Summary
Telomerase comprises a reverse transcriptase and an internal RNA template that maintains telomeres in many eukaryotes, and it is a well-validated cancer target. However, there is a dearth of small molecules with efficacy against human telomerase in vivo. We developed a surrogate yeast high-throughput assay to identify human telomerase inhibitors. The reversibility of growth arrest induced by active human telomerase was assessed against a library of 678 compounds preselected for bioactivity in S. cerevisiae. Four of eight compounds identified reproducibly restored growth to strains expressing active human telomerase, and three of these four compounds also specifically inhibited purified human telomerase in vitro. These compounds represent probes for human telomerase function, and potential entry points for development of lead compounds against telomerase-positive cancers.
Graphical Abstract
Highlights
► Growth arrest induced by human telomerase in yeast is chemically reversible ► Readout is sensitive to telomerase catalytic activity and telomere recruitment ► Three cell-permeable compounds also inhibit purified human telomerase ► Yeast can be successfully used to screen for human telomerase inhibitors
Ability of baker's yeast to arrest cell division reversibly in response to the expression of human telomerase was used to screen for compounds that impaired the enzyme. Wong et al. found several compounds with desired activity, establishing yeast as a model system to identify new probes for telomerase function.
Introduction
The reactivation of telomerase is important for cancer cell survival. While primary somatic cells do not normally express telomerase and undergo progressive telomere shortening that leads subsequently to cell senescence, many malignant cell types activate telomerase expression to counteract telomere attrition and support tumor growth. Given its selective expression in cancer and requirement for cell immortality, telomerase in principle represents an ideal therapeutic cancer target (Harley, 2008). Mounting preclinical and clinical data strongly suggest that telomerase inhibition is a viable approach to suppress cancer progression, particularly in cancer cells with shorter telomeres, low telomerase expression, and rapid turnover (Harley, 2008). Furthermore, telomerase inhibition augments the sensitivity of carcinoma cell lines to radiation therapy and cytotoxic drugs (Gomez-Millan et al., 2007; Röth et al., 2010; Ward and Autexier, 2005).
Approaches to inhibit telomerase in cancer cells include direct or indirect enzyme inhibition though gene therapy, oligonucleotides that are complementary to the RNA template, and small molecule antagonists of enzyme activity (Harley, 2008). The most potent known in vitro telomerase inhibitor, BIBR1532 is a nonnucleosidic compound that interferes with the processivity of telomere elongation; however, BIBR1532 has not been taken into clinical trials due to issues with solubility and bioavailability (Damm et al., 2001; El-Daly et al., 2005; Pascolo et al., 2002). Imetelstat (GRN163L) is an oligonucleotide template antagonist that targets the active site of human telomerase through the telomerase RNA (hTR) template region and is the only telomerase inhibitor of its class to have progressed to phase II clinical trial (Röth et al., 2010; Harley, 2008). Thus, there remains a pressing need for the development of additional candidate telomerase inhibitors for cancer treatment.
Here, we report a surrogate genetic screening assay for human telomerase activity based on conditional heterologous expression of the two core catalytic subunits of the human ribonucleoprotein (RNP), hTR and telomerase reverse transcriptase (hTERT) in budding yeast (Bachand and Autexier, 1999; Bah et al., 2004). When hTERT is tethered to the yeast telomere via fusion with a yeast telomeric DNA binding protein, Cdc13, growth arrest occurs upon coexpression with hTR (Bachand and Autexier, 1999; Bah et al., 2004). We exploited this inducible growth arrest phenotype in a high-throughput assay to identify three compounds that reverse growth suppression via the specific inhibition of human telomerase.
Results
Induction of Active Human Relomerase in Budding Yeast
To reconstitute functional human telomerase in S. cerevisiae, we introduced phTR, for constitutive expression of hTR, and pGAL1-CDC13-hTERT-FLAG, to permit galactose-inducible expression of CDC13-TERT-FLAG (Bah et al., 2004; Evans and Lundblad, 1999) (Figure S1A available online). Expression of hTR and Cdc13-hTERT-FLAG in galactose-containing medium was confirmed by reverse transcriptase (RT)-PCR and western blot (Figures 1A and 1B). Galactose induction of CDC13-TERT-FLAG resulted in microcolony formation and growth suppression by 48 hr, and persisted up to 96 hr (Figures 1C and S1C). No growth delay was observed during the first 24 hr, likely due to a lag in the assembly and recruitment of active human telomerase to levels sufficient to induce a response (Figure S2A; data not shown). As expected, strains expressing hTR alone, which is insufficient for human telomerase activity in yeast (Bah et al., 2004), exhibited no growth delay (Figure 1C). The growth impedance caused by human telomerase expression depended upon the presence of the ATM-like kinase Mec1, which is the predominant DNA damage checkpoint kinase in budding yeast (d’Adda di Fagagna et al., 2004) (Figures 1D and S1E). The arrest did not depend on Esc4, a key factor in DNA replication restart that is dispensable for the intra-S-phase checkpoint arrest (Rouse, 2004) (Figure 1E). Expression of human telomerase did not interfere with endogenous yeast telomerase function, since there were no changes in the terminal telomere DNA-containing restriction fragment (TRF) length and no human telomeric repeats were detected at yeast telomeres (Figure S1D; data not shown) (Bah et al., 2004). Unlike the Mec1-dependent, irreversible arrest in response to a double-strand break at a native yeast telomere (Sandell and Zakian, 1993), the growth inhibition induced by human telomerase was reversible, and growth resumed if glucose was added to the medium to suppress CDC13-TERT-FLAG (Figure S1H).
