Abstract
The myelodysplastic syndrome (MDS) is a clonal disorder characterized by increased stem cell proliferation coupled with aberrant differentiation resulting in a high rate of apoptosis and eventual symptoms related to bone marrow failure. Cellular differentiation is an epigenetic process that requires specific and highly ordered DNA methylation and histone modification programs. Aberrant differentiation in MDS can often be traced to abnormal DNA methylation (both gains and losses of DNA methylation genome wide and at specific loci) as well as mutations in genes that regulate epigenetic programs (TET2 and DNMT3a, both involved in DNA methylation control; EZH2 and ASXL1, both involved in histone methylation control). The epigenetic nature of MDS may explain in part the serendipitous observation that it is the disease most responsive to DNA methylation inhibitors; other epigenetic-acting drugs are being explored in MDS as well. Progression in MDS is characterized by further acquisition of epigenetic defects as well as mutations in growth-controlling genes that seem to tip the proliferation/apoptosis balance and result in the development of acute myelogenous leukemia. Although MDS is clinically and physiologically heterogeneous, a case can be made that subsets of the disease can be largely explained by disordered stem cell epigenetics.
MDS is a disease of disordered differentiation
Myelodysplastic syndrome (MDS) carries in its name the fundamental pathognomonic defect that characterizes it: dysplasia, a catchy description of what is essentially abnormal differentiation.1,2 There are many histologic hallmarks of aberrant hematopoietic cell differentiation in MDS: nuclear/cytoplasmic ratio, nuclear shape, agranularity (or persistence of granules when they should be absent at that particular stage of differentiation), etc. There are also functional defects characteristic of abnormal differentiation: patients with MDS are prone to serious infections even when the neutrophil count is apparently preserved and they can have serious bleeding episodes despite reasonable platelet counts. In vitro, one can demonstrate altered differentiation through clonogenic assays3 and, in vivo, MDS cells have gene expression defects that are often in differentiation-related pathways.4 Thus, while the characteristic bone marrow picture suggests abnormal proliferation (hypercellular marrow), the actual defect in MDS appears more traceable to abnormal differentiation, perhaps itself a trigger of compensatory proliferation. Indeed, a major difference between MDS and more classically proliferative neoplasms (such as acute myelogenous leukemia [AML]) is that myelodysplastic cells have a high rate of apoptosis, presumably as a result of the differentiation defects.5 These properties account for one of the striking paradoxes in the disease: clinically, it often behaves as a bone marrow failure syndrome, even though it has many of the hallmarks of a classical neoplasm (clonality, hypercellularity, progression to more advanced stages, etc). It is also well established that “MDS” is a catch phrase for different diseases that have distinct etiologies. For example, in a mouse model of the 5q− syndrome, the pathologic abnormalities could be traced to high expression of P53 triggered by haploinsufficiency of several genes including the ribosomal protein gene Rps14.6 Thus, there may also be instances of MDS without direct differentiation defects.
Differentiation is an epigenetic process
The molecular mechanisms underlying differentiation were quite mysterious when the era of DNA began in the middle of the 20th century. How could one go from a single genome to over 200 tissues without substantial alterations in the underlying DNA sequence? The developmental biologist Waddington coined the term epigenetics to account for this phenomenon.7 In the past 2 decades, there has been much progress in understanding the mechanisms underlying epigenetic regulation and, while “epigenetics” is now a catch word for a myriad of phenomena, differentiation remains central to understanding the core principle of epigenetics: stable, long-term regulation of gene expression that is unrelated to variation in the DNA coding sequence and that can survive across numerous rounds of cell division.8
Details of the molecular mechanisms of epigenetic regulation can be found in other chapters in this series. Suffice it to say that much of the focus has been on three mechanisms: DNA methylation, posttranslational histone tail modifications, and micro-RNA expression. DNA methylation of CpG-rich promoters (CpG islands) is the purest epigenetic mechanism in that, once established, it can be perpetuated without the initial regulatory signal.9 Indeed, such DNA methylation is sufficient to maintain 2 of the best-recognized allele-specific epigenetic phenomena: imprinting and X-chromosome inactivation. DNA methylation in other parts of the genome (eg, CpG-poor gene bodies, enhancers, etc) can contribute to gene regulation but changes dynamically with gene activation/inactivation and is thus less stable than promoter CpG island methylation and less clearly involved in epigenetic regulation.9 Posttranslational modifications of histone tails are closely linked to particular gene expression patterns and have been proposed as an epigenetic code.10 However, many of these histone modifications dynamically respond to the presence of enhancers or repressors, and the short half-life of modified histones have led some to question whether they can serve as an epigenetic code independent of transcription factors.11 The issue of stable vs dynamic histone modifications remains an important research area. Finally, micro-RNAs are often discussed in the context of epigenetic regulation, and they clearly influence gene expression.12 Micro-RNA expression can also be dynamically regulated; thus, their involvement in epigenetics is limited to the instances when their own transcription is subject to epigenetic regulation such as promoter DNA methylation. In that context, micro-RNAs can serve as useful mediators of fine-tuning gene expression in differentiated cells.
