Abstract
Mutations of the isocitrate dehydrogenase 1 and 2 genes (IDH1 and IDH2) are commonly found in primary brain cancers. We previously reported that a novel enzymatic activity of these mutations results in the production of the putative oncometabolite, R(–)-2-hydroxyglutarate (2-HG). Here we investigated the ability of magnetic resonance spectroscopy (MRS) to detect 2-HG production in order to non-invasively identify patients with IDH1 mutant brain tumors. Patients with intrinsic glial brain tumors (n = 27) underwent structural and spectroscopic magnetic resonance imaging prior to surgery. 2-HG levels from MRS data were quantified using LC-Model software, based upon a simulated spectrum obtained from a GAMMA library added to the existing prior knowledge database. The resected tumors were then analyzed for IDH1 mutational status by genomic DNA sequencing, Ki-67 proliferation index by immunohistochemistry, and concentrations of 2-HG and other metabolites by liquid chromatography–mass spectrometry (LC–MS). MRS detected elevated 2-HG levels in gliomas with IDH1 mutations compared to those with wild-type IDH1 (P = 0.003). The 2-HG levels measured in vivo with MRS were significantly correlated with those measured ex vivo from the corresponding tumor samples using LC–MS (r2 = 0.56; P = 0.0001). Compared with wild-type tumors, those with IDH1 mutations had elevated choline (P = 0.01) and decreased glutathione (P = 0.03) on MRS. Among the IDH1 mutated gliomas, quantitative 2-HG values were correlated with the Ki-67 proliferation index of the tumors (r2 = 0.59; P = 0.026). In conclusion, water-suppressed proton (1H) MRS provides a non-invasive measure of 2-HG in gliomas, and may serve as a potential biomarker for patients with IDH1 mutant brain tumors. In addition to 2-HG, alterations in several other metabolites measured by MRS correlate with IDH1 mutation status.
Keywords: 2 hydroxyglutarate (2-HG), Glioma, Magnetic resonance spectroscopy, Brain tumor, Isocitrate dehydrogenase, LC-Model
Introduction
Somatic mutations of the isocitrate dehydrogenase 1 and 2 genes (IDH1 and IDH2) have recently been implicated in gliomagenesis and are found in approximately 80% of World Health Organization (WHO) grade II–III gliomas and secondary glioblastomas (WHO grade IV) in humans [1–5]. The vast majority of IDH1 mutant, high-grade gliomas have evolved from lower grade lesions [1, 4–6]. IDH1 mutations have recently been found to be associated with a distinct gene expression profile (in particular, the “proneural” subset of glioblastomas) [7, 8] and are considered to be an independent prognostic indicator in these patients [7, 9, 10]. When analyzed in relation to other genes implicated in brain tumors, the compiled evidence suggests that IDH1 is often an early mutation identified in the neoplastic transformation of primary brain cancers [6].
Isocitrate dehydrogenase 1 mutations almost always occur at a single amino acid residue of the IDH1 active enzymatic site, namely arginine 132. We previously demonstrated that, when arginine 132 is mutated to histidine, residues at the IDH1 active site are shifted, thereby producing structural changes that result in a new ability of the enzyme to catalyze the NADPH-dependent reduction of α-ketoglutarate to R(–)-2-hydroxyglutarate (2-HG) [11]. Excess accumulation of the enantiomer L2-HG has been shown to lead to an elevated risk of malignant brain tumors in patients with inborn errors of 2-HG metabolism [12]. Given that 2-HG accumulates in vivo and is thought to contribute to the formation and malignant progression of gliomas, the ability to non-invasively measure this putative oncometabolite could be of potential clinical utility. Noninvasive detection of 2-HG in glioma patients with IDH1 mutant tumors may be of benefit in the management of these patients by detecting disease recurrence and/or monitoring treatment effects when future interventions targeting IDH1 mutations are clinically proven. Therefore, we developed a magnetic resonance spectroscopic (MRS) protocol, with LC-Model post-processing, to evaluate whether 2-HG was measurable in a series of patients with gliomas.
Methods
Patients
Between November 2009 and March 2010, we prospectively studied 27 consecutive adult patients with intracranial gliomas who presented to the UCLA Department of Neurosurgery for surgical resection of their brain tumors. This research protocol was approved by the UCLA Institutional Review Board (UCLA IRB #09-09-094), and all subjects gave written informed consent. All patients had measurable disease on magnetic resonance imaging (MRI) for which surgical resection was warranted. Clinical classification and grading of the tissue was performed by a board-certified neuropathologist (WHY).
Magnetic resonance spectroscopy (MRS) studies
Within 1 week prior to surgery, patients with newly diagnosed or recurrent gliomas underwent pre-operative MRI and MRS. A 3T MRI/MRS scanner (Trio-Tim, Siemens Medical Systems, Erlangen, Germany) was used to acquire the MR spectral data. A 12-channel head MRI coil was used for receiving the signal and the quadrature body MRI coil for transmitting the radio-frequency (RF) pulses. Single-voxel localized MR spectra were acquired using the double-echo PRESS sequence [13]. Voxels were localized to representative areas of solid tumor, as determined by a board-certified neuro-radiologist (WBP or NS). Regions of probable necrosis, hemorrhage, or edema, if present, were excluded from the interrogated area. For large lesions, the spectral voxel was placed in the center of the lesion over a majority of the solid tumor area, excluding the surrounding regions of peri-tumoral vasogenic edema. As a control, voxels were also placed in the same anatomical location on the contralateral (non-tumor) side of the brain to obtain control spectra. The MRS acquisition parameters were as follows: 2 × 2 × 2 cm3, TR/TE = 2 s/30 ms, 128 averages and 2,048 complex points for the spectral data. Water enhanced through T1 (WET)-based global pre-suppression of water was achieved using three frequency-selective RF pulses, each RF pulse followed by dephasing B0-crusher gradient pulses [14]. Unsuppressed water signal was also acquired using the same parameters as above, except with four averages only. Patient spectra were assessed for artifacts and signal to noise ratio (SNR). Spectra with the full width at half maximum (FWHM) of NAA of above 30 Hz or poor water suppression were excluded based on these objective criteria.