Figure 1.
Reconstitution of Active Human Telomerase in S. cerevisiae via Coexpression of Wild-Type Cdc13-hTERT-FLAG and hTR
(A) RT-PCR analysis of hTR expression from total cellular RNA (30 ng) prepared from a W303-1a strain containing pCDC13-hTERT-FLAG and phTR plasmids in media containing galactose (gal; lanes 5–7) or glucose (glc; lanes 8–10), and, as a control, hTR synthesized in vitro (lanes 1–3; 0.5 ng, 0.2 ng, 0.05 ng). Irrelevant lanes between lanes 7–8 and 9–10 were omitted. RT, reverse transcriptase; Taq, Taq polymerase; M, DNA markers.
(B) Immunoprecipitation (IP) of 500 μg crude lysate onto anti-FLAG resin followed by detection with anti-FLAG (Oulton and Harrington, 2004) after growth in noninducible (raffinose, raf), repressive (glc), and galactose-containing (gal) media. The predicted mass of Cdc13-hTERT-FLAG is 232 kDa, indicated by the arrow at right. Asterisk indicates immunoglobulin G heavy chain (53 kDa) of anti-FLAG antibody.
(C) Cell number during an 8-day growth period of W303-1a in galactose (gal) or glucose (glc) or W303-1a in galactose containing an empty GAL1 plasmid (empty vector), hTR alone (hTR), or Cdc13-hTERT-FLAG and hTR. Error bars indicate SD, n = 3.
(D) Growth analysis as in (C) in strains expressing Cdc13-hTERT-FLAG + hTR in a mec1Δ sml1Δ strain background. Error bars indicate SD, n = 3.
(E) Growth analysis as in (C) in strains expressing Cdc13-hTERT-FLAG + hTR in an esc4Δ strain background. Error bars indicate SD, n = 3.
(F) Cell number after 2 days of growth of W303-1a strains expressing an inactive hTR mutant (hTR190), truncated, inactive hTERT (TERT1–677), inactive hTERT (containing an amino acid substitution in the catalytic site, D868A), or lacking the DNA-binding domain (DBD) of Cdc13. Error bars indicate SD, n = 3.
To confirm the specificity of the inducible growth suppression, we assessed the effect of mutations in the cDNAs encoding hTR, hTERT, and Cdc13 upon growth after confirmation that the mutant mRNA and proteins were expressed at levels comparable to their wild-type counterparts (Figures S1F and S1G). A 10-nucleotide (nt) substitution between nts 190 and 199 was introduced into hTR (hTR190), rendering it catalytically inactive (Autexier et al., 1996). Two mutations of hTERT were also introduced: the first (TERT1–677) lacks RT motifs essential for activity (Banik et al., 2002), and the second (hTERT-D868A-FLAG) contains an alanine substitution at D868, a residue essential for activity (Harrington et al., 1997). Finally, the telomere DNA binding domain within Cdc13, which is essential for its telomere recruitment (Evans and Lundblad, 1999), was removed from the hTERT fusion protein. All four mutations abolished the growth arrest (Figure 1F). Thus, the arrest depended on telomere recruitment and the catalytic activity of human telomerase conferred by hTR and hTERT.
