of ampicillin/ml, 50 μg of kanamycin/ml, 50 μg of rifamycin/ml, and 10 μg of tetracycline/ml. Chemicals were obtained from Sigma (St. Louis, MO).
Table 1.
M. extorquens strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| CM 253.1 | ΔmtdB | 16 |
| CM 258.1 | ΔmptG | 16 |
| NM 132 | ΔmgsA | This study |
| NM 133 | ΔmgsA ΔmtdB | This study |
| AM1 | Rifr derivative | 17 |
| Plasmids | ||
| pCM62 | M. extorquens expression vector (Plac) | 21 |
| pCM80 | M. extorquens expression vector (PmxaF) | 21 |
| pCM106 | pCM80 with fghA flhA | 16 |
| pAP774 | M. extorquens expression vector (Pmeta1_2136) | E. Skovran, unpublished data |
| pAP776 | M. extorquens expression vector (Pmeta1_002) | E. Skovran, unpublished data |
| pAP775 | M. extorquens expression vector (Pmeta1_3616) | E. Skovran, unpublished data |
| pNM1 | pAP774 with mtdA | This study |
| pNM2 | pAP776 with mtdA | This study |
| pNM3 | pAP775 with mtdA | This study |
| pNM125 | pCM80 with mch | This study |
Generation of mutant strains.
M. extorquens deletion mutants lacking mgsA (NMG synthase) were generated with the allelic exchange suicide vector pCM184 (20). Approximately 0.5-kb regions upstream and downstream of mgsA were amplified by PCR and directly cloned into pCM184 (donor) with the primers nmgsyn AM1 Rev KpnI up, 5′ ACGGTACCCGACCACGAAGGTGAAGAAG 3′; nmgsyn AM1 For EcoRI up, 5′ GCGAATTCCGGCATTACCTGCACCTG 3′; nmgsyn AM1 Rev SacI down, 5′ GCGAGCTCTCATCACCGCGAGGTAGTTC 3′; and nmgsyn AM1 For HpaI down, 5′ CAGTTAACGCAAGGATTCCCGCGAGAC 3′. Mutant strains of M. extorquens were generated by conjugation of the plasmid from an E. coli S17-1 donor as previously described (21). Unmarked deletion strains were generated with the cre-lox-expressing plasmid pCM157 (20). Mutant strains were confirmed by diagnostic PCR analysis.
Phenotypic analyses of mutant strains.
Growth of M. extorquens strains was assessed for 2 biological replicates grown in liquid medium containing the carbon source described and with monitoring of the optical density at 600 nm (OD600). The strains were grown in minimal medium with succinate (15 mM) at 30°C to late exponential phase and subcultured into flasks containing 100 ml of minimal medium containing methylamine (35 mM) and the appropriate antibiotic if needed.
Generation of plasmids overexpressing mtdA.
The coding region of mtdA was amplified by PCR and cloned into pCM62 (21) with the primers MtdA SacI For, 5′ GCGAGCTCATGTCCAAGAAGCTGCTCTTCCAGTTCG 3′, and MtdA HindIII Rev, 5′ CGCGAATTCTCAGGCCATTTCCTTGGCCAGC 3′, containing different promoter regions: Meta1_2136 for low expression, Meta1_0002 for medium expression, and Meta1_3616 for high expression. Relative expression was based on published microarray results (18, 22).
Purification and characterization of dH4MPT species.
M. extorquens AM1 wild type was grown at 30°C on a minimal salts medium containing methanol (125 mM) until it reached an OD600 of 2.2. Cells were harvested by centrifugation. The cell paste (30 g) was introduced into an anaerobic chamber (Coy, Grass Lake, MI) containing 95% N2 and 5% H2. All experiments were performed in the dark. Cells were resuspended in 30 ml of anoxic buffer A (5 mM potassium phosphate buffer, pH 4.8, 10 mM β-mercaptoethanol) and broken by boiling (15 min). Cell extracts were cleared by centrifugation in a sealed tube outside the anaerobic glove box (28,000 × g, 45 min, 4°C) and transferred back into the glove box. Forty milliliters of supernatant was applied to an Oasis weak anion exchange (WAX) extraction cartridge (6 ml, 500 mg) (Waters, Milford, MA) previously activated with 1% (vol/vol) formic acid and equilibrated with methanol. After loading, the column was washed with 1 column volume of distilled water. dH4MPT was eluted with 1 column volume of elution solution 1 (5% [vol/vol] NH4OH, 80% [vol/vol] methanol, 15% [vol/vol] H2O). The elution fraction was analyzed under UV-visible light to confirm the characteristic maximal peaks of the species. The fraction was also tested by monitoring NADP+-dependent MtdB activity (15). After corroborating activity, the active fraction was lyophilized under anoxic conditions. The lyophilized powder was transferred into the glove box and resuspended with anoxic buffer A. The active fraction (approximately 3 ml) was applied to an Oasis mixed-mode anion exchange (MAX) cartridge (3 ml, 60 mg) (Waters, Milford, MA) previously activated with water and equilibrated with methanol. After loading, the column was washed with 1 column volume of distilled water. While partially purified dH4MPT species did not bind the sorbent, some contaminants did bind and were discarded. The flowthrough fraction and washes were pooled (2 fractions of 3 ml each) and lyophilized under anoxic conditions. The powder was transferred into the glove box and resuspended with anoxic buffer A. The fraction was analyzed by UV-visible analysis and activity as described above. The process was repeated once more to further purify the cofactor with an Oasis mixed-mode cation exchange (MCX) cartridge (3 ml, 60 mg), and the resulting active fraction was lyophilized. The powder was transferred into the glove box and resuspended in anoxic buffer B (100 mM potassium phosphate buffer, pH 6.0; 2 ml). The UV-visible spectrum and NADP+-dependent MtdB activity were corroborated in the final fraction. Methenyl-dH4MPT was purified by the same protocol with the following differences. The initial cell pellet was derived from an mtdB mutant strain grown on minimal salts medium containing methylamine (35 mM). Methenyl-dH4MPT did not bind the WAX resin. The flowthrough from this step was lyophilized and resuspended with anoxic buffer A (approximately 12 ml). The fraction containing methenyl-dH4MPT was further purified with the MAX and MCX sorbents, and although methenyl-dH4MPT did not bind the resins, other contaminants bound. Analysis by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) corroborated the m/z typical of dH4MPT and methenyl-dH4MPT, respectively. The mass spectrometer (Quatto Micro API; Micromass, Manchester, United Kingdom) was operated in the positive (3.5-kV) electrospray ionization (ESI) mode. Nitrogen was used as the desolvation gas at 300 liters/h and as cone gas at 50 liters/h. The syringe pump was used to infuse the purified sample at a flow rate of 5 μl/ml for MS and tandem MS (MS/MS) analysis. The mass spectrometer was first operated in Q1MS mode to detect the parent ions of interest (targeted ions). It was then operated in MS/MS mode to look for the product ions for the selected parent. The collision energy was optimized to obtain a good signal-to-noise ratio of the product ions.