Differentiation can now be understood as an interplay between transcription programs that initiate the process and epigenetic changes that stabilize gene expression and limit plasticity.8 As predicted, DNA methylation is exquisitely tissue specific,13 though the details of genes involved and mechanisms of methylation initiation are only now becoming clear. There are also dynamic switches in histone modifications that accompany differentiation; one of the most characteristic of these is a progressive loss in the bivalent state.14 Bivalent genes are those characterized by the simultaneous presence of activating and inactivating marks in embryonic stem cells. These contribute to the plasticity of embryonic stem cells in that their expression state can go either way depending on the differentiation path. As cells differentiate, bivalent genes are steered toward an irreversible gene expression state (active or inactive) and, as the number of bivalent genes declines, cells acquire the characteristic of terminal differentiation. Finally, micro-RNA expression is also exquisitely tissue specific, which undoubtedly contributes to the final identify of the differentiated cell.12
MDS cells carry an abnormal epigenome
Every aspect of epigenetic regulation can be shown to be abnormal in neoplastic cells,15 and MDS is no exception. This includes methylation abnormalities, histone code changes and micro-RNA expression (Figure 1). It is important but somewhat tricky to tease out epigenetic changes that are truly abnormal from those that simply reflect the differentiation state of the cells under study. Even more difficult is the task of separating driver events from passenger events that arise as a function of time/neoplasia. Still, much progress has been made in these areas, and our knowledge base is accelerating thanks to the emergence of genome-wide epigenetic profiling technologies.
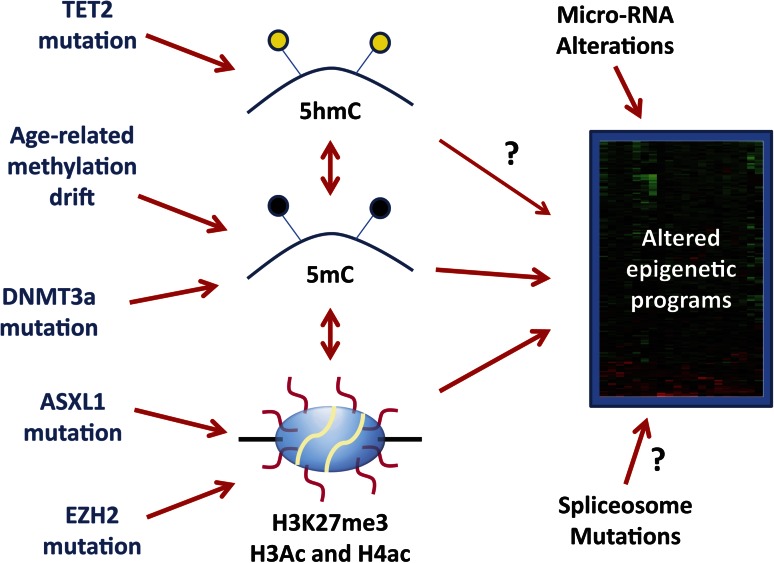
Figure 1.
Origin of aberrant epigenetic programs in MDS. MDS carries an altered epigenome that results in stable gene expression changes represented by a heatmap on the right side of the figure. These can be influenced by DNA methylation (5mC) and histone code posttranslational modifications such as histone H3 lysine 27 trimethylation (H3K27me3) and acetylation (Ac) of multiple residues on histone H3 and H4. Cytosine hydroxymethylation (5hmC) influences 5mC content and may have direct effects on gene expression (arrow with a question mark). There are also complex correlations between DNA methylation and histone modifications. Molecularly, 5hmC can be altered by TET2 mutations, 5mC is altered by age-related drift and possibly by DNMT3a mutations, and H3K27me3 is potentially influenced by ASXL1 and EZH2 mutations. However, the precise links between TET2 mutations, DNMT3a mutations, and DNA methylation in MDS remain somewhat uncertain (illustrated by dotted lines). Changes in micro-RNA expression (due to genetic or epigenetic lesions) also influence the final gene expression patterns and it is possible (though speculative) that spliceosome mutations also do this. It remains unclear how much of the final MDS gene expression patterns are driven by the described epigenetic alterations, and the heterogeneity of the disease implies that these mechanisms may be more important in some cases than in others.
DNA methylation
Promoter-associated CpG islands are largely unmethylated in normal tissues, regardless of differentiation state.9 Dozens to hundreds of these become aberrantly hypermethylated in MDS.16 Just like the rare instances where this is seen in normal cells9,17 (imprinting, X-inactivation, differentiation), aberrant promoter methylation is stable, clonally propagated, and invariably associated with silencing of the involved gene. Such aberrant methylation was first seen in myeloid leukemias years ago in a series of studies that are still relevant today.18 We now know that abnormal methylation can occur early, is often independent of cytogenetic changes, and is associated with more rapid disease progression to AML.16,19 Updated studies using genome-wide technologies showed that 3% to 5% of promoter CpG islands are hypermethylated in MDS,20 and that at least some of the genes involved are potential driver events because (1) they are expressed in normal hematopoietic cells, (2) they are silenced when methylated, and (3) silencing of the genes has well-defined functional consequences to the neoplastic cells. The P15ink4b cell-cycle regulator is emblematic of these potential driver events: it is hypermethylated in 10% to 30% of MDS cases, methylation is associated with a poor outcome, and mouse studies suggest that this gene behaves as a tumor suppressor in hematopoietic cells.16,19,21 But P15 is but one of many genes that behave this way and multiple pathways are involved. It is likely that many of the hallmarks of MDS can be traced to functional pathway alterations due to aberrant promoter CpG island methylation.