For post-processing of MRS, raw data files were processed using the LC-Model algorithm [15]. LC-Model was performed in exactly the same way for each spectrum, with no changes in pre-processing or modifications to the control file. The water-suppressed spectral domain data were analyzed between the range of 0.5 and 4.0 ppm. Using the GAMMA library [16], the basis set for 2-HG was developed, assuming a pH value of 7.0. In addition, the basis set provided by the vendor was used and then scaled to a consistent transmitter gain. Assuming a water concentration of 35 M (15), absolute values of 2-HG, glutamate (Glu), glutamine (Gln), choline (Cho), creatine (Cr), N-acetylaspartate (NAA), lactate (Lac), and other metabolites were reported in mM per Kg of wet tissue uncorrected for T1 and T2 saturation. All metabolites used in the full basis set are listed in Supplemental Table 1. Metabolite ratios were also calculated with respect to the total Cr concentrations. Using LC-Model to process the MRS data allows for calculation of both ratios of metabolites with respect to total Cr, as well as absolute concentrations using known concentrations of tissue water [15]. However, fluctuating SNR with changes in FWHM, could result in low or high concentrations of 2-HG systematically in the LC-Model analysis [17]. Therefore, Cramer–Rao lower bounds (CLRB) values provide a lower bound on the variance of the MRS fitting estimate. It represents the precision of the estimate but not the accuracy, and reports to us how close repeated measurements will be to the current one under similar experimental conditions [18]. The CRLB values were calculated from the residual error and the Fisher matrix of the partial derivatives of the concentrations.
Clinical specimens and sequencing of genomic tumor DNA
The pre-operative MRS scans were fused to the anatomical T1-weighted MRI and registered to a BrainLAB™ intra-operative navigational system. At the time of tumor resection, brain tumor samples corresponding to the voxel localized by the pre-operative MRS were stereotactically localized, biopsied/resected, and immediately snap-frozen in isopentane cooled by dry ice, and stored at −80°C. Stereotactic intra-operative navigation fused to the pre-op MRS was used to make certain that the area investigated by MRS was also the tissue collected for examination of IDH1 mutations. To determine the mutation status, genomic DNA was isolated from frozen tumor tissue and amplified by PCR. Exon 4 of IDH1 was sequenced and analyzed for somatic mutations, as previously described [11].
Metabolite extraction and analysis
Metabolite extraction from the clinical specimens was accomplished by adding a 10× volume (m/v ratio) of −80°C methanol:water mix (80:20%) to the brain tumor tissue (approximately 100 mg) followed by 30 s homogenization at 4°C. For analysis, a 2× volume of tributylamine (10 mM), acetic acid (10 mM) pH 5.5 was added to cleared tissue supernatants and analyzed by LC–MS as follows. Sample extracts were injected onto a reverse-phase high-performance liquid chromatography (HPLC) column (Synergi 150 × 2 mm, Phenomenex Inc.) and eluted using a linear gradient LC–MS-grade methanol. Eluted metabolite ions were detected using a triple-quadrupole mass spectrometer, tuned to detect in negative mode with multiple-reaction-monitoring mode transition set according to the molecular weights and fragmentation patterns for eight known central metabolites, including 2-HG as described above. Data was processed using Analyst Software (Applied Biosystems, Inc.) and metabolite signal intensities were obtained by standard peak integration methods.
Immunohistochemistry
Paraffin sections of intracranial tumor specimens were cut to 5 μm thickness and stained with an anti-human antibody against Ki-67 (1:100, clone MIB-1, DAKO, Carpinteria, CA). Immunostaining for Ki-67 was performed using the Ventana medical Systems BenchMark® XT (Tucson, Arizona) automated immunostainer by sequentially incubating in primary antibody for 30–60 min, then mouse anti-rabbit secondary immunoglobulins (DAKO Corp.) for 30 min. Diaminobenzidine and hydrogen peroxide were used as the substrates for the peroxidase enzyme. For the negative controls, mouse isotype or rabbit immunoglobulins (DAKO Corp.) were used in place of the primary antibodies. At 200× magnification, the percentage of Ki-67+ cells was quantitatively counted in areas of highest labeling. This Ki-67 labeling index was determined by a neuropathologist (WHY) in an unbiased fashion, with no prior knowledge of IDH1 mutational status or metabolic analysis data.
Statistical analysis
Variations in quantitative data are represented as the standard error of the mean. Significant differences in metabolite concentrations were calculated using a non-parametric, Mann–Whitney test. Generated P-values are two-tailed, and P < 0·05 was considered statistically significant. Best-fit linear regression lines are shown for data comparison, calculated by the Goodness-of-fit of this model (SStot), and represented as r2. P-values for linear regressions are calculated from an F test. All statistical analyses and graphs were constructed using Graph Pad software.