Preparation of extracts.
M. extorquens strains, wild type and mutants, were grown on minimal medium with methylamine (100 ml) and harvested at an OD600 of 0.4 to 0.5. Cell pellets were harvested by centrifugation, and the supernatant was removed and immediately transferred to the anaerobic chamber (Coy, Grass Lake, MI) containing 95% N2 and 5% H2. Further experiments were performed under strictly anoxic conditions and in the dark. Cell pellets were resuspended in anoxic buffer (100 mM potassium phosphate buffer, pH 6.0; 1 ml). Lysozyme was added and incubated on ice for 10 min. Cells were broken by sonication (9 cycles of intermittent pulses for 45 s each), with monitoring of the temperature to ensure that it remained below 10°C. The cell extracts were centrifuged (10 min, 28,000 × g, 25°C), and the supernatant was set on ice until used for assays. When indicated, the extracts were desalted with a PD-10 gel filtration column (8.3-ml bed volume, 5-cm bed height) previously equilibrated with 100 mM anoxic potassium phosphate buffer, pH 6.0, and further concentrated with microconcentrators (Amicon-Ultra; Millipore, Billerica, MA).
Purification of MtdA.
For high-level expression of MtdA, mtdA was amplified and cloned into pQE30Xa with chromosomal DNA (purified according to the MoBio protocol [Carlsbad, CA]) from wild-type M. extorquens AM1 as the template. The construct was transformed into M15/prep4 cells. This strain was grown at 30°C in Superbroth medium with kanamycin (50 μg/ml) and ampicillin (50 μg/ml). IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM) was added to induce expression of MtdA when the culture reached an OD600 of 0.5. Cultures were grown after induction for 4 h at 30°C, and cells were harvested by centrifugation (4,800 × g, 10 min, 4°C). The cell paste (30 g) was resuspended in 30 ml of buffer A (50 mM Tris-HCl, pH 8.0, 5 mM imidazole, and 15% [vol/vol] glycerol), and cells were broken with a French press. Cell extracts were cleared by centrifugation (28,000 × g, 45 min, 4°C), and the supernatant was applied to a Ni-charged chelating Sepharose column (Qiagen, Germantown, MD) (8 ml) previously equilibrated with buffer A. After loading, the column was washed with 5 column volumes of buffer B (buffer A with 200 mM NaCl). MtdA-His6 protein was eluted off the column by running an imidazole gradient (0 to 500 mM) over 50 ml in buffer A. Fractions (4 fractions of 3 ml each) were pooled and desalted with a PD-10 gel filtration column (8.3-ml bed volume, 5-cm bed height) equilibrated with buffer C (50 mM morpholineethanesulfonic acid [MES]-NaOH buffer, pH 5.5). The protein was concentrated with centrifugal filter devices (Amicon-Ultra 10K; 4,000 × g, 15 min, 4°C). The concentrated protein sample (100 μM) was purified further with a HiTrap Sepharose S column (GE Healthcare, Pittsburgh, PA) (5 ml) previously equilibrated with buffer C. After loading the column, MtdA-His6 was eluted off the column with a salt gradient (0 to 0.2 M NaCl) over 25 ml in buffer C. Two fractions of 2 ml each were pooled and desalted with buffer D (120 mM potassium phosphate buffer, pH 6.0) and the gel filtration PD-10 column previously described. Proteins were concentrated to a final concentration of 100 μM and used as indicated. The protein was stable when stored at −80°C. Protein concentration was determined by the bicinchoninic acid method (Pierce).
MtdA activity.
Activity of MtdA was measured with NADP+ as a cosubstrate, and the dehydrogenation of methylene-dH4MPT to methenyl-dH4MPT was followed by monitoring production of NADPH as described previously (13) with the following differences: 200 mM potassium phosphate buffer, purified methylene-H4MPT dehydrogenase MtdA (100 μM, 50 μl), and formaldehyde (2 mM). All assays were performed under anoxic conditions and in the dark. Formaldehyde was prepared by autoclaving paraformaldehyde for 10 min (Sigma, St. Louis, MO) in distilled water (4.6 mg/ml). The reaction mixture was incubated at room temperature for 20 min. The total volume of the reaction was 420 μl. Activity was monitored at 340 nm (i.e., NADPH production) after addition of the cosubstrate NADP+ (125 μM). When indicated, hydrogenation of methenyl-H4F was also measured with NADPH as a cosubstrate and methenyl-H4F (Schircks Laboratories, Switzerland) as a substrate. Extinction coefficients were 6.22 cm−1 mM−1 for NADP+ and 21.68 cm−1 mM−1 for methenyl-H4F. For inhibition experiments, two concentrations of methenyl-H4F were used, 10 and 50 μM, and five different concentrations of methenyl-dH4MPT were used, 2.5, 5, 10, 20, and 50 μM. The concentration of pure MtdA used was 100 ng.
Mch activity.
The activity of Mch was followed photometrically by monitoring the decrease in absorbance at 335 nm (ε = 21.6 mM−1 cm−1) as described previously (23) with the following differences: the assay mixture contained 50 mM potassium phosphate buffer (pH 8.0), 0.3 M NaCl, and 125 μM methenyl-dH4MPT. The reaction was started with extract.
NMGDH activity.
NMG dehydrogenase (NMGDH) activity included (per 0.5 ml): 50 mM sodium phosphate buffer, pH 7.6, 5 mM NMG, 0.5 mM NAD+, and 0.5 mg protein extract. The reaction was initiated by addition of NMG and, after incubation of the reaction mixture at room temperature for 15 to 20 min, terminated by addition of 0.5 ml Nash reagent. Accumulation of formaldehyde was recorded spectrophotometrically at 412 nm (11).
RESULTS
The NMG pathway contributes to methylamine oxidation in the mtdB mutant but not in the wild type.