In addition to promoter CpG island hypermethylation, one can see multiple DNA methylation changes in MDS when compared with normal hematopoietic cells. These include both gains and losses of methylation in CpG-poor promoters, nonpromoter CpG islands, gene bodies, enhancer regions, and intergenic areas (mostly composed of repeat sequences including retrotransposons). Because each of these genomic compartments can dynamically change with gene transcription and differentiation, it can be difficult to establish those events that are primary (and responsible for altered gene expression) from those that are secondary, either physiologic (ie, reflect the unique differentiation state of the MDS clone) or pathologic (eg, a downstream result of gene activation by a nonepigenetic mechanism). Regardless of etiology, it is likely that these unique methylation states mediate the abnormal differentiation behavior of MDS cells in some cases and thus could be useful as biomarkers as well as targets for therapeutic intervention. Finally, a global loss of 5-methylcytosine characterizes many cancer cells22 (and this was actually the first cancer-specific DNA methylation change described23), but it is relatively rare in MDS.
A currently debated question is the origin of DNA methylation changes in MDS. There are 2 not-mutually-exclusive hypotheses. In the first one, drift, it is postulated that epigenetic patterns drift somewhat randomly over time (and age), creating diversity in epigenomic patterns. In turn, this diversity provides the fodder for Darwinian forces to select those patterns most permissive of neoplastic growth (and aberrant differentiation) in MDS. This hypothesis was first developed in solid tumor models where age-related epigenomic diversity can be easily appreciated in normal and neoplastic tissues,24-26 but it equally applies to hematopoietic malignancies. Indeed, DNA methylation and epigenetic drift can be seen in aging blood cells in both humans and mice.27-29 A second hypothesis links DNA methylation alterations to specific genomic defects. Particular patterns of DNA methylation can be seen in leukemias with distinct cytogenetic abnormalities,20 and mutations in genes that control DNA methylation can also be seen in MDS.30 Although there are compelling reasons to suspect that some of the methylation patterns in MDS reflect underlying genetic damage, it appears that some of the most common changes (eg, promoter CpG island methylation) are not unique to any genetic or cytogenetic group.16 Even if such associations were seen, it remains to be directly demonstrated that the genetic changes actually cause DNA methylation damage as opposed to coevolve during clonal selection. For example, one of the most striking associations between a genetic event and aberrant methylation is the link between BRAF (and KRAS) mutations and the CpG island methylator phenotype in colon cancer.31 However, there are no convincing data showing that those mutations lead to aberrant methylation of the affected genes, and the association is most likely an example of coselection. Thus, it will be important to clearly determine which genetic events directly contribute to aberrant DNA methylation in MDS.
Histone modifications and micro-RNAs
Genome-wide studies of histone modifications have not yet been reported in MDS, though studies in AML show distinct patterns of histone posttranslational modifications between leukemia and normal hematopoietic cells.32 Similarly, gene expression profiles reveal differences in micro-RNA expression between MDS and normal hematopoietic cells.33 These studies need to be interpreted with caution. As discussed earlier, there are large-scale switches in chromatin regulation and gene expression associated with differentiation. Thus, one needs to carefully consider the comparator when evaluating the significance of these studies. Another complicating issue is technical: whether measured by microarrays or by deep sequencing, the precision and reproducibility of histone modifications is around 90%, which introduces significant difficulties when comparing samples. A commonly used strategy to overcome these problems is to compare sorted CD34+ leukemic cells to CD34+ normal cells both studied simultaneously; distinct differences are found in these well-conducted studies but it is likely that only large differences are reliably detected in this way. Ultimately, it will be most interesting to link these altered patterns to underlying defects that could account for them, be it mutations in histone modifiers or genetic changes at micro-RNA loci.
MDS shows frequent epigenetic effector mutations
Epigenomic anomalies in MDS coexist with cytogenetic changes in more than half the patients and with somatic mutations in virtually every case. Thus, the full manifestations of the disease appear to require concurrent genetic and epigenetic damage. The case for MDS as an epigenetic disease (in some instances) received a strong boost when genome-wide sequencing studies revealed frequent mutations in epigenetic effectors.30 These include mutations in DNA methylation controllers (DNMT3a, a de novo DNA methylase and TET2, a DNA demethylase), histone modifiers (EZH2, a methyltransferase that catalyzes histone H3 lysine 27 [H3K27] trimethylation and ASXL1, a member of the polycomb group proteins that are also regulators of H3K27). Much remains to be learned about the specific effects of these mutations but it is very likely that their transforming ability is linked to specific epigenetic regulation (Figure 1).