A Yeast-Based HTS for Human Telomerase Inhibitors
We used a Tecan shaker-reader platform to establish conditions in a 96-well format (Figure 2A) that recapitulated the growth delay specific to active human telomerase (compare Figure 2B and Figure 1F). Expression of hTERT and hTR was confirmed, and, as anticipated, no growth delay was observed during the first 39 hr of growth (this preassay phase is hereinafter referred to as time course 1) (Figures S2A and S2B). Following an additional 86 hr (the experimental growth assay phase, hereinafter referred to as time course 2), strains expressing catalytically active human telomerase (Cdc13-hTERT-FLAG + hTR) exhibited a delay in exponential phase growth when compared to a strain expressing inactive telomerase (hTERT-D868/A868) (Figure 2B). Using an optical density at 595 nm (OD595) of 0.62 as a reference for the midpoint of exponential phase (see Figure S2B), cells expressing Cdc13-hTERT-FLAG + hTR reached this OD an average of 8.75 hr later during time course 2 than strains expressing inactive hTERT [Cdc13-hTERT(D868A)-FLAG + hTR]. We next screened two independent replicates of a library of 678 bioactive compounds against the query strain expressing active human telomerase and compared growth to the same strain treated with 2% v/v dimethyl sulfoxide (DMSO). These 678 compounds were selected from a 50,000 member Maybridge library based upon a spectrum of growth inhibitory bioactivity in a drug pump-deficient yeast strain (Ishizaki et al., 2010). This bioactive library yields a hit rate of 12% in a HeLa cell proliferation assay, such that most of the compounds are nontoxic to human cells at the screening concentration used (M.S., Jan Wildenhain, Sonam Dolma, David Bellows, and M.T., unpublished data). In each 96-well assay plate, growth of the query strain in the presence of compound (20 μM), beginning at the onset of the experimental growth phase (i.e., time course 2, from 43 hr onward), was normalized against DMSO treatment (Figures 2C and 2D; Figures S2D and S2E). As expected, the growth rate of the query strain was consistent between both screens and individual assay plates upon treatment with DMSO (Figure S2D); however, the growth rates upon treatment with compounds between the two screens was more variable, with a Pearson correlation coefficient of 0.4 (Figure S2E). Relative to DMSO, a total of eight compounds rescued the time to reach an OD595 of 0.62 by at least 8 hr (Figure 2C), with a predicted false-positive rate of 1.4% (see Experimental Procedures). Follow-up screens that included the eight putative hits (and 43 other compounds that did not initially meet the 8-hr threshold) confirmed that three of the eight high-throughput screen (HTS) hits reproducibly and specifically advanced the growth rate of strains expressing active human telomerase (Table 1; Figure 2E; see Supplemental Experimental Procedures for details). One of the rescreened compounds, SEW05920, did not exhibit significant growth recovery in the initial HTS but exhibited a growth recovery of ∼8 hr in follow-up screens and inhibited telomerase activity in vitro (Table 1; Figures 2E and 3). In contrast, the known in vitro telomerase inhibitor, BIBR1532 (Figures 3A and S2F), had no effect upon the growth of control strains or yeast expressing human telomerase (Figure 2F). Furthermore, in a hyperpermeable strain, pdr1DBD-cyc8 (Stepanov et al., 2008), BIBR1532 was toxic (Figure S2G).
Figure 2.
High-Throughput Chemical Screens of W303-1a Expressing Cdc13-hTERT-FLAG + hTR
(A) Schematic of HTS design. Cells induced with active human telomerase were dispensed in assay plates with media containing galactose and compounds, and OD595 was assessed throughout two serial time courses that totaled ∼128 elapsed hr (see Experimental Procedures for details).
(B) Growth profiles in a 96-well format, obtained with a Tecan shaker-reader, of W303-1a cells expressing wild-type Cdc13-hTERT-FLAG + hTR or a catalytically inactive hTERT mutant (D868A) + hTR during time course 2 (commencing at 43 hr in culture, labels spaced every 4.5 hr and rounded up or down accordingly). Horizontal double-sided arrow indicates the relative growth delay of 8.75 hr between the two strains at an OD595 of 0.62. Error bars, in black, indicate SD, n = 8.
(C) Histogram of the number of compounds in categories of time difference (hr) to reach an OD595 of 0.62 relative to DMSO treatment (screen 1, light gray; screen 2, dark gray). Compounds that rescued relative growth delay by between 8 and 16 hr are shown in pink or red.
(D) Heatmap analysis of time difference (hr) to reach an OD595 of 0.62 relative to DMSO in a representative 96-well plate during the assay phase time course 2. C, cycloheximide; D, DMSO. Wells containing compounds (or cycloheximide) that impeded growth by more than 8 hr relative to DMSO appear white. Wells in which the time to reach an OD595 of 0.62 relative to DMSO was advanced by 8 hr or more are red (e.g., CD11359, outlined in black).
(E) Fold change in time difference (hr) to reach an OD595 of 0.62 relative to DMSO of strains expressing active hTERT (Cdc13-hTERT + hTR), an inactive hTERT truncation (Cdc13-TERT1–677 + hTR), or Cdc13 lacking its DNA binding domain [Cdc13(-DBD)-hTERT + hTR] in the presence of repurchased candidate compounds (20 μM). Values were normalized to the time rescue observed in strains expressing catalytically inactive Cdc13-hTERT(D868A)-FLAG + hTR treated with DMSO. Error bars indicate SD, n = 4.