Previous work (14, 16) demonstrated that the mtdB mutant strain was unable to grow in methanol liquid medium. However, in this study, it was found that the mtdB mutant strain is able to grow poorly on the C1 compound methylamine as a sole source of carbon and energy, with a doubling time of 9.5 h and a final OD600 of 0.7 compared to a doubling time of 4 h and a final OD600 of 1.3 for the wild type (Fig. 2A). It has been shown that M. extorquens uses methylamine dehydrogenase to grow on methylamine, and this enzyme catalyzes the conversion of methylamine to formaldehyde (12). Once formaldehyde is formed and transported into the cytoplasm, it undergoes the same dH4MPT-dependent oxidation to generate formate as it does when methanol is the carbon source (Fig. 1). In order to understand the relative roles of MtdA and MtdB in C1 metabolism, it is important to understand why MtdB appears to have different roles for growth on methanol and methylamine. Recently, Latypova et al. defined the genetics of the NMG pathway (an alternative pathway for methylamine oxidation) in another methylotroph, Methyloversatilis universalis FAM5, and defined a cluster of sox-like, gltB-like, and gsIII-like genes as encoding the enzymes necessary for an operational NMG (11). Homologs of this alternative pathway were identified in M. extorquens—Meta1_1545, Meta1_1546, Meta1_1547, and Meta1_1548 for the sox-like cluster; Meta1_1550, Meta1_1551, and Meta1_1552 for the gltB-like cluster; and Meta1_1553 for the gamma-glutamylmethylamide synthetase (GMA synthetase) homolog (Fig. 2B)—suggesting that M. extorquens has the potential for a functional NMG pathway. NMG dehydrogenase was detected in wild-type cell extracts at 0.2 ± 0.1 nmol/min/mg protein, supporting the existence of this pathway. However, its contribution is not sufficient to support growth, as a mau mutant (missing MaDH activity) is not able to grow on methylamine (12). It has been suggested that the final product of the NMG pathway is not formaldehyde but methylene-H4F (11). If the NMG pathway is functional in the mtdB mutant strain, it could allow contribution of carbon flux to the assimilation pathways bypassing the dH4MPT-dependent oxidation pathway and, consequently, MtdB activity. No equivalent alternative is available for methanol oxidation, and so the difference in phenotypes of the mtdB mutant strain on methanol and methylamine may be due to this alternative pathway. In order to test this possibility, NMG dehydrogenase activity was measured in cell extracts from the mtdB mutant strain grown in methylamine medium. This activity was found to be 17.8 ± 2 nmol/min/mg protein, 89-fold higher than that in the wild-type strain, suggesting that when MtdB is not present, the NMG pathway for methylamine oxidation is upregulated. Phenotypic studies of mutants were used to corroborate the involvement of this pathway for methylamine growth of the mtdB mutant strain. As shown in Fig. 2A, the mgsA mutant strain (mgsA encodes NMG synthase) showed no growth defect in methylamine medium, while the double mtdB mgsA mutant strain showed a growth defect compared to the mgsA mutant (final OD600 of 0.2 to 0.3 of 2 biological replicates). Together, these data demonstrate the contribution of the NMG pathway for methylamine growth in a strain lacking mtdB.
Fig 2.
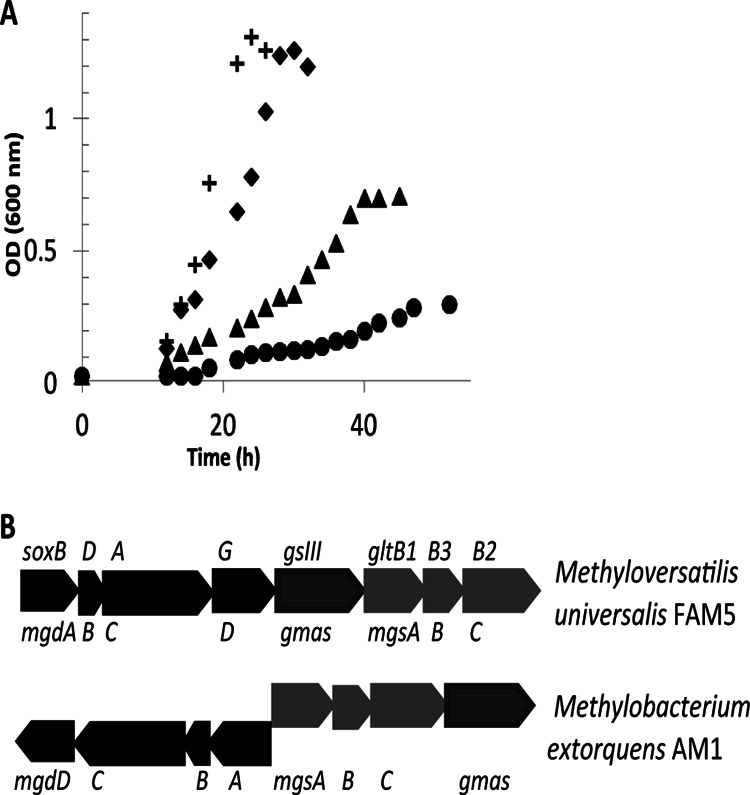
Contribution of the NMG pathway to methylamine growth. (A) Growth of wild-type M. extorquens (crosses), mtdB mutant strain (triangles), an msgA (encoding N-methyl glutamate synthase) mutant strain (diamonds), and a double mtdB msgA mutant strain (circles) pregrown on succinate and inoculated in medium containing methylamine (35 mM). (B) Comparison of the gene clusters encoding the N-methyl glutamate pathway in Methylobacterium extorquens AM1 and Methyloversatilis universalis FAM5.
Reduced growth of the mtdB mutant strain on methylamine is due in part to formaldehyde toxicity.
Each of the mutants that affect formaldehyde consumption has two possible components to growth defects, the change in flux to later steps in metabolism and the toxicity of formaldehyde, if it accumulates. Formaldehyde toxicity is manifested as decreased growth in the presence of a cosubstrate in combination with methanol (methanol sensitivity) or formaldehyde compared to that of the wild type (16). The glutathione (GSH)-dependent formaldehyde oxidation pathway (FghA and FlhA [Fig. 1]) alleviates both methanol and formaldehyde sensitivity in M. extorquens mutants (including the mtdB mutant), due to decreased formaldehyde accumulation (16, 24, 25), and it also likely provides increased flux to formate, which might increase assimilation. In order to test the formaldehyde toxicity component of growth defects in the mtdB mutant strain, phenotypic studies were carried out comparing this strain to mptG, mgsA, and mgsA mtdB mutant strains with and without the GSH-dependent formaldehyde oxidation pathway provided in trans. mptG is the gene that encodes the first enzyme in the dH4MPT biosynthesis pathway, β-ribofuranosylaminobenzene 5′-phosphate synthase (26). A null mutant in mptG generates no dH4MPT (27) and therefore is unable to synthesize the substrate for MtdB and MtdA activity. Thus, this mutant can assimilate methylamine only via the NMG pathway. This mutant is unable to grow on either methanol or methylamine and is extremely methanol sensitive (16). The lack of growth on methylamine of a null mutant lacking MptG was confirmed (Fig. 3A). It had previously been shown that an mptG mutant containing the heterologous GSH-dependent formaldehyde oxidation pathway was no longer methanol sensitive but grew only poorly on methanol (16). Thus, the GSH-dependent formaldehyde oxidation pathway alleviates formaldehyde toxicity and allows only slow growth.