DNMT3a is a de novo DNA methylase that is expressed at high levels in hematopoietic stem cells.34 Mutations in DNMT3a can be seen in both MDS35 and AML.36 The mutation distribution (a hotspot present in about half the cases and mutations throughout the gene in the other half) and in vitro studies37 suggest that some of the mutations reduce its methyltransferase catalytic activity, but the ultimate functional consequences of these mutations remain incompletely understood. Indeed, there are few well-characterized specific methylation targets of DNMT3a that are altered as a result of these mutations. In a mouse model, hematopoietic-specific deletion of DNMT3a results in stem cell expansion and reduced differentiation, along with DNA methylation changes in an unexpectedly narrow subset of genes.34 It is likely that this mouse model is relevant to understanding the defects in DNMT3a mutant myeloid leukemias.
TET2 is an enzyme that catalyzes the production of 5-hydroxy-methylcytosine from 5-methyl-cytosine, in a reaction that ultimately leads to DNA demethylation. Inactivating mutations in TET2 are frequent in MDS, and are present in almost half of all cases of chronic myelomonocytic leukemia (CMML).38 The effects of these mutations on genomic DNA methylation remain controversial. Although TET2 mutant cases clearly show decreased 5-hmC39 and increased 5-mC,40 effects on gene-specific methylation have been inconsistent, with 1 study reporting mostly CpG island hypomethylation39 (comparison of MDS with/without mutation), another reporting CpG island hypermethylation41 (comparison of AML with mutation to normal cells), and a third showing no effect on CpG islands40 (comparison of CMML with/without mutations). TET2 is part of a family of 3 proteins.42 TET1 and TET3 have a CXXC domain that is characteristic of proteins that bind to CpG islands.43 By contrast, TET2 lacks a CXXC domain, and it is likely that its effects on DNA methylation lie outside of CpG islands. Deletion of TET2 in mice results in a myelomonocytic expansion reminiscent of CMML,44 and it is likely an excellent example of a genetic defect leading to aberrant differentiation through specific epigenetic changes (that remain to be conclusively delineated).
The polycomb pathway consists of 2 complexes, PRC1 and PRC2, that read and write the unique histone modification H3K27 trimethylation. This methylation is generally associated with repression of gene transcription, and is key to the transition from stem cell to differentiated cell.45 Thus, it is not surprising that polycomb alterations characterize MDS, the quintessential differentiation disease. EZH2, a member of the PRC2 complex, writes the H3K27 trimethylation signal and inactivating mutations in EZH2 can be found in a subset of MDS cases.46 These mutations would presumably be associated with loss of H3K27 trimethylation at specific gene targets, though this remains to be determined. Interestingly, these mutations are the exact opposite of what can be seen in some lymphomas that are characterized by activating EZH2 mutations and increased H3K27 trimethylation.47 Thus, it is really the balance of the H3K27 mark that matters in neoplasia. A second gene in the polycomb pathway is also mutated in MDS, namely ASXL1.30 This gene is part of the PRC1 complex, and mutations are common in MDS and very frequent in CMML. The mutations lead to loss of H3K27 trimethylation and promote leukemic transformation in a mouse model.48 There are also rare mutations in other epigenetic modifiers in MDS. For example, mutations in the metabolic enzymes IDH1 and IDH2 can affect the function of both DNA demethylases (such as TET241) and histone demethylases (such as KDM4C), and can be seen in some MDS cases,30 though they are more frequent in AML. Similarly, mutations in the histone H3K27 demethylase UTX and in the H3K4 methylase MLL family have been reported in some cases.
It is also worth mentioning mutations in the spliceosome in this section. These mutations are very frequent in MDS subsets49 and, while specific gene targets remain unknown, their likely consequences are aberrant gene expression, particularly in differentiation programs. The general control of gene elongation and splicing remains incompletely understood, but there are intriguing links between DNA methylation, histone modifications, and splicing programs.50-52 Thus, it may well be that some epigenetic alterations and spliceosome mutations converge pathophysiologically on similar pathways that affect hematopoietic cell differentiation, but this remains speculative at this point.