(F) Growth profiles of W303-1a cells alone (left) or expressing wild-type Cdc13-hTERT-FLAG + hTR (right) commencing at 35 hr after induction (i.e., following 32 hr of treatment during time course 1, then 3 hr in galactose only, and readdition of compound or DMSO at 35 hr) containing 500, 150, 50, 10, or 1 μM BIBR1532 or 2% v/v DMSO. Error bars indicate SD, n = 3.
See also Figure S2.
Table 1.
Screen Hits and Corresponding Z Score Values
| Compounds | Time (hr) Difference at an OD595 of 0.62 Relative to DMSO | Z Score |
|---|---|---|
| CD11359 | 8.3 | 1.7 |
| SPB03924 | 12 | 2.3 |
| RJC00417 | 10 | 2.0 |
| SEW05920 | 8.3a | 1.7 |
SEW05920 did not significantly rescue growth in the initial HTS analysis but was validated as a hit based on significant growth recovery in two follow-up subscreens and upon repurchase of the compound from the commercial supplier. Please refer to Supplemental Experimental Procedures.
Figure 3.
Assessment of Human Telomerase Inhibition In Vitro
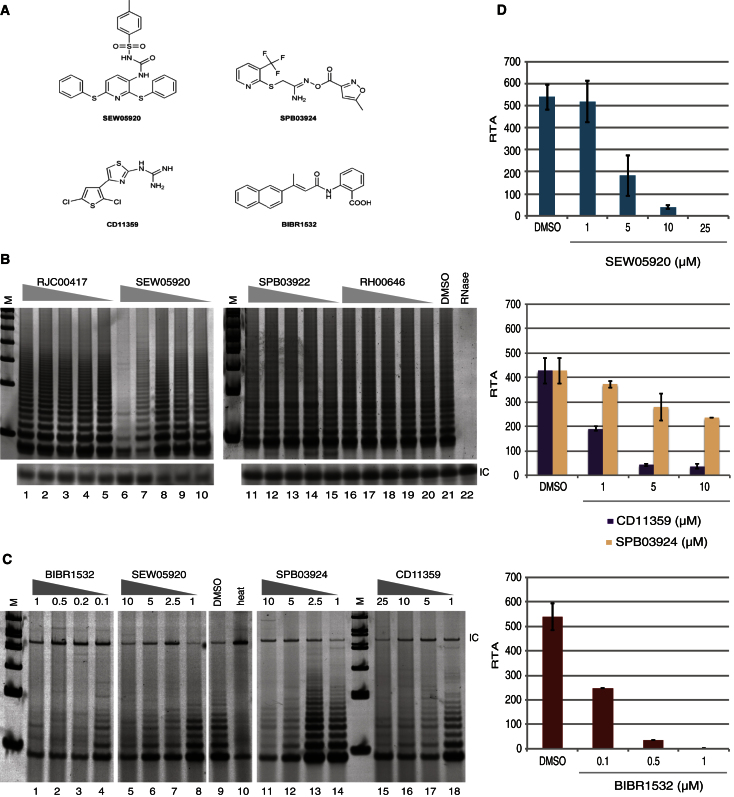
(A) Chemical structures of BIBR1532 and hit compounds SEW05920, SPB03924, and CD11359.
(B) Representative gel of the telomerase repeat addition protocol (TRAPeze, Millipore) after addition of RJC00417, SEW05920, SPB03922, and RH00646 to recombinant human telomerase purified in vitro (Beattie et al., 1998; Oulton and Harrington, 2004) (n = 3). Lanes 1–5, treatment with 200, 100, 20, 10, or 2 μM RJC00417; lanes 6–10, treatment with 200, 100, 20, 10, or 2 μM SEW05920; lanes 11–15, treatment with 200, 100, 20, 10, or 2 μM SPB03922; lanes 16–20, treatment with 200, 100, 20, 10, or 2 μM RH00646; lane 21, treatment with 2% v/v DMSO; lane 22, pretreatment with ribonuclease A. IC; internal PCR control (36 base pairs [bp]).
(C) Representative gel of the relative telomerase activity (RTA) by ELISA-TRAP (TeloTAGGG Telomerase PCR ELISA-PLUS, Roche Diagnostics) of purified human telomerase upon incubation with BIBR1532 (lanes 1–4), SEW05920 (lanes 5–8), SPB03924 (lanes 11–14), CD11359 (lanes 15–18) (concentration indicated in micromolar), 2% v/v DMSO (lane 9), or heat (lane 10). IC; internal PCR control (216 bp).
(D) Levels of RTA (%) after normalization to a 216 bp internal control, as shown in (C). Error bars indicate SD, n = 3.
For synthesis of BIBR1532, see Figure S2.