Results
Patients
Of the 27 patients enrolled in this study, 24 (89%) were found to have high quality MRS data based on objective criteria defined in the “Methods” section. Of note, the three excluded cases were all recurrent tumors post-treatment, which may be a factor to consider in determining types of cases for which 2-HG MRS would be useful. Comprehensive, quantitative metabolomic analyses were performed on tumor tissues collected from all 24 of the evaluable cases using LC–MS. These patients were then divided into wild-type and mutant IDH1 genotypes, based on genomic sequence analysis of their tumor DNA. Nine of the patients had IDH1 mutant tumors, and 15 were wild-type for IDH1. Of the IDH1 mutant cases (n = 9), there were four females and five males, and the average age was 43 years (range 22–67 years old). Of the patients with IDH1 wild-type tumors (n = 15), eight were females and seven were males, with an average age of 59 years (range 40–87 years old). The patient characteristics and findings are summarized in Table 1. The age difference between the two groups is reflective of the fact that IDH1 mutations tend to be found in lower grade gliomas (WHO grade II and III) and secondary glioblastomas, which are typically diagnosed in younger patients. Since this a consecutive series of patients, we did not select for subgroups based on histology, which is a limitation of this study.
Table 1.
Patient characteristics and findings
| Case # | Gender | Age | Diagnosis | Clinical status* |
IDH1 status |
[2HG] (ng/ml)—LC–MS |
2HG/Cr ratio—MRS |
|---|---|---|---|---|---|---|---|
| 2117 | Female | 22 | Anaplastic astrocytoma, WHO grade III | ND | R132C | 48700 | 1.044 |
| 2128 | Male | 61 | Anaplastic oligoastrocytoma, WHO grade III | Rec. | R132H | 166000 | 1.424 |
| 2140 | Female | 67 | Anaplastic oligodendroglioma, WHO grade III | ND | R132H | 58700 | 1.494 |
| 2147 | Male | 30 | Anaplastic astrocytoma, WHO grade III | Rec. | R132H | 64350 | 0.358 |
| 2153 | Male | 38 | Oligoastrocytoma, WHO grade II | Rec. | R132H | 13500 | 0.146 |
| 2155 | Male | 66 | Anaplastic astrocytoma, WHO grade III | Rec. | R132H | 58400 | N/A |
| 2160 | Male | 55 | Anaplastic oligodendroglioma, WHO grade III | ND | R132H | 148500 | 1.368 |
| 2166 | Female | 36 | Anaplastic oligoastrocytoma, WHO grade III | Rec. | R132C | 179000 | 0.810 |
| 2169 | Male | 41 | Anaplastic oligodendroglioma WHO grade III | ND | R132H | 117500 | 0.882 |
| 2199 | Female | 41 | Oligoastrocytoma (WHO grade III) | Rec. | R132H | 50200 | 0.330 |
| 2132 | Male | 62 | Glioblastoma, WHO grade IV | ND | WT | 2330 | 0.089 |
| 2133 | Male | 66 | Glioblastoma, WHO grade IV | Rec. | WT | 990.5 | 0 |
| 2139 | Male | 40 | Glioblastoma, WHO grade IV | Rec. | WT | 3860 | 0 |
| 2145 | Male | 85 | Glioblastoma, WHO grade IV | ND | WT | 2880 | N/A |
| 2146 | Male | 57 | Anaplastic astrocytoma, WHO grade III | ND | WT | 460 | 0 |
| 2149 | Female | 87 | Glioblastoma, WHO grade IV | ND | WT | 739.5 | 0 |
| 2163 | Male | 76 | Glioblastoma, WHO grade IV | Rec. | WT | 2630 | N/A |
| 2164 | Female | 63 | Glioblastoma, WHO grade IV | Rec. | WT | 175.5 | 0 |
| 2165 | Female | 44 | Glioblastoma, WHO grade IV | ND | WT | 1135 | 0 |
| 2172 | Female | 65 | Glioblastoma, WHO grade IV | ND | WT | 789 | 0.793 |
| 2173 | Female | 55 | Glioblastoma, WHO grade IV | Rec. | WT | 1540 | 0.047 |
| 2179 | Female | 60 | Glioblastoma, WHO grade IV | Rec. | WT | 340 | 0.509 |
| 2182 | Female | 66 | Glioblastoma, WHO grade IV | Rec. | WT | 631 | 0.412 |
| 2183 | Female | 45 | Glioblastoma, WHO grade IV | ND | WT | 787 | 0.030 |
| 2191 | Male | 56 | Glioblastoma, WHO grade IV | Rec. | WT | 853 | 0 |
| 2195 | Male | 59 | Glioblastoma, WHO grade IV | ND | WT | 403 | 0.424 |
| 2201 | Male | 63 | Glioblastoma, WHO grade IV | Rec. | WT | 4500 | 0 |
ND newly diagnosed; Rec. recurrent
MRI and MRS studies
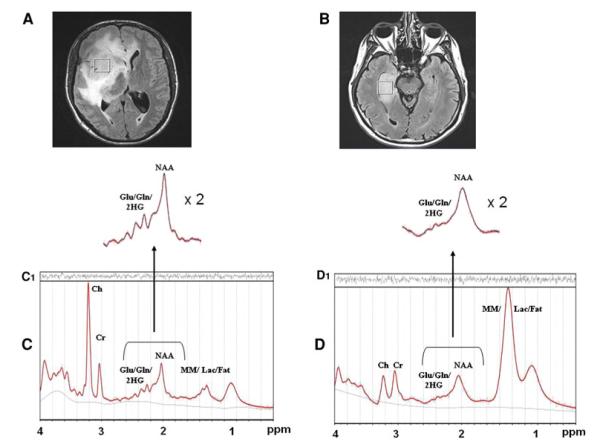