MDS responds to epigenetic therapy
The final part of the argument for MDS being a disorder of abnormal epigenetic regulation (in some cases) comes from its remarkable sensitivity to drugs that modify the epigenome (epigenetic therapy). Two nucleoside analog DNA methylation inhibitors, 5-azacytidine (azacitidine) and 5-aza-2’-deoxycytidine (decitabine), have shown significant activity in all stages of MDS.53 The clinical data suggest responses in about half of patients54,55 (including complete responses in 10%-30% of patients and partial remissions such as transfusion independence in an additional 20%-30% cases), delays in time to progression to AML, and prolonged survival compared with supportive care or traditional chemotherapy in randomized studies54,56 or in case-control studies.57 The responses follow an unusual pattern with little measurable effect in the first few weeks to months of therapy and eventual clinical responses that coincide with clonal elimination (including cytogenetic remissions). This pattern of delayed responses along with a lack of toxicities classically associated with cytotoxic agents has been argued as evidence that the responses are mediated by epigenetic modulation.53
The mechanisms of sensitivity and resistance to hypomethylating drugs are a matter of ongoing investigations. Some studies reported that early and sustained DNA hypomethylation of multiple genes after decitabine correlate with subsequent achievement of a clinical response16,55,58 but this was not seen with azacitidine.59 These studies are difficult to do and interpret because clonal elimination is indistinguishable from demethylation of aberrantly methylated loci, and patients with MDS have a limited number of circulating cells for such analyses. Clearly, the issue deserves more precise investigation in well-controlled studies. Interestingly, gene activation independent of demethylation (eg, the aforementioned P15 gene55 and the P53R2 gene60) also correlates with response and it is not known whether this reflects pathway activation by demethylation or a truly unrelated mechanism of action. In some cases, clinical improvement (eg, platelet and hemoglobin increases) is seen without clonal elimination, suggesting that the drugs help overcome a differentiation block. In the best responses, however, clonal elimination can be documented61 (and correlates with improved survival) and it remains to be seen whether this is due to delayed cell death/senescence, effects on neoplastic stem cell renewal, activation of an immune response, or a combination of mechanisms. Moreover, the mechanisms of resistance to hypomethylating agents remain poorly understood. Some data suggest that primary resistance could be due to pharmacologic factors that limit incorporation of the drugs into nucleic acids, for example, high expression of CDA62,63 which metabolizes the drugs and/or, in the case of decitabine, low expression of DCK which is rate limiting for DNA incorporation.62 However, it is well documented that hypomethylation is not sufficient for a response59 and thus other mechanisms should be investigated. Secondary resistance also develops commonly and was found to occur despite DNA demethylation,62,64 suggesting nonpharmacologic mechanisms in these cases.
Patients with MDS also respond to drugs that inhibit multiple histone deacetylases.65 Single-agent response to this class of drugs is relatively low, and it is worthwhile noting that activation of hypermethylated genes by decitabine in vitro is much more sustained than that seen with histone deacetylase inhibitors.66 However, combinations of DNA methylation and histone deacetylase inhibition have shown impressive responses in phase 2 studies,67 though randomized studies have yet to document a response or survival advantage to the combinations.
Oncogene activation at the transition from MDS to AML
Much can be learned from studying the transition from MDS to AML. One way to approach the problem is to consider those MDS-specific changes associated with more rapid progression to AML. These include some chromosomal subsets2 (notably monosomy 7), specific mutations (eg, DNMT3a35), and a high degree of CpG island methylation abnormalities.16 Another way is to consider those changes that are exclusive to very advanced MDS, or AML derived from MDS. Notably, some of the frequent epigenetic pathway anomalies in MDS occur quite early and, while sometimes associated with more rapid progression, mutations in epigenetic effectors (TET2, ASXL1, etc) can also be seen years before progression to full-blown AML. Indeed, TET2 mutations have been reported in older individuals with skewed hematopoiesis but no overt MDS.68 By contrast, mutations in growth- and/or apoptotic-controlling genes tend to be restricted to very advanced MDS or at the (somewhat arbitrary) MDS/AML boundary. Examples of these include NRAS, P53, FLT3, CBL, etc69,70 though this list is undoubtedly incomplete. Whole genome sequencing studies of AML derived from MDS will be important to reveal the whole spectrum of these anomalies. Nevertheless, it is possible to advance a speculative model whereby MDS pathogenesis (in some cases) is largely attributable to aberrant epigenetic regulation, and MDS progression is a consequence of acquired abnormalities in growth- and apoptosis-controlling genes (Figure 2).
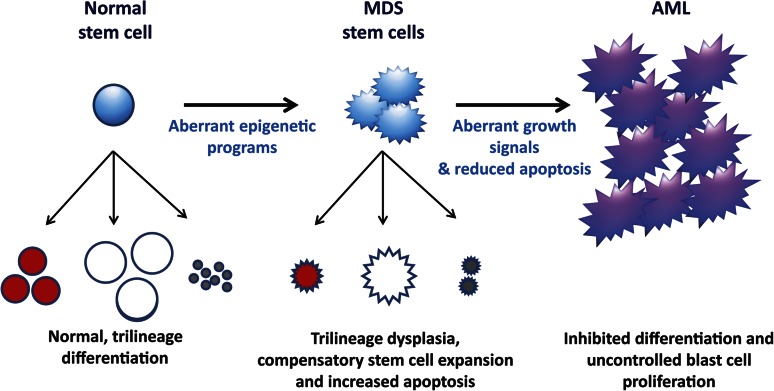
Figure 2.
A model of MDS formation and progression. It is hypothesized that the altered differentiation programs and dysplasias pathognomonic of MDS are due to aberrant epigenetic regulation (summarized in Figure 1). These differentiation defects signal compensatory stem cell growth but also result in increased apoptosis, which explains the paradox of hypercellular marrows but peripheral cytopenias in MDS. With time, MDS cells acquire mutations that confer uncontrolled growth signals (eg, NRAS) and/or inhibited apoptosis (eg, P53). These mutations (and, possibly additional epigenetic defects) lead to the blast expansion and inhibited differentiation characteristic of the transition from MDS to AML.