In Vitro Characterization of Inhibitors Specific to Human Telomerase
We found that three of the four compounds (SEW05920, SPB03924, and CD11359) exhibited a dose-dependent inhibition of both purified recombinant human telomerase and telomerase activity in crude HeLa cell extracts using a telomerase repeat amplification protocol (TRAP) assay (Figures 3A–3C). The fourth compound, RJC00417, did not inhibit telomerase activity in vitro and thus may inhibit human telomerase in yeast through a mechanism distinct from catalytic inhibition (Figure 3B). As a control, we observed no inhibition of the Taq polymerase used for amplification of telomerase extension products in the TRAP (data not shown). In addition, as negative controls, two other compounds that were not identified as hits (SPB03922 and RH00646) were tested and confirmed to exert no effect upon telomerase activity in vitro (Figure 3B). Using a semiquantitative assay for the detection of telomerase elongation products (ELISA-TRAP) the in vitro half maximal inhibitory concentration (IC50) values of the compounds fell between 1 and 6.5 μM (Figures 3C and 3D). Similar IC50 values were also observed against purified recombinant murine telomerase (data not shown). These values are higher than that of BIBR1532 (IC50 = 0.1 μM) (Damm et al., 2001; Pascolo et al., 2002) but within a concentration range typical of first hits from commercial libraries. SEW05920, SPB03924, and CD11359 are heteroaromatic small molecules that differ structurally from BIBR1532 (Figures 3A and S2F). A search of PubChem revealed that SEW05920 had not been registered in a previous bioassay; similarly, CD11359 and SPB03924 were screened in nine and 14 bioassays, respectively, against diverse targets including toll-like receptor 4, Sphingosine 1-Phosphate receptor 1, NADPH oxidase 1, phosphatase methylesterase 1, and NOTCH1 but were not identified as hits.
Discussion
Analysis of human enzymatic activity as a direct output of a high-throughput screen in yeast has been used previously (Ceyhan et al., 2012; Griffioen et al., 2006; Perkins et al., 2001). Our results extend the utility of budding yeast as a model system to identify and validate compounds, in particular, its utility for a large multi-subunit RNP enzyme (383 kDa, including hTERT-Cdc13 and hTR). This surrogate genetic system possesses several advantages over conventional chemical screening approaches. The yeast-based assay obviates the need for large quantities of purified human telomerase in vitro and allows the facile incorporation of mutational controls. The heterologous expression system enables human telomerase to be specifically targeted, unlike human cell line-based assays, and therefore allows rapid counterselection against compounds that exhibit off-target effects on cell growth. The yeast system also selects for bioactive small molecules with well-balanced hydrophilic-lipophilic properties due to the presence of two chemically different barriers, namely, the hydrophilic polysaccharide wall and the lipophilic phospholipid membrane. Notably, the compound library used in this study was already highly enriched for yeast cell bioavailability, which allows a substantially higher hit rate that conventional library screens (Ishizaki et al., 2010).
Aside from the general features of planar, aromatic groups that typify the Maybridge compound collection, the three putative telomerase inhibitors span diverse chemical classes that do not coincide with known telomerase inhibitors (Harley, 2008). The compounds were nontoxic in isogenic human telomerase-positive or telomerase-negative cell lines with long telomeres (Sealey et al., 2010; Taboski et al., 2012) over a concentration range of 0.5–5.0 μM after 28–35 days of continuous treatment (data not shown). Although nontoxic, longer treatment periods will be required to detect telomere shortening in human cells, since significant shortening in the presence of GRN163L can take up to 3 months (Uziel et al., 2010). The chemical properties of SEW05920, SPB03924, and CD11359 appear amenable to structure-activity-relationship analysis, which will enable the design and synthesis of more potent derivatives to be tested for efficacy in telomerase-positive human cancer cell lines, and for determination of their precise mechanism of action in vitro.
Significance
The specificity of yeast growth suppression for active human telomerase will also enable the screening of active hTERT variants capable of drug resistance. We note that acquired drug resistance to single-drug regimes against HIV reverse transcriptase is a prevalent concern in the treatment of HIV infection (Penazzato and Giaquinto, 2011). The diverse compound structures recovered in the screen suggest the possibility that more than one site can be targeted on active telomerase. Combinations of inhibitors with different mechanisms of action may exhibit additive or synergistic inhibition and impede the emergence of single-point mutations in hTERT that are drug resistant. The surrogate budding yeast system thus provides a sensitive and cost-effective assay for the identification of human telomerase inhibitors and will accelerate classical hit-to-lead chemistry in parallel with testing in human cell lines and animal models.