For patients with the same tumor pathology and grade, pre-operative structural MRI imaging characteristics were generally indistinguishable between IDH1 mutant and wild-type tumors as shown in Fig. 1A, B, respectively. Tumors of larger size had more surrounding edema and mass effect compared to smaller tumors, irrespective of IDH1 mutational status. However, localized MR spectroscopy of the areas of tumor revealed distinct MR spectra patterns between IDH1 mutant and wild-type gliomas. MRS of the tumors with IDH1 mutations revealed prominent extra peaks resonating at approximately 2.25 ppm (Fig. 1C) in contrast to that of the wild-type gliomas (Fig. 1D). As shown in Fig. 1, the area under the peaks at around 2.25 ppm is greater in the IDH1 mutant patients. This was attributable to 2-HG based on the LC-Model and phantom measurements (Suppl. Fig. 1), and was not seen in the majority (11/15, 73%) of the wild-type tumors. Of note, MR spectra from the contralateral (non-tumor) hemisphere of patients were obtained in IDH1 mutated tumors and did not reveal elevated 2-HG peaks (Suppl. Fig. 2), suggesting that this metabolic finding was specific to the tumor area itself.
Fig. 1.
Magnetic Resonance Imaging and MRS. Top panel (A and B): Axial MRI scans of two anaplastic astrocytomas (WHO grade III) show focal regions of fluid-attenuated inversion recovery (FLAIR) hyperintensity corresponding to areas of tumor. Other than the surrounding edema related to the size of the tumors, pre-operative structural MRI imaging characteristics were generally indistinguishable between IDH1 mutant (A) and wild-type (B) gliomas of the same grade and histopathology. Voxels of interest for MR spectroscopic analysis were localized by two neuroradiologists. Spectral voxels were placed in the center of the area of solid tumor, excluding regions of probable necrosis or vasogenic edema. Comparison of representative MR spectra from IDH1 mutant gliomas (C) versus wild-type spectra (D). Note the extra peaks in the region of Glu/Gln/2-HG (centered at 2.25 ppm) that are increased in the IDH1mutant tumors, compared to the wild-type MR spectra
Out of the 15 wild-type tumors analyzed using LC-Model, 11 patients had very low to undetectable levels of 2-HG. High CRLB values in the other four samples strongly suggest that the measurements of 2-HG generated by LC-Model for these samples were uncertain, and likely false positive calculations. The nine mutant tumors all had measurable levels of 2-HG by MRS, with low CRLB values, suggesting that the measurement of 2-HG was accurate. In addition, there were no obvious differences in the observed spectra due solely to differences in tumor histology or grade, which was confirmed by LC–MS.
Quantitative measure of 2-HG and other metabolites
Liquid chromatography–mass spectrometry quantitation of tumor-derived 2-HG demonstrated log-fold increases in 2-HG in IDH1 mutant tumor tissues ex vivo compared to wild-type (P < 0.0001) (Fig. 2A), validating our previous results [11]. Furthermore, in vivo MRS data from the same tumors also demonstrated that 2-HG was elevated in gliomas with the IDH1 mutation compared to wild-type tumors (P = 0.003) (Fig. 2B). All IDH1 mutant gliomas had measurable levels of 2-HG by MRS (100%), whereas only 4 of the 15 wild-type tumors did (26%). Tumor tissues from these four cases revealed low levels of 2-HG by LC–MS. Together with the high CRLB values, the compiled evidence suggests a false positive detection of 2-HG in these four wild-type tumors by the LC-Model algorithm. Since there is only a slight deviation in the chemical structure of 2-HG from that of Glu and Gln, the multiplets of 2-HG appear very close to that of Glu and Gln (Suppl. Fig. 1), which may account for a false positive MRS detection of 2-HG in wild-type tumors. Of note, the four false positive cases had relatively higher Glu/Gln levels than the other wild-type tumors (data not shown), and two of these four had very high Lac levels on MRS (Fig. 4D). Overall, there was a statistically significant correlation between 2-HG levels measured in glioma patients in vivo with MRS and those measured within resected tumor tissue samples ex vivo using LC–MS (correlation coefficient r2 = 0.56; P < 0.0001) (Fig. 2C).
Fig. 2.
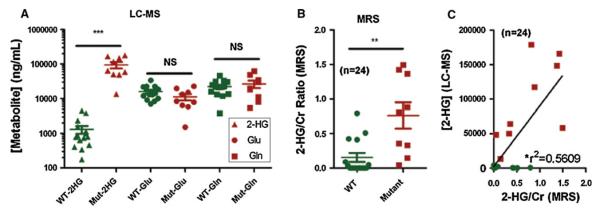
2-hydroxyglutarate, Glu and Gln levels in IDH1 wild-type versus IDH1 mutant gliomas. Quantitative 2-HG levels were calculated ex vivo using LC–MS (A) and in vivo using MRS (B). Good correlation was found between 2-HG levels measured by in vivo MRS and ex vivo LC–MS of tumor samples from the same patients (P < 0.0001) (C). IDH1 mutant cases are highlighted in red. ***P < 0.0001; **P = 0.003
Fig. 4.