Conclusions: MDS as an epigenetic disease
The data reviewed above provide a compelling if speculative argument for MDS as an epigenetic disease. The MDS phenotype is one of altered differentiation accompanied by profound epigenomic alterations and mutations in epigenetic controllers. These mutations are sufficient to recapitulate some of the MDS characteristics in mouse models,34,44 and drugs that reprogram the epigenome induce in vivo differentiation, clonal elimination, clinical remissions, and improved survival.53 Progression to AML is characterized by accumulation of epigenetic defects and emergence of new mutations, particularly in growth-controlling genes. It has been proposed that myeloproliferative neoplasms require alterations in 2 classes of genes: growth controllers and transcription factors.71 The accumulating data on epigenetic abnormalities suggest an adaptation of this model (Figure 2) whereby the MDS phenotype is proposed to be due to aberrant epigenetic programs that result in altered differentiation but limited growth potential through increased apoptosis. In turn, progression to AML is likely due to genetic and additional epigenetic lesions that override the apoptotic signals and result in the characteristic blast cell proliferation typical of acute leukemias. This concept suggests that epigenetic modulation will remain key to treatment of MDS, while abrogation of proliferative signals may be a particularly effective treatment at the transition to AML. The outlined model does not discount the role of genetic lesions (which are essential) or the possibility that aspects of the MDS phenotype can be unrelated to epigenetic changes, but it provides a useful framework to define research questions.
The argument proposed here has interesting implications for research directions. One of the central assumptions, that some MDS cases represent essentially an epigenetic disease that responds to epigenetic therapy, triggers many questions. Can one get to MDS without overt epigenetic deregulation (as suggested by the 5q− mouse model discussed earlier)? And would response to epigenetic interventions be limited in those cases? Are genetic, cytogenetic, and epigenetic lesions mostly linked or mostly independent of each other? The latter would suggest that a molecular staging of MDS must involve simultaneous genetic and epigenetic profiling. Do many of the genetic/epigenetic anomalies in MDS converge on a common set of altered pathways (Figure 2) and does this intersection shed unique light on the driver lesions in this disease? Finally, the excitement in the field owes a substantial debt to the activity of epigenetic drugs in MDS. There are many clinical/translational questions there: is it really epigenetic therapy? What are the pathways to response downstream of hypomethylation or histone acetylation induction? Are there subsets of patients that benefit particularly from this approach and others where more classical treatment is indicated? What are the mechanisms of primary and secondary resistance to epigenetic therapy? And, perhaps most intriguingly of all, can effective epigenetic modulation eventually contribute to a higher cure rate in MDS?
Acknowledgments
Research in the author’s laboratory was supported by National Institutes of Health grants CA100632, CA046939, CA121104, CA158112, and CA108631, and by grants from the Ellison Medical Foundation and the Stand Up to Cancer Foundation. J.-P.J.I. is an American Cancer Society Clinical Research professor supported by a generous gift from the F. M. Kirby Foundation.
Authorship
Contribution: J.-P.J.I. is the sole author of the manuscript.
Conflict-of-interest disclosure: J.-P.J.I. is a consultant for Astex and Janssen and receives research support from Astex.
Correspondence: Jean-Pierre J. Issa, Fels Institute for Cancer Research & Molecular Biology, 3307 North Broad St, Room 154, PAHB, Philadelphia, PA 19140; e-mail: jpissa@temple.edu.
References
- 1.Komrokji RS, Zhang L, Bennett JM. Myelodysplastic syndromes classification and risk stratification. Hematol Oncol Clin North Am. 2010;24(2):443–457. doi: 10.1016/j.hoc.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Foran JM, Shammo JM. Clinical presentation, diagnosis, and prognosis of myelodysplastic syndromes. Am J Med. 2012;125(suppl 7):S6–S13. doi: 10.1016/j.amjmed.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 3.Richert-Boe KE, Bagby GC., Jr In vitro hematopoiesis in myelodysplasia: liquid and soft-gel culture studies. Hematol Oncol Clin North Am. 1992;6(3):543–556. [PubMed] [Google Scholar]
- 4.Theilgaard-Mönch K, Boultwood J, Ferrari S, et al. Gene expression profiling in MDS and AML: potential and future avenues. Leukemia. 2011;25(6):909–920. doi: 10.1038/leu.2011.48. [DOI] [PubMed] [Google Scholar]
- 5.Kerbauy DB, Deeg HJ. Apoptosis and antiapoptotic mechanisms in the progression of myelodysplastic syndrome. Exp Hematol. 2007;35(11):1739–1746. doi: 10.1016/j.exphem.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barlow JL, Drynan LF, Hewett DR, et al. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q- syndrome. Nat Med. 2010;16(1):59–66. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- 8.Cedar H, Bergman Y. Epigenetics of haematopoietic cell development. Nat Rev Immunol. 2011;11(7):478–488. doi: 10.1038/nri2991. [DOI] [PubMed] [Google Scholar]
- 9.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 10.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 11.Ahmad K, Henikoff S. Epigenetic consequences of nucleosome dynamics. Cell. 2002;111(3):281–284. doi: 10.1016/s0092-8674(02)01081-4. [DOI] [PubMed] [Google Scholar]