Fig 3.
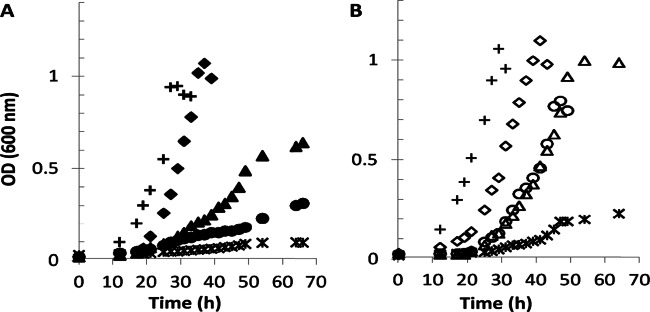
Formaldehyde accumulation reduces growth of the mtdB mutant strain in methylamine medium. (A) Growth of wild-type M. extorquens (crosses), mtdB mutant (triangles), msgA mutant (diamonds), mptG (encoding the first gene product necessary for dH4MPT biosynthesis) mutant (asterisks), and mtdB msgA double mutant (circles), all carrying the plasmid pCM80 as the vector control. All strains were pregrown on succinate and inoculated in medium containing methylamine (35 mM). (B) Growth of the same strains (represented by the same symbols but open instead of solid) under the same conditions, carrying the heterologous GSH-dependent formaldehyde oxidation system [pCM106 (fghA flhA)].
When the GSH-dependent formaldehyde oxidation pathway was introduced into the mptG mutant, the strain grew better on methylamine than did the mptG strain carrying the empty plasmid but still not at the wild-type level (doubling time of 11.5 h; final OD600 of 0.22) (Fig. 3), similar to the results reported previously with methanol (16). This amount of growth reflects the contributions of formate assimilation via the GSH-dependent formaldehyde oxidation pathway plus assimilation of methylene-H4F via the NMG pathway. The difference in growth between the mgsA mtdB double mutant and the mtdB mutant reflects assimilation via the NMG pathway. Since that growth difference is similar to the growth of the mptG mutant containing the GSH-dependent formaldehyde oxidation pathway, it suggests that the main impact of the GSH-dependent formaldehyde oxidation pathway is in detoxification. Therefore, the difference in growth between the mptG mutant expressing FghA and FlhA and other mutants containing this pathway is an indicator of the relative contribution of formaldehyde toxicity to the mutant phenotype. However, when the GSH-dependent formaldehyde oxidation pathway was expressed in the mtdB mutant, the strain was able to grow in methylamine medium with a growth rate (4 h) and final optical density (OD600 of 1.02) similar to those of the wild type (Fig. 3B).
fghA and flhA were also introduced in the double mtdB mgsA mutant, and the growth of the strain was measured. Significant increases in growth rate and final optical density were observed compared to those of the same double mutant without the pathway (Fig. 3). However, the growth of the double mutant containing the GSH-dependent formaldehyde oxidation pathway was clearly defective compared to those of the wild-type strain and the mtdB mutant strain containing the same heterologous pathway. Together, these results suggest that part but not all of the methylamine growth defect of an mtdB mutant strain is due to formaldehyde toxicity.
Increasing MtdA levels inhibit growth on methylamine.
Previous studies on methanol medium (16) have shown that increasing levels of MtdA in an mtdB mutant strain relieved methanol and formaldehyde sensitivity but did not allow growth on methanol. However, the effect of MtdA overexpression on methylamine growth was not tested. In this study, the region encompassing mtdA was cloned and introduced into a set of plasmids with different promoters (pAP774, low expression; pAP775, medium expression; pAP776, high expression [Table 1]) of the expression vector pCM62 (21). These plasmids were used to overexpress mtdA at different levels in the mtdB mutant strain during growth on methylamine. Increasing levels of MtdA inhibited growth on methylamine (Fig. 4). One plausible explanation is that increased levels of MtdA resulted in the accumulation of a downstream metabolite affecting growth. Since the mtdB mutant strain is able to grow on formate (16), an intermediate downstream from formate would be an unlikely candidate for this inhibitor.
Fig 4.
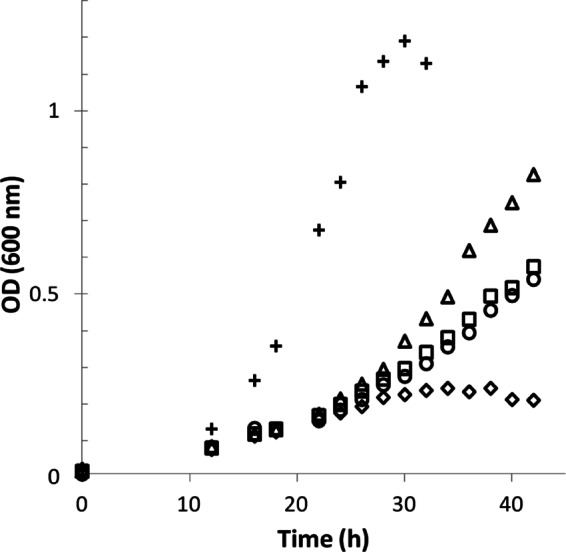
Increased levels of MtdA inhibit growth of the mtdB mutant strain. Growth of wild-type M. extorquens (crosses) in methylamine medium with empty plasmid (pCM62) and mtdB mutant strains overexpressing different levels of mtdA. Higher levels of mtdA are denoted by different symbols (triangles, empty vector; squares, pNM1; circles, pNM2; diamonds, pNM3).
An inhibitor of MtdA accumulates in the mtdB mutant strain.