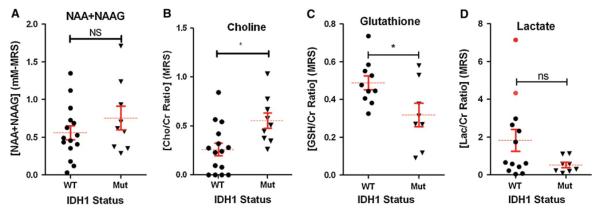
Metabolite levels in IDH1 wild-type vs. IDH1 mutant gliomas. Levels of A N-acetyl-aspartate (NAA), B choline (Cho), C glutathione (GSH), and D Lactate (Lac) were determined by MRS and quantified using LC-Model software. Metabolite ratios are represented with respect to the total concentrations of Cr. *P = 0.01 for Cho and P = 0.03 for GSH. Red dots represent wild-type samples with artifactually detectable 2-HG on MRS and high lactate levels, showing the metabolic profile of the “false positive” cases
To further validate that the increases in the spectral region of 2-HG peaks from IDH1 mutant tumors were due specifically to increases in 2-HG, we measured the concentrations of Glu and Gln in all the tumor tissue samples by LC–MS. As shown in Fig. 2A, no differences in the concentrations of either Glu or Gln were observed between IDH1 mutant and wild-type tumor tissues, which strongly suggests that the increases in the area of the Glu/Gln/2-HG peaks detected by MRS were attributable to 2-HG.
Furthermore, to correct for the impact of tumor grade, given that all but one of the IDH1 wild-type tumors in our consecutive series were glioblastoma and none of the IDH1 mutant tumors were glioblastoma, we examined 2-HG levels by LC–MS in an additional set of 43 IDH1-mutated gliomas of varying grades. As shown in Fig. 3, there was no significant difference in quantitative values of 2-HG based solely on tumor grade.
Fig. 3.
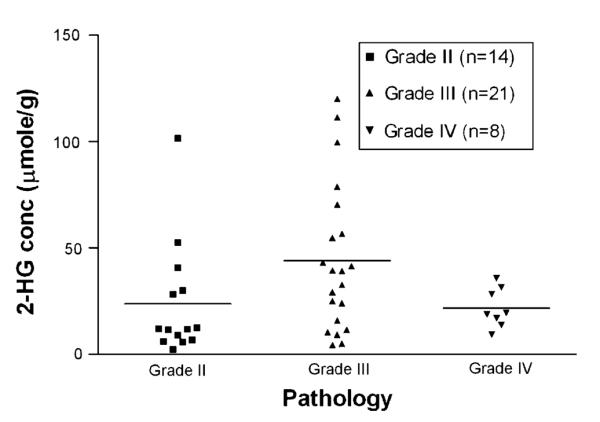
Tumor grade does not correlate with 2-HG concentrations in IDH1 mutant gliomas. Surgically resected glioma tissues were tested by LC–MS for the concentration of 2-HG in confirmed cases of IDH1 mutated gliomas. No significant differences in quantitative 2-HG levels were found due to differences in histology (p < 0.05 by ANOVA test)
Magnetic resonance spectroscopy demonstrated no statistical difference in total NAA levels in mutant versus wild-type IDH1 gliomas (Fig. 4A), but significantly elevated Cho ratios were seen in the IDH1 mutants (P = 0.01) (Fig. 4B). Wild-type tumors, on the other hand, revealed elevated levels of glutathione (GSH) compared to those with IDH1 mutations (P = 0.03) (Fig. 4C). Such findings are consistent with a recent report detailing metabolic differences between wild-type and mutant tumors [19]. Although there was a trend of higher Lac levels in some of the wild-type tumors, the difference in Lac ratios was not significant between the two groups (Fig. 4D). MR spectra from the wild-type tumors also revealed a very prominent lipid/Lac peak at around 1.3 ppm (not shown). However, because of the overlap of the MR spectral peaks due to lipid, Lac, alanine (Ala), and macromolecules (MM), no statistically significant difference in these individual compounds could be detected between IDH1 mutant and wild-type tumors by the MRS LC-Model algorithm.
Interestingly, among the IDH1 mutant tumors, a significant correlation was also found between the Ki-67 proliferation index of the tumor tissues and the quantified 2-HG levels measured in vivo by MRS (Fig. 5). When Cho levels detected by MRS were compared with proliferation in these tumors, there was a trend towards increasing Cho with a higher Ki-67 proliferation index, although the correlation was not statistically significant. This was most likely due to the small sample size (data not shown).
Fig. 5.