- 12.Amiel J, de Pontual L, Henrion-Caude A. miRNA, development and disease. Adv Genet. 2012;80:1–36. doi: 10.1016/B978-0-12-404742-6.00001-6. [DOI] [PubMed] [Google Scholar]
- 13.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128(4):669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 15.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen L, Kantarjian H, Guo Y, et al. DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes. J Clin Oncol. 2010;28(4):605–613. doi: 10.1200/JCO.2009.23.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen L, Kondo Y, Guo Y, et al. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3(10):2023–2036. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baylin SB, Fearon ER, Vogelstein B, et al. Hypermethylation of the 5′ region of the calcitonin gene is a property of human lymphoid and acute myeloid malignancies. Blood. 1987;70(2):412–417. [PubMed] [Google Scholar]
- 19.Uchida T, Kinoshita T, Nagai H, et al. Hypermethylation of the p15INK4B gene in myelodysplastic syndromes. Blood. 1997;90(4):1403–1409. [PubMed] [Google Scholar]
- 20.Figueroa ME, Skrabanek L, Li Y, et al. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood. 2009;114(16):3448–3458. doi: 10.1182/blood-2009-01-200519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosu-Myles M, Taylor BJ, Wolff L. Loss of the tumor suppressor p15Ink4b enhances myeloid progenitor formation from common myeloid progenitors. Exp Hematol. 2007;35(3):394–406. doi: 10.1016/j.exphem.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Estécio MR, Gharibyan V, Shen L, et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS ONE. 2007;2(5):e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lapeyre JN, Becker FF. 5-Methylcytosine content of nuclear DNA during chemical hepatocarcinogenesis and in carcinomas which result. Biochem Biophys Res Commun. 1979;87(3):698–705. doi: 10.1016/0006-291x(79)92015-1. [DOI] [PubMed] [Google Scholar]
- 24.Issa JP. Epigenetic variation and cellular Darwinism. Nat Genet. 2011;43(8):724–726. doi: 10.1038/ng.897. [DOI] [PubMed] [Google Scholar]
- 25.Hansen KD, Timp W, Bravo HC, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43(8):768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7(4):536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 27.Maegawa S, Hinkal G, Kim HS, et al. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20(3):332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rakyan VK, Down TA, Maslau S, et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010;20(4):434–439. doi: 10.1101/gr.103101.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chambers SM, Shaw CA, Gatza C, Fisk CJ, Donehower LA, Goodell MA. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5(8):e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12(9):599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 31.Shen L, Toyota M, Kondo Y, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci U S A. 2007;104(47):18654–18659. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller-Tidow C, Klein HU, Hascher A, et al. Study Alliance Leukemia. Profiling of histone H3 lysine 9 trimethylation levels predicts transcription factor activity and survival in acute myeloid leukemia. Blood. 2010;116(18):3564–3571. doi: 10.1182/blood-2009-09-240978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dostalova Merkerova M, Krejcik Z, Votavova H, Belickova M, Vasikova A, Cermak J. Distinctive microRNA expression profiles in CD34+ bone marrow cells from patients with myelodysplastic syndrome. Eur J Hum Genet. 2011;19(3):313–319. doi: 10.1038/ejhg.2010.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Challen GA, Sun D, Jeong M, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2012;44(1):23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walter MJ, Ding L, Shen D, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25(7):1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan XJ, Xu J, Gu ZH, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43(4):309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 38.Langemeijer SM, Kuiper RP, Berends M, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41(7):838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 39.Ko M, Huang Y, Jankowska AM, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468(7325):839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamazaki J, Taby R, Vasanthakumar A, et al. Effects of TET2 mutations on DNA methylation in chronic myelomonocytic leukemia. Epigenetics. 2012;7(2):201–207. doi: 10.4161/epi.7.2.19015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139(11):1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song J, Rechkoblit O, Bestor TH, Patel DJ. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science. 2011;331(6020):1036–1040. doi: 10.1126/science.1195380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moran-Crusio K, Reavie L, Shih A, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci. 2010;35(6):323–332. doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 46.Nikoloski G, Langemeijer SM, Kuiper RP, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42(8):665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 47.McCabe MT, Graves AP, Ganji G, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc Natl Acad Sci U S A. 2012;109(8):2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abdel-Wahab O, Adli M, LaFave LM, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22(2):180–193. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maciejewski JP, Padgett RA. Defects in spliceosomal machinery: a new pathway of leukaemogenesis. Br J Haematol. 