A likely target for inhibition is the methenyl-H4F reduction activity by MtdA, since methylene-H4F is the entry metabolite for the assimilatory pathways. To test the hypothesis that an inhibitor of the H4F-dependent activity of MtdA accumulates in the mtdB mutant strain, methenyl-H4F oxidation activity by MtdA was measured in cell extracts from cells grown on methylamine of the wild type, the mtdB mutant strain, and the mtdB mutant strain overexpressing mtdA. The extracts were generated under strict anoxic conditions and in the dark to minimize degradation of dH4MPT derivatives. As shown in Table 2, the activities of MtdA with limiting amounts of methylene-H4F were similar for the wild-type extract and the mtdB extract and decreased slightly when MtdA was overexpressed. However, when the extracts were desalted to remove a potential small-molecule inhibitor, a 4-fold increase of the MtdA activity catalyzing the oxidation of methylene-H4F to methenyl-H4F was found (from 0.3 mU/mg to 1.2 mU/mg enzyme) from the mtdB extract and a 10-fold increase was found in the mtdB extract overexpressing MtdA (from 0.13 mU/mg to 1.4 mU/mg), while the activity from the wild-type extract remained the same. These results confirmed the presence of an inhibitor of H4F-dependent activity of MtdA in the mtdB mutant strain.
Table 2.
Specific activity of the methylene-H4MPT dehydrogenase MtdA in cell extracts with methylene-H4F as the substrate and NADP+ as the cosubstrate
| Strain | Sp act (mU/mg) with 20 μM H4F |
Fold change | |
|---|---|---|---|
| Normal extracts | Desalted extracts | ||
| WT | 0.23 ± 0.09 | 0.36 ± 0.09 | 1.5 |
| mtdB mutant | 0.34 ± 0.1 | 1.23 ± 0.1 | 3.6 |
| mtdB mutant/pNM3 | 0.13 ± 0.12 | 1.38 ± 0.15 | 10 |
Methenyl-dH4MPT accumulates when MtdB is absent.
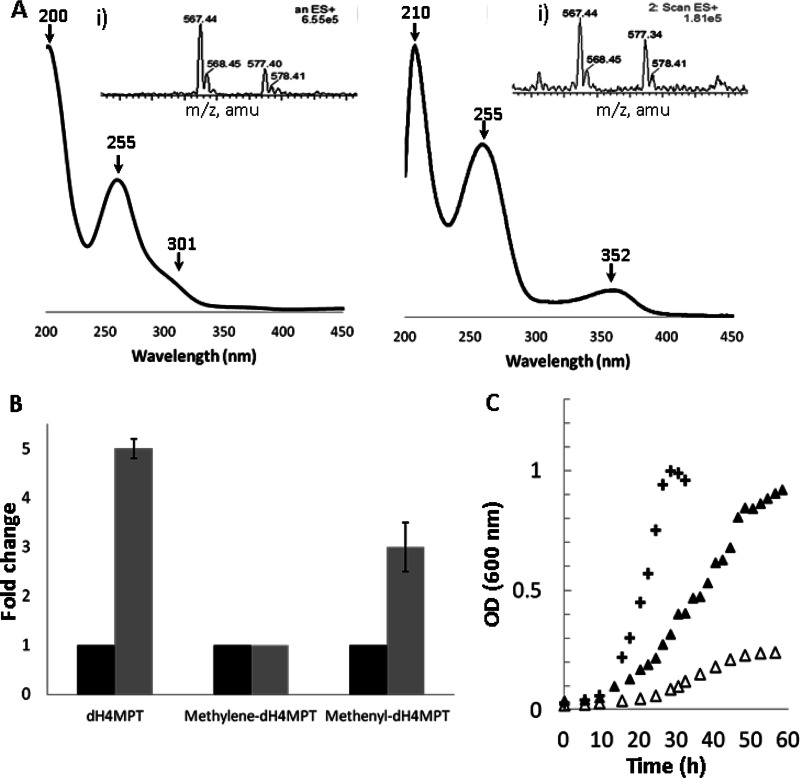
Since MtdA has dual specificity for H4F and dH4MPT intermediates, a possible candidate for the inhibitor of H4F-dependent activity is a dH4MPT species. To test for accumulation of dH4MPT species in the mtdB mutant strain compared to the wild type, dH4MPT and its derivatives were purified from the wild-type and mtdB mutant strains grown on methylamine and analyzed as described in Materials and Methods. Two fractions showed higher absorbance in the extract derived from the mtdB mutant than in that from the wild type, and each fraction was further purified. UV-visible maximal peaks of 200 nm, 255 nm, and 301 nm were observed in the purified fractions, along with maximal absorbance peaks of 210 nm, 255 nm, and 352 nm, consistent with dH4MPT and methenyl-dH4MPT, respectively (14, 28) (Fig. 5A). Each of these fractions was then analyzed by electrospray ionization mass spectrometry along with standards. The m/z for the two species (567 and 577 [Fig. 5A, inset]) corroborated the identification of dH4MPT and methenyl-dH4MPT, respectively, suggesting that these two species accumulated in the mtdB mutant strain when growing in methylamine medium. Quantification of the accumulation of both compounds with respect to the wild-type strain showed a 5-fold increase for dH4MPT (by UV-visible analysis and mass spectrometry analysis) and a 2- to 3-fold increase for methenyl-dH4MPT (Fig. 5B). In keeping with accumulation of methenyl-dH4MPT, the methenyl-H4MPT cyclohydrolase (Mch) activity decreased in an mtdB mutant strain compared to the wild type, with activities in extracts of 0.4 ± 0.1 μmol/min/mg protein and 1 ± 0.2 μmol/min/mg protein, respectively. Overexpression of mch should decrease the methenyl-dH4MPT pool and thereby alleviate inhibition. mch in an overexpression plasmid was introduced into the mtdB mutant strain, and the cells were grown on methylamine. Increasing the levels of Mch (from 0.4 μmol/min/mg protein in the mtdB mutant to 1.2 μmol/min/mg protein with the overexpression plasmid) allowed the mutant to grow to higher densities (OD600 of 1.2 versus 0.6 [Fig. 5C]) and partially rescued the defect in growth rate. This result is consistent with an inhibitory effect of methenyl-dH4MPT on the assimilation step.
Fig 5.
Isolation and identification of methenyl-dH4MPT in the mtdB mutant during growth on methylamine. (A) Identification of the maximal absorbance points of the purified compounds and MS analysis of reaction products (insets: left, dH4MPT; right, methenyl-dH4MPT). Both species were compared to standards. amu, atomic mass units. (B) Accumulation of dH4MPT and methenyl-dH4MPT in the mtdB mutant strain (gray bars) compared to the wild-type strain (black bars). (C) Growth of wild-type M. extorquens (crosses) and mtdB mutant strain with empty vector (open triangles) and overexpressing mch (filled triangles). All strains were pregrown on succinate and inoculated in medium containing methylamine (35 mM).
Methenyl-dH4MPT inhibits MtdA H4F-dependent activity.