Experimental Procedures
Construction of pGAL1-CDC13-hTERT-FLAG and phTR Plasmids
Human TERT-FLAG cDNA was PCR amplified from pCR3-FLAG-hTERT-FLAG (Beattie et al., 1998) and inserted downstream of the 3′ terminal 1.2 kilobase pairs (kbp) of CDC13 in an intermediate StrataClone vector, to which the terminator sequence of the ADH1 gene (ADHter) was subsequently inserted downstream of the fusion protein. This fragment, CDC13SphI-hTERT-ADHter, was inserted into pGAL1-pVL1091 (Evans and Lundblad, 1999). The hTR sequence was amplified from pUC19-hTR (Beattie et al., 2000), digested with EcoRI, and inserted into pIII426 (Good and Engelke, 1994). The final constructs were verified by DNA sequencing.
Galactose Induction and Growth Assay
W303-1a haploid cells, MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 can1-100 (Thomas and Rothstein, 1989), or a mec1Δ sml1Δ strain, MATa ura3-52 trp1-289 leu2-3,112 bar1::LEU2 (Makovets et al., 2004; Zhao et al., 1998), were transformed with pCDC13-hTERT-FLAG and phTR plasmids (Gietz and Woods, 2002) and were propagated on synthetic dropout liquid medium lacking uracil and leucine and containing 3% (w/v) raffinose. At an OD 600 (OD600) of 0.3, cells were pelleted, washed, and resuspended in selective media containing 2% (w/v) galactose (to induce expression of Cdc13-hTERT-FLAG) or 2% (w/v) glucose.
Cdc13-hTERT-FLAG and hTR Expression by RT-PCR Analysis
Total cellular RNA was isolated as described by Burke et al. (2000) and amplified using the Sensiscript RT Kit protocol (QIAGEN); hTR, forward: 5′-GGGTTGCGGAGGGTGGGCC-3′, reverse: 5′-GCATGTGTGAGCCGAGTCCTGG-3′; hTERT, forward 5′-GCGAGCTGCGGTCACCCC-3′, reverse 5′- AGCTCCTGCAGCGAGAGC-3′; Act1, forward 5′- ATGGATTCTGAGGTTGCTGCTTTGGTTA-3′, reverse 5′- TGTTCTTCTGGGGCAACTCTCAATT-3′.
High-Throughput Chemical Screens
W303-1a cells bearing pCDC13-hTERT-FLAG and phTR plasmids were grown in selective medium containing 3% (w/v) raffinose until an OD600 of 0.3, followed by a 2 hr pregrowth in medium containing 2% (w/v) galactose prior to addition of compounds. To conduct the HTS (Figure 2A), 98 μl (3 × 105 cells per milliliter) was dispensed into 96-well clear flat-bottomed microplates (Corning Costar) using a Biomek FX (Beckman Coulter). A small molecule bioactive library of 678 compounds was utilized for the screen, consisting of compounds selected from the Maybridge collection (Thermo Fisher Scientific). Two microliters of each compound from the master library plate was dispensed into the assay plate at a final concentration of 20 μM, 2% (v/v) DMSO. DMSO at 2% (v/v) and 20 μM cycloheximide (Sigma) final concentration were added as controls to each assay plate, into alternate wells in columns 1 and 12 using the Span 8 module of the Biomek FX. Nine plates were analyzed, 80 compounds per plate, with 16 wells for controls. After compound addition, the plates were sealed and followed for time course 1 (up to 39 hr) on a Sunrise plate reader (Tecan) at absorbance 595 nm, maintained at 30°C and shaking at 564 rpm. After time course 1, cells in assay plates were diluted to 3 × 105 cells per milliliter in fresh media containing galactose using the Biomek FX. The plates were preincubated in galactose-containing media at 30°C for 3 hr (to allow a period of recovery from the stationary phase in the absence of compound), 2 μl of each compound or control was added to plates as described earlier, returned to the plate readers, and followed for time course 2 (up to 86 hr), for a total of ∼128 elapsed hours. In follow-up experiments, we confirmed that the ability of a validated hit (SPB03924) to accelerate growth recovery did not differ significantly when the compound was added immediately at the outset of time course 2 versus its readdition after a 3 hr period in galactose only (data not shown).
Identification of Human Telomerase Inhibitors
Growth curves were analyzed using a growth curve analysis tool implemented in R. After smoothing of growth curves, the time required for cells induced with human telomerase to reach an OD595 of 0.62 in the presence of each compound during time course 2 was derived using a data-based approach (Warringer and Blomberg, 2003). The time to reach an OD595 of 0.62 for each compound was normalized to the average DMSO value from each assay plate (to arrive at a normalized value, x, for each compound) and used to generate heatmaps as shown in Figure 2C. Z scores were calculated based on the equation: z = (x − μ)/σ, where μ is the mean time difference for all compounds in the bioactive library and σ is the SD for all compounds in the bioactive library. The rate of false-positives (1.4%) was estimated based on the number of DMSO-treated wells that were able to accelerate the time to reach an OD595 of 0.62 by 8 hr or more, using the following equation: 1 − (atrue-positives/btotal) × 100%, where a corresponds to the number of DMSO controls that exhibited a rescue in growth equal to or above 8 hr and b corresponds to the total number of DMSO controls included in the screen. Four follow-up subscreens were performed after the high-throughput screen, leading to the identification of the validated hit compounds listed in Table 1. These compounds were also analyzed for their ability to inhibit human telomerase activity in vitro (see Supplemental Experimental Procedures for details).