2012;158(2):165–173. doi: 10.1111/j.1365-2141.2012.09158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ameyar-Zazoua M, Rachez C, Souidi M, et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol. 2012;19(10):998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- 51.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13(2):115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oberdoerffer S. A conserved role for intragenic DNA methylation in alternative pre-mRNA splicing. Transcription. 2012;3(3):106–109. doi: 10.4161/trns.19816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Issa JP, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res. 2009;15(12):3938–3946. doi: 10.1158/1078-0432.CCR-08-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fenaux P, Mufti GJ, Hellstrom-Lindberg E, et al. International Vidaza High-Risk MDS Survival Study Group. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009;10(3):223–232. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kantarjian H, Oki Y, Garcia-Manero G, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109(1):52–57. doi: 10.1182/blood-2006-05-021162. [DOI] [PubMed] [Google Scholar]
- 56.Lübbert M, Suciu S, Baila L, et al. Low-dose decitabine versus best supportive care in elderly patients with intermediate- or high-risk myelodysplastic syndrome (MDS) ineligible for intensive chemotherapy: final results of the randomized phase III study of the European Organisation for Research and Treatment of Cancer Leukemia Group and the German MDS Study Group. J Clin Oncol. 2011;29(15):1987–1996. doi: 10.1200/JCO.2010.30.9245. [DOI] [PubMed] [Google Scholar]
- 57.Kantarjian HM, O’Brien S, Huang X, et al. Survival advantage with decitabine versus intensive chemotherapy in patients with higher risk myelodysplastic syndrome: comparison with historical experience. Cancer. 2007;109(6):1133–1137. doi: 10.1002/cncr.22508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Metzeler KH, Walker A, Geyer S, et al. DNMT3A mutations and response to the hypomethylating agent decitabine in acute myeloid leukemia. Leukemia. 2012;26(5):1106–1107. doi: 10.1038/leu.2011.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fandy TE, Herman JG, Kerns P, et al. Early epigenetic changes and DNA damage do not predict clinical response in an overlapping schedule of 5-azacytidine and entinostat in patients with myeloid malignancies. Blood. 2009;114(13):2764–2773. doi: 10.1182/blood-2009-02-203547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Link PA, Baer MR, James SR, Jones DA, Karpf AR. p53-inducible ribonucleotide reductase (p53R2/RRM2B) is a DNA hypomethylation-independent decitabine gene target that correlates with clinical response in myelodysplastic syndrome/acute myelogenous leukemia. Cancer Res. 2008;68(22):9358–9366. doi: 10.1158/0008-5472.CAN-08-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oki Y, Jelinek J, Shen L, Kantarjian HM, Issa JP. Induction of hypomethylation and molecular response after decitabine therapy in patients with chronic myelomonocytic leukemia. Blood. 2008;111(4):2382–2384. doi: 10.1182/blood-2007-07-103960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qin T, Castoro R, El Ahdab S, et al. Mechanisms of resistance to decitabine in the myelodysplastic syndrome. PLoS ONE. 2011;6(8):e23372. doi: 10.1371/journal.pone.0023372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mahfouz RZ, Jankowska A, Ebrahem Q, et al. Increased CDA expression/activity in males contributes to decreased cytidine analog half-life and likely contributes to worse outcomes with 5-azacytidine or decitabine therapy. Clin Cancer Res. 2013;19(4):938–948. doi: 10.1158/1078-0432.CCR-12-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Issa JP, Gharibyan V, Cortes J, et al. Phase II study of low-dose decitabine in patients with chronic myelogenous leukemia resistant to imatinib mesylate. J Clin Oncol. 2005;23(17):3948–3956. doi: 10.1200/JCO.2005.11.981. [DOI] [PubMed] [Google Scholar]
- 65.Batty N, Malouf GG, Issa JP. Histone deacetylase inhibitors as anti-neoplastic agents. Cancer Lett. 2009;280(2):192–200. doi: 10.1016/j.canlet.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 66.Raynal NJ, Si J, Taby RF, et al. DNA methylation does not stably lock gene expression but instead serves as a molecular mark for gene silencing memory. Cancer Res. 2012;72(5):1170–1181. doi: 10.1158/0008-5472.CAN-11-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia-Manero G, Kantarjian HM, Sanchez-Gonzalez B, et al. Phase 1/2 study of the combination of 5-aza-2′-deoxycytidine with valproic acid in patients with leukemia. Blood. 2006;108(10):3271–3279. doi: 10.1182/blood-2006-03-009142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Busque L, Patel JP, Figueroa ME, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44(11):1179–1181. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dicker F, Haferlach C, Sundermann J, et al. Mutation analysis for RUNX1, MLL-PTD, FLT3-ITD, NPM1 and NRAS in 269 patients with MDS or secondary AML. Leukemia. 2010;24(8):1528–1532. doi: 10.1038/leu.2010.124. [DOI] [PubMed] [Google Scholar]
- 70.Schwaab J, Ernst T, Erben P, et al. Activating CBL mutations are associated with a distinct MDS/MPN phenotype. Ann Hematol. 2012;91(11):1713–1720. doi: 10.1007/s00277-012-1521-3. [DOI] [PubMed] [Google Scholar]
- 71.Gilliland DG. Molecular genetics of human leukemias: new insights into therapy. Semin Hematol. 2002;39(4 suppl 3):6–11. doi: 10.1053/shem.2002.36921. [DOI] [PubMed] [Google Scholar]