The methylene-H4MPT dehydrogenase MtdA was purified to assess the inhibitory effect of methenyl-dH4MPT on methenyl-H4F reduction activity by MtdA. As shown in Table 3, when methenyl-dH4MPT was present in the assay, the Km value of MtdA activity for methenyl-H4F increased more than 3-fold and the Vmax changed slightly, suggesting a competitive inhibition. The estimated Ki was 10 μM.
Table 3.
Kinetic parameters of the methylene-H4MPT dehydrogenase MtdA with NADPH as a cosubstrate in the presence and absence of methenyl-dH4MPTa
| Substrate | Km (μM) | Vmax (U/mg) |
|---|---|---|
| Methenyl-H4F | 45 | 103 |
| Methenyl-H4F + methenyl-dH4MPT | 187 | 135 |
Concentrations of methenyl-H4F varied from 10 μM to 100 μM; the concentration of methenyl-dH4MPT was 100 μM.
DISCUSSION
MtdA is a well-characterized methylene-dH4MPT dehydrogenase (13, 15, 29) known to efficiently catalyze the oxidation of methylene-dH4MPT to methenyl-dH4MPT during methanol and methylamine metabolism and also to catalyze the reversible reaction with methenyl-H4F. However, the significance of its activity under physiological conditions in this step has been unclear, considering that MtdA is unable to replace the alternative methylene-dH4MPT dehydrogenase MtdB. One plausible explanation concerns differential pyridine nucleotide usage, since MtdA is able to use only NADP+ as a cosubstrate, while MtdB can use both NAD+ and NADP+. However, the intracellular concentrations of both species are on the order of millimolar concentrations and should be saturating (30). Furthermore, deuterium experiments have shown that the majority of flux during methanol oxidation to formate generates NADPH (8).
In methanol medium, two problems are associated with the lack of MtdB in the cell: the accumulation of formaldehyde in the presence of methanol and the decreased production of formate for further metabolism (16). If the methylene-dH4MPT-dependent activity of MtdA in vivo is not sufficient for the flux to methenyl-dH4MPT, formaldehyde would accumulate in an mtdB mutant strain. However, when MtdA was overexpressed and its activity increased 7-fold, it was observed that MtdA was able to avoid the accumulation of formaldehyde but the mtdB mutant strain was still unable to grow on methanol (16). This result suggests that the normal level of MtdA is insufficient to handle the full formaldehyde flux through the dH4MPT-dependent oxidative pathway. When MtdA is overexpressed in the mtdB mutant strain, formaldehyde flux is sufficient and should allow growth on methanol. The lack of growth under this condition suggests a block in assimilation.
We showed that the mtdB mutant strain is able to grow on methylamine, although with a lower growth rate than that of the wild type. The growth of the mutant on methylamine is dependent on the presence of an alternative methylamine oxidation pathway (the NMG pathway), which is proposed to generate methylene-H4F directly without involvement of the dH4MPT-dependent oxidative pathway. The lower growth rate and lower final cell density of the mtdB mutant strain than those of the wild type suggest that MaDH provides a higher in vivo flux of methylamine than does the NMG synthase/GMA synthetase. Although this finding did not illuminate the reason why MtdA cannot substitute for MtdB, it did provide a system for studying the role of MtdA in one-carbon utilization. However, this finding does provide evidence of a functional NMG pathway for methylamine oxidation in M. extorquens as previously hypothesized but not shown.
It is clear that MtdB alone at its normally expressed level is sufficient to accommodate the flux from formaldehyde, since an mtdA mutant is not methanol sensitive (31). In this study, we demonstrate that the activity of MtdA alone allows growth on methylamine in the presence of alternate pathways such as the NMG pathway and the heterologous GSH-dependent formaldehyde oxidation pathway. Each of these pathways contributes to both the detoxification of formaldehyde and the contribution of carbon to assimilation in a dH4MPT-independent manner, thus at least partially alleviating both problems that arise when MtdB is lacking. In addition, the difference in biochemistry between the GSH-dependent formaldehyde oxidation pathway and the NMG pathway allows us to further propose mechanistic details that were unknown. These results show that the NMG pathway is functional and inducible and that when it is upregulated it is essential for growth of an mtdB mutant strain on methylamine. In addition, the data presented here further suggest that the final product of the pathway is not formaldehyde but methylene-H4F. We also demonstrate partial growth in the absence of an NADH-generating step both in the dH4MPT-dependent oxidative pathway and in the NMG pathway, consistent with the lack of NADH production found in previous labeling studies. Finally, although both pathways contribute to assimilation (generating formate or methylene-H4F [11, 16]), neither pathway allows significant growth by itself. It is only when MtdA activity is present and carbon flux occurs through the dH4MPT-dependent oxidative pathway that the mtdB mutant strain is able to grow. We have demonstrated that accumulation of dH4MPT occurs in an mtdB mutant. This accumulation is consistent with the accumulation of formaldehyde found in the mtdB mutant grown on methanol plus succinate, suggesting that the increase in MtdA is still not sufficient to allow full carbon flux to formate, as it occurs when both MtdA and MtdB are present. On the other hand, the product, methenyl-dH4MPT, also accumulates. Concomitantly, the methenyl-dH4MPT-consuming activity (Mch) drops in the mtdB mutant strain to only 40% of that of the wild-type strain, which would be expected to contribute to methenyl-dH4MPT accumulation. This result suggests that the lack of MtdB affects Mch activity. It is possible that Mch and MtdB physically associate and that Mch activity is higher in the complex than alone. Another possibility is that alterations in the pools of one or more small molecules (e.g., formaldehyde, pyridine nucleotides, etc.) in the mtdB mutant strain affect Mch activity, either directly or through either transcriptional or posttranscriptional mechanisms. These hypotheses will require further investigation.
Methenyl-dH4MPT, methylene-H4F, and methenyl-H4F are proposed to bind the same active site (29), and therefore, it is feasible to suggest that accumulation of methenyl-dH4MPT might affect MtdA activity in both the methenyl-H4F-dependent assimilation step and the methylene-H4F-dependent oxidation step. Desalting of extracts with elevated methenyl-dH4MPT relieved inhibition, as would be expected for reversible inhibition. The estimated Km of 45 μM and Ki of 10 μM are also consistent with this hypothesis. dH4MPT total cellular concentration (all species) in wild-type M. extorquens is approximately 400 μM, while H4F total cellular concentration is approximately 150 μM (15). The in vivo concentrations of each of the four species for each compound are not known; however, the concentration of methenyl-H4F is predicted to be at or below the Km. It is feasible to suggest that the intracellular concentration of methenyl-dH4MPT is higher than the intracellular concentration of methenyl-H4F under normal conditions and that therefore the assimilatory MtdA activity is already partially inhibited. Thus, the calculated Ki value suggests that even a 3-fold increase in the normal concentration of methenyl-dH4MPT could have a dramatic effect on assimilatory flux.