Acknowledgments
We thank Dr. Svetlana Makovets for providing the yeast strains NK399 and mec1Δ sml1Δ; Larissa Christian, Jennifer Dorrens, and Dr. Fiona Pryde for experimental assistance; Dr. David Kelly for microscope assistance; and Prof. David Finnegan, Dr. Maurice Gallagher, Dr. Heidrun Interthal, and Prof. David Lydall for helpful input. The research was funded by Cancer Research UK (A9299 to L.H.), the Wellcome Trust (084637 to L.H.), the European Research Council (AdG-N°233457 to M.T.), and the Medical Research Council (G0401194, to Prof. Mark Bradley, in whose laboratory the synthesis of BIBR1532 was conducted).
Published: March 21, 2013
Footnotes
Supplemental Information includes two figures, Supplemental Experimental Procedures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.chembiol.2012.12.008.
Supplemental Information
References
- Autexier C., Pruzan R., Funk W.D., Greider C.W. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 1996;15:5928–5935. [PMC free article] [PubMed] [Google Scholar]
- Bachand F., Autexier C. Functional reconstitution of human telomerase expressed in Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:38027–38031. doi: 10.1074/jbc.274.53.38027. [DOI] [PubMed] [Google Scholar]
- Bah A., Bachand F., Clair E., Autexier C., Wellinger R.J. Humanized telomeres and an attempt to express a functional human telomerase in yeast. Nucleic Acids Res. 2004;32:1917–1927. doi: 10.1093/nar/gkh511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banik S.S., Guo C., Smith A.C., Margolis S.S., Richardson D.A., Tirado C.A., Counter C.M. C-terminal regions of the human telomerase catalytic subunit essential for in vivo enzyme activity. Mol. Cell. Biol. 2002;22:6234–6246. doi: 10.1128/MCB.22.17.6234-6246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie T.L., Zhou W., Robinson M.O., Harrington L. Reconstitution of human telomerase activity in vitro. Curr. Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- Beattie T.L., Zhou W., Robinson M.O., Harrington L. Polymerization defects within human telomerase are distinct from telomerase RNA and TEP1 binding. Mol. Biol. Cell. 2000;11:3329–3340. doi: 10.1091/mbc.11.10.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D., Dawson D., Stearns T. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. [Google Scholar]
- Ceyhan O., Birsoy K., Hoffman C.S. Identification of biologically active PDE11-selective inhibitors using a yeast-based high-throughput screen. Chem. Biol. 2012;19:155–163. doi: 10.1016/j.chembiol.2011.12.010. [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F., Teo S.H., Jackson S.P. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 2004;18:1781–1799. doi: 10.1101/gad.1214504. [DOI] [PubMed] [Google Scholar]
- Damm K., Hemmann U., Garin-Chesa P., Hauel N., Kauffmann I., Priepke H., Niestroj C., Daiber C., Enenkel B., Guilliard B. A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J. 2001;20:6958–6968. doi: 10.1093/emboj/20.24.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Daly H., Kull M., Zimmermann S., Pantic M., Waller C.F., Martens U.M. Selective cytotoxicity and telomere damage in leukemia cells using the telomerase inhibitor BIBR1532. Blood. 2005;105:1742–1749. doi: 10.1182/blood-2003-12-4322. [DOI] [PubMed] [Google Scholar]
- Evans S.K., Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Woods R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Gomez-Millan J., Goldblatt E.M., Gryaznov S.M., Mendonca M.S., Herbert B.S. Specific telomere dysfunction induced by GRN163L increases radiation sensitivity in breast cancer cells. Int. J. Radiat. Oncol. Biol. Phys. 2007;67:897–905. doi: 10.1016/j.ijrobp.2006.09.038. [DOI] [PubMed] [Google Scholar]
- Good P.D., Engelke D.R. Yeast expression vectors using RNA polymerase III promoters. Gene. 1994;151:209–214. doi: 10.1016/0378-1119(94)90658-0. [DOI] [PubMed] [Google Scholar]
- Griffioen G., Duhamel H., Van Damme N., Pellens K., Zabrocki P., Pannecouque C., van Leuven F., Winderickx J., Wera S. A yeast-based model of alpha-synucleinopathy identifies compounds with therapeutic potential. Biochim. Biophys. Acta. 2006;1762:312–318. doi: 10.1016/j.bbadis.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Harley C.B. Telomerase and cancer therapeutics. Nat. Rev. Cancer. 2008;8:167–179. doi: 10.1038/nrc2275. [DOI] [PubMed] [Google Scholar]