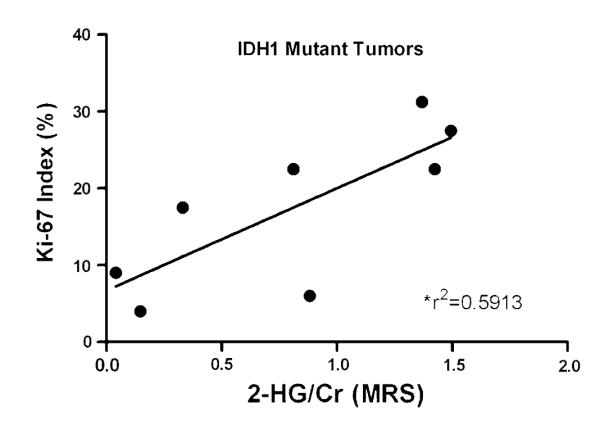
Relationship between Ki-67 proliferation index and 2-HG concentration. For the mutant IDH1 tumors (n = 9), the Ki-67 index for each tumor sample was determined by immunohistochemistry and compared to 2-HG concentrations calculated by MRS analysis. Correlation coefficients were calculated and are shown with a linear regression line drawn (P = 0.026)
Discussion
High-throughput sequencing previously revealed that the metabolic enzymes, IDH1 and IDH2, are frequently mutated in a majority of several types of malignant gliomas [4, 5]. Subsequently, we found that human malignant gliomas harboring R132 IDH1 mutations produce elevated levels of 2-HG, a finding recently confirmed by other groups [19, 20]. All mutant IDH1 tumors that we have examined to date, including those reported in this study, had between 5 and 35 micromol of 2-HG per gram of tumor tissue, whereas gliomas with wild-type IDH1 had over 100-fold less 2-HG [11]. Herein, we discovered that MRS could sensitively detect 2-HG within patient tumors in vivo, which correlated reproducibly with ex vivo quantitation of 2-HG via LC–MS. The non-invasive detection of a novel putative oncometabolite (2-HG) that is directly linked with IDH1 mutations potentially provides a unique and clinically applicable imaging biomarker associated with this major subset of gliomas.
All the IDH1 mutant gliomas we tested had measurable levels of 2-HG by MRS, suggesting the potential for a high level of sensitivity (100%) in the detection of 2-HG using this MRS algorithm. The specificity of detection of 2-HG may be challenging, however, as the structure of 2-HG is similar to Glu and Gln, resulting in overlap of the resultant multiplets. For the wild-type case cohort, there was a 26% (4/15) false positive rate in the detection of 2-HG, presumably due to the overlap of the 2-HG peak with those of Glu and Gln in the one-dimensional (1D) MR spectra (Suppl. Fig. 1). Nevertheless, the Cramer-Rao lower bounds calculations support a true difference in the detection of 2-HG between IDH1 mutant and wild-type tumors. Overall, there was good correlation between 2-HG as measured by MRS in vivo compared to ex vivo quantitation. One avenue to potentially reduce system noise is to perform two-dimensional (2D) MRS, which may help separate 2-HG from adjacent peaks, thereby increasing the accuracy of its quantification. Future refinements notwithstanding, our report demonstrates the feasibility of non-invasively tracking metabolic changes that are associated with mutant IDH1 enzymes in clinical practice.
Since we did not see any significant differences in the concentrations of Glu and Gln between wild-type and mutant IDH1 tumors, our data provide substantial evidence that we are specifically detecting differences in 2-HG. Detection of 2-HG by MRS may provide a method to follow disease progression in patients with IDH1 mutant tumors and/or to monitor treatment effects once specific molecular inhibitors of IDH1 are available clinically. In a recent large study of acute myeloid leukemia (AML), IDH mutational status was serially analyzed in 130 patients and found to correlate with disease remission and relapse [21]. Two other independent studies have associated elevated levels of 2-HG in AML samples harboring IDH mutations, suggesting a correlation of 2-HG levels to disease status [22, 23]. Conceivably, MRS of 2-HG in brain tumors could be used to track and monitor glioblastoma patients in the same way.
Furthermore, our data suggest that gliomas harboring IDH1 mutations exhibit additional metabolic alterations beyond elevated levels of 2-HG. For instance, we found that Cho levels, as measured by MRS, were elevated in IDH1-mutated tumors compared to wild-type gliomas in vivo. In a recent study profiling the effects of IDH on the cellular metabolome in vitro, elevated Cho derivatives were also found in human oligodendroglioma (HOG) cells engineered to express mutant IDH1 and IDH2 [19]. Total Cho is a marker for membrane turnover, and is elevated in a variety of pathologic conditions, including brain tumors [24]. For gliomas, Cho has been shown to increase in concentration in lower grade tumors that underwent malignant degeneration into higher grades, in comparison to non-progressing tumors, [25] and the concentration of Cho is correlated with tumor cell density [26]. Therefore, increased Cho in IDH1 mutants could reflect increased cell density due to IDH1-mutation-mediated cellular proliferation. Although previous studies have indicated that IDH1 mutations identify a subgroup of gliomas with improved survival compared to those with wild-type IDH1 [9, 27–29], this does not preclude the possibility that IDH1 mutant-mediated 2-HG production leads to increased cell proliferation, whereas the ultimate survival of patients is dictated by additional oncogenic processes (i.e., migration, angiogenesis, necrosis, etc.). Since mutations in IDH1 are known to occur early in gliomagenesis, [5, 6, 27] the quantitative levels of 2-HG within IDH1 mutant tumors may provide some mechanistic insight into the role of IDH1 mutations in gliomagenesis and tumor progression.