The results presented in the current study suggest an explanation for why both MtdA and MtdB are required for methylotrophic growth. The apparent biochemical redundancy of MtdA and MtdB underlies a simple but elegant mechanism for balancing oxidative and assimilatory metabolism, based on the dual H4F/dH4MPT specificity of MtdA. A previous study had shown that when succinate-grown cells are exposed to methanol, one-carbon flux is directed to oxidative metabolism until the entire assimilatory machinery is induced and functional (22). This strategy is one mechanism to prevent formaldehyde from accumulating during shifts in formaldehyde flux. Our results suggest a possible model for a mechanism of this block in assimilatory flux (Fig. 6) that is consistent with our results, in which MtdB is the dehydrogenase involved in constitutively removing formaldehyde while MtdA is dynamically changed to increase or decrease levels of methenyl-dH4MPT. The levels of methenyl-dH4MPT modulate the assimilatory activity of MtdA. Thus, the dual H4F/dH4MPT specificity of MtdA couples oxidative flux to assimilatory flux and ensures that assimilation does not occur under conditions of imbalance in the oxidative flux. Interestingly, FtfL and Fch, the enzymes that complete the H4F pathway that generates the methylene H4F precursor for assimilatory metabolism, both catalyze reversible reactions. This would suggest that the entire H4F pathway would respond to MtdA activity inhibition, consistent with our hypothesis for flux regulation. In this model, the extra formate produced while assimilation is blocked is oxidized to CO2, as was shown to occur previously in metabolic imbalance in M. extorquens AM1 (22). It is likely that the one-step oxidation of formate to CO2 provides a relatively simple mechanism for dynamic adjustment to keep formaldehyde from accumulating during formaldehyde flux changes, as opposed to the alternative of maintaining and regulating activity of the 22 enzymes of the combined serine cycle and ethylmalonyl coenzyme A (CoA) pathways, the machinery of assimilatory metabolism.
Fig 6.
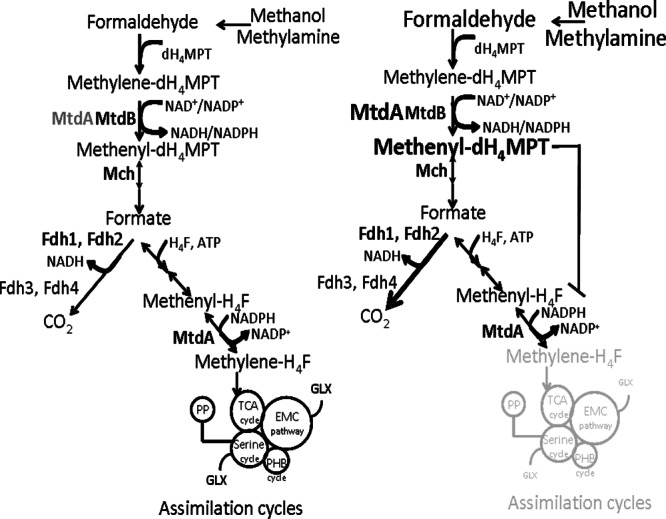
Proposed model for the role of MtdA in methylotrophic metabolism in M. extorquens AM1. Under moderate formaldehyde flux, the dH4MPT-dependent activity of MtdA is relatively low, while the H4F-dependent activity is normal. When the flux upshifts, methenyl-dH4MPT accumulates, inhibiting production of methylene-H4F and thus assimilatory metabolism. Formate utilization shifts to oxidative metabolism, producing more CO2. TCA, tricarboxylic acid; PHB, poly-β-hydroxybutyrate; EMC, ethylmalonyl-CoA; GLX, glyoxylate.
Likewise, we suggest that the inhibitory effect on MtdA by accumulation of methenyl-dH4MPT when the strain lacks MtdB could also regulate distribution of carbon flux from the NMG pathway when methylamine is the carbon substrate. In this scenario, the inhibition of MtdA catalyzing the oxidation of methylene-H4F to methenyl-H4F ensures that the predicted product of the NMG pathway, methylene-H4F, is incorporated into the assimilatory cycles and not further oxidized to CO2. This is particularly important considering that our in vivo data suggest that the GMA synthetase and NMG synthase are considerably less efficient than MaDH in utilizing methylamine.
In summary, the results presented here suggest that one role of MtdA during one-carbon compound metabolism is to regulate carbon flux between assimilation and oxidation when the metabolic network is resetting after perturbation of formaldehyde flux. Further, the identification of methenyl-dH4MPT as a small-molecule regulator for one-carbon compound metabolism is shown. The inhibitory effect of methenyl-dH4MPT and MtdA might not be the only regulatory mechanism that contributes to the lack of assimilation during this transition. Decreased Mch activity strongly suggests an additional regulatory mechanism that could potentially involve additional small regulators. The growth rate differences between the wild-type strain and the mtdB mutant strain might shift the nucleotide ratios, so we cannot rule an effect due to nucleotides.
This growing insight into how the metabolic network is controlled at the level of small molecules will facilitate the manipulation of methylotrophic metabolic networks for a variety of applications, including the production of valued-added chemicals from methanol.
ACKNOWLEDGMENTS
This work was funded by a grant from the NIH (GM58933). N.C.M.-G. was supported with award no. F32GM096705 from the National Institute of General Medical Sciences.
We thank Mila Chistoserdova, Elizabeth Skovran, and Nathan Good for critical reading of the manuscript; Alexander Palmer for the construction of the pAP plasmids; and Martin Sadilek for assistance with the mass spectroscopy analysis.