- Harrington L., Zhou W., McPhail T., Oulton R., Yeung D.S., Mar V., Bass M.B., Robinson M.O. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki H., Spitzer M., Wildenhain J., Anastasaki C., Zeng Z., Dolma S., Shaw M., Madsen E., Gitlin J., Marais R. Combined zebrafish-yeast chemical-genetic screens reveal gene-copper-nutrition interactions that modulate melanocyte pigmentation. Dis. Model Mech. 2010;3:639–651. doi: 10.1242/dmm.005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makovets S., Herskowitz I., Blackburn E.H. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol. Cell. Biol. 2004;24:4019–4031. doi: 10.1128/MCB.24.9.4019-4031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulton R., Harrington L. A human telomerase-associated nuclease. Mol. Biol. Cell. 2004;15:3244–3256. doi: 10.1091/mbc.E04-03-0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascolo E., Wenz C., Lingner J., Hauel N., Priepke H., Kauffmann I., Garin-Chesa P., Rettig W.J., Damm K., Schnapp A. Mechanism of human telomerase inhibition by BIBR1532, a synthetic, non-nucleosidic drug candidate. J. Biol. Chem. 2002;277:15566–15572. doi: 10.1074/jbc.M201266200. [DOI] [PubMed] [Google Scholar]
- Penazzato M., Giaquinto C. Role of non-nucleoside reverse transcriptase inhibitors in treating HIV-infected children. Drugs. 2011;71:2131–2149. doi: 10.2165/11597680-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Perkins E., Sun D., Nguyen A., Tulac S., Francesco M., Tavana H., Nguyen H., Tugendreich S., Barthmaier P., Couto J. Novel inhibitors of poly(ADP-ribose) polymerase/PARP1 and PARP2 identified using a cell-based screen in yeast. Cancer Res. 2001;61:4175–4183. [PubMed] [Google Scholar]
- Röth A., Harley C.B., Baerlocher G.M. Imetelstat (GRN163L)—telomerase-based cancer therapy. Recent Results Cancer Res. 2010;184:221–234. doi: 10.1007/978-3-642-01222-8_16. [DOI] [PubMed] [Google Scholar]
- Rouse J. Esc4p, a new target of Mec1p (ATR), promotes resumption of DNA synthesis after DNA damage. EMBO J. 2004;23:1188–1197. doi: 10.1038/sj.emboj.7600129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell L.L., Zakian V.A. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- Sealey D.C., Zheng L., Taboski M.A., Cruickshank J., Ikura M., Harrington L.A. The N-terminus of hTERT contains a DNA-binding domain and is required for telomerase activity and cellular immortalization. Nucleic Acids Res. 2010;38:2019–2035. doi: 10.1093/nar/gkp1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanov A., Nitiss K.C., Neale G., Nitiss J.L. Enhancing drug accumulation in Saccharomyces cerevisiae by repression of pleiotropic drug resistance genes with chimeric transcription repressors. Mol. Pharmacol. 2008;74:423–431. doi: 10.1124/mol.107.044651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taboski M.A., Sealey D.C., Dorrens J., Tayade C., Betts D.H., Harrington L. Long telomeres bypass the requirement for telomere maintenance in human tumorigenesis. Cell Rep. 2012;1:91–98. doi: 10.1016/j.celrep.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B.J., Rothstein R. The genetic control of direct-repeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics. 1989;123:725–738. doi: 10.1093/genetics/123.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel O., Beery E., Dronichev V., Samocha K., Gryaznov S., Weiss L., Slavin S., Kushnir M., Nordenberg Y., Rabinowitz C. Telomere shortening sensitizes cancer cells to selected cytotoxic agents: in vitro and in vivo studies and putative mechanisms. PLoS ONE. 2010;5:e9132. doi: 10.1371/journal.pone.0009132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward R.J., Autexier C. Pharmacological telomerase inhibition can sensitize drug-resistant and drug-sensitive cells to chemotherapeutic treatment. Mol. Pharmacol. 2005;68:779–786. doi: 10.1124/mol.105.011494. [DOI] [PubMed] [Google Scholar]
- Warringer J., Blomberg A. Automated screening in environmental arrays allows analysis of quantitative phenotypic profiles in Saccharomyces cerevisiae. Yeast. 2003;20:53–67. doi: 10.1002/yea.931. [DOI] [PubMed] [Google Scholar]
- Zhao X., Muller E.G., Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.