We found that IDH1 mutant tumors have decreased levels of GSH compared to gliomas with the wild-type allele. Because IDH is an important NADPH-producing node, somatic mutation of this enzyme could alter the cellular NADPH/NADP+ ratio. Importantly, NADPH is essential for the regeneration of reduced GSH and maintenance of redox homeostasis. GSH is indeed a major antioxidant that neutralizes reactive oxygen species and free radicals. Furthermore, it contributes to DNA synthesis and repair, mediates xenobiotic metabolism, and participates in numerous metabolic reactions [30, 31]. Thus, alterations in cellular concentration, as well as the ratio of reduced to oxidized forms of GSH, may contribute to malignancy. Other studies have also shown lower GSH levels in mutant IDH-expressing glioma cell cultures in vitro [19]. The decreased levels of GSH seen in IDH1 mutated gliomas may provide a potential clue to the role of this putative oncogene in the development of human brain cancers.
Future studies will help to further define whether IDH1 mutant gliomas exhibit a more global and distinct metabolic fingerprint, which would shed additional light on the potential link between metabolic alterations and mutations in IDH1. Recent evidence suggests that 2-HG accumulation in IDH1- and IDH2-mutated tumors may lead to genome-wide histone and DNA methylation alterations [32]. Our present study establishes an MRS-based method to non-invasively monitor 2-HG levels in gliomas in the clinical setting. Development of this methodology provides an opportunity to investigate this putative oncometabolite in relationship to gliomagenesis, global metabolic alterations, and disease progression in situ. In addition to a diagnostic application, it could provide a means to monitor the activity of inhibitors of mutant IDH1/IDH2 enzymes once available. Since IDH mutations appear to be early and key events in the pathogenesis of glioma, have prognostic implications, and present attractive targets for future pharmacologic inhibition, such non-invasive methods of clinical assessment may be of potential value and clinical utility.
Supplementary Material
Acknowledgments
Supported in part by grants from the National Cancer Institute (K01-CA111402 and R01-CA123396 to Dr. Prins; R01-CA112358 and R01-CA121131 to Dr. Liau), and research grants from the Neidorf Family Foundation, the Phillip R. Jonsson & Kenneth A. Jonsson Foundations, the Brad Kaminsky Foundation, and the Accelerate Brain Cancer Cure Foundation. Scientific support of Mr. Brian Burns is also gratefully acknowledged.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11060-011-0737-8) contains supplementary material, which is available to authorized users.
Contributor Information
Whitney B. Pope, Department of Radiological Sciences, David Geffen School of Medicine at UCLA, University of California at Los Angeles, Los Angeles, CA 90095, USA
Robert M. Prins, Department of Neurosurgery, David Geffen School of Medicine at UCLA, University of California at Los Angeles, Gonda Building, Room 3357, 695 Charles Young Drive, Los Angeles, CA 90095-1761, USA LLIAU@mednet.ucla.edu
M. Albert Thomas, Department of Radiological Sciences, David Geffen School of Medicine at UCLA, University of California at Los Angeles, Los Angeles, CA 90095, USA.
Rajakumar Nagarajan, Department of Radiological Sciences, David Geffen School of Medicine at UCLA, University of California at Los Angeles, Los Angeles, CA 90095, USA.
Noriko Salamon, Department of Radiological Sciences, David Geffen School of Medicine at UCLA, University of California at Los Angeles, Los Angeles, CA 90095, USA.
Arthur P. Chou, Department of Neurosurgery, David Geffen School of Medicine at UCLA, University of California at Los Angeles, Gonda Building, Room 3357, 695 Charles Young Drive, Los Angeles, CA 90095-1761, USA LLIAU@mednet.ucla.edu
William H. Yong, Department of Pathology & Laboratory Medicine, David Geffen School of Medicine at UCLA, University of California at Los Angeles, Los Angeles, CA 90095, USA
Horacio Soto, Department of Neurosurgery, David Geffen School of Medicine at UCLA, University of California at Los Angeles, Gonda Building, Room 3357, 695 Charles Young Drive, Los Angeles, CA 90095-1761, USA LLIAU@mednet.ucla.edu.
Neil Wilson, Department of Radiological Sciences, David Geffen School of Medicine at UCLA, University of California at Los Angeles, Los Angeles, CA 90095, USA.
Hyun G. Jang, Agios Pharmaceuticals, 38 Sidney Street, Cambridge, MA 02139, USA michael.su@agios.com
Shinsan M. Su, Agios Pharmaceuticals, 38 Sidney Street, Cambridge, MA 02139, USA michael.su@agios.com
David P. Schenkein, Agios Pharmaceuticals, 38 Sidney Street, Cambridge, MA 02139, USA michael.su@agios.com
Albert Lai, Department of Neurosurgery, David Geffen School of Medicine at UCLA, University of California at Los Angeles, Gonda Building, Room 3357, 695 Charles Young Drive, Los Angeles, CA 90095-1761, USA; Department of Neurology, David Geffen School of Medicine at UCLA, University of California at Los Angeles, Los Angeles, CA 90095, USA.
Timothy F. Cloughesy, Department of Neurology, David Geffen School of Medicine at UCLA, University of California at Los Angeles, Los Angeles, CA 90095, USA
Harley I. Kornblum, Department of Psychiatry & Behavioral Sciences, David Geffen School of Medicine at UCLA, University of California at Los Angeles, Los Angeles, CA 90095, USA
Hong Wu, Department of Molecular & Medical Pharmacology, David Geffen School of Medicine at UCLA, University of California at Los Angeles, Los Angeles, CA 90095, USA.