Footnotes
Published ahead of print 15 March 2013
REFERENCES
- 1. Anthony C. 1982. The biochemistry of methylotrophs. Academic Press, London, United Kingdom [Google Scholar]
- 2. Chistoserdova L, Chen S, Lapidus A, Lidstrom ME. 2003. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J. Bacteriol. 185:2980–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Naqvi SW, Bange HW, Gibb SW, Goyet C, Hatton AD, Upstill-Goddard RC. 2005. Biogeochemical ocean-atmosphere transfers in the Arabian Sea. Prog. Oceanogr. 65:116–144 [Google Scholar]
- 4. Schäfer H, Miller LG, Oremland RS, Murrell JC. 2007. Bacterial cycling of methyl halides. Adv. Appl. Microbiol. 61:307–3046 [DOI] [PubMed] [Google Scholar]
- 5. Trotsenko YA, Murrell JC. 2008. Metabolic aspects of aerobic obligate methanotrophy. Adv. Appl. Microbiol. 63:183–229 [DOI] [PubMed] [Google Scholar]
- 6. Singh BK, Bardgett RD, Smith P, Reay DS. 2010. Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat. Rev. Microbiol. 8:779–790 [DOI] [PubMed] [Google Scholar]
- 7. Chistoserdova L. 2011. Modularity of methylotrophy, revisited. Environ. Microbiol. 13:2603–2622 [DOI] [PubMed] [Google Scholar]
- 8. Crowther GJ, Kosály G, Lidstrom ME. 2008. Formate as the main branch point for methylotrophic metabolism in Methylobacterium extorquens AM1. J. Bacteriol. 190:5057–5062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chistoserdova L, Crowther GJ, Vorholt JA, Skovran E, Portais JC, Lidstrom ME. 2007. Identification of a fourth formate dehydrogenase in Methylobacterium extorquens AM1 and confirmation of the essential role of formate oxidation in methylotrophy. J. Bacteriol. 189:9076–9081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peyraud R, Schneider K, Kiefer P, Massou S, Vorholt JA, Portais JC. 2011. Genome-scale reconstruction and system level investigation of the metabolic network of Methylobacterium extorquens AM1. BMC Syst. Biol. 5:189 doi:10.1186/1752-0509-5-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Latypova E, Yang S, Wang YS, Wang T, Chavkin TA, Hackett M, Schäfer H, Kalyuzhnaya MG. 2010. Genetics of the glutamate-mediated methylamine utilization pathway in the facultative methylotrophic beta-proteobacterium Methyloversatilis universalis FAM5. Mol. Microbiol. 75:426–439 [DOI] [PubMed] [Google Scholar]
- 12. Chistoserdov AY, Chistoserdova LV, McIntire WS, Lidstrom ME. 1994. Genetic organization of the mau gene cluster in Methylobacterium extorquens AM1: complete nucleotide sequence and generation and characteristics of mau mutants. J. Bacteriol. 176:4052–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vorholt JA, Chistoserdova L, Lidstrom ME, Thauer RK. 1998. The NADP-dependent methylene tetrahydromethanopterin dehydrogenase in Methylobacterium extorquens AM1. J. Bacteriol. 180:5351–5356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chistoserdova L, Vorholt JA, Thauer RK, Lidstrom ME. 1998. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic archaea. Science 281:99–102 [DOI] [PubMed] [Google Scholar]
- 15. Hagemeier CH, Chistoserdova L, Lidstrom ME, Thauer RK, Vorholt JA. 2000. Characterization of a second methylene tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1. Eur. J. Biochem. 267:3762–3769 [DOI] [PubMed] [Google Scholar]
- 16. Marx CJ, Chistoserdova L, Lidstrom ME. 2003. Formaldehyde-detoxifying role of the tetrahydromethanopterin-linked pathway in Methylobacterium extorquens AM1. J. Bacteriol. 185:7160–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nunn DN, Lidstrom ME. 1986. Isolation and complementation analysis of 10 methanol oxidation mutant classes and identification of the methanol dehydrogenase structural gene of Methylobacterium sp. J. Bacteriol. 166:581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Okubo Y, Skovran E, Guo X, Sivam D, Lidstrom ME. 2007. Implementation of microarrays for Methylobacterium extorquens AM1. OMICS 11:325–340 [DOI] [PubMed] [Google Scholar]
- 19. Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1:784–791 [Google Scholar]
- 20. Marx CJ, Lidstrom ME. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. Biotechniques 33:1062–1067 [DOI] [PubMed] [Google Scholar]
- 21. Marx CJ, Lidstrom ME. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147:2065–2075 [DOI] [PubMed] [Google Scholar]
- 22. Skovran E, Crowther GJ, Guo X, Yang S, Lidstrom ME. 2010. A systems biology approach uncovers cellular strategies used by Methylobacterium extorquens AM1 during the switch from multi- to single-carbon growth. PLoS One 5:e14091 doi:10.1371/journal.pone.0014091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pomper BK, Vorholt JA, Chistoserdova L, Lidstrom ME, Thauer RK. 1999. A methenyl tetrahydromethanopterin cyclohydrolase and a methenyl tetrahydrofolate cyclohydrolase in Methylobacterium extorquens AM1. Eur. J. Biochem. 261:475–480 [DOI] [PubMed] [Google Scholar]
- 24. Ras J, Van Ophem PW, Reijnders WN, Van Spanning RJ, Duine JA, Stouthamer AH, Harms N. 1995. Isolation, sequencing, and mutagenesis of the gene encoding NAD- and glutathione-dependent formaldehyde dehydrogenase (GD-FALDH) from Paracoccus denitrificans, in which GD-FALDH is essential for methylotrophic growth. J. Bacteriol. 177:247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harms N, Ras J, Reijnders WN, van Spanning RJ, Stouthamer AH. 1996. S-formylglutathione hydrolase of Paracoccus denitrificans is homologous to human esterase D: a universal pathway for formaldehyde detoxification? J. Bacteriol. 178:6296–6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scott JW, Rasche ME. 2002. Purification, overproduction, and partial characterization of β-RFAP synthase, a key enzyme in the methanopterin biosynthesis pathway. J. Bacteriol. 184:4442–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rasche ME, Havemann SA, Rosenzvaig M. 2004. Characterization of two methanopterin biosynthesis mutants of Methylobacterium extorquens AM1 by use of a tetrahydromethanopterin bioassay. J. Bacteriol. 186:1565–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Escalante-Semerena JC, Rinehart KL, Wolfe RS. 1984. Tetrahydromethanopterin, a carbon carrier in methanogenesis. J. Biol. Chem. 259:9447–9455 [PubMed] [Google Scholar]
- 29. Ermler U, Hagemeier CH, Roth A, Demmer U, Grabarse W, Warkentin E, Vorholt JA. 2002. Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1. Structure 10:1127–1137 [DOI] [PubMed] [Google Scholar]
- 30. Guo X, Lidstrom ME. 2006. Physiological analysis of Methylobacterium extorquens AM1 grown in continuous and batch cultures. Arch. Microbiol. 186:139–149 [DOI] [PubMed] [Google Scholar]
- 31. Marx CJ, Lidstrom ME. 2004. Development of an insertional expression vector system for Methylobacterium extorquens AM1 and generation of null mutants lacking mtdA and/or fch. Microbiology 150:9–19 [DOI] [PubMed] [Google Scholar]