References
- 1.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116(6):597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 2.Bleeker FE, Lamba S, Leenstra S, Troost D, Hulsebos T, Vandertop WP, et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30(1):7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- 3.De Carli E, Wang X, Puget S. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(21):2248. doi: 10.1056/NEJMc090593. author reply 9. [DOI] [PubMed] [Google Scholar]
- 4.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174(4):1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ducray F, Marie Y, Sanson M. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(21):2248. author reply 9. [PubMed] [Google Scholar]
- 8.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubbink HJ, Taal W, van Marion R, Kros JM, van Heuvel I, Bromberg JE, et al. IDH1 mutations in low-grade astrocytomas predict survival but not response to temozolomide. Neurology. 2009;73(21):1792–1795. doi: 10.1212/WNL.0b013e3181c34ace. [DOI] [PubMed] [Google Scholar]
- 10.Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1, 010 diffuse gliomas. Acta Neuropathol. 2009;118(4):469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 11.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aghili M, Zahedi F, Rafiee E. Hydroxyglutaric aciduria and malignant brain tumor: a case report and literature review. J Neurooncol. 2009;91(2):233–236. doi: 10.1007/s11060-008-9706-2. [DOI] [PubMed] [Google Scholar]
- 13.Bottomley PA. Spatial localization in NMR spectroscopy in vivo. Ann N Y Acad Sci. 1987;508:333–348. doi: 10.1111/j.1749-6632.1987.tb32915.x. [DOI] [PubMed] [Google Scholar]
- 14.Ogg RJ, Kingsley PB, Taylor JS. WET, a T1- and B1-insensitive water-suppression method for in vivo localized 1H NMR spectroscopy. J Magn Reson B. 1994;104(1):1–10. doi: 10.1006/jmrb.1994.1048. [DOI] [PubMed] [Google Scholar]
- 15.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30(6):672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 16.Smith SA, Levante TO, Meier BH, Ernst RR. Computer simulations in magnetic resonance.An object oriented programming approach. J Magn Reson. 1994;A106:75–105. [Google Scholar]
- 17.Helms G. The principles of quantification applied to in vivo proton MR spectroscopy. Eur J Radiol. 2008;67(2):218–229. doi: 10.1016/j.ejrad.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 18.Cavassila S, Deval S, Huegen C, van Ormondt D, Graveron-Demilly D. Cramer-Rao bounds: an evaluation tool for quantitation. NMR Biomed. 2001;14(4):278–283. doi: 10.1002/nbm.701. [DOI] [PubMed] [Google Scholar]
- 19.Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci U S A. 2011;108(8):3270–3275. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin G, Reitman ZJ, Spasojevic I, Batinic-Haberle I, Yang J, Schmidt-Kittler O, et al. 2-Hydroxyglutarate production, but not dominant negative function, is conferred by glioma-derived NADP-dependent isocitrate dehydrogenase mutations. PLoS One. 2011;6(2):e16812. doi: 10.1371/journal.pone.0016812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou WC, Hou HA, Chen CY, Tang JL, Yao M, Tsay W, et al. Distinct clinical and biologic characteristics in adult acute myeloid leukemia bearing the isocitrate dehydrogenase 1 mutation. Blood. 2010;115(14):2749–2754. doi: 10.1182/blood-2009-11-253070. [DOI] [PubMed] [Google Scholar]
- 22.Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207(2):339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Bisbal MC, Celda B. Proton magnetic resonance spectroscopy imaging in the study of human brain cancer. Q J Nucl Med Mol Imaging. 2009;53(6):618–630. [PubMed] [Google Scholar]
- 25.Tedeschi G, Lundbom N, Raman R, Bonavita S, Duyn JH, Alger JR, et al. Increased choline signal coinciding with malignant degeneration of cerebral gliomas: a serial proton magnetic resonance spectroscopy imaging study. J Neurosurg. 1997;87(4):516–524. doi: 10.3171/jns.1997.87.4.0516. [DOI] [PubMed] [Google Scholar]
- 26.Dowling C, Bollen AW, Noworolski SM, McDermott MW, Barbaro NM, Day MR, et al. Preoperative proton MR spectroscopic imaging of brain tumors: correlation with histopathologic analysis of resection specimens. AJNR Am J Neuroradiol. 2001;22(4):604–612. [PMC free article] [PubMed] [Google Scholar]
- 27.Labussiere M, Wang XW, Idbaih A, Ducray F, Sanson M. Prognostic markers in gliomas. Future oncology (London, England) 2010;6(5):733–739. doi: 10.2217/fon.10.25. [DOI] [PubMed] [Google Scholar]
- 28.Toedt G, Barbus S, Wolter M, Felsberg J, Tews B, Blond F, et al. Molecular signatures classify astrocytic gliomas by IDH1 mutation status. Int J Cancer. 2011;128(5):1095–1103. doi: 10.1002/ijc.25448. [DOI] [PubMed] [Google Scholar]
- 29.van den Bent MJ, Dubbink HJ, Marie Y, Brandes AA, Taphoorn MJ, Wesseling P, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res. 2010;16(5):1597–1604. doi: 10.1158/1078-0432.CCR-09-2902. [DOI] [PubMed] [Google Scholar]
- 30.Dang L, Jin S, Su SM. IDH mutations in glioma and acute myeloid leukemia. Trends Mol Med. 2010;16(9):387–397. doi: 10.1016/j.molmed.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Wiseman H, Halliwell B. Damage to DNA by reactive oxygen and nitrogen species: role in inflammatory disease and progression to cancer. Biochem J. 1996;313(Pt 1):17–29. doi: 10.1042/bj3130017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim S-H, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of [alpha